Abstract
Background
Liver metastasis is the primary cause of colorectal cancer (CRC)-associated death. Aryl-hydrocarbon receptor-interacting protein (AIP), a putative positive intermediary in aryl-hydrocarbon receptor-mediated signalling, is overexpressed in highly metastatic human KM12SM CRC cells and other highly metastatic CRC cells.
Methods
Meta-analysis and immunohistochemistry were used to assess the relevance of AIP. Cellular functions and signalling mechanisms mediated by AIP were assessed by gain-of-function experiments and in vitro and in vivo experiments.
Results
A significant association of high AIP expression with poor CRC patients’ survival was observed. Gain-of-function and quantitative proteomics experiments demonstrated that AIP increased tumorigenic and metastatic properties of isogenic KM12C (poorly metastatic) and KM12SM (highly metastatic to the liver) CRC cells. AIP overexpression dysregulated epithelial-to-mesenchymal (EMT) markers and induced several transcription factors and Cadherin-17 activation. The former induced the signalling activation of AKT, SRC and JNK kinases to increase adhesion, migration and invasion of CRC cells. In vivo, AIP expressing KM12 cells induced tumour growth and liver metastasis. Furthermore, KM12C (poorly metastatic) cells ectopically expressing AIP became metastatic to the liver.
Conclusions
Our data reveal new roles for AIP in regulating proteins associated with cancer and metastasis to induce tumorigenic and metastatic properties in colon cancer cells driving liver metastasis.
Subject terms: Metastasis, Colorectal cancer
Introduction
Metastasis is the final step of malignant transformation and is the main responsible for morbidity and mortality in cancer. Indeed, more than 90% of the mortality associated with cancer is due to metastasis [1]. Metastasis cannot be understood as a unique process but as a collection of different events with unique molecular characteristics, where cancer cells need to interact with different microenvironments, affecting and being affected by the cell and extracellular matrix of such tissues [2, 3]. There, cancer cells have to adapt to each situation, altering the expression, localisation and activation of proteins to generate a metastasis.
Identifying metastasis-associated proteins and altered pathways underlying cancer metastasis would contribute to discover new targets of intervention and to improve patients’ survival. Quantitative proteomic analysis of cell lines with the same genetic background, but differing in their metastatic capabilities, has an immense potential to unveil clinically relevant underlying mechanisms [4–7]. Recently, we have quantitatively studied by in-depth proteomics the secretome and spatial proteome of the KM12 cell system to study the biology of CRC liver metastasis [8, 9]. This cell system derived from a CRC patient classified as Duke’s B (actual T3, N0 of TNM classification) allows for the study of late metastatic events to the liver in CRC, including liver colonisation and survival [10, 11]. It is composed of isogenic KM12C and KM12SM epithelial cells, which differ only in their metastatic properties [10, 11]. KM12SM CRC cell line with high metastatic capacity was isolated from liver metastases in nude mice after five cycles of intrasplenic injection of the poorly metastatic KM12C cell line. Numerous studies support a good correlation between the findings observed in the KM12 cell system and patient samples, indicating that these isogenic cell lines recapitulate quite effectively critical issues in CRC liver metastasis [12–16]. In these proteomics studies using isogenic cell lines with the same genetic background, liver metastatic KM12SM cells were analysed in comparison to KM12C cells [10, 11]. In such analyses, many identified proteins had been previously described as key molecules in CRC, as VEGFA, ERBB2, EGFR, MMP7, FGFR4, Cadherin-17 (CDH17) or IL13Rα2. These results encouraged us to continue characterising these cell lines and gain further insights into proteins dysregulated between KM12C and KM12SM cells [8, 9]. In this sense and taking into account that the function of about 20% of all human proteins remains unknown [17, 18], we focused this work on the analysis of one of these barely known proteins in CRC: aryl-hydrocarbon receptor-interacting protein (AIP).
Here, we observed a clear association of AIP expression with increased liver metastasis and a worse prognosis of CRC patients. At the molecular level, gain-of-function experiments in the KM12 cell system of CRC liver metastasis showed EMT dysregulation and a strong increase in cell adhesion, invasion, colony formation, migration, in vivo liver homing and liver metastasis. Proteomics analysis pointed out to an AIP-associated expression of transcription factors, EGFR and CDH17, among others, as the driving force for the observed changes promoting liver metastasis.
Materials and methods
In silico analysis of prognostic value
GSE17538 database, containing 244 tumour samples with clinicopathological description from colorectal cancer patients, was used for AIP prognostic analysis. Data were normalised using Bioconductor’s Affymetrix package and transformed into z-scores. The prognostic value of AIP was assessed by Kaplan–Meier curves using the best cut-off method for separating high and low-expression populations. The significance of the difference in survival between both populations was estimated by log-rank test. To validate the results with a different cohort of patients, the GEPIA2 tool (http://gepia2.cancer-pku.cn/) was employed with colon adenocarcinoma TCGA dataset (270 tumour samples) [19].
Tissue microarrays and immunohistochemistry
Core tumour tissue samples from 50 metastatic and non-metastatic CRC patients and core tumour tissue samples from 94 recurrent or non-recurrent CRC Stage II patients composed the two tissue microarrays (TMAs) used in the study. Immunohistochemical staining using optimised antibody dilutions and visualisation and immunoreactivity was conducted according to established protocols [20–22].
Cell lines, vectors and transfection of KM12 cells
KM12C and KM12SM human colon cancer cells were obtained directly from Dr. Fidler’s lab (MD Anderson Cancer Center, Houston, TX, USA). KM12 cells were expanded in the laboratory to prepare a large batch of working aliquots for liquid nitrogen storage. For each experiment, cells were thawed and cultured for a maximum of ten passages. These two cell lines were not authenticated in our laboratory. Other CRC cell lines were purchased directly from the ATCC. All these cell lines were authenticated by ATCC and passaged after purchase for all the experiments. Parental cells, and their derivatives, were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco-Life Technologies) containing 10% foetal calf serum and antibiotics at 37 °C in a 5% CO2-humidified atmosphere and continuously monitored for mycoplasma contamination.
AIP gene and pcDNA3.1(+) were obtained from the DNASU plasmid repository and Thermo Fisher Scientific. AIP cloning into pcDNA3.1 vector was performed according to established procedures and sequence verified prior use. Stably transfected cells were obtained according to established protocols (see Supplementary Methods for full methodology) [23–26].
Cell adhesion, invasion, apoptosis detection, proliferation, soft agar colony formation and wound-healing assays
Functional cell-based assays were performed at least in triplicate according to established protocols [23–26]. For cell-adhesion assays, 96-well plates were coated with Matrigel (0.4 µg/mm2) (BD Biosciences) in coating buffer (0.1 M NaHCO3 pH 8.8) overnight at 4 °C and, then, incubated with adhesion medium (0.5% bovine serum albumin (BSA) in serum-free DMEM) for 2 h at 37 °C to block unspecific binding. Cells were starved without serum for 5 h and labelled with BCECF-AM (Molecular Probes) for 30 min at 37 °C, detached with 4 mM EDTA in PBS and resuspended in adhesion medium. Then, 105 cells were added in triplicate to plates and incubated for 30 min. Plates were washed with PBS 1× to remove non-adherent cells. Bound cells were then lysed using 1% SDS in PBS and the fluorescence was quantified in The Spark multimode microplate reader (Tecan Trading AG). For Matrigel invasion assays, 8 × 105 cells were resuspended in invasion medium (serum-free DMEM containing 0.5% BSA) and loaded onto 8-µm pore-size Transwells filters (Costar) coated with 35–50 µl of 1:3 dilution of Matrigel (BD Biosciences). The lower compartments of invasion chambers were filled with medium containing 10% FBS (Gibco). After 22 h of incubation at 37 °C, non-invading cells were removed from the filter’s upper surface, and cells that migrated through the filter were fixed with 4% paraformaldehyde (Sigma), stained with crystal violet and the invading cells counted under a microscope. For wound healing, cells were seeded inside IBIDI silicone wound-healing inserts (IBIDI, #80209) in 24-well plates at a density of 1.5 × 105 cells per insert well. The day after, the insert was removed to uncover a cell-free area of 500 ± 100 µm. Fresh cell growth medium was added, and time-lapse imaging of the cells was recorded. For imaging, a confocal microscope (TCS-SP5-AOBS-UV, Leica-Microsystems) imaging position was set using the Mark and Find Leica imaging software tool, and 1500 × 908 µm (2048 × 1200 pixels) images were acquired every 90 min. Temperature (37 °C) and CO2 concentration (5%) were kept constant throughout the whole imaging time (48 h). The cell-free area was measured with the MRI’s Wound Healing tool for Image J software (NIH) (https://github.com/MontpellierRessourcesImagerie/imagej_macros_and_scripts/wiki/Wound-Healing-Tool) [27], and the calculated areas were visually inspected to verify all the time points were correct.
