Abstract
Background
Immunotherapy has revolutionised the field of cancer therapy and immunology, but has demonstrated limited therapeutic efficacy in high-grade serous ovarian cancer (HGSOC).
Methods
Multi-omics data of 495 TCGA HGSOC tumours and RNA-seq data of 1708 HGSOC tumours were analyzed. Multivariate Cox regression analysis and meta-analyses were used to identify prognostic genes. The immune microenvironment was characterised using the ssGSEA methods for 28 immune cell types. Immunohistochemistry staining of tumour tissues of 14 patients was used to validate the key findings further.
Results
A total of 1142 genes were identified as favourable prognostic genes, which are prevailing in immune-related pathways and the infiltration of most immune subpopulations was observed to be associated with a favourable prognosis suggesting that tumour immunogenicity was the most prominent factor associated with improved clinical outcomes and response to chemotherapy of HGSOC. We identified multiple genomic and transcriptomic determinants of immunogenicity, including the copy loss of chromosome 4q and deficiencies of the homologous recombination pathway. Finally, an immunological subtype characterised by increased infiltration of activated CD8 T cells and decreased Tregs was associated with favourable prognosis and improved therapeutic efficacy.
Conclusions
Our study characterised the immunogenomic landscape and refined the immunological classifications of HGSOC. This may improve the selection of patients with HGSOC who are suitable candidates for immunotherapy.
Subject terms: Predictive medicine, Prognostic markers
Introduction
High-grade serous ovarian cancer (HGSOC) is the most common histological subtype of ovarian cancer, and accounts for the majority (~70–80%) of ovarian cancer deaths [1, 2]. HGSOC is the most aggressive gynecologic cancer, and is generally diagnosed at advanced stages. Despite a high initial response rate to surgery and chemotherapy, most HGSOC patients will likely relapse or develop metastases, leading to poor outcomes, and thus, HGSOC has a 5-year survival of ~30% [3, 4].
Recently, cancer treatment has been revolutionised by the clinical successes of immunotherapy (including anti-PD-L1/PD-1, anti-CTLA-4 and CAR-T-based therapies) by harnessing the power of the immune system [5, 6]. Immunotherapy with immune checkpoint inhibitors (ICI) has been widely reported to enable unprecedented long-term survival in several types of cancer, including non-small cell lung cancer, melanoma and head and neck cancer [7–9]. However, malignant tumour cells can evade immune surveillance through complex and diverse immune evasion mechanisms [10]. Therefore, immunotherapy response rates vary greatly, even in patients with the same type of tumour [11]. Traditionally, HGSOCs are considered ‘immune-cold’ tumours characterised by lower immune infiltrates and a low response rate to immunotherapy. However, there is increasing evidence showing that HGSOC is one of the few types of cancer in which a spontaneous anti-tumour immune response is significantly associated with prolonged survival, and the immune evasion mechanisms are associated with poor survival [12], indicating the immunogenic nature of the HGSOC. Therefore, it is hypothesised that there is a solid rationale for using immunotherapy in HGSOC patients. However, clinical trials and resulting data from recent studies have demonstrated the low efficacy of ICI immunotherapy [13], which could be attributed to the complex tumour evasion mechanisms present in HGSOC. Furthermore, despite the limited efficacy in ICI immunotherapy of HGSOC, patients who exhibit a long-term response have also been observed in some trials, suggesting the existence of an immunogenic HGSOC subgroup of patients who are more likely to benefit from immunotherapy. Thus, a comprehensive understanding of immune evasion mechanisms and their relationship with prognosis and immunotherapy response is urgently required to enable improved prognostication, response prediction and immunotherapy strategies for HGSOC.
In the present study, a systematic analysis of immunogenomic features was performed in a large cohort of 2203 HGSOC tumours across 16 multicenter studies to gain a comprehensive insight into the immune evasion mechanisms and refine the classification of the HGSOC immunologic subtypes.
Materials and methods
Patients and samples
Transcriptional profiles and clinical annotations of HGSOC tumours were collected from Gene Expression Omnibus (GEO; ncbi.nlm.nih.gov/geo), The Cancer Genome Atlas (TCGA) (xenabrowser.net/), ArrayExpress (ebi.ac.uk/arrayexpress/) and published literature according to the following inclusion criteria: (1) tumours with FIGO stage III and IV and grade ≥ 2; (2) included overall survival (OS) information and (3) death events ≥20. After manual curation, a total of 2203 HGSOC tumours from 16 multicenter studies were included in the present study, and the detailed description of the 16 cohorts is summarised in supplementary table 1. Except for the tumours we indicated specifically in the manuscript, all the samples we used are primary tumours prior to systemic therapy. Raw data from 16 cohorts were pre-processed and normalised according to our previous studies [14]. A workflow of this study was shown in Supplementary Fig. 1.
Prognostic genes and functional analysis
A Multivariate Cox proportional hazards model with stage, grade and age were used to evaluate the association of each gene with OS time in each cohort. Then prognostic genes were identified by meta-analysis with a fixed-effects model across the 16 cohorts with a Benjamini-Hochberg false discovery rate-corrected P ≤ 0.2. Gene ontology (GO)-based enrichment analyses for prognostic genes were conducted and visualised using Cytoscape with the ClueGO plug-in [15, 16].
Multi-omics data and related analysis of TCGA
Amplification and deletion of genes were determined using GISTIC 2.0 software based on TCGA segmented copy number variation (CNV) files derived from Genomic Data Commons (GDC) (portal.gdc.cancer.gov) [17]. CNVs were categorised as amplification if the GISTIC value was 2 or 1, or a deletion if the GISTIC value was −2 or −1. Cytoband enrichment analyses were performed using an EASE score (modified Fisher’s exact maximum probability) ≤0.1 for identifying statistically significant cytoband overrepresentation. The number of predicted MHC Class I-associated neoantigens for TCGA HGSOC tumours were retrieved from Rooney et al. [18]. Tumour samples with BRCA1/2 mutation, BRCA1 hypermethylation, EMSY amplification, defects of PTEN, RAD, FA, ATR, ATM, CHEK1/2 or PALB2 were considered as homologous recombination (HR)-defective tumours according to previous studies [14, 19, 20].
