Abstract
5-hydroxytryptamine receptor 5A (5-HT5A) belongs to the 5-HT receptor family and signals through the Gi/o protein. It is involved in nervous system regulation and an attractive target for the treatment of psychosis, depression, schizophrenia, and neuropathic pain. 5-HT5A is the only Gi/o-coupled 5-HT receptor subtype lacking a high-resolution structure, which hampers the mechanistic understanding of ligand binding and Gi/o coupling for 5-HT5A. Here we report a cryo-electron microscopy structure of the 5-HT5A–Gi complex bound to 5-Carboxamidotryptamine (5-CT). Combined with functional analysis, this structure reveals the 5-CT recognition mechanism and identifies the receptor residue at 6.55 as a determinant of the 5-CT selectivity for Gi/o-coupled 5-HT receptors. In addition, 5-HT5A shows an overall conserved Gi protein coupling mode compared with other Gi/o-coupled 5-HT receptors. These findings provide comprehensive insights into the ligand binding and G protein coupling of Gi/o-coupled 5-HT receptors and offer a template for the design of 5-HT5A-selective drugs.
Subject terms: Cryoelectron microscopy, Hormone receptors
Introduction
5-hydroxytryptamine (5-HT) receptors are widely expressed in the central and peripheral nervous systems and are involved in a variety of psychiatric disorders. They are one of the most promising drug targets for the treatment of nervous system diseases1. There are seven distinct types (5-HT1–7), comprised of 14 subtypes in the 5-HT receptor family, of which 13 are G protein-coupled receptors (GPCRs) (Fig. 1a). So far, 26 structures of 5-HT receptors have been reported, including crystal structures of 5-HT1B2,3, 5-HT2A4,5, 5-HT2B6–9, and 5-HT2C10, as well as cryo-electron microscopy (cryo-EM) structures of all members of 5-HT1, including 5-HT1A11, 5-HT1B12, 5-HT1D11, 5-HT1E11, and 5-HT1F13, in complex with Gi/o protein. These structures provide a basis for understanding ligand recognition and functional regulation of these 5-HT receptors. Besides 5-HT1, 5-HT5 is another type of Gi/o-coupled 5-HT receptor and also remains the last type of Gi/o-coupled 5-HT receptor without a reported structure.
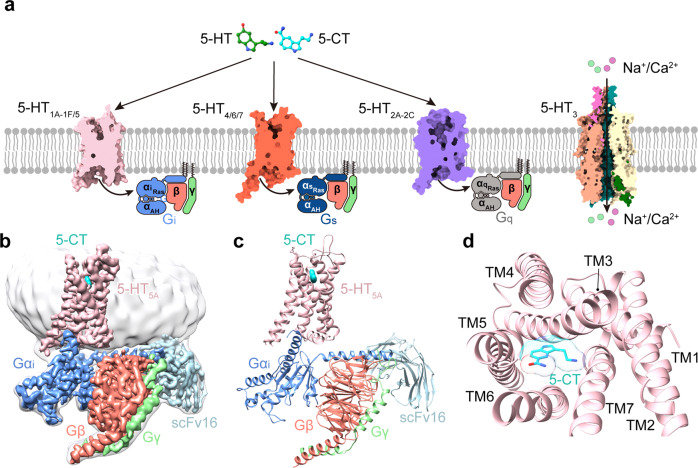
Fig. 1. Cryo-EM structure of the 5-CT–5-HT5A–Gi–scFv16 complex.
a Schematic illustration of 5-HT receptors coupling to distinct G proteins upon stimulation by 5-HT and 5-CT. 5-HT1 and 5-HT5 belong to Gi/o-coupled 5-HT receptors. 5-HT1 subfamily includes 5-HT1A, 5-HT1B, 5-HT1D, 5-HT1E, and 5-HT1F. Different from other 5-HT receptor subtypes, 5-HT3 is an ion channel. b, c Orthogonal views of the density map (b) and model (c) for the 5-CT–5-HT5A–Gi–scFv16 complex. The EM density of 5-CT is shown as cyan surface presentation. d The extracellular view of the 5-HT5A. The complex is colored by subunits. Light pink, 5-HT5A; cyan, 5-CT; blue, Gαi; salmon, Gβ; light green, Gγ; light blue, scFv16.
The 5-HT5 subfamily consists of two members, designated as 5-HT5A and 5-HT5B, which share 69% sequence identity with each other and have 23%–34% homology with other 5-HT receptors14. Of note, 5-HT5B is the first example of a brain-specific receptor that is absent in humans, of which the coding sequence is interrupted by stop codons14,15. Thus, 5-HT5A stands out as the only 5-HT5 subtype expressed in human brain regions, including the cerebral cortex, hippocampus, and raphe nuclei16,17. 5-HT5A shows an antinociceptive role and is involved in the regulation of memory, learning, and food intake18,19. Its specific ligands have shown potential in the treatment of psychosis, depression, schizophrenia, and neuropathic pain20. Thus, the development of 5-HT5A-selective drugs will offer a new opportunity for the treatment of these nervous system diseases.
However, the selective ligands for 5-HT5A are still lacking. 5-Carboxamidotryptamine (5-CT) is a synthetic agonist for 5-HT5A and also activates other Gi/o-coupled 5-HT receptors with distinct affinities. It shows a moderate affinity for 5-HT5A with a pKi value of 7.7 and displays high affinities for 5-HT1A, 5-HT1B, and 5-HT1D (pKi = 8.9–9.0)21–25. In contrast, 5-CT displays negligible affinities for 5-HT1E (pKi = 5.4) and 5-HT1F (pKi = 6.1) (https://pdsp.unc.edu/pdspweb/)26,27. SB699551 and ASP5736 stand out as two selective antagonists, which have been widely used for functional studies of 5-HT5A20,28. The availability of the structure of ligand-bound 5-HT5A may accelerate the design of 5-HT5A-targeting drugs by providing an accurate structure template.
