Abstract
Mitogenomes have been widely used for phylogenetic reconstruction of various Dipteran groups, but specifically for chironomid, they have not been carried out to resolve the relationships. Diamesinae (Diptera: Chironomidae) are important bioindicators for freshwater ecosystem monitoring, but its evolutionary history remains uncertain for lack of information. Here, coupled with one previously published and 30 new mitogenomes of Diamesinae, we carried out comparative mitogenomic analysis and phylogenetic analysis. Mitogenomes of Diamesinae were conserved in structure, and all genes arranged in the same order as the ancestral insect mitogenome. All protein‐coding genes in Diamesinae were under stronger purifying selection than those of other nonbiting midge species, which may exhibit signs of adaptation to life at cold living conditions. Phylogenetic analyses strongly supported the monophyly of Diamesinae, with Boreheptagyiini deeply nested within Diamesini. In addition, phylogenetic relationship of selected six genera was resolved, except Sympotthastia remained unstable. Our study revealed that the mitogenomes of Diamesinae are highly conserved, and they are practically useful for phylogenetic inference.
Keywords: Diamesinae, mitogenome, phylogeny
Here, coupled with one previously published and 30 new mitogenomes of Diamesinae, we carried out comparative mitogenomic analysis and phylogenetic analysis. Mitogenomes of Diamesinae were conserved in structure, and all genes arranged in the same order as the ancestral insect mitogenome. Phylogenetic analyses strongly supported the monophyly of Diamesinae, with Boreheptagyiini deeply nested within Diamesini.

1. INTRODUCTION
Dipteran family Chironomidae have the most abundant species richness among freshwater macroinvertebrates, including more than 6300 species worldwide, even in Antarctica (Kelley et al., 2014; Kim et al., 2016). Since their great species diversity and ability to inhabit different types of water body, chironomid larvae are key bioindicators for freshwater ecosystem monitoring. Several phylogenetic studies have been conducted based on morphological characters or combining genetic markers to reconstruct the evolutionary history of Chironomidae (Brundin, 1966; Cranston et al., 2012; Cranston & Krosch, 2015; Ekrem, 2003; Krosch & Cranston, 2013; Lin et al., 2018; Qi et al., 2019; Sæther, 1977, 2000; Serra‐Tosio, 1973; Silva et al., 2015), but few has attempted to use mitogenomes. Diamesinae (Figure 1) is a relatively small subfamily within Chironomidae, containing over 100 species of six tribes: Boreheptagyiini, Diamesini, Harrisoniini, Heptagyiini, Lobodiamesini, and Protanypini (Ashe & O'Connor, 2009; Brundin, 1966; Sæther, 2000). At present, Boreheptagyiini includes three genera (Boreoheptagyia Brundin, Palatovia Makarchenko & Semenchenko, and Shilovia Makarchenko) distributed in Holarctic and Oriental regions (Makarchenko et al., 2017). Diamesini contains 11 genera: Arctodiamesa Makarchenko, Diamesa Meigen, Kaluginia Makarchenko, Lappodiamesa Serra‐Tosio, Pagastia Oliver, Potthastia Kieffer, Pseudodiamesa Goetghebuer, Pseudokiefferiella Zavrel, Sasayusurika Makarchenko, Sympotthastia Pagast, and Syndiamesa Kieffer (Ashe & O'Connor, 2009) distributed in Afrotropical, Holarctic, and Oriental regions. Phylogenetic relationships within Diamesinae are still controversial despite more than 50 years of research. Traditionally, the phylogenetic relationships of Diamesinae were inferred by morphological characters (Brundin, 1966; Sæther, 1977, 2000). Until last decade, the phylogenetic relationship of very limited sets of Diamesinae subgroups has been explored based on a few molecular loci (Cranston et al., 2012; Lencioni et al., 2021; Montagna et al., 2016). However, Boreheptagyiini and Protanypini were missing, and Diamesini taxa were undersampled in their study. Therefore, the phylogenetic relationship within Diamesini and Boreheptagyiini was recovered by morphological characters is misleading.
FIGURE 1.

An adult male of Diamesa loeffleri Reiss, 1968 on the ice in Qinghai, China. Photo: Qing‐Bo Huo
In general, mitogenomes of most insects is a double‐strand circular DNA molecule ranging from 14 kb to 20 kb in size, encoding 37 genes (13 protein‐coding genes, two ribosomal RNA genes, and 22 transfer RNA genes) and a control region (Boore, 1999; Cameron, 2014; Wolstenholme, 1992). Since its small genome size, maternal inheritance, low sequence recombination, and fast evolutionary rates (Brown et al., 1979; Curole & Kocher, 1999), the mitogenome is considered as powerful marker for phylogenetic and evolutionary analysis in many insect groups (Condamine et al., 2018; Crampton‐Platt et al., 2015; Jacobsen et al., 2012; Tang, Zhu, et al., 2019; Yan et al., 2019). Benefiting from the advances of high‐throughput sequencing technology, an increasing number of complete mitogenomes have been sequenced among the Diptera (Kang et al., 2016; Li et al., 2020; Miao et al., 2020; Ramakodi et al., 2015; Tang, Yan, et al., 2019; Wang et al., 2021; Yan et al., 2021; Zhang et al., 2022), and have been widely used for mitochondrial structure comparison and phylogenetic analysis at different taxonomic levels (Chen et al., 2018; de Oliveira Aragão et al., 2019; Yan et al., 2019; Zhang et al., 2016; Zhang, Kang, et al., 2019). Prior to this study, rare mitogenomes of Chironomidae were available (Beckenbach, 2012; Deviatiiarov et al., 2017; Fang et al., 2022; Jiang et al., 2022; Kim et al., 2016; Kong et al., 2021; Lei et al., 2021; Park et al., 2020; Zhang, Xu, et al., 2019; Zheng et al., 2022; Zheng et al., 2021), limiting our understanding of their mitochondrial structure and phylogenetic pattern. Besides, it is still unknown whether mitogenomes can effectively resolve phylogenetic relationships at different levels within Chironomidae. To date, only one mitogenome of Diamesinae was available, representing Diamesini (Zheng et al., 2021).
In this study, we provide 30 newly sequenced (nearly) complete mitogenomes from 30 species representing Boreheptagyiini (four species of one genus) and Diamesini (26 species of five genera) using next‐generation sequencing. We analyzed the genomic structure, base composition, substitution, and evolutionary rates among Diamesinae, expanding our knowledge of its diversity of mitogenomes. Coupled with published data, we carried out phylogenetic analysis of Boreheptagyiini and Diamesini based on 31 mitogenomes.
2. MATERIALS AND METHODS
2.1. Taxon sampling and dna extraction
Field collection of 30 species were conducted in China during 2014–2020, using classical insect collection techniques such light traps, sweep traps, Malaise traps, and D‐nets. Specimens were preserved in ethanol (85% for adults, 95% for immature), and stored at dark at −20°C before morphological and molecular analyses. The total genomic DNA was extracted from thorax of adult and middle larval bodies using a Qiagen DNA Blood and Tissue Kit (Qiagen) following the manufacturer's protocol. After DNA extraction, the cleared exoskeleton of thorax was mounted in Euparal on microscopy slides together with the corresponding wings, legs, and antennae following the procedures outlined by Sæther (1969). The DNA and vouchers of the species are deposited at the college of Life Sciences, Nankai University, Tianjin, China. Specimens were identified morphologically using relevant taxonomic revisions and species descriptions (Lin, Chang, et al., 2021; Lin, Yu, et al., 2021; Makarchenko et al., 2008, 2021; Makarchenko & Wang, 2017; Moubayed‐Breil & Orsini, 2016; Oliver, 1983, 1989; Reiss, 1968; Sun et al., 2019), belonging to two tribes of Diamesinae.
Thirty mitogenomes were newly sequenced in this study, representing four species of Boreheptagyiini (four Boreoheptagyia species) and 26 species of Diamesini (15 Diamesa species, four Pagastia species, four Potthastia species, two Pseudodiamesa species and one Sympotthastia species). Since mitogenomes of another four tribes were not available for current molecular study, we could not reconstruct the phylogeny of the whole subfamily Diamesinae. Therefore, by integrating one public Potthastia species (GenBank accession: MW373523), a total of 31 species of Boreheptagyiini and Diamesini were selected as in‐groups. In addition, we selected one Prodiamesinae species (Prodiamesa olivacea [Meigen, 1818], GenBank accession: MW373525) and one Orthocladiinae species (Propsilocerus akamusi [Tokunaga, 1938], GenBank accession: MW846253) as outgroups for phylogenetic analyses. Detailed information could be found in Table 1. Each sample ID in Table 1 represents the voucher unique identifier.
