Antigen stimulation induces a subset of CD4+ and CD8+ T cells to differentiate into CD4+CD8+ T cell subsets gaining in polyfunctional characteristics within the tumor. Monitoring CD4+CD8+ T cells may thus favor the identification and selection of antigen-reactive T cells to drive clinical benefit.
Abstract
Transcription factors ThPOK and Runx3 regulate the differentiation of “helper” CD4+ and “cytotoxic” CD8+ T cell lineages respectively, inducing single positive (SP) T cells that enter the periphery with the expression of either the CD4 or CD8 co-receptor. Despite the expectation that these cell fates are mutually exclusive and that mature CD4+CD8+ double positive (DP) T cells are present in healthy individuals and augmented in the context of disease, yet their molecular features and pathophysiologic role are disputed. Here, we show DP T cells in murine and human tumors as a heterogenous population originating from SP T cells which re-express the opposite co-receptor and acquire features of the opposite cell type’s phenotype and function following TCR stimulation. We identified distinct clonally expanded DP T cells in human melanoma and lung cancer by scRNA sequencing and demonstrated their tumor reactivity in cytotoxicity assays. Our findings indicate that antigen stimulation induces SP T cells to differentiate into DP T cell subsets gaining in polyfunctional characteristics.
Introduction
Mature CD4+CD8+ T cells are considered an anomaly, generally overlooked as cell doublets or cells having escaped faulty thymic selection. The presence of these cells in the periphery refutes the understanding that CD4 and CD8 expression is fixed and mutually exclusive in mature T cells (Egawa, 2015). This concept is derived from the fact that key transcription factors ThPOK and Runx3 negatively regulate one another during T cell development in the thymus to induce functionally distinct “helper” CD4+ or “cytotoxic” CD8+ single positive (SP) T cell fates. MHC class II–restricted CD4+ T cells rely on ThPOK-mediated suppression of Runx3 to prevent CD8αβ expression and the cytotoxic lineage program (Wang et al., 2008; Wildt et al., 2007; Egawa and Littman, 2008). Runx3 binds ThPOK’s upstream silencer, directing commitment of MHC class I–restricted thymocytes toward the CD8 lineage (Mucida et al., 2013; Setoguchi et al., 2008). Expression of these transcription factors is further maintained in the periphery, suggesting that their lineage-defining roles remain active outside the thymus (Vacchio and Bosselut, 2016). Despite the current understanding that T cells are programmed to commit to and maintain a SP fate, a heterogenous population of circulating CD4+CD8+ double positive (DP) T cells have been observed in the peripheral blood and tissues of numerous species, including humans, suggesting a degree of plasticity in ThPOK and Runx3 regulation (Blue et al., 1985; Ghia et al., 2007). While considerable variability exists between individuals, DP T cells typically represent <1% of circulating T cells in healthy individuals and localize at sites of inflammatory responses, including viral infections and malignant diseases (Clenet et al., 2017; Overgaard et al., 2015; Nascimbeni et al., 2004).
Several attempts have been made to characterize extrathymic CD4+CD8+ T cells in mice and humans based on their function, expression of surface markers, and TCR repertoire, but the results are contradictory (Overgaard et al., 2015). In renal cell carcinoma patients, CD4+CD8+ T cells can make up over 30% of the tumor-infiltrating lymphocyte (TIL) population with phenotypic analysis, pointing their identity toward an effector memory CD8+ T cell subset with cytotoxic activity (Menard et al., 2018). In HIV patients, CD4+CD8+ T cells in circulation are highly proliferative and demonstrate polyfunctionality following specific HIV antigen stimulation, resembling CD4+ T cell responses (Frahm et al., 2012). In contrast, suppressive IL-4– and IL-10–producing CD4+CD8+ T cells have been identified in the context of inflammatory bowel disease, urological cancer, and systemic sclerosis (Parel et al., 2007; Das et al., 2003; Bohner et al., 2019). These studies demonstrate the unique heterogeneity and plasticity of the CD4+CD8+ T cell pool, indicating that the disease type and/or local environmental stimuli may substantially contribute to shaping the differentiation and function of these cells.
Of note, a commonality in the CD4+CD8+ T cell literature is data supporting antigen-driven expansion and evidence for TCR clonality observed within CD4+CD8+ T cells, highlighting the potential therapeutic relevance of this population, especially in the setting of cancer and cancer immunotherapy (Clenet et al., 2017; Overgaard et al., 2015; Frahm et al., 2012; Menard et al., 2018; Nishida et al., 2020; Bohner et al., 2019). However, with the origin and function(s) of CD4+CD8+ T cells unclear, a comprehensive understanding of this cell type and its mechanism of differentiation in the tumor microenvironment is necessary to harness the full potential of these cells.
Here, we defined the molecular underpinning of the unique polyfunctional features of DP T cell states in open-repertoire and transgenic TCR T cells within murine and human melanoma and lung cancer. Upon TCR signaling, CD4-derived DP T cells (CD4+cd8+) upregulate CD8 lineage regulator Runx3 and cytolytic genes, Gzmb and Prf1, to become potent cytotoxic T cells. In contrast, murine CD8-derived DP T cells (CD8+cd4+) express Foxp3 and display dual cytotoxic and mild suppressive properties. Transcriptional analysis of CD4+ and CD8+ T-cell-derived DP cells revealed distinct modulation of the epigenetic landscape over time, regulating the differentiation from SP to DP T cells.
We identified distinct clonally expanded DP T cell states in human melanoma and lung cancer samples by single-cell RNA (scRNA) and TCR sequencing and identified a unique overlap of DP TCRs matching CD4 and CD8 SP T cells. Further, we observed an enrichment in cytolytic markers in CD4+CD8+ T cells among patients who ultimately responded to ICB and demonstrated the potent tumor reactivity of CD4+CD8+ T cells in ex vivo cytotoxicity assays where we observed cell lysis to be MHC dependent.
Our findings indicate that antigen stimulation induces SP T cells to differentiate into DP T cell subsets gaining in polyfunctional characteristics. Monitoring DP T cells may thus favor identification of T cells enriched for antigen specificity and allow for determination of signaling pathways underlying their functional plasticity. These findings can be exploited to reprogram T cells to maximize polyfunctionality for clinical application, such as adoptive T cell and immune checkpoint blockade (ICB)–based therapies.
Results
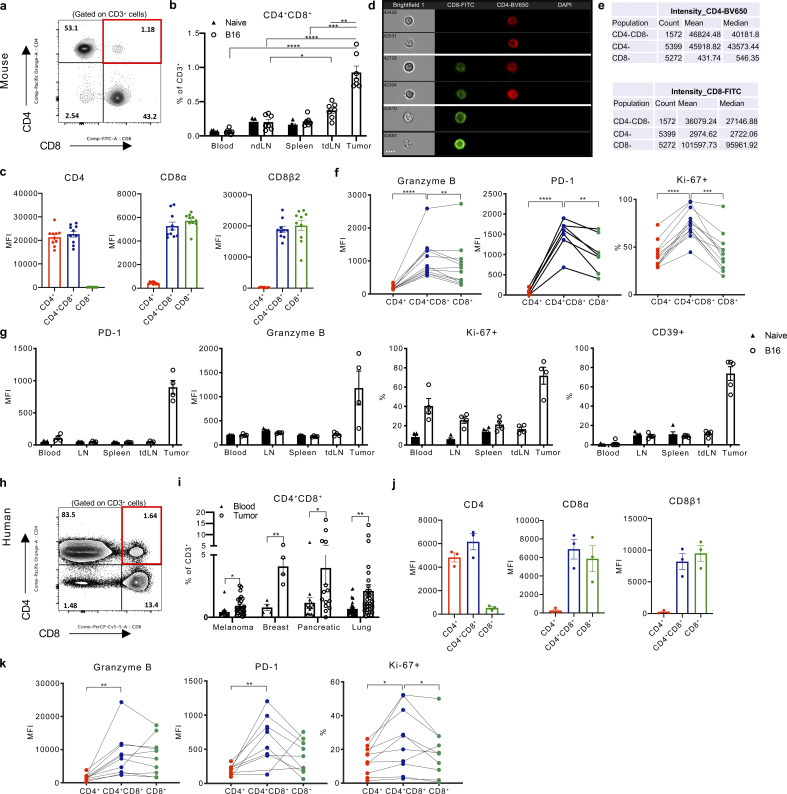
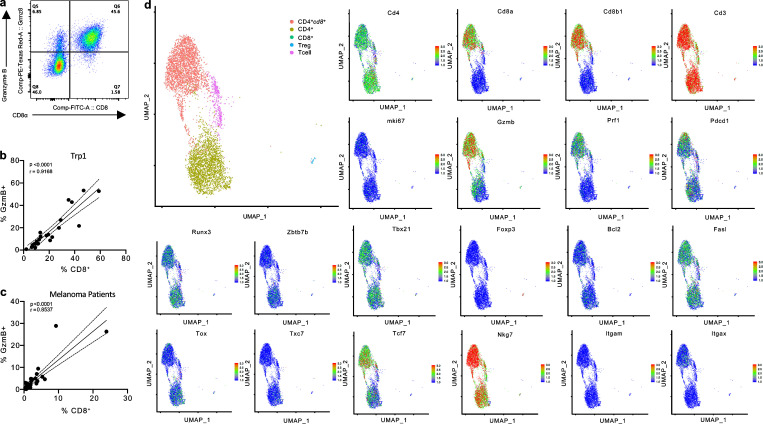
Previous studies suggested that CD4+CD8+ (DP) T cells are polyfunctional, possessing cytotoxic, proliferative, or suppressive features (Overgaard et al., 2015). However, a uniform consensus regarding the distinct contribution of CD4+CD8+ T cells to anti-tumor immunity is lacking (Bohner et al., 2019; Menard et al., 2018; Nishida et al., 2020). To evaluate the role of CD4+CD8+ T cells during tumor progression, we studied these cells in naive and B16F10 (referred to as B16) melanoma-bearing C57BL/6J mice. CD4+CD8+ T cells comprised <0.5% of CD3+ T cells in circulation and the lymphoid organs of naive mice and accumulated within the tumor microenvironment (Fig. 1, a and b; and Fig. S1 a). Importantly, CD4+CD8+ T cells expressed both the CD8α and CD8β co-receptors (Fig. 1 c and Fig. S1 c) and confirmed that CD4+CD8+ T cells were singlet T cells expressing both CD4 and CD8 on their surface by imaging cells in suspension using Image Stream flow cytometry (Fig. 1, d and e). Intra-tumoral CD4+CD8+ T cells expressed increased the levels of granzyme B (Gzmb) and PD-1 and comprised a higher fraction of proliferating Ki-67+ cells compared with SP CD4+ or CD8+ T cells, displaying an activated and proliferative effector phenotype (Fig. 1 f and Fig. S1 e). Intriguingly, the elevated expression of these activation and cytolytic markers, including CD39, a marker reportedly upregulated in tumor-reactive T cells (Duhen et al., 2018), was unique to T cells isolated from the tumor and was not observed in CD4+CD8+ T cells in naive mice or in the periphery of tumor-bearing mice (Fig. 1 g).
Figure 1.
CD4+CD8+ DP T cells with a cytotoxic profile reside in mouse and human melanoma tumors. (a) Representative expression of CD4 and CD8 detected by surface staining on CD3+ T cells in B16 tumors. (b) Percentage of endogenous CD4+CD8+ T cells among CD3+ cells present in tissues (peripheral blood, non-tumor draining lymph node [LN], spleen, tumor-draining lymph node [tdLN]), and tumor of naive or B16 tumor-bearing mice. (c) CD4, CD8α, and CD8β expression detected by flow cytometry in CD4+, CD4+CD8+, or CD8+ T cells isolated from B16 tumors implanted in mice. (d) Representative images of CD4+, CD8+, and CD4+CD8+ T cells that were FACS-sorted from B16 tumors and immediately analyzed by an ImageStream cytometer for staining with CD8 (FITC), CD4 (BV650), and DAPI. Scale bar, 7 μm. (e) Count of gated CD4+, CD4+CD8+, and CD8+ T cells. Mean and median expression intensity of CD4 (BV650) and CD8 (FITC). (f) Granzyme B, PD-1 expression (mean fluorescence intensity [MFI]) and percentage of Ki-67+ cells detected by flow cytometry in CD4+, CD4+CD8+, and CD8+ T cells isolated from B16 tumors. (g) Granzyme B, PD-1 expression (MFI), and percentage of Ki-67+ and CD39+ cells detected by flow cytometry in CD4+CD8+ T cells isolated from peripheral blood, non-tumor draining lymph node (LN), spleen, tumor-draining lymph node (tdLN), and tumor of naive and B16 tumor-bearing mice. (h) Representative expression of CD4 and CD8 by surface staining on CD3+ T cells and (i) percentage of CD4+CD8+ T cells isolated from melanoma (n = 25), breast (n = 4), pancreatic (n = 15), and lung (n = 33) patient tumor resections or peripheral blood from independent cohorts of patients with the respective tumors. (j) CD4, CD8α, and CD8β expression detected by flow cytometry in CD4+, CD4+CD8+, or CD8+ T cells isolated from melanoma patient tumor resections. (k) Granzyme B, PD-1 expression (MFI), and percentage of Ki-67+ cells detected by flow cytometry in CD4+, CD4+CD8+, and CD8+ T cells isolated from melanoma patient resections. Data are representative of three independent experiments (a, b, and e–g) or representative images of >1,500 cells (g) are displayed. Data with error bars are represented as mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, two-sided unpaired (b and i) or paired (e, f, and k) Student’s t test.
