Abstract
Background
Systemic inflammation is associated with survival outcomes in colon cancer. However, it is not well-known which systemic inflammatory marker is a powerful prognostic marker in patients with colon cancer.
Methods
A total of 4535 colon cancer patients were included in this study. We developed a novel prognostic index using a robust combination of seven systemic inflammation-associated blood features of the discovery set. The predictability and generality of the novel prognostic index were evaluated in the discovery, validation and replication sets.
Results
Among all combinations, the combination of albumin and monocyte count was the best candidate expression. The final formula of the proposed novel index is named the Prognostic Immune and Nutritional Index (PINI). The concordance index of PINI for overall and progression-free survival was the highest in the discovery, validation and replication sets compared to existing prognostic inflammatory markers. PINI was found to be a significant independent prognostic factor for both overall and progression-free survival.
Conclusions
PINI is a novel prognostic index that has improved discriminatory power in colon cancer patients and appears to be superior to existing prognostic inflammatory markers. PINI can be utilised for decision-making regarding personalised treatment as the complement of the TNM staging system.
Subject terms: Prognostic markers, Colon cancer
Background
Pathologic characteristics of the tumour are the major factors in prognosis of colorectal cancer. However, the survival outcomes can vary even in colorectal cancer patients with similar pathologic characteristics. Tumour-related inflammation may contribute to these differences in outcomes. Similar to tumour-related inflammation, systemic inflammation is associated with tumour progression [1]. It is mediated by cytokines and immune cells. Since these mediators can be detected in the blood, these may be related to long-term outcomes [2].
Several systemic inflammatory markers have been suggested as prognostic markers in patients with colorectal cancer [3]. These markers consist of systemic inflammation-associated blood features, like immune cell enumeration or inflammatory-associated proteins from routine blood tests. Each of these blood features, such as monocyte, lymphocyte, neutrophil and albumin, are also related to prognosis in colorectal cancer [4, 5]. As systemic inflammatory markers, the lymphocyte-to-monocyte ratio (LMR), neutrophil-to-lymphocyte ratio (NLR), platelet-to-lymphocyte ratio (PLR) and prognostic nutritional index (PNI) have been investigated for the prediction of survival outcomes in patients with colorectal cancer [6–9]. These are generated using two blood features by basic mathematical operators, such as addition and division.
However, the existing systemic inflammatory markers are not derived from an extensive comparison in terms of long-term outcomes. It is still not well-known which combination of blood features is a powerful prognostic marker in patients with colon cancer. The aim of this study was to develop a novel strong prognostic index for colon cancer through a robust combination of blood features for systemic inflammation.
Methods
Study population
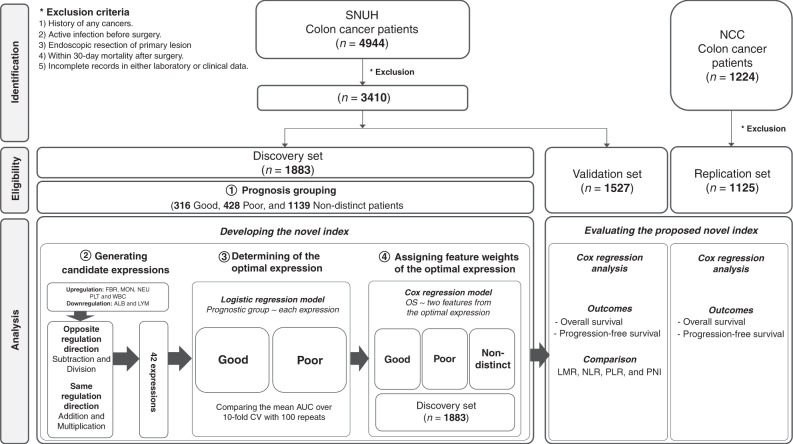
A total of 4944 patients, who underwent surgical resection for primary colon cancer between January 1, 2002 and December 31, 2015, were collected at the Seoul National University Hospital (SNUH). These patients were included in the discovery and validation sets. For the replication set, a total of 1224 colon cancer patients between January 1, 2008 and December 31, 2010, who underwent primary tumour resection, were collected from the National Cancer Center Korea (NCC). All eligible patients underwent colonic resection and lymph node dissection. Adjuvant chemotherapy was administered after surgery based on the tumour stage. Patients were excluded if one of the following criteria was met: (1) with a history of colorectal cancer or other types of cancers; (2) with an active infection before surgery; (3) had undergone endoscopic resection of the primary lesion; (4) within 30-day mortality after surgery and (5) with incomplete records in both laboratory and clinical data. Out of 4944 patients from the SNUH, 3410 patients were included for the discovery and validation sets. Discovery and validation sets were split on the basis of a cut-off date, set on January 1, 2011. Out of 1224 patients from the NCC, 1125 patients were included in the replication set after exclusion (Fig. 1). Details on characteristics of the three sets are described in Table 1. This study was approved by the Institutional Review Board (no. 2101-042-1187, NCC2021-0057). The Institutional Review Board granted waivers of informed consent for retrospective medical chart review and data analysis.
Fig. 1. The workflow of developing a novel prognostic inflammatory index in colon cancer.
SNUH Seoul National University Hospital, NCC National Cancer Center Korea, FBR fibrinogen, MON monocyte, NEU neutrophil, PLT platelet, WBC white blood cell, ALB albumin, LYM lymphocyte, LMR lymphocyte to monocyte ratio, NLR neutrophil to lymphocyte ratio, PLR platelet-to-lymphocyte ratio, PNI prognostic nutritional index.
Table 1.
