Abstract
Background and purpose
Radiomics offers great potential in improving diagnosis and treatment for patients with glioblastoma multiforme. However, in order to implement radiomics in clinical routine, the features used for prognostic modelling need to be stable. This comprises significant challenge in multi-center studies. The aim of this study was to evaluate the impact of different image normalization methods on MRI features robustness in multi-center study.
Methods
Radiomics stability was checked on magnetic resonance images of eleven patients. The images were acquired in two different hospitals using contrast-enhanced T1 sequences. The images were normalized using one of five investigated approaches including grey-level discretization, histogram matching and z-score. Then, radiomic features were extracted and features stability was evaluated using intra-class correlation coefficients. In the second part of the study, improvement in the prognostic performance of features was tested on 60 patients derived from publicly available dataset.
Results
Depending on the normalization scheme, the percentage of stable features varied from 3.4% to 8%. The histogram matching based on the tumor region showed the highest amount of the stable features (113/1404); while normalization using fixed bin size resulted in 48 stable features. The histogram matching also led to better prognostic value (median c-index increase of 0.065) comparing to non-normalized images.
Conclusions
MRI normalization plays an important role in radiomics. Appropriate normalization helps to select robust features, which can be used for prognostic modelling in multicenter studies. In our study, histogram matching based on tumor region improved both stability of radiomic features and their prognostic value.
Keywords: Image normalization, Glioblastoma multiforme, Radiomics, Features stability, Prognostic modelling
1. Introduction
Glioblastoma multiforme (GBM) is the most common primary brain tumor in adults with poor median overall and long-term survival rates below 10%[1]. Recently introduced disease phenotyping using isocitrate dehydrogenase (IDH) and methylation status of O6-methylguanine-methyltransferase (MGMT) as prognostic biomarkers has improved disease classification and selection of the optimal treatment strategy [2], [3]. However, these biomarkers alone have limited predictive power and better patient stratification is needed. Radiomics refers to the extraction of imaging characteristics using automated data-mining algorithms. Numerous studies have shown that these characteristics known as radiomic features have the potential to decode tumor phenotypes and predict treatment outcome [4]. Magnetic resonance imaging (MRI)-based radiomics have already demonstrated their promising abilities to predict disease progression [5], [6], methylation status [7], tumor grade [8] and overall survival [9], [10] for patients with central nervous system malignancies. Magnetic resonance (MR) images are routinely acquired for patients with GBM at baseline and for follow-up. Combined with radiomic analysis these images may enable further tumor profiling. On the other hand, radiomic features used for prognostic modelling need to be stable in terms of any variability excluding intratumoral heterogeneity. This comprises significant challenges in MRI-based radiomics, particularly in multi-centric data collections.
Most of the existing studies investigated MRI-based radiomic features reproducibility on datasets collected from a single institution [11], [12], [13]; however, robustness of radiomic features in a multi-center framework is a prerequisite for successful translation of radiomic biomarkers into the clinical workflow [14].
A particular challenge in the multi-centric setting is the lack of harmonization between MR imaging protocols, which results in large variability in image intensities among inter-patient and intra-patient acquisitions. Unlike computed tomography (CT) and positron emission tomography (PET) imaging, where intensities are calibrated to Hounsfield units (HU) and standard uptake values (SUV), respectively, MR intensities have arbitrary values. This fact can significantly affect the absolute values of radiomic features, compromising their robustness. In order to mitigate the aforementioned issues, intensity normalization of the MR images should be performed prior to any quantitative analysis.
The aim of this study was to evaluate the intra-patient stability of radiomic features for patients with GBM derived from MR images sequentially acquired in multi-centric study and to investigate the impact of different types of intensity normalization methods on radiomics reproducibility. The hypothesis was that normalization of MR intensities leads to reduction of discrepancy in the dataset and thus improves prognostic value of MR-based biomarkers. To that end we analyzed the prognostic value of features extracted from contrast-enhanced T1-weighted images before and after intensity normalization.