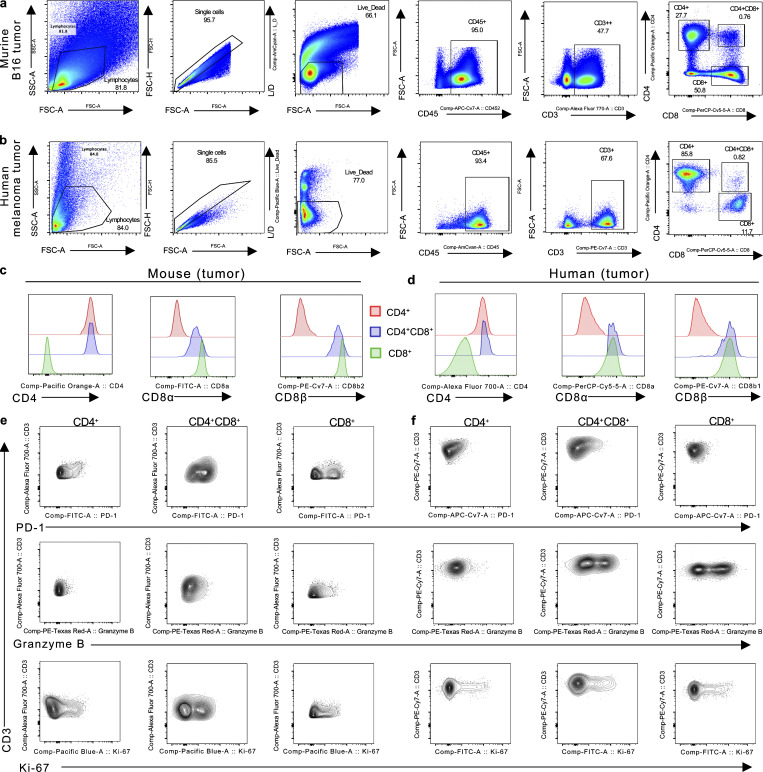
Figure S1.
Gating strategy and representative FACS plots of CD4+, CD4+CD8+, and CD8+ T cells. (a and b) Representative gating strategy of murine (a) and human (b) CD4+, CD4+CD8+, and CD8+ T cell identification by flow cytometry. (c and d) Representative histogram of CD4, CD8α, and CD8β expression in CD4+, CD4+CD8+, or CD8+ T cells isolated from B16 tumor-bearing mice (c) or human melanoma (d) tumors. (e and f) Representative FACS plot of PD-1, Granzyme B, and Ki-67 staining in CD4+, CD4+CD8+, and CD8+ T cells from mouse (e) and human (f) tumors. Data are representative of three independent experiments (a, c, and e).
We next investigated if the observation of CD4+CD8+ T cells in the murine setting is relevant in humans by assessing the presence and phenotype of CD4+CD8+ T cells in the peripheral blood and within melanoma tumors resected from 25 patients. CD4+CD8+ T cells expressing the CD8α and CD8β co-receptors were present in all samples evaluated and comprised up to 3% of the total CD3+ population in tumors, in contrast to ∼0.5% of circulating CD3+ T cells displaying a CD4+CD8+ phenotype (Fig. 1, h–j; and Fig. S1, b and d). Intra-tumoral CD4+CD8+ T cells were also observed at increased frequencies in patients with breast, pancreatic, and non-small cell lung cancer (NSCLC) relative to CD4+CD8+ T cells in circulation (Fig. 1 i). Consistent with the phenotype of CD4+CD8+ T cells in murine tumors, the protein expression of GzmB, PD-1, and Ki-67 was increased in CD4+CD8+ T cells isolated from melanoma resections (Fig. 1 k and Fig. S1 f).
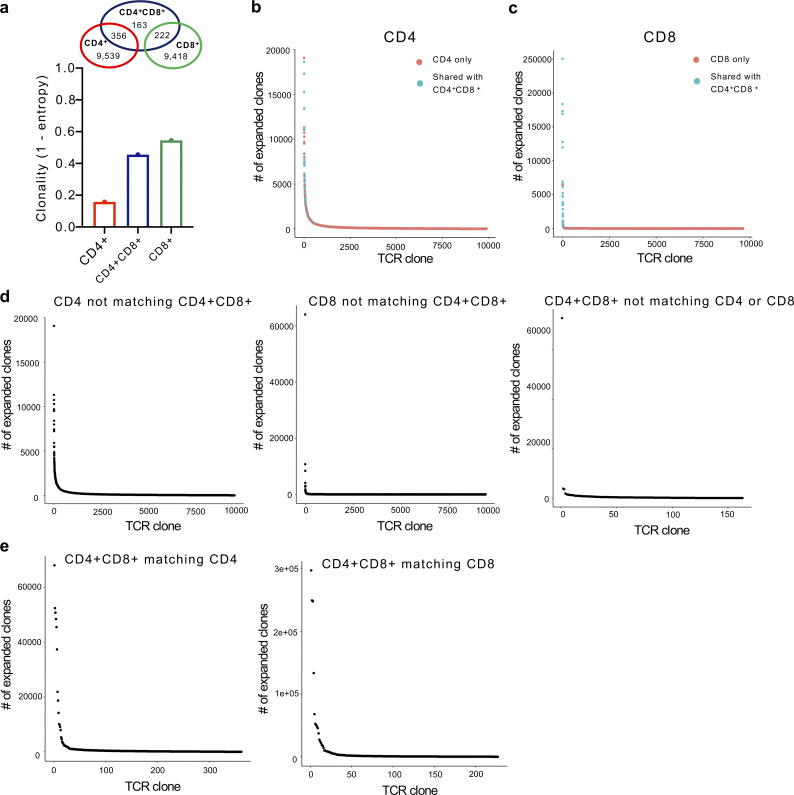
Based on the prominent activation phenotype of CD4+CD8+ T cells within the tumor, we hypothesized that DP T cells were differentiating into polyfunctional effector cells following antigen stimulation. To assess the clonality of these T cell populations, we implanted C57BL/6 mice with B16 cells and FACS-sorted CD4+, CD8+, and CD4+CD8+ T cells from the tumor for TCR sequencing. Reconstructed CDR3 sequences revealed enriched TCRβ transcripts within the intra-tumor CD4+CD8+ T cell population compared with tumor-infiltrating SP CD4+ T cells and similar to tumor-infiltrating SP CD8+ T cells, further suggesting antigen-driven expansion (Fig. 2 a). Surprisingly, we also observed a number of shared TCRs between CD4+CD8+ T cells and SP CD4+ or CD8+ TCRs (Fig. 2 a), where the majority of the expanded SP CD4+ or CD8+ TCRs were those shared with CD4+CD8+ T cells (Fig. 2, b and c). Notably, among the CD4+CD8+ TCRs that were not shared with SP CD4+ or CD8+ TCRs, expanded TCRβ transcripts were significantly reduced (Fig. 2, d and e). These data suggest that the observed heterogeneity of the CD4+CD8+ T cell pool may be derived from both CD4+ and CD8+ SP T cells re-expressing the opposite co-receptor upon antigen priming in the periphery.
Figure 2.
Expanded and shared TCRβ transcripts among CD4+CD8+ T cells. (a) Clonality of reconstructed CDR3 regions as assessed by TCR diversity score and normalized by Shannon-Weiner index of CD4+, CD4+CD8+, and CD8+ T cells FACS-sorted out of 10 pooled B16 tumors. Shared CDR3 regions among CD4+, CD4+CD8+, and CD8+ T cells are represented in a Venn diagram. (b and c) Frequency of expanded CDR3 clones among CD4+ (b) and CD8+ (c) T cells. Clones present among CD4+CD8+ T cells are colored in blue. (d and e) Frequency of unique expanded CDR3 clones among CD4+, CD4+CD8+, and CD8+ T cells (d) or CD4+CD8+ TCRs shared with CD4+ or CD8+ T cells (e).
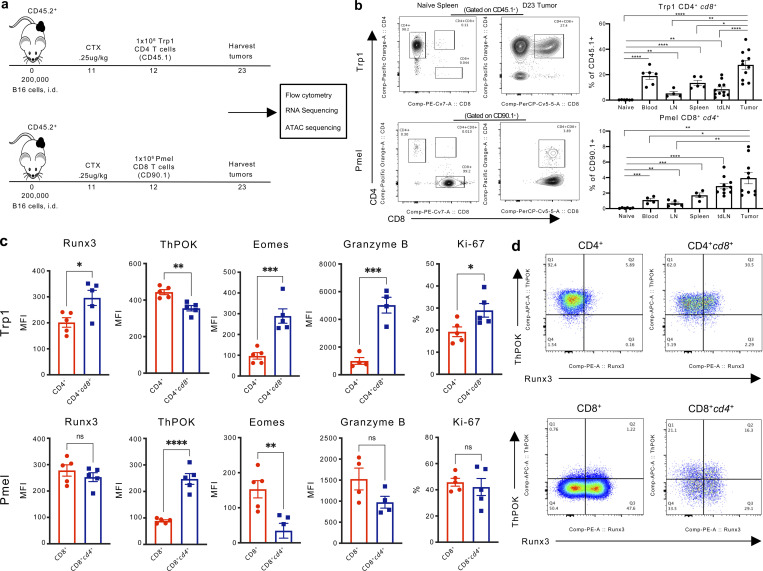
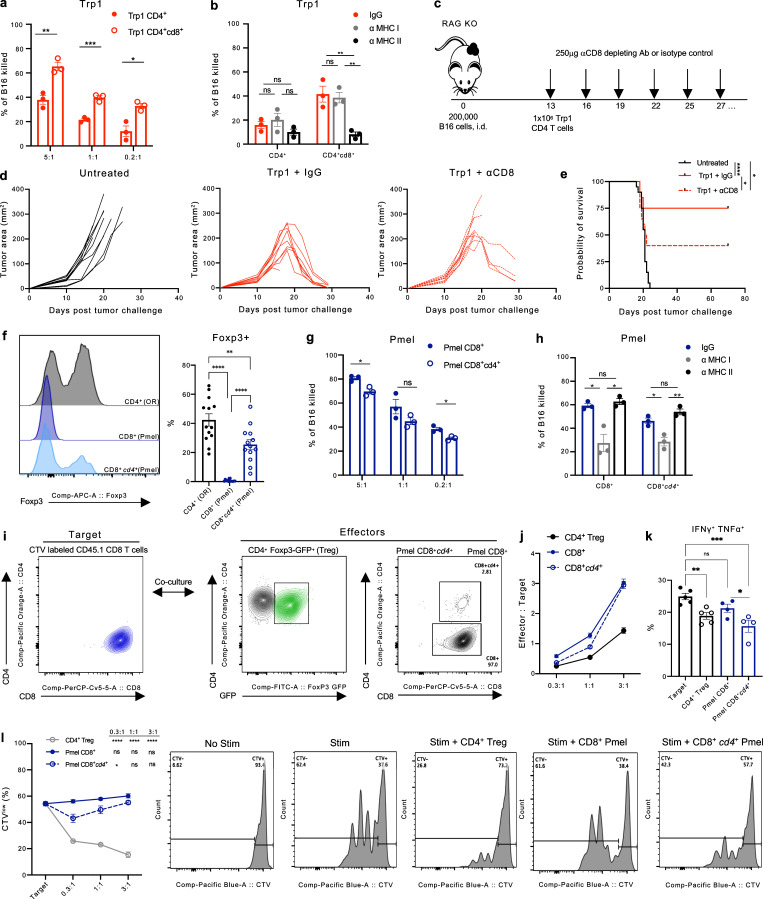
To investigate the hypothesis that TCR signaling drives ectopic re-expression of the opposite CD4 or CD8 co-receptor in tumor-infiltrating T cells, we utilized CD4+ and CD8+ T cells from TCR transgenic mice specific to common melanosomal antigens expressed by B16 and normal melanocytes. Specifically, we utilized transgenic TCR CD4+ T cells recognizing tyrosinase-related protein 1 (trp1, gp75) in the context of MHC class II (Trp1 T cells), and transgenic TCR CD8+ T cells responding to premelanosome matrix glycoprotein (pmel, gp100) presented by MHC class I (Pmel T cells; Muranski et al., 2008; Overwijk et al., 2003). Using these congenically labeled T cells (CD45.1 and CD90.1 respectively), we monitored the development of DP T cells from naive SP CD4+CD45.1+ Trp1 and CD8+CD90.1+ Pmel T cells upon transfer into B16-bearing CD45.2 recipient mice pre-conditioned with cyclophosphamide (Fig. 3 a). Importantly, both naive CD4+ Trp1 and CD8+ Pmel T cells were able to acquire a CD4+CD8+ (CD4+cd8+ Trp1 or CD8+cd4+ Pmel) phenotype in the blood, lymphoid organs, and tumors in the recipient mice (Fig. 3 b). Similar to our observations with open-repertoire T cells, the transgenic DP T cell populations were enriched in the tumor, albeit more so for Trp1-derived CD4+cd8+ T cells (Fig. 3 b). Flow cytometric analysis of transcription factors regulating CD4+ and CD8+ T cell fate and effector function revealed that CD8 re-expression in CD4+ Trp1 T cells was accompanied by increased expression of CD8 lineage regulator Runx3, GzmB, and eomesdermin (Eomes), a transcription factor implicated in regulating granzyme B (GzmB; Glimcher et al., 2004; Cruz-Guilloty et al., 2009; Fig. 3, c and d). Further, CD4+cd8+ Trp1 T cells displayed downregulation of the CD4 lineage commitment regulator ThPOK and increased the proliferative capacity by Ki-67 staining relative to SP CD4+ Trp1 T cells (Fig. 3, c and d). In contrast, CD8+cd4+ Pmel T cells had reduced protein expression of both Eomes and GzmB and increased expression of ThPOK relative to SP CD8+ Pmel T cells (Fig. 3, c and d).
Figure 3.
CD4+ and CD8+ derived tumor-antigen-specific CD4+CD8+ T cells acquire features of the opposite co-receptor. (a) Experimental scheme of adoptive T cell transfer of antigen-specific CD4+ Trp1 or CD8+ Pmel T cells from naive mice into B16 tumor-bearing mice following cyclophosphamide treatment. T cells were isolated from subcutaneous tumors and peripheral tissues 11 d after transfer for flow cytometry and transcriptomic analyses. (b) Representative plots and percentage of CD4+CD8+ among CD45.1+ (Trp1) or CD90.1+ (Pmel) T cells isolated from a naive spleen of the respective transgenic donor mice, or on day 23 from the blood, non-tumor draining lymph node (LN), spleen, tumor-draining lymph node (tdLN) and tumor of the recipient mice after transfer. (c) Runx3, ThPOK, Eomes, and Granzyme B expression (MFI) and percentage of Ki-67+ detected in Trp1 CD4+ (SP, red), Trp1 CD4+cd8+ (DP, blue), or Pmel CD8+ (SP, red) and Pmel CD8+cd4+ (DP, blue) T cells isolated from B16 tumors 11 d after adoptive transfer. (d) Representative FACS plots of ThPOK and Runx3 expression in CD4+ and CD4+cd8+ Trp1 T cells (top), and CD8+ and CD8+cd4+ Pmel T cells (bottom) isolated from the tumor of B16 tumor-bearing mice. Data are representative of at least three independent experiments. Data are mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, two-sided paired Student’s t test.