Clinicopathologic characteristics of the discovery, validation and replication sets.
| SNUH | NCC | ||
|---|---|---|---|
| Total | Discovery set | Validation set | Replication set |
| (N = 4535) | (N = 1883) | (N = 1527) | (N = 1125) |
| Age, years (mean ± SD) | 63.2 ± 11.2 | 62.8 ± 11.4 | 61.3 ± 11.5 |
| Sex | |||
| Male | 1062 (56.4%) | 941 (61.6%) | 643 (57.2%) |
| Female | 821 (43.6%) | 586 (38.4%) | 482 (42.8%) |
| Body mass index, kg/m2 (mean ± SD) | 23.5 ± 3.3 | 23.1 ± 3.0 | 23.8 ± 3.1 |
| <30 kg/m2 | 1823 (96.8%) | 1494 (97.8%) | 1081 (97.3%) |
| ≥30 kg/m2 | 60 (3.2%) | 33 (2.2%) | 30 (2.7%) |
| Carcinoembryonic antigen | |||
| <5 μg/ml | 1414 (75.1%) | 1049 (68.7%) | 735 (65.6%) |
| ≥5 μg/ml | 469 (24.9%) | 478 (31.3%) | 385 (34.4%) |
| Tumour location | |||
| Proximal colon | 758 (40.3%) | 540 (35.4%) | 316 (28.1%) |
| Distal colon | 1115 (59.2%) | 964 (63.1%) | 785 (69.8%) |
| Mixed | 10 (0.5%) | 23 (1.5%) | 23 (2.1%) |
| Tumour stage | |||
| I | 264 (14.0%) | 194 (12.7%) | 201 (17.9%) |
| II | 624 (33.1%) | 493 (32.3%) | 331 (29.4%) |
| III | 643 (34.1%) | 501 (32.8%) | 493 (43.8%) |
| IV | 352 (18.7%) | 339 (22.2%) | 100 (8.9%) |
| Tumour grade | |||
| Low | 1732 (92.0%) | 1371 (89.8%) | 950 (90.5%) |
| High | 151 (8.0%) | 156 (10.2%) | 100 (9.5%) |
| Lymphovascular invasion | |||
| Absent | 1178 (62.6%) | 906 (59.3%) | 125 (11.8%) |
| Present | 705 (37.4%) | 621 (40.7%) | 931 (88.2%) |
| Adjuvant chemotherapy | |||
| No | 563 (29.9%) | 445 (29.1%) | 348 (36.98%) |
| Yes | 1320 (70.1%) | 1082 (70.9%) | 700 (63.1%) |
SD standard deviation, SNUH Seoul National University Hospital, NCC National Cancer Center Korea.
The following clinicopathologic characteristics of patients were analysed retrospectively: age, sex, body mass index (BMI), carcinoembryonic antigen (CEA) levels, tumour location, tumour stage, tumour grade, lymphovascular invasion and adjuvant chemotherapy. With regard to tumour location, proximal tumours were defined as tumours in the cecum, ascending colon and transverse colon; distal tumours were defined as those in the descending and sigmoid colon. Seven blood features were considered for our analysis, which included albumin (ALB) level, fibrinogen (FBR) level, lymphocyte count (LYM), monocyte count (MON), neutrophil count (NEU), platelet count (PLT) and white blood cell count (WBC). All of these were routine blood measurements that are related to systemic inflammation. These blood features are the components of systemic inflammatory markers, such as the leukocyte count, LMR, NLR, PLR, PNI and plasma FBR, which have been investigated previously as prognostic biomarkers in colorectal cancer [6–11]. Although the serum level of C-reactive protein (CRP) was investigated in the modified Glasgow prognostic score (mGPS), it was excluded since it was not a routine blood test in the cohort study [12]. The values of these blood features were collected retrospectively from preoperative blood test results, which were obtained within 4 weeks before surgery.
Patient follow-up was conducted every 3 or 6 months for 5 years and every year thereafter. Physical examination, chest X-ray, and tests for serum CEA levels were performed in every follow-up visit. An abdominopelvic computed tomography was performed every 6 months. Colonoscopy was performed every 1 or 2 years. Cancer recurrence or progression was detected through imaging studies, pathologic examinations or both.
Developing a novel index
Defining the distinctive prognosis group in the discovery set
The patients were classified into good, poor and non-distinct prognosis groups based on their cancer progression status. The distinctive prognosis groups, which comprised patients with either good or poor prognosis, was represented as the explicit ground-truth [13]. Hence, we determined a novel index using good and poor prognosis groups from the discovery set. In our study, the good prognosis group was composed of patients with no recurrence or any cancer-related death during a 5-year follow-up after surgery, while the poor prognosis group was composed of patients with either recurrence or cancer-related death between 30 days and 3 years after surgery. The remaining patients were in the non-distinct prognosis group.
Generating candidate expressions of laboratory features
First, an association analysis was conducted to divide seven laboratory features into those involved in upregulation (that is, presenting a positive association with cancer progression: FBR, MON, NEU, PLT and WBC) and downregulation (that is, presenting a negative association with cancer progression: ALB and LYM). Next, we generated candidate expressions of a pair of laboratory features using the four basic mathematical operators, which were addition, subtraction, multiplication and division. For the laboratory features from the same regulation direction, we considered addition and multiplication operators. For instance, the candidate expressions between MON and FBR were MON + FBR and MON × FBR. The subtraction and division operators were used for the laboratory features from the opposite regulation direction. Additionally, the min-max normalisation was used in order to acquire the range of features on the same scale. In this study, we considered a combination of two laboratory features.
The optimal expression of two weighted laboratory features
The novel index was defined as the optimal expression of two weighted laboratory features. The optimal expression was determined by the best model performance for predicting the prognosis group. The weights of the two laboratory features in the optimal expression were set as the estimated coefficients obtained by Cox regression models for overall survival (OS) in the entire discovery set. The workflow of developing a novel index is described in Fig. 1.
