Abstract
SARS-CoV-2-infected subjects are generally asymptomatic during initial viral replication but may suffer severe immunopathology after the virus has receded and monocytes have infiltrated the airways. In bronchoalveolar lavage fluid from severe COVID-19 patients, monocytes express mRNA encoding inflammatory mediators and contain SARS-CoV-2 transcripts. We leverage a human small airway model of infection and inflammation, whereby primary blood monocytes transmigrate across SARS-CoV-2-infected lung epithelium to characterize viral burden, gene expression, and inflammatory mediator secretion by epithelial cells and monocytes. In this model, lung-infiltrating monocytes acquire SARS-CoV-2 from the epithelium and upregulate expression and secretion of inflammatory mediators, mirroring in vivo data. Combined use of baricitinib (Janus kinase inhibitor) and remdesivir (nucleoside analog) enhances antiviral signaling and viral clearance by SARS-CoV-2-positive monocytes while decreasing secretion of proneutrophilic mediators associated with acute respiratory distress syndrome. These findings highlight the role of lung-infiltrating monocytes in COVID-19 pathogenesis and their importance as a therapeutic target.
Keywords: RNA-seq, cytokine release syndrome, inflammation, interferon, Janus kinase, therapy
Graphical abstract
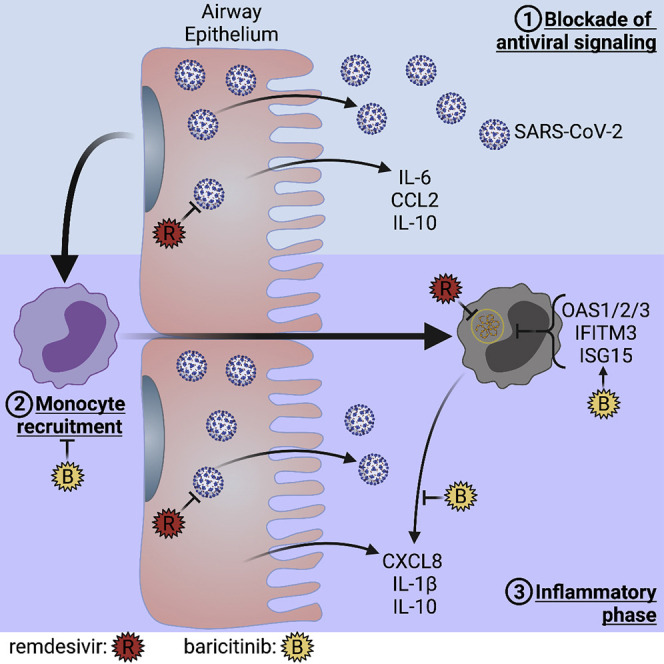
Dobosh et al. recapitulate infection by SARS-CoV-2 and subsequent inflammation by blood monocytes entering the lung in a human small airway model. Lung-recruited monocytes acquire SARS-CoV-2 from the infected epithelium but are unable to propagate virus. The immunomodulatory drug baricitinib promotes antiviral signaling in monocytes, which enhances clearance of SARS-CoV-2.
Introduction
Coronavirus disease 2019 (COVID-19) continues to be a rapidly evolving pandemic caused by the lung-tropic severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) (Weston and Frieman, 2020). COVID-19 symptoms range from asymptomatic to mild to severe, and the course of lung disease can be broadly categorized into three phases (Blanco-Melo et al., 2020). In the first phase of immune avoidance, the virus infects epithelial cells and replicates rapidly for about 3–6 days while avoiding activation of canonical antiviral responses like type I interferon (IFN) (Acharya et al., 2020; Tay et al., 2020). Respiratory symptoms occur in those patients who may progress to a second phase of “cytokine release syndrome (CRS), involving the infiltration of blood monocytes into the lung (Henderson et al., 2020; Merad and Martin, 2020), during which the viral titers decline. Eventually, in the second week post-infection, some patients will enter into a third, life-threatening, phase of acute respiratory distress syndrome (ARDS), featuring high levels of CXCL8, interleukin-1β (IL-1β), and other proinflammatory mediators, resulting in neutrophil recruitment (Coperchini et al., 2020; Tang et al., 2020). Considering the distinct phases of COVID-19 (Gordon et al., 2020), antiviral drugs like remdesivir may be particularly effective if given early (epithelial and monocytic stages), while immunomodulators, such as dexamethasone and the JAK1/2 inhibitor baricitinib, may be impactful later (monocytic and neutrophilic stages) (Beigel et al., 2020; Kalil et al., 2021; Perez-Alba et al., 2021).
Severe cases of COVID-19 have a dysfunctional immune response largely reflected by the CRS and decreases in most immune cells in the systemic compartment, including T, B, and natural killer (NK) cells (Tan et al., 2020), as well as monocytes, eosinophils, and basophils (Qin et al., 2020). Meanwhile, neutrophil counts and the neutrophil-to-lymphocyte ratio are elevated in the blood of patients with severe COVID-19 (Qin et al., 2020; Zhang et al., 2020). It is not yet known what underlying circumstances cause such changes in some patients but not others. As a result, the timing of therapeutics, in particular, host-directed treatments targeting the immune system, needs to be paid close attention to in relation to the course of disease to avoid suppressing antiviral responses in early stages or promoting further damage in late stages. Understanding mechanistic effects of antiviral and immunomodulatory drugs during the course of COVID-19 requires models that can recapitulate the complex interactions between SARS-CoV-2, host epithelial cells, and lung-infiltrating leukocytes (Mason, 2020). To this end, we adapted a small airway model of infection and inflammation (Dobosh et al., 2021) previously developed and validated by our group to study lung-infiltrating leukocytes in cystic fibrosis (CF) and ARDS (Forrest et al., 2018; Grunwell et al., 2019). This model features a human lung epithelium (HLE) monolayer differentiated at the air-liquid interface (ALI) that enables luminal infection with SARS-CoV-2 and subsequent infiltration by primary human leukocytes.
Mimicking the early, epithelium-restricted stage of SARS-CoV-2 infection in humans (Ravindra et al., 2021), HLE-ALI cells infected with SARS-CoV-2 in vitro did not upregulate canonical antiviral pathways or express inflammatory cytokines. Next, we observed, by reanalysis of a previously published dataset of single-cell RNA-seq (scRNA-seq) of bronchoalveolar lavage fluid (BALF) from patients with severe COVID-19 (Liao et al., 2020), that lung-infiltrating monocytes express CXCL8 and IL-1β and contain transcripts from SARS-CoV-2; both features were recapitulated by lung-infiltrating monocytes in vitro. CXCL8 and IL-1β are critical to the development of ARDS via recruitment and activation of neutrophils (Donnelly et al., 1993; Zhou et al., 2020). We next leveraged this model to investigate the effects on HLE-ALI cells and lung-infiltrating monocytes of remdesivir and baricitinib, two drugs that have been granted emergency use authorization by the US Food and Drug Administration as single and combined therapies. While remdesivir blocked progression of viral replication in both cell types, baricitinib selectively enhanced the expression of antiviral genes in lung-infiltrating monocytes. Both drugs used alone or in combination enhanced viral clearance in monocytes. Taken together, our findings confirm that lung-infiltrating monocytes are a critical target for COVID-19 treatment.