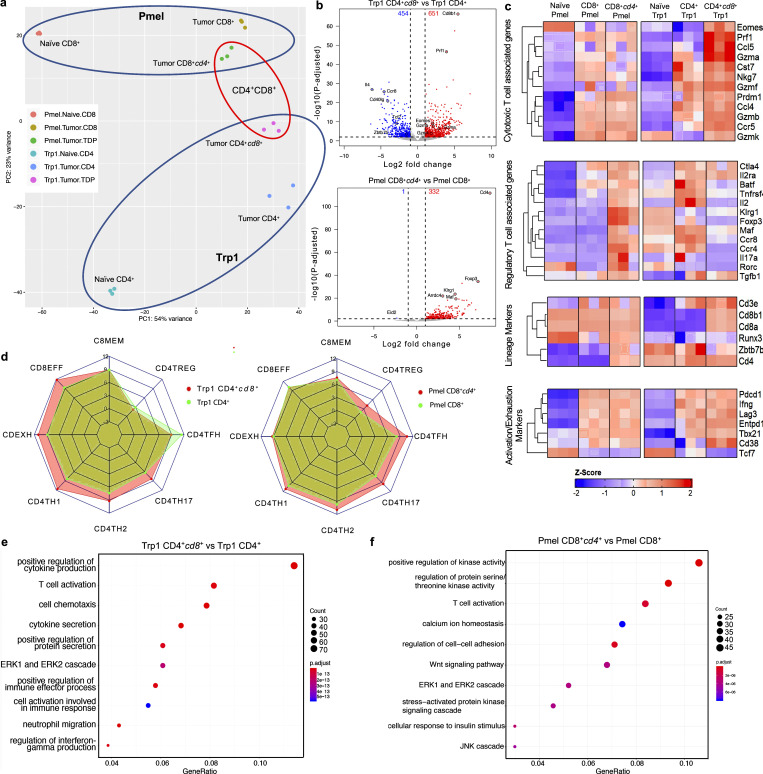
To gain a deeper understanding of DP T cells derived from CD4+ vs. CD8+ SP T cells, we FACS-sorted SP Trp1 and Pmel T cells from spleens of naive transgenic mice, and SP and DP Trp1 and Pmel T cells from tumors of mice treated as in Fig. 3 a for transcriptomic profiling (Table S1). Principal component analysis (PCA) of the top 500 variably expressed genes demonstrated that Trp1 and Pmel naive, SP, and DP subsets are transcriptionally distinct populations (Fig. 4 a). We observed 1,105 (651 up, 454 down) differentially expressed genes (DEGs) between tumor-infiltrating CD4+ and CD4+cd8+ Trp1 T cells, with cytotoxicity related genes Prf1, Gzma, and Gzmb upregulated in CD4+cd8+ Trp1 T cells, while Il4, Ccr8, Cd40lg, and Tcf7 were preferentially expressed in CD4+ Trp1 T cells (Fig. 4, b and c). In contrast, 333 DEGs were observed between tumor-infiltrating CD8+ and CD8+cd4+ Pmel T cells, with all but 1, Eid2, being preferentially expressed in CD8+cd4+ Pmel T cells. Intriguingly, among the DEGs upregulated in Pmel CD8+cd4+ T cells was Foxp3 and genes associated with regulatory T cell (Treg) signatures including Klrg1, Maf, Ccr4, and Il2ra (CD25; Fig. 4, b and c). Of note, intra-tumor Trp1 and Pmel SP and DP cells expressed Cd3, Cd4, Cd8a, and Cd8b1 lineage markers as expected based on our gating strategy for sorting, and similar levels of activation and exhaustion genes (Fig. 4 c). Gene set enrichment analysis (GSEA) further demonstrated that tumor-infiltrating CD4+cd8+ Trp1 T cells showed extensive similarity with gene sets representing known effector and exhausted CD8 T cell states, while tumor-infiltrating CD8+cd4+ Pmel T cells more closely resembled CD4 T cell states than CD8+ Pmel T cells (Fig. 4 d). Additionally, Gene Ontology enrichment analysis of the DEGs between CD4+ and CD4+cd8+ Trp1 T cells revealed enrichment in signatures associated with T cell activation, leukocyte proliferation, and cytokine secretion, while DEGs between CD8+ and CD8+cd4+ Pmel T cells were enriched in signatures associated with the positive regulation of MAP kinase activity (Fig. 4, e and f).
Figure 4.
Transcriptional Characterization of antigen-specific CD4 and CD8 derived CD4+CD8+ T cells. (a) Principal component analysis (PCA) of top 500 variably expressed genes in Pmel naive, Pmel CD8+, Pmel CD8+cd4+, Trp1 Naive, Trp1 CD4+, Trp1 CD4+cd8+ T cells. (b) Volcano plots of RNA-seq dataset (top: Trp1 CD4+cd8+ vs. Trp1 CD4+; bottom: Pmel CD8+cd4+ vs. Pmel CD8+). DEGs are shown in red (positive) or blue (negative). (c) Heatmap of bulk RNA-seq expression of genes associated with T cell lineage, activation, cytotoxic, and Treg signatures in Pmel naive, Pmel CD8+, Pmel CD8+cd4+, Trp1 naive, Trp1 CD4+, and Trp1 CD4+cd8+ cells. (d) Spider plot depicting GSEA normalized enrichment scores of prior defined gene sets between Trp1 CD4+ and Trp1 CD4+cd8+ (left) and Pmel CD8+ and CD8+cd4+ T cells (right). Gene sets for CD4TH1, CD4TH2, CD4TH17, and CD4TREG are from GSE14308, CD4TFH is from GSE85316, and CD8EFF, CD8EXH, CD8MEM are from GSE30431 and are all relative to naive T cells. (e and f) Gene Ontology enrichment analysis of signatures across differentially expressed genes between Trp1 CD4+cd8+ and Trp1 CD4+ (e) and Pmel CD8+cd4+ and Pmel CD8+ T cells (f).
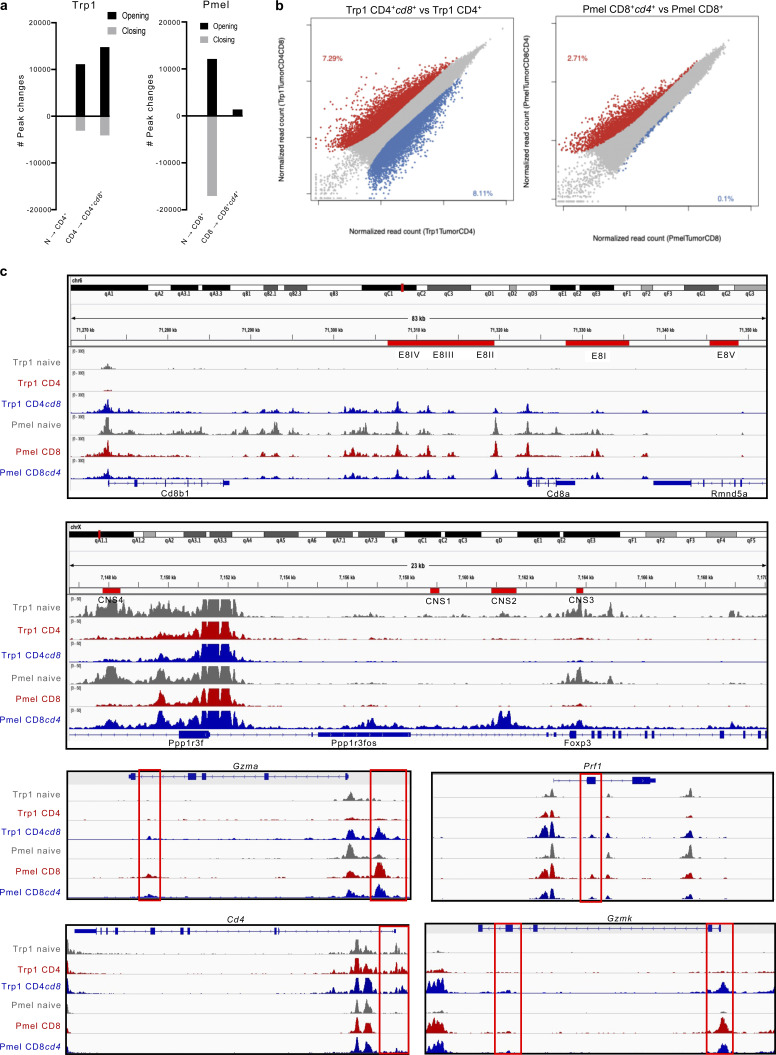
We next investigated the chromatin state dynamics of Trp1 and Pmel T cells following their differentiation from a SP naive state in the spleen of naive mice to intra-tumor SP and DP T cells, as depicted in Fig. 3 a. Consistent with previous reports (Philip et al., 2017), we identified substantial chromatin remodeling between T cells from naive spleen and SP effector T cells isolated from tumor, as well as between intra-tumoral SP and DP Trp1 and Pmel T cells (Fig. S2, a and b). We observed a broad modulation in peak accessibility with CD4+cd8+ Trp1 T cells demonstrating increased accessibility in the E8I-V enhancer regions along the CD8a and Cd8b1 gene loci, as well as specifically along the loci of genes associated with cytotoxicity including Gzma and Prf1 (Fig. S2 c). In contrast, CD8+cd4+ Pmel T cells had distinct openings in the CD4 and Foxp3 gene loci, with accessibility along the Foxp3 locus distinctly in all four CNS regions known to induce and sustain Foxp3 expression (Chakraborty et al., 2017; Fig. S2 c). These findings suggest differential epigenetic regulation of these unique cytotoxic and suppressive DP T cell states originating respectively from SP CD4 or CD8 T cells.
Figure S2.
Epigenetic states of Trp1 and Pmel T cells. (a) Number of chromatin peaks whose accessibility changes during transition from naive Trp1 CD4+ to tumor-infiltrating CD4+ and CD4+cd8+ T cells (right) or from naive Pmel CD8+ to tumor-infiltrating CD8+ and CD8+cd4+ T cells (left). (b) Scatter plot of differentially accessible peaks between Trp1 CD4+cd8+ and Trp1 CD4+ (left) and Pmel CD8+cd4+ and Pmel CD8+ (right). Increased accessibility is colored in red, and reduced accessibility is colored in blue. (c) Genome browser view of ATAC-seq signal intensities of Trp1 naive, Trp1 CD4+, Trp1 CD4+cd8+, Pmel naive, Pmel CD8+, and Pmel CD8+cd4+ cells across Cd8a, Cd8b1, Foxp3, Gzma, Gzmk, Prf1, and Cd4 loci. Red boxes indicate peaks that become more accessible in Trp1 CD4+cd8+ vs. Trp1 CD4 or Pmel CD8+cd4+ vs. Pmel CD8, respectively. ATAC-seq peaks from naive Trp1 and Pmel are shown in gray as a control. E8I-V enhancer regions along the CD8ab locus and CNS I-V regions along the Foxp3 locus are annotated.
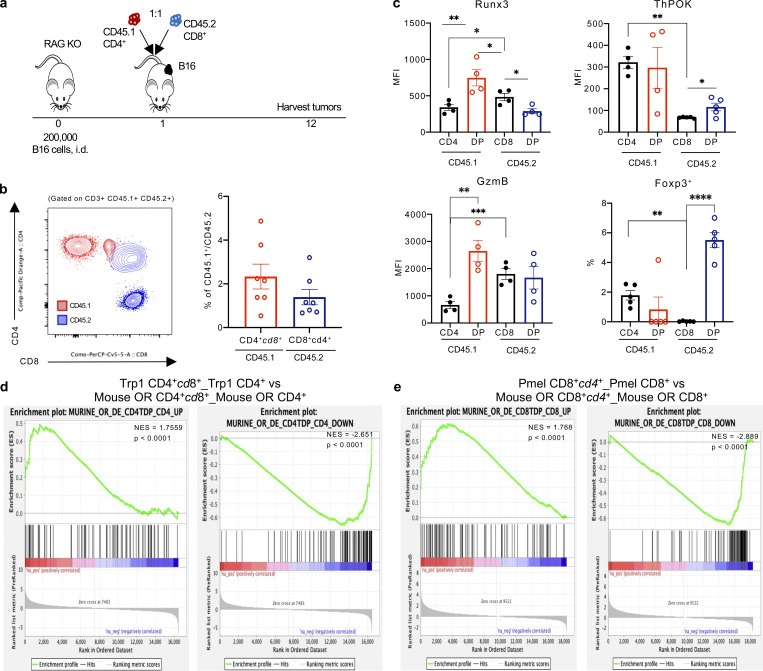
Notably, the phenotype of Trp1 and Pmel DP T cells was not unique to our transgenic model and was consistent with DP T cells derived from adoptively transferred open-repertoire CD4+ (CD45.1) or CD8+ (CD45.2) T cells within the tumor microenvironment of B16-bearing RAG KO mice (Fig. 5, a and b). CD4-derived open-repertoire CD4+cd8+ T cells expressed elevated levels of Runx3 and GzmB, while CD8-derived CD8+cd4+ T cells had an increased expression of ThPOK and Foxp3 relative to SP CD8+ T cells (Fig. 5 c). Additionally, we FACS-sorted the four T cell populations (as identified in Fig. 5 b) for 10× scRNA sequencing (scRNA-seq) and compared the transcriptional signatures to our transgenic CD4+cd8+ Trp1 and CD8+cd4+ Pmel T cells. GSEA analysis revealed significant similarities of gene expression changes between the open repertoire and transgenic CD4+cd8+ and CD8+cd4+ T cell populations compared with matched CD4+ and CD8+ T cells (Fig. 5, d and e).
Figure 5.
CD4+ and CD8+ derived open repertoire CD4+CD8+ T cells acquire features of the opposite co-receptor. (a) Experimental schema of adoptive transfer of naive FACS-sorted CD4+ CD45.1+ and CD8+ CD45.2+ T cells into B16 tumor-bearing RAG KO mice. Tumors were harvested for analysis 11 d after T cell transfer. (b) Representative plot of CD4+ CD45.1+ (red) and CD8+ CD45.2+ (blue) derived CD4+CD8+ T cells in tumor (left), frequency of CD4+ CD45.1+ (red) and CD8+ CD45.2+ (blue) derived CD4+CD8+ T cells of parent population (right). (c) Runx3, ThPOK, and Granzyme B expression (MFI), and percentage of Foxp3+ cells among CD4+, CD4 derived CD4+cd8+ (red), CD8+, and CD8 derived CD8+cd4+ (blue) T cells in the tumor. (d) GSEA of DEGs in murine open repertoire CD4+cd8+ vs. CD4+ T cells (left, UP; right, DOWN) shows significantly concordant differences in the same direction as murine Trp1 CD4+cd8+ vs. Trp1 CD4+ T cells. (e) GSEA of DEGs in murine open repertoire CD8+cd4+ vs. CD8+ T cells (left, UP; right, DOWN) shows significantly concordant differences in the same direction as murine Pmel CD8+cd4+ vs. Pmel CD8+ T cells. Data are representative of at least three independent experiments (b and c). Data with error bars are represented as mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, two-sided paired Student’s t test.