Immune microenvironment characterisation
Our study does not require evaluating relative fractions of immune infiltrations that might be biased by the deconvolution method. Therefore, the infiltration levels for 28 immune cell types were quantified using the single sample gene set enrichment analysis (ssGSEA) implemented in the GSVA package based on 782 genes that specifically expressed in particular immune cell types [21]. The resulting ssGSEA enrichment scores were then used to cluster the HGSOC tumours using the unified manifold approximation and projection (UMAP) analysis, which reduced the 28-dimensional space to a two-dimensional space and coloured each sample based on the molecular subtype. The cytolytic immune activity (CYT) was quantified using the geometric mean of transcript levels of GZMA and PRF1 genes, as previously reported [18].
Immunohistochemistry (IHC) and cell quantification
The ovarian cancer tissues of 14 patients obtained from Harbin Medical University were cut into 4 μm sections using a microtome, fixed on a glass slide, dried on the 42 °C grill surface for 2 min, and then placed in an oven at 65 °C for 1 h. The tissue sections were dewaxed three times with xylene (30 min each) and gradually hydrated using a decreasing gradient of alcohol solutions. Slides were then soaked three times in distilled water (3 min each), and then immersed in pH6.0 citrate buffer solution and heated for antigen retrieval. The slides were removed after 10 min of boiling and then allowed to cool passively, washed three times with PBS (3 min each), and incubated with 0.3% H2O2 for 20 min at room temperature to quench endogenous peroxidase activity. The slides were subsequently incubated overnight at 4 °C in a wet box with anti-CD3 antibodies (1:100, Rabbit, clone 2GV6, Roche Diagnostics) and anti-CD8 antibodies (1:100, Rabbit, clone SP57, Roche Diagnostics). The specimens treated with PBS solution or primary antibody diluent without the addition of the antibody were used as the negative controls. After washing several times with PBS, the sections were incubated with the biotinylated secondary antibodies (PV-6000D, ZSGB-BIO) for 1 h at room temperature. Freshly prepared DAB chromogenic solution (1 ml DAB substrate buffer and 50 μl DAB concentrated solution) was added to the tissues to develop the signals (~1 min), and tissues were subsequently observed under a microscope. When the cytoplasm turned brown and the background had no obvious staining, slides were immersed in tap water to stop staining. Next, tissues were counterstained with hematoxylin for 4 min at room temperature, rinsed with tap water, immersed in 1% hydrochloric acid alcohol solution for differentiation for 2–3 sec, and washed under flowing tap water for 15 min. The slides were further dehydrated, cleared using xylene and sealed with neutral resin. The positive control group was a positive immune-stained slide obtained from the Department of Pathology, Harbin Medical University. Three pathologists blinded to the conditions independently evaluated and recorded the IHC results in all cases for analysis. Data are presented as the absolute number of positive cells/mm2 (for CD3+ T cells and CD8+ T cells), as previously described [22, 23].
Statistical analysis
A one-way ANOVA, a Wilcoxon rank-sum test or a Fisher’s exact test were used to compare differences between the groups. Survival analysis was performed using the Kaplan–Meier method and statistically compared using a log-rank test. All statistical analyses were performed using R version 3.3.1, with two-sided P-values.
Results
The immune microenvironment is a crucial determinant of cancer prognosis
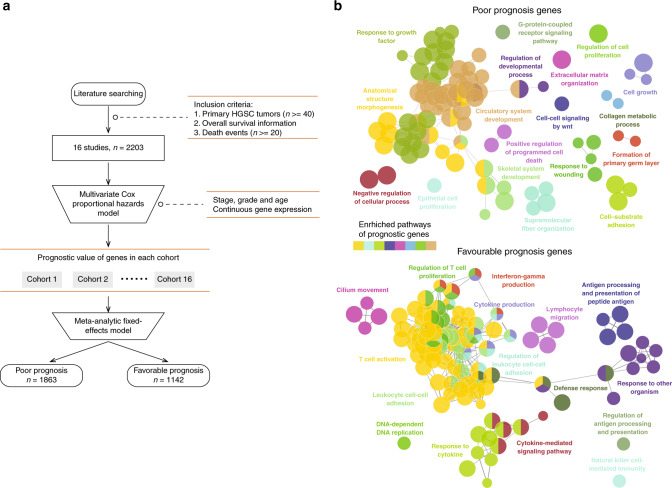
To determine the transcriptional features associated with clinical outcomes, we first built a compendium of genes related to OS using the gene expression profiles of 2203 HGSOC samples from 16 studies. An overview of the analytical strategy for identifying reliable prognostic genes is shown in Fig. 1a. We performed multivariate Cox regression analysis for gene expression adjusting for stage, grade and age in each dataset independently, and then integrated the results using meta-analyses to leverage the prognostic value of all 16 studies. A total of 1142 genes were identified as favourable prognostic genes, and 1863 genes were considered risky prognostic transcriptional features associated with a poor prognosis (Supplementary Table 2). We then conducted pathway enrichment analysis for these favourable and risky prognostic genes, which showed functional heterogeneity between favourable and risky prognostic genes. As shown in Fig. 1b, the results of the functional characterisation showed that risky prognostic genes were enriched in cancer invasion and metastasis-related pathways, such as cell proliferation and cell death, whereas favourable prognostic genes were prevailing in immune-related pathways, such as T cell activation, T cell proliferation, lymphocyte migration and cytokine-mediated signaling pathways (Fig. 1b). These results suggest that the immune microenvironment context is a crucial determinant contributing to a favourable prognosis.
Fig. 1. Functional characterisation of prognostic genes.
a Analytical pipeline for the indentation of prognostic genes. b Visualisation of functionally grouped networks with enriched GO terms as nodes, where the node size represents the enrichment significance; the label of the most significant term per group is shown. Functionally related groups partially overlap. GO Gene Ontology.