Results
Antiviral signaling in human lung epithelium differentiated at the air-liquid interface is blocked by SARS-CoV-2 and is not rescued by baricitinib and/or remdesivir
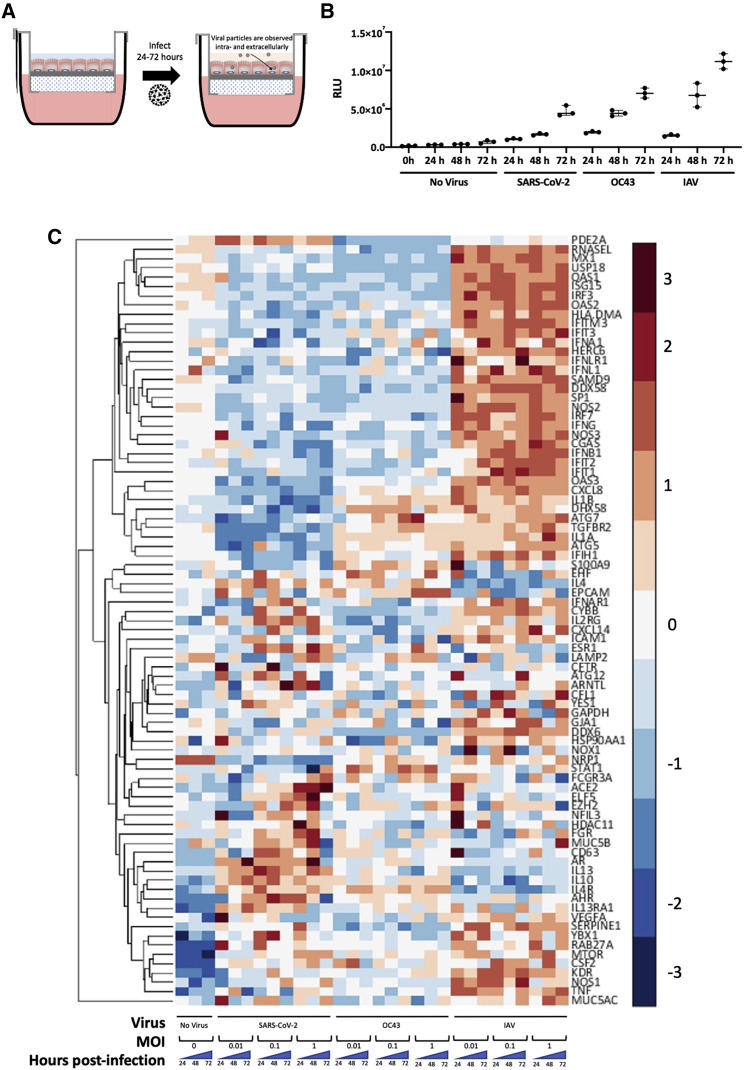
Critical aspects of virus-host interactions in the lung are species-specific, which makes models of the human lung particularly relevant for mechanistic investigations. Here, we differentiated an HLE for two weeks at ALI on a scaffold enabling en masse transmigration of leukocytes to simultaneously investigate the dynamics of luminal infection by viruses and leukocyte infiltration following epithelial infection (Figure 1A). SARS-CoV-2-infected HLE-ALI cells showed minimal amounts of extracellular ATP compared with OC43- and influenza strain A/PR8/1934 (IAV)-infected cells up to 48 h post-infection at an MOI of 0.1 (Figure 1B). This suggests that the 48-h time point and MOI may be appropriate conditions to study SARS-CoV-2 infection in the absence of substantial epithelial cell toxicity. Next, we conducted a multiplexed qRT-PCR assay of infected HLE-ALI cells at different input concentrations of virus over 72 h of infection and observed that HLE-ALI cells infected with SARS-CoV-2 and OC43 downregulated antiviral genes in contrast to IAV-infected cells (Figure 1C). For example, core IFN pathway genes IFNA1, IFNLR1, IFNL1, IFNG, and IFNB1, and to a lesser extent IFNAR1 were all lowered in cells infected with coronaviruses, while the same genes were dramatically increased in IAV-infected cells. Similarly, expression of inflammatory mediators, such as IL1α, IL-1β, and CXCL8, was consistently downregulated in SARS-CoV-2-infected cells, although IL1α expression was observed in HLE-ALI cells infected with OC43. Of note, a subset of allergy-associated genes, such as IL4, IL13, IL4R, IL13RA1, and the common γ-chain IL2RG, were upregulated in most of the SARS-CoV-2 infection conditions. Also of interest, infection with SARS-CoV-2 at an MOI of 1 showed upregulation of ACE2 throughout the course of infection, suggesting that the virus may be able to enhance its own infection efficiency.
Figure 1.
HLE-ALI cells grown at the air-liquid interface can be infected by OC43, SARS-CoV-2, and influenza and exhibit different transcriptional responses
(A) Experimental scheme illustrating the infection of human lung epithelium (HLE) differentiated at the air-liquid interface (ALI) with influenza A strain A/PR8/1934 (IAV), betacoronaviruses OC43, or SARS-CoV-2, isolate USA-WA1/2020, for 24–72 h.
(B) Extracellular ATP generated by HLE-ALI cells infected with IAV, OC43, or SARS-CoV-2 at an MOI of 0.1 for 24–72 h was measured using a luciferase assay. Plotted is the median with interquartile range. No statistics were calculated due to low sample number.
(C) Multiplexed qRT-PCR of HLE-ALI cells infected with either no virus or influenza A strain A/PR8/1934, betacoronaviruses OC43, or SARS-CoV-2, isolate USA-WA1/2020, for 24–72 h at MOIs of either 0.01, 0.1, or 1. Data were normalized using the delta delta Ct method relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the no virus condition at time 0, and then Z score normalized. Values shown are the average of biological triplicates.
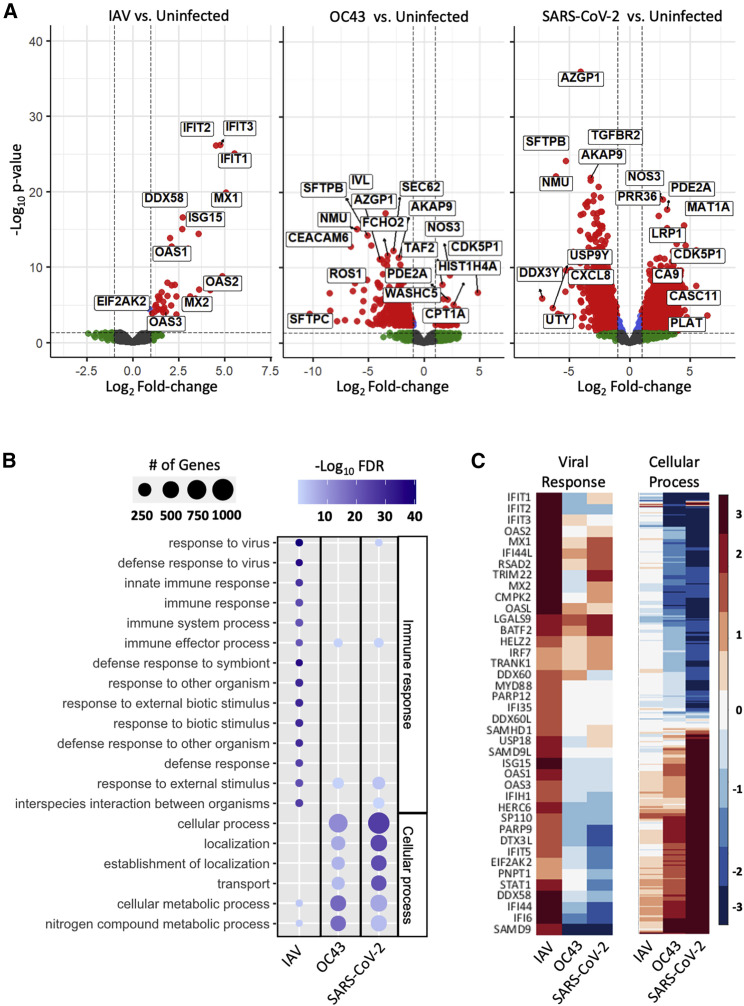
In order to better understand the global changes in the infected HLE-ALI cells, we then sequenced their entire transcriptome. As before, infection with IAV/PR8/1934 (IAV; MOI 0.1) induced canonical antiviral signaling in HLE-ALI cells, most notably IFITM1/2/3, ISG15, and OAS1/2/3, while infection with betacoronaviruses (MOI 0.1) OC43 and SARS-CoV-2, isolate USA-WA1/2020, did not (Figure 2A; Table S1). Across all infections, 50 gene ontology (GO) terms were significantly enriched (Table S2). Most genes enriched in HLE-ALI cells infected with IAV related to the immune response, particularly the response to virus and immune effector processes. In contrast, the enrichment profiles of HLE-ALI cells infected with OC43 and SARS-CoV-2 related to cellular processes, such as localization, transport, and metabolism (Figure 2B). Many of the antiviral genes upregulated in IAV-infected HLE-ALI cells were not upregulated in OC43 and SARS-CoV-2 infections (Figure 2C). Additionally, the genes associated with cellular processes in SARS-CoV-2-infected cells were similarly downregulated or remained unchanged in IAV-infected cells (Figure 2C; Table S2, row 9).
Figure 2.
SARS-CoV-2 prevents induction of an antiviral response in differentiated human lung epithelium
HLE-ALI cells were infected at an MOI of 0.1 with IAV, OC43, or SARS-CoV-2 for 48 h, after which total RNA was extracted and library prepared following the protocol of the TruSeq RNA Sample Preparation Kit.
(A) Volcano plots of differentially expressed genes (DEGs) in HLE-ALI cells infected with IAV, OC43, and SARS-CoV-2 compared with uninfected cells.