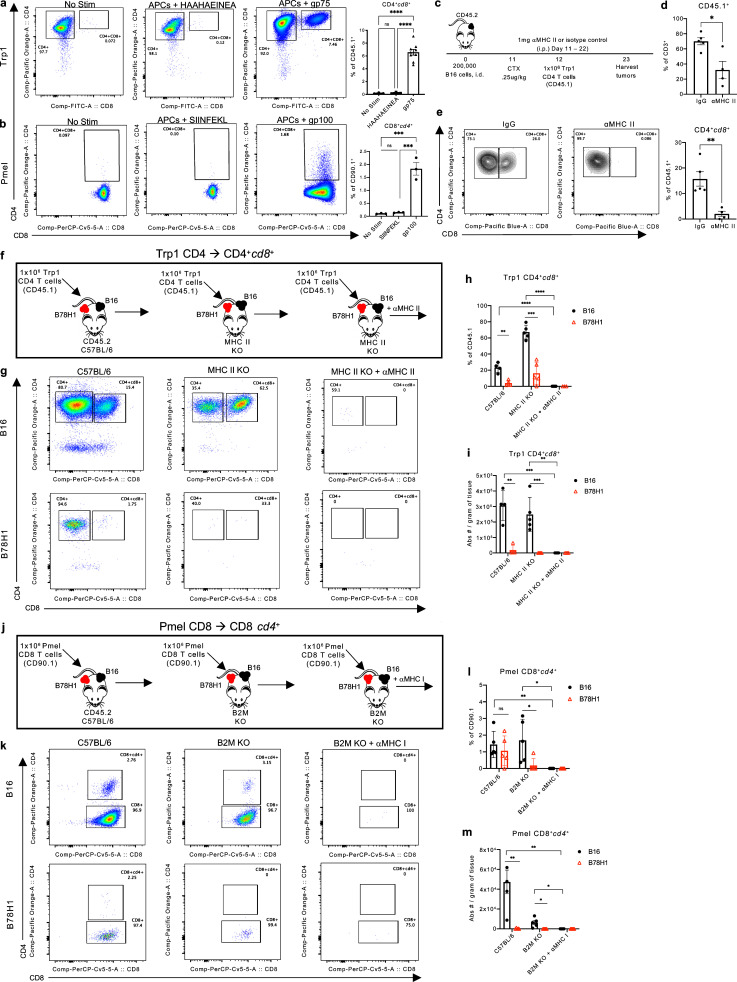
Based on the tumor-specific phenotype and increased number of TCRβ transcripts among CD4+CD8+ T cells, we next assessed TCR signaling as a driving force regulating co-receptor re-expression and CD4+CD8+ differentiation from SP T cells. Following stimulation in vitro with peptide-pulsed APCs, naive CD4+ Trp1 and CD8+ Pmel T cells were capable of inducing the opposite co-receptor when cultured in the presence of their cognate antigen (Fig. 6, a and b). CD4+CD8+ differentiation was not observed when T cells were cultured in the presence of APCs pulsed with an irrelevant peptide. Further, in vivo differentiation of CD4+ Trp1 and CD8+ Pmel T cells to DP T cell states was abrogated when B16 tumor-bearing C57BL/6 mice were treated with anti-MHC class I or MHC class II antibodies (Fig. 6, c–m). Trp1 CD4+cd8+ and Pmel CD4+cd8+ T cell populations were also significantly reduced in B78H1 tumors (a B16-derived cell line lacking MHC expression and gp75 and gp100 antigens) in C57BL/6, MHC class II KO, and B2M KO mice with bilateral B16 and B78H1 tumors (Fig. 6, f–m). Of note, Trp1 CD4+cd8+ T cells were present at an increased frequency within B16 tumors of MHC class II KO compared with C57BL/6 mice (Fig. 6, g–i), suggesting direct stimulation of CD4+ Trp1 T cells by antigen presented on MHC class II in B16 cells (Quezada et al., 2010; Xie et al., 2010) and increased frequency potentially due in part to reduced competition for cytokine with open-repertoire CD4+ T cells. These data point to TCR signaling as a primary mechanism by which antigen-specific T cells re-express the opposite co-receptor in the periphery.
Figure 6.
TCR signaling induces CD4+CD8+ T cell differentiation. (a) Representative plots and percentage of Trp1 CD4+cd8+ T cells following activation with OT-II or gp75-pulsed APCs. (b) Representative plots and percentage of Pmel CD8+cd4+ T cells following activation with SIINFEKL or gp100-pulsed APCs. (c) Experimental schema, as in Fig. 3 a, with daily treatment of 1 mg αMHC class II blocking antibody (M5/114) or isotype control (IgG). (d) Percentage of CD45.1+ T cells in the tumor of IgG and αMHC class II mice. (e) Representative plots and percentage of Trp1 CD4+cd8+ T cells in the tumor of IgG and αMHC class II treated mice 11 d after T cell transfer. (f) Experimental schema of adoptive transfer of antigen-specific CD4 Trp1 T cells into B78H1 or B16 melanoma tumor-bearing C57BL/6 or MHC class II KO mice treated with αMHC class II. (g) Representative plot of CD4+ and CD8+ expression in B16 and B78H1 tumors of mice of each group. (h and i) Percentage (h) and absolute number (i) of Trp1 CD4+cd8+ T cells isolated from the tumor 11 d after T cell transfer. (j) Experimental schema of adoptive transfer of antigen specific CD8 Pmel T cells into B78H1 or B16 melanoma tumor-bearing C57BL/6 or B2M KO mice treated with αMHC class I. (k) Representative plot of CD4+ and CD8+ expression in B16 and B78H1 tumors of mice of each group. (l and m) Percentage (l) and absolute number (m) of Pmel CD8+cd4+ T cells isolated from the tumor 11 d after T cell transfer. Data are representative of three (a–e) or one (f–m) independent experiments. Data are mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, two-sided paired (h, i, l, and m) or unpaired (a, b, and d) Student’s t test.
To clarify the role of CD8 re-expression in determining the acquisition of cytotoxic potential by CD4+ T cells, we further investigated Trp1 T cells in the tumor microenvironment, where these cells displayed maximum induction of the cytotoxic phenotype together with the acquisition of a DP state (Fig. 3 b and Fig. 4 c). Upon transfer into lymphodepleted B16-bearing mice, as in Fig. 3 a, ∼50% of Trp1 CD4+ T cells expressed GzmB within the tumor (Fig. 7 a), where GzmB expression correlated with surface expression of CD8 (Fig. 7 b). Co-expression of GzmB and CD8 was also observed in intra-tumor CD4+ T cells isolated from melanoma patient resections (Fig. 7 c), suggesting that CD8 expression may identify CD4+ T cells gaining in cytotoxic potential. We further investigated the correlation between re-expression of CD8 in CD4+ T cells and the acquisition of cytotoxic potential using single-cell transcriptomic analysis of CD4+ and CD4+cd8+ Trp1 T cells sorted from B16 tumors. Unsupervised clustering of 5,840 cells revealed five distinct clusters, with the majority of cells belonging to either cluster CD4+cd8+ or cluster CD4+ (Fig. 7 d). Both CD4+cd8+ and CD4+ clusters were enriched for Cd4 expression, while CD4+cd8+ was exclusively enriched in Cd8a, Cd8b1, along with cytotoxic markers GzmB, Prf1, and Nkg7 (Fig. 7 d).
Figure 7.
scRNA-seq of Trp1 T cells. (a) Representative expression of CD8 and Granzyme B (GzmB) in CD45.1+ CD4+ Trp1 T cells isolated from B16 tumors 11 d after transfer. (b and c) Pearson correlation analyses of % Gzmb+ and % CD8+ among CD45.1+ CD4+ Trp1 T cells (n = 22; b), or intra-tumor CD4+ T cells isolated from melanoma patient resections (n = 36; c). (d) UMAP (uniform manifold approximation and projection) plot of single cell transcriptomic profiles of sorted CD4+ and CD4+cd8+ Trp1 T cells isolated from 10 pooled B16 tumors 11 d after T cell transfer, with cells colored based on five clusters found by k-means clustering or based on expression of stated gene. Data are representative of three independent experiments (a) or pooled from three independent experiments (b).
To functionally assess the tumoricidal potential of Trp1 T cells re-expressing CD8 in the tumor microenvironment, we compared CD4+cd8+ with CD4+ Trp1 T cells FACS-sorted from B16 tumors in ex vivo cytotoxicity assays using B16 as target cells. CD4+cd8+ Trp1 T cells consistently exerted superior cytotoxicity against B16 melanoma at multiple E:T ratios (Fig. 8 a); however, the cytolytic function remained MHC class II-dependent (Fig. 8 b). We next examined the in vivo contributions of CD4+cd8+ Trp1 T cells in the tumor microenvironment by adoptively transferring naive CD4+ Trp1 T cells into RAG KO mice bearing B16 tumors and by treating with anti-CD8 antibody (2.43) or isotype control (IgG) to selectively deplete CD4+cd8+ Trp1 T cells (Fig. 8 c). Mice lacking CD4+cd8+ Trp1 T cells had diminished capacity to control tumor growth following T cell transfer and had significantly reduced survival (Fig. 8, d and e). Taken together, these results point to CD8 expression in CD4+ T cells as a marker of cytotoxic function.
Figure 8.
Ectopic re-expression of CD8 and CD4 co-receptors drives respective acquisition of cytotoxic and suppressive properties in murine CD4+CD8+ T cells. (a and b) Percentage of target B16 tumor cells lysed ex vivo after 48 h incubation with Trp1 CD4+ T cells or Trp1 CD4+cd8+ T cells isolated from subcutaneous B16 tumors and cultured at the indicated E:T ratios (a), and cultured in the presence of αMHC class I or II blocking antibodies or matched isotype controls (b; n = 3). (c) Experimental schema of adoptive T cell transfer of antigen-specific CD4+ Trp1 T cells from naive mice into B16 tumor-bearing RAG KO mice. Recipient mice were treated with αCD8 depleting antibodies or isotype control IgG starting on day 13 after tumor implantation (n = 10). (d) Tumor area of B16-bearing RAG KO mice following the adoptive transfer of 5 × 105 Trp1 CD4+ T cells and treated with αCD8 depleting antibodies or isotype control IgG (250 μg/2× weekly) starting on day 13 after tumor implantation (n = 10). (e) Overall survival of B16 tumor-bearing RAG KO mice. (f) Representative flow cytometry histogram (left) and quantification (right) of Foxp3 expression in open repertoire (OR) CD4+ T cells and Pmel CD8+ and CD8+cd4+ T cells isolated from the tumor of B16-bearing mice. (g and h) Percentage of target B16 tumor cells lysed ex vivo after 48 h incubation with Pmel CD8+ T cells or Pmel CD8+cd4+ T cells isolated from subcutaneous B16 tumors and cultured at the indicated E:T ratios (g), and cultured in the presence of anti-MHC class I or II blocking antibodies or matched isotype controls at 1:1 E:T ratio (h). (i) Schema of in vitro suppression assay with CTV-labeled CD8+ CD45.1+ target T cells co-cultured with CD8+ Pmel or CD8+cd4+ Pmel T cells FACS-sorted from B16 tumors, and with CD4+ Foxp3+ (Tregs) FACS-sorted from spleens of naive Foxp3-GFP mice as controls. (j) Quantification of E:T ratios following 48 h of incubation. (k) Percentage of IFN-γ+ TNF-α+ and expression of CD44 (MFI) in CD45.1+ CD8+ target T cells in the indicated co-cultures at 1:1 E:T ratio after 48 h incubation. (l) CTV dilution quantification (left) and representative histograms (right) in total CD45.1 CD8+ target T cells following culture with OR CD4+ Tregs, Pmel CD8+, or Pmel CD8+cd4+ T cells at the indicated E:T ratios after 48-h incubation. Data are representative of three independent experiments. Data with error bars are mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, two-sided unpaired Student’s t test.
We next sought to determine the functional role of ectopic expression of CD4 and Treg profiles in CD8+ Pmel T cells upon antigen encounter in B16 tumors (Fig. 3 b and Fig. 4 c), where, in our model of adoptive transfer (Fig. 3 a), ∼30% of Pmel CD8+cd4+ T cells express Foxp3 protein detectable by flow cytometry (Fig. 8 f). In parallel, cytotoxicity assays with the Trp1 cell subsets, CD8+cd4+ and CD8+ Pmel T cells, and CD8+cd4+ Pmel T cells had a modest decline in cytotoxic potential and remained MHC class I restricted (Fig. 8, g and h). Using ex vivo suppression assays with CTV-labeled CD8 T cells as targets, we compared tumor-derived Pmel CD8+ and Pmel CD8+cd4+, with bona fide Tregs (Foxp3+CD4+) as a positive control for suppression (Fig. 8, i and j). CD8+cd4+ Pmel T cells significantly reduced the production of inflammatory cytokines TNF-α and IFN-γ (Fig. 8 k) and suppressed target cell proliferation (Fig. 8 i).