2. Materials and methods
2.1. Robustness study
2.1.1. Patient cohort
To perform robustness analysis we identified patients with GBM diagnosed between September 2012 and December 2019. Using local picture archiving and communication system (PACS), we selected patients, who underwent preoperative brain MRI scans in two different hospitals. The inclusion criteria were the following: a) pathologically confirmed GBM, b) availability of two preoperative contrast-enhanced T1-weighted (T1c) MRI examinations, c) without significant tumor growth between two scans (<30% change in tumor volume) d) without surgical treatment/biopsy between two examinations. The study was approved by the local ethics committee (BASEC Nr.2020-00859).
2.1.2. Image acquisition
For all patients the T1c images were sequentially acquired in our healthcare institution (internal) and elsewhere (external). The MRI data was acquired using either 1.5 or 3 Tesla scanners, following standard local protocols of each healthcare institution, the detailed technical information is shown in Table 1.
Table 1.
Technical characteristics and imaging protocol details for internal and external patient cohorts.
| Robustness study |
Prognostic study | |||
|---|---|---|---|---|
| Characteristics | Internal cohort | External cohort | Overall survival cohort | |
| MRI scanner | Siemens | |||
| Skyra | N = 10 (91%) | N = 1 (9%) | ||
| Aera | N = 1 (9%) | |||
| Avanto | N = 1 (9%) | N = 3 (5%) | ||
| Verio | N = 4 (7%) | |||
| TrioTim | N = 1 (1%) | |||
| Symphony | N = 2 (3%) | |||
| GE Medical Systems | ||||
| Signa | N = 1 (9%) | |||
| Genesis Sigma | N = 1 (9%) | N = 15 (25%) | ||
| Signa Excite | N = 8 (13%) | |||
| Philips | ||||
| Ingenia | N = 3 (27%) | |||
| Achieva | N = 4 (36%) | N = 5 (8%) | ||
| Intera | N = 2 (3%) | |||
| Unknown | N = 20 (33%) | |||
| Magnetic field strength | 1.16 T | |||
| 1.5 T | N = 4 (36%) | N = 33 (55%) | ||
| 3 T | N = 11 (100%) | N = 7 (64%) | N = 22 (37%) | |
| unknown | N = 5 (8%) | |||
| Slice thickness | Median (range) | 0.88 (0.7–0.9) mm | 1.17 (0.6–3.0) mm | 5.0 (1.0 – 12.5) mm |
| Repetition time (TR) | Median (range), s | 1622 (8.9–1900) s | 441.9 (5.5–2300) s | 586.3 (34.0 – 3285.6) s |
| Echo time (TR) | Median (range) | 2.65 (2.54–3.42) s | 3.78 (2.31–9) s | 8.00 (2.48 – 17.00) s |
| Flip angle | Median (range) | 9.54 (9–15) ° | 15.6 (8–90) ° |
|
2.1.3. Segmentation
For each patient, the volume of interest (VOI) represented by the enhanced tumour region was manually delineated by a radiologist with 5 years of experience in medical imaging using MIM VISTA (Version 6.7.9., MIM software Inc., Cleveland, USA). All segmented images were validated by an experienced radiation oncologist with more than 5 years of experience in oncologic imaging.
2.1.4. Image postprocessing
All images were resampled using trilinear interpolation to 3 mm voxel size, which represented the largest voxel size (3 × 3 × 3 mm) among both internal and external cohorts. None of the voxels in the acquired datasets exceeded 3 mm in any dimension. Image normalization was performed prior to the radiomics analysis, using five different methods described below (Table 2).
Table 2.
Short description of the image normalization approaches considered in this study.