Evaluation of proposed novel index
To evaluate the predictability and generality of the newly developed index proposed in this study, we investigated the association between the proposed index and the prognosis of colon cancer patients in the discovery, validation and external replication sets. First, we compared the predictive performance of the proposed index in each of the discovery set and validation set with the existing systemic inflammatory markers with the C-index from the univariate Cox regression analysis. In addition, we investigated whether the proposed novel index affects the prognosis independently of the clinicopathologic confounders through the adjusted multivariate Cox regression analysis. Finally, we checked whether the proposed novel index is statistically significant through the external replication set.
Statistical analysis
As for the clinicopathologic and laboratory features, categorical variables were presented as count (%), and continuous variables as mean (standard deviation, SD). The t-test was used for continuous variables and the chi-square test for categorical variables.
Two survival outcomes, including OS and progression-free survival (PFS), were considered for the following survival analysis. PFS was defined as the duration of patient survival without any recurrence or progression of colon cancer or death from any cause. OS was defined as the duration of patient survival with regard to any possible cause of death.
To define the novel index, univariate logistic regression with each candidate expression was performed, and the area under the receiver operating characteristic curve (AUC) was used to evaluate the predictive performance. The optimal expression was determined by the highest AUC among all candidate expressions. Delong’s tests [14] were performed pairwise to compare AUC between the Top 1 ranked expression and each candidate expression using pROC R package, respectively. The estimated coefficients from univariate Cox proportional hazard regression models for OS were set as the weights of the two laboratory features in the optimal expression. The dichotomous novel index was defined by its optimal cut-off value, which was determined using the maximally selected rank statistics. The cut-off points of each systemic inflammatory marker were determined based on the existing literature [6, 7, 15, 16].
Univariate Cox proportional hazard regression models for OS and PFS were performed to evaluate the risk prediction of the novel index. We then conducted multivariate Cox proportional hazard regression models in order to adjust the impact of clinicopathologic features. To compare the predictive performance of the novel index, existing prognostic inflammatory markers were considered as benchmarks. Harrell’s C-index (that is, the concordance index) was used to assess the predictive performance of the survival models. To estimate the improvement of the novel index in the univariate and multivariate models, we calculated the continuous net reclassification index (NRI) for 5-year reclassification improvement for OS and PFS with 1000 bootstrapping using the uricens R package, which is used for time-to-event NRI analysis. For further survival analyses, Kaplan–Meier curves and log-rank tests were conducted to cheque if there were significant differences (adjusted by false discovery rate) in survival between groups, which were divided by the novel index. Subgroup analyses were performed to evaluate the prognostic value of the novel index in stage IIa (T3N0) and low-risk (T1-3N1) and high-risk stage III (T4 and/or N2) colon cancer from all datasets.
The Reporting Recommendations for Tumour Marker Prognostic Studies (REMARK) criteria were considered in this study report (Supplementary Table 1) [17]. Although formal estimation of sample size was not performed in advance, the number of events (438 deaths and 551 progressions in the discovery set) compared with the number of Cox model variables (7) implied that “a minimum of 10 events per predictor” rule was surpassed, signifying the precision and accuracy of the regression estimates [18]. A p-value less than 0.05 was considered statistically significant. All analyses were carried out with R 4.0.0.
Results
Patient characteristics
Out of 3410 patients in this study, 1883 patients were included in the discovery set and 1527 patients were included in the validation set on the basis of a cut-off date, set on January 1, 2011. The average age was 63.2 years (SD: 11.2) in the discovery set and 62.8 years (SD: 11.4) in the validation set. The mean BMI was 23.5 kg/m2 (SD: 3.3) in the discovery set and 23.1 kg/m2 (SD: 3.0) in the validation set. Detailed clinicopathologic characteristics of both discovery and validation sets are described in Table 1. In the discovery set, ~40% of the patients belonged to the distinctive prognosis group, with 316 patients in the good prognosis group and 428 in the poor prognosis group. As hypothesised, the poor prognosis group had a significantly higher level of CEA, a larger proportion of late tumour stage and high tumour grade, had more records of lymphovascular invasion and adjuvant chemotherapy than the good prognosis group (Supplementary Table 2).
In the replication set, there were 1125 colon cancer patients from the NCC to cross-validate the performance of the proposed index. The average age was 61.3 years (SD: 11.5) and the mean BMI was 23.8 kg/m2 (SD: 3.1), exhibiting a similar distribution as both the discovery set and validation set. Further detailed information on the replication set is shown in Table 1.
Development of the novel index
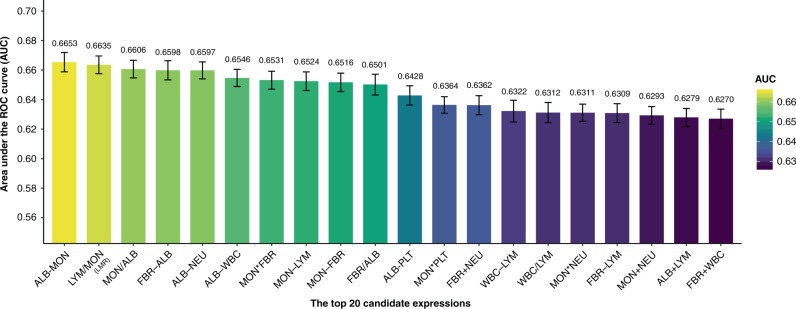
First, association analyses were conducted between OS and seven laboratory features. FBR, MON, NEU, PLT and WBC were positively associated with OS, while ALB and LYM were negatively associated (Supplementary Table 3). With respect to the association direction per feature, 42 candidate expressions were generated following the workflow described in Section Developing a novel index. The 10-fold cross-validation (CV) repeated 100 times produced results that showed that the expression of ALB − MON achieved the highest average AUC of 0.6653 (Fig. 2 and Supplementary Table 4). The second highest AUC was achieved with the expression of LYM /MON, which is one of the known existing systemic inflammatory markers (i.e. LMR). The third highest AUC’s expression was MON/ALB (MAR, monocyte-to-albumin ratio). We defined these three expressions, identified as no significant difference with Delong’s test, as candidate expressions (Supplementary Table 4). We have confirmed that ALB and MON were captured most frequently in the top-ranked indices among the 42 expressions as influential factors (Supplementary Table 5). To obtain the weights of ALB and MON, we performed the Cox regression analysis with ALB and MON as variables and OS as the outcome in the discovery set. The weights were 0.9000 for ALB and 0.0007 for MON. Hence, (ALB × 0.9) − (MON × 0.0007) was defined as the novel index and named the Prognostic Immune and Nutritional Index (PINI). Dichotomous PINI was defined by the cut-off value of 3.0 in order to maximise the OS prediction. The high PINI group (≥3.0) was associated with a young age, high BMI, low level of CEA, and a small proportion of late tumour stage in the discovery set (Supplementary Table 6).