(B) Gene ontology enrichment profiles of DEGs in HLE-ALI cells infected with IAV, OC43, or SARS-CoV-2. Gene names associated with GO terms are in Table S2C. Heatmaps of targeted analysis of genes associated with the viral response or cellular process pathways. Gene names associated with the cellular process terms are in Table S2, row 9, and raw expression values in Table S1.
Further analysis of the RNA-seq dataset revealed that at 48 h post-infection with SARS-CoV-2 at an MOI of 0.1, HLE-ALI cells had the greatest number of uniquely differentially expressed genes (DEGs) compared with the uninfected time = 0 control (Figure S1A). At both 24 and 48 h post-infection, these DEGs showed dramatic changes in pathways associated with metabolic processes, showing that the HLE-ALI cells were affected by infection with SARS-CoV-2 despite the avoidance of canonical antiviral signaling (Figure S1B; Table S3).
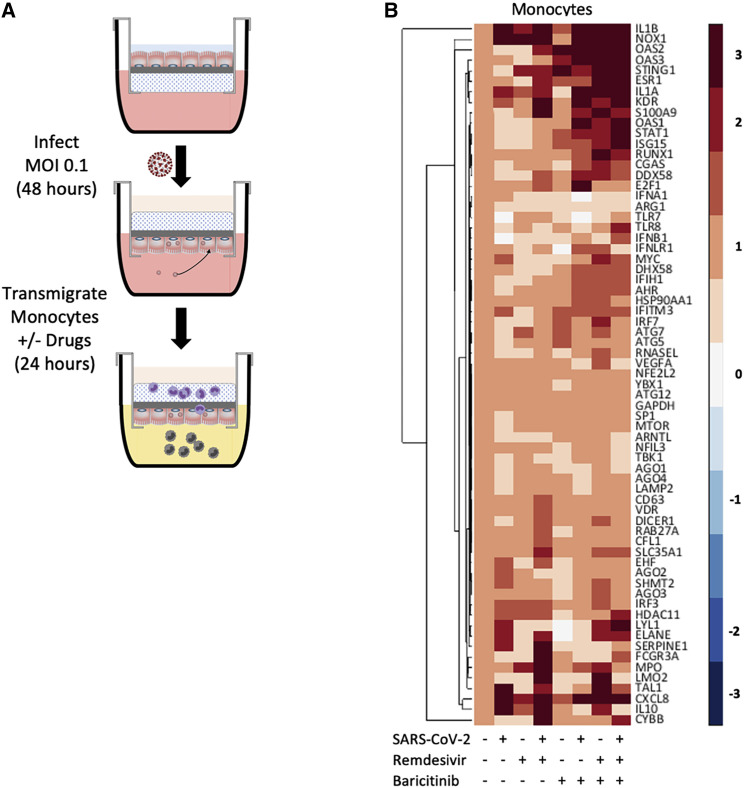
Following SARS-CoV-2 infection of the lung epithelium, subjects may remain asymptomatic or progress toward COVID-19. In vivo studies suggest that the pivotal response distinguishing asymptomatic from symptomatic subjects is the induction of a proinflammatory phase driven by lung-infiltrating monocytes (Falasca et al., 2020; Li et al., 2021). To investigate this key process in our model, we applied primary human blood monocytes to the basolateral side of SARS-CoV-2-infected HLE-ALI cells and allowed them to transmigrate across. To mimic in vivo conditions, the epithelial phase of infection was allowed to proceed for 48 h without any drug treatment, while the monocytic phase was studied for another 24 h in vehicle control (no drug condition) or in the presence of baricitinib (1 μM) and/or remdesivir (1 μM) (Figure 3A). The chemoattractants, CCL2 (250 pg/mL) and LTB4 (100 nM), were used in all conditions.
Figure 3.
Lung-recruited primary human monocytes induce proinflammatory signaling after transmigration across a SARS-CoV-2-infected epithelium
(A) Schematic diagram of the monocyte transmigration process: HLE-ALI cells were differentiated at ALI for two weeks and infected with SARS-CoV-2 at an MOI of 0.1 for 48 h, after which 106 monocytes were transmigrated across the infected epithelium toward leukotriene B4 (LTB4) (100 nM) and C-C motif chemokine ligand 2 (CCL2) (250 pg/mL) for 24 h, in the absence or presence of remdesivir (1 μM) and/or baricitinib (1 μM). All conditions contained 0.1% v/v DMSO.
(B) Heatmaps of selected genes from each treatment condition. Monocyte mRNA was analyzed by multiplexed qRT-PCR and first normalized to GAPDH using the delta delta Ct method and then Z score normalized.
To understand the impact of monocyte transmigration on the epithelium, we identified DEGs in HLE-ALI cells pre- versus post-transmigration. From these DEGs, we listed the top 10 GO enrichment profiles (Figure S2; Table S4). Following transmigration by monocytes, HLE-ALI cells showed an enrichment in genes related to the regulation of cell communication, signaling, and multicellular organismal processes, which was abrogated when HLE-ALI cells had been infected by SARS-CoV-2 prior to monocyte transmigration (Figure S2). Moreover, in the absence of SARS-CoV-2, transmigration of monocytes showed many fewer DEGs in the HLE-ALI cells (Figure S3A) than when monocyte transmigration occurred in the presence of virus (Figure S1A). Approximately 55% of the upregulated DEGs were shared in all conditions, suggesting that these genes are commonly expressed by HLE-ALI cells during the course of transmigration. Furthermore, most of the uniquely downregulated genes were observed in the no drug condition, suggesting that remdesivir and baricitinib have minimal effects on HLE-ALI cells, unless they are infected by SARS-CoV-2 (Figure S3A). Many of the GO terms observed were related to changes in cellular morphology and metabolic remodeling (Figure S3B; Table S5). Similarly, about 59% of the upregulated DEGs and 34% of the downregulated genes were shared by all conditions (Figure S4A), and many of the same metabolic process GO terms were observed. Notably however, the no drug condition showed an enrichment of GO terms associated with MHC class II antigen presentation (Figure S4B; Table S6).
Moreover, in concordance with the biological processes identified in SARS-CoV-2-infected HLE-ALI cells pre-transmigration by monocytes (Figure 2B), SARS-CoV-2-infected HLE-ALI cells post-transmigration still failed to induce genes included under the response to virus or immune system processes GO terms (Figure S5). Taken together, these findings suggest that during early SARS-CoV-2 infection, epithelial antiviral responses are delayed or blocked while favoring cellular and metabolic processes and regulation of communication. Remdesivir treatment did not induce an enrichment of these pathways when SARS-CoV-2 was present. Baricitinib treatment, however, promoted an enrichment in GO terms related to the regulation of cell communication and signaling in the SARS-CoV-2-infected epithelium post-transmigration, suggesting that this drug can affect epithelial signaling (Figures S2 and S4).
After transmigration across SARS-CoV-2-infected HLE-ALI cells, human monocytes induce proinflammatory signaling, which is attenuated by baricitinib treatment
To investigate the expression profile of lung-infiltrating monocytes during SARS-CoV-2 infection (compared with uninfected conditions), a set of 67 genes was measured by multiplexed qRT-PCR and grouped by unsupervised clustering (Figure 3B). Nucleic acid signaling sensor (CGAS and STING1, DDX58, and OAS1/2/3), kinase (TBK1), effector (RNAse L), and transcription factor (IRF3 and IRF7) genes as well as critical neutrophil chemoattractant genes, such as CXCL8 and IL-1β, were upregulated in monocytes recruited across SARS-CoV-2-infected HLE-ALI cells. The same set of genes were extracted from the RNA-seq dataset of the HLE-ALI cells pre- and post-transmigration. Although the post-transmigration HLE-ALI cells did not show global enrichment of antiviral pathways by analysis of GO terms compared with the pre-transmigration control, specific antiviral genes, such as DDX58 and OAS1/2/3, and the proneutrophilic chemokine CXCL8 were also upregulated (Figure S5). This response was not affected by treatment with baricitinib and/or remdesivir.