For cell proliferation assays, the growth medium was changed 24 h after seeding (day 0), and cells were further incubated for three days. Then, medium was removed, and cells were stained with 100 µl of the chromogenic dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) at a final concentration of 1 mg/ml in DMEM. The cells were further incubated for 1 h at 37 °C and 5% CO2. Then, medium was carefully aspirated, and cells disrupted with 100 µl of DMSO (Sigma) prior to reading absorbance at 570 nm. All the experiments were done three times in duplicate. Colony soft agar assay was performed as previously described [26].
Proteomic analysis by isobaric TMT, peptide separation and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
For identification and quantification by TMT of the AIP-associated proteome, 25 μg of protein extracts of each sample in 100 μL RIPA buffer were reduced with 10 μL 100 mM tris(2-carboxyethyl)phosphine (TCEP, Sigma Aldrich) for 45 min at 37 °C and 600 rpm and alkylated with 11 μL of 0.4 M chloroacetamide (Sigma Aldrich) for 30 min at room temperature and 600 rpm and in darkness. Next, samples were incubated with 100 µL of Sera-Mag magnetic beads mix (50% hydrophilic beads—50% hydrophobic beads, GE Healthcare) and 200 μL of acetonitrile 100% for 35 min at room temperature and 600 rpm to allow protein binding to beads. Then, supernatants were discarded, and magnetic beads were washed twice with ethanol 70% and once with acetonitrile 100%. Finally, supernatants were discarded and protein digested overnight at 37 °C and 600 rpm by incubating each sample with 100 μl 20 mM HEPES pH 8.0 supplemented with 1 μg porcine trypsin (Thermo Fisher Scientific). The next day, samples were sonicated and the supernatant was collected. All peptides from the four samples and both pools were separately labelled with ten different Tandem Mass Tags (Thermo Scientific, San Jose, CA, lot UG288073) according to the manufacturer’s instructions. Finally, the content of the tubes was pooled and dried under speed-vacuum before separation using High pH Reversed-Phase Peptide Fractionation Kit (Pierce). In brief, dehydrated samples were reconstituted in 300 μl H2O, TFA 0.1% and applied to the columns and peptides eluted according to the manufacturer’s instructions. Six fractions were collected in total, and directly dried under speed-vacuum and stored at −80 °C until LC-MS/MS analysis using a Q Exactive mass spectrometer (Thermo Fisher Scientific).
Peptide separations were carried out on an Easy-nLC 1000 nanosystem (Thermo Fisher Scientific). For each analysis, samples were loaded into a precolumn Acclaim PepMap 100 (Thermo Fisher Scientific) and eluted in an RSLC PepMap C18, 15-cm long, 50-μm inner diameter and 2-μm particle size (Thermo Fisher Scientific). The mobile phase flow rate was 300 nl/min using 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The gradient profile was set as follows: 0%-35% solvent B for 90 min, 35–100% solvent B for 4 min, 100% solvent B for 8 min. Four microliters of each sample were injected.
For ionisation, 2000 V of liquid junction voltage and 270 °C capillary temperature were used. The full scan method employed an m/z 400–1500 mass selection, an Orbitrap resolution of 70,000 (at m/z 200), a target automatic gain control (AGC) value of 3e6, and maximum injection times of 100 ms. After the survey scan, the 15 most intense precursor ions were selected for MS/MS fragmentation. Fragmentation was performed with a normalised collision energy of 27 and MS/MS scans were acquired with a starting mass of m/z 100, AGC target was 2e5, resolution of 17,500 (at m/z 200), intensity threshold of 8e3, isolation window of 2 m/z units and maximum IT was 100 ms. Charge state screening was enabled to reject unassigned, singly charged, and greater than or equal to seven protonated ions. A dynamic exclusion time of 20 s was used to discriminate against previously selected ions.
MS data analysis
MS data were analysed with MaxQuant (version 1.6.6.0) using standardised workflows. Mass spectra *.raw files were searched against Uniprot UP000005640_9606.fasta Homo sapiens (human) 2019 database (20962 protein entries) using Reporter ion MS2 type. Precursor and reporter mass tolerance were set to 4.5 ppm and 0.003 Da, respectively, allowing two missed cleavages. Carbamidomethylation of cysteines was set as a fixed modification, and methionine oxidation, acetylation N-terminal and Ser, Thr and Tyr phosphorylation were set as variable modifications. Reporter ion intensities were bias-corrected for the overlapping isotope contributions from the TMT tags according to the manufacturer’s certificate. Unique and Razor peptides were considered for quantification. Minimal peptide length and maximal peptide mass were fixed to 7 amino acids and 4600 Da, respectively. Identified peptides were filtered by their precursor intensity fraction with a FDR threshold of 0.01. Proteins identified with at least one peptide and an ion score above 99% were considered for evaluation, whereas proteins identified as potential contaminants were excluded from the analysis. The protein sequence coverage was estimated for specific proteins by the percentage of matching amino acids from the identified peptides having confidence greater than or equal to 95% divided by the total number of amino acids in the sequence.
As the same amount of protein was labelled in each TMT sample, differences in each channel’s total sum of signals were corrected by computing normalisation factors to equal these sums. For data normalisation, sample loading (SL) normalisation was carried out with R Studio (version 3.6.2) according to established protocol (https://github.com/pwilmart), using “tidyverse”, “psych”, “gridExtra” and “scales” packages [28]. Data were then exported to Microsoft Excel 2019 and Perseus (version 1.6.10) for the subsequent analysis. Proteins identified with a fold change of 1.5 for each ratio were selected as potential proteins dysregulated in AIP-overexpressed cells (upregulated = ratio≥1.5 or downregulated=ratio≤0.67).
In vivo animal experiments
The Ethical Committee of the Instituto de Salud Carlos III approved the protocols used for experimental work with mice (Proex 285/19). Liver metastasis, in vivo homing and subcutaneous experiments were performed in Nude mice according to established protocols [23].
Statistical analysis
Statistical analyses were performed with Microsoft Office Excel and Graphpad Prism 8. Data were analysed by one-way analysis of variance followed by Tukey–Kramer multiple comparison test. In both analyses, the minimum acceptable level of significance was P values < 0.05. For discrete variable data, as presence or absence of metastasis, we used χ2 test. For continuous variable data with not Gaussian distribution, as overall survival, we performed a Mann–Whitney U test. For immunohistochemistry analysis, data distribution using the Shapiro–Wilk test and variance homogeneity using the Bartlett test was first evaluated. Since data normality was discarded in all cases, we then assessed whether each indicated group’s means were statistically different from each other using the non-parametric Mann–Whitney U test assuming unequal variances. P values < 0.05 were considered statistically significant.
Results
AIP overexpression in colorectal cancer patients correlates with lower overall survival and liver metastasis
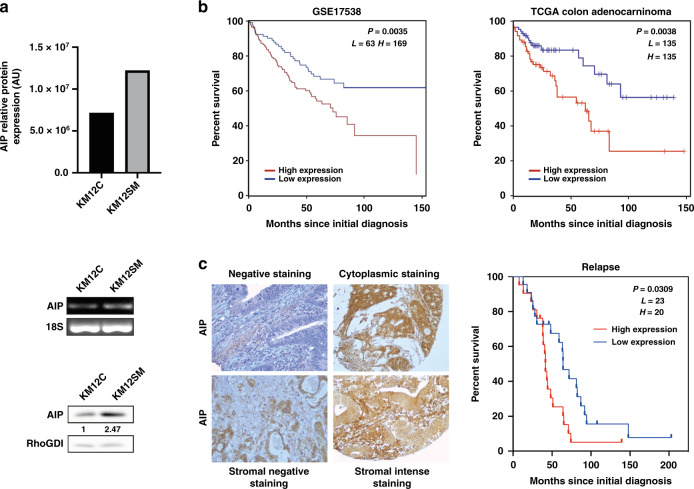
The overexpression of AIP was previously observed by multidimensional proteomics in highly metastatic KM12SM compared to isogenic poorly metastatic KM12C CRC cells (Fig. 1a) [9]. Here, we further assessed the differential expression of AIP by semi-quantitative PCR and WB analysis (Fig. 1a). In addition, we also investigated whether AIP dysregulation could be associated with other CRC cells. mRNA expression of AIP was observed in five out of the eight tested cell lines by semi-quantitative PCR analyses (Supplementary Fig. 1a). qPCR analysis confirmed that the highest AIP expression mRNA levels were observed in the metastatic SW620 and Lim1215 colon cancer cell lines, together with Colo320 cells (Supplementary Fig. 1b). Cell lines with lower metastatic capacity, like SW480 cells from the SW480/SW620 isogenic pair [29], RKO or HT29, showed the lowest mRNA expression levels of AIP (Supplementary Fig. 1). SW480/SW620 results were in concordance with KM12 cells results, where metastatic cells showed higher AIP expression than the poorly or non-metastatic cells from the isogenic pair.