| Normalization | Bins | ROIs for intensity interpolation |
|---|---|---|
| Fixed bin number (FBN) | Number of bins equal to 32 | no |
| Fixed bin size (FBS1) | Bin size 30 | vitreous body of an eye, white matter |
| Fixed bin size (FBS2) | Bin size 30 | vitreous body of an eye, contrast-enhanced confluence of sinuses |
| Histogram matching (HM) | Fixed bin size of 20 | delineated VOI |
| Fixed bin size of 20,000 | brain mask | |
| Z-score (ZS) | Fixed bin size of 0.15 | delineated VOI |
| Fixed bin size of 0.2 | brain mask |
In the first approach, the entire range of intensities was divided into a series of 32 bins (fixed bin number, FBN) usually defined in the literature as a relative grey-level discretization [15], [16]. In the second approach, absolute discretization with fixed bin size of 30 and linear intensity interpolation using two small ROIs positioned in the white matter of the contralateral brain and vitreous body of one eye were applied (FBS1). In the third approach, we also used linear intensity interpolation and a fixed bin size of 30, but in this case the vitreous body of one eye and the contrast-enhanced region of torcular Herophili (confluence of sinuses) were selected as reference regions (FBS2). In both FBS1 and FBS2, the intensities outside of the selected range were extrapolated.
Additionally to the methods based on the gray-level discretization, we used two less common approaches: histogram matching and z-score, which were recently suggested for radiomics analysis based on MRI collected from several institutions [17], [18]. In histogram matching (HM) approach we used one intensity histogram as a reference and linearly mapped the intensities of other images to it. In the z-score (ZS) method, the mean intensity of the entire image or a region of interest was subtracted from each voxel value followed by division by the corresponding standard deviation. In both HM and ZS methods we considered two possible regions of interest a) delineated tumor (VOI) and b) whole brain. In latter, brain segmentation was performed using automated approach described by Isensee et al. [19].
In the approaches with fixed bin size (FBS1, FBS2, HM and ZS), the size of the bins was adapted so that on median 32 bins were analyzed. This has been done to match the noise in the FBN normalization.
2.1.5. Feature extraction and statistical analysis
Radiomic features were extracted from each VOI using the in-house developed software Z-Rad. This software performs three-dimensional image analysis allowing the extraction of four feature types: shape (n = 18), intensity (n = 17), texture (n = 137) and wavelet (n = 1232). In total, 1404 features were extracted. The mathematical description of the features was previously published [20], [21]. To compare the radiomic features from two different images of a single subject, the one-way random intra-class correlation coefficients (ICC) were calculated using R-package version 3.3.2. Features with ICC ≥ 0.90 were considered to be robust, ICC < 0.90 non-robust. With respect to the literature, this level of ICC allows to keep the error below 0.05 even for a small patient cohort [22]. The ICC calculations were performed separately for each of the investigated normalization techniques. Features with ICC ≥ 0.9 were examined for their correlations with the tumor volume using Spearman’s rho statistic to estimate a rank-based measure of association. Additionally the features robustness was tested using Lin’s concordance correlation coefficients (CCCs) as previously described [23], [24]. Stable features were defined as CCC ≥ 0.90.
2.2. Prognostic study
2.2.1. Dataset for evaluation
To investigate improvement in the prognostic performance of features extracted from MR images after intensity normalization a publicly available dataset of patients with GBM was analyzed [25]. This multicenter collection of data provided large heterogeneity in imaging parameters (Table 1). For 110 cases tumor segmentation masks were available based on the results of the International multimodal BRAin Tumor Segmentation challenge (BRATS 2015) [26], [27].
For this analysis, we selected region of interest comprising the contrast-enhanced part of the tumors similarly to the robustness study.
For 60 patients both tumor segmentation and follow-up data were available. Patients were treated with surgery (resection n = 53, excisional biopsy n = 2, data not available n = 5) combined with radiotherapy (yes n = 56, no n = 3, data not available n = 1)) and/or chemotherapy (yes n = 50, no n = 5, data not available n = 5). The analyzed endpoint was overall survival at 18 months (35 events). The median overall survival in the cohort was 13 months and median follow-up time was 42 months.
2.2.2. Value of the robust features
Based on the robustness analysis the normalization method resulting in the largest number of stable features was selected for further prognostic performance investigation. For this part of the study, we decided to exclude shape features, since different normalization methods have very little influence on absolute value of shape features. The prognostic value of the stable intensity, texture and wavelet features was tested in the univariable logistic regression a) when simple grey-level discretization was used (fixed bin size = 32, no ROI), b) when the normalization technique providing the maximum robust features was applied. The number of prognostic features (p-value < 0.05) between those two scenarios was compared. Additionally, the increase in area under receiver operating characteristic (AUC) after application of the image normalization was reported. Statistical analysis was performed using R (Version 3.3.2).