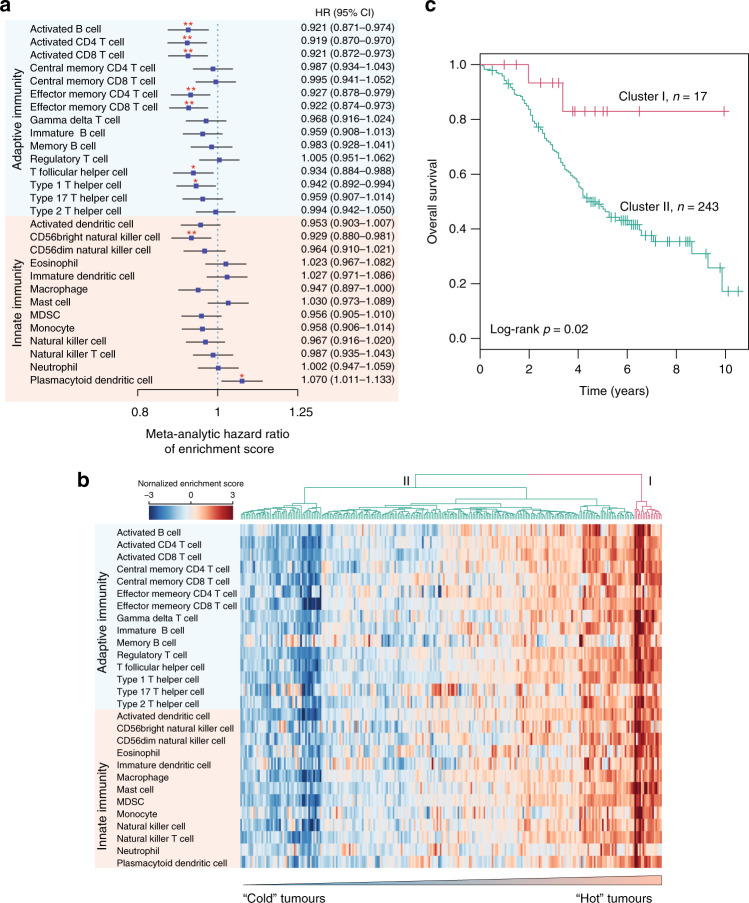
To further examine which cellular composition of the immune infiltrate in the tumour immune microenvironment (TIME) affected patient prognosis, we estimated the correlation of 28 infiltrating immune subpopulations with prognosis by performing univariate Cox regression analysis for a fraction of the individual infiltrating immune cell types with OS within each dataset, and then conducted a meta-analysis to leverage the 16 datasets (Fig. 2a, Supplementary Fig. 2 and Supplementary Table 3). The results of the meta-analysis revealed associations between specific infiltrating immune subpopulations with prognosis. As shown in Fig. 2a, the infiltration of most immune subpopulations was observed to be associated with a favourable prognosis. Of these, seven immune subpopulations related to adaptive immunity, including activated B cells, activated CD4 T cells, activated CD8 T cells, effector memory CD4 T cells, effector memory CD8 T cells, T follicular helper cells and type 1 T helper cells, were significantly associated with a favourable prognosis. On the other hand, only one of the immune subpopulations of innate immunity (CD56 bright natural killer cells) was significantly associated with a favourable prognosis (Fig. 2a). Plasmacytoid dendritic cells (pDC) were significantly associated with a poor prognosis (Fig. 2a), reminiscent of its tumour-promoting function and poor prognosis in breast cancer [24]. To further characterise the heterogeneity of immune infiltrates within HGSOC tumours, we performed hierarchical clustering of the HGSOC tumours based on the infiltrating immune cell composition, which identified two patient clusters with distinct immunophenotypes. As shown in Fig. 2b, tumours in cluster II, accounting for the majority of HGSOC tumours, tended to be immune-cold and exhibited low levels of immune infiltrates, whereas tumours in cluster I, accounting for a small fraction of HGSOC tumours, tended to be immune-hot and were associated with higher levels of immune infiltrates. Patients in the immune-hot-like cluster were observed to experience significantly longer survival times compared with those in the immune-cold-like cluster [hazard ratio = 0.61, 95% confidence interval (CI), 0.38–0.97, P = 0.02; log-rank test] (Fig. 2c). The high immunogenicity in a small subset of patients suggests that immunotherapy may be beneficial for some HGSOC patients that can be identified by tumour immunogenomic features.
Fig. 2. The landscape of the tumour immune microenvironment.
a Forest plot visualising the meta-analytic hazard ratios (95% confidence interval) of univariate Cox regression analyses of each infiltrating immune subpopulation in the 16 cohorts. b Hierarchical clustering of intratumoural immune states based on the cellular composition of the immune infiltrates calculated using ssGSEA. The two predominant clusters are referred to as ‘Cluster I’ and ‘Cluster II’. c Kaplan–Meier survival curves of overall survival between Cluster I and Cluster II. Statistical analysis was performed using a log-rank test. *P < 0.05, **P < 0.01. ssGSEA single sample gene set enrichment analysis.
Genomic states contribute to immunophenotypes of HGSOC tumours
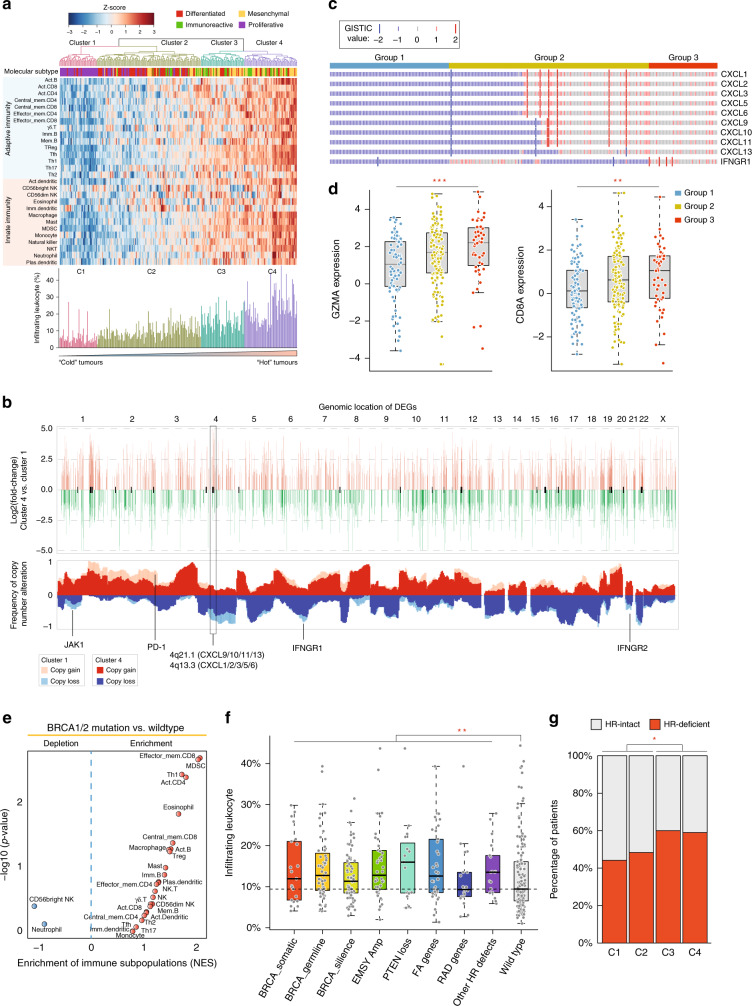
To characterise the intratumoural immune states, we first performed unsupervised hierarchical clustering for TCGA HGSOC tumours with genomic data based on the cellular composition of the immune infiltrate evaluated using the GSEA method. In total, four clusters with different immune phenotypes were identified (Fig. 3a). As shown in Fig. 3a, the immune infiltrate increases from cluster C1 to C4. Tumours in the C1 cluster displayed the lowest levels of immune infiltrate, and therefore were designated as immune-cold HGSOC tumours, whereas tumours in the C4 cluster exhibited the highest levels of immune infiltrates and therefore were designated as immune-hot HGSOC tumours. Tumours in C2 and C3 clusters were observed to have intermediate levels of immune infiltrates, and therefore were combined and designated as immune-medium HGSOC tumours. We found a distinct distribution of molecular subtypes that characterised the four clusters by analysing the association of molecular subtypes and the four clusters. Cluster 1 was composed almost entirely of proliferative subtypes, and Cluster 4 was dominated by mesenchymal and immunoreactive subtypes.