In this study, we report the structure of Gi-coupled 5-HT5A complex bound to 5-CT at a resolution of 3.1 Å. This structure clarified the feature of 5-CT recognition by 5-HT5A and identified a determinant for 5-CT affinities against Gi/o-coupled 5-HT receptors, thus providing a rationale for designing drugs targeting 5-HT5A. Structural comparison of the 5-HT5A–Gi with other Gi/o-coupled 5-HT receptor complexes deepens our understanding of the mechanism underlying ligand recognition and Gi/o coupling.
Results
Cryo-EM structure of the 5-CT–5-HT5A–Gi–scFv16 complex
We used the full-length human 5-HT5A for structural studies. A BRIL was fused to the N-terminus of 5-HT5A to improve expression. The NanoBiT tethering strategy was applied to stabilize the 5-HT5A–Gi complex, which had been widely used in the structure determination of several GPCR–G protein complexes29–31 (Supplementary Fig. S1). The C-terminus of the receptor and the Gβ1 subunit were connected to the LgBiT and HiBiT, respectively. A dominant-negative form of the human Gαi1 mutant containing four mutations (S47N, G203A, E345A, and A326S), referred to as Gαi1(4DN), was applied32. The 5-HT5A–Gi complex was assembled by co-expressing the engineered receptor with Gαi1(4DN), Gβ1, Gγ2 subunits, and scFv16 in High Five (Hi5) cells in the presence of 5-CT.
The structure of the 5-CT–5-HT5A–Gi–scFv16 complex was determined with an overall resolution of 3.1 Å (Fig. 1b, c; Supplementary Fig. S2 and Table S1). The high-quality density maps are clear for modeling 5-HT5A from residue 31 to residue 353, with the exception of residues 237–275 in the intracellular loop 3 (ICL3). The majority of the residue side chains in the seven-transmembrane helical domain (TMD), three extracellular loops (ECL1–ECL3), and two ICLs (ICL1 and ICL2) of 5-HT5A were well-defined. 5-CT, scFv16, and the three subunits of Gi protein are also well-fitted in the EM map. The entire model provides detailed structural information on the 5-CT-binding pocket and 5-HT5A–Gi interaction interface (Fig. 1c, d; Supplementary Fig. S3).
The recognition of Gi/o-coupled 5-HT receptors by agonists
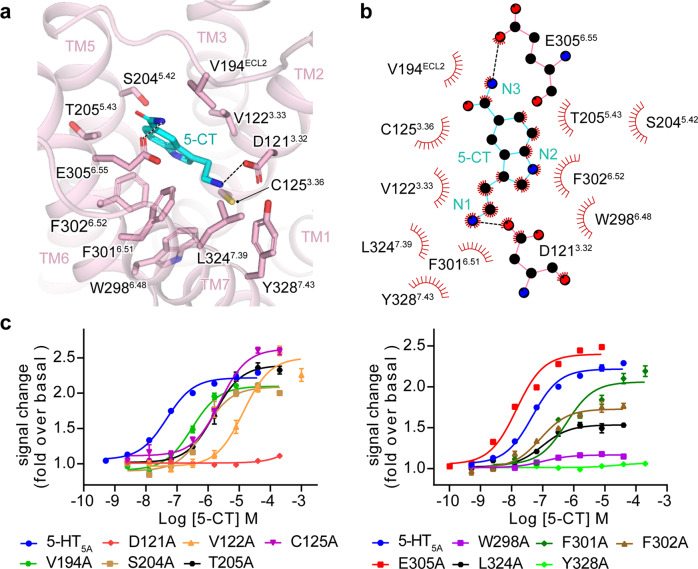
The binding pocket in 5-HT5A is largely overlapped with that in other Gi/o-coupled 5-HT receptors, sharing 11 of 16 identical residues. 5-CT is embedded deep into the pocket constituted by TM3, ECL2, and TM5–TM7 of 5-HT5A (Fig. 2a). Compared with other ligands bound to Gi/o-coupled 5-HT receptors, 5-CT adopts a similar binding pose in 5-HT5A (Supplementary Fig. S4a). Its indole scaffold is anchored through a salt bridge between its positively charged nitrogen at the 3-aminoethyl group and the carboxylate of D1213.32 (Fig. 2a, b). This salt bridge is highly conserved across ligand-bound 5-HT receptors with known structures (Supplementary Fig. S4b). Mutating D1213.32 to alanine abolished the 5-CT-induced 5-HT5A activation, highlighting its importance to 5-CT activity (Fig. 2c). In addition, the side chain of D1213.32 is further stabilized by an intramolecular hydrogen bond between D1213.32 and Y3287.43, which is supported by the alanine mutagenesis data. On the other side, the nitrogen at the 5-carboxamide of 5-CT forms a hydrogen bond with the side chain of E3056.55. Besides polar interactions, the indole scaffold of 5-CT tightly packs against a hydrophobic cleft comprising side chains of V1223.33, F3016.51, F3026.52, and L3247.39. These hydrophobic residues substantially contribute to 5-CT-induced 5-HT5A activation (Fig. 2c; Supplementary Table S2).