3. Results
3.1. Stable features
In total, MR images from eleven patients (7-male, 4-female, mean age 66.1 ± 14.8 years old) were identified and included in the analysis (Table 3).
Table 3.
The patient characteristics and the time gap between two MR acquisitions for each patient.
| Patient | Gender | MGMT mutation status | IDH mutation status | Age, years | Time between scans, days |
|---|---|---|---|---|---|
| 1 | male | positive | not confirmed | 72 | 5 |
| 2 | female | not confirmed | not confirmed | 60 | 2 |
| 3 | male | not confirmed | positive | 52 | 8 |
| 4 | female | negative | negative | 70 | 6 |
| 5 | male | negative | negative | 76 | 36 |
| 6 | male | negative | negative | 72 | 9 |
| 7 | male | not confirmed | negative | 69 | 4 |
| 8 | male | positive | positive | 30 | 1 |
| 9 | female | negative | not confirmed | 75 | 17 |
| 10 | female | negative | negative | 66 | 9 |
| 11 | male | negative | negative | 86 | 11 |
| Mean: | 66 | 9 | |||
| Median: | 70 | 8 | |||
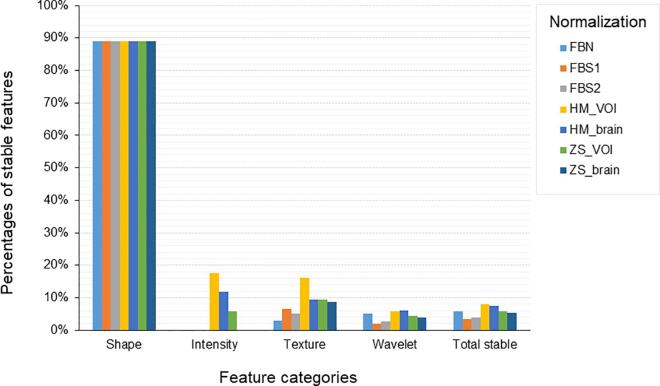
The number of stable radiomic features (ICC ≥ 0.9) in each category (all, shape, intensity, texture and wavelets) for the investigated normalization methods are shown in Table 4. The proportion of stable features by category for all investigated normalizations is shown in Fig. 1. The overall stability was low (at most 8%) and stability of certain features was dependent on normalization method. The ICC values for features stable in at least one normalization are shown in Supplementary Fig. S1. Some features were stable regardless of the normalization method (mostly shape). The HM_VOI showed superior stability for texture features, but there was also a subgroup of texture feature solely stable for the HM_brain normalization. HM_brain normalization also resulted in a higher number of stable features in wavelet features LLH/LHL/HLL similarly to ZS_brain. These results suggested that combining different normalization methods could be beneficial to retain larger number of stable features. For instance, combination of HM_VOI (the one with superior features stability) with other normalizations has shown that the maximum number of stable features (169 / 1404) can be achieved by combination of HM_VOI and HM_brain normalization. The number of stable features that can be achieved from pairwise combination of investigated normalization methods are shown in Supplementary Fig. S2. The complete list of ICCs for all features is provided in supplementary table (Supplementary Table S1-S4). Lin’s concordance coefficients have shown similar trend in features stability (Supplementary Table S5).
Table 4.
Number of robust features using different normalization approaches.
| Normalization | Stable features, ICC > 0.9 |
||||
|---|---|---|---|---|---|
| Shape | Intensity | Texture | Wavelets | All | |
| FBN | 16/18 | 0/17 | 4/137 | 63/1232 | 83/1404 (5.9%) |
| FBS1 | 16/18 | 0/17 | 9/137 | 23/1232 | 48/1404 (3.4%) |
| FBS2 | 16/18 | 0/17 | 7/137 | 32/1232 | 55/1404 (3.9%) |
| HM_VOI | 16/18 | 3/17 | 22/137 | 72/1232 | 113/1404 (8.0%) |
| HM_brain | 16/18 | 2/17 | 13/137 | 74/1232 | 104/1404 (7.4%) |
| ZS_VOI | 16/18 | 1/17 | 13/137 | 53/1232 | 83/1404 (5.9%) |
| ZS_brain | 16/18 | 0/17 | 12/137 | 49/1232 | 77/1404 (5.4%) |
| Regardless of the normalization | 16/18 | 0/17 | 2/137 | 8/1232 | 26/1404 (1.9%) |
Fig. 1.
