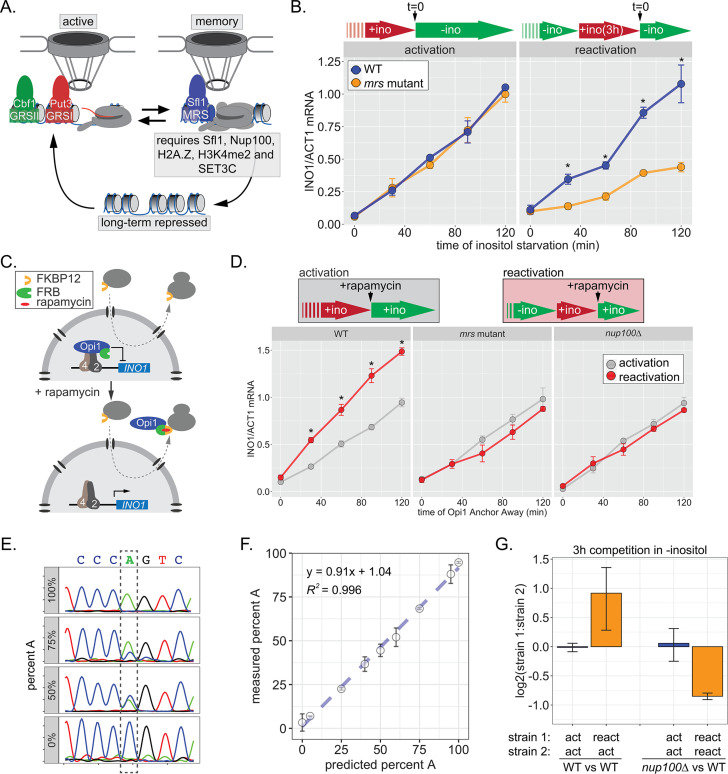
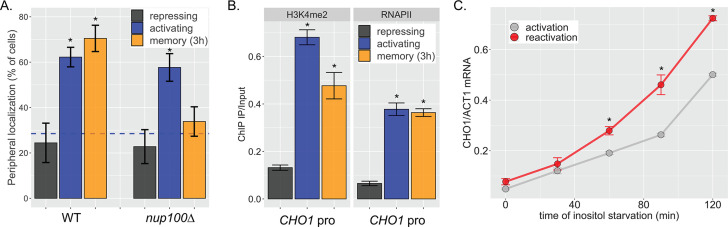
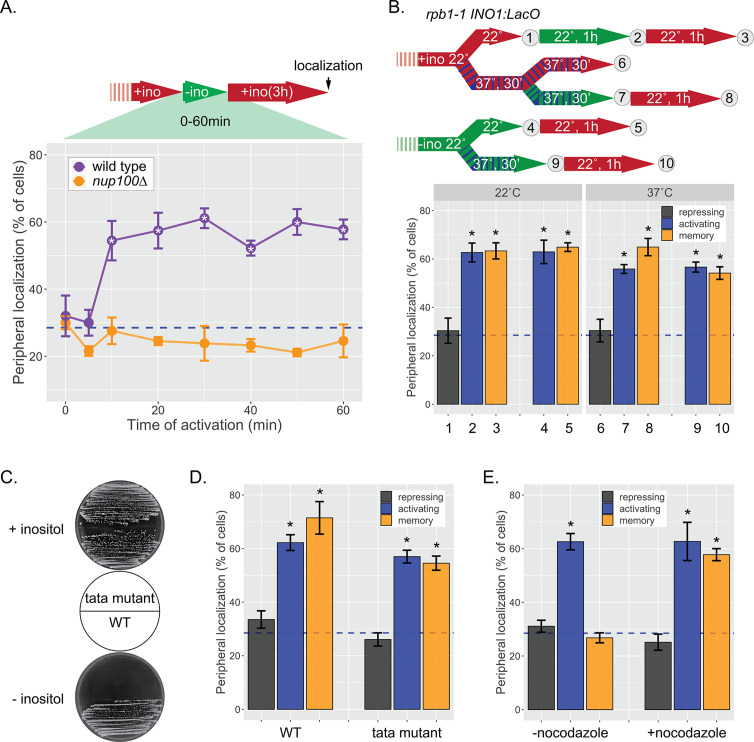
Figure 1. INO1 transcriptional memory stimulates faster transcription and provides a competitive fitness advantage.
(A) Model of INO1 in the active, memory, and long-term repressed states, highlighting factors that are specifically required for memory. (B) Activation (left) and reactivation (right) of INO1 in wild type (WT) and mrs mutant strains upon starvation of inositol. Cells were harvested at indicated time points and the INO1 mRNA was quantified relative to ACT1 mRNA by real time quantitative PCR (RT-qPCR) (*p-value<0.05 from one-tailed t-test comparing WT and mrs mutant, alternative = greater). (C) Schematic of Anchor Away of Opi1 to induce INO1. (D) Top: experimental scheme for synthetic activation and reactivation of INO1. Activation and reactivation of INO1 in WT (left), mrs mutant (middle), and nup100∆ (right) strains upon removal of Opi1 by Anchor Away in the presence of inositol. Bottom: Cells were harvested at indicated time points and INO1 mRNA was quantified relative to ACT1 mRNA by RT-qPCR (*p-value<0.05 from one-tailed t-test comparing reactivation and activation, alternative = greater). (E) Chromatograms resulting from sequencing mixtures of strains having either ‘A’ or ‘C’ SNP within an integrated plasmid, as indicated (dashed box). (F) Standard curve comparing the predicted percentage of strain A (as estimated by O.D.600) with the measured percentage of A (as quantified by the relative area under the peaks, as shown in E). (G) Relative abundance of competing strains, as indicated. The log2 ratio of the abundance of the two strains after 3 hr of competition in media lacking inositol is shown. For panels B, D, F, and G, data are averages of three biological replicates ± SEM.