Fig. 1. AIP overexpression is associated with colorectal cancer KM12SM liver metastatic cells and to poor survival of colorectal cancer patients.
a AIP protein expression levels depicted as bar graph were higher in KM12SM liver metastatic cells than in the poorly metastatic KM12C colon cancer cells by spatial proteomics [9]. AIP mRNA and protein expression levels were assessed by semi-quantitative PCR and WB analyses using 18S and RhoGDi as controls, respectively. Protein abundance was quantified by densitometry using Image J. AU, arbitrary units. b Kaplan–Meier analyses of overall survival of patients with colon cancer with the log-rank test according to the expression of AIP. Significant association of AIP expression gene with lower overall survival was found by comparing differences between high- versus low-expression groups with the log-rank test. The publicly available GSE17538 cohort containing colorectal cancer samples with clinicopathological data was used for the prognostic study. The prognostic value of AIP was independently assessed with a different cohort of patients: colon adenocarcinoma (COAD) TCGA dataset containing 270 tumour samples. c Immunohistochemical analysis of AIP expression in tissue microarrays showing representative images of weak, moderate or intense staining of different colon carcinomas. Counterstaining was made with hematoxylin. Pictures were taken at ×100 or ×200 magnification. Significant association of AIP tumoral stromal overexpression with poor survival was found with the log-rank test.
Then, to investigate the clinical relevance in human CRC, we analysed AIP mRNA expression in tumour tissue samples using two public cohorts containing 508 CRC patients (Fig. 1b). Kaplan–Meier analysis showed a strong significant association between high AIP expression and lower overall survival in the GSE17538 cohort (P = 0.0035). These results were validated with the COAD TCGA dataset containing 270 colon adenocarcinoma samples (P = 0.0038). Moreover, AIP protein expression was analysed using tissue microarrays containing 144 core tissue samples from CRC patients followed for more than 5 years and retrospectively selected (Fig. 1b) [20]. AIP high expression significantly correlated with CRC lower survival (P = 0.03) (Fig. 1c).
These results demonstrated an association between AIP expression and poor prognosis of CRC patients, besides its association with liver metastasis observed by proteomics.
AIP overexpression promotes adhesion, colony formation, migration and invasion of colorectal cancer cells
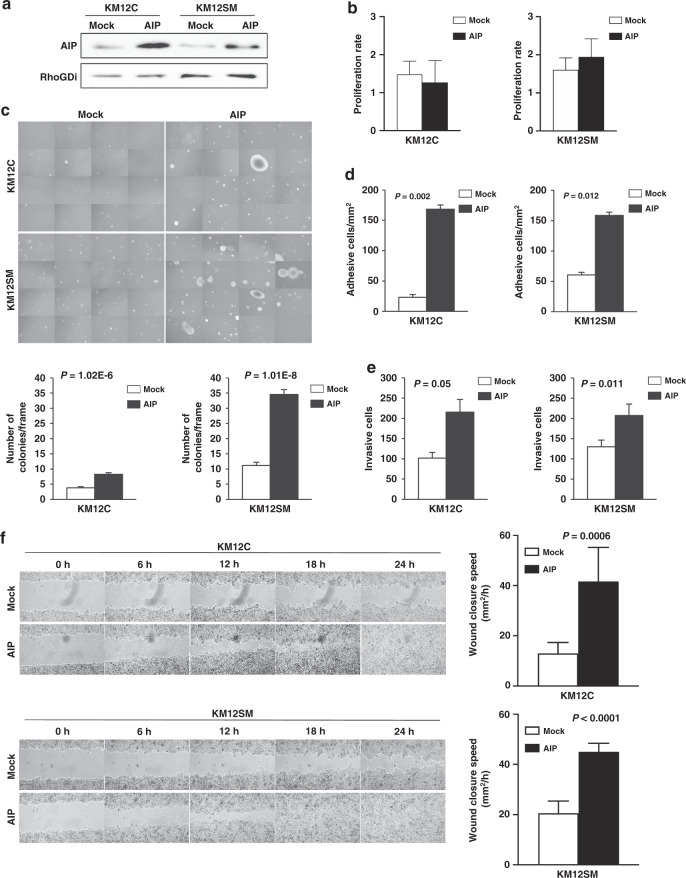
To address the role of AIP overexpression in tumorigenesis and metastasis, we studied the effect of stably overexpressing AIP in the KM12 cell model of CRC liver metastasis in comparison to Mock-stably transfected control cells. A significant increase in AIP expression was observed in KM12 cells stably transfected with AIP by WB (Fig. 2a), semi-quantitative PCR, and immunofluorescence (IF; Supplementary Fig. 2). AIP-stable transfection effect was more pronounced on KM12C, which showed less AIP protein expression by proteomics, than on KM12SM cells.
Fig. 2. AIP-ectopic expression in colorectal cancer cells increases tumorigenic and metastatic properties of colorectal cancer cells.
a WB analysis of AIP in KM12C and KM12SM cells stably expressing AIP and Mock confirmed the ectopic expression of AIP in both cell lines. b Proliferation was determined by MTT assays after 96 h of culture. A non-significant slightly decreased optical density in AIP-overexpressing KM12C and KM12SM cells was observed in comparison to Mock control cells. c AIP-ectopic expression induces the formation of soft agar colonies in KM12C and KM12SM cells in comparison to Mock control cells. Single-cell suspensions of AIP- and Mock-stably transfected KM12C and KM12SM cells were seeded in soft agar and allowed to form colonies for 21 days in six-well plates. Then, colonies were visualised by microscopy by taking photographs of 16 random fields. The average colony number per frame was represented by bar graphs. d Cell adhesion to Matrigel of AIP- or Mock-stably transfected, after starving cells for 5 h in medium alone. e KM12C and KM12SM ectopically expressing AIP showed approximately twofold higher invasion than Mock-stably transfected cells. f AIP and Mock control cells were grown until confluence and their migratory capabilities were analysed in a wound-healing assay every 90 min until confluence. KM12C and KM12SM cells migratory capabilities were significantly enhanced by AIP-ectopic expression. Representative images of the wound-healing assay are shown. Migration speed (mm2/h) of AIP and Mock control cells was calculated as the distance covered every 24 h. Data for all the experiments represent the mean ± SEM of three independent experiments. P values of all the experiments are shown.
Next, the tumorigenic (proliferation and colony formation) and metastatic (adhesion, migration and invasion) properties of the AIP- and Mock-stably transfected KM12 cells were investigated. AIP-overexpressing KM12 cells showed non-significant changes in proliferation (Fig. 2b). Regarding colony-forming ability, AIP-stably transfected KM12C and KM12SM cells showed a 2.5-fold increase and about 3.5-fold, respectively, with respect to Mock cells. Similar colony formation ability for AIP-stably transfected KM12C cells was observed in comparison to Mock KM12SM cells (Fig. 2c).
Then, the adhesive properties of the cell lines were analysed using Matrigel assays. AIP induced a fivefold higher adhesion capacity on KM12C cells. In KM12SM cells, a striking 155% increase in their adhesion capacity was observed (Fig. 2d). Remarkably, AIP-ectopic expression induced KM12C and KM12SM cells to have a similar adhesion capacity.
AIP-stably transfected cells also showed increased migration and invasive capacities. A significant increase in the invasive properties induced by AIP, more pronounced on KM12C cells, was found on KM12 cells (Fig. 2e). Changes in migration, assessed via wound-healing assays in 24-well plates coated with Matrigel, showed that KM12 Mock controls were unable to close the wound (Fig. 2f). On the other hand, AIP-overexpressing cells showed a steep increase in migration speed, more accentuated in KM12C cells. Collectively, these results demonstrated that AIP significantly augments the tumorigenic and metastatic properties of KM12 CRC cells. Effects were more evident for the poorly metastatic KM12C cells, which upon AIP-stable transfection increased their tumoral and metastatic properties nearby to KM12SM Mock cells or surpassed them regarding invasive and migration capacities.