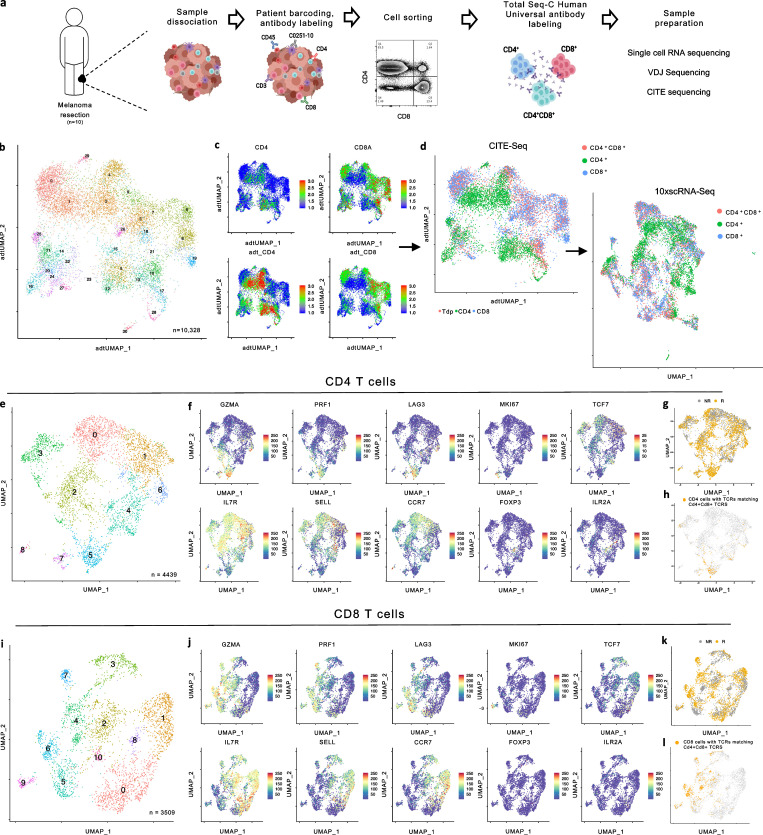
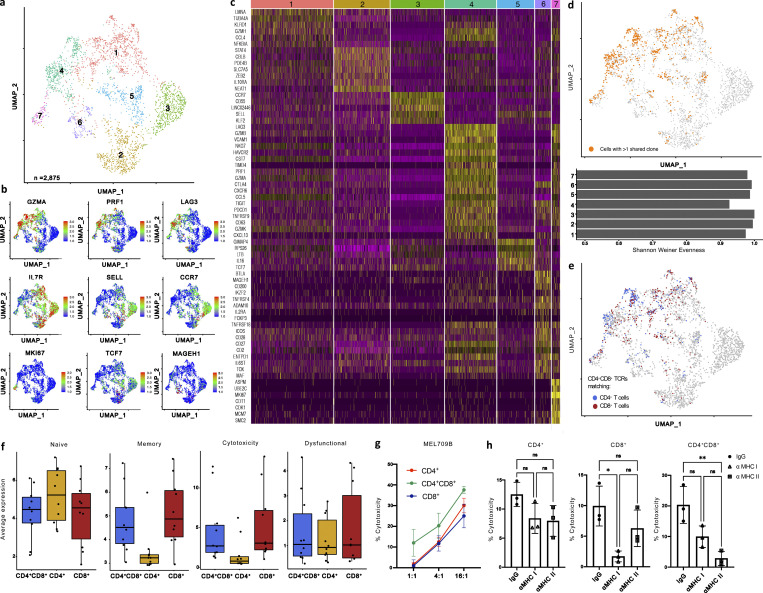
With unique gene signatures for CD4-derived CD4+cd8+ and CD8-derived CD8+cd4+ T cells, we next evaluated whether we could distinguish similar subsets of DP T cells among patient TILs. To assess this, CD4+, CD8+, and CD4+CD8+ T cells were FACS-sorted in equal numbers from 10 immunotherapy naive melanoma tumor samples (Table S2) for 10× scRNA, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq), and VDJ sequencing analyses (Fig. 9 a). Initial unsupervised clustering of 10,328 cells by CITE-seq surface markers revealed 29 distinct clusters of T cells, which were then mapped back to a scRNA-based UMAP projection. We identified and labeled CD4+CD8+ T cells as having >0.5 scaled protein expression for both Cd4 and Cd8a, and assessed the resulting 2,875 CD4+CD8+, 4,439 CD4+, and 3,509 CD8+ T cells by re-clustering each cell type independently (Fig. 9, b–l and Fig. 10 a). After re-clustering the CD4+CD8+ T cells, seven DP subclusters were identified. Cytotoxic markers Prf1, Gzmk, Gzmb, Gzma, and Nkg7 were enriched in clusters 1 and 4 specifically, while cluster 4 simultaneously expressed a number of exhaustion markers, including Lag3, Pdcd1 (PD-1), Havcr2 (Tim-3), and Entpd1 (CD39; Fig. 10, b and c). Clusters 2 and 3 were enriched for tissue-resident memory and naive T cell markers, respectively, and Cluster 6 had elevated expression of costimulatory markers, including Tnfrsf4 (OX40), Btla, Cd28, Icos, and Tnfrsf18 (GITR), as well as the Treg markers Foxp3 and Il2ra.
Figure 9.
Paired CITE-seq and scRNA-seq based analysis of melanoma patient TILs. (a) Experimental schema for sequencing of CD4+, CD8+, and CD4+CD8+ T cells collected from 10 ICB treatment naive melanoma patient single cell suspension samples. (b) UMAP plot of CITE-seq profiles of sorted T cells collected from 10 melanoma patient single cell suspension samples, cells colored based on 29 clusters found by k-means clustering. (c) UMAP projection of Cd4 and Cd8a expression by scRNA (top) or CITE-seq (bottom). (d) UMAP projection of cells labeled as CD8+ (blue), CD4+ (green), or CD4+CD8+ (red; left) and UMAP projection by scRNA preserving CITE-seq identify labels (right). (e) UMAP plot of single-cell transcriptomic profiles of sorted CD4+T cells collected from 10 ICB treatment naive melanoma patient single-cell suspension samples; cells colored based on eight clusters found by k-means clustering. (b) UMAP projections of CD4+ T cells with coloring based on expression for Gzma, Prf1, Lag3, Il7r, Sell, Ccr7, Mki67, Tcf7, Mageh1, and Foxp3. (g and h) UMAP projection of CD4+ T cells with cells colored based on response to immunotherapy (g) or with cells colored based on shared TCRs with CD4+CD8+ T cells (h). (i) UMAP plot of single-cell transcriptomic profiles of sorted CD8+ T cells collected from 10 ICB treatment naive melanoma patient single-cell suspension samples; cells colored based on eight clusters found by k-means clustering. (j) UMAP projections of CD8+ T cells with coloring based on expression for Gzma, Prf1, Lag3, Il7r, Sell, Ccr7, Mki67, Tcf7, Mageh1, and Foxp3. (k and l) UMAP projection of CD8+ T cells with cells colored based on response to immunotherapy (k) or with cells colored based on shared TCRs with CD4+CD8+ T cells (l).
Figure 10.
Human CD4+CD8+ T cells are clonally expanded and cytotoxic. (a) UMAP plot of single cell transcriptomic profiles of sorted CD4+CD8+ T cells collected from 10 ICB treatment naive stage IV melanoma patient single-cell suspension samples; cells colored based on seven clusters found by k-means clustering. (b) UMAP of CD4+CD8+ T cells displaying scaled expression for Gzma, Prf1, Lag3, Il7r, Sell, Ccr7, Mki67, Tcf7, and Mageh1. (c) Heatmap displaying scaled expression values of discriminative gene sets per cluster as defined in a. (d) UMAP projection of CD4+CD8+ T cells with cells colored based on >1 shared TCRs among CD4+CD8+ T cells (top); Shannon-Weiner evenness score measuring evenness of TCR composition among cell clusters (bottom). (e) UMAP projection of CD4+CD8+ T cells with cells colored based on shared TCRs with CD4+ T cells (blue) or CD8+ T cells (red). (f) Average expression of naive, memory, cytotoxicity, and dysfunctional gene signatures in CD4+, CD4+CD8+, and CD8+ T cells. (g and h) Cytotoxicity of a melanoma cell line derived from MEL 709B after 36 h incubation with FACS-sorted CD4+, CD8+, or CD4+CD8+ T cells obtained from the same cryopreserved tumor. Different E:T ratios were evaluated (g) and cytotoxicity in the presence of αMHC class I or II blocking antibodies or matched isotype controls were assessed (h). Data with error bars are represented as mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01, two-sided unpaired Student’s t test (g).
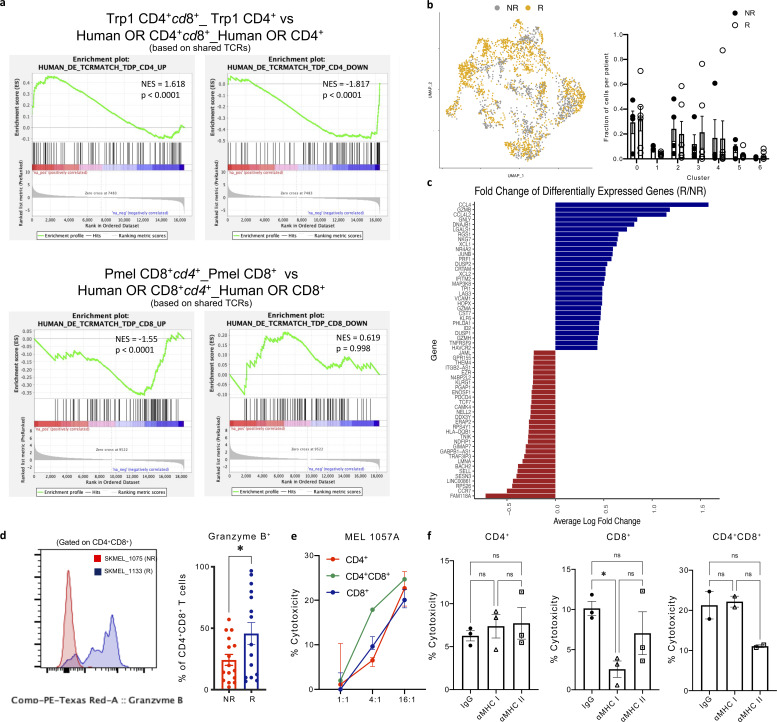
We reconstructed TCR sequences to assess TCR clonality based on shared CDR3 ⍺ and β chains and observed enriched representation of the same TCRs (detected in more than one T cell in a single patient sample; Sade-Feldman et al., 2019) predominantly among clusters 1 and 4 (Fig. 10 d), which is consistent with our hypothesis of antigen-driven CD4+CD8+ T cell differentiation that is associated with a highly cytotoxic state. Additionally, clusters 1 and 4 had reduced evenness of their TCR composition as measured by the Shannon-Weiner index (Fig. 10 d). Further, CD4+CD8+ TCRs shared with SP CD4+, and CD8+ T cells were present predominantly within clusters 1 and 4 (Fig. 10 e). To assess whether the CD4- and CD8-derived CD4+CD8+ T cell states we identified in the murine setting were consistent with human CD4+CD8+ phenotypes, we examined the DEGs between CD4+ and CD4+cd8+ T cells as well as CD8+ and CD8+cd4+ T cells in our data set based on shared TCRs between the SP and DP T cell populations. The GSEA analysis revealed significant similarities of gene expression changes between murine transgenic and human CD4+cd8+ T cell populations compared with matched CD4+ T cells (Fig. S3 a). However, we did not observe any significant similarity between CD8+cd4+ T cells compared with matched CD8+ T cells in our murine transgenic and human T cell populations (Fig. S3 a), suggesting that the regulatory T cell signature induced in the murine CD8-derived CD8+cd4+ T cells may be differentially regulated in mice and humans.
Figure S3.
Identification of CD4+CD8+ T cell states associated with clinical outcome. (a) GSEA of DEGs in human open repertoire CD4+cd8+ vs. CD4+ T cells (left, UP; right, DOWN) shows significantly concordant differences in the same direction as murine Trp1 CD4+cd8+ vs. Trp1 CD4+ T cells (top); GSEA of DEGs in human open repertoire CD8+cd4+ vs. CD8+ T cells (left, UP; right, DOWN) shows significantly concordant differences in the same direction as murine Pmel CD8+cd4+ vs. Pmel CD8+ T cells (bottom). (b) UMAP projection of CD4+CD8+ T cells with cells colored based on response to immunotherapy (left), percentage of cells in each cluster per patient (right) by response status: non-responder (NR), responder (R). (c) Average log fold change of top 20 DEGs (P < 0.05) among CD4+CD8+ T cells from responders and non-responders. (d) Representative protein expression of GzmB in CD4+CD8+ T cells isolated from SKMEL_1075 (NR) and SKMEL_1133 (R); frequency of GzmB+ CD4+CD8+ T cells within melanoma patient resections (n = 33 patients: 16 NR, 17 R). (e and f) Percentage of target patient melanoma cells lysed ex vivo after 36 h incubation with CD4+, CD8+, or CD4+CD8+ T cells FACS-sorted from frozen single cell suspension and cultured at the stated E:T ratios (e) and in the presence of αMHC class I or II blocking antibodies or matched isotype controls (f). Data are mean ± SEM. *, P ≤ 0.05, two-sided unpaired Student’s t test (d and f).
While there was no correlation between the presence of specific CD4+CD8+ T cell clusters with patients’ response to ICB therapy (Fig. S3 b), we observed an enrichment in cytotoxic genes including Gzmb, Gnly, Nkg7, Prf1, and Gzma in CD4+CD8+ T cells isolated from responders, whereas CD4+CD8+ T cells from non-responders had increased expression of genes linked to T cell memory markers, including Sell and Ccr7 (Fig. S3, c and d). To evaluate the phenotype of CD4+CD8+ T cells relative to SP CD4+ and CD8+ T cells, we assessed the expression of genes associated with previously published T cell states (van der Leun et al., 2020) and found that CD4+CD8+ T cells had similar memory and cytotoxic signatures to CD8+ T cells, while CD4+ T cells were enriched for naive markers (Fig. 10 f).
We next investigated whether CD4+CD8+ T cells in melanoma tumors were enriched for tumor-reactive T cells by assessing their functional relevance in ex vivo cytotoxicity assays. CD4+, CD4+CD8+, and CD8+ T cells were FACS-sorted from frozen single-cell suspensions generated from patients’ resected tumors. T cells were cultured in the presence of IL-2 and CTV-labeled autologous tumor cells in an imaging-based cytotoxicity assay, where cell death was assessed by cytotox red (Oh et al., 2020). CD4+CD8+ mediated tumor cell death was observed in two patient samples at rates comparable with or superior to CD4+ and CD8+ T cell-mediated killing (Fig. 10 g and Fig. S3 e). Furthermore, the cytotoxic activity of CD4+CD8+ T cells was at least partially dependent on MHC class I and II recognition as cell death was reduced in the presence of pan anti-MHC class I (HLA-A, B, C) and anti-MHC II (HLA-DR, DP, DQ) antibodies (Fig. 10 h and Fig. S3 f), whereas CD4+ T cell killing was less dependent on MHC recognition. These data demonstrate that CD4+CD8+ TILs have heightened cytolytic capacity and are enriched for tumor reactive TCRs.