Fig. 2. The 5-CT-binding pocket of 5-HT5A.
a Detailed interactions of 5-CT with residues in the binding pocket of 5-HT5A. b 2D schematic representation of interactions between 5-CT and residues in the ligand-binding pocket of 5-HT5A, analyzed by LigPlot+ program. The polar interactions are indicated as black dashed lines. c Effects of alanine mutation of pocket residues of 5-HT5A on 5-CT-induced Gi protein recruitment. Three independent NanoBiT assays in triplicates were performed. Each data point presents mean ± SEM from a representative experiment.
It has been thought that the ligand-binding pocket of each Gi/o-coupled 5-HT receptor is comprised of two subpockets, the orthosteric binding pocket (OBP) and the extended binding pocket (EBP)2,8. OBP of Gi/o-coupled 5-HT receptors locates deep into the core of the TMD pocket, whereas the EBP approaches the extracellular surface of the entire binding pocket (Supplementary Fig. S4a). The 11 conserved residues in the binding pocket across Gi/o-coupled 5-HT receptors are located in the OBP, including D3.32, C3.36, T3.37, I4.56, S5.42, T5.43, A5.46, W6.48, F6.51, F6.52, and Y7.43 (Supplementary Fig. S4b), of which D3.32 is thought critical to ligand binding for 5-HT and other monoamine receptors by forming a conserved salt bridge with the basic cyclic amine of ligands2. The featured benzene ring of the ligand is surrounded by conserved hydrophobic residues in Gi/o-coupled 5-HT receptors, including W6.48, F6.51, F6.52, and Y7.43. This hydrophobic environment is crucial for ligand-induced receptor activation. Inspection of the binding poses of ligands in Gi/o-coupled 5-HT receptors reveals that 5-HT, 5-CT, and BRL54443 only occupy the OBP. In contrast, Donitriptan and Lasmiditan, two anti-migraine drugs selectively targeting 5-HT1B/1D and 5-HT1F, respectively, are relatively bulky and occupy both OBP and EBP of specific receptors (Supplementary Fig. S4a). These structural observations are consistent with the contention that OBP is critical to the binding potency of ligands, whereas the EBP plays a predominant role in determining ligand selectivity2. Together, these findings provide insights into the 5-CT recognition for 5-HT5A and deepen our understanding of ligand selectivity for 5-HT receptors.
Role of the residue at 6.55 in the determination of 5-CT selectivity for Gi/o-coupled 5-HT receptors
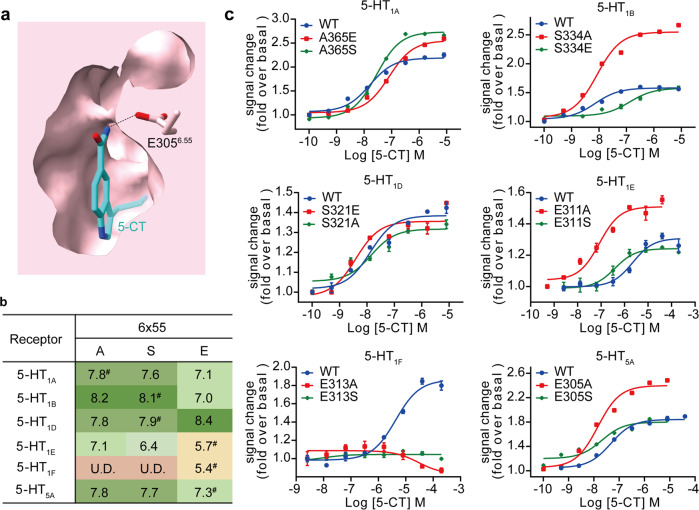
5-CT shows different selectivity for Gi/o-coupled 5-HT receptors. It exhibits high affinities for 5-HT1A, 5-HT1B, and 5-HT1D (pKi = 7.9–8.1) and relatively weak affinities for 5-HT1E, 5-HT1F, and 5-HT5A (pKi = 5.4–7.0) (Fig. 3b; Supplementary Table S3). Sequence comparison of residues in the EBP of Gi/o-coupled 5-HT receptors reveals a low sequence identity at position 6.55 (Supplementary Fig. S4b). The residue at 6.55 is alanine in 5-HT1A and serine in 5-HT1B and 5-HT1D. In contrast, the cognate residue in 5-HT1E, 5-HT1F, and 5-HT5A is glutamic acid. The difference in residue composition raises a hypothesis that the residue at 6.55 is involved in 5-CT selectivity for Gi/o-coupled 5-HT receptors.
Fig. 3. Role of the residue at 6.55 in determining 5-CT selectivity of Gi/o-coupled 5-HT receptors.
a Cross-section of the orthosteric binding pocket of 5-HT5A. The hydrophobic interaction between 5-CT and E3056.55 is indicated as a black dashed line. b 5-CT activity (pEC50 values) for wild-type (WT) and mutants of Gi/o-coupled 5-HT receptors. #5-CT activity (pEC50 values) for WT receptors. U.D., undetectable. c Effects of mutations at 6.55 on 5-CT-induced Gi protein recruitment of Gi/o-coupled receptors. Three independent NanoBiT assays in triplicates were performed. Each data point presents mean ± SEM from a representative experiment.