Fig. 3. Genomic states associated with the immunological phenotypes.
a Heatmap of the cellular composition of the immune infiltrate. TCGA HGSOC tumours were hierarchically clustered into four primary clusters. b Visual representation of the statistically enriched chromosome locus and CNV pattern for differentially expressed genes between immune-cold and immune-hot tumours. c Oncoplot showing the copy number variation pattern of CXC chemokines and IFNGR1. Copy number loss or gain was determined using GISTIC 2.0. d Boxplot showing mRNA expression levels of GZMA and CD8A amongst the different patient groups. Statistical analysis was performed using a one-way ANOVA. e Volcano plots showing the enrichment of infiltrating immune subpopulations for tumours with and without BRCA1/2 mutations calculated based on the NES score from GSEA. f Boxplot showing the relative abundance of infiltrated leukocytes in HR-defective and HR-intact tumours. Statistical analysis was performed using a Wilcoxon rank-sum test. g Distribution of tumours with and without HRD across different patient groups. Statistical analysis was performed using a Fisher’s exact test. *P < 0.05, **P < 0.01, ***P < 0.001. TCGA The Cancer Genome Atlas, HGSOC high-grade serous ovarian cancer, CNV copy number variation, IFNGR1 interferon-γ receptor 1, NES normalised enrichment score, GSEA gene set enrichment analysis, HR homologous recombination, HRD homologous recombination deficiency.
To characterise the genomic features of immune-hot HGSOC tumours, we compared gene expression patterns between immune-cold and immune-hot HGSOC tumours, to identify the upregulated and downregulated genes (Fig. 3b). Cytoband enrichment analysis showed that these immune phenotype-related DEGs were significantly enriched in several chromosomal bands in which specific essential immune genes, such as JAK1, PD-1 and IFNGR1/2, were located. We plotted the copy number status of C1 and C4 clusters across the genome and revealed a significant association between CNVs with the immune phenotype. As shown in Fig. 3b, frequent copy number deletion or amplification was observed in enriched chromosomal bands. On chromosomes 1, 4, 5, 9, 21 and 22, genes tended to have copy number deletions in the immune-cold HGSOC tumours, whereas on chromosomes 2, 3, 13 and 20, genes were likely to be amplified in the immune-cold HGSOC tumours relative to the immune-hot HGSOC tumours. In particular, copy number deletions frequently occurred at the chromosomal loci 4q21.1 and 4q13.3 in immune-cold HGSOC tumours, consistent with the above observation that upregulated genes in immune-hot HGSOC tumours tended to be enriched in these chromosomal regions.
Furthermore, many important members of the CXC chemokine family are located in chromosomal bands 4q21.1 (including CXCL9/10/11 and CXCL13) and 4q13.3 (including CXCL1–3, CXCL5–6 and CXCL9). By sorting tumours according to the GISTIC values of the members of the CXC chemokine family and IFNGR1, we found that the CNV pattern of the CXC chemokines could be used to classify tumours into three groups: Tumours with complete deletion, partial deletion or no deletion. It has been previously shown that one of the functions of CXC chemokines is the recruitment of effector cells (such as monocytes and neutrophils) into the tumour microenvironment (TME) [25]. Therefore, we examined the expression patterns of GZMA and CD8A amongst the three groups and revealed a significant difference in the expression distribution of GZMA (P < 0.001, one-way ANOVA) and CD8A (P < 0.006, one-way ANOVA) (Fig. 3d). The expression levels of GZMA and CD8A are generally associated with a decrease in the deletion of genes encoding the CXC chemokines. As shown in Fig. 3d, the expression of GZMA and CD8A in tumours with no deletions of the CXC chemokines genes was significantly higher compared with those classed as complete and partial deletion. These results suggested that the copy number loss of these genes might be a reason why these tumours have relatively lower immune infiltration.
To define whether homologous recombination deficiency (HRD) was related to the immunophenotype, we examined whether the BRCA1/2 genotype of tumours was associated with the immunophenotype by estimating the infiltration of different immune subpopulations. As shown in Fig. 3e, most immune subpopulations were enriched in BRCA1/2 mutated tumours compared with wild-type tumours. Additionally, with the BRCA1/2 mutations, similar results were observed for other HR-defective tumours, which showed significantly higher levels of infiltrated leukocytes than the HR-intact tumours (P = 0.01, Wilcoxon rank-sum test) (Fig. 3f). Besides, HR-defective tumours tended to frequently occur in immune-hot clusters relative to immune-cold clusters (p < 0.05, Fisher exact test) (Fig. 3g). These results demonstrated that the HRD genotype was associated with a highly active immune phenotype and the non-HRD genotype was associated with the immunosuppressive phenotype.