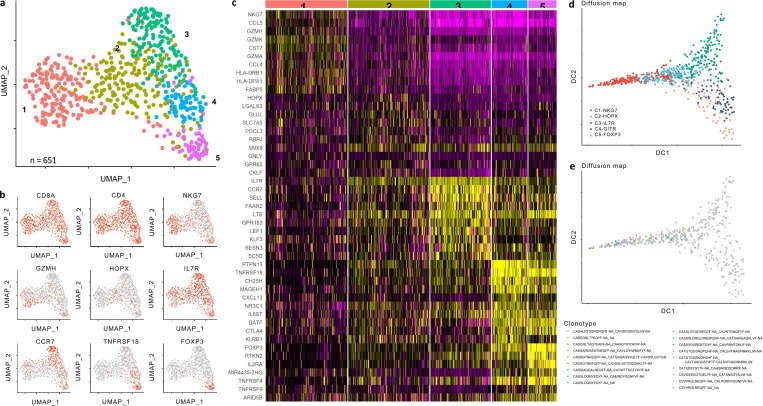
In an independent cohort of scRNA data generated from TILs isolated from nine NSCLC patients previously treated with neoadjuvant nivolumab (αPD-1) prior to resection, unsupervised analysis of 651 DP T cells revealed five distinct clusters (Fig. S4 a). Consistent with the phenotypes of intra-tumor DP T cells from melanoma patients, we observed enrichment in cytotoxic molecules (Cluster 1) and naive markers (Cluster 2). In addition, we identified co-stimulatory and Treg markers enriched in Clusters 4 and 5, which were not abundant in our melanoma data and likely can be attributed to heterogeneity arising from the patient, tumor, and treatment variety (Fig S4 c). Further, pseudotime analysis projected diffusion of Cluster 1 and Clusters 4 and 5 in opposite components from central Clusters 2 and 3, suggesting distinct differentiation pathways of cytotoxic and suppressive DP T cell phenotypes (Fig. S4 d). In addition, 16 of the 18 clonally expanded TCRs were predominantly observed in Cluster 1, further suggesting antigen-driven expansion among this cytotoxic cluster (Fig. S4 e). Taken together, these data demonstrate the unique and diverse functional states of DP T cells, highlighting the multiple modes of CD4+CD8+ T cell differentiation within the tumor microenvironment.
Figure S4.
CD4+CD8+ T cell analysis in NSCLC patient tumors. (a) UMAP plot of single cell transcriptomic profiles of sorted CD4+CD8+ T cells collected from nine NSCLC patients treated with two doses of nivolumab prior to surgery, with cells colored based on five clusters found by k-means clustering. (b) UMAP projections of CD4+CD8+ T cells with coloring based on expression of Cd8a, Cd4, Nkg7, Gzmh, Hopx, Il7r, Ccr7, Tnfrsr18, and Foxp3. (c) Heatmaps displaying scaled expression values of discriminative gene sets per cluster as defined in a. (d) Diffusion component plot displaying clusters over diffusion component 1 (x axis) or diffusion component 2 (y axis), with cells colored based on cluster identity. (e) Clonal TCRs (defined as clonotypes with n > 2) overlaid on diffusion map projection from (d). Of the 18 clonotypes, 16 clones were present in the C1-NKG7 cluster.
Discussion
In summary, our findings reveal divergent differentiation and functions of CD4+CD8+ T cells from SP CD4 and CD8 T cell states specifically in the tumor setting, highlighting the plasticity of lineage commitment mechanisms in the periphery. We find that ectopic expression of CD8 in CD4+ T cells is mediated through distinct transcriptional and epigenetic modulation induced via TCR signaling that correlates with activation of the cytotoxic program in conventional CD4+ T cells to drive a potent cytolytic population of MHC class II–restricted CD4+cd8+ T cells. Notably, enrichment of clonally expanded CD4+ T cells with cytotoxic effector function has previously been reported in human melanoma and urothelial bladder cancer tumors, with a gene signature predictive of the response to PD-1 blockade (Sledzinska et al., 2020; Oh et al., 2020). Our findings suggest that this population of clinically relevant cytotoxic CD4+ T cells can be identified by co-expression of CD8. In parallel, we identified a population of CD8-derived CD8+cd4+ T cells in mice that significantly overexpressed genes associated with suppressive CD4 T cell states, including the Treg maker Foxp3, and displayed unique polyfunctionality, maintaining cytotoxic properties while limiting effector T cell functions and proliferation.
While we demonstrated TCR signaling to be a primary mechanism influencing co-receptor re-expression in the periphery, our data do not exclude the possibility of non-antigen reactive bystander CD4+CD8+ T cells within an open-repertoire population. In fact, our TCR sequencing analysis of CD4+, CD8+, and CD4+CD8+ T cells revealed that clonal expansion was only observed among the CD4+CD8+ TCRs shared with CD4+ or CD8+ T cells, and it was seemingly absent among unique CD4+CD8+ TCR clones. Further, patient-derived CD4+CD8+ TILs had clonally expanded TCRs shared with CD4+ and CD8+ TCRs specifically in Clusters 1 and 4, enriched for activation and cytotoxicity markers, while few to no clonal or shared TCRs were observed in Clusters 2 or 3 with heightened expression of naive and memory markers. These data are supportive of three distinct populations of CD4+CD8+ T cells: antigen-specific CD4+ T cells re-expressing CD8, antigen-specific CD8+ T cells re-expressing CD4, and antigen-irrelevant bystander CD4+CD8+ T cells that may have escaped thymic selection or persisted in a memory state from a prior immune response.
Our findings further illustrate the functional relevance of DP T cells specifically within melanoma patient tumors where CD4+CD8+ T cells displayed enhanced cytotoxic potential against patient-derived target cell lines in an MHC-restricted manner. While murine CD8-derived CD8+cd4+ T cells had increased expression of Foxp3 and exhibited modest suppressive features in vitro, human CD8+cd4+ T cells (identified by shared TCRs with CD8+ T cells) did not demonstrate this phenotype, but rather displayed a profile commensurate with traditional cytotoxic CD8 T cells. This differential regulation of Foxp3 between a mouse and a human suggests that the bulk CD4- and CD8-derived CD4+CD8+ T cell population in humans is highly cytolytic and uniquely capable of recognizing antigens presented on both MHC class I and II. Despite their low frequency, these cells may play a pivotal role in tumor rejection as previous reports have demonstrated that very low levels of cytolytic T cells are influential in tumor eradication (Budhu et al., 2010). Further, the exclusive expression of activation markers within this clonally expanded population serves as an identity marker of T cells with enhanced tumor specificity that may allow for the selection of antigen reactive clones or whose frequency may inform the response to therapy. However, further analysis into the longevity of these cells and special attention to their inclusion during the sequencing and analysis of TILs is required.
The present study demonstrates the plasticity and variability of lymphocyte lineages in response to disease and environmental stimuli, suggesting that the investigation of T cell fates should not consider the expression of immune cell markers as fixed, but rather take into consideration their transcriptional and epigenetic modulation during differentiation in response to antigen stimulation. Our data provide a novel framework for improving the identification and selection of antigen-reactive T cells in the context of immune-based therapies such as adoptive cell transfer, and the definition of relevant pathways for genetically engineering T cells with the desired functionality for the treatment of cancer and other immune-related diseases.
Materials and methods
Mice and tumor cell lines
All mouse procedures were performed in accordance with institutional protocol guidelines at Memorial Sloan Kettering Cancer Center (MSK). C57BL/6J, CD45.1+ C57BL/6J, C57BL/6J Rag1−/−, Nur77-GFP (stock no: 016617), MHC class II KO (stock no: 003584), and B2M KO (stock no: 002087) 8–10 wk males were obtained from the Jackson Laboratory. Pmel-1/gp100-specific CD8 TCR and Trp1/gp-specific CD4 TCR transgenic mice were generously provided by Dr. Nicholas Restifo (National Cancer Institute, Bethesda, MD; Overwijk et al., 2003; Mucida et al., 2013). Trp1 CD4+ TCR transgenic mice were backcrossed to Rag1−/− Trp1−/− CD45.1 background. Foxp3-GFP mice were generously provided by Dr. Alexander Rudensky (MSK, New York, NY) and backcrossed to C57BL/6. All mice were bred at MSK. Mice were maintained according to the National Institutes of Health Animal Care guidelines under a protocol approved by the MSK Institutional Animal Care Committee. Littermates of the same age (8–10 wk-old) and the same sex were randomly assigned to experimental groups. The B16F10 mouse melanoma cell line was originally obtained from I. Fidler (M.D. Anderson Cancer Center, Houston, TX), B78H1 mouse melanoma cell line was originally obtained from A. Albino (Sloan Kettering Institute, New York, NY) and cultured in RPMI 1640 medium supplemented with 7.5% inactivated FBS, 1× nonessential amino acids, and 2 mM L-glutamine. We confirmed the expression of melanoma differentiation antigens in B16F10 melanoma cells. Cells were routinely screened to avoid mycoplasma contamination and maintained in a humidified chamber with 5% CO2 at 37°C for up to 1 wk after thawing before injection in mice.
Patient material
All the patients signed an approved informed consent before providing tissue samples. Patient samples were collected on a tissue-collection protocol approved by the MSK Institutional Review Board. Patients’ peripheral blood mononuclear cells were isolated from whole blood collected in cell preparation tubes containing sodium heparin (BD Vacutainer) according to the manufacturer’s instruction and cryopreserved in 10% DMSO FBS until use. Single-cell suspensions from patients’ tumors were obtained by digesting the tumor samples with type I collagenase (2 mg/ml), type V hyaluronidase (2 mg/ml), and type IV deoxyribonuclease I (200 U/ml) in serum-free RPMI 1640 using a GentleMACs Octo Dissociator (Miltenyi Biotech) and viably frozen at −80°C prior to use. Sorted TILs were maintained in a human-complete medium (RPMI, 10% human serum, 1% penicillin with streptomycin, 2 mM L-glutamine, 0.1% amphotericin) + 100 IU/ml of human recombinant IL-2 (#200-02; Peprotech). The patient radiographical response was determined by RECIST v1.1.
In vivo tumor implantation and treatments
B16F10 melanoma cells (2 × 105 cells) were implanted subcutaneously in 0.2 ml of matrigel (Matrigel Matrix Growth Factor Reduced; Becton Dickinson) for immune infiltrate analyses or implanted intradermally in 0.1 ml PBS for tumor growth and survival analyses. Cyclophosphamide monohydrate (Sigma-Aldrich) mixed in sterile PBS was administered intraperitoneally as a single dose at 250 mg/kg once the tumors were established (1–2 wk later). 1 d after cyclophosphamide injection, Trp1 CD4+ T cells (CD45.1) and Pmel-1 CD8+ T cells (CD90.1) were purified from the lymph nodes and spleen of male mice by positive selection magnetic cell sorting using CD4 beads (L3T4) or CD8 (Ly-2; Miltenyi Biotech) according to the manufacturer’s instructions. 1 × 106 purified Trp1 CD4+ T cells or Pmel-1 CD8+ T cells were intravenously injected into the tail vein in 100 μl PBS. For the adoptive transfer of polyclonal T cells, 1 × 106 FACS-sorted CD4 T cells (from CD45.1 C57BL/6) and 1 × 106 FACS-sorted CD8+ T cells (from CD45.2 C57BL/6) were mixed at a 1:1 ratio and intravenously injected via the tail vein into RAG−/− mice with established B16 tumors. Treatment was with αCD8 depleting antibodies (clone 2.43; BioXcell, 250 µg/injection i.p., 2× weekly), αMHC class II (clone M5/114; BioXcell, 1 mg/injection i.p., daily), αMHC class I (clone H2; BioXcell, 1 mg/injection i.p., daily), or matched isotype control IgGs (BioXcell). Tissues were harvested at indicated time points.
Flow cytometry and cell sorting
Tumors were dissociated after 30 min incubation with Liberase TL and DNAse I (Roche) and passed through a 40-µM filter to obtain single-cell suspensions. Lymphocytes were enriched by Percoll (GE Healthcare) gradient centrifugation. Cells from lymph nodes and spleens were prepared by mechanical dissociation on 40-µM filters and RBC lysis (ammonium-chloride-potassium [ACK] buffer; Lonza). Mouse peripheral blood was collected by retro-orbital puncture and the RBCs were lysed with ACK buffer. Surface staining of mouse cells was performed on ice in staining buffer (2% BSA and 2 mM EDTA in PBS) for 20 min after 15 min of pre-incubation with anti-mouse CD16/CD32 Ab (clone 2.4G2; BD Biosciences) to block Fcγ receptor binding, with panels of appropriately diluted fluorochrome-conjugated Abs (from BD Biosciences, eBioscience, or Invitrogen) against the following mouse proteins in different combinations: CD45.2 (clone 104), CD45.1 (clone A20), CD3 (clone 145-2C11), CD4 (clone RM4-5), CD8 (53-6.7), CD90.1 (clone OX-7), Runx3 (R3-5G4), ThPok (clone T43-94), CD44 (clone IM7), CD25 (clone PC61.5), PD-1 (clone J43), and an eFlour506 fixable viability dye. For intracellular staining, mouse cells were fixed and permeabilized (Foxp3 fixation/permeabilization buffer; eBioscience) and incubated with appropriately diluted PE-labeled anti-Runx3 (R3-5G4), Alexa647-labeled anti-ThPok (clone T43-94), FITC-labeled anti-Ki67 (clone SolA15), PE Texas Red-labeled anti-Granzyme B, PerCP-Cyanine5.5-labeled anti-T-bet (clone 4B10), eFlour450-labeled anti-Eomes (clone Dan11mag), and AF700-labeled anti-Foxp3 (clone FJK-16s). Surface staining of human immune cells was performed in the presence of the Fcγ receptor blocking reagent (Miltenyi Biotech) in staining buffer with proper dilutions of fluorochrome-conjugated Abs (from BD Pharmingen or BioLegend) against the following human proteins in different combinations: CD45 (clone 2D1), CD3 (clone SK7), CD4 (clone OKT4), CD8 (clone SK1), CD39 (clone A1), PD-1 (clone EH12.2H7), and an eFlour506 fixable viability dye. For intracellular staining, human cells were fixed and permeabilized (Foxp3 fixation/permeabilization buffer; eBioscience) according to the manufacturer’s instructions and then incubated on ice for 20 min with appropriately diluted FITC-labeled anti-Ki67, PE-labeled anti-Foxp3 (clone 259D/C7), and PE Texas Red-labeled anti-Granzyme B. Flow cytometry was performed on an LSRII (BD). FlowJo software (version 10.7.1) was used for all flow cytometry analyses.