To prove this hypothesis, we introduced swap mutations to residues at 6.55 across Gi/o-coupled 5-HT receptors. Our Gi protein recruitment data support two-facet roles of the residue at 6.55 in 5-CT-induced receptor activation. One is the steric hindrance arising from the side chain of the residue at position 6.55 (Fig. 3c). For 5-HT1A, A3656.55E and A3656.55S mutations, which increase the size of the side chain, reduced 5-CT-induced receptor activation relative to the WT receptor. S3346.55E mutation of 5-HT1B decreased 5-CT activity, while substituting the serine with a smaller side chain residue alanine dramatically increased 5-CT potency. Similarly, mutating glutamic acid of 5-HT1E and 5-HT5A to alanine or serine, two residues with a relatively small side chain, notably promoted receptor activation. These findings corroborate the idea that the bulkier side chain of glutamic acid relative to alanine and serine may prevent the binding of 5-CT and receptor activation through steric hindrance. On the other hand, 5-CT may form hydrogen bonds with the glutamic acid at 6.55 in Gi/o-coupled 5-HT receptors, which may dominate the ligand–receptor interaction over the hindrance effects of the side chain (Fig. 3c). This point is supported by functional analysis of 5-HT1D and 5-HT1F. S3216.55E mutation of the 5-HT1D enhanced 5-CT activity, despite the increased side-chain size. Consistently, E3136.55S and E3136.55A mutations in 5-HT1F almost abolished 5-CT activity. Thus, residues at position 6.55 modulate the activity of Gi/o-coupled 5-HT receptors through two aspects of roles: the steric hindrance effects for 5-HT1A, 5-HT1B, 5-HT1E, and 5-HT5A and the hydrogen bond-forming capacity as an acceptor for 5-HT1D and 5-HT1F. Together, our findings provide further evidence for the previous speculation that the residue at 6.55 is responsible for the ligand-recognition specificity of 5-HT receptors and offer a new opportunity for the design of drugs selectively targeting 5-HT receptors11.
General features of the activation and G protein coupling of Gi/o-coupled 5-HT receptors
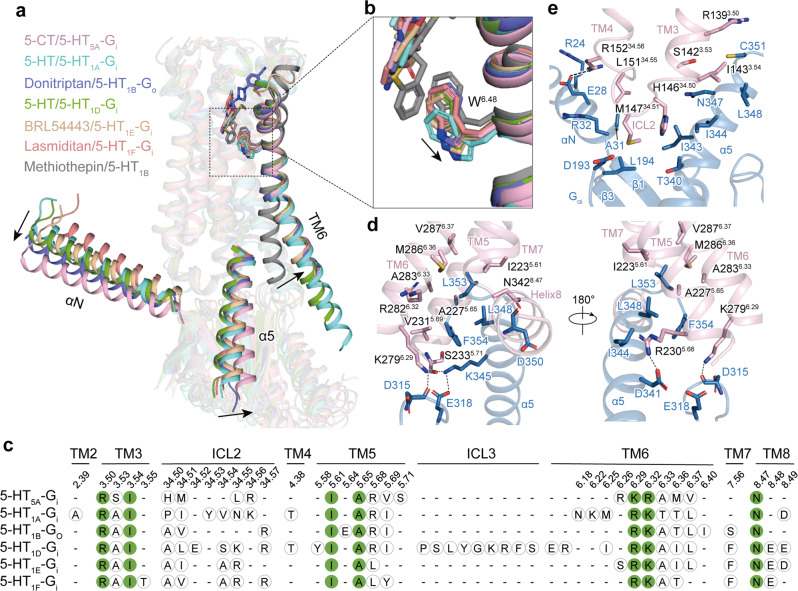
Similar to agonists bound to other Gi/o-coupled 5-HT receptors, 5-CT directly contacts the toggle switch residue W6.48 of 5-HT5A and triggers its rotameric switch. The change of W6.48 initiates the rotation and outward movement of the TM6 cytoplasmic end of 5-HT5A relative to inverse agonist-bound 5-HT1B (PDB: 5V54), the hallmark of class A GPCR activation (Fig. 4a, b).
Fig. 4. Structural features of the activation and Gi/o coupling of 5-HT5A and other Gi/o-coupled 5-HT receptors.
a The activation of 5-HT5A and other Gi/o-coupled 5-HT receptors. The outward movement of TM6 of agonist-bound Gi/o-coupled 5-HT receptors compared with methiothepin-bound 5-HT1B in the inactive state is shown as a black arrow. The movements of the αN and α5 helix of Gαi protein in the 5-HT5A–Gi complex relative to that in other Gi/o-coupled 5-HT receptors are indicated as black arrows. The structures of the active 5-HT5A (light pink), 5-HT1A (cyan, PDB: 7E2Y), 5-HT1B (purple, PDB: 6G79), 5-HT1D (green, PDB: 7E32), 5-HT1E (wheat, PDB: 7E33), 5-HT1F (coral, PBD: 7EXD) and inactive-state 5-HT1B (gray, PDB: 4IAQ) complexes were superimposed based on TM2, TM3 and TM4. b Structural comparison of ligands and the W6.48 residues of active 5-HT receptors with that of inactive 5-HT1B. Ligands directly contact W6.48. Compared with methiothepin, the inverse agonist of 5-HT1B, agonists trigger the rotameric switch of W6.48, which leads to the outward movement of TM6. c The sequence alignment of receptor residues at receptor–Gi/o protein interfaces. The conserved residues are highlighted in solid green circles. d, e Detailed interactions between 5-HT5A and the Gαi subunit. Detailed Interactions between TM5, TM6, and the TM7–helix 8 junction of 5-HT5A and the α5 helix of the Gαi subunit (d). Detailed interactions between ICL2 of 5-HT5A and the α5 and αN helices, β1 and β3 strands of the Gαi subunit (e).