TABLE 1.
Taxonomic information, sampling metadata, GenBank accession numbers, and references of mitochondrial genomes used in the study
| Sample ID | Subfamily | Species | Sampling metadata | Life stage | Accession no | Reference |
|---|---|---|---|---|---|---|
| XL3275 | Prodiamesinae | Prodiamesa olivacea | Jiuzhaigou Valley Scenic and Historic Interest Area, Sichuan, China, 33.1928°N, 103.8942°E, 12‐Jul‐2019, leg. X.‐Y. Ge | Larva | MW373525 | Lin et al. (2022) |
| XL3436 | Orthocladiinae | Propsilocerus akamusi | Yuqiao Reservoir, Jizhou, Tianjin, China, 40.0197°N, 117.6389°E, 21‐Nov‐2019, leg. H.‐J. Yu | Adult male | MW846253 | Lin et al. (2022) |
| XL1177 | Diamesinae | Boreoheptagyia alulasetosa | Cangshan Mountain, Dali, Yunnan, China, 25.6475°N, 100.1426°E, 20‐May‐2018, leg. X.‐L. Lin | Adult male | MZ043574 | This study |
| ZJ837 | Diamesinae | Boreoheptagyia brevitarsis | Lingdi, Wenzhou, Zhejiang, China, 28.3276°N, 120.8774°E, 5‐May‐2019, leg. X.‐L. Lin | Adult male | MZ043575 | This study |
| LGS62 | Diamesinae | Boreoheptagyia kurobebrevis | Leigongshan Natural Reserve, Guizhou, China, 26.3960°N, 108.2609°N, 20‐Jan‐2020, leg. H.‐J. Yu | Adult female | MZ043576 | This study |
| XL3519 | Diamesinae | Boreoheptagyia zhengi | Gaoligongshan National Nature Reserve, Baoshan, Yunnan, China, 25.3106°N, 98.7950°E, 22‐May‐2018, X.‐L. Lin | Adult male | OM302508 | This study |
| XL4059 | Diamesinae | Diamesa loeffleri | Shoule town, Haidong, Qinghai, China, 36.7707°N, 102.4887°E, 26‐Nov‐2020, leg. Q.‐B. Huo | Adult male | MZ127838 | This study |
| CHMIT19 | Diamesinae | Diamesa qiangi | Lulang, Xizang, China, 29.77°N, 94.74°E, 14‐Aug‐2013, leg. Q. Wang | Adult male | MZ127839 | This study |
| XL4057 | Diamesinae | Diamesa sp. 1XL | Shoule town, Haidong, Qinghai, China, 36.7707°N, 102.4887°E, 26‐Nov‐2020, leg. Q.‐B. Huo | Larva | MZ048035 | This study |
| XL3288 | Diamesinae | Diamesa sp. 2 XL | Huanglong Scenic and Historic Interest Area, Sichuan, China, 30.72538°N, 103.8331°E, 17‐Jul‐2019, leg. X.‐Y. Ge | Larva | MZ048036 | This study |
| XL2214 | Diamesinae | Diamesa sp. 3XL | Shangchayuzhen, Zayu, Xizang, China, 28.73868694°N, 96.76293611°E, 24‐Mar‐2016, leg. Z.‐Y. Liu | Larva | MZ048037 | This study |
| XL1967 | Diamesinae | Diamesa sp. 4XL | Zhongshacun, Mainling, Xizang, 29.1873°N, 93.9954°E, 16‐Jul‐2014, leg. X.‐L. Lin | Larva | MZ048038 | This study |
| XL3464 | Diamesinae | Diamesa sp. 5XL | Erdaobaihezhen, Antu, Jilin, China, 42.4011°N, 128.1008°E, 22‐Aug‐2019, leg. S. Qiu | Larva | MZ231027 | This study |
| XL2212 | Diamesinae | Diamesa sp. 6XL | Shangchayuzhen, Zayu, Xizang, China, 28.7387°N, 96.76294°E, 24‐Mar‐2016, leg. Z.‐Y. Liu | Larva | MZ158293 | This study |
| XL1930 | Diamesinae | Diamesa sp. 7XL | Bomi, Xizang, China, 29.8035°N, 95.8672°E, 11‐Jul‐2014, leg. X.‐L. Lin | Larva | MZ158294 | This study |
| XL2216 | Diamesinae | Diamesa sp. 8XL | Shangchayuzhen, Zayu, Xizang, China, 28.7387°N, 96.7629°E, 24‐Mar‐2016, leg. Z.‐Y. Liu | Larva | MZ231028 | This study |
| XL3286 | Diamesinae | Diamesa sp. 9XL | Huanglong Scenic and Historic Interest Area, Sichuan, China, 30.7253°N, 103.8331°E, 17‐Jul‐2019, leg. X.‐Y. Ge | Larva | MZ231029 | This study |
| XL2133 | Diamesinae | Diamesa sp. 10XL | Baiyanggou, Qinghai, China, 38.2283°N, 100.2674°E, 24‐Jul‐2019, leg. X.‐J. Zhu | Adult male | MZ043577 | This study |
| XL1929 | Diamesinae | Diamesa sp. 11XL | Bomi, Xizang, China, 29.8035°N, 95.8672°E, 11‐Jul‐2014, leg. X.‐L. Lin | Larva | MZ043578 | This study |
| XL1907 | Diamesinae | Diamesa sp. 12XL | Ranwu Lake, Chamdo, Xizang, China, 29.5050°N, 96.7489°E, 10‐Jul‐2014, leg. X.‐L. Lin | Larva | MZ158295 | This study |
| XL2121 | Diamesinae | Diamesa tonsa | Qihan, Qinghai, China, 37.1555°N, 102.0238°E, 17‐Apr‐2019, leg. X.‐J. Zhu | Adult male | MZ158292 | This study |
| XL877 | Diamesinae | Pagastia lanceolata | Gaoligongshan National Nature Reserve, Baoshan, Yunnan, China, 25.3106°N, 98.7950°E, 22‐May‐2018, leg. X.‐L. Lin | Adult male | OM302510 | This study |
| XL3361 | Diamesinae | Pagastia sp. 1XL | Sangzhuziqu, Xizang, 12‐Aug‐2019, leg. J. Jiang | Larva | OM302507 | This study |
| XL3290 | Diamesinae | Pagastia sp. 2XL | Huanglong Scenic and Historic Interest Area, Sichuan, China, 30.7253°N, 103.8331°E, 17‐Jul‐2019, leg. X.‐Y. Ge | Larva | OM302505 | This study |
| XL3460 | Diamesinae | Pagastia tianmumontana | Erdaobaihezhen, Antu, Jilin, China, 42.4011°N, 128.1008°E, 22‐Aug‐2019, leg. S. Qiu | Larva | MZ231025 | This study |
| XL3152 | Diamesinae | Potthastia gaedii | Fanjingshan National Nature Reserve, Tongren, Guizhou, China, 27.7392°N, 108.8212°E, 7‐Oct‐2019, leg. H.‐J. Yu | Larva | OM302504 | This study |
| LGS11 | Diamesinae | Potthastia sp. 1XL | Xianlvtang, Leigongshan Natural Reserve, Guizhou, China, 20‐Dec‐2019, leg. H.‐J. Yu | Adult male | OM302509 | This study |
| XL1347 | Diamesinae | Potthastia sp. 2XL | Juma River, Baoding, Hebei, China, 39.4280°N, 115.1701°E, 8‐May‐2018, leg. X.‐L. Lin | Adult male | MZ064641 | This study |
| XL1561 | Diamesinae | Potthastia sp. 3XL | Wuying River, Yichun, Heilongjiang, China, 48.0869°N, 129.2468°E, 27‐Jul‐2016, leg. C. Song | Adult male | MW373523 | Zheng et al. (2021) |
| XL1623 | Diamesinae | Potthastia sp. 4XL | Erdaobaihezhen, Antu, Jilin, China, 42.4567°N, 128.1442°E, 12‐Jul‐2016, leg. C. Song | Adult male | OM302503 | This study |
| XL2282 | Diamesinae | Pseudodiamesa sp. 1XL | Songduo Mian Steam, Xizang, China, 29.9067°N, 92.3981°E, 14‐Oct‐2016, leg. Z.‐Y. Liu | Larva | MZ064643 | This study |
| XL3334 | Diamesinae | Pseudodiamesa sp. 2XL | Huanglong Scenic and Historic Interest Area, Sichuan, China, 30.7253°N, 103.8331°E, 16‐Jul‐2019, leg. X.‐Y. Ge | Larva | OM302506 | This study |
| ZJ283 | Diamesinae | Sympotthastia takatensis | Lingdi, Wenzhou, Zhejiang, China, 28.3044°N, 120.9295°E, 7‐Apr‐2019, leg. X.‐L. Lin | Pupa | MZ231026 | This study |
2.2. Sequencing and mitogenome assembly
The whole genomes were sequenced using the Illumina NovaSeq 6000 platform with 150‐bp paired‐end reads at Novogene Co., Ltd. (Beijing, China). The raw sequencing reads were trimmed with Trimmomatic (Bolger et al., 2014), and then about two Gb of clean data were obtained for each sample. The clean data were assembled using IDBA‐UD (Peng et al., 2012) with minimum and maximum k values of 40 and 120 bp, respectively, and the similarity was set as 98%.