Fig. 2. The average predictive performance of the top 20 candidate expressions in the distinctive prognosis group over 10-fold CV with 100 repeats.
WBC white blood cell count, NEU neutrophil count, LYM lymphocyte count, MON monocyte count, PLT platelet count, ALB albumin, FBR fibrinogen.
Comparing PINI with the existing systemic inflammatory markers
Table 2 shows the univariate Cox regression performance of PINI and the existing systemic inflammatory markers (that is, LMR, NLR, PLR and PNI) and MAR for OS and PFS in the discovery set. PINI outperformed in both OS and PFS. For OS, the highest C-index of 0.677 (standard error [SE]: 0.013) was achieved with PINI; LMR obtained the second rank of C-index, which was 0.659. For PFS, PINI obtained the highest C-index of 0.649 (SE: 0.012), and a C-index of 0.632 for LMR was obtained.
Table 2.
Univariate survival analysis of candidate indices and systemic inflammatory markers for OS and PFS in the discovery set.
| Overall survival | Progression-free survival | |||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | C-index (SE) | HR (95% CI) | P-value | C-index (SE) | |
| Continuous | ||||||
| PINI | 0.371 (0.320–0.431) | <0.001 | 0.677 (0.013) | 0.424 (0.369–0.486) | <0.001 | 0.649 (0.012) |
| LMR | 0.716 (0.670–0.764) | <0.001 | 0.659 (0.014) | 0.764 (0.723–0.808) | <0.001 | 0.632 (0.013) |
| MAR | 1.004 (1.003–1.005) | <0.001 | 0.628 (0.014) | 1.004 (1.003–1.004) | <0.001 | 0.611 (0.013) |
| NLR | 1.123 (1.096–1.151) | <0.001 | 0.648 (0.014) | 1.111 (1.084–1.139) | <0.001 | 0.618 (0.013) |
| PLR | 1.003 (1.002–1.003) | <0.001 | 0.612 (0.014) | 1.565 (1.323–1.851) | <0.001 | 0.559 (0.011) |
| PNI | 0.922 (0.910–0.934) | <0.001 | 0.666 (0.014) | 0.933 (0.923–0.944) | <0.001 | 0.635 (0.013) |
| Dichotomous | ||||||
| PINI | ||||||
| <3.0 | 1 | 1 | ||||
| ≥3.0 | 0.346 (0.287–0.418) | <0.001 | 0.628 (0.012) | 0.412 (0.348–0.487) | <0.001 | 0.603 (0.011) |
| LMR | ||||||
| <2.4 | 1 | 1 | ||||
| ≥2.4 | 0.338 (0.278–0.412) | <0.001 | 0.601 (0.011) | 0.404 (0.336–0.484) | <0.001 | 0.586 (0.010) |
| MAR | ||||||
| <192 | 1 | 1 | ||||
| ≥192 | 2.874 (2.326–3.550) | <0.001 | 0.582 (0.011) | 2.296 (1.883–2.799) | <0.001 | 0.562 (0.009) |
| NLR | ||||||
| <5.0 | 1 | 1 | ||||
| ≥5.0 | 2.399 (1.849–3.112) | <0.001 | 0.545 (0.009) | 2.013 (1.577–2.571) | <0.001 | 0.536 (0.008) |
| PLR | ||||||
| <150 | 1 | 1 | ||||
| ≥150 | 1.831 (1.515–2.215) | <0.001 | 0.580 (0.012) | 1.565 (1.323–1.851) | <0.001 | 0.559 (0.011) |
| PNI | ||||||
| <45 | 1 | 1 | ||||
| ≥45 | 0.365 (0.302–0.441) | <0.001 | 0.618 (0.012) | 0.430 (0.362–0.510) | <0.001 | 0.597 (0.011) |
CI confidence interval, SE standard error, HR hazard ratio, PINI prognostic immune and nutritional index, LMR lymphocyte-to-monocyte ratio, MAR monocyte-to-albumin ratio, NLR neutrophil-to-lymphocyte ratio, PLR platelet-to-lymphocyte ratio, PNI prognostic nutritional index.
In case of multivariate analysis for survival, age, CEA level, tumour stage, tumour grade, lymphovascular invasion and adjuvant chemotherapy were adjusted as clinicopathologic confounders (Supplementary Table 7). After adjusting for the above confounders, PINI achieved the competitive C-index of 0.846 (SE: 0.009) for OS and a slightly high C-index of 0.788 (SE: 0.010) for PFS (Supplementary Table 8). PINI divulged as a significant independent prognostic factor for OS and PFS in the discovery set (hazard ratio [HR] = 0.553, 95% confidence interval [CI] = 0.463–0.659; HR = 0.627, 95% CI = 0.536–0.734, respectively).
The predictive performance of PINI was evaluated with the internal validation set. Table 3 illustrates that PINI outperformed the other four markers for OS and PFS in the univariate Cox analyses. The C-index of PINI for OS was 0.637 (SE = 0.013) and 0.617 (SE = 0.012) for PFS. In case of multivariate Cox analyses, PINI obtained slightly higher C-index values of 0.789 for OS and 0.757 for PFS (Supplementary Table 9). PINI was also found to be a significant independent prognostic factor for OS and PFS in the validation set (HR = 0.644, 95% CI = 0.545–0.761; HR = 0.687, 95% CI = 0.583–0.808, respectively).