The percentages of stable features by categories for FBN, FBS1, FBS2, HM_VOI, HM_brain, ZS_VOI and ZS_brain normalizations.
3.1.1. Gray-level discretization and linear interpolation
For FBN, FBS1 and FBS2 normalizations, the ICC analysis showed that only 5.9% (83/1404), 3.4% (48/1404) and 3.9% (55/1404) of the total number of radiomic features were stable, respectively. Among all feature types extracted from the MR images performed in two different healthcare centers shape features have shown the highest reproducibility (89%), while only 7% of the texture features and 5% of the wavelet features satisfied this criterion. The intensity features did not show any significant reproducibility while using these normalizations.
3.1.2. Z-score
Z-score normalization with brain and VOI selected as a region for intensity normalization showed similar results to FBN, with 5.9% and 5.4% robust features for ZS_VOI and ZS_brain, respectively. However, unlike FBN both ZS_VOI and ZS_brain resulted in a higher proportion of stable texture features and a lower proportion of wavelet features.
3.1.3. Histogram matching
Histogram matching image normalization method showed higher stability of the features with total number of 113/1404 (8.0%) and 104/1404 (7.4%) stable features for HM_VOI and HM_brain, respectively. This approach also identified few stable features in intensity domain.
3.2. Correlations between features and tumor volume
Certain features are known to be volume correlated and thus their stability might be independent of the MR intensities. Across all normalization types, the mean absolute of Spearman’s |r| between stable features and volume was 0.78 ± 0.06. The number of features which correlated highly (|r| > 0.8), strongly (0.6 ≤ |r| ≤ 0.8) and weakly (|r| < 0.6) with the tumor volume for each normalization type is shown in Table 5.
Table 5.
Correlations between features and tumor volume.
| Normalization | Highly correlated features, |r| >0.8 | Strongly correlated features, |r| >0.6 | Weakly correlated features, |r| <0.6 |
|---|---|---|---|
| FBN | 8/83 | 67/83 | 9/83 |
| FBS1 | 37/48 | 8/48 | 3/48 |
| FBS2 | 40/55 | 7/55 | 8/55 |
| HM_VOI | 57/113 | 19/113 | 37/113 |
| HM_brain | 54/104 | 11/104 | 39/104 |
| ZS_VOI | 59/83 | 11/83 | 13/83 |
| ZS_brain | 53/77 | 12/77 | 12/77 |
3.3. Prognostic value of the robust features
The radiomic features for 60 patients from publicly available dataset were extracted using basic discretization and histogram matching based on the tumor region. The stable intensity, texture and wavelet features were analyzed for their value to predict overall survival at 18 months in univariable logistic regression. Without normalization only 3 features were found to be prognostic, whereas in case of histogram matching (HM_VOI) 7 out of 97 analyzed features were prognostic (p-value < 0.05). Features, which were identified as prognostic after histogram normalization had in general higher AUC (AUC range 0.65 – 0.70) in comparison to features significant in the non-processed images (AUC range 0.57 – 0.73). For the 3 features already significant prior tonormalization, histogram matching resulted in additional increase in AUC (median = 0.065). The increase in AUC was not significant for this relatively small patient cohort.
4. Discussion
This study aimed to identify stable MRI-based radiomic features in multi-center study.. The results showed that only 8% (113 out of 1404 investigated features) remain stable. The analysis also demonstrated that image normalization has significant impact on radiomics robustness. Depending on the normalization the number of robust features was between 48 and 113 (out of 1404). A similar study performed recently on diffusion-weighted magnetic resonance imaging (DWI) for patients with lung, liver and ovarian cancer has shown about that 122 features out of 1322 remain robust [18]. Interestingly, Shiri at al showed that if the acquisition parameters remain the same in test–retest MR data separated with two days interval as much as 53.8% of features can remain stable (out of 107 investigated features) [12].