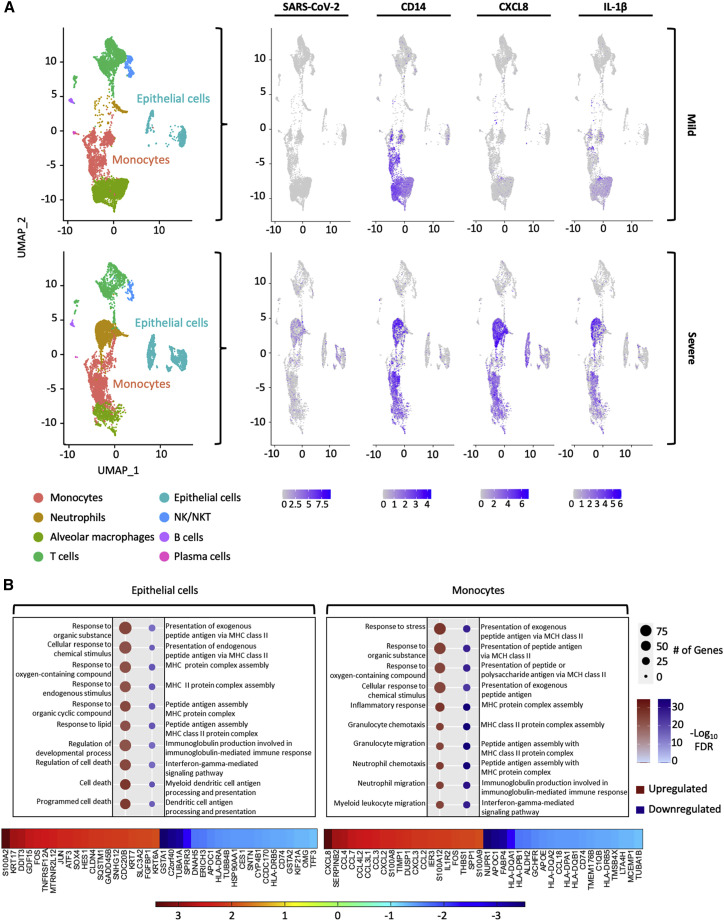
Computational analysis of a publicly available scRNA-seq dataset from the BALF of COVID-19 patients hospitalized with COVID-19 (Liao et al., 2020) confirmed that lung-infiltrating monocytes (differentiated from macrophages by exclusion of the macrophage gene MARCO) displayed elevated CXCL8 and IL-1β in patients with severe rather than mild disease (Figures 4A and S6). To our knowledge, this is the only dataset that has obtained scRNA-seq from BALF of hospitalized patients spanning mild and severe COVID-19, pre-remdesivir. Another study, which analyzed BALF from patients with severe COVID-19, found that there was expression of numerous inflammatory cytokines in BALF cells from a variety of cell types, particularly neutrophils (Bost et al., 2021). However, that study was conducted later during the course of the COVID-19 pandemic and did not report on BALF from relatively milder COVID-19.
Figure 4.
scRNA-seq of BALF from patients hospitalized with mild or severe COVID-19 shows that monocytes harbor SARS-CoV-2 genomes and express CXCL8 and IL-1β among other chemokines
(A) Combined Uniform Manifold Approximation and Projection (UMAP) plots of scRNA-seq from n = 3 mild patients and n = 3 severe patients. Seurat was used to normalize gene barcodes and generate UMAP clustering plots. Expression values for SARS-CoV-2, CD14, CXCL8, and IL-1β were overlaid onto the UMAP plot.
(B) DEGs in cells identified as epithelial cells or monocytes between the severe and mild patient groups were investigated. Enriched pathways for each cell type were plotted based on whether they were up- or down-regulated (severe versus mild). The top 20 up- and downregulated genes were listed for both cell types and plotted as a heatmap.
Epithelial cells recovered in BALF of severe COVID-19 patients upregulated cell-death-associated pathways and downregulated genes associated with antigen presentation, both of which may promote inflammation by innate immune cells (Figure 4B; Table S7). In addition to CXCL8, mentioned above, infiltrated monocytes present in the BALF of severe COVID-19 patients upregulated many chemokines, such as CCL4, CCL7, CCL4L2, CCL3, CXCL2, CXCL3, and CCL2, most of which are potent monocyte and macrophage chemoattractants and actually downregulated pathways associated with antigen presentation, similar to the epithelial cells (Figure 4B; Table S8). In addition, patients with severe disease showed a greater number of neutrophils in BALF compared with those with mild COVID-19, in whom monocytes and T cells were more abundant. These responses are consistent with those observed in lung-infiltrating monocytes produced in vitro (Figure 3B).
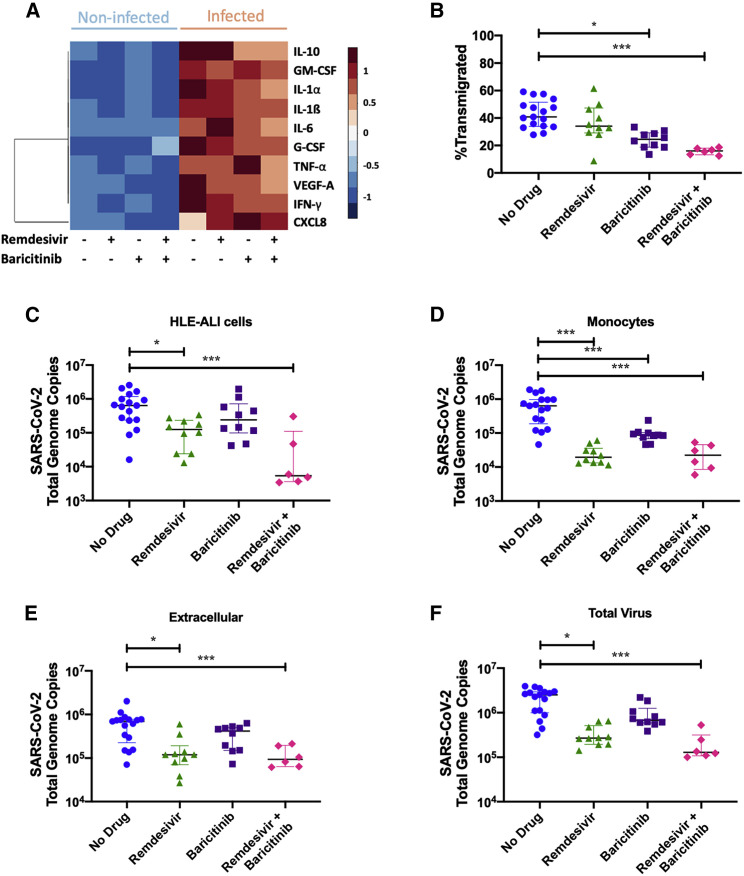
Next, we sought to confirm that changes in the expression of immune mediator genes was reflected in protein levels present in the apical aspect of our small airway model. Monocyte transmigration following SARS-CoV-2 infection of the epithelium led to increased extracellular levels of CRS-associated mediators, such as CXCL8, IL-1β, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-1α, IL-6, granulocyte-colony-stimulating factor (G-CSF), tumor necrosis factor alpha (TNF-α), vascular endothelial growth factor A (VEGFA), and IFN-γ, in the fluid compared with conditions where monocyte transmigration was conducted in the absence of virus (Figure 5A). While remdesivir treatment did not alter mediator levels, baricitinib treatment alone and in combination with remdesivir decreased extracellular IL-10 levels. This coincided with a decrease in IL-10 transcripts in lung-infiltrating monocytes but not epithelial cells (Figures 3B and S5). Baricitinib treatment also increased extracellular CXCL8 levels, while its combination with remdesivir decreased IL-1α, IL-1β, IL-6, TNF-α, VEGFA, and IFN-γ levels (Figure 5A).
Figure 5.
Lung-infiltrating monocytes release inflammatory mediators and harbor replicative SARS-CoV-2
(A) Inflammatory mediators in the apical fluid following transmigration were quantified by a multiplexed electrochemiluminescent assay. The Z score for each mediator in each condition was calculated and plotted.
(B) Transmigration efficiency of monocytes was calculated by dividing the number of monocytes in the apical fluid after 24 h by the input number of cells.
(C–E) RNA was extracted from each component of the model (epithelium, lung-infiltrating monocytes, and extracellular fluid) and reverse transcribed. Total SARS-CoV-2 genome copies were calculated.
(F) The sum of each of the components of the model was calculated to depict the total amount of virus remaining in the system after monocytes were allowed to transmigrate for 24 h.
All statistics were calculated using the Mann-Whitney U-test in Prism between the “no drug” and each treatment group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Shown are median and interquartile range.
Treatment with baricitinib and remdesivir decreases total viral burden in lung-infiltrating monocytes, which acquire SARS-CoV-2 from infected epithelium
Importantly, the JAK1/2 pathway can promote leukocyte recruitment in the lung and, in our model treatment with the JAK1/2 inhibitor baricitinib, decreased the number of lung-infiltrating monocytes (Figure 5B), which we determined by a qPCR-based DNA quantitation method (Figure S7A). As expected, remdesivir treatment decreased the viral burden in the HLE-ALI cells (Figure 5C), as measured by qRT-PCR (Figures S7B and S7C), despite the 48 h afforded to the virus to infect epithelial cells prior to treatment.