Table 3.
Univariate survival analysis of candidate indices and systemic inflammatory markers for OS and PFS in the validation set.
| Overall survival | Progression-free survival | |||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | C-index (SE) | HR (95% CI) | P-value | C-index (SE) | |
| Continuous | ||||||
| PINI | 0.402 (0.320–0.431) | <0.001 | 0.637 (0.013) | 0.478 (0.412–0.554) | <0.001 | 0.617 (0.012) |
| LMR | 0.795 (0.756–0.836) | <0.001 | 0.629 (0.012) | 0.846 (0.808–0.885) | <0.001 | 0.614 (0.012) |
| MAR | 1.005 (1.004–1.006) | <0.001 | 0.629 (0.011) | 1.935 (1.630–2.298) | <0.001 | 0.566 (0.009) |
| NLR | 1.059 (1.041–1.078) | <0.001 | 0.593 (0.012) | 1.084 (1.062–1.106) | <0.001 | 0.590 (0.012) |
| PLR | 1.002 (1.001–1.003) | <0.001 | 0.562 (0.012) | 1.002 (1.001–1.002) | <0.001 | 0.555 (0.012) |
| PNI | 0.940 (0.910–0.934) | <0.001 | 0.616 (0.012) | 0.953 (0.942–0.964) | <0.001 | 0.599 (0.012) |
| Dichotomous | ||||||
| PINI | ||||||
| <3.0 | 1 | 1 | ||||
| ≥3.0 | 0.470 (0.401–0.551) | <0.001 | 0.597 (0.010) | 0.563 (0.483–0.655) | <0.001 | 0.582 (0.010) |
| LMR | ||||||
| <2.4 | 1 | 1 | ||||
| ≥2.4 | 0.471 (0.397–0.560) | <0.001 | 0.573 (0.009) | 0.552 (0.467–0.652) | <0.001 | 0.568 (0.009) |
| MAR | ||||||
| <192 | 1 | 1 | ||||
| ≥192 | 2.299 (1.928–2.741) | <0.001 | 0.575 (0.009) | 1.935 (1.630–2.298) | <0.001 | 0.566 (0.009) |
| NLR | ||||||
| <5.0 | 1 | 1 | ||||
| ≥5.0 | 1.963 (1.530–2.519) | <0.001 | 0.531 (0.007) | 1.822 (1.430–2.321) | <0.001 | 0.533 (0.007) |
| PLR | ||||||
| <150 | 1 | 1 | ||||
| ≥150 | 1.429 (1.219–1.674) | <0.001 | 0.547 (0.010) | 1.288 (1.107–1.499) | <0.001 | 0.542 (0.011) |
| PNI | ||||||
| <45 | 1 | 1 | ||||
| ≥45 | 0.499 (0.425–0.585) | <0.001 | 0.585 (0.010) | 0.574 (0.492–0.669) | <0.001 | 0.573 (0.010) |
CI confidence interval, SE standard error, HR hazard ratio, PINI prognostic immune and nutritional index, LMR lymphocyte-to-monocyte ratio, MAR monocyte-to-albumin ratio, NLR neutrophil-to-lymphocyte ratio, PLR platelet-to-lymphocyte ratio, PNI prognostic nutritional index.
Survival analysis for dichotomous PINI
The predictive performance of dichotomous PINI was compared with those of LMR, NLR, PLR, PNI and MAR with respect to the provided cut-off values. In the univariate analysis, dichotomous PINI achieved the highest C-index of 0.628 (SE: 0.012) for OS and 0.603 (SE: 0.011) for PFS in the discovery set (Table 2). Moreover, dichotomous PINI outperformed in the univariate analysis for OS and PFS in the validation set (Table 3). As for the multivariate analysis, dichotomous PINI retained the competitive predictive performance for OS and PFS in both discovery and validation sets (Supplementary Tables 8-9). After adjustment for covariates, a high dichotomous PINI was significantly associated with good OS (HR = 0.567, 95% CI = 0.466–0.690) and PFS (HR = 0.650, 95% CI = 0.544–0.776) in the discovery set. In the validation set, a high dichotomous PINI was also significantly associated with good OS (HR = 0.695, 95% CI = 0.587–0.821) and PFS (HR = 0.780, 95% CI = 0.666–0.915) after adjustment.
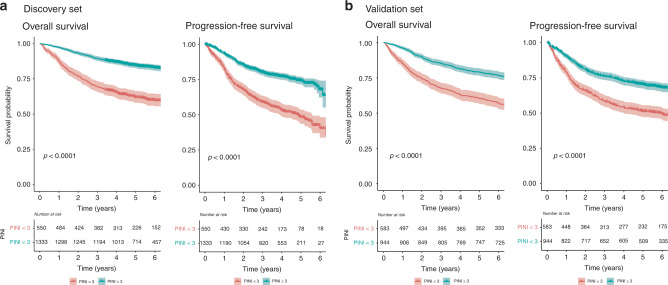
With regards to dichotomous PINI, the Kaplan–Meier curves in Fig. 3 shows that the patients in the high PINI group (≥3.0) had better OS than those in the low PINI group (<3.0; 5-year OS in the discovery set: 84.4% vs 62.0%, p < 0.001; 5-year OS in the validation set: 79.1% vs 60.4%, p < 0.001, respectively). Analogously, Fig. 3 also indicates that the high PINI group had better PFS than the low PINI group (5-year PFS in the discovery set: 74.3% vs 48.3%, p < 0.001; 5-year PFS in the validation set: 70.3% vs 51.2%, p < 0.001, respectively). Furthermore, the finding that a higher PINI score is associated with a better survival was consistent with the patients categorised into four groups based on the quantiles of PINI (Supplementary Fig. 1).