The cytochrome c oxidase I (COI) barcode sequence for each species was obtained by Sanger sequencing herein and from previous study (Lin, Yu, et al., 2021), and served as the “bait” references to acquire the best‐fit and targeted mitochondrial contigs by BLAST (Altschul et al., 1990) search in Geneious 2020.2.1 (Kearse et al., 2012). Moreover, clean reads were mapped onto the obtained mitogenome using Geneious to check the accuracy of the assembly.
2.3. Genome annotation, composition, and substitution rate
Genome annotation was conducted following previous study (Zheng et al., 2020). Transfer RNA (tRNA) genes and their secondary structures were identified on MITOS2 webserver (available at http://mitos2.bioinf.uni‐leipzig.de/index.py). Ribosomal RNA (rRNA) genes and protein‐coding genes (PCGs) were annotated by aligning with homologous genes of Potthastia sp. in Geneious. Newly sequenced mitogenomes were submitted to GenBank (accession numbers: pending). The mitogenome maps were drawn by the CG View server V 1.0 (Grant & Stothard, 2008). The base composition, codon usage, and relative synonymous codon usage (RSCU) values were calculated in MEGA X (Kumar et al., 2018). The bias of the nucleotide composition was measured by AT‐skew [(A − T)/(A + T)] and GC‐skew [(G − C)/(G + C)]. The ratio (ɷ) of nonsynonymous substitution rates (Ka) to Synonymous substitution rates (Ks) was an excellent estimator of evolutionary selection pressure. Synonymous substitution rates (Ks) and nonsynonymous substitution rates (Ka) of mitochondrial PCGs were calculated using DnaSP 6.12.03 (Rozas et al., 2017).
2.4. Substitution rate and phylogenetic analyses
The level of base substitution saturation for each gene and each position of the PCGs was assessed using DAMBE 5.6.14 (Xia, 2013). Substitution of each of the three codon positions are generally not saturated, except for the transition of 3rd codon positions (Figure S1). Therefore, the 3rd codon positions of PCGs were excluded for the phylogenetic analyses. Each gene was aligned using MAFFT 7.402 (Katoh & Standley, 2013) with algorithm G‐INS‐i strategy. Gap in each matrix was treated as the fifth character and was retained in this study. Alignments of individual genes were then concatenated using SequenceMatrix v1.7.8 (Vaidya et al., 2011), after which three datasets were prepared for phylogenetic analyses: PCG12 (the 1st and 2nd codon positions of the 13 PCGs), PCG12R (the 1st and 2nd codon positions of the 13 PCGs and two rRNAs), and third AA (amino acid sequences of the 13 PCGs). The best partitioning scheme and best‐fit substitution model for each partition was tested using PartitionFinder 2.0 (Lanfear et al., 2017) with the Bayesian Information Criterion (BIC). Phylogenetic analyses were conducted with Maximum likelihood (ML) reconstruction and Bayesian inference (BI). The ML analysis was performed using IQ‐TREE 1.6.10 (Nguyen et al., 2015) with the best‐fit substitution model and 1000 bootstrap replicates. BI analysis was performed using MrBayes 3.2.7a (Ronquist et al., 2012) with substitution model in Table S1. Two simultaneous Markov chain Monte Carlo (MCMC) runs of 10,000,000 generations were conducted, trees were sampled every 1000 generations, and the first 25% of trees discarded as burn‐in. Tracer 1.7 (Rambaut et al., 2018) was used to check convergence of runs.
3. RESULTS
3.1. Mitogenome features of Diamesinae
The mitogenomes of 31 Diamesinae species were included in this study, 21 of which are complete, with the entire length ranging from 15,913 bp to 16,411 bp (Table S2). Each mitogenome contains 37 genes (13 PCGs, two rRNAs, and 22 tRNAs) and one control region. Nine PCGs, 14 tRNAs, and 2 rRNAs are coded on the majority strand (J‐strand), while the other genes are coded on the minority strand (N‐strand). The A + T content of the whole mitogenomes ranged from 72% in Pseudodiamesa sp. 1XL to 77.6% in Potthastia gaedii (Meigen, 1838) (Figure 2). Among the mitogenomes of Diamesinae, the control region and the 3rd codon of PCGs have the highest A + T content, while the 1st and 2nd codons of PCGs exhibit the lowest A + T content. The A + T content of rRNA genes is slightly higher than that in the whole mitogenomes, PCGs, and tRNA genes (Figure 2). In all selected species, the AT‐skew value of tRNA genes is positive while that of PCGs is negative. The GC‐skew values of rRNA genes and the 1st codon of PCGs are positive, while negative in the whole mitogenomes and the 2nd codon of PCGs (Figure S2). The start codons in most PCGs of the mitogenomes among Diamesinae are ATN (N represents one of four nucleotides, A, T, C, G), while COI and ND1 start with TTG. In addition, ND5 start with GTG in most mitogenomes of Diamesinae (Figure 3). The most prevalent termination codon used in mitogenomes of Diamesinae is TAA, with a small number of PCGs terminate with TAG, TGA, and T‐ (Figure 3). The total codon numbers, except the termination codons in mitogenomes of Diamesinae range from 3565 to 3735. Leu2, Phe, and Ile are the three most frequently used codon families, each with a number of more than 300. The least frequently used codon family is Cys, with a number less than 50 (Figure S3).
FIGURE 2.
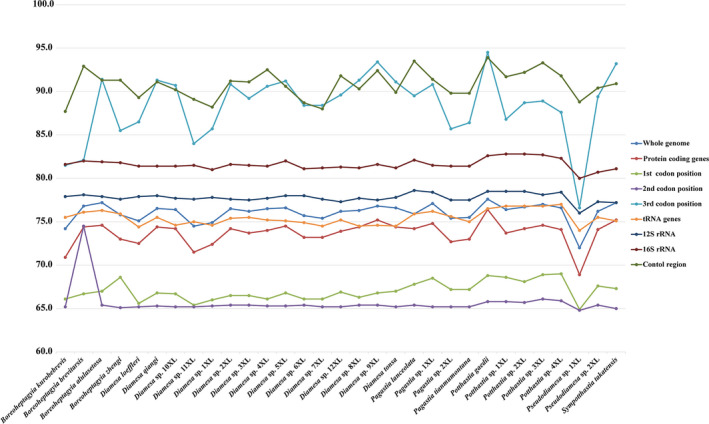
A+T content of mitochondrial genes of Diamesinae species. The X‐axis shows the species names and the Y‐axis shows the percentage of A+T content
FIGURE 3.
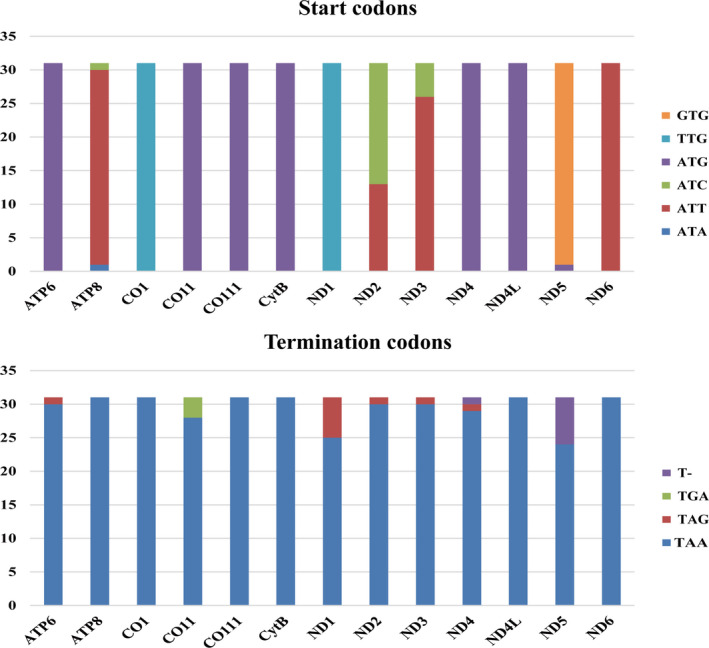
Start and termination codons of PCGs among Diamesinae species. The X‐axis shows the names of PCGs and the Y‐axis shows the codon numbers
For the entire Diamesinae, the ratio of Ka/Ks (ω) of all the 13 PCGs is less than 0.35, and the ATP8 exhibits the largest Ka/Ks value while the COI has the lowest Ka/Ks value (Figure 4, Table S3). To better understand the evolutionary pattern and the role of selection in Diamesinae species, the values of Ka/Ks were also calculated at congeneric level. The Ka/Ks value was quite heterogeneous at congeneric level. For individual genes, ATP6 showed a lower Ka/Ks value in Boreoheptagyia, ND1 and ND4L showed a lower Ka/Ks value in Boreoheptagyia and in Pagastia, and the remaining ten showed a lower Ka/Ks value in Diamesa and in Pagastia (Figure 4, Table S3). We also provided the Ka/Ks values of mitochondrial PCGs of Orthocladiinae and Stenochironomus that we previous reported in Table S3, which are higher than that in Diamesinae.