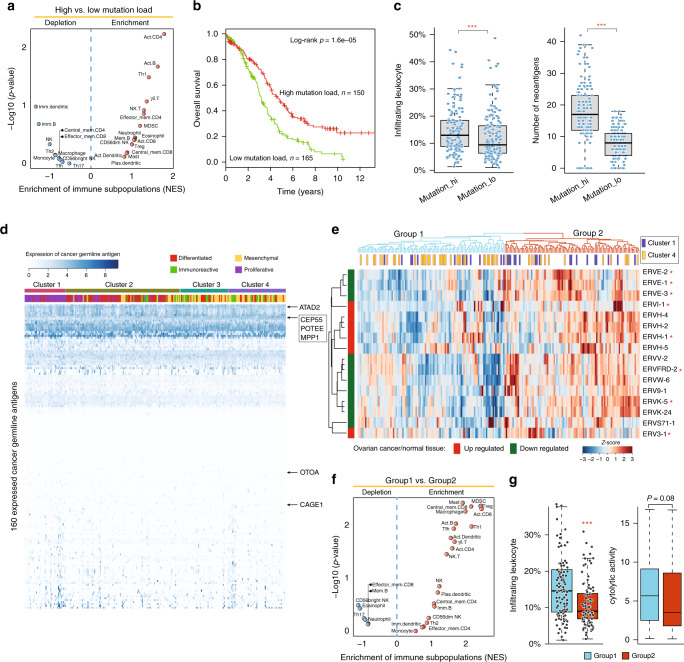
In has been reported that tumour immunogenicity is a critical aspect of intrinsic immune escape [26]. Therefore, we further investigated the associations of mutational load and neoantigens, which are two important factors determining tumour immunogenicity, with immunophenotypes of HGSOC tumours. The HGSOC tumours were first classified into two groups: Tumours with high and low mutational loads. As shown in Fig. 4a, effector cells, such as activate CD4 and T cells, were significantly overrepresented in the TME of tumours with a high mutational load compared with those with a low mutational load, consistent with the observation that a high mutational load was associated with improved patient survival (hazard ratio = 0.55, 95% CI, 0.41–0.72, P < 0.001; log-rank test) (Fig. 4b). In addition, tumours with a high mutational load exhibited significantly higher levels of lymphocytic infiltrates and neoantigen burden than those with a low mutational load (P < 0.001, Wilcoxon rank-sum test) shown in Fig. 4c. However, there are no significant differences in the expression of cancer-germline antigens among the different immunophenotypes of HGSOC tumours (Fig. 4d). Further investigation of the expression pattern of endogenous retroviruses (ERVs) suggested that ERV expression could be used to classify HGSOC tumours into two groups, as shown in Fig. 4e. HGSOC tumours in group 2 with high levels of ERV expression were observed to have a lower enrichment of immune cells, tumour-infiltrating lymphocytes (TILs) and cytolytic activity (Fig. 4f, g).
Fig. 4. Tumour immunogenicity is associated with the different immunological phenotypes.
a Volcano plots showing the enrichment of infiltrating immune subpopulations of tumours with a high or low mutational load calculated based on the NES score from the GSEA. b Kaplan–Meier curves of overall survival between patients with a high and low mutational load. Statistical analysis was performed using a log-rank test. c Boxplot showing the relative abundance of infiltrated leukocytes and neoantigens in patients with a high and low mutational load. d Heatmap of the expression pattern of 160 cancer-germline antigens amongst the different immunophenotypes of HGSOC tumours. e Hierarchical clustering of HGSOC tumours based on the expression pattern of ERVs. f Volcano plots showing the enrichment of infiltrating immune subpopulations of tumours in two different groups derived from ERVs expression. g Boxplot showing the infiltrated leukocytes abundance and cytolytic activity. ***P < 0.001. ERVs endogenous retroviruses, NES normalised enrichment score, GSEA gene set enrichment analysis.
Chemotherapy modulates the immune microenvironment of HGSOC tumours
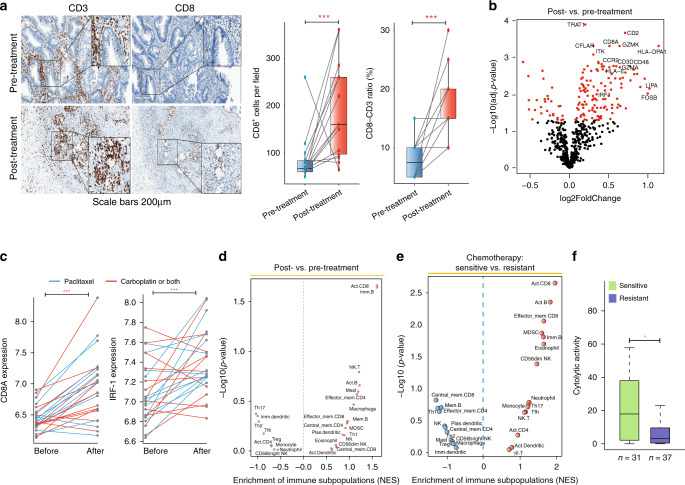
To investigate the impact of chemotherapy on the immune microenvironment of HGSOC tumours, we first collected paired samples before and after neoadjuvant chemotherapy of 14 HGSOC patients. Results of IHC staining of the 14 patients showed a significant increase in CD8+ T cell densities and in the CD8/CD3 ratio of patients following neoadjuvant chemotherapy relative to their treatment naïve counterparts (Fig. 5a), suggesting that chemotherapy-induced the infiltration of effector immune cells. To further validate the finding, we compared the expression changes of immune marker genes in 25 paired HGSOC tumours before and after three cycles of either carboplatin- or paclitaxel-based chemotherapy derived from the GSE15622 cohort [27]. As shown in Fig. 5b, immune marker genes, especially those from effector cells, are significantly upregulated following chemotherapy. For example, the T-effector gene CD8A and IRF-1 exhibited significantly higher expression levels in post-chemotherapy tumours relative to pre-chemotherapy (P < 0.001, paired t-test) (Fig. 5c). Similar results were also observed for non-paired HGSOC tumours either pre- or post-neoadjuvant treatment from the GSE71340 cohort [28]. HGSOC tumours received neoadjuvant treatment revealed significant enrichment of immune subpopulations, especially effector cells, such as activated CD8+ T cells, NK T cells and activated B cells (Fig. 5d).