Fig. 3. Kaplan–Meier curves of the high and low PINI groups for OS and PFS.
a Discovery and b validation sets. P-values were calculated using the log-rank tests. OS overall survival, PFS progression-free survival, PINI prognostic immune and nutritional index.
NRI analysis for 5-year reclassification improvement of PINI
To compare the predictive performance of the univariate and multivariate cox models, we performed continuous NRI analysis for 5-year OS and PFS in the validation set (Supplementary Table 10). In the univariate analysis for OS and PFS, PINI provided significant incremental value compared to existing indices and MAR (Candidate index). In the multivariate analysis, even though performance increase is relatively small, improvements remained in the multivariate analysis for OS and PFS.
In particular, PINI improved 18.07% (95% CI 9.79–27.05) and 11.03 (95% CI 4.01–15.58) for OS and PFS compared with NLR, the most widely validated prognostic index in the univariate analysis, and PINI improved 2.23% (95% CI 0.48–6.89) and 3.64% (95% CI 0.93–5.13) for OS and PFS compared with NLR in multivariate analysis, respectively.
The association between PINI and survival outcomes in the replication set
A high PINI score was significantly related to a good OS and PFS in the external replication set (Supplementary Table 11). After adjustment with the clinical factors, a high PINI score was significantly associated with better OS and PFS (HR = 0.714, 95% CI = 0.558–0.917; HR = 0.766, 95% CI = 0.617–0.951, respectively). The high dichotomous PINI group had better OS and PFS than the low dichotomous PINI group (5-year OS: 82.8% vs 70.0%, p < 0.001; 5-year PFS: 72.2% vs 58.2%, p < 0.001; Supplementary Fig. 2). Dichotomous PINI was found to be a significant independent prognostic factor for OS and PFS in the replication set (HR = 0.763, 95% CI = 0.595–0.979; HR = 0.801, 95% CI = 0.644–0.996, respectively).
Subgroup analysis
To investigate the benefits of PINI as a prognostic novel index for clinical utility, we evaluated PINI score in stage IIa (T3N0) and low (T1-3N1) and high-risk stage III (T4 and/or N2) colon cancer subgroups among the 4535 (SNUH 3410 and NCC 1125) eligible study participants. In the stage IIa (T3N0) subgroup (N = 1197), patients with high PINI had better OS and PFS than patients with low PINI (Supplementary Fig. 3). PINI was an independently significant factor for OS (HR = 0.606, 95% CI = 0.442–0.832) and PFS (HR = 0.702, 95% CI = 0.530–0.930) in the multivariate survival analysis adjusted for age, sex, BMI, CEA levels, tumour location, tumour grade, lymphovascular invasion and adjuvant chemotherapy.
For the stage III subgroup analysis, patients with colon cancer of stage III high (T4 and/or N2, N = 587) and low (T1-3N1, N = 855) patients were identified. Low-risk stage III patients with low PINI and high-risk stage III patients with high PINI had a better prognosis than high-risk stage III patients with low PINI for OS and DFS. These patients had a worse prognosis than low-risk stage III patients with high PINI for OS and DFS (Supplementary Fig. 4).
Discussion
In this study, a novel prognostic index using systemic inflammation-associated blood features, termed PINI, was developed for colon cancer. This index had better predictive performance than other existing prognostic inflammatory markers, such as LMR, NLR, PLR and PNI. There were few studies to investigate which markers outperform including the existing prognostic inflammatory markers. However, to the best of our knowledge, most previous studies investigated only a small set of combinations of blood features as prognostic markers. The present study evaluated 42 candidate expressions generated by four basic mathematical operators. PINI was discovered through the robust analyses of 42 candidates. With respect to OS and PFS, PINI consistently outperformed in both the discovery and validation sets. PINI presented as an independent prognostic factor in the replication set. Moreover, dichotomous PINI provided promising outcomes in the discovery, validation and replication sets.
PINI is composed of two components (that is, ALB and MON) reflecting nutrition and inflammation. Although ALB is known as a nutritional marker, its concentrations are affected by inflammation regardless of malnutrition [19]. A high tumour burden can produce a large number of inflammatory cytokines, which in turn suppresses ALB synthesis from the liver [20]. A recent study showed that a low ALB concentration was related to the activation of systemic inflammation, the risk of malnutrition, low BMI, low subcutaneous and visceral obesity and low skeletal muscle mass in colorectal cancer [21]. Several other studies also demonstrated that ALB was associated with body composition in colorectal cancer [22, 23]. Colorectal cancer patients with hypoalbuminaemia are more likely to have unhealthy body composition and poor long-term outcomes. Several studies have demonstrated that serum ALB is an independent prognostic factor for survival in colorectal cancer [5, 24]. Independent of systemic inflammatory markers, a low blood level of ALB was associated with worse survival outcomes in colorectal cancer [21]. Monocytes are one of the major inflammatory components in cancer, and promote tumour progression through angiogenesis and invasion [25]. Circulating monocytes infiltrate the sites of inflammation and differentiate into macrophages in the tumour microenvironment [26]. These macrophages promote tumour cell migration and extravasation. Consequently, elevated MON was associated with poor survival in patients with colorectal cancer. PINI is formulated as (ALB × 0.9) − (MON × 0.0007), which has sign of coefficients consistent with their direction of regulation.
Among 42 candidate expressions, the predictive performance of the LMR was ranked second. The LMR has been suggested as an independent prognostic factor in patients with colorectal cancer [27]. In a meta-analysis study, a high LMR was associated with good survival in colorectal cancer patients [16]. In this study, the LMR was also an independent prognostic factor for OS and PFS. In the univariate analysis for the validation set, the LMR achieved the next highest C-index after PINI. Owing to this, LMR can be seen as a promising prognostic biomarker in colorectal cancer.