Our re-analysis of publicly available scRNA-seq data from lung-infiltrating monocytes in the BALF of patients with severe COVID-19 revealed that some of them harbored SARS-CoV-2 transcripts (Figure 4A). In severe COVID-19 patients included in another BALF study, infiltrating monocytes also contained SARS-CoV-2, although a vast majority of viral transcripts were found in the neutrophil compartment (Bost et al., 2021). Similarly, lung-infiltrating monocytes in our model, which were not directly infected but rather were made to cross an infected epithelium, also contained SARS-CoV-2 genome copies, as determined by qRT-PCR (Figures S5D and S7D). The total viral burden in lung-infiltrating monocytes was decreased in all treatment conditions, with the greatest effect being contributed by remdesivir. Absolute quantification of SARS-CoV-2 in both epithelial and monocytic compartments showed similar results as relative quantification of SARS-CoV-2 to GAPDH and 18S rRNA transcripts (Figures S8A–S8D). SARS-CoV-2 genome copies were also detectable in the extracellular fluid and were decreased by treatment with remdesivir alone and combined with baricitinib (Figure 5E). Altogether, remdesivir decreased total virus in all compartments of the transmigration model, while baricitinib did not have a noticeable effect despite lowering the number of genome copies in the recruited monocytes (Figure 5F).
In the 72 h during which the virus was applied to the HLE-ALI cells from start to finish of transmigration experiments, HLE-ALI cells had a median of 6.3 × 105 viral genome copies in the absence of drugs, which is about 5× the initial input (Figure 5C). To determine if viral replication occurred, we assessed SARS-CoV-2 N-subgenome relative to 18S rRNA transcripts in HLE-ALI cells. There was a detectable amount of N-subgenome that remained unchanged in conditions including remdesivir. However, treatment with baricitinib resulted in an ∼24-fold increase in the N-subgenome relative amount (Figure S7E). Relative to 18S rRNA transcript, all treatment conditions increased the amount of N-subgenome compared with the untreated control in monocytes (remdesivir: ∼21; baricitinib: ∼23; and remdesivir + baricitinib: ∼21) (Figure S7F).
In order to gain a better understanding of how SARS-CoV-2 enters monocytes, we analyzed the expression of surface angiotensin-converting enzyme 2 (ACE2) and also included various treatments during transmigration known to affect SARS-CoV-2 entry into cells. Blood monocytes were found to express surface ACE2 as measured by flow cytometry, and ACE2 appeared to be slightly upregulated upon transmigration (Figure S9A). The observation that immune cells, including monocytes and macrophages, express ACE2 and thus could be susceptible to direct SARS-CoV-2 infection has been validated by many groups (Abassi et al., 2020). Accordingly, the inclusion of soluble ACE2 (200 μg/mL, also contained a C-terminal 10xHis-tag) lowered the amount of SARS-CoV-2 observed in the monocytes, as other groups have shown (Krishnamurthy et al., 2021; Monteil et al., 2020). Next, the use of the antibiotic dalbavancin (1 μM), which has been shown to bind to the ACE2 receptor (Wang et al., 2021), also decreased the amount of SARS-CoV-2 in transmigrated monocytes. As a control, the use of either soluble ACE2 or dalbavancin did not decrease the amount of OC43 in the monocytes (Figure S9B). The dynamin GTPase inhibitor and pinocytosis blocker dynasore (80 μM) and endocytosis inhibitor pitstop2 (15 μM) also inhibited SARS-CoV-2 entry into monocytes. Taken together, these findings indicate that SARS-CoV-2 likely enters monocytes through a variety of mechanisms, including ACE2-receptor-mediated endocytosis, either with or without the aid of clathrin (Bayati et al., 2021), as well as pinocytosis.
Treatment with baricitinib and remdesivir decreases the rate of viral clearance and replication in lung-infiltrating monocytes harboring SARS-CoV-2
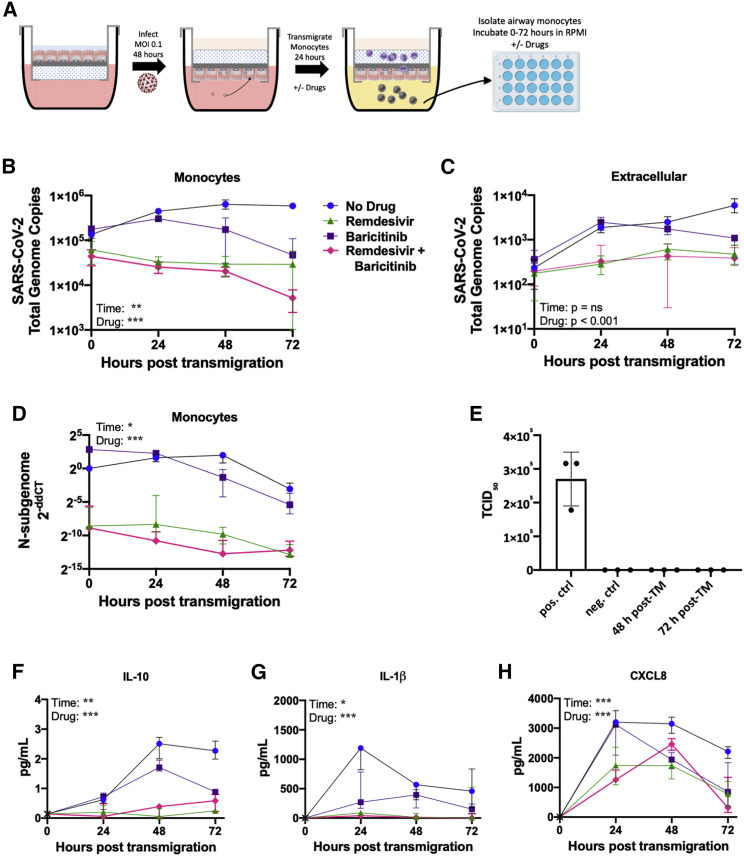
Since SARS-CoV-2 genome copies were detectable in lung-infiltrating monocytes, we examined whether these genomes were replication-competent. To this end, we isolated and washed lung-infiltrating monocytes via negative selection with anti-CD326 beads to remove contaminating epithelial cells and incubated the purified monocytes in fresh medium for 72 h (Figure 6A). Every 24 h, RNA and protein were isolated from the monocytes and the extracellular fluid. In the absence of drug, viral genome copies in monocytes increased from 0 to 48 h and then plateaued (Figure 6B) while extracellular viral genome copies steadily increased across the 72 h (Figure 6C). Since baricitinib induced the expression of several antiviral genes and remdesivir reduced viral genome copies in lung-infiltrating monocytes in our model, we next tested the effect of these drugs on viral propagation upon an additional 72 h of culture. As expected, treatment with remdesivir abrogated the replication of the virus and few genome copies were detected in monocytes. Treatment with baricitinib decreased viral genome copies in monocytes at 48- and 72-h time points, suggesting that this drug may also promote clearance of virus by lung-infiltrating monocytes. Furthermore, treatment with remdesivir alone or in combination with baricitinib resulted in a stark decrease in levels of N-subgenome in monocytes at all time points (Figure 6D). Treatment with baricitinib also lowered N-subgenome in monocytes compared with the untreated control at 48- and 72-h time points.
Figure 6.
Treatment with baricitinib and remdesivir increases the rate of viral clearance in lung-infiltrating monocytes
(A) Schematic of the experimental setup to quantify replication of the virus in monocytes. HLE-ALI cells were infected with SARS-CoV-2 at an MOI 0.1 for 48 h, after which 106 monocytes were transmigrated across the infected epithelium toward LTB4 (100 nM) and CCL2 (250 pg/mL) for 24 h, in the absence/presence of remdesivir (1 μM) and/or baricitinib (1 μM). Monocytes were washed and purified by negative depletion using anti-CD326 beads to remove contaminating epithelial cells and placed into medium with no drug, remdesivir (1 μM), and/or baricitinib (1 μM) for 0–72 h. All conditions contained 0.1% v/v DMSO.
(B) Quantification of total SARS-CoV-2 genome copies in the monocytes.
(C) Quantification of the amount of virus in the extracellular fluid at each time point.
(D) Quantification of the N-subgenome in monocytes at each time point. Data were normalized to 18S rRNA in the untreated condition at 0 h using the delta delta Ct method.
(E) The extracellular fluid was layered onto VeroE6 cells to perform a plaque assay from which a TCID50 was calculated. The positive control was the direct application of 2.5 × 104 genome copies of SARS-CoV-2 to the cells.