Among four investigated feature types the shape features have demonstrated the highest stability with only 2 non-robust features (fractal dimension and center of mass shift). For both of these features it can be explained by small variation in contours performed manually and the distribution of contrast which might be different in two consequent examinations. The study from Peerlings et al. [18] has demonstrated similar rate of shape features in DWI scans. The test–retest study by Shiri et al. has demonstrated the superior robustness of the shape features, however that study did not include the above-mentioned features in the analysis.
Texture features, which describe intra-tumour heterogeneity [28], have shown lower percentage of stable features. This can be explained by the fact that texture features are sensitive to acquisition parameters which influence spatial resolution, unless the spatial resolution is sufficiently high [29]. In our study, the median slice thickness was 0.88 mm and 1.17 mm for internal and external cohorts, respectively (Table 1). However, since one patient in the external cohort had thicker slices of 3 mm, we resampled all datasets to the common resolution of 3 mm. In prospective studies this problem can be solved by harmonization of the imaging protocols [5], [6], however harmonizing between machines of different generations often comes with degrading the image quality achieved by the most recent one[30]. In retrospective studies, one can reduce the variability in voxel size by applying low-pass filtering in the frequency domain [31], however this also reduces the quality of images acquired by the most recent device [32]. Alternatively, one can select the images to match spatial resolution, however, this will strongly decrease the amount of investigated subjects.
Our findings indicated that histogram matching and z-score normalizations improved the stability of the texture features compared to gray-level discretization. Similar improvement in texture features reproducibility has been previously shown by several groups investigating other cancers [33] or collected in a single institution [33], [34].
Intensity features showed the lowest stability for all investigated normalizations. Despite our effort the highest number of stable intensity features was equal to 3, for HM_VOI normalization. Other researchers have also reported, that MR-based intensity features generally show lower stability [18].
Potentially the amount of stable radiomic features can be increased by combining histogram matching based on VOI and brain (see Supplementary Fig. S12).
Despite the low number of stable features, our results showed that relevant numbers of informative features were preserved. In the overall survival prediction task, we were also able to show that our selected normalization technique (histogram matching based on the tumor region) reduced noise in dataset and improved the prognostic value.
While several researchers have already investigated the prognostic power of radiomics in multi-center studies, the robustness of the MRI-based features were mainly investigated in test–retest studies, when MR images were acquired under similar conditions. In order to generalize prognostic models, the features used for modelling need to be stable irrespective of image acquisition and reconstruction parameters. To the best of our knowledge, our study is the first one aiming to investigate intra-patient robustness of the radiomic features for GBM patients, using MRI collected in different institutions.
Our study has several limitations. First of all, only few patients were included into this study due to strict inclusion criteria. Similar test–retest studies rarely exceed our population number. Second, we have only investigated contrast-enhanced T1-weighted sequence, since it was available for all patients in our cohort and this sequence is often reported as the most informative one for GBM patients [35], [36]. Additionally, in this study the tumor segmentation was controlled by two independent observers, however such study could potentially benefit from automatic segmentation. Last, but not least, we only considered the images collected retrospectively in real clinical routine, however it would be interesting to compare results of our work with similar investigations performed on MR images acquired in standard test–retest environment.