FIGURE 4.
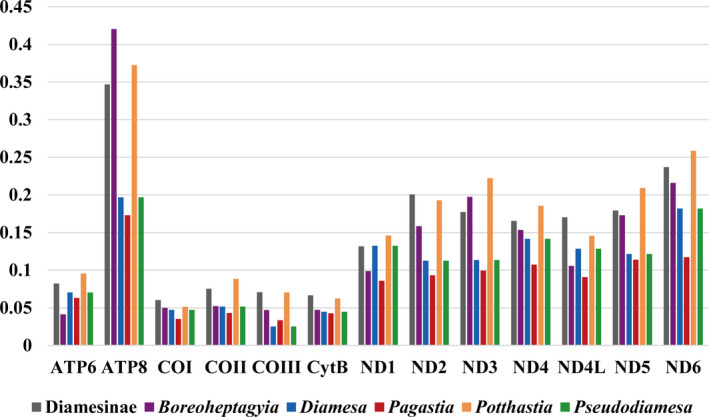
Evolution rate of each PCG of Diamesinae species. The X‐axis shows the names of PCGs and the Y‐axis shows the Ka/Ks value
Each mitogenome of Diamesinae contains 22 typical tRNA genes, with A+T content ranging from 74.0% to 77%. The nucleotide skew of tRNA genes among Diamesinae is consistent, exhibiting positive AT‐skew and negative GC‐skew (Figure 2, Figure S2). Both 12S and 16S rRNA genes transcribe from the minority strand (N‐strand). Among the mitogenomes of Diamesinae, the length of 12S rRNA gene varies from 794 to 815 bp, and the length of 16S rRNA gene varies from 1345 to 1374 bp (Table S2). The A+T content of 12S and 16S rRNA genes ranges from 76% to 78.6% and 80% to 82.8%, respectively. Both 12S and 16S rRNA genes exhibit positive GC‐skew in the mitogenomes of Diamesinae (Figure 2, Figure S2). A total of 21 mitogenomes in the present study have the complete control region, varying from 907 to 1309 bp (Table S2). The A+T content of the control region among the mitogenomes of Diamesinae ranges from 87.7% to 93.9% (Figure 2), extremely higher than the whole mitogenomes.
3.2. Phylogenetic relationships
Generally, six phylogenetic trees constructed by BI and ML analyses are similar in topology, only with the position of Sympotthastia was unstable (Figure 5). The monophyly of the Diamesinae is fully supported across all analyses using different datasets (Figure 5). Within the Diamesinae, two genera‐level topologies were inferred from three datasets: (i) (Potthastia + ((Boreoheptagyia +Sympotthastia) + (Diamesa + (Pagastia +Pseudodiamesa)))) was inferred from the PCG12 and PCG12R datasets; (ii) (Potthastia + (Boreoheptagyia + (Sympotthastia + (Diamesa + (Pagastia +Pseudodiamesa))))) was inferred from the AA dataset. The topology inferred from the AA dataset had the strongest nodal support. Based on three different datasets, Boreheptagyiini was deeply nested within Diamesini. The monophyly of Boreoheptagyia, Diamesa, Pagastia, Potthastia, and Pseudodiamesa was well supported by mitogenomes.
FIGURE 5.
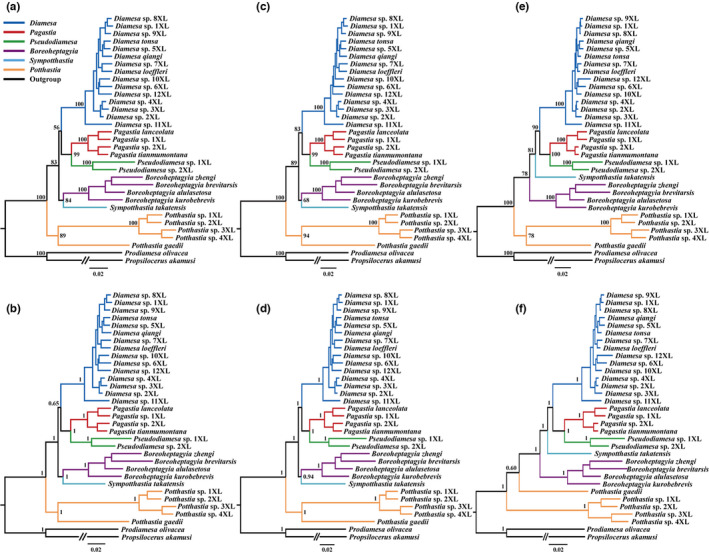
Phylogenetic relationships of Diamesinae inferred from mitogenomes. (a) ML tree obtained based on PCG12; (b) BI tree obtained based on PCG12; (c) ML tree obtained based on PCG12R; (d) BI tree obtained based on PCG12R; (e) ML tree obtained based on AA; (f) BI tree obtained based on AA. Numbers at the nodes are ML bootstrap values (a, c, e) and BI posterior probabilities (b, d, f)
4. DISCUSSION
4.1. Mitogenome features
A total of 31 mitogenomes of Diamesinae are included in this study, of which 10 mitogenomes have incomplete control region by the highly gene duplication (Cameron, 2014; Zhang & Hewitt, 1997). The lengths of 21 complete mitogenomes of Diamesinae range from 15,913 bp to 16,411 bp due to the variation of the control region. The gene number and arrangement of these mitogenomes are conserved, and all genes arranged in the same order as the ancestral insect mitogenome (Clary & Wolstenholme, 1985). The nucleotide composition of the mitogenomes of Diamesinae is biased toward A+T, which is consistent with other published chironomid species (Beckenbach, 2012; Deviatiiarov et al., 2017; Zheng et al., 2021). The mitogenomes of Diamesinae exhibit positive or negative AT‐skew and negative GC‐skew, the nucleotide bias of these mitogenomes may be related to the asymmetric mutation processes during replication (Hassanin et al., 2005). Most PCGs of mitogenomes of Diamesinae terminated with complete termination codons, except ND4 and ND5 in a few mitogenomes, terminated with a single T that may be completed by post‐transcriptional polyadenylation (Ojala et al., 1981). The ratio of Ka/Ks (ω) is used to assess the evolutionary rate of PCGs in mitogenome (Cheng et al., 2018; Li et al., 2020). The lengths of rRNA genes are inconsistent among Diamesinae species, indicating a relatively high level of variation in these genes. The A+T content of the control region is significantly higher than the whole mitogenome and other regions in mitogenome in Diamesinae, indicating a strong A+T bias in this region.
4.2. Evolutionary rate
We compared the Ka/Ks value between Diamesinae and other subfamilies of Chironomidae. Previous studies reported the Ka/Ks values of mitochondrial PCGs of Orthocladiinae and Stenochironomus (Lin et al., 2022; Zheng et al., 2022), and the Ka/Ks values of each PCG in these chironomids are higher than that in Diamesinae (Table S3), indicating that the mitochondrial genomes of Diamesinae are under stronger purifying selection than other nonbiting midge species (Hurst, 2002). Mitochondrial genome played a central role in animal energy production, and stronger purification selection could enhance their conserved role in energy production (Hassanin et al., 2009; Yuan et al., 2020). The existence of stronger purifying selection in Diamesinae species may exhibit signs of adaptation to life at cold living conditions (high latitude and high altitude) (Makarchenko et al., 2017). Severe habitats generally accumulate more deleterious mutations, and the stronger purifying selection of mitochondrial PCGs in Diamesinae species may help against these deleterious mutations (Sarkar et al., 2020; Wang et al., 2019). In addition, Diamesinae species lives in the cold environment (Lencioni & Rossaro, 2005; Montagna et al., 2016; Sun et al., 2019) and have a small range of activities, which could lead to a lower metabolic rate. Given the correlation between purification selection and metabolic rate has been reported in several species (Chong & Mueller, 2013; Shen et al., 2009; Wang et al., 2019), we hypothesized that the stronger purifying selection in Diamesinae species may also be associated with metabolic requirement.