Structural comparison of the Gi-coupled 5-HT5A with other Gi/o-coupled 5-HT receptors whose structures had been solved revealed an almost overlapped receptor activation conformation. However, the α5 helices of Gαi/o subunits in these 5-HT receptor complexes showed slight tilts to different extents, which cause rotation of the entire Gi/o proteins, leading to the most noticeable translational movement of the αN helices (Fig. 4a). The global coupling interface profile analysis showed that the majority of the Gαi subunit-interacting residues in TM3, ICL2, TM5, TM6, TM7, and helix 8 are conserved across Gi/o-coupled 5-HT receptors, including R3.50, I3.54, I5.61, A5.65, R/K6.29, R/K6.32, and N8.47. Differently, no substantial interactions were seen between ICL3 of Gi/o-coupled 5-HT receptors and Gi protein, with an exception of 5-HT1D, which shows an additional EM density of ICL3 and a more extensive ICL3–Gi interaction11 (Fig. 4c).
Two major interfaces exist between 5-HT5A and Gi protein. The cytoplasmic receptor cavity constituted by TM3, TM5, TM6, and the TM7–helix 8 junction accommodates the distal C-terminal end of the α5 helix of the Gαi subunit, forming a primary interface (Fig. 4d). The residues of α5 helix hydrophobically contact the receptor cavity. L348, C351, L353, and F354 in the α5 helix of the Gαi subunit contact a hydrophobic patch comprised of residues in TM3 (I1433.54), TM5 (I2235.61, A2275.65, and V2315.69), and TM6 (A2836.33 and V2876.37). In addition, residues R2305.68 and S2335.71 of TM5 form well-defined hydrogen bonds with D341 and K345 from the α5 helix, respectively (Fig. 4d). The ICL2 also interacts with Gαi, constituting the other major interface. M14734.51 of ICL2 inserts into the groove constituted by hydrophobic residues in the α5 helix, the β1 and β3 strands, and αN of the Gαi subunit. A hydrogen bond present between R15234.56 and E28 may further stabilize the ICL2–Gαi interface (Fig. 4e). Together, these findings clarify the activation and Gi coupling features of 5-HT5A and provide a comprehensive understanding of the G protein coupling mechanism of Gi/o-coupled 5-HT receptors.
Discussion
5-HT5A is a Gi/o-coupled 5-HT receptor subtype and is involved in nervous system disorders, thus serving as an important drug target. It is the only Gi/o-coupled 5-HT receptor subtype lacking a high-resolution structure to date. In this paper, we report a 3.1 Å-resolution cryo-EM structure of the 5-HT5A–Gi complex bound to a synthetic agonist, 5-CT, which is also the first 5-CT-bound 5-HT receptor structure. Our structure reveals the recognition mechanism of 5-HT5A by 5-CT and adds to the pool of the structures for deepening our understanding of the ligand-binding mode of 5-HT receptors. Furthermore, structural comparison and functional analysis of the ligand-binding pockets reveal that the residue at 6.55 serves as a determinant for the 5-CT specificity for Gi/o-coupled 5-HT receptors. This ligand specificity is partly attributed to the steric hindrance arising from the side chain of the residue at 6.55 or its potential polar interaction with ligands. In addition, our structure reveals a similar activation mechanism and an overall conserved Gi protein coupling mode for 5-HT5A compared with other Gi/o-coupled 5-HT receptors. These findings broaden our understanding of ligand recognition in the 5-HT system.
Although 5-HT5A has been cloned for ~3 years, it is still one of the less well-characterized receptors in the 5-HT receptor family. The lack of selective ligands has delayed the functional studies on 5-HT5A until the discovery of the selective antagonists SB699551 and ASP5736, which have improved our understanding of the localization of 5-HT in the brain and its function. However, it should be noted that we are still far from fully understanding the pharmacological characteristics of 5-HT5A. Meanwhile, no drugs targeting 5-HT5A have been registered for clinical trials or approved. Recently, virtual screening based on a homology model identified UCSF678, a 42 nM new chemical probe with partial agonism activity for 5-HT5A. USCF678 exhibits enhanced selectivity for 5-HT5A and a more restricted off-target profile than the existing 5-HT5A antagonist SB699551. Unlike the promiscuous ligand 5-HT, molecular docking reveals that USCF678 extends into the upper region of the binding pocket, known as EBP. W1173.28 in EBP of 5-HT5A is further proved to be responsible for the high-affinity binding of UCSF638, and the tryptophan at position 3.28 may contribute to the off-target binding of UCSF638 analogs33. These findings are consistent with the two pockets (OBP and EBP) binding model2,8 and highlight the importance of EBP to the discovery of selective ligands. Consequently, the 5-HT5A structure provides an accurate template for the rational design of drugs targeting 5-HT5A and may offer a new opportunity for the treatment of nervous system diseases, including psychosis, depression, schizophrenia, and neuropathic pain.
Materials and methods
Constructs
The human full-length 5-HT5A was cloned into the pFastBac with an N-terminal haemagglutinin (HA) sequence followed by a Flag-tag, 15× His-tag, BRIL-tag, and a LgBiT sequence at the C-terminus to facilitate the protein expression and purification. The human Gαi with four dominant-negative mutations, S47N, G203A, E245A, and A326S was applied. Human Gβ1, human Gγ2, and scFv16 were cloned into pFastBac vector using homologous recombination (ClonExpress One Step Cloning Kit, Vazyme).