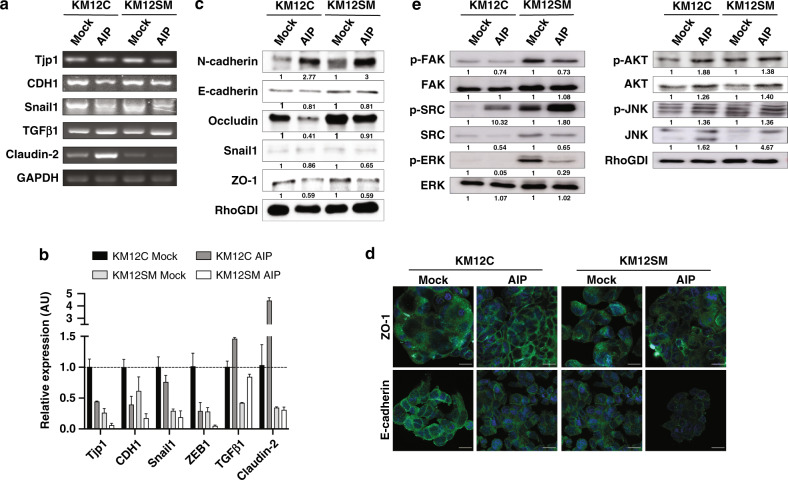
AIP overexpression modified the expression of inducers of the mesenchymal phenotype
Since cell adhesion, migration and invasive capacity of epithelial cells correlate with the epithelial-to-mesenchymal transition (EMT), we investigated for alterations in EMT inducers. We studied changes in the mRNA expression levels of Snail1 (SNAI1), ZEB1, TGFβ1, Claudin-2, ZO-1 and E-cadherin (CDH1) by semi-quantitative PCR and/or qPCR analyses. In KM12C and KM12SM cells, AIP overexpression caused a significant alteration in the EMT inducers TGFβ1, Snail1, and ZEB1, accompanied by a large decrease in the epithelial marker CDH1 (Fig. 3a). Semi-quantitative PCR results for Tjp1 (ZO-1), CDH1, Snail1, TGFβ1 and Claudin-2 were further assessed by qPCR analysis (Fig. 3b). Collectively, AIP overexpression induced in KM12 cells a large decrease in ZEB1, Snail1 and CDH1, and an increase in TGFβ1. Furthermore, AIP-ectopic expression altered the expression of the mRNAs of the tight junctions’ proteins Tjp1 (ZO-1) and Claudin-2.
Fig. 3. Alterations in EMT inducers after AIP-ectopic expression in colorectal cancer cells.
a cDNA synthesised from total RNA from AIP- and Mock-stably transfected cells was subjected to semi-quantitative PCR using specific primers (Supplementary Table 3) for the EMT inducers Tjp1 (ZO-1), CDH1, Snail1, TGFβ1, and Claudin-2 using GAPDH for normalisation. b cDNA synthesised from total RNA from AIP- and Mock-stably transfected cells were subjected to qPCR analysis using specific primers (Supplementary Table 3) for the EMT inducers Tjp1 (ZO-1), CDH1, Snail1, ZEB1, TGFβ1 and Claudin-2 using 18 S for normalisation. Data are shown as mean ± SD. c KM12C and KM12SM Mock and AIP cells were lysed and subjected to WB analysis using specific antibodies against the indicated proteins and EMT markers. The abundance of each protein was quantified by densitometry. RhoGDI was used as a loading control. d Immunofluorescence analysis of ZO-1 and E-cadherin (green) in Mock and AIP-stably transfected KM12C and KM12SM cells. DAPI was used for counterstaining the nucleus in blue. e Mock and AIP KM12C and KM12SM cells were starved, lysed and subjected to WB analysis using specific antibodies against phosphorylated p-AKT, p-ERK1/2, p-FAK, p-JNK and p-SRC and total AKT, ERK1/2, FAK, JNK and SRC. RhoGDi was used as a loading control. Protein abundance was quantified by densitometry using Image J. AU arbitrary units. Scale bar: 20 μm.
At the protein level, Snail1, ZO-1 and E-cadherin confirmed mRNA results (Fig. 3c). We observed a considerable decrease of E-cadherin together with a noticeable increase in N-cadherin supporting their opposite dysregulation [30], indicative of a reduction of the mesenchymal phenotype. Observed changes in E-cadherin and adherens junction protein ZO-1 upon AIP-ectopic expression were also confirmed by IF (Fig. 3d). Differences in E-cadherin expression were more evident in the cell membrane, suggesting that AIP facilitated N-cadherin expression and the suppression of functional E-cadherin on the cell surface. Collectively, these data confirm that AIP induces a significant alteration on EMT effectors.
Signalling analysis in AIP-stably transfected colorectal cancer cells
Then, the effect of AIP-ectopic expression on signalling pathways associated with effects on tumorigenic and metastatic properties was analysed. We observed a significant activation of phospho-SRC, phospho-JNK, and phospho-AKT in AIP-overexpressed KM12 cells (Fig. 3e). In contrast, we detected a noticeable reduction of phospho-ERK1/2, correlating with the non-significant changes in proliferation of AIP-stably transfected cells, and phospho-FAK. These changes suggest an effect mediated by AIP through AKT on EMT and cell survival, and SRC and JNK on cell migration, adhesion, and invasion, which may play a role in advanced CRC facilitating liver metastatic colonisation.
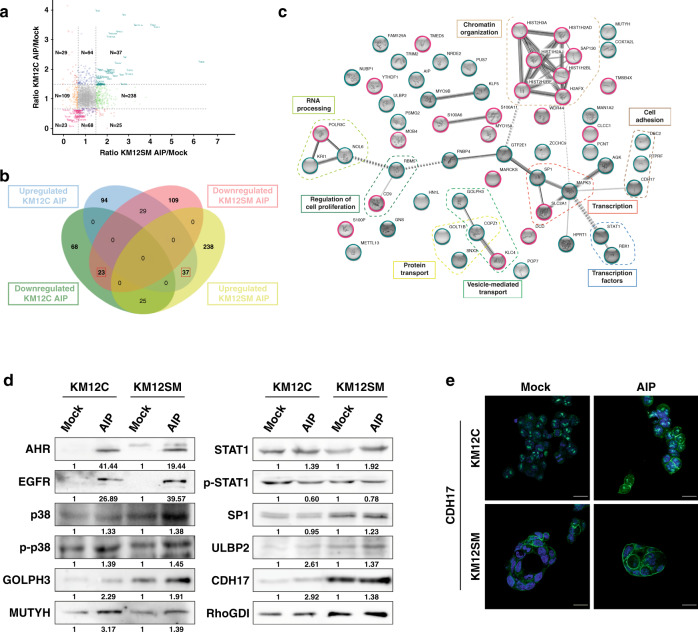
Identification of proteins affected by AIP overexpression on KM12 colorectal cancer cells by proteomics
We then carried out a proteomic approach to identify AIP-modulated proteins and characterise their interaction network (Supplementary Fig. 3a). AIP-stably transfected KM12 lysed cells were analysed by quantitative TMT proteomics analysis. As control, we included in the assay Mock and parental cells. After data normalisation (Supplementary Fig. 3b), a total of 3124 proteins were identified and quantified with at least one peptide (Supplementary Table 1). Among them, 569 proteins identified and quantified with two or more peptides showed upregulation or downregulation because of AIP-overexpression with a fold change ≥1.5 or ≤0.67, respectively (Fig. 4a, b and Supplementary Table 2). To identify proteins highly specifically modulated by AIP, we focused on those proteins commonly dysregulated in KM12C and KM12SM cells. In total, we found 60 proteins up- or downregulated (Fig. 4b and Table 1). As expected, AIP was among the overexpressed proteins.