To conclude, our study assessed the intra-patient stability of radiomic features based on sequential MR images of patients with GBM collected from different healthcare centers. This methodology allowed the selection of stable radiomic features which can be further investigated as reliable biomarker for prognostic modelling, phenotyping and treatment strategy selection in realistic clinical conditions. Histogram matching based on tumor region was identified as the most reliable normalization method, which improved both stability of radiomic features and their prognostic value.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2022.05.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Weller M., van den Bent M., Hopkins K., Tonn J.C., Stupp R., Falini A., et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014;15:e395–e403. doi: 10.1016/S1470-2045(14)70011-7. [DOI] [PubMed] [Google Scholar]
- 2.Li H., Li J., Cheng G., Zhang J., Li X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clin Neurol Neurosurg. 2016;151:31–36. doi: 10.1016/j.clineuro.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Suh C.H., Kim H.S., Jung S.C., Choi C.G., Kim S.J. Clinically relevant imaging features for MGMT promoter methylation in multiple glioblastoma studies: a systematic review and meta-analysis. AJNR Am J Neuroradiol. 2018;39:1439–1445. doi: 10.3174/ajnr.A5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogowicz M., Vuong D., Huellner M.W., Pavic M., Andratschke N., Gabrys H.S., et al. CT radiomics and PET radiomics: ready for clinical implementation? Q J Nucl Med Mol Imaging. 2019;63:355–370. doi: 10.23736/S1824-4785.19.03192-3. [DOI] [PubMed] [Google Scholar]
- 5.Le Fevre C., Lhermitte B., Ahle G., Chambrelant I., Cebula H., Antoni D., et al. Pseudoprogression versus true progression in glioblastoma patients: A multiapproach literature review: Part 1 - Molecular, morphological and clinical features. Crit Rev Oncol Hematol. 2021;157 doi: 10.1016/j.critrevonc.2020.103188. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y.Z., Yan L.F., Han Y., Nan H.Y., Xiao G., Tian Q., et al. Differentiation of pseudoprogression from true progressionin glioblastoma patients after standard treatment: a machine learning strategy combinedwith radiomics features from T1-weighted contrast-enhanced imaging. BMC Med Imaging. 2021;21:17. doi: 10.1186/s12880-020-00545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vils A., Bogowicz M., Tanadini-Lang S., Vuong D., Saltybaeva N., Kraft J., et al. Radiomic analysis to predict outcome in recurrent glioblastoma based on multi-center MR imaging from the prospective DIRECTOR trial. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.636672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zacharaki E.I., Wang S., Chawla S., Soo Yoo D., Wolf R., Melhem E.R., et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabowski M.M., Recinos P.F., Nowacki A.S., Schroeder J.L., Angelov L., Barnett G.H., et al. Residual tumor volume versus extent of resection: predictors of survival after surgery for glioblastoma. J Neurosurg. 2014;121:1115–1123. doi: 10.3171/2014.7.JNS132449. [DOI] [PubMed] [Google Scholar]
- 10.Chaddad A., Daniel P., Desrosiers C., Toews M., Abdulkarim B. Novel radiomic features based on joint intensity matrices for predicting glioblastoma patient survival time. IEEE J Biomed Health Inform. 2019;23:795–804. doi: 10.1109/JBHI.2018.2825027. [DOI] [PubMed] [Google Scholar]
- 11.Baessler B., Weiss K., Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol. 2019;54:221–228. doi: 10.1097/RLI.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 12.Shiri I, Hajianfar G, Sohrabi A, Abdollahi H, S PS, Geramifar P, et al. Repeatability of radiomic features in magnetic resonance imaging of glioblastoma: Test-retest and image registration analyses. Med Phys. 2020;47:4265-80. [DOI] [PubMed]
- 13.Suter Y., Knecht U., Alao M., Valenzuela W., Hewer E., Schucht P., et al. Radiomics for glioblastoma survival analysis in pre-operative MRI: exploring feature robustness, class boundaries, and machine learning techniques. Cancer Imaging. 2020;20:55. doi: 10.1186/s40644-020-00329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier L., Costaridou L., Bidaut L., Michoux N., Lecouvet F.E., de Geus-Oei L.F., et al. Incorporating radiomics into clinical trials: expert consensus endorsed by the European Society of Radiology on considerations for data-driven compared to biologically driven quantitative biomarkers. Eur Radiol. 2021;31:6001–6012. doi: 10.1007/s00330-020-07598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duron L., Balvay D., Vande Perre S., Bouchouicha A., Savatovsky J., Sadik J.C., et al. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS ONE. 2019;14:e0213459. doi: 10.1371/journal.pone.0213459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crombe A., Kind M., Fadli D., Le Loarer F., Italiano A., Buy X., et al. Intensity harmonization techniques influence radiomics features and radiomics-based predictions in sarcoma patients. Sci Rep. 2020;10:15496. doi: 10.1038/s41598-020-72535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaddad A., Kucharczyk M.J., Daniel P., Sabri S., Jean-Claude B.J., Niazi T., et al. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front Oncol. 2019;9:374. doi: 10.3389/fonc.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peerlings J., Woodruff H.C., Winfield J.M., Ibrahim A., Van Beers B.E., Heerschap A., et al. Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci Rep. 2019;9:4800. doi: 10.1038/s41598-019-41344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isensee F., Schell M., Pflueger I., Brugnara G., Bonekamp D., Neuberger U., et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum Brain Mapp. 2019;40:4952–4964. doi: 10.1002/hbm.24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwanenburg A LS, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv preprint arXiv:161207003. 2013.