Expression and complex purification
5-HT5A-LgBiT, DN_Gαi, Gβ1-SmBiT, Gγ2, and scFv16 were co-expressed in Hi5 insect cells (Invitrogen) using the Bac-to-Bac baculovirus expression system (ThermoFisher). Cell cultures were grown in ESF 921 medium (Expression Systems) to a density of 3 × 106 cells/mL, and then infected with five separate baculoviruses, respectively, at the ratio of 1:1:1:1:1.5. The culture was harvested by centrifugation 48 h post-infection and stored at −80 °C for further usage.
Cell pellets were lysed in 20 mM HEPES, pH 7.4, 20 mM KCl, 10 mM MgCl2, 5 mM CaCl2, and 10% glycerol supplemented with Protease Inhibitor Cocktail (TargetMol). The 5-HT5A–Gi complex was formed on the membrane for 1.5 h at room temperature by addition of 10 µM 5-CT and 25 mU/mL apyrase (Sigma), and then solubilized from the membrane by using 0.5% (w/v) n-dodecyl-β-d-maltoside (DDM) (Anatrace) and 0.1% (w/v) cholesteryl hemisuccinate (CHS) (Anatrace) for 2 h at 4 °C, followed by centrifugation at 85,000 × g for 30 min to extract the solubilized complex. The supernatant was subsequently incubated by nickel affinity chromatography (Ni Smart Beads 6FF, SMART Lifesciences) at 4 °C for 3 h. The resin was washed with 20 column volumes of the buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 25 mM imidazole, 0.01% (w/v) LMNG (Anatrace), 0.005% GDN (Anatrace), 0.004% (w/v) CHS (Anatrace), and 5 µM 5-CT. The complex was then eluted with six column volumes of the same buffer containing 300 mM imidazole. The protein was concentrated and subjected onto a Superdex 200 Increase 10/300 column (GE Healthcare) in the buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 0.00075% (w/v) LMNG (Anatrace), 0.00025% (w/v) GDN (Anatrace), 0.0002% (w/v) CHS (Anatrace) and 10 mM 5-CT. The purified complex fractions were collected and concentrated for cryo-EM experiments.
Cryo-EM grid preparation and data collection
For the cryo-EM grid preparation, 3 μL of the purified 5-CT–5-HT5A–Gi complex at a final concentration of 25 mg/mL was applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, 300 mesh), and vitrified using a Vitrobot Mark IV (ThermoFisher Scientific) subsequently. Grids were plunge-frozen in liquid ethane using Vitrobot Mark IV (Thermo Fischer Scientific). Frozen grids were transferred to liquid nitrogen and stored for data acquisition. Cryo-EM images were collected by an FEI Titan Krios at 300 kV accelerating voltage equipped with a Gatan K3 Summit direct electron detector at the Center of Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China). A total of 5303 movies were automatically acquired using SerialEM10 in super-resolution counting mode at a pixel size of 1.071 Å. The images were recorded at a dose rate of about 26.7 e/Å2/s with a defocus ranging from –1.2 to –2.2 μm. The total exposure time was 3 s, and intermediate frames were recorded in 0.083-s intervals, resulting in a total of 36 frames per micrograph.
Image processing and 3D reconstruction
Image stacks were subjected to beam-induced motion correction using MotionCor2.134, while contrast transfer function (CTF) parameters were determined by Gctf35. Automated particle selection and data processing were performed using Relion 3.036. Automated particle selection yielded 3,767,450 particles. The particles were subjected to reference-free 2D classification, producing 1,327,660 particles with well-defined averages. The map of 5-HT1E–Gi–scFv16 complex (EMDB-30975)11 low-pass filtered to 40 Å was used as an initial reference model for 3D classification, which produced two good subsets showing clear structural features accounting for 754,854 particles. These particles were subsequently subjected to Bayesian polishing, CTF refinement, and 3D refinement, which generated a map with an indicated global resolution of 3.1 Å at a Fourier shell correlation of 0.143. Local resolution was determined using the Resmap37 with half maps as input maps.
Model building and refinement
The cryo-EM structure of the 5-CT–5-HT5A–Gi complex (PDB: 7E2Y) and the Gi protein model (PDB: 6DDE) were used to generate the initial model and refinement against the electron microscopy map. The model was docked into the EM density map using UCSF Chimera38, followed by iterative manual adjustment and rebuilding in COOT39 and ISOLDE40 according to side-chain densities. Real-space refinement was performed using Phenix programs41. The model statistics were validated using MolProbity42. Structural figures were prepared in Chimera, ChimeraX43, and PyMOL (https://pymol.org/2/). The final refinement statistics are provided in Supplementary Table S1.
NanoBiT G protein recruitment assay
NanoBiT, a NanoLuc luciferase-based method, is used to detect the interaction between receptor and G protein in living cells44. The full-length 5-HT5A was fused with a LgBiT fragment (17.6 kDa) at its C-terminus via a 15-amino acid flexible linker. SmBiT, a 13-amino acid peptide, was C-terminally fused to the Gβ subunit using the same linker. The cDNAs of 5-HT5A-LgBiT, Gαi1, Gβ1-SmBiT, and Gγ2 were cloned into pFastBac vector (Invitrogen). The baculoviruses were prepared using the bac-to-bac system (Invitrogen). Hi5 cells were cultured in ESF 921 medium (Expression Systems) to a density of 2.5–3 million cells per mL and then infected with four separate baculoviruses at the ratio of 1:1:1:1. After 48 h infection, the culture was collected by centrifugation, and the cell pellet was resuspended with PBS. The cell suspension was seeded onto 384-well microtiter plates (40 μL per well) and loaded with 5 μL of 50 μM coelenterazine (Yeasen) diluted in the assay buffer. 5 μL of ligands were added and incubated for 3–5 min at room temperature before measurement. Luminescence counts were normalized to the initial count to show the G protein binding response.