Fig. 4. Mass spectrometry analysis of protein alterations modulated by AIP overexpression in CRC cells.
a Correlation scatter plot of proteins identified and quantified as altered in AIP-overexpressed KM12C and KM12SM cells in comparison to Mock control cells with at least one high-confidence peptide in the different TMT channels (q value<0.01). The x axis shows the ratio of protein expression differences between AIP- and Mock-transfected KM12SM cells, and the y axis the ratio of protein expression differences between AIP- and Mock-transfected KM12C cells. Coloured dots represent differentially expressed proteins upregulated (green) and downregulated (orange) in AIP-overexpressed KM12SM, and differentially expressed proteins upregulated (blue) and downregulated (purple) in AIP-overexpressed KM12C with 1.5-fold expression difference (represented by the four black lines). Proteins differentially expressed with ≥1.5-fold change difference in both AIP-overexpressed KM12C and KM12SM cells are represented in bluish-green (upregulated) and pink (downregulated) dots, whereas proteins showing an opposite dysregulation in AIP-overexpressed KM12C and KM12SM cells are represented in red. b Venn Diagram of the 569 proteins dysregulated in AIP-overexpressed KM12C and KM12SM cells identified with two or more peptides and ≥1.5-fold expression difference. Red squares, the 60 proteins commonly dysregulated in both AIP-overexpressed cell lines. c Protein interactome map of the proteins significantly dysregulated by AIP in both KM12C and KM12SM cells. STRING analysis of known and predicted protein-protein interactions of upregulated and downregulated gene-products differentially expressed in KM12C and KM12SM cells by AIP. STRING Version 11.0 was used. The MCL algorithm set to 2 was used to define the clusters of interaction. Upregulated proteins by AIP overexpression are circled in bluish-green and downregulated proteins in pink. d SDS-PAGE 10% gels were run with protein extracts from the indicated conditions and transferred onto nitrocellulose membranes. Then, membranes were incubated with specific antibodies against the indicated dysregulated proteins by AIP at optimised dilutions and the signal developed with HRP-labelled secondary antibodies (Supplementary Table 4). RhoGDI was used as loading control in the same gels and for normalisation using protein abundance in Mock-stably transfected cells as basal protein levels. p-p38, phosphorylated p38. p-STAT1, phosphorylated STAT1. Protein abundance was quantified by densitometry using Image J. AU arbitrary units. e Immunofluorescence analysis of CDH17 (green) in Mock- and AIP-stably transfected KM12C and KM12SM cells. DAPI was used for counterstaining of the nucleus in blue.
Table 1.
Proteins upregulated or downregulated in both AIP-overexpressed KM12C and KM12SM cells.
| Protein IDs | Protein names | Gene names | KM12C and KM12SM AIP-transfected cells* |
|---|---|---|---|
| P06703 | Protein S100-A6 | S100-A6 | Down |
| P11166 | Solute carrier family 2, facilitated glucose transporter member 1 | SLC2A1 | Down |
| P16104 | Histone H2AX | H2AFX | Down |
| P20671 | Histone H2A type 1-D | HIST1H2AD | Down |
| P21926 | CD9 antigen | CD9 | Down |
| P25815 | Protein S100-P | S100-P | Down |
| P29966 | Myristoylated alanine-rich C-kinase substrate | MARCKS | Down |
| P31949 | Protein S100-A11; Protein S100-A11, N-terminally processed | S100-A11 | Down |
| P62328 | Thymosin beta-4; Hematopoietic system regulatory peptide | TMSB4X | Down |
| P81605 | Dermcidin; Survival-promoting peptide; DCD-1 | DCD | Down |
| Q16778 | Histone H2B type 2-E | HIST2H2BE | Down |
| Q5JSH3 | WD repeat-containing protein 44 | WDR44 | Down |
| Q71DI3 | Histone H3.2 | HIST2H3A | Down |
| Q96S66 | Chloride channel CLIC-like protein 1 | CLCC1 | Down |
| Q99878 | Histone H2A type 1-J | HIST1H2AJ | Down |
| Q99880 | Histone H2B type 1-L | HIST1H2BL | Down |
| Q9BUI4 | DNA-directed RNA polymerase III subunit RPC3 | POLR3C | Down |
| Q9BYJ9 | YTH domain-containing family protein 1 | YTHDF1 | Down |
| Q9H0E3 | Histone deacetylase complex subunit SAP130 | SAP130 | Down |
| Q9NSK0 | Kinesin light chain 4 | KLC4 | Down |
| Q9UKN7 | Unconventional myosin-XV | MYO15A | Down |
| Q9Y3A3 | MOB-like protein phocein | MOB4 | Down |
| Q9Y3A6 | Transmembrane emp24 domain-containing protein 5 | TMED5 | Down |
| O00170 | AH receptor-interacting protein | AIP | Up |
| O14548 | Cytochrome c oxidase subunit 7A-related protein, mitochondrial | COX7A2L | Up |
| O60476 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB | MAN1A2 | Up |
| O60493 | Sorting nexin-3 | SNX3 | Up |
| O75817 | Ribonuclease P protein subunit p20 | POP7 | Up |
| O95613 | Pericentrin | PCNT | Up |
| P00492 | Hypoxanthine-guanine phosphoribosyltransferase | HPRT1 | Up |
| P08047 | Transcription factor Sp1 | SP1 | Up |
| P10586 | Receptor-type tyrosine-protein phosphatase F | PTPRF | Up |
| P15586 | N-acetylglucosamine-6-sulfatase | GNS | Up |
| P27361 | Mitogen-activated protein kinase 3 | MAPK3 | Up |
| P29083 | General transcription factor IIE subunit 1 | GTF2E1 | Up |
| P42224 | Signal transducer and activator of transcription 1-alpha/beta | STAT1 | Up |
| P52756 | RNA-binding protein 5 | RBM5 | Up |
| P53384 | Cytosolic Fe-S cluster assembly factor NUBP1 | NUBP1 | Up |
| P61923 | Coatomer subunit zeta-1 | COPZ1 | Up |
| P62877 | E3 ubiquitin-protein ligase RBX1 | RBX1 | Up |
| Q02487 | Desmocollin-2 | DSC2 | Up |
| Q12864 | Cadherin-17 | CDH17 | Up |
| Q13459 | Unconventional myosin-IXb | MYO9B | Up |
| Q13887 | Krueppel-like factor 5 | KLF5 | Up |
| Q53H12 | Acylglycerol kinase, mitochondrial | AGK | Up |
| Q8N3X1 | Formin-binding protein 4 | FNBP4 | Up |
| Q8N567 | Zinc finger CCHC domain-containing protein 9 | ZCCHC9 | Up |
| Q8N6R0 | Methyltransferase-like protein 13 | METTL13 | Up |
| Q8N9T8 | Protein KRI1 homolog | KRI1 | Up |
| Q969U7 | Proteasome assembly chaperone 2 | PSMG2 | Up |
| Q96PZ0 | Pseudouridylate synthase 7 homolog | PUS7 | Up |
| Q9BZM5 | NKG2D ligand 2 | ULBP2 | Up |
| Q9BZQ8 | Protein Niban | FAM129A | Up |
| Q9C040 | Tripartite motif-containing protein 2 | TRIM2 | Up |
| Q9H4A6 | Golgi phosphoprotein 3 | GOLPH3 | Up |
| Q9H6R4 | Nucleolar protein 6 | NOL6 | Up |
| Q9H7Z3 | Protein NRDE2 homolog | NRDE2 | Up |
| Q9H910 | Haematological and neurological expressed 1-like protein | HN1L | Up |
| Q9UIF7 | A/G-specific adenine DNA glycosylase | MUTYH | Up |
| Q9Y3E0 | Vesicle transport protein GOT1B | GOLT1B | Up |
*Down, ≤0.67-fold change. Up, ≥1.5-fold change.
Using STRING and data mining [31], proteins were classified into eight clusters of interaction, including proteins related to cell adhesion, cell cycle and regulation of proliferation, transport, transcription, chromatin organisation, transcription factors and RNA processing (Fig. 4c). Using Reactome [32], altered processes due to AIP-ectopic expression were observed (Supplementary Fig. 4). Among them, AIP was observed to induce changes in DNA repair (P value = 4.08E-4), cell cycle (P value = 1.34E-2), gene expression (P value = 4.57E-2) and metabolism of proteins (mainly protein deubiquitination (P value = 2.26E-2) and asparagine N-glycosylation (P value = 2.81E-2)). Moreover, the dysregulation of all the cell-adhesion proteins upregulated by AIP—CDH17, DSC2 and PTPRF—has been described to increase metastasis, in vivo homing and proliferation, while also contributing to adherens junctions redistribution and cytoskeletal rearrangement [23, 33–35]. Furthermore, AIP was found to upregulate the transcription factors SP1 and STAT1, which play a major role in cancer and metastatic progression [36–38].
WB and IF analyses of selected targets confirmed the dysregulation of indicated proteins (Fig. 4d, e). The overexpression observed by proteomics of EGFR, p38, GOLPH3, MUTYH, STAT1 and CDH17 due to AIP-ectopic expression in KM12C and KM12SM cells was confirmed, whereas SP1 overexpression could be only validated in KM12SM cells. In addition, AIP was able to activate phospho-p38, in contrast to phospho-STAT1, whose expression decreased in parallel with AIP overexpression (Fig. 4d). It was also observed that the ectopic expression of AIP-induced Aryl-hydrocarbon receptor (AHR) expression in KM12C and KM12SM cells (Fig. 4d).