(F–H) Inflammatory mediators were measured using an electrochemiluminescent assay.
All statistics were calculated with a two-way ANOVA, main effects model in Prism with Geisser-Greenhouse correction applied. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Shown are median and interquartile range.
Because the virus replicated its genome in lung-infiltrating monocytes, we next sought to determine whether infectious particles could be formed from these cells. To this end, the extracellular fluid from monocyte cultures collected at 48- and 72-h time points were applied to virus-permissive VeroE6 cells. Remarkably, no plaques or cytopathic effects were observed (Figures 6E and S10), an observation repeated with a foci assay. These findings suggest that although lung-infiltrating monocytes in our in vitro model are amenable to SARS-CoV-2 genome replication, a blockade exists in these cells that results in abortive virus.
Finally, to better understand the inflammatory state of lung-infiltrating monocytes harboring viral genome copies, we performed a multiplexed electrochemiluminescent assay to quantify mediators released by these cells. While extracellular IL-10 levels steadily increased from 0 to 48 h in the absence of drugs, treatment with baricitinib alone or in combination with remdesivir kept those levels low (Figure 6F). Treatment with remdesivir alone increased extracellular IL-10 levels, although not to the same levels as in the untreated control conditions. Extracellular levels of CXCL8 and IL-1β were higher in the absence of drug at 0- and 24-h time points than in all treatment groups. However, treatment with baricitinib lowered the magnitude of IL-1β increase, suggesting that this drug may promote lower grade neutrophilic inflammation. Although treatment with remdesivir resulted in lower levels of CXCL8 and IL-1β (Figures 6G and 6H), it elevated levels of IL-1α compared with the untreated control condition at 24- and 48-h time points (Figure S11A). IFN-γ increased over time in the untreated control condition but remained low in all treatment groups (Figure S11B), while no effects were observed with IL-6, TNF-α, G-CSF, GM-CSF, and VEGF-A (Figures S11C–S11G).
Discussion
COVID-19 includes an early viral phase and a later immune phase. Upon infection by SARS-CoV-2, differentiated epithelial cells in the small airway model show enhanced proapoptotic signaling and attenuated type I IFN signaling, which may allow for enhanced viral propagation during early disease and prevent mounting of an effective innate immune response by epithelial and myeloid cells. An ineffective innate immune response with suboptimal type I IFN signaling can impede development of subsequent type II IFN (IFN-γ)-dependent adaptive immune responses (Zuniga et al., 2015), as occurs in severe COVID-19 (Rao et al., 2020). Prior studies noted that SARS-CoV-2-infected human lung tissue failed to upregulate IFN genes during infection and that, in patients with severe COVID-19, lung-infiltrating monocytes showed a reduced IFN signature compared with patients with mild disease (Chu et al., 2020; Schulte-Schrepping et al., 2020). In contrast with prior studies that did not combine differentiated epithelial cells with infiltrating leukocytes (Vanderheiden et al., 2020), our small airway model did show increased IFN-γ release upon SARS-CoV-2 infection.
As viral burden wanes, host responses may devolve into CRS, featuring proinflammatory mediators supporting monocyte infiltration combined with high IL-10 levels, which act to initially limit neutrophil recruitment (Eroshenko et al., 2020). To overcome the barrier posed by IL-10, lung-infiltrating virus-infected monocytes may synthesize additional CXCL8, which eventually results in neutrophil infiltration, leading to ARDS. Moreover, high levels of monocyte-derived IL-1β can also overcome IL-10 blockade and promote neutrophilic inflammation (Dinarello, 2018). Lung-infiltrating monocytes in our model were infected by SARS-CoV-2 from the epithelium. They subsequently increased expression of proinflammatory genes, including CXCL8 and IL-1β, among other changes in their transcriptional poise, likely to contribute to further inflammation. This mirrors in vivo findings that infiltrating monocytes isolated from the BALF of severe COVID-19 patients contain SARS-CoV-2 transcripts and increased expression of CXCL8 and IL-1β and multiple other chemokines, including CCL4, CCL7, CCL3, and CCL2. These findings implicate lung-infiltrating monocytes as key contributors to COVID-19 progression. Thus, our model recapitulates the distinct phases in the clinical progression of COVID-19, including initial immune avoidance in epithelial cells, recruitment of airway monocytes, and the release of inflammatory molecules reflective of CRS and, later, ARDS.
Although the viral products detected in lung-infiltrating monocytes were abortive, these cells may yet act as a reservoir of virus or as a continuing source of inflammatory cytokines and contribute not only to fueling the CRS but also to paving the way for neutrophil-driven ARDS. Furthermore, it is possible that replicative SARS-CoV-2 in airway monocytes serves as a continual source of dsRNA-stimulation, resulting in inflammatory cytokine release contributing to symptomatology, seen in so-called “long-haulers,” i.e., patients who have relatively mild disease weeks or even months after the initial infection. Indeed, it was previously observed that CD16+ blood monocytes expressed SARS-CoV-2 S1 spike protein up to 15 months after initial SARS-CoV-2 infection, despite the inability to isolated full-length viral RNA (Patterson et al., 2021). The finding that monocytes harbor cell-associated SARS-CoV-2 copies has been observed by others and poses a key therapeutic opportunity for early host-directed disease intervention. In one study, blood monocytes were able to be infected by SARS-CoV-2 in an antibody-dependent manner, triggering inflammasome activation and contributing to severe COVID-19 pathology (Junqueira et al., 2021). In another study, circulating lymphomononuclear cells, including monocytes as well as T and B cells were susceptible to SARS-CoV-2 infection (Pontelli et al., 2020). A third study described that blood monocytes and differentiated macrophages could be infected by SARS-CoV-2, resulting in abortive viral products (Boumaza et al., 2021). However, these prior instances were in blood monocytes, not lung-infiltrating cells, as used in our study, which we previously showed to acquire a distinct transcriptional poise (Ford et al., 2021). Recently, another group showed that tissue-resident alveolar macrophages take up and replicate SARS-CoV-2, after which a type I IFN response ensued (Sefik et al., 2022).
In the Adaptive COVID-19 Treatment Trial 1 (ACTT-1), the antiviral remdesivir showed greatest clinical benefits (improved time to recovery) in patients with an ordinal oxygen score of 4 or 5 (not receiving or receiving low-flow oxygen, respectively) compared with a placebo, suggesting that antivirals will work best in early-stage disease (Beigel et al., 2020). ACTT-2 evaluated baricitinib in combination with remdesivir against baricitinib alone (Kalil et al., 2021). The combined treatment shortened total time to recovery and had more improvement in symptoms and fewer adverse events than the control group. The efficacy of combined baricitinib and remdesivir treatment was greatest in patients who had an ordinal score of 6 at baseline (receiving high-flow oxygen or a non-invasive ventilation), indicating the profound effects of baricitinib on more severe late-stage disease. In the subset of patients who started treatment at an ordinal score of 6, time to recovery was shorted from 18 days in the group to 10 days in the combination group (Kalil et al., 2021).
In rhesus macaques infected with SARS-CoV-2, baricitinib given alone from 2 to 10 days post-infection suppressed proinflammatory cytokine production in lung macrophages, reduced recruitment of neutrophils, and resulted in reduced lung pathology compared with the control group. However, viral load in nasal and throat swabs as well as BALF was unchanged (Hoang et al., 2021). Other groups have proposed that, in addition to its immunomodulatory effects via JAK1/2 inhibition, baricitinib treatment may have a direct effect on SARS-CoV-2 viral entry via ACE2 by inhibiting numb-associated kinases to prevent clathrin-mediated endocytosis (Stebbing et al., 2020). Our study supports the notion that treatment with baricitinib (alone and in combination with remdesivir) reduces viral load in lung-infiltrating monocytes and that baricitinib (alone and in combination with remdesivir) reduces viral replication and the release of inflammatory mediators. Meanwhile, an exploratory trial evaluating baricitinib plus standard of care versus a placebo plus standard of care found that the treatment group had a lower mortality rate at both 28 and 60 days, although ventilator-free days and length of hospital duration were unchanged (Ely et al., 2022). At such late time points post-infection, viral titers are low and baricitinib is likely functioning primarily as an immunomodulator.
Findings presented here are made relevant to the clinical prevention and resolution of COVID-19 by, first, comparing our in vitro data to in vivo scRNA-seq data from patients with severe COVID-19 and, second, by using the approved drugs remdesivir and baricitinib. These drugs may now be tested in combination with or as comparators for emerging therapies to bring to trial. Findings here suggest that candidate drugs can affect the epithelium and lung-infiltrating leukocytes in different and sometimes divergent ways with regard to their impact on antiviral and immune signaling, notably. For this reason, the use of our small airway model as a testing platform for candidate COVID-19 drugs overcomes some limitations associated with using non-human/non-lung in vitro models.