Fig. 5. Chemotherapy modulates the immune microenvironment of HGSOC tumours.
a Representative immunohistochemical images of formalin-fixed paraffin-embedded HGSOC samples and related statistics show that neoadjuvant chemotherapy increased the presence of tumour-infiltrating lymphocytes. Scale bar: 200 μm. b Differentially expressed marker genes of the different immune subpopulations in the paired HGSOC tumours before and after three cycles of either carboplatin- or paclitaxel-based chemotherapy. c Pairwise comparison of the expression levels of CD8A and IRF-1 in tumours before and after chemotherapy. Statistical analysis was performed using a paired t-test. d Volcano plots showing the enrichment of infiltrating immune subpopulations for tumours before and after chemotherapy calculated based on the NES score from GSEA. e Volcano plots showing the enrichment of infiltrating immune subpopulations of sensitive and resistant tumours. f Boxplot showing the distribution of cytolytic activity in chemotherapy-resistant and -sensitive tumours. Statistical analysis was performed using a Wilcoxon rank-sum test. *P < 0.05, ***P < 0.001. HGSOC high-grade serous ovarian cancer.
By comparing the cellular composition of immune infiltrates of patients with resistant and sensitive information from Patch et al. [29], we found that chemotherapy-sensitive HGSOC tumours were characterised by significant enrichment of infiltrating immune effector cells compared with chemotherapy-resistant tumours (Fig. 5e). Furthermore, cytolytic activity was observed to be significantly higher in chemotherapy-sensitive tumours compared with chemotherapy-resistant tumours (P = 0.02, Wilcoxon rank-sum test) (Fig. 5f). These results indicated that chemotherapy is involved in modulating the immune microenvironment and stimulating the intratumoural infiltration of immune effector cells, both of which are associated with chemotherapy sensitivity.
The interplay between immunophenotypes and molecular subtypes of HGSOC tumours
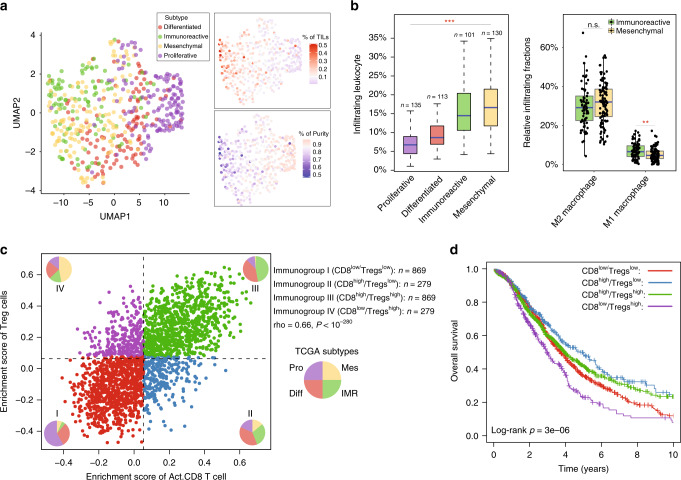
We first clustered HGSOC tumours based on the infiltration levels of 28 immune subpopulations and projected to the 2-dimensional UMAP. The distribution of subtypes in the UMAP (Fig. 6a) shows that molecular subtypes have different immunophenotypes. The UMAP1 dimension is associated with immune infiltration, and the mesenchymal and immunoreactive molecular subtypes tended to be distinguished by the UMAP2 dimension. Although both mesenchymal and immunoreactive molecular subtypes revealed significantly higher levels of TILs compared with the proliferative and differentiated subtypes, there was still a difference in the compositions of TILs between the mesenchymal and immunoreactive molecular subtypes (Fig. 6b). For example, there was a significant difference in the infiltration of M1 macrophages (P = 0.03, Wilcoxon rank-sum test), but no difference in M2 macrophages (P = 0.2, Wilcoxon rank-sum test) between the mesenchymal and immunoreactive molecular subtypes. These observations highlight the heterogeneity of the cellular composition of the immune infiltrates in the TME of HGSOC tumours.
Fig. 6. The interplay between immunophenotypes and intrinsic molecular subtypes.
a UMAP plot visualising spatial distributions of intrinsic molecular subtypes based on the infiltration levels of 28 immune subpopulations. b Boxplot showing the distribution of tumour-infiltrating lymphocytes between the different intrinsic molecular subtypes. Statistical analysis was performed using a one-way ANOVA and a Wilcoxon rank-sum test. c Visualisation of the four novel immunophenotypes based on the crosstalk between CD8+ T and regulatory T cells (Tregs) for HGSOC tumours. d Kaplan–Meier curves of overall survival between the different immunophenotypes. Statistical analysis was performed using a log-rank test. **P < 0.01, ***P < 0.001. n.s. not significant, mUC metastatic urothelial cancer.
Immunologic subtype refinement based on a combination of activated CD8+ T and regulatory T cells
It is well established that activated CD8 T cells serve a critical role in the anti-tumour immune responses, whereas regulatory T cells (Tregs) are capable of forming immunological barriers against CD8+ T-cell-mediated anti-tumour immune responses [30, 31]. However, a significantly positive correlation was observed between the activated CD8 T cells and Tregs was observed in the TME (Fig. 6c). Therefore, we further examined whether the crosstalk between activated CD8 T cells and Tregs could implicate clinically different behaviours. We defined high and low activated CD8 T infiltration as values above and below the median density, and similarly defined high and low Treg levels based on the median, and found that the combination of activated CD8 T and Tregs could yield four different immunological subtypes of HGSOC. As shown in Fig. 6c, the immunogroup I was characterised by a low infiltration of both activated CD8 T cells and Tregs (referred to as CD8low/Tregslow), whereas immunogroup III was defined by strong activation of both activated CD8 T cells and Tregs (referred to as CD8high/Tregshigh). Immunogroup II was characterised by high infiltration of activated CD8 T cells and low infiltration of Tregs (referred to as CD8high/Tregslow), and immunogroup IV was classed as high levels of infiltration of Tregs and low levels of infiltration of activated CD8 T cells (referred to as CD8low/Tregshigh) (Fig. 6c). By examining the strength of the association between four immunogroups and four existing molecular subtypes, we found that the mesenchymal subtype was enriched in CD8low/Tregshigh subtype, whereas the immunoreactive subtype was enriched in CD8high/Tregslow and CD8high/Tregshigh subtypes. The CD8low/Tregslow subtype had the highest representation of the proliferative subtype, and no obvious different distribution was observed for the differentiated subtype across the four immunological subtypes (Fig. 6c). Next. we assessed the survival of patients across the four immunological subtypes. As shown in Fig. 6d, patients in the CD8high/Tregslow subtype exhibited the longest survival times, whereas CD8low/Tregshigh subtype was associated with the worst overall survival. Although TILs between CD8low/Tregslow and CD8high/Tregshigh differed significantly, no significant difference in overall survival was found between these two groups.