Surface expression analysis
Cell surface expression for each mutant was monitored using flow cytometry. The expressed Hi5 cells (10 μL) were incubated with 10 μL anti-FLAG-FITC antibody (Sigma), which is diluted with PBS containing 4% BSA at a final ratio of 1:1000, at 4 °C for 15 min, and 180 μL 1× PBS was then added to the cells. The surface expression of each mutant was monitored by detecting the fluorescent intensity of FITC using a BD ACCURI C6.
Supplementary information
Acknowledgements
The Cryo-EM data of the 5-CT–5-HT5A–Gi complex were collected at the Cryo-Electron Microscopy Research Center, Shanghai Institute of Material Medica. We thank all the staff at the cryo-EM facilities for their technical support. This work was partially supported by the National Natural Science Foundation (32171187 to Y.J., 82121005 to Y.J., and H.E.X., 32130022 to H.E.X.); the Ministry of Science and Technology (China) grants (2018YFA0507002 to H.E.X.); the Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to H.E.X.); Shanghai Municipal Science and Technology Major Project (H.E.X.); the CAS Strategic Priority Research Program (XDB37030103 to H.E.X.); Shandong University (Qilu Young Scholar 62450082163091 to H.M.); Shanghai Municipal Science and Technology Commission (19ZR1467500 to H.M.); the Young Innovator Association of CAS Enrollment (H.M.).
Author contributions
Y.T. and P.X. optimized the conditions of protein samples for final structure determination; Y.T. and S.H. generated the expression constructs and optimized the 5-HT5A–Gi protein complex; Y.T. purified the protein samples for final structure determination, participated in cryo-EM grid inspection, data collection, model building, executed the functional assay, and prepared the figures and manuscript draft; P.X. evaluated the specimen by negative-stain EM, screened the cryo-EM conditions, prepared the cryo-EM grids, collected cryo-EM images, and refined the structures; G.Y. participated in the functional assay; F.Z. analyzed the interactions between 5-HT5A and 5-CT by LigPlot+ program; X.H. and H.M. participated in structure analysis and model building. Y.J. participated in the supervision of Y.T., S.H., P.X., X.H., and F.Z., analyzed the structures, prepared the figures, and edited the manuscript. H.E.X. and Y.J. conceived and supervised the project, and wrote the manuscript with inputs from all authors.
Data availability
The atomic coordinate and the electron microscopy map for the 5-CT–5-HT5A–Gi–scFv16 complex have been deposited in the Protein Data Bank (PDB) under accession number 7X5H and Electron Microscopy Data Bank (EMDB) under accession number EMD-33014, respectively.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yangxia Tan, Peiyu Xu, Sijie Huang
Contributor Information
H. Eric Xu, Email: eric.xu@simm.ac.cn.
Yi Jiang, Email: yijiang@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-022-00412-3.
References
- 1.Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Annu. Rev. Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340:610–614. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin W, et al. Crystal structure of the human 5-HT1B serotonin receptor bound to an inverse agonist. Cell Discov. 2018;4:12. doi: 10.1038/s41421-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura KT, et al. Structures of the 5-HT2A receptor in complex with the antipsychotics risperidone and zotepine. Nat. Struct. Mol. Biol. 2019;26:121–128. doi: 10.1038/s41594-018-0180-z. [DOI] [PubMed] [Google Scholar]
- 5.Kim K, et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell. 2020;182:1574–1588.e19. doi: 10.1016/j.cell.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishchenko A, et al. Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody. Proc. Natl. Acad. Sci. USA. 2017;114:8223–8228. doi: 10.1073/pnas.1700891114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCorvy JD, et al. Structural determinants of 5-HT2B receptor activation and biased agonism. Nat. Struct. Mol. Biol. 2018;25:787–796. doi: 10.1038/s41594-018-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wacker D, et al. Crystal structure of an LSD-bound human serotonin receptor. Cell. 2017;168:377–389.e12. doi: 10.1016/j.cell.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y, et al. 5-HT2C receptor structures reveal the structural basis of GPCR polypharmacology. Cell. 2018;172:719–730.e14. doi: 10.1016/j.cell.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu P, et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature. 2021;592:469–473. doi: 10.1038/s41586-021-03376-8. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Nafria J, Nehme R, Edwards PC, Tate CG. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature. 2018;558:620–623. doi: 10.1038/s41586-018-0241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, et al. Structural basis for recognition of anti-migraine drug lasmiditan by the serotonin receptor 5-HT1F-G protein complex. Cell Res. 2021;31:1036–1038. doi: 10.1038/s41422-021-00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grailhe R, Grabtree GW, Hen R. Human 5-HT(5) receptors: the 5-HT(5A) receptor is functional but the 5-HT(5B) receptor was lost during mammalian evolution. Eur. J. Pharm. 2001;418:157–167. doi: 10.1016/S0014-2999(01)00933-5. [DOI] [PubMed] [Google Scholar]