In conclusion, we showed that HLE cells differentiated at ALI propagate SARS-CoV-2 and fail to induce antiviral signaling but can recruit monocytes. These lung-infiltrating monocytes are in turn infected by SARS-CoV-2 yet successfully induce host antiviral pathways while also activating IL-1β and CXCL8 transcription. Critically, we show that virus-laden, IL-1β- and CXCL8-positive monocytes are also present in vivo in the lung fluid of patients with severe, but not mild, COVID-19. Finally, we show that treatment with the JAK1/2 inhibitor baricitinib combined with the antiviral drug remdesivir decreases monocyte recruitment through the virus-infected epithelium and viral burden in both cell types and also alters signaling pathways in monocytes. Taken together, our findings establish that lung-infiltrating monocytes retain virus and contribute to the pathogenesis of COVID-19 and thus must be appropriately targeted for successful resolution of the disease.
Limitations of the study
Although the human small airway model leveraged here can be generated with primary epithelial cells (Laucirica et al., 2022), the present study relies on a club cell line. In addition, blood monocytes transmigrated in the model are from healthy subjects. Use of primary monocytes from patients may shed information on disease course in light of the observations that they may behave abnormally in severe COVID-19 (Qin et al., 2020). Finally, this study uses the Washington strain of SARS-CoV-2. Follow-up studies combining primary airway epithelial cells and monocytes from infected subjects and other SARS-CoV-2 variants of interest (e.g., Delta [B.1.617.2] or Omicron [B.1.1.529]) may help identify host-virus interactions causing the divergence between asymptomatic, mild, and severe cases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACE2 | R&D Systems | RRID: AB_2305177; BAF933 |
| CD45 biotin | Biolegend | RRID: AB_314392; 304004 |
| CD115 Biotin | Biolegend | RRID: AB_2650968; 347314 |
| Bacterial and virus strains | ||
| OC43 | ATCC | Betacoronavirus 1 Strain OC43 |
| IAV | ATCC | IAV A/PR/8/34 |
| SARS-CoV-2 | BEI Resources | Isolate USA-WA1/2020 |
| Biological samples | ||
| Blood | Blood was drawn by a trained phlebotomist from healthy donors | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dynasore | Selleckchem | S8047 |
| Tripure | Millipore Sigma | 11667165001 |
| Recombinant Human ACE-2 | R&D Systems | 933-ZN |
| Leukotriene B4 (LTB4) | Millipore Sigma | Cat#L0517-10UG |
| CCL2 | Biolegend | 571402 |
| Dalbavancin | Selleckchem | S4848 |
| Pitstop2 | Selleckchem | S9670 |
| Critical commercial assays | ||
| Mesoscale U-PLEX Biomarker Group 1 (hu) Assays | MSD | Kit Catalog Numer: K15067L-1 |
| RealTime-Glo™ Extracellular ATP Assay | Promega | GA5010 |
| Deposited data | ||
| RNA-seq | GEO | GSE186460 |
| scRNAseq | Liao et al., 2020; GEO | GSE145926 |
| Experimental models: Cell lines | ||
| NCI-H441 | ATCC | RRID: CVCL_1561; HTB-174 |
| Oligonucleotides | ||
| Primers | IDT | Table S9 |
| gBlocks | IDT | Table S10 |
| Software and algorithms | ||
| SAMtools | Danecek et al., 2021 | N/A |
| FeatureCounts | Liao et al., 2014 | N/A |
| DESeq2 | Love et al., 2014 | N/A |
| HISAT2 | Siren et al., 2014 | N/A |
| Other | ||
| Streptavidin Coated Magnetic Particles 1.0% w/v 8.0–9.9 μm | Spherotech | SVM-80-5 |
Resource availability
Lead contact
Requests for further information should be directed to and will be fulfilled by the lead contact, Rabindra Tirouvanziam (tirouvanziam@emory.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Maintenance of submerged H441 Club cells
The H441 Club cell line was acquired from ATCC and grown in 50/50 DMEM/F12 supplemented with 1% v/v penicillin/streptomycin and 10% FBS.
In vitro transmigration experiments and infection with virus
The H441 Club cell line is grown on Alvetex scaffolds (ReproCELL, Glasgow, UK) coated with rat-tail collagen (Sigma) for 2 weeks at air liquid interface with 2% v/v Ultroser G (Crescent Chemical, Islandia, NY) in 50/50 DMEM/F12 supplemented with 1% v/v penicillin/streptomycin (Forrest et al., 2018; Grunwell et al., 2019). The filters are then flipped and placed into fresh media in the bottom of the well. Virus (PR8: A/Puerto Rico/8/1934; OC43; or NR-52281, SARS-CoV-2 Isolate USA-WA1/2020) is added to the media such that that the multiplicity of infection (MOI) is 0.1 and incubated for 24 h (unless otherwise indicated) (Figure 1A). This setup requires manual flipping of filters prior to transmigration (Dobosh et al., 2021), a delicate process to perform in BSL3 conditions. Thus, the epithelial cells must be infected while the cells are submerged and no longer at ALI, which may introduce artifacts reminiscent of pneumonia. The filters are transferred to RPMI media with LTB4 (100 nM) and CCL2 (250 pg/mL) with or without additional drugs. Baricitinib and remdesivir were each used at a final concentration of 1 μM. The untreated condition contained 0.1% v/v DMSO as a vehicle control. Blood was collected by venipuncutre in K2-EDTA tubes from healthy donord and monocytes were purified using RosetteSep (StemCell) as described previously (Ford et al., 2021). A total of 106 cells is loaded onto the Alvetex scaffold for transmigration which was allowed to occur for 24 h (Figure 3A). After transmigration, TriPure (Roche) was added to epithelial cells and frozen at −80°C.
Method details
Isolation of monocytes from transmigration fluid
To purify transmigrated cells in a low volume-manner and without the use of a centrifuge, we conjugated 8 μm magnetic beads coated in streptavidin (Spherotech) with biotinylated antibodies targeting CD45 and CD115. Beads and cells were incubated at room temperature for 15 min and the supernatant was removed. The bead precipitate was resuspended in TriPure and frozen at −80°C.
Isolation of nucleic acids
For isolation of RNA (and DNA) from epithelial cells, after thawing, chloroform (Sigma) was added to the TriPure and RNA was extracted using the standard purification procedure in the manufacturer’s protocol followed by a sodium acetate precipitation to further clean the RNA. For isolation of RNA (and DNA) from monocytes, the tubes were thawed and placed on a magnet to remove the beads from solution. The TriPure supernatant was transferred to a clean tube and chloroform was added and spun following the manufacturer’s protocol. Due to the small amount of expected RNA yield, the aqueous phase (containing the RNA) was mixed 1:1 with 100% ethanol (Sigma) and loaded onto an RNA clean and concentrator-5 column (Zymo). RNA was isolated following the manufacturer’s protocol. DNA was isolated from the organic phase following the manufacturer’s protocol.
Cell counting by DNA quantification
Traditional cell counting by hemocytometer was not available in the BSL3. To solve this problem, an estimate of cell number was achieved by quantifying the amount of DNA in the sample. The extracted genomic DNA is amplified using a primer and probe pair which bind an exon-intron junction of AP endonuclease 1 (APEX1; Table S9). These sequences appear once in the genome and the primers and probe do not exhibit off-target amplification. Luna Universal Probe qPCR Master Mix (New England Biolabs) was used to amplify the gDNA. A standard curve to calculate copy number was generated using a double stranded gBlock from IDT which contains the expected binding sequence (Table S10). The copy number was then divided by two to account for diploidy and multiplied by the dilution factor to estimate the total number of cells. Naturally, this number is an estimate as dividing cells as well as dead cells, which have not yet fully degraded their DNA, will elevate the count resulting in an overestimate of the total cell yield.
Flow cytometric analysis of monocytes
Purified blood monocytes from blood before and after transmigration across HLE-ALI cells with or without infection with SARS-CoV-2 were stained (R&D Systems) for the presence of surface ACE2 and then measured by flow cytometry on a Cytoflex S (Beckman Coulter).
Extracellular ATP assay
100 μL of extracellular fluid was centrifuged at 800 xg for 10 min and then used with the Promega Realtime-Glo Extracellular ATP Assay following the manufacturer’s instructions. Luminescence was measured on a SpectraMax iD3 (Molecular Devices).