The evolutionary rate analyses of Diamesinae also provided new insights for the study of species delimitation. The evolutionary rate of COI was generally considered to be consistent with the evolutionary rate of the species itself, so it has been widely used in species delimitation and phylogenetic studies (Havill et al., 2021; Jones et al., 2021). However, for species with lower evolution rate of mitochondrial PCGs, COI barcodes sometimes failed to accurately define the species boundary of Diamesa (Montagna et al., 2016) (E. Stur, pers. comm.). The mitochondrial genome or individual genes with higher evolution rate may be better choices for species delimitation.
4.3. Phylogenetic analyses
Previous study has revealed that mitogenomes have poor phylogenetic signals at the subfamily level of Chironomidae (Zheng et al., 2021). However, our study reveals that the mitogenomes of Diamesinae are practically useful for phylogenetic inference. In our study, we applied a variety of strategies to explore the phylogenetic relationships of six genera of the Diamesinae using mitogenomic data, and confirmed the monophyly of Diamesinae. According to traditional morphological systematics, Boreheptagyiini could be separated from other tribes of Diamesinae by having distinct pubescence, low antennal ratio, and usual dorsocentral and prealar setae in adults (Brundin, 1966; Sæther, 1977; Serra‐Tosio, 1973). According the morphological phylogeny, tribe Boreheptagyiini is sister to Heptagyini + Lobodiamesini, and Diamesini is sister to Protanypini. However, in the dated molecular phylogeny of the Chironomidae (Cranston et al., 2012), Boreheptagyiini was not sampled, and only one Diamesa species was selected. Our result gives a new insight for the systematic status of Boreheptagyiini. Serra‐Tosio (1973) presented a simplistic analysis of the tribe Diamesini, indicating that the clade Pagastia + Pseudodiamesinae is sister to the clade (Sympatthastia + Potthastia) + (Pseudokiefferiella + (Parapotthastia + (Onychodiamesa + (Diamesa +Lappodiamesa)))). Willassen (2011) presented an unpublished study based on two mitochondrial genetic markers (COX2 and 16S) of all Diamesini genera except Arctodiamesa and Sympotthastia in the 18th International Symposium on Chironomidae, and found that the tribe Diamesini is not monophyletic unless Potthastia is removed, and Boreoheptagyia (and Sasayusurika) are sister to the remaining Diamesini. In our study, Potthastia is placed as the oldest of all Diamesinae genera studied here. Our result corresponds very well by Willassen (2011), supporting that Potthastia is not a Diamesini which is contradictory with traditional morphology‐based systematics. Moreover, the phylogenetic position of Sympotthastia remain unstable based on mitogenomic phylogeny. In general, missing taxa and lacking of informative genetic characters can give a wrong picture of phylogeny estimation (Xi et al., 2016). Therefore, to finally explore the evolutionary history of Diamesinae, a complete resolution will require a comprehensive taxa sampling with the most informative mitochondrial and nuclear markers.
5. CONCLUSION
In this study, we sequenced 30 mitogenomes representing 30 species of six genera of Boreheptagyiini and Diamesini by whole genome sequencing technologies, and did the first comparative analysis of mitogenome base composition and evolutionary history in Diamesinae. This study showed that mitogenomes of Diamesinae were conserved in structure, gene order, and nucleotide composition. All protein‐coding genes in Diamesinae were under stronger purifying selection than those of other nonbiting midge species, which may exhibit signs of adaptation to life at cold living conditions. Mitogenomes could provide new insight into evolutionary history of Diamesinae based on the dated molecular phylogeny.
AUTHOR CONTRIBUTIONS
Xiao‐Long Lin: Data curation (lead); Investigation (equal); Methodology (equal); Software (equal); Writing – original draft (lead). Zheng Liu: Funding acquisition (lead); Writing – review & editing (equal). Li‐Ping Yan: Investigation (equal); Methodology (equal); Writing – review & editing (equal). Xin Duan: Formal analysis (equal); Software (equal). Wen‐Jun Bu: Supervision (equal); Validation (equal). Xin‐Hua Wang: Investigation (equal); Supervision (equal); Validation (equal). Chen‐Guang Zheng: Formal analysis (equal); Investigation (equal); Writing – original draft (equal); Writing – review & editing (equal).
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
Financial support from National Natural Science Foundation of China (31900344, 41502021), Technological Innovation Talent Training Project of the Ministry of Natural Resources of China (12110600000018003910), and the China Postdoctoral Science Foundation (2018M640227) are acknowledged with thanks. The authors thank Prof. Endre Willassen and another two anonymous reviewers for their suggestions and comments. The authors sincerely thank Hai‐Jun Yu, Qing‐Bo Huo, Xin‐Yu Ge, Zhen‐Yuan Liu, Xiao‐Ju Zhu, Dr. Chao Song, Dr. Shuang Qiu, Dr. Yu Peng, and Prof. Xiao‐Li Tong for their collecting material.
Lin, X.‐L. , Liu, Z. , Yan, L.‐P. , Duan, X. , Bu, W.‐J. , Wang, X.‐H. , & Zheng, C.‐G. (2022). Mitogenomes provide new insights of evolutionary history of Boreheptagyiini and Diamesini (Diptera: Chironomidae: Diamesinae). Ecology and Evolution, 12, e8957. 10.1002/ece3.8957
Xiao‐Long Lin and Zheng Liu contributed equally to this work as first authors.
DATA AVAILABILITY STATEMENT
The new mitogenomes of Boreoheptagyia alulasetosa, Boreoheptagyia brevitarsis, Boreoheptagyia kurobebrevis, Boreoheptagyia zhengi, Diamesa loeffleri, Diamesa qiangi, Diamesa sp. 1XL, Diamesa sp. 2 XL, Diamesa sp. 3XL, Diamesa sp. 4XL, Diamesa sp. 5XL, Diamesa sp. 6XL, Diamesa sp. 7XL, Diamesa sp. 8XL, Diamesa sp. 9XL, Diamesa sp. 10XL, Diamesa sp. 11XL, Diamesa sp. 12XL, Diamesa tonsa, Pagastia lanceolata, Pagastia sp. 1XL, Pagastia sp. 2XL, Pagastia tianmumontana, Potthastia gaedii, Potthastia sp. 1XL, Potthastia sp. 2XL, Potthastia sp. 4XL, Pseudodiamesa sp. 1XL, Pseudodiamesa sp. 2XL, and Sympotthastia takatensis are deposited in GenBank of NCBI under accession numbers MZ043574, MZ043575, MZ043576, OM302508, MZ127838, MZ127839, MZ048035, MZ048036, MZ048037, MZ048038, MZ231027, MZ158293, MZ158294, MZ231028, MZ231029, MZ043577, MZ043578, MZ158295, MZ158292, OM302510, OM302507, OM302505, MZ231025, OM302504, OM302509, MZ064641, OM302503, MZ064643, OM302506, and MZ231026, respectively.