- 15.Nelson DL. 5-HT5 receptors. Curr. Drug Targets CNS Neurol. Disord. 2004;3:53–58. doi: 10.2174/1568007043482606. [DOI] [PubMed] [Google Scholar]
- 16.Kassai F, et al. Effect of 5-HT5A antagonists in animal models of schizophrenia, anxiety and depression. Behav. Pharm. 2012;23:397–406. doi: 10.1097/FBP.0b013e3283565248. [DOI] [PubMed] [Google Scholar]
- 17.Thomas DR. 5-ht5A receptors as a therapeutic target. Pharm. Ther. 2006;111:707–714. doi: 10.1016/j.pharmthera.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Vidal-Cantu GC, et al. Role of 5-HT5A and 5-HT1B/1D receptors in the antinociception produced by ergotamine and valerenic acid in the rat formalin test. Eur. J. Pharm. 2016;781:109–116. doi: 10.1016/j.ejphar.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Esparza S, Berumen LC, Padilla K, Miledi R, Garcia-Alcocer G. Expression of hippocampal serotonin receptors 5-HT2C and 5-HT5A in a rat model of diet-induced obesity supplemented with tryptophan. Int J. Dev. Neurosci. 2015;42:80–85. doi: 10.1016/j.ijdevneu.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki M, et al. ASP5736, a novel 5-HT5A receptor antagonist, ameliorates positive symptoms and cognitive impairment in animal models of schizophrenia. Eur. Neuropsychopharmacol. 2014;24:1698–1708. doi: 10.1016/j.euroneuro.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Leysen JE, et al. Alniditan, a new 5-hydroxytryptamine1D agonist and migraine-abortive agent: ligand-binding properties of human 5-hydroxytryptamine1D alpha, human 5-hydroxytryptamine1D beta, and calf 5-hydroxytryptamine1D receptors investigated with [3H]5-hydroxytryptamine and [3H]alniditan. Mol. Pharm. 1996;50:1567–1580. [PubMed] [Google Scholar]
- 22.Newman-Tancredi A, et al. Agonist activity of antimigraine drugs at recombinant human 5-HT1A receptors: potential implications for prophylactic and acute therapy. Naunyn Schmiedebergs Arch. Pharm. 1997;355:682–688. doi: 10.1007/PL00005000. [DOI] [PubMed] [Google Scholar]
- 23.Newman-Tancredi A, et al. Agonist and antagonist actions of antipsychotic agents at 5-HT1A receptors: a [35S]GTPgammaS binding study. Eur. J. Pharm. 1998;355:245–256. doi: 10.1016/S0014-2999(98)00483-X. [DOI] [PubMed] [Google Scholar]
- 24.Parker EM, Izzarelli DG, Lewis-Higgins L, Palmer D, Shapiro RA. Two amino acid differences in the sixth transmembrane domain are partially responsible for the pharmacological differences between the 5-HT1D beta and 5-HT1E 5-hydroxytryptamine receptors. J. Neurochem. 1996;67:2096–2103. doi: 10.1046/j.1471-4159.1996.67052096.x. [DOI] [PubMed] [Google Scholar]
- 25.Rees S, et al. Cloning and characterisation of the human 5-HT5A serotonin receptor. FEBS Lett. 1994;355:242–246. doi: 10.1016/0014-5793(94)01209-1. [DOI] [PubMed] [Google Scholar]
- 26.McAllister G, et al. Molecular cloning of a serotonin receptor from human brain (5HT1E): a fifth 5HT1-like subtype. Proc. Natl. Acad. Sci. USA. 1992;89:5517–5521. doi: 10.1073/pnas.89.12.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adham N, et al. Cloning of another human serotonin receptor (5-HT1F): a fifth 5-HT1 receptor subtype coupled to the inhibition of adenylate cyclase. Proc. Natl. Acad. Sci. USA. 1993;90:408–412. doi: 10.1073/pnas.90.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas, D. R. et al. SB-699551-A (3-cyclopentyl-N-[2-(dimethylamino)ethyl]-N-[(4’-{[(2-phenylethyl)amino]methyl}-4 -biphenylyl)methyl]propanamide dihydrochloride), a novel 5-ht5A receptor-selective antagonist, enhances 5-HT neuronal function: evidence for an autoreceptor role for the 5-ht5A receptor in guinea pig brain. Neuropharmacology51, 566–577 (2006). [DOI] [PubMed]
- 29.Duan J, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun. 2020;11:4121. doi: 10.1038/s41467-020-17933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin YL, et al. Molecular basis for kinin selectivity and activation of the human bradykinin receptors. Nat. Struct. Mol. Biol. 2021;28:755–761. doi: 10.1038/s41594-021-00645-y. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor. Nat. Commun. 2021;12:5064. doi: 10.1038/s41467-021-25364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu P, et al. The structural basis of the dominant negative phenotype of the Galphai1beta1gamma2 G203A/A326S heterotrimer. Acta Pharm. Sin. 2016;37:1259–1272. doi: 10.1038/aps.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levit Kaplan, A. et al. Structure-based design of a chemical probe set for the 5-HT5A serotonin receptor. J. Med. Chem.10.1021/acs.jmedchem.1c02031 (2022). [DOI] [PMC free article] [PubMed]
- 34.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heymann JB. Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 2018;27:159–171. doi: 10.1002/pro.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Croll TI. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu P, et al. Structures of the human dopamine D3 receptor-Gi complexes. Mol. Cell. 2021;81:1147–1159.e44. doi: 10.1016/j.molcel.2021.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinate and the electron microscopy map for the 5-CT–5-HT5A–Gi–scFv16 complex have been deposited in the Protein Data Bank (PDB) under accession number 7X5H and Electron Microscopy Data Bank (EMDB) under accession number EMD-33014, respectively.