Mouse T cell subsets were sorted from B16 tumors harvested from C57BL/6 mice following enrichment by Percoll gradient centrifugation. Naive CD4 and CD8 T-cell subsets were sorted from CD45.1 and CD45.2 C57BL/6 and Foxp3-GFP mice by using pre-enriched splenocytes (CD4 or CD8 Microbeads; Miltenyi Biotech). Following incubation with anti-mouse CD16/CD32 Abs, the samples were stained with anti-CD45.1, anti-CD90.1, anti-CD3, anti-CD4, and anti-CD8 in different combinations depending on the populations to isolate. DAPI was added to the stained samples immediately before acquisition. Human CD4+, CD8+, and CD4+CD8+ T cells were sorted directly from thawed single-cell suspensions following incubation with Fcγ receptor blocking reagent (Miltenyi Biotech) and staining with anti-CD45, anti-CD3, anti-CD4, and anti-CD8 Abs, and DAPI immediately before acquisition. FACS sorting was conducted on a FACSAria II cell sorter (BD Biosciences). After gating according to lymphocyte morphology, excluding doublets and dead cells, CD45+ CD3+ cells were sub-gated into CD4+, CD8+, and CD4+CD8+ to sort the indicated populations from mouse and human tissues.
In vitro assays
Murine cytotoxicity assays were performed ex vivo following a previously described clonogenic assay (Budhu et al., 2010). Briefly, TRP1 CD4+ and TRP1 CD4+CD8+ T cells were FACS-sorted from B16 tumors harvested from C57BL/6 mice that had previously received adoptive transfer of naive TRP1 CD4 T cells and were incubated at the indicated ratios in a 24-well plate with 10,000 B16F10 cells (target) for 48 h at 37°C in RPMI 1640 medium supplemented with 10% inactivated FBS, 1× nonessential amino acids, 2 mM L-glutamine, and 100 U/ml recombinant murine IL-2 (Millipore). Where indicated, MHC class I (clone Y-3), MHC class II (clone M5/114), or isotype control (Clone LTF-2) was added to the culture at 10 µg/ml. After 48 h, the T cells and dead target cells were removed by collecting the supernatant, the plate was washed 1× with PBS, and 250 μl of trypsin was added to each well to detach the remaining targets. Then, 750 μl of T cell culture medium was added to each well and the resulting solution was diluted 1:100 in a T-cell culture medium. 100 μl of the final dilution was plated in each well of a 6-well tissue culture plate containing 5 ml T cell culture medium. The plates were incubated at 37°C for 7 d in a 95% air 5% CO2 humidified atmosphere to allow B16 cells to form macroscopic colonies, washed with PBS, treated with 2 ml of 3.7% formaldehyde in PBS for 15 min to fix the B16 cells, washed again with PBS, incubated for 2 h in 2 ml of 4% wt/vol methylene blue in H2O at room temperature, washed with distilled water to remove excess methylene blue, dried, and the blue-stained colonies counted manually as previously described (Budhu et al., 2010).
Activation assays with mouse T cells were performed by incubating naive Trp1 CD4+ or Pmel CD8+ T cells at a 1:1 ratio with peptide-pulsed mouse splenocytes in vitro for 3 d in the presence of IL-2. Briefly, a mouse spleen (CD45.1+ C57BL/6) was homogenized to generate a single-cell suspension, and the released cells were pelleted and resuspended in 3 ml of ACK lysis buffer (Lonza) to lyse RBCs. The splenocytes were washed, resuspended at 1 × 106 cell/ml in T-cell growth medium containing gp75 peptide, gp100 peptide, or OVA peptide (0.75 µg/ml). 100 μl of peptide-pulsed APCs were added to 100 μl of Trp1 CD4+ (CD45.1) or Pmel CD8+ (CD90.1) T cells (1 × 106 cell/ml) and incubated in a 96-well plate (Corning) at 37°C for 3 d in a 95% air 5% CO2 humidified atmosphere. On day 3, cells were collected and stained for analysis of double-positive T cells by flow cytometry.
Suppression assays with mouse T cells were performed by incubating 10,000 CD4+ Foxp3-, CD4+Foxp3+, PMEL CD8+, or PMEL CD4+CD8+ T cells at a 1:1 ratio with CellTrace Violet (CTV; Invitrogen)–labeled target T cells that were magnetic-activated cell sorting (MACS)–purified (CD4 or CD8 Microbeads) and FACS-sorted from spleens of CD45.1+ C57BL/6 congenic mice or Foxp3-GFP mice. PMEL populations were FACS-sorted from Percoll-enriched B16 tumors harvested from C56BL/6 mice that had previously received adoptive transfer of naive PMEL CD8 T cells. Cultures were stimulated for 48–72 h with 0.5 µg/ml soluble anti-CD3 Ab and irradiated splenocytes before analysis of target CTV dilution (proliferation) and CD25 and CD44 upregulation (activation) by flow cytometry.
Human cytotoxicity assays were adapted from Oh et al. (2020) and carried out with samples from two independent patients. Primary cancer cells from paired patient samples were freshly thawed from frozen aliquots and expanded in culture. Cells were labeled with CTV (Invitrogen) prior to plating. To achieve various effector-to-target (E:T) ratios, 1,000 CTV labeled target cells were suspended in a complete human medium and seeded into each well of a 384-well plate (#781946; Greiner). Different ratios of freshly thawed and sorted CD4+, CD8+, and CD4+ CD8+ TILs were serially diluted and added to the corresponding wells in triplicate. Each well contained 100 μl of medium supplemented with 2 µM Cytotox Red (#4632; Essen Bioscience). For MHC class I and MHC class II blockade, 10 µg of blocking antibody (MHC class II: clone Tu39; BD Biosciences, MHC class I: clone W6/32, MSKCC Core) was added to wells containing cancer cells and cultured at 37°C for 1 h prior to co-culture with TILs. Cell culture was monitored by Celigo Imaging Cytometer at 6, 18, 24, and 36 h. Cell viability was assessed as the percentage of CTV+ Cytotox-target cells of the total CTV+ population based on a minimum area threshold of 75 µm2, and background cell death (cell death observed in target cells cultured alone) was subtracted from the total.
Single cell imaging
Approximately 200,000 CD4+, CD8+, and CD4+CD8+ T cells were sorted by a FACSAria II and pooled in 50 μl PBS/2% FBS. Images were taken on an Amnis ImageStreamX Mark II Imaging flow cytometry and analyzed in IDEAS software.
Sample preparation for RNA sequencing (RNA-seq) and ATAC sequencing (ATAC-seq)
Samples were isolated from three independent experiments of pooled spleens (n = 3) or tumors (n = 5–8) as follows: (i) Naive TRP1 CD4+ T cells and naive PMEL CD8+ T cells were sorted by flow cytometry from spleens of Trp1 and Pmel-1 transgenic mice. (ii) TRP1 CD4+ and TRP1 CD4+CD8+ were sorted from B16 tumors of C57BL/6 mice that had been adoptively transferred with naive TRP1 CD4+ T cells 11 d prior. (iii) PMEL CD8+ and PMEL CD4+CD8+ were sorted from B16 tumors of C57BL/6 mice that had been adoptively transferred with naive PMEL CD8+ T cells 11 d prior. Cells were gated on congenic labels (TRP1 – CD45.1, PMEL – CD90.1), CD3+, CD4+CD8− (TRP1), CD4+CD8+ (TRP1 and PMEL), or CD4−CD8+ (PMEL). After flow sorting, all samples for downstream ATAC-seq analysis were frozen in 10% DMSO and stored at −80°C; samples for RNA-seq were directly sorted into Trizol (Invitrogen), adjusted to 500 μl total volume and frozen and stored at −80°C.
Single-cell transcriptome sequencing
Cryopreserved melanoma patient samples were individually hash-tagged and labeled with the TotalSeq-C human universal cocktail to enable protein detection of genes by cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq). Sorted T cells were stained with Trypan blue, and Countess II Automated Cell Counter (Thermo Fisher Scientific) was used to assess both cell number and viability. Following QC, the single-cell suspension was loaded onto Chromium Chip A (10X Genomics; PN 230027), and GEM generation, cDNA synthesis, cDNA amplification, and library preparation of 5,800–7,000 cells with 57% viability proceeded using the Chromium Single Cell 5′ Reagent Kit (10X Genomics PN 1000006) according to the manufacturer’s protocol. cDNA amplification included 13 cycles and 50 ng of the material was used to prepare sequencing libraries with 14 cycles of PCR. Indexed libraries were pooled equimolar and sequenced on a NovaSeq 6000 in a 28 bp/91 bp paired end run using the NovaSeq 6000 SP Reagent Kit (100 cycles; Illumina). An average of 273 million paired reads was generated per sample.
Single-cell V(D)J analysis from RNA
An aliquot of cDNA generated using the methods described above was used to enrich for V(D)J regions using the Chromium Single Cell V(D)J Enrichment Kit Human T Cell (10X Genomics; PN 1000005) according to the manufacturer’s protocol with 10 cycles of PCR during enrichment and nine cycles during library preparation. Indexed libraries were pooled equimolar and sequenced on a NovaSeq 6000 in a 150 bp/150 bp paired-end run using the NovaSeq 6000 S4 Reagent Kit (300 cycles; Illumina). An average of 107 million paired reads was generated per sample.
Cell surface protein feature barcode analysis
Amplification products generated using the methods described above included both cDNA and feature barcodes tagged with cell barcodes and unique molecular identifiers. Smaller feature barcode fragments were separated from longer amplified cDNA using a 0.6× cleanup using aMPure XP beads (catalog # A63882; Beckman Coulter). Libraries were constructed using the Chromium Single Cell 5′ Feature Barcode Library Kit (10X Genomics; PN 1000080) according to the manufacturer’s protocol with nine cycles of PCR. Indexed libraries were pooled equimolar and sequenced on a NovaSeq 6000 in a 28 bp/91 bp paired end run using the NovaSeq 6000 SP Reagent Kit (100 cycles; Illumina). An average of 106 million paired reads was generated per sample.
TCR sequencing
RNA was extracted from purified open repertoire mouse CD4+, CD8+, and CD4+CD8+ TILs FACS-sorted from B16 tumors. Purified RNA samples were quantified using Qubit RNA HS Assay Kit (Cat. No. Q32852; Thermo Fisher Scientific). The Agilent 2100 Bioanalyzer and Agilent RNA 600 Nano Kit were used to quantify and evaluate RNA integrity. 25 ng of total RNA was reverse transcribed using the Ion Torrent NGS Reverse Transcription Kit (Cat. No. A45003; Thermo Fisher Scientific). For each sample, 25 ng cDNA was amplified using Ion AmpliSeq Mouse TCR Beta SR Assay (RNA; Cat. No. A45489; Thermo Fisher Scientific), and the protocol was described in the Ion AmpliSeq Mouse Immune Repertoire User Guide (MAN0018524 rev. B.0). Libraries were purified with Agencourt AMPure XP beads (Cat. No. A63880; Beckman Coulter), washed with 70% ethanol, and eluted in 50 μl Low TE buffer. The resulting library samples were diluted 1:100 and quantified using the Ion Library Quantification Kit (Cat. No. 4468802; Thermo Fisher Scientific) and then diluted to 25 pM with Low TE buffer. Samples were pooled and sequenced using the Ion 540 chip to a target depth between 2 and 3M reads, followed by analysis via Ion Reporter version 5.12. TCR sequencing the Illumina-based platform was performed by Sequenta as described in Klinger et al. (2013), the ImmunoSeq assay (Adaptive Biotechnologies) or Archer Immunoverse HS TCRB assay, per manufacturer instructions.
Transcriptome sequencing
RNA was extracted using RNeasy mini kit (Qiagen) per instructions provided by the manufacturer. After ribogreen quantification and quality control of Agilent BioAnalyzer, total RNA underwent amplification using the SMART-seq V4 (Clonetech) ultralow input RNA kit for sequencing (12 cycles of amplification for 2–10 ng of total RNA). Subsequently, 10 ng of amplified cDNA was used to prepare Illumina Hiseq libraries with the Kapa DNA library preparation chemistry (Kapa Biosystems). Samples were run on a Hiseq 4000, in a 50-bp/50-bp paired-end run, using the TruSeq SBS Kit v3 (Illumina).
ATAC-seq
Profiling of chromatin was performed by assay for transposase-accessible chromatin (ATAC)–seq as described by Buenrostro et al. (2015). Briefly, 8–50,000 viably frozen T cells were thawed, washed in cold PBS, and lysed. The transposition reaction containing TDE1 Tagment DNA Enzyme (catalog # 20034198; Illumina) was incubated at 37°C for 30 min. The DNA was cleaned with the MinElute PCR Purification Kit (catalog # 28004; Qiagen), and the material was amplified for five cycles using NEBNext High-Fidelity 2× PCR Master Mix (catalog #M0541L; New England Biolabs). After evaluation by real-time PCR, 7–13 additional PCR cycles were done. The final product was cleaned by aMPure XP beads (catalog # A63882; Beckman Coulter) at a 1× ratio, and size selection was performed at a 0.5× ratio. Libraries were sequenced on a HiSeq4000 in a PE50 run, using the HiSeq 3000/4000 SBS Kit (Illumina). An average of 57 million paired reads were generated per sample.
Bioinformatics
TCR-seq analysis
Mouse TCR-seq data was preprocessed using the MiXCR (v. 3.0.9, https://github.com/milaboratory/mixcr) “analyze” pipeline. Tables of subsequent TCR clones were analyzed for various metrics using VDJtools (v. 1.2.1, https://github.com/mikessh/vdjtools). Additional visualizations were generated using the R statistical platform.
RNA-seq analysis
RNA-seq data were aligned to the mm9 reference genome using STAR 2.3.0e. Read counts were derived from featureCounts 1.4.6-p5. Differentially expressed genes were generated by DESeq2 1.14.1 in R. Upregulated genes were decided by log2 fold change >1 and the adjusted P value <0.01 while downregulated genes were decided by log2 fold change < −1 and adjusted P value <0.01.