REFERENCES
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Ashe, P. , & O'Connor, J. P. (2009). A world catalogue of Chironomidae (Diptera), Part 1: Buchonomyiinae, Chilenomyiinae, Podonominae, Aphroteniinae, Tanypodinae, Usambaromyiinae, Diamesinae, Prodiamesinae and Telmatogetoninae (445 pp). Irish Biogeographical Society. [Google Scholar]
- Beckenbach, A. T. (2012). Mitochondrial genome sequences of Nematocera (lower Diptera): evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biology and Evolution, 4(2), 89–101. 10.1093/gbe/evr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Research, 27(8), 1767–1780. 10.1093/nar/27.8.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. M. , George, M. , & Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America, 76(4), 1967–1971. 10.1073/pnas.76.4.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundin, L. (1966). Transantarctic relationships and their significance, as evidenced by chironomid midges with a monograph of the subfamilies Podonominae and Aphroteniinae and the austral Heptagynae. Kongliga Svenska Vetenskaps Academiens nya Handlingar, 11, 1–472. [Google Scholar]
- Cameron, S. L. (2014). Insect mitochondrial genomics: implications for evolution and phylogeny. Annual Review of Entomology, 59, 95–117. 10.1146/annurev-ento-011613-162007 [DOI] [PubMed] [Google Scholar]
- Chen, J.‐Y. , Chang, Y.‐W. , Zheng, S.‐Z. , Lu, M.‐X. , & Du, Y.‐Z. (2018). Comparative analysis of the Liriomyza chinensis mitochondrial genome with other Agromyzids reveals conserved genome features. Scientific Reports, 8(1), 1–10. 10.1038/s41598-018-27213-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. C. , Chen, M. Y. , Wang, J. F. , Liang, A. P. , & Lin, C. P. (2018). Some mitochondrial genes perform better for damselfly phylogenetics: Species‐and population‐level analyses of four complete mitogenomes of Euphaea sibling species. Systematic Entomology, 43(4), 702–715. 10.1111/syen.12299 [DOI] [Google Scholar]
- Chong, R. A. , & Mueller, R. L. (2013). Low metabolic rates in salamanders are correlated with weak selective constraints on mitochondrial genes. Evolution, 67(3), 894–899. 10.1111/j.1558-5646.2012.01830.x [DOI] [PubMed] [Google Scholar]
- Clary, D. O. , & Wolstenholme, D. R. (1985). The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. Journal of Molecular Evolution, 22(3), 252–271. 10.1007/BF02099755 [DOI] [PubMed] [Google Scholar]
- Condamine, F. L. , Nabholz, B. , Clamens, A.‐L. , Dupuis, J. R. , & Sperling, F. A. (2018). Mitochondrial phylogenomics, the origin of swallowtail butterflies, and the impact of the number of clocks in Bayesian molecular dating. Systematic Entomology, 43(3), 460–480. 10.1111/syen.12284 [DOI] [Google Scholar]
- Crampton‐Platt, A. , Timmermans, M. J. , Gimmel, M. L. , Kutty, S. N. , Cockerill, T. D. , Vun Khen, C. , & Vogler, A. P. (2015). Soup to tree: The phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Molecular Biology and Evolution, 32(9), 2302–2316. 10.1093/molbev/msv111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranston, P. S. , Hardy, N. B. , & Morse, G. E. (2012). A dated molecular phylogeny for the Chironomidae (Diptera). Systematic Entomology, 37(1), 172–188. 10.1111/j.1365-3113.2011.00603.x [DOI] [Google Scholar]
- Cranston, P. S. , & Krosch, M. N. (2015). DNA sequences and austral taxa indicate generic synonymy of Paratrichocladius Santos‐Abreu with Cricotopus Wulp (Diptera: Chironomidae). Systematic Entomology, 40(4), 719–732. 10.1111/syen.12130 [DOI] [Google Scholar]
- Curole, J. P. , & Kocher, T. D. (1999). Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends in Ecology & Evolution, 14(10), 394–398. 10.1016/S0169-5347(99)01660-2 [DOI] [PubMed] [Google Scholar]
- de Oliveira Aragão, A. , Neto, J. P. N. , Cruz, A. C. R. , Casseb, S. M. M. , Cardoso, J. F. , da Silva, S. P. , & Ishikawa, E. A. Y. (2019). Description and phylogeny of the mitochondrial genome of Sabethes chloropterus, Sabethes glaucodaemon and Sabethes belisarioi (Diptera: Culicidae). Genomics, 111(4), 607–611. 10.1016/j.ygeno.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Deviatiiarov, R. , Kikawada, T. , & Gusev, O. (2017). The complete mitochondrial genome of an anhydrobiotic midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA Part A, 28(2), 218–220. 10.3109/19401736.2015.1115849 [DOI] [PubMed] [Google Scholar]
- Ekrem, T. (2003). Towards a phylogeny of Tanytarsus van der Wulp (Diptera: Chironomidae). Is morphology alone sufficient to reconstruct the genealogical relationship? Insect Systematics & Evolution, 34(2), 199–219. [Google Scholar]
- Fang, X. , Li, X. , Lu, T. , Fu, J. , Shen, M. , Xiao, Y. , & Fu, Y. (2022). Complete mitochondrial genome of Limnophyes minimus (Diptera: Chironomidae). Mitochondrial DNA Part B, 7(1), 280–282. 10.1080/23802359.2022.2029604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, J. R. , & Stothard, P. (2008). The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Research, 36(Suppl 2), 181–184. 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanin, A. , Leger, N. , & Deutsch, J. (2005). Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Systematic Biology, 54(2), 277–298. 10.1080/10635150590947843 [DOI] [PubMed] [Google Scholar]
- Hassanin, A. , Ropiquet, A. , Couloux, A. , & Cruaud, C. (2009). Evolution of the mitochondrial genome in mammals living at high altitude: New insights from a study of the tribe Caprini (Bovidae, Antilopinae). Journal of Molecular Evolution, 68(4), 293–310. 10.1007/s00239-009-9208-7 [DOI] [PubMed] [Google Scholar]
- Havill, N. P. , Griffin, B. P. , Andersen, J. C. , Foottit, R. G. , Justesen, M. J. , Caccone, A. , D'Amico, V. , & Elkinton, J. S. (2021). Species delimitation and invasion history of the balsam woolly adelgid, Adelges (Dreyfusia) piceae (Hemiptera: Aphidoidea: Adelgidae), species complex. Systematic Entomology, 46(1), 186–204. 10.1111/syen.12456 [DOI] [Google Scholar]
- Hurst, L. D. (2002). The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends in Genetics, 18(9), 486. 10.1016/S0168-9525(02)02722-1 [DOI] [PubMed] [Google Scholar]
- Jacobsen, M. W. , Hansen, M. M. , Orlando, L. , Bekkevold, D. , Bernatchez, L. , Willerslev, E. , & Gilbert, M. T. P. (2012). Mitogenome sequencing reveals shallow evolutionary histories and recent divergence time between morphologically and ecologically distinct European whitefish (Coregonus spp.). Molecular Ecology, 21(11), 2727–2742. 10.1111/j.1365-294X.2012.05561.x [DOI] [PubMed] [Google Scholar]
- Jiang, Y.‐W. , Zhao, Y.‐M. , & Lin, X.‐L. (2022). First report of the complete mitogenome of Tanypus punctipennis Meigen, 1818 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA Part B, 7(1), 215–216. 10.1080/23802359.2021.2022544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L. , Twyford, A. D. , Ford, C. R. , Rich, T. C. G. , Davies, H. , Forrest, L. L. , Hart, M. L. , McHaffie, H. , Brown, M. R. , Hollingsworth, P. M. , & Vere, N. (2021). Barcode UK: A complete DNA barcoding resource for the flowering plants and conifers of the United Kingdom. Molecular Ecology Resources, 21(6), 2050–2062. 10.1111/1755-0998.13388 [DOI] [PubMed] [Google Scholar]
- Kang, Z. , Li, X. , & Yang, D. (2016). The complete mitochondrial genome of Dixella sp. (Diptera: Nematocera, Dixidae). Mitochondrial DNA Part A, 27(2), 1528–1529. 10.3109/19401736.2014.953123 [DOI] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , Buxton, S. , Cooper, A. , Markowitz, S. , Duran, C. , Thierer, T. , Ashton, B. , Meintjes, P. , & Drummond, A. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, J. L. , Peyton, J. T. , Fiston‐Lavier, A.‐S. , Teets, N. M. , Yee, M.‐C. , Johnston, J. S. , Bustamante, C. D. , Lee, R. E. , & Denlinger, D. L. (2014). Compact genome of the Antarctic midge is likely an adaptation to an extreme environment. Nature Communications, 5, 4611. 10.1038/ncomms5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Kim, H. , & Shin, S. C. (2016). Complete mitochondrial genome of the Antarctic midge Parochlus steinenii (Diptera: Chironomidae). Mitochondrial DNA Part A, 27(5), 3475–3476. 10.3109/19401736.2015.1066355 [DOI] [PubMed] [Google Scholar]