Remarkably, IF analysis showed that the ectopic expression of AIP-induced changes in the abundance and the localisation of CDH17, with the recruitment of CDH17 to the plasmatic membrane (Fig. 4e).
AIP induces in vivo tumour growth, liver metastasis and decreases mice survival
Finally, we investigated the in vivo effects of AIP-ectopic expression. First, we examined its effects on the capacity of KM12 cells inoculated in the spleen of nude mice for liver homing. As a surrogate marker for homing, human GAPDH was highly detected in the livers of mice inoculated with AIP-stably transfected KM12 cells in comparison to Mock cells (Fig. 5a). More importantly, AIP-ectopic expression induced in KM12C cells ability to colonise the liver.
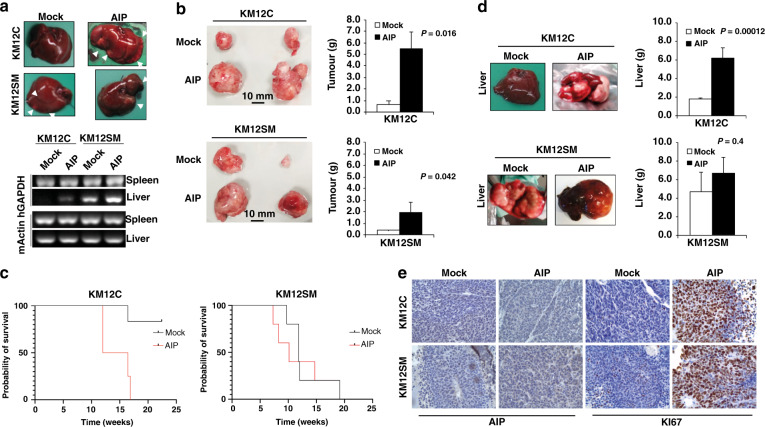
Fig. 5. AIP-ectopic expression induces tumour growth and liver metastasis in KM12 cells.
a Nude mice intrasplenically inoculated with indicated KM12 cells were sacrificed 24 h after inoculation for analysis of in vivo homing. RNA was isolated from the liver and subjected to RT-PCR to amplify human GAPDH (hGAPDH). Representative experiments out of three are shown. Murine β-actin (mβ-actin) was amplified as a control. b KM12 transfectants were inoculated subcutaneously in nude mice (n = 6 per group). Tumour size was measured every day for 4 weeks and mean ± SEM of the endpoint represented by bar graphs. Representative tumour images from KM12 cell stably transfectants are also depicted. Tumours were significantly higher in AIP-transfected cells (P < 0.05) and in AIP-transfected KM12C cells vs KM12SM cells (P = 0.013). c Kaplan–Meier survival assay of nude mice inoculated intrasplenic with the indicated KM12 cell transfectants (n = 5 per group). Survival of mice inoculated with AIP-stably transfected cells significantly decreased (**P < 0.01) when compared with those inoculated with Mock control cells. d Mice were examined for macroscopic metastases in liver. (Left) representative images of macroscopic metastases in the liver significantly induced in mice inoculated intra-spleen with AIP-stably transfected KM12C and KM12SM cells are shown. (Right) Livers were weighed at the endpoint and mean ± SEM represented (n = 5 per group). Livers were significantly heavier in mice inoculated intra-spleen with AIP-stably transfected KM12C and KM12SM cells because of tumour metastasis. e Representative IHC images of FFPE tumours developed in the mice injected with either KM12C or KM12SM Mock or AIP-stably expressing cells. Tumour samples were stained with anti-KI67 or anti-AIP antibodies. A more intense AIP staining was observed in tumours from mice inoculated with AIP-overexpressing cells compared to Mock controls. Likewise, a clearly more intense nuclear KI67 staining was observed in tumours from mice subcutaneously inoculated with AIP-overexpressing cells.
Then, the role of AIP in CRC tumour growth, metastasis and survival was investigated by subcutaneous and intrasplenic cell inoculations. AIP-stably transfected cells developed significantly higher measurable tumours after subcutaneous inoculation than Mock KM12 cells (Fig. 5b). In addition, mice inoculated intrasplenically with AIP-stably transfected KM12C cells showed shorter survival than those inoculated with Mock control cells (Fig. 5c). Importantly, Mock KM12C control cells did not develop any metastasis to the liver as opposed to AIP-stably transfected KM12C cells. This reduced survival was associated with the AIP-induced higher capacity for liver colonisation, as visually inspected and depicted by the mice’s liver weight at the endpoint (Fig. 5d). Regarding KM12SM cells, although no significant differences were observed in survival because AIP-stably transfected and Mock KM12SM cells develop liver metastasis (Fig. 5c), liver metastasis were considerably higher in AIP-stably transfected KM12SM cells (Fig. 5d). Finally, to confirm that the observed differences were due to the ectopic expression of AIP in the injected cells, we analysed AIP protein abundance on tumour sections by immunohistochemistry (IHC; Fig. 5e). We could observe a more intense AIP staining in the tumours of mice injected with AIP-stably expressing cells. In addition, AIP-stably expressing cells showed a clear increase in the marker for cellular proliferation KI67 [39] compared to Mock conditions (Fig. 5e), indicating that tumours overexpressing AIP were more proliferative than their Mock counterparts.
Discussion
We here found that AIP is highly overexpressed in CRC metastatic cell lines, and liver metastatic KM12SM CRC cells. AIP is associated to poor prognosis in CRC patients, and acts as a novel key player promoting CRC metastasis. AIP affected a plethora of transcription factors, cell-adhesion molecules and signalling cascades leading to SRC, JNK and PI3K/AKT pathways’ activation. Associated with these changes, AIP altered adhesion, colony formation, migration, and invasion capacity of cells. Remarkably, these effects together with AIP-associated dysregulation of multiple EMT factors induced non-metastatic KM12C cells to become metastatic to the liver.
Germline AIP mutations have been strongly associated with familial isolated pituitary adenoma [40]. However, AIP’s mutation analysis in colorectal, breast and prostate cancer showed that the presence of somatic mutations is not a common finding [41]. In contrast, AIP overexpression in tumors has been strongly associated with a poor outcome in gastric [42], pancreatic [43] and colorectal cancer patients (here presented data). Notably, AIP-induced alterations in the EMT process have been previously reported. Gene expression analysis of AIP germline-mutated pituitary adenomas showed that the EMT pathway was altered with 16 upregulated and 31 downregulated genes [44]. Here, we observed that AIP-induced alterations in EMT, as depicted by the downregulation of Snail1, Zeb1 and E-cadherin and an increase of N-cadherin and TGFβ1 accompanied by CDH17 overexpression. Altogether, these observations suggest that AIP-mediated EMT dysregulation is a common event on AIP-mutated or AIP-overexpressing tumors.
Previous clinical data supported the role of AIP as a tumour suppressor since AIP germline-mutated gene was associated with familial isolated pituitary adenoma [45]. Germline mutational analysis of these adenomas detected 50 different pathogenic mutations leading to AIP disruption. Moreover, wild-type AIP overexpression in human fibroblast and pituitary cell lines reduced cell proliferation in vitro, whereas the mutant AIP loses this ability compared to the wild-type AIP [46]. However, AIP overexpression in gastrointestinal—colorectal, gastric and pancreatic—cancers was associated with a worse prognosis, and in CRC cells produced a significant increase in overall metastatic capacities. In addition, this work showed the first association of AIP overexpression with liver homing and liver metastasis in CRC. Therefore, our results indicate that beyond its role as a tumour suppressor in pituitary adenomas, AIP acts as an oncogene in CRC.
Twenty interaction partners have been described for AIP, of which fourteen were confirmed to interact directly with AIP. These include viral proteins (HBV X and EBNA-3), chaperones (hsp90 and hsc70), PDEs (PDE4A5 and PDE2A3), nuclear (AHR, PPARα and TRβ1) and transmembrane (RET) receptors, G proteins (Gα13 and Gαq), survivin and a mitochondrial import receptor (TOMM20) [47]. Besides the initial interest drawn upon AIP and AHR in the 2000s, the interactome or the cellular pathways in which AIP is involved remain obscure. Here, we have shed some light on these processes by quantitative proteomics and orthogonal techniques. In this sense, AIP-ectopic expression induced a vast protein dysregulation in KM12 cells, associated to chromatin organisation, DNA repair, cell cycle, or signal transduction, among others. Notably, adhesion proteins—as CDH17—and transcription factors—as SP1 and STAT1—known to play central roles in cancer and metastasis were also dysregulated by AIP [23, 36–38]. In addition to the vast number of proteins dysregulated by AIP, we found AIP in the cytoplasm and nucleus by IF and IHC from AIP-overexpressing cells and CRC tumoral tissue in Nude mice, respectively. Thus, it is plausible to think that beyond its interaction with AHR and the translocation of this receptor to the nucleus where it functions as a transcription factor [48, 49], AIP might act independently to AHR as a (direct or indirect) transcription factor itself when overexpressed. Two of the most interesting proteins upregulated by AIP were the atypical cadherin CDH17 and EGFR. CDH17 facilitates cell clustering via homotypic cadherin interactions and cell adhesion through integrin activation to initiate micrometastasis formation [23]. Therefore, AIP-mediated increase of CDH17 should facilitate liver adhesion and in vivo homing of CRC cells ectopically expressing AIP. On the other hand, previous reports have already established the role of EGFR in tumour onset and progression [50–52]. Mutation or overexpression of EGFR leads to altered EGFR signalling, which in turn induces abnormal trafficking and contributes to increased signalling and tumour development [50].