ATAC-seq analysis
ATAC-seq data was aligned to the mm9 reference genome. Alignment and peak calling were performed by atac-seq-pipeline v1.7.1 from ENCODE Data Coordination Center (ENCODE DCC). Differential binding analysis was performed by DiffBind 2.2.12 in R. Gain differentially bound sites were decided by log2 fold change >1 and adjusted P value <0.01, while loss differentially bound sites were decided by log2 fold change < −1 and adjusted P value <0.01. Normalized density was calculated using bamCoverage module from deeptools 3.1.3 with normalize using RPGC effective Genome Size 2620345972 ignore duplicates.
scRNA-seq analysis
Single-cell sequencing data were aligned to the Genome Reference Consortium Human Build 38 (GRCh38) using Cell Ranger (v3.1.0; 10X Genomics) to obtain T cell clonotypes, feature barcoding, CITE-seq antibody detection, and gene expression profiles associated with individual single-cells. Each datatype was matched to create a UMI matrix and cells were filtered out based on three metrics: (1) cells with fewer than 200 detectable genes; (2) cells with more than 3,000 detectable genes; (3) cells that had fewer than 5% of counts related to mitochondrial genes. Data normalization, Principal Component Analysis, and subsequent Uniform Manifold Approximation and Projections (UMAP) were performed on the dataset using the R package Seurat v.3.1.1 (https://github.com/satijalab/seurat). The differential expression comparisons were generated using the DESeq2 package with selected genes (FDR < 0.05). After filtering, we created subclusters of cells using the Louvain algorithm (Blondel et al., 2008; Lancichinetti and Fortunato, 2009). Double positive T cells were selected by a cutoff of 0.5 coexpression in CITEseq data for both CD4 and CD8A.
Online supplemental material
Fig. S1 shows the flow cytometric detection of CD4+CD8+ T cells and representative flow cytometry plots and histograms of cell surface markers on CD4+, CD8+, and CD4+CD8+ T cells. Fig. S2 shows the epigenetic signature of SP and CD4 and CD8 derived CD4+CD8+ T cells. Fig. S3 shows the similarity in gene signatures among CD4+CD8+ T cell states between a mouse and a human as well as CD4+CD8+ T cell signatures associated with clinical response in melanoma patient samples. Fig. S4 shows CD4+CD8+ T cell states detected in lung cancer patient samples. Table S1 shows the Trp1 and Pmel RNA and ATAC-seq samples. Table S2 shows the clinical characteristics of melanoma patients analyzed.
Supplementary Material
shows the Trp1 and Pmel RNA and ATAC-seq samples.
shows the clinical characteristics of melanoma patients analyzed.
Acknowledgments
We would like the thank the melanoma DMT tissue repository team at MSK, specifically Mohsen Abu-Akeel and the MSK Flow Cytometry Core. We thank the New York Blood Center for use of the ImageStream.
We acknowledge the use of the Integrated Genomics Operation Core, funded by the National Cancer Institute Cancer Center Support Grant (P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. The research was funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748/NCI R01 CA056821, the Swim Across America, Ludwig Institute for Cancer Research, Ludwig Center for Cancer Immunotherapy at Memorial Sloan Kettering, Cancer Research Institute, and Parker Institute for Cancer Immunotherapy. The research was also supported in part by a Cancer Immunology Translational Cancer Research Grant (SU2C-AACR-DT1012) from Cancer Research Institute-Stand Up 2 Cancer, John Hopkins Bloomberg-Kimmel Institute for Cancer Immunotherapy, and Bristol-Myers Squibb International Immuno-Oncology Network. Stand Up 2 Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Author contributions: conceptualization: S.E. Schad, T. Merghoub, and J.D. Wolchok; methodology: S.E. Schad, A. Chow, R. Zappasodi, M. Gigoux, D. Higoux, and S. Budhu; investigation and resources: S.E. Schad, A. Chow, R. Zappasodi, M. Gigoux, D. Higoux, N. Malandro, S. Budhu, L. Mangarin, J. Zhang, H. Pan, M. Arniella, and D. Redmond; formal analysis: S.E. Schad; computation analysis: L. Mangarin, J. Zhang, H. Pan, D. Redmond, M. Arniella, and O. Elemento; human samples: J.D. Wolchok, T. Merghoub, J.E. Chaft, P.M. Forde, J.F. Gainor, M.D. Hellmann, V.P. Balachandran, K.N. Smith, and D.M. Pardoll; writing: S.E. Schad; review and editing: S.E. Schad, R. Zappasodi, J.D. Wolchok, and T. Merghoub; supervision and funding acquisition: J.D. Wolchok and T. Merghoub.
Data availability
All data generated and supporting the findings of this study are available within the paper. Datasets have been deposited to the Gene Expression Omnibus database under accession numbers GSE203187 (TCR-seq), GSE203182, GSE203183, GSE203184 (scRNA-seq), GSE203186 (RNA-seq), and GSE203188 (ATAC-seq). Additional information and materials will be made available upon request.
References
- Blondel, V.D., Guillaume J.-L., Lambiotte R., and Lefebvre E.. 2008. Fast unfolding of communities in large networks. J. Stat. Mech. P10008. 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Blue, M.L., Daley J.F., Levine H., and Schlossman S.F.. 1985. Coexpression of T4 and T8 on peripheral blood T cells demonstrated by two-color fluorescence flow cytometry. J. Immunol. 134:2281–2286. [PubMed] [Google Scholar]
- Bohner, P., Chevalier M.F., Cesson V., Rodrigues-Dias S.-C., Dartiguenave F., Burruni R., Tawadros T., Valerio M., Lucca I., Nardelli-Haefliger D., et al. 2019. Double positive CD4+CD8+ T cells are enriched in urological cancers and favor T helper-2 polarization. Front. Immunol. 10:622. 10.3389/fimmu.2019.00622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu, S., Loike J.D., Pandolfi A., Han S., Catalano G., Constantinescu A., Clynes R., and Silverstein S.C.. 2010. CD8+ T cell concentration determines their efficiency in killing cognate antigen-expressing syngeneic mammalian cells in vitro and in mouse tissues. J. Exp. Med. 207:223–235. 10.1084/jem.20091279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109:21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S., Panda A., Sayantan B., Roy D., Kajal K., Guha D., and Sa G.. 2017. Transcriptional regulation of FOXP3 requires integrated activation of both promoter and CNS regions in tumor-induced CD8 + Treg cells. Sci. Rep. 7:1628. 10.1038/s41598-017-01788-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clénet, M.L., Gagnon F., Moratalla A.C., Viel E.C., and Arbour N.. 2017. Peripheral human CD4+CD8+ T lymphocytes exhibit a memory phenotype and enhanced responses to IL-2, IL-7 and IL-15. Sci. Rep. 7:11612. 10.1038/s41598-017-11926-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty, F., Pipkin M.E., Djuretic I.M., Levanon D., Lotem J., Lichtenheld M.G., Groner Y., and Rao A.. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206:51–59. 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, G., Augustine M.M., Das J., Bottomly K., Ray P., and Ray A.. 2003. An important regulatory role for CD4+CD8 alpha alpha T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 100:5324–5329. 10.1073/pnas.0831037100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen, T., Duhen R., Montler R., Moses J., Moudgil T., de Miranda N.F., Goodall C.P., Blair T.C., Fox B.A., McDermott J.E., et al. 2018. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9:2724. 10.1038/s41467-018-05072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa, T. 2015. Regulation of CD4 and CD8 coreceptor expression and CD4 versus CD8 lineage decisions. Adv. Immunol. 125:1–40. 10.1016/bs.ai.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Egawa, T., and Littman D.R.. 2008. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9:1131–1139. 10.1038/ni.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm, M.A., Picking R.A., Kuruc J.D., Mcgee K.S., Gay C.L., Eron J.J., Hicks C.B., Tomaras G.D., and Ferrari G.. 2012. CD4+CD8+ T cells represent a significant portion of the anti-HIV T cell response to acute HIV infection. J. Immunol. 188:4289–4296. 10.4049/jimmunol.1103701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghia, P., Prato G., Stella S., Scielzo C., Geuna M., and Caligaris-Cappio F.. 2007. Age-dependent accumulation of monoclonal CD4+CD8+ double positive T lymphocytes in the peripheral blood of the elderly. Br. J. Haematol. 139:780–790. 10.1111/j.1365-2141.2007.06867.x [DOI] [PubMed] [Google Scholar]
- Glimcher, L.H., Townsend M.J., Sullivan B.M., and Lord G.M.. 2004. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 4:900–911. 10.1038/nri1490 [DOI] [PubMed] [Google Scholar]
- Klinger, M., Kong K., Moorhead M., Weng L., Zheng J., and Faham M.. 2013. Combining next-generation sequencing and immune assays: A novel method for identification of antigen-specific T cells. PLoS One. 8:e74231. 10.1371/journal.pone.0074231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard, L.C., Fischer P., Kakrecha B., Linsley P.S., Wambre E., Liu M.C., Rust B.J., Lee D., Penhallow B., Manjarrez Orduno N., and Nadler S.G.. 2018. Renal cell carcinoma (RCC) tumors display large expansion of double positive (DP) CD4+CD8+ T cells with expression of exhaustion markers. Front. Immunol. 9:2728. 10.3389/fimmu.2018.02728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida, D., Husain M.M., Muroi S., van Wijk F., Shinnakasu R., Naoe Y., Reis B.S., Huang Y., Lambolez F., Docherty M., et al. 2013. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 14:281–289. 10.1038/ni.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski, P., Boni A., Antony P.A., Cassard L., Irvine K.R., Kaiser A., Paulos C.M., Palmer D.C., Touloukian C.E., Ptak K., et al. 2008. Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood. 112:362–373. 10.1182/blood-2007b11-120998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimbeni, M., Shin E.-C., Chiriboga L., Kleiner D.E., and Rehermann B.. 2004. Peripheral CD4(+)CD8(+) T cells are differentiated effector memory cells with antiviral functions. Blood. 104:478–486. 10.1182/blood-2003blo12-4395 [DOI] [PubMed] [Google Scholar]
- Nishida, K., Kawashima A., Kanazawa T., Kidani Y., Yoshida T., Hirata M., Yamamoto K., Yamamoto Y., Sawada M., Kato R., et al. 2020. Clinical importance of the expression of CD4+CD8+ T cells in renal cell carcinoma. Int. Immunol. 32:347–357. 10.1093/intimm/dxaa004 [DOI] [PubMed] [Google Scholar]
- Oh, D.Y., Kwek S.S., Raju S.S., Li T., McCarthy E., Chow E., Aran D., Ilano A., Pai C.-C.S., Rancan C., et al. 2020. Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 181:1612–1625.e13. 10.1016/j.cell.2020.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard, N.H., Jung J.-W., Steptoe R.J., and Wells J.W.. 2015. CD4+/CD8+ double-positive T cells: More than just a developmental stage? J. Leukoc. Biol. 97:31–38. 10.1189/jlb.1RU0814-382 [DOI] [PubMed] [Google Scholar]
- Overwijk, W.W., Theoret M.R., Finkelstein S.E., Surman D.R., de Jong L.A., Vyth-Dreese F.A., Dellemijn T.A., Antony P.A., Spiess P.J., Palmer D.C., et al. 2003. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 198:569–580. 10.1084/jem.20030590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parel, Y., Aurrand-Lions M., Scheja A., Dayer J.-M., Roosnek E., and Chizzolini C.. 2007. Presence of CD4+CD8+ double-positive T cells with very high interleukin-4 production potential in lesional skin of patients with systemic sclerosis. Arthritis Rheum. 56:3459–3467. 10.1002/art.22927 [DOI] [PubMed] [Google Scholar]
- Philip, M., Fairchild L., Sun L., Horste E.L., Camara S., Shakiba M., Scott A.C., Viale A., Lauer P., Merghoub T., et al. 2017. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 545:452–456. 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada, S.A., Simpson T.R., Peggs K.S., Merghoub T., Vider J., Fan X., Blasberg R., Yagita H., Muranski P., Antony P.A., et al. 2010. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 207:637–650. 10.1084/jem.20091918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman, M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. 2019. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 176:404. 10.1016/j.cell.2018.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi, R., Tachibana M., Naoe Y., Muroi S., Akiyama K., Tezuka C., Okuda T., and Taniuchi I.. 2008. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 319:822–825. 10.1126/science.1151844 [DOI] [PubMed] [Google Scholar]
- Sledzinska, A., Vila de Mucha M., Bergerhoff K., Hotblack A., Demane D.F., Ghorani E., Akarca A.U., Marzolini M.A.V., Solomon I., Vargas F.A., et al. 2020. Regulatory T cells restrain interleukin-2- and Blimp-1-dependent acquisition of cytotoxic function by CD4+ T cells. Immunity. 52:151–166.e6. 10.1016/j.immuni.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacchio, M.S., and Bosselut R.. 2016. What happens in the thymus does not stay in the thymus: How T cells recycle the CD4+-CD8+ lineage commitment transcriptional circuitry to control their function. J. Immunol. 196:4848–4856. 10.4049/jimmunol.1600415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancichinetti, A., and Fortunato S.. 2009. Community detection algorithms: A comparative analysis. Phys. Rev. E. 80:056117. 10.1103/PhysRevE.80.056117 [DOI] [PubMed] [Google Scholar]
- van der Leun, A.M., Thommen D.S., and Schumacher T.N.. 2020. CD8(+) T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer. 20:218–232. 10.1038/s41568-019s4150235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Wildt K.F., Castro E., Xiong Y., Feigenbaum L., Tessarollo L., and Bosselut R.. 2008. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 29:876–887. 10.1016/j.immuni.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt, K.F., Sun G., Grueter B., Fischer M., Zamisch M., Ehlers M., and Bosselut R.. 2007. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J. Immunol. 179:4405–4414. 10.4049/jimmunol.179.7.4405 [DOI] [PubMed] [Google Scholar]
- Xie, Y., Akpinarli A., Maris C., Hipkiss E.L., Lane M., Kwon E.K., Muranski P., Restifo N.P., and Antony P.A.. 2010. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J. Exp. Med. 207:651–667. 10.1084/jem.20091921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the Trp1 and Pmel RNA and ATAC-seq samples.
shows the clinical characteristics of melanoma patients analyzed.
Data Availability Statement
All data generated and supporting the findings of this study are available within the paper. Datasets have been deposited to the Gene Expression Omnibus database under accession numbers GSE203187 (TCR-seq), GSE203182, GSE203183, GSE203184 (scRNA-seq), GSE203186 (RNA-seq), and GSE203188 (ATAC-seq). Additional information and materials will be made available upon request.