- 21.USZ Medical Physics (medical-physics-usz.github.io) [Available from: https://medical-physics-usz.github.io/.
- 22.Walter S.D., Eliasziw M., Donner A. Sample size and optimal designs for reliability studies. Stat Med. 1998;17:101–110. doi: 10.1002/(sici)1097-0258(19980115)17:1<101::aid-sim727>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 23.Lin L.I. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 24.Balagurunathan Y., Kumar V., Gu Y., Kim J., Wang H., Liu Y., et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27:805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedano N., Flanders A.E., Scarpace L., Mikkelsen T., Eschbacher J.M., Hermes B., Ostrom Q. Radiology data from the cancer genome atlas low grade glioma [TCGA-LGG] collection. The Cancer Imaging Archive. 2016 [Google Scholar]
- 26.Bakas S., Akbari H., Sotiras A., Bilello M., Rozycki M., Kirby J.S., et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4 doi: 10.1038/sdata.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C. Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-LGG collection. The Cancer Imaging Archive. 2017. [DOI] [PMC free article] [PubMed]
- 28.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G., Granton P., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayerhoefer M.E., Szomolanyi P., Jirak D., Materka A., Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: An application-oriented study. Med Phys. 2009;36:1236–1243. doi: 10.1118/1.3081408. [DOI] [PubMed] [Google Scholar]
- 30.RBE for deterministic effects. A report of a Task Group of Committee I of the International Commission on Radiological protection. Ann ICRP. 1989;20:1-57. [PubMed]
- 31.Mackin D., Fave X., Zhang L., Yang J., Jones A.K., Ng C.S., et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS ONE. 2017;12:e0178524. doi: 10.1371/journal.pone.0178524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlhac F., Lecler A., Savatovski J., Goya-Outi J., Nioche C., Charbonneau F., et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur Radiol. 2021;31:2272–2280. doi: 10.1007/s00330-020-07284-9. [DOI] [PubMed] [Google Scholar]
- 33.Chatterjee A., Vallières M., Dohan A., Levesque I., Ueno Y., Saif S., Reinhold C., Seuntjens J. Creating robust predictive radiomic models for data from independent institutions using normalization. IEEE Trans Radiat Plasma Med Sci. 2019 [Google Scholar]
- 34.Carre A., Klausner G., Edjlali M., Lerousseau M., Briend-Diop J., Sun R., et al. Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Sci Rep. 2020;10:12340. doi: 10.1038/s41598-020-69298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingrisch M., Schneider M.J., Norenberg D., Negrao de Figueiredo G., Maier-Hein K., Suchorska B., et al. Radiomic analysis reveals prognostic information in T1-weighted baseline magnetic resonance imaging in patients with glioblastoma. Invest Radiol. 2017;52:360–366. doi: 10.1097/RLI.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 36.Nakamoto T., Takahashi W., Haga A., Takahashi S., Kiryu S., Nawa K., et al. Prediction of malignant glioma grades using contrast-enhanced T1-weighted and T2-weighted magnetic resonance images based on a radiomic analysis. Sci Rep. 2019;9:19411. doi: 10.1038/s41598-019-55922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.