In the subgroup analyses, this study demonstrated that the high-risk group among patients with stage IIa can be identified using PINI. The absolute survival benefits of adjuvant chemotherapy in stage IIa cancer seem to be smaller than those in stage III cancer. In patients with stage IIa cancer, the high-risk group, who has a higher risk of recurrence, might obtain potential benefits from adjuvant chemotherapy. PINI can be utilised for stratifying the patients with stage IIa cancer for adjuvant therapy. In another subgroup analyses of the present study, PINI classified patients into more detailed prognostic groups in stage III colon cancer. For stage III cancer, the optimal duration of adjuvant oxaliplatin chemotherapy has been investigated. On the basis of the results of the International Duration Evaluation of Adjuvant Chemotherapy (IDEA) collaboration, 6 months of therapy is suggested for patients with high-risk stage III cancer and three months of therapy for those with low-risk stage III cancer [28]. This study presented that high-risk stage III patients with low PINI had worse survival than those with high PINI or low-risk stage III patients. Considering the results of PINI about the oncological outcome, evaluation of preoperative PINI could select high-risk groups who require intensive follow-up surveillance, adjuvant chemotherapy and proper duration of adjuvant therapy after resection.
The present study has several strengths for developing a novel prognostic index. Compared with other existing systemic inflammatory markers, PINI was developed and validated based on a relatively large cohort. Importantly, PINI was further validated with an externally independent cohort. Moreover, our approach can be seen as a comprehensive study, which investigated all simple, but effective combinations of any two systemic inflammatory factors, as well as four existing systemic inflammatory markers. Moreover, an optimal combination was determined by the distinctive prognostic group only, which has been successfully used to reduce false-positive errors [13, 29]. The non-distinctive prognostic group can be included as well to estimate the weights of factors in the optimal combination thoroughly. In addition, PINI is composed of MON and ALB, which are routinely measured for preoperative blood testing. Therefore, PINI is easily accessible for daily clinical application.
Meanwhile, there are several possible limitations regarding the current study. First, we mainly investigated seven laboratory features, which were considered as systemic inflammatory factors. However, other laboratory blood features, such as the CRP level, may also contribute to the prognosis of colon cancer [12, 30]. With regard to board investigation of the prognostic index for colon cancer patients, all laboratory features in the routine complete blood count test need to be considered in future works. Next, driven by the simplicity principle, we focused only on a combination of two laboratory features to develop a novel index. Combinations of more than two laboratory features should be considered in further studies. Although PINI was cross-validated using an external cohort, it would be helpful to further explore the generalisation of our proposed PINI in a multi-centre cohort.
In conclusion, PINI is a novel prognostic index that has improved the discriminatory power in colon cancer patients, including the influence of markers on both axes of acute-phase protein and inflammation. PINI may be utilised for predicting the prognosis of patients with colon cancer in daily clinical practise. This prognostic index may help to refine patients’ stratification for individualised therapy and tailored follow-up surveillance as the complement of the TNM staging system.
Supplementary information
Acknowledgements
We appreciate Mi Ae Lee, R.N. for assisting the collection of clinical data.
Author contributions
S-HJ, DK and JWP conceived and designed the study. MJK, S-BR, S-YJ, KJP, SCP, DKS, JHO and JWP collected the data. S-HJ, JH, DK and JWP analysed and interpreted the data. S-HJ did the statistical analysis. S-HJ, JH and JWP wrote the manuscript. MS, YN, JK, EKC, MJK, S-BR, S-YJ, KJP, SCP, DKS, JHO, H-HW and DK read and provided critical revision of the manuscript for intellectual contents; and all authors read and approved the final manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1F1A1063000).
Data availability
The datasets generated during and/or analysed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (no. 2101-042-1187, NCC2021-0057).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sang-Hyuk Jung, Jie Hao.
Contributor Information
Dokyoon Kim, Email: dokyoon.kim@pennmedicine.upenn.edu.
Ji Won Park, Email: sowisdom@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01767-w.
References
- 1.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 2.Park JW, Chang HJ, Yeo HY, Han N, Kim BC, Kong S-Y, et al. The relationships between systemic cytokine profiles and inflammatory markers in colorectal cancer and the prognostic significance of these parameters. Br J Cancer. 2020;123:610–8. doi: 10.1038/s41416-020-0924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sylman JL, Mitrugno A, Atallah M, Tormoen GW, Shatzel JJ, Tassi Yunga S, et al. The predictive value of inflammation-related peripheral blood measurements in cancer staging and prognosis. Front Oncol. 2018;8:78. doi: 10.3389/fonc.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanio A, Saito H, Uejima C, Takaya S, Yamamoto M, Tokuyasu N, et al. A prognostic index for colorectal cancer based on preoperative absolute lymphocyte, monocyte, and neutrophil counts. Surg Today. 2019;49:245–53. doi: 10.1007/s00595-018-1728-6. [DOI] [PubMed] [Google Scholar]
- 5.Heys S, Walker L, Deehan D, Eremin O. Serum albumin: a prognostic indicator in patients with colorectal cancer. J R Coll Surg Edinb. 1998;43:163–8. [PubMed] [Google Scholar]
- 6.Li MX, Liu XM, Zhang XF, Zhang JF, Wang WL, Zhu Y, et al. Prognostic role of neutrophil‐to‐lymphocyte ratio in colorectal cancer: a systematic review and meta‐analysis. Int J Cancer. 2014;134:2403–13. doi: 10.1002/ijc.28536. [DOI] [PubMed] [Google Scholar]