Discussion
One of the primary roles of the immune system is to monitor and eliminate malignant transformation, which is referred to as tumour immune surveillance [32]. However, malignant tumour cells can evade immune surveillance. Although considerable efforts have been made in identifying markers of prognosis in HGSOC involved in expression patterns, gene amplification, ERVs and TILs [14, 33–37], to date, there have been no studies that have comprehensively characterised the immune evasion mechanisms and its therapeutic implication in HGSOC from a systems biology viewpoint, to the best of our knowledge. An improved understanding of tumour immune evasion mechanisms promises to develop novel immunotherapeutic strategies and guide personalised treatments.
In the present study, we conducted an extensive immune and genomic characterisation of 2203 HGSOC tumour samples from 16 multicenter studies to investigate the immunogenomic features and their influence on prognosis and therapeutic response. Functional analysis of prognostic genes highlighted that favourable prognostic genes are involved in immune-related pathways and genes associated with a worse prognosis were enriched in known cancer invasion and metastasis-related pathways. A recently published meta-analysis for HGSOC also found that the best performing prognostic genes were enriched in pathways involved in the immune system [37]. The intratumoural cellular characterisation of immune infiltrates identified four novel patient clusters with heterogeneous immunophenotypes of low, intermediate and high levels of immune infiltrate, termed ‘immune-cold’, ‘immune-medium’ and ‘immune-hot’ tumours, respectively (Fig. 3a). We revealed a cluster of’immune-hot tumors’ in HGSOC (Cluster 4), which was indicative of an improved outcome compared with the other three patient clusters. As reported in urothelial cancer, which is generally considered an immune-cold tumour, patients with high levels of immune infiltrates are most responsive to checkpoint blockade [38]. Therefore, identifying an immune-hot subset supports the rationale for selecting HGSOC patients suitable for ICI-based treatment.
Previous studies on the association between genotypes and immunophenotypes have revealed that the genotypes of the tumours determine the immunophenotypes and thus the tumour escape mechanisms [21]. Here, by comparing the genomic states from a gene-specific view between immune-cold and immune-hot tumours, our results revealed enrichment of copy number deletion of chromosome arm 4q, which includes genes encoding for the CXC chemokine family members (CXCL1–13) in the immune-cold tumours, which tended to be associated with immunosuppressive features. Therefore, it is reasonable to speculate that constitutive copy number alteration in chromosome arm 4q may be a mechanism underlying immune evasion that allows HGSOC tumours to shift towards immunosuppressive phenotypes. For example, the CXC chemokine family from a cluster located on chromosomal band 4q serves a critical role in the recruitment of effector cells [25]. Therefore, the deletion of CXC chemokines may contribute to tumour immune evasion, implying an opportunity to design combinatorial approaches for cold tumours with a heterozygous loss or normal copies of chemokine genes.
Similarly, focal amplification on chromosome segment 4q21 in the favourable immune phenotype was also observed in colon cancer and breast cancer [39, 40]. A previous study has demonstrated the association of genetic and environmental properties of tumours with local immune cytolytic activity [18]. Therefore, we further characterised the cancer antigenome among different immunophenotypes of HGSOC tumours, and revealed that neoantigens arising from somatic mutations and ERVs were heterogeneous across the different HGSOC immunophenotypes. In contrast, the cancer-germline antigens were independent of the immunophenotype of the HGSOC tumours, which was largely due to the high-frequency occurrence of HR pathway alterations, particularly for BRCA mutations in HGSOC tumours, which can significantly increase genomic mutations and produce an increased quantity of neoantigens (Fig. 3e), thus enhancing the immune response. Viruses, another class of germline-encoded elements, have been reported as inducers of immune responses [18]. Our results revealed that ERVs in HGSOC tumours demnonstrated significant association with immune microenvironment, with high ERV-expressing tumours showing a higher ability to evade immune surveillance, consistent with the recently reported association between ERVs and anti-tumour immunity in epithelial ovarian cancer (EOC) [36].
We further examined a relationship between IMS and immunophenotypes of HGSOC tumours, and revealed heterogeneity in tumour immune microenvironments across four molecular subtypes. Therefore, based on the above observations, we hypothesised an immunological classification system based on the crosstalk between the CD8+ T cells and Tregs, and separated HGSOC patients into four immunological subtypes: CD8low/Tregslow, CD8high/Tregshigh, CD8high/Tregslow and CD8low/Tregshigh. There is an imbalance in the distribution of IMS across the four immunologic subtypes. Immunogroup IV, which has high Tregs and low levels of CD8 + T cells, was enriched in the mesenchymal subtype, which had the highest amount of TILs. Conversely, immunogroup I with low infiltration of both CD8+ T cells and Tregs were enriched in the proliferative subtype, in which the quantity of TILs was relatively low. Although the number of patients did not differ in terms of IMS, the OS of patients in the CD8high/Tregslow group was better than those in the other immunological groups.
Furthermore, there was no significant difference in OS between CD8low/Tregslow and CD8high/Tregshigh, even if CD8high/Tregshigh exhibited significantly higher numbers of TILs compared with CD8low/Tregslow. These observations suggest complex immunomodulatory interactions between intrinsic molecular programs and intratumoural immune response, and show that the relative cellular composition between immune infiltrates in the immune microenvironment has a more significant influence on survival than the absolute abundance. Therefore, we reasoned that the four immunological subtypes based on the infiltration of CD8+ T cells and Tregs in the TME not only significantly expand on gene expression-based subgrouping but may also facilitate the selection of patients suitable for immunotherapy.
This study has several limitations that should be acknowledged. First, considering the limitations of measurement technology and intratumoural heterogeneity, we chose a lenient FDR during the discovery phase of a biomarker which has limitations in the more rigorous and might contain biases. Second, the definition of OS in previously published literature was not used in the same manner. Third, the criteria of HGSOC were not validated by pathology, immunohistochemistry or mutation/gene expression status, but come from the previously published literature that is not always consistent. Therefore, to keep it consistent among different datasets and to include as many samples as possible, we defined HGSOC as the tumours with both high-grade (≥2) and high-stage (≥III). The fourth is the retrospective nature of our study. Although our retrospective study tried to include as many patient cohorts as possible for more rigorous discovery, biomarkers and immunological classifications need to be prospectively validated in independent well-designed clinical trials following REMARK criteria.