- Kong, F.‐Q. , Zhao, Y.‐C. , Chen, J.‐L. , & Lin, X.‐L. (2021). First report of the complete mitogenome of Microchironomus tabarui Sasa, 1987 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA Part B, 6(10), 2845–2846. 10.1080/23802359.2021.1970638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosch, M. , & Cranston, P. S. (2013). Not drowning, (hand) waving? Molecular phylogenetics, biogeography and evolutionary tempo of the ‘Gondwanan’ midge Stictocladius Edwards (Diptera: Chironomidae). Molecular Phylogenetics and Evolution, 68(3), 595–603. 10.1016/j.ympev.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R. , Frandsen, P. B. , Wright, A. M. , Senfeld, T. , & Calcott, B. (2017). PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34(3), 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Lei, T. , Song, C. , Zhu, X.‐D. , Xu, B.‐Y. , & Qi, X. (2021). The complete mitochondrial genome of a non‐biting midge Polypedilum unifascium (Tokunaga, 1938) (Diptera: Chironomidae). Mitochondrial DNA Part B, 6(8), 2212–2213. 10.1080/23802359.2021.1945977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencioni, V. , Rodriguez‐Prieto, A. , & Allegrucci, G. (2021). Congruence between molecular and morphological systematics of Alpine non‐biting midges (Chironomidae, Diamesinae). Zoologica Scripta, 50(4), 455–472. 10.1111/zsc.12480 [DOI] [Google Scholar]
- Lencioni, V. , & Rossaro, B. (2005). Microdistribution of chironomids (Diptera: Chironomidae) in Alpine streams: an autoecological perspective. Hydrobiologia, 533(1), 61–76. 10.1007/s10750-004-2393-x [DOI] [Google Scholar]
- Li, X.‐Y. , Yan, L.‐P. , Pape, T. , Gao, Y.‐Y. , & Zhang, D. (2020). Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. International Journal of Biological Macromolecules, 149, 371–380. 10.1016/j.ijbiomac.2020.01.249 [DOI] [PubMed] [Google Scholar]
- Lin, X.‐L. , Chang, T. , Yan, C.‐C. , Wang, B. , & Liu, W.‐B. (2021). Redescription of Diamesa loeffleri Reiss, 1968 (Diptera, Chironomidae) and new record from China. Annales Zoologici Fennici, 58(1–3), 109–113. 10.5735/086.058.0110 [DOI] [Google Scholar]
- Lin, X.‐L. , Stur, E. , & Ekrem, T. (2018). Molecular phylogeny and temporal diversification of Tanytarsus van der Wulp (Diptera: Chironomidae) support generic synonymies, a new classification and centre of origin. Systematic Entomology, 43, 659–677. 10.1111/syen.12292 [DOI] [Google Scholar]
- Lin, X.‐L. , Yu, H.‐J. , Wang, X.‐H. , Bu, W.‐J. , Wang, X.‐H. , Yan, C.‐C. , & Liu, W.‐B. (2021). New or little‐known Boreoheptagyia (Diptera: Chironomidae) in China inferred from morpholgy and DNA barcodes. ZooKeys, 1040, 187–200. 10.3897/zookeys.1040.66527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X.‐L. , Zhao, Y.‐M. , Yan, L.‐P. , Liu, W.‐B. , Bu, W.‐J. , Wang, X.‐H. , & Zheng, C.‐G. (2022). Mitogenomes provide new insights into the evolutionary history of Prodiamesinae (Diptera: Chironomidae). Zoologica Scripta, 51(1), 119–132. 10.1111/zsc.12516 [DOI] [Google Scholar]
- Makarchenko, E. A. , Endo, K. , Wu, J.‐Y. , & Wang, X.‐H. (2008). A review of Boreoheptagyia Brundin, 1966 (Chironomidae: Diamesinae) from East Asia and bordering territories, with the description of five new species. Zootaxa, 1817, 1–17. 10.11646/zootaxa.1817.1.1 [DOI] [Google Scholar]
- Makarchenko, E. A. , Semenchenko, A. A. , & Palatov, D. M. (2017). Review of subfamily Diamesinae (Diptera, Chironomidae) from Tien Shan and Pamir mountains. Paper presented at the 20th International Symposium on Chironomidae. Abstract Book of the 20th International Symposium on Chironomidae MUSE—Museo delle Scienze, Trento, Italy, 2–8 July 2017. [Google Scholar]
- Makarchenko, E. A. , Semenchenko, A. A. , & Palatov, D. M. (2021). New species and findings of Pagastia Oliver (Diptera: Chironomidae: Diamesinae) from Central Asia, with DNA barcoding of known species of the genus. Zootaxa, 4951(3), 559–570. 10.11646/zootaxa.4951.3.8 [DOI] [PubMed] [Google Scholar]
- Makarchenko, E. A. , & Wang, X.‐H. (2017). Pagastia tianmumontana sp. n.‐a new species of chironomids (Diptera: Chironomidae: Diamesinae) from South China. Far Eastern Entomologist, 336, 13–15. [Google Scholar]
- Miao, X. , Huang, J. , Menzel, F. , Wang, Q. , Wei, Q. , Lin, X.‐L. , & Wu, H. (2020). Five mitochondrial genomes of black fungus gnats (Sciaridae) and their phylogenetic implications. International Journal of Biological Macromolecules, 150, 200–205. 10.1016/j.ijbiomac.2020.01.271 [DOI] [PubMed] [Google Scholar]
- Montagna, M. , Mereghetti, V. , Lencioni, V. , & Rossaro, B. (2016). Integrated taxonomy and DNA barcoding of alpine midges (Diptera: Chironomidae). PLoS One, 11(3), e0149673. 10.1371/journal.pone.0149673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayed‐Breil, J. , & Orsini, A. (2016). On the genus Potthastia Kieffer, 1922 from Corsica and continental France with description of three new species [Diptera, Chironomidae, Diamesinae]. Ephemera, 17, 1–36. [Google Scholar]
- Nguyen, L.‐T. , Schmidt, H. A. , Von Haeseler, A. , & Minh, B. Q. (2015). IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32(1), 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala, D. , Montoya, J. , & Attardi, G. (1981). tRNA punctuation model of RNA processing in human mitochondria. Nature, 290(5806), 470–474. [DOI] [PubMed] [Google Scholar]
- Oliver, D. R. (1983). The larvae of Diamesinae (Diptera: Chironomidae) of the Holarctic region– Keys and diagnoses. Entomologica Scandinavica, Supplement, 19, 115–138. [Google Scholar]
- Oliver, D. (1989). The adult males of Diamesinae (Diptera. Chironomidae) of the Holarctic region – keys and diagnoses. Entomologica Scandinavica, Supplement, 34, 129–154. [Google Scholar]
- Park, K. , Jo, H. , Choi, B. , & Kwak, I.‐S. (2020). Complete mitochondrial genome of Stictochironomus akizukii (Tokunaga) (Chironomidae, Diptera) assembled from next‐generation sequencing data. Mitochondrial DNA Part B, 5(3), 2310–2311. 10.1080/23802359.2020.1750320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Leung, H. C. , Yiu, S.‐M. , & Chin, F. Y. (2012). IDBA‐UD: A de novo assembler for single‐cell and metagenomic sequencing data with highly uneven depth. Bioinformatics, 28(11), 1420–1428. 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- Qi, X. , Lin, X.‐L. , Ekrem, T. , Beutel, R. G. , Song, C. , Orlov, I. , Chen, C.‐T. , & Wang, X.‐H. (2019). A new surface gliding species of Chironomidae: An independent invasion of marine environments and its evolutionary implications. Zoologica Scripta, 48(1), 81–92. 10.1111/zsc.12331 [DOI] [Google Scholar]
- Ramakodi, M. P. , Singh, B. , Wells, J. D. , Guerrero, F. , & Ray, D. A. (2015). A 454 sequencing approach to dipteran mitochondrial genome research. Genomics, 105(1), 53–60. 10.1016/j.ygeno.2014.10.014 [DOI] [PubMed] [Google Scholar]
- Rambaut, A. , Drummond, A. J. , Xie, D. , Baele, G. , & Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, F. (1968). Neue Chironomiden‐Arten (Diptera) aus Nepal. Khumbu Himal, 3, 55–73. [Google Scholar]
- Ronquist, F. , Teslenko, M. , van der Mark, P. , Ayres, D. L. , Darling, A. , Höhna, S. , Larget, B. , Liu, L. , Suchard, M. A. , & Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3), 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J. , Ferrer‐Mata, A. , Sánchez‐DelBarrio, J. C. , Guirao‐Rico, S. , Librado, P. , Ramos‐Onsins, S. E. , & Sánchez‐Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12), 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- Sæther, O. A. (1969). Some Nearctic Podonominae, Diamesinae, and Orthocladiinae (Diptera: Chironomidae). Bulletin of the Fisheries Research Board of Canada, 170, 1–154. [Google Scholar]