- 7.Lu C, Gao P, Yang Y, Chen X, Wang L, Yu D, et al. Prognostic evaluation of platelet to lymphocyte ratio in patients with colorectal cancer. Oncotarget. 2017;8:86287. doi: 10.18632/oncotarget.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song W, Wang K, Zhang R-J, Zou S-B. Prognostic value of the lymphocyte monocyte ratio in patients with colorectal cancer: a meta-analysis. Medicine. 2016;95:49. doi: 10.1097/MD.0000000000005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun G, Li Y, Peng Y, Lu D, Zhang F, Cui X, et al. Impact of the preoperative prognostic nutritional index on postoperative and survival outcomes in colorectal cancer patients who underwent primary tumor resection: a systematic review and meta-analysis. Int J Colorectal Dis. 2019;34:681–9. doi: 10.1007/s00384-019-03241-1. [DOI] [PubMed] [Google Scholar]
- 10.Hu X, Li Y-Q, Li Q-G, Ma Y-L, Peng J-J, Cai S-J. Baseline peripheral blood leukocytosis is negatively correlated with T-cell infiltration predicting worse outcome in colorectal cancers. Front Immunol. 2018;9:2354. doi: 10.3389/fimmu.2018.02354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Son H-J, Park JW, Chang HJ, Kim DY, Kim BC, Kim SY, et al. Preoperative plasma hyperfibrinogenemia is predictive of poor prognosis in patients with nonmetastatic colon cancer. Ann Surg Oncol. 2013;20:2908–13. doi: 10.1245/s10434-013-2968-8. [DOI] [PubMed] [Google Scholar]
- 12.Proctor M, Morrison D, Talwar D, Balmer S, O’reilly D, Foulis A, et al. An inflammation-based prognostic score (mGPS) predicts cancer survival independent of tumour site: a Glasgow Inflammation Outcome Study. Br J Cancer. 2011;104:726–34. doi: 10.1038/sj.bjc.6606087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrede O-J, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–60. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 14.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;8:37–45. [PubMed] [Google Scholar]
- 15.Yang Y, Gao P, Chen X, Song Y, Shi J, Zhao J, et al. Prognostic significance of preoperative prognostic nutritional index in colorectal cancer: results from a retrospective cohort study and a meta-analysis. Oncotarget. 2016;7:58543. doi: 10.18632/oncotarget.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Hu T, Zheng E, Deng X, Wang Z. Prognostic role of the lymphocyte-to-monocyte ratio in colorectal cancer: an up-to-date meta-analysis. Medicine. 2017;96:20. doi: 10.1097/MD.0000000000007051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst. 2018;110:803–11. doi: 10.1093/jnci/djy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peduzzi P, Concato J, Feinstein AR, Holford TR. Importance of events per independent variable in proportional hazards regression analysis II. Accuracy and precision of regression estimates. J Clin Epidemiol. 1995;48:1503–10. doi: 10.1016/0895-4356(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 19.McMillan DC, Watson WS, O’Gorman P, Preston T, Scott HR, McArdle CS. Albumin concentrations are primarily determined by the body cell mass and the systemic inflammatory response in cancer patients with weight loss. Nutr Cancer. 2001;39:210–3. doi: 10.1207/S15327914nc392_8. [DOI] [PubMed] [Google Scholar]
- 20.Cengiz O, Kocer B, Sürmeli S, Santicky M-J, Soran A. Are pretreatment serum albumin and cholesterol levels prognostic tools in patients with colorectal carcinoma? Med Sci Monit. 2006;12:CR240–CR247. [PubMed] [Google Scholar]
- 21.Almasaudi AS, Dolan RD, Edwards CA, McMillan DC. Hypoalbuminemia reflects nutritional risk, body composition and systemic inflammation and is independently associated with survival in patients with colorectal cancer. Cancers. 2020;12:1986. doi: 10.3390/cancers12071986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feliciano EMC, Kroenke CH, Meyerhardt JA, Prado CM, Bradshaw PT, Kwan ML, et al. Association of systemic inflammation and sarcopenia with survival in nonmetastatic colorectal cancer: results from the C SCANS study. JAMA Oncol. 2017;3:e172319–e172319. doi: 10.1001/jamaoncol.2017.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okugawa Y, Toiyama Y, Yamamoto A, Shigemori T, Kitamura A, Ichikawa T, et al. Close relationship between immunological/inflammatory markers and myopenia and myosteatosis in patients with colorectal cancer: a propensity score matching analysis. J Parenter Enter Nutr. 2019;43:508–15. doi: 10.1002/jpen.1459. [DOI] [PubMed] [Google Scholar]
- 24.Boonpipattanapong T, Chewatanakornkul S. Preoperative carcinoembryonic antigen and albumin in predicting survival in patients with colon and rectal carcinomas. J Clin Gastroenterol. 2006;40:592–5. doi: 10.1097/00004836-200608000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Chittezhath M, Dhillon MK, Lim JY, Laoui D, Shalova IN, Teo YL, et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity. 2014;41:815–29. doi: 10.1016/j.immuni.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Qian B-Z, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan JC, Chan DL, Diakos CI, Engel A, Pavlakis N, Gill A, et al. The lymphocyte-to-monocyte ratio is a superior predictor of overall survival in comparison to established biomarkers of resectable colorectal cancer. Ann Surg. 2017;265:539. doi: 10.1097/SLA.0000000000001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grothey A, Sobrero AF, Shields AF, Yoshino T, Paul J, Taieb J, et al. Duration of adjuvant chemotherapy for stage III colon cancer. N Engl J Med. 2018;378:1177–88. doi: 10.1056/NEJMoa1713709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danielsen H, Hveem T, Domingo E, Pradhan M, Kleppe A, Syvertsen R, et al. Prognostic markers for colorectal cancer: estimating ploidy and stroma. Ann Oncol. 2018;29:616–23. doi: 10.1093/annonc/mdx794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki S, Akiyoshi T, Oba K, Otsuka F, Tominaga T, Nagasaki T, et al. Comprehensive comparative analysis of prognostic value of systemic inflammatory biomarkers for patients with stage II/III colon cancer. Ann Surg Oncol. 2020;27:844–52. doi: 10.1245/s10434-019-07904-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available, but are available from the corresponding author on reasonable request.