In summary, our study identified and characterised heterogeneous immune phenotypes associated with differential clinical outcomes and therapeutic effects. These immunophenotypes could be characterised by specific cellular, molecular, genetic and environmental factors contributing to the distinct potential immune escape mechanisms evoked. With an increasing understanding of immune evasion mechanisms, the characterisation of immunogenomic features holds important promise for allowing a more precise categorisation of patients who may benefit from immunotherapy.
Supplementary information
Author contributions
MZ and DPH conceived and designed the study. JS, DPH, CCY, DDX, ZCZ and KL prepared and carried out all analyses, including the development of their statistical framework, and interpreting the data. XBL collected the patient samples and performed immunohistochemical analysis. MZ and DPH drafted the manuscript. All authors participated in the interpretation and discussion of the results and in the version of the manuscript.
Funding
This study was supported by the Zhejiang Provincial Natural Science Foundation of China (Grant No. LY22C060001). The funders had no roles in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Data availability
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Harbin Medical University and written informed consent was obtained from all the participants.
Consent for publication
All authors agree with the content of the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jie Sun, Congcong Yan, Dandan Xu.
Contributor Information
Meng Zhou, Email: zhoumeng@wmu.edu.cn.
Dapeng Hao, Email: dapeng.hao@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01692-4.
References
- 1.Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ, Bast RC, Jr., Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–79. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 3.Olivier RI, van Beurden M, Lubsen MA, Rookus MA, Mooij TM, van de Vijver MJ, et al. Clinical outcome of prophylactic oophorectomy in BRCA1/BRCA2 mutation carriers and events during follow-up. Br J Cancer. 2004;90:1492–7. doi: 10.1038/sj.bjc.6601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945–56.. doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–68. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akinleye A, Rasool Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J Hematol Oncol. 2019;12:92. doi: 10.1186/s13045-019-0779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadal E, Massuti B, Domine M, Garcia-Campelo R, Cobo M, Felip E. Immunotherapy with checkpoint inhibitors in non-small cell lung cancer: insights from long-term survivors. Cancer Immunol Immunother. 2019;68:341–52. doi: 10.1007/s00262-019-02310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szturz P, Vermorken JB. Immunotherapy in head and neck cancer: aiming at EXTREME precision. BMC Med. 2017;15:110. doi: 10.1186/s12916-017-0879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss SA, Wolchok JD, Sznol M. Immunotherapy of melanoma: facts and hopes. Clin Cancer Res. 2019;25:5191–201. doi: 10.1158/1078-0432.CCR-18-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35:S185–S98. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kandalaft LE, Motz GT, Duraiswamy J, Coukos G. Tumor immune surveillance and ovarian cancer: lessons on immune mediated tumor rejection or tolerance. Cancer Metastasis Rev. 2011;30:141–51. doi: 10.1007/s10555-011-9289-9. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Martin A, Sanchez-Lorenzo L. Immunotherapy with checkpoint inhibitors in patients with ovarian cancer: still promising? Cancer. 2019;125:4616–22. doi: 10.1002/cncr.32520. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Bao S, Xu D, Zhang Y, Su J, Liu J, et al. Large-scale integrated analysis of ovarian cancer tumors and cell lines identifies an individualized gene expression signature for predicting response to platinum-based chemotherapy. Cell Death Dis. 2019;10:661. doi: 10.1038/s41419-019-1874-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Wu D, Li C, Zhou M, Hao D. Correlation between gene expression and mutator phenotype predicts homologous recombination deficiency and outcome in ovarian cancer. J Mol Med (Berl) 2014;92:1159–68.. doi: 10.1007/s00109-014-1191-9. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Truxova I, Kasikova L, Hensler M, Skapa P, Laco J, Pecen L, et al. Mature dendritic cells correlate with favorable immune infiltrate and improved prognosis in ovarian carcinoma patients. J Immunother Cancer. 2018;6:139. doi: 10.1186/s40425-018-0446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goc J, Germain C, Vo-Bourgais T, Lupo A, Klein C, Knockaert S, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014;74:705–15. doi: 10.1158/0008-5472.CAN-13-1342. [DOI] [PubMed] [Google Scholar]
- 24.Le Mercier I, Poujol D, Sanlaville A, Sisirak V, Gobert M, Durand I, et al. Tumor promotion by intratumoral plasmacytoid dendritic cells is reversed by TLR7 ligand treatment. Cancer Res. 2013;73:4629–40. doi: 10.1158/0008-5472.CAN-12-3058. [DOI] [PubMed] [Google Scholar]
- 25.Gorbachev AV, Fairchild RL. Regulation of chemokine expression in the tumor microenvironment. Crit Rev Immunol. 2014;34:103–20. doi: 10.1615/CritRevImmunol.2014010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–27. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohm S, Montfort A, Pearce OM, Topping J, Chakravarty P, Everitt GL, et al. Neoadjuvant chemotherapy modulates the immune microenvironment in metastases of tubo-ovarian high-grade serous carcinoma. Clin Cancer Res. 2016;22:3025–36. doi: 10.1158/1078-0432.CCR-15-2657. [DOI] [PubMed] [Google Scholar]
- 29.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 30.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–8. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol. 2019;234:8509–21. doi: 10.1002/jcp.27782. [DOI] [PubMed] [Google Scholar]
- 32.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–46. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Gu S, Bao S, Yan C, Zhang Z, Hou P, et al. Mechanistically derived patient-level framework for precision medicine identifies a personalized immune prognostic signature in high-grade serous ovarian cancer. Brief Bioinformatics. 2021;22:bbaa069. doi: 10.1093/bib/bbaa069. [DOI] [PubMed] [Google Scholar]
- 34.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macintyre G, Goranova TE, De Silva D, Ennis D, Piskorz AM, Eldridge M, et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet. 2018;50:1262–70. doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natoli M, Gallon J, Lu H, Amgheib A, Pinato DJ, Mauri FA, et al. Transcriptional analysis of multiple ovarian cancer cohorts reveals prognostic and immunomodulatory consequences of ERV expression. J Immunother Cancer. 2021;9:e001519. doi: 10.1136/jitc-2020-001519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millstein J, Budden T, Goode EL, Anglesio MS, Talhouk A, Intermaggio MP, et al. Prognostic gene expression signature for high-grade serous ovarian cancer. Ann Oncol. 2020;31:1240–50. doi: 10.1016/j.annonc.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–8. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrickx W, Simeone I, Anjum S, Mokrab Y, Bertucci F, Finetti P, et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology. 2017;6:e1253654. doi: 10.1080/2162402X.2016.1253654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.