In summary, this work demonstrates the value of AIP, previously identified from a spatial proteomic analysis of metastatic cells [9], and multidimensional proteomics for identifying relevant proteins in metastasis with actual value in CRC patients. Although AIP depletion was not explored in CRC cells as reciprocal phenotypes—as a limitation of the study—we report here a new role for AIP in adhesion, invasion, migration, colony formation and liver metastasis in CRC as depicted from AIP overexpression. Our conclusions were based on the following observations: (i) AIP high expression was associated with liver metastasis and poor overall survival, (ii) AIP-ectopic overexpression increased cell adhesion, migration, invasion and colony formation in KM12 cells, particularly in non-metastatic KM12C cells, (iii) AIP overexpression induced the dysregulation of proteins related to the EMT transition or involved in cancer and metastasis, suggesting a transcription factor role for AIP, (iv) AIP overexpression also induced an increase in the phosphorylation levels of JNK, SRC and AKT, (v) AIP overexpression increased in vivo tumour growth and decreased mice survival and (vi) AIP overexpression induced KM12C cells to acquire the ability for liver colonisation. Together, these data confirm that AIP has a key role in CRC and liver metastasis, possessing oncogene features.
Supplementary information
Legend to Supplementary Figures and Tables
Author contributions
GSF: data curation; formal analysis, investigation, writing—original draft and writing—review and editing. AMC: data curation, investigation, writing—original draft and writing—review and editing. MSM: data curation and investigation. APG: formal analysis, investigation, methodology, supervision and writing—review and editing. MJFA: formal analysis, methodology, supervision and writing—review and editing. PP: investigation and writing—review. MAN: investigation and writing—review. MM: methodology, supervision and writing—review. J Hendrix: supervision and writing—review. DH: methodology, supervision and writing—review. RAB: conceptualisation, investigation, supervision and writing—review & editing. J Hofkens: conceptualisation, investigation, supervision, writing—review & editing and funding. SR: conceptualisation, formal analysis, investigation, writing—original draft, writing—review and editing and funding. RB: conceptualisation, formal analysis, investigation, writing—original draft, writing—review and editing and funding.
Funding
This work was supported by the financial support of the PI17CIII/00045 and PI20CIII/00019 grants from the AES-ISCIII program to RB. J Hendrix acknowledges funding by UH-BOF (BOF20TT06). J Hofkens acknowledges financial support from the Research Foundation-Flanders (FWO, Grant No. ZW15_09-G0H6316N), the Flemish government through long-term structural funding Methusalem (CASAS2, Meth/15/04) and the MPI as MPI fellow. S.R. acknowledges the financial support of the KU Leuven through the internal C1 funding (KU Leuven (C14/16/053)). GSF is the recipient of a predoctoral contract (grant number 1193818 N) supported by The Flanders Research Foundation (FWO). The FPU predoctoral contract to AMC is supported by the Spanish Ministerio de Educación, Cultura y Deporte.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files. The Mass Spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031119. Other data are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All animal experiments were performed under the guidelines of the Instituto de Salud Carlos III Institutional Animal Care Committee. All experimental protocols were approved by the Committee (Proex 285/19). This article does not contain any studies with human participants by any of the authors out of public databases. However, immunohistochemical analyses of human tumour specimens were in compliance with ethical standards and approved by the Hospital Clínico San Carlos Ethics Board; and informed consent was obtained from all individual participants included in the study.
Consent to publish
All authors of this article gave consent for the publication of the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ana Montero-Calle, Maricruz Sánchez-Martínez.
These authors jointly supervised this work: Rubén A. Bartolomé, Rodrigo Barderas.
Contributor Information
Susana Rocha, Email: susana.rocha@kuleuven.be.
Rodrigo Barderas, Email: r.barderasm@isciii.es.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01762-1.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Torres S, Bartolome RA, Mendes M, Barderas R, Fernandez-Acenero MJ, Pelaez-Garcia A, et al. Proteome profiling of cancer-associated fibroblasts identifies novel proinflammatory signatures and prognostic markers for colorectal cancer. Clin Cancer Res. 2013;19:6006–19. doi: 10.1158/1078-0432.CCR-13-1130. [DOI] [PubMed] [Google Scholar]
- 3.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Z, Chiu JF, He QY. Application of proteomics in the study of tumor metastasis. Genomics Proteom Bioinforma. 2004;2:152–66. doi: 10.1016/S1672-0229(04)02021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aleckovic M, Wei Y, LeRoy G, Sidoli S, Liu DD, Garcia BA, et al. Identification of Nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev. 2017;31:1439–55. doi: 10.1101/gad.301937.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song P, Bao H, Yu Y, Xue Y, Yun D, Zhang Y, et al. Comprehensive profiling of metastasis-related proteins in paired hepatocellular carcinoma cells with different metastasis potentials. Proteom Clin Appl. 2009;3:841–52. doi: 10.1002/prca.200780131. [DOI] [PubMed] [Google Scholar]
- 7.Sun B, Zhang S, Zhang D, Li Y, Zhao X, Luo Y, et al. Identification of metastasis-related proteins and their clinical relevance to triple-negative human breast cancer. Clin Cancer Res. 2008;14:7050–9. doi: 10.1158/1078-0432.CCR-08-0520. [DOI] [PubMed] [Google Scholar]
- 8.Barderas R, Mendes M, Torres S, Bartolome RA, Lopez-Lucendo M, Villar-Vazquez R, et al. In-depth characterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion. Mol Cell Proteom. 2013;12:1602–20. doi: 10.1074/mcp.M112.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendes M, Pelaez-Garcia A, Lopez-Lucendo M, Bartolome RA, Calvino E, Barderas R, et al. Mapping the spatial proteome of metastatic cells in colorectal cancer. Proteomics. 2017;17:1700094. [DOI] [PubMed]
- 10.Morikawa K, Walker SM, Jessup JM, Fidler IJ. In vivo selection of highly metastatic cells from surgical specimens of different primary human colon carcinomas implanted into nude mice. Cancer Res. 1988;48:1943–8. [PubMed] [Google Scholar]
- 11.Morikawa K, Walker SM, Nakajima M, Pathak S, Jessup JM, Fidler IJ. Influence of organ environment on the growth, selection, and metastasis of human colon carcinoma cells in nude mice. Cancer Res. 1988;48:6863–71. [PubMed] [Google Scholar]
- 12.Li A, Varney ML, Singh RK. Constitutive expression of growth regulated oncogene (gro) in human colon carcinoma cells with different metastatic potential and its role in regulating their metastatic phenotype. Clin Exp Metastasis. 2004;21:571–9. doi: 10.1007/s10585-004-5458-3. [DOI] [PubMed] [Google Scholar]
- 13.Kuniyasu H, Ohmori H, Sasaki T, Sasahira T, Yoshida K, Kitadai Y, et al. Production of interleukin 15 by human colon cancer cells is associated with induction of mucosal hyperplasia, angiogenesis, and metastasis. Clin Cancer Res. 2003;9:4802–10. [PubMed] [Google Scholar]
- 14.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–84. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barderas R, Bartolome RA, Fernandez-Acenero MJ, Torres S, Casal JI. High expression of IL-13 receptor alpha2 in colorectal cancer is associated with invasion, liver metastasis, and poor prognosis. Cancer Res. 2012;72:2780–90. doi: 10.1158/0008-5472.CAN-11-4090. [DOI] [PubMed] [Google Scholar]
- 16.Hegde P, Qi R, Gaspard R, Abernathy K, Dharap S, Earle-Hughes J, et al. Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray. Cancer Res. 2001;61:7792–7. [PubMed] [Google Scholar]