- Sæther, O. A. (1977). Female genitalia in Chironomidae and other Nematocera: Morphology, phylogenies, keys. Bulletin of the Fisheries Research Board of Canada, 197, 1–209. [Google Scholar]
- Sæther, O. A. (2000). Phylogeny of the subfamilies of Chironomidae (Diptera). Systematic Entomology, 25(3), 393–403. 10.1046/j.1365-3113.2000.00111.x [DOI] [Google Scholar]
- Sarkar, I. , Dey, P. , Sharma, S. K. , Ray, S. D. , Kochiganti, V. H. S. , Singh, R. , Pramod, P. , & Singh, R. P. (2020). Turdoides affinis mitogenome reveals the translational efficiency and importance of NADH dehydrogenase complex‐I in the Leiothrichidae family. Scientific Reports, 10(1), 1–11. 10.1038/s41598-020-72674-4 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Serra‐Tosio, B. (1973). Ecologie et biogéographie des Diamesini d’Europe (Diptera, Chironomidae). Travaux du Laboratoire d’hydrobiologie et de pisciculture de l’université de Grenoble, 63, 5–175. [Google Scholar]
- Shen, Y.‐Y. , Shi, P. , Sun, Y.‐B. , & Zhang, Y.‐P. (2009). Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Research, 19(10), 1760–1765. 10.1101/gr.093138.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, F. L. , Ekrem, T. , & Fonseca‐Gessner, A. A. (2015). Out of South America: phylogeny of non‐biting midges in the genus Labrundinia suggests multiple dispersal events to Central and North America. Zoologica Scripta, 44(1), 59–71. 10.1111/Zsc.12089 [DOI] [Google Scholar]
- Sun, B.‐J. , Lin, X.‐L. , Wang, X.‐H. , & Makarchenko, E. A. (2019). New or little‐known Diamesinae (Diptera: Chironomidae) from Oriental China. Zootaxa, 4571(4), 544–550. 10.11646/zootaxa.4571.4.6 [DOI] [PubMed] [Google Scholar]
- Tang, L. , Yan, L. , Gao, Y. , & Zhang, D. (2019). First report of mitochondrial genome from the subfamily Bengaliinae (Diptera: Calliphoridae). Mitochondrial DNA Part B, 4(1), 1560–1561. 10.1080/23802359.2019.1601037 [DOI] [Google Scholar]
- Tang, P. , Zhu, J.‐C. , Zheng, B.‐Y. , Wei, S.‐J. , Sharkey, M. , Chen, X.‐X. , & Vogler, A. P. (2019). Mitochondrial phylogenomics of the Hymenoptera. Molecular Phylogenetics and Evolution, 131, 8–18. 10.1016/j.ympev.2018.10.040 [DOI] [PubMed] [Google Scholar]
- Vaidya, G. , Lohman, D. J. , & Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics, 27(2), 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Huang, J. , & Wu, H. (2021). Mitogenomes provide insights into the phylogeny of Mycetophilidae (Diptera: Sciaroidea). Gene, 783, 145564. 10.1016/j.gene.2021.145564 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Zhang, Y. , Zhang, H. , Qin, G. , & Lin, Q. (2019). Complete mitochondrial genomes of eight seahorses and pipefishes (Syngnathiformes: Syngnathidae): Insight into the adaptive radiation of syngnathid fishes. BMC Evolutionary Biology, 19(1), 1–11. 10.1186/s12862-019-1430-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willassen, E. (2011). Phylogeny of Diamesinae inferred from mtDNA sequences. Paper presented at the 18th International Symposium on Chironomidae. Scientific Program and Abstracts (p. 50). https://www.ntnu.no/c/document_library/get_file?uuid=515831d0‐116b‐4dd6‐a5d7‐03de9f235816&groupId=10476 [Google Scholar]
- Wolstenholme, D. R. (1992). Animal mitochondrial DNA: Structure and evolution. International Review of Cytology, 141, 173–216. 10.1016/S0074-7696(08)62066-5 [DOI] [PubMed] [Google Scholar]
- Xi, Z. X. , Liu, L. , & Davis, C. C. (2016). The impact of missing data on species tree estimation. Molecular Biology and Evolution, 33(3), 838–860. 10.1093/molbev/msv266 [DOI] [PubMed] [Google Scholar]
- Xia, X. H. (2013). DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Molecular Biology and Evolution, 30(7), 1720–1728. 10.1093/molbev/mst064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L. , Pape, T. , Elgar, M. A. , Gao, Y. , & Zhang, D. (2019). Evolutionary history of stomach bot flies in the light of mitogenomics. Systematic Entomology, 44(4), 797–809. 10.1111/syen.12356 [DOI] [Google Scholar]
- Yan, L. , Xu, W. , Zhang, D. , & Li, J. (2021). Comparative analysis of the mitochondrial genomes of flesh flies and their evolutionary implication. International Journal of Biological Macromolecules, 174, 385–391. 10.1016/j.ijbiomac.2021.01.188 [DOI] [PubMed] [Google Scholar]
- Yuan, M.‐L. , Zhang, L.‐J. , Zhang, Q.‐L. , Zhang, L. I. , Li, M. , Wang, X.‐T. , Feng, R.‐Q. , & Tang, P.‐A. (2020). Mitogenome evolution in ladybirds: Potential association with dietary adaptation. Ecology and Evolution, 10(2), 1042–1053. 10.1002/ece3.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.‐X. , & Hewitt, G. M. (1997). Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochemical Systematics and Ecology, 25(2), 99–120. 10.1016/S0305-1978(96)00042-7 [DOI] [Google Scholar]
- Zhang, D. , Yan, L. , Zhang, M. , Chu, H. , Cao, J. , Li, K. , Hu, D. , & Pape, T. (2016). Phylogenetic inference of calyptrates, with the first mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). International Journal of Biological Sciences, 12(5), 489–504. 10.7150/ijbs.12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Xu, W. , Peng, K. , Zou, L. , Li, Y. , Chen, Y. , Cai, Y. , & Gong, Z. (2019). The complete mitochondrial genome of Propsilocerus akamusi (Diptera, Chironomidae). Mitochondrial DNA Part B, 4(2), 3983–3984. 10.1080/23802359.2019.1688703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Kang, Z. , Ding, S. , Wang, Y. , Borkent, C. , Saigusa, T. , & Yang, D. (2019). Mitochondrial genomes provide insights into the phylogeny of Culicomorpha (Insecta: Diptera). International Journal of Molecular Sciences, 20(3), 747. 10.3390/ijms20030747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Yang, D. , & Kang, Z. (2022). New data on the mitochondrial genome of Nematocera (lower Diptera): Features, structures and phylogenetic implications. Zoological Journal of the Linnean Society, zlac012. 10.1093/zoolinnean/zlac012 [DOI] [Google Scholar]
- Zheng, C.‐G. , Liu, Z. , Zhao, Y.‐M. , Wang, Y. , Bu, W.‐J. , Wang, X.‐H. , & Lin, X.‐L. (2022). First report on mitochondrial gene rearrangement in non‐biting midges, revealing a Synapomorphy in Stenochironomus Kieffer (Diptera: Chironomidae). Insects, 13(2), 115. 10.3390/insects13020115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C. , Ye, Z. , Zhu, X. , Zhang, H. , Dong, X. , Chen, P. , & Bu, W. (2020). Integrative taxonomy uncovers hidden species diversity in the rheophilic genus Potamometra (Hemiptera: Gerridae). Zoologica Scripta, 49(2), 174–186. 10.1111/zsc.12401 [DOI] [Google Scholar]
- Zheng, C.‐G. , Zhu, X.‐X. , Yan, L.‐P. , Yao, Y. , Bu, W.‐J. , Wang, X.‐H. , & Lin, X.‐L. (2021). First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ, 9, e11294. 10.7717/peerj.11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The new mitogenomes of Boreoheptagyia alulasetosa, Boreoheptagyia brevitarsis, Boreoheptagyia kurobebrevis, Boreoheptagyia zhengi, Diamesa loeffleri, Diamesa qiangi, Diamesa sp. 1XL, Diamesa sp. 2 XL, Diamesa sp. 3XL, Diamesa sp. 4XL, Diamesa sp. 5XL, Diamesa sp. 6XL, Diamesa sp. 7XL, Diamesa sp. 8XL, Diamesa sp. 9XL, Diamesa sp. 10XL, Diamesa sp. 11XL, Diamesa sp. 12XL, Diamesa tonsa, Pagastia lanceolata, Pagastia sp. 1XL, Pagastia sp. 2XL, Pagastia tianmumontana, Potthastia gaedii, Potthastia sp. 1XL, Potthastia sp. 2XL, Potthastia sp. 4XL, Pseudodiamesa sp. 1XL, Pseudodiamesa sp. 2XL, and Sympotthastia takatensis are deposited in GenBank of NCBI under accession numbers MZ043574, MZ043575, MZ043576, OM302508, MZ127838, MZ127839, MZ048035, MZ048036, MZ048037, MZ048038, MZ231027, MZ158293, MZ158294, MZ231028, MZ231029, MZ043577, MZ043578, MZ158295, MZ158292, OM302510, OM302507, OM302505, MZ231025, OM302504, OM302509, MZ064641, OM302503, MZ064643, OM302506, and MZ231026, respectively.