- 17.Ellens KW, Christian N, Singh C, Satagopam VP, May P, Linster CL. Confronting the catalytic dark matter encoded by sequenced genomes. Nucleic Acids Res. 2017;45:11495–514. doi: 10.1093/nar/gkx937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perdigao N, Heinrich J, Stolte C, Sabir KS, Buckley MJ, Tabor B, et al. Unexpected features of the dark proteome. Proc Natl Acad Sci USA. 2015;112:15898–903. doi: 10.1073/pnas.1508380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garranzo-Asensio M, San Segundo-Acosta P, Poves C, Fernandez-Acenero MJ, Martinez-Useros J, Montero-Calle A, et al. Identification of tumor-associated antigens with diagnostic ability of colorectal cancer by in-depth immunomic and seroproteomic analysis. J Proteom. 2020;214:103635. doi: 10.1016/j.jprot.2020.103635. [DOI] [PubMed] [Google Scholar]
- 21.Garranzo-Asensio M, San Segundo-Acosta P, Martinez-Useros J, Montero-Calle A, Fernandez-Acenero MJ, Haggmark-Manberg A, et al. Identification of prefrontal cortex protein alterations in Alzheimer’s disease. Oncotarget. 2018;9:10847–67. doi: 10.18632/oncotarget.24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barderas R, Villar-Vazquez R, Fernandez-Acenero MJ, Babel I, Pelaez-Garcia A, Torres S, et al. Sporadic colon cancer murine models demonstrate the value of autoantibody detection for preclinical cancer diagnosis. Sci Rep. 2013;3:2938. doi: 10.1038/srep02938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartolome RA, Barderas R, Torres S, Fernandez-Acenero MJ, Mendes M, Garcia-Foncillas J, et al. Cadherin-17 interacts with alpha2beta1 integrin to regulate cell proliferation and adhesion in colorectal cancer cells causing liver metastasis. Oncogene. 2014;33:1658–69. doi: 10.1038/onc.2013.117. [DOI] [PubMed] [Google Scholar]
- 24.Pelaez-Garcia A, Barderas R, Torres S, Hernandez-Varas P, Teixido J, Bonilla F, et al. FGFR4 role in epithelial-mesenchymal transition and its therapeutic value in colorectal cancer. PLoS ONE. 2013;8:e63695. doi: 10.1371/journal.pone.0063695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barderas R, Shochat S, Timmerman P, Hollestelle MJ, Martinez-Torrecuadrada JL, Hoppener JW, et al. Designing antibodies for the inhibition of gastrin activity in tumoral cell lines. Int J Cancer. 2008;122:2351–9. doi: 10.1002/ijc.23395. [DOI] [PubMed] [Google Scholar]
- 26.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. J Vis Exp. 2014;92:e51998. [DOI] [PMC free article] [PubMed]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Exploring data normalization and analysis in large TMT experimental designs [https://pwilmart.github.io/IRS_normalization/understanding_IRS.html].
- 29.Kubens BS, Zanker KS. Differences in the migration capacity of primary human colon carcinoma cells (SW480) and their lymph node metastatic derivatives (SW620) Cancer Lett. 1998;131:55–64. doi: 10.1016/S0304-3835(98)00201-8. [DOI] [PubMed] [Google Scholar]
- 30.Araki K, Shimura T, Suzuki H, Tsutsumi S, Wada W, Yajima T, et al. E/N-cadherin switch mediates cancer progression via TGF-beta-induced epithelial-to-mesenchymal transition in extrahepatic cholangiocarcinoma. Br J Cancer. 2011;105:1885–93. doi: 10.1038/bjc.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griss J, Viteri G, Sidiropoulos K, Nguyen V, Fabregat A, Hermjakob H. ReactomeGSA—efficient Multi-Omics comparative pathway analysis. Mol Cell Proteom. 2020;19:2115–25. doi: 10.1074/mcp.TIR120.002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian X, Yang C, Yang L, Sun Q, Liu N. PTPRF as a novel tumor suppressor through deactivation of ERK1/2 signaling in gastric adenocarcinoma. Onco Targets Ther. 2018;11:7795–803. doi: 10.2147/OTT.S178152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang WK, Liao LD, Li LY, Xie YM, Xu XE, Zhao WJ, et al. Down-regulated desmocollin-2 promotes cell aggressiveness through redistributing adherens junctions and activating beta-catenin signalling in oesophageal squamous cell carcinoma. J Pathol. 2013;231:257–70. doi: 10.1002/path.4236. [DOI] [PubMed] [Google Scholar]
- 35.Bujko M, Kober P, Mikula M, Ligaj M, Ostrowski J, Siedlecki JA. Expression changes of cell-cell adhesion-related genes in colorectal tumors. Oncol Lett. 2015;9:2463–70. doi: 10.3892/ol.2015.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bajpai R, Nagaraju GP. Specificity protein 1: its role in colorectal cancer progression and metastasis. Crit Rev Oncol Hematol. 2017;113:1–7. doi: 10.1016/j.critrevonc.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Li K, Mei Y, Huang X, Li Z, Yang Q, et al. Sp1 suppresses miR-3178 to promote the metastasis invasion cascade via upregulation of TRIOBP. Mol Ther Nucleic Acids. 2018;12:1–11. doi: 10.1016/j.omtn.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, Shao MY, et al. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS ONE. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basak O, van de Born M, Korving J, Beumer J, van der Elst S, van Es JH, et al. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014;33:2057–68. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coopmans EC, Muhammad A, Daly AF, de Herder WW, van Kemenade FJ, Beckers A, et al. The role of AIP variants in pituitary adenomas and concomitant thyroid carcinomas in the Netherlands: a nationwide pathology registry (PALGA) study. Endocrine. 2020;68:640–9. doi: 10.1007/s12020-020-02303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georgitsi M, Karhu A, Winqvist R, Visakorpi T, Waltering K, Vahteristo P, et al. Mutation analysis of aryl hydrocarbon receptor interacting protein (AIP) gene in colorectal, breast, and prostate cancers. Br J Cancer. 2007;96:352–6. doi: 10.1038/sj.bjc.6603573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diaz Del Arco C, Estrada Munoz L, Barderas Manchado R, Pelaez Garcia A, Ortega Medina L, Molina Roldan E, et al. Prognostic role of Aryl hydrocarbon receptor interacting protein (AIP) immunohistochemical expression in patients with resected gastric carcinomas. Pathol Oncol Res. 2020;26:2641–50. doi: 10.1007/s12253-020-00863-7. [DOI] [PubMed] [Google Scholar]
- 43.Fernández-Aceñero MJ, Barderas R, Peláez-García A, Martínez-Useros J, Díez-Valladares L, Ortega-Medina L, et al. Aryl hydrocarbon receptor interacting protein (AIP) significantly influences prognosis of pancreatic carcinoma. Annal Diagnostic Pathol. 2021;53:151742. [DOI] [PubMed]
- 44.Barry S, Carlsen E, Marques P, Stiles CE, Gadaleta E, Berney DM, et al. Tumor microenvironment defines the invasive phenotype of AIP-mutation-positive pituitary tumors. Oncogene. 2019;38:5381–95. doi: 10.1038/s41388-019-0779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasilev V, Daly AF, Trivellin G, Stratakis CA, Zacharieva S, Beckers A. Hereditary endocrine tumours: current state-of-the-art and research opportunities: the roles of AIP and GPR101 in familial isolated pituitary adenomas (FIPA) Endocr Relat Cancer. 2020;27:T77–T86. doi: 10.1530/ERC-20-0015. [DOI] [PubMed] [Google Scholar]
- 46.Leontiou CA, Gueorguiev M, van der Spuy J, Quinton R, Lolli F, Hassan S, et al. The role of the aryl hydrocarbon receptor-interacting protein gene in familial and sporadic pituitary adenomas. J Clin Endocrinol Metab. 2008;93:2390–401. doi: 10.1210/jc.2007-2611. [DOI] [PubMed] [Google Scholar]
- 47.Trivellin G, Korbonits M. AIP and its interacting partners. J Endocrinol. 2011;210:137–55. doi: 10.1530/JOE-11-0054. [DOI] [PubMed] [Google Scholar]
- 48.Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 2019;19:184–97. doi: 10.1038/s41577-019-0125-8. [DOI] [PubMed] [Google Scholar]
- 49.Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol Cell Biol. 1998;18:978–88. doi: 10.1128/MCB.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12:3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010;51:71–119. doi: 10.1016/S0065-2423(10)51004-7. [DOI] [PubMed] [Google Scholar]
- 52.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–19. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legend to Supplementary Figures and Tables
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files. The Mass Spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031119. Other data are available from the corresponding author on reasonable request.