Quantification of viral RNA via qRT-PCR
Total RNA is reverse-transcribed using SuperScript IV (ThermoFisher) and an anchored oligo-d(T)20 primer followed by amplification with a primer probe pair which targets the N gene (Table S9) and Luna Universal Probe qPCR Master Mix (New England Biolabs) was used to amplify the cDNA. A standard curve to calculate copy number was also generated using a double stranded gBlock from IDT which contains the expected binding sequence (Table S10).
qRT-PCR
RNA is reverse-transcribed using SuperScript IV and anchored oligo-d(T)20 primers to make a cDNA library that can be used multiple times. cDNA is amplified using relevant primer pairs (Table S9) and SYBR Green in an Applied Biosystems 7500. Data are analyzed via the ddCt method for 18S rRNA and GAPDH (reference controls).
RNA quantification and quality control
RNA was initially quantified by Nanodrop 1000 spectrophotometer (ThermoScientific). For samples that would be analyzed by RNA-seq, an aliquot was further quality controlled by an Agilent Bioanalyzer 2100.
Transcriptomic analysis of monocytes
Isolated RNA was reverse transcribed using Superscript IV Reverse Transcriptase (ThermoFisher Scientific) and an anchored oligo-d(T)20 primer following the manufacturer’s protocol. A Fluidigm Biomark machine and Delta Gene Assay chip (96 × 96) was used to conduct multiplexed qPCR of 96 genes and 96 samples. Data was analyzed using a modified version of the jpouch script available in GitHub (https://github.com/jpouch/qPCR-Biomark; Date of Access: May 31st, 2020).
RNA-sequencing of epithelial cells
For RNA-seq, purified RNA was given to the Yerkes Genomics Core for processing. Briefly, rRNA is depleted and the TruSeqRNA Sample Preparation v2 kit and LS protocol is used. All the produced Fastq files from single end reads were aligned to the human reference genome (GRCh38.p13- Ensembl) using the alignment tool HISAT2 (version 2.1.0), using the default settings (Siren et al., 2014). Then, BAM files reads were sorted using SAMtools (Li et al., 2009). The resulting BAM files were used as input to determine read counts using FeatureCounts (1.5.2) (Liao et al., 2014). All processed counts were used to conducted differential expression analysis using DEseq2 (Love et al., 2014), considering as differentially expressed genes, genes with fold changes >2 folds, and a False discovery rate <0.1. Finally, to understand the functions of the differentially expressed genes Metacore server is used against pathway maps and networks. RNA-seq data has been deposited at GEO and are available as of the date of publication. It can be accessed under accession: GEO: GSE186460.
Protein extraction
Once the DNA was removed from the organic phase, the supernatant was mixed 1:1 with 1% v/v SDS and loaded onto a 3,000 kDa MWCO column (Sartorius) and spun at 3,000 xg for 2 min. The column was buffer exchanged with 500 μL of 1% v/v SDS five times.
Quantification of inflammatory mediators
Ten mediators (CXCL8, G-CSF, GM-CSF, IFNγ, IL-1α, IL1β, IL-6, IL-10, IL-18, TNFα) were measured using a chemiluminescent assay according to the manufacturer’s protocol (U-plex; Meso Scale Diagnostics). In general, to measure CXCL8, samples had to be diluted 1:10, but for all other cytokines we did not need to dilute the samples.
Single-cell RNA sequencing
The scRNA-seq FASTQs data from BALF of mild and severe patients were acquired from the GEO database under accession code GEO: GSE145926 (Liao et al., 2020): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145926. The Cell Ranger Software (v.3.1.0) was used to perform barcode processing and single-cell 5′ unique molecular identifier (UMI) counting. To detect SARS-CoV-2 reads, a customized reference was built by integrating human GRCh38 and SARS-CoV-2 genome (severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome, GenBank MN908947.3). Specifically, splicing-aware aligner STAR (Dobin et al., 2013) was used in FASTQs alignment. Cell barcodes were then determined based on the distribution of UMI counts automatically. The filtered gene-barcode matrices were first normalized using ‘LogNormalize’ methods in Seurat v.3 (Stuart et al., 2019) with default parameters. The top 2,000 variable genes were then identified using the ‘vst’ method by the FindVariableFeatures function. To remove the batch effect between the mild and severe datasets, the standard integration workflow of Seurat v.3 was used with the first 30 dimensions from canonical correlation analysis as the input of the ‘FindTransferAnchors()’ function. PCA was performed using the top 2,000 variable genes of the integrated matrix. Then UMAP was performed on the top 30 principal components for visualizing the cells. Meanwhile, graph-based clustering was performed on the PCA-reduced data for clustering analysis with Seurat v.3. The resolution was set to 0.8 to obtain the UMAP.
Quantification and statistical analysis
All statistics were calculated using GraphPad Prism 9.2.0. Details of each test number of including the n, where n is the independent biological replicates, plotted data always indicate the median with the interquartile range, as well as threshold cutoffs for statistical significance are indicated in the legends accompanying the figures.
Acknowledgments
This study was supported by NSF EAGER award 2032273 (to R.T., R.F.S., and K.Z.) and a Woodruff Health Science Center COVID-19 CURE award (to R.T., R.F.S., K.Z., M.B., and V.S.). R.F.S. is also funded in part by NIH grant R01MH116695. We thank Dr. Matt Frieman (University of Maryland) for sharing primer sequences targeting the N-subgenome of SARS-CoV-2. We thank Dalia Gulick, Naima Djeddar, and Gregory Gibson (Georgia Tech) for assisting with multiplexed qRT-PCR. Research reported in this publication was supported in part by the Pediatrics/Winship Flow Cytometry Core of Winship Cancer Institute of Emory University, Children's Healthcare of Atlanta. The graphical abstract was generated in Biorender. SARS-CoV-2 isolate USA-WA1/2020 was obtained from BEI Resources, Manassas, VA.
Author contributions
B.D., K.Z., D.M.G., S.L.G., K.M., M.A., J.L., V.D.G., J.K., D.E., and J.Y. performed and analyzed experiments. M.B., V.S., and E.G. provided conceptualization and input for the work. B.D., K.Z., D.M.G., R.T., and R.F.S. wrote the original manuscript. R.F.S. and R.T. conceptualized the experiments. All authors reviewed and edited the manuscript.
Declaration of interests
R.F.S. is the inventor of the use of baricitinib for coronavirus infections and receives royalties from Eli Lilly. His conflict of interest has been reviewed and approved by Emory University. All other authors have declared that no conflict of interest exists. Contents within this manuscript are included in patent USPTO 10670594.
Published: May 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110945.
Supplemental information
Row 9 contains the list of all genes related to the cellular process GO term relevant to Figure 1C. Please refer to Table S1 for expression values.
All sequences are written from 5’->3’.
All sequences are written as the top strand 5’->3’.
Data and code availability
-
•
RNA-seq data has been deposited at Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. It can be accessed under accession: GEO: GSE186460 with full link here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186460.
This paper analyzes existing, publicly available scRNA-seq data (Liao et al., 2020) and can be accessed from the GEO database under accession code GEO: GSE145926 with full link here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145926.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abassi Z., Knaney Y., Karram T., Heyman S.N. The lung macrophage in SARS-CoV-2 infection: a friend or a foe? Front. Immunol. 2020;11:1312. doi: 10.3389/fimmu.2020.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayati A., Kumar R., Francis V., McPherson P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., De Sanctis F., Cane S., Ugel S., Donadello K., Castellucci M., Eyal D., Fiore A., Anselmi C., Barouni R.M., et al. Deciphering the state of immune silence in fatal COVID-19 patients. Nat. Commun. 2021;12:1428. doi: 10.1038/s41467-021-21702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumaza A., Gay L., Mezouar S., Bestion E., Diallo A.B., Michel M., Desnues B., Raoult D., La Scola B., Halfon P., et al. Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: the clue for coronavirus disease 2019 immunoparalysis. J. Infect. Dis. 2021;224:395–406. doi: 10.1093/infdis/jiab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018;281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobosh B., Giacalone V.D., Margaroli C., Tirouvanziam R. Mass production of human airway-like neutrophils via transmigration in an organotypic model of human airways. STAR Protoc. 2021;2:100892. doi: 10.1016/j.xpro.2021.100892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly S.C., Strieter R.M., Kunkel S.L., Walz A., Robertson C.R., Carter D.C., Grant I.S., Pollok A.J., Haslett C. Interleukin-8 and development of adult respiratory distress syndrome in at-risk patient groups. Lancet. 1993;341:643–647. doi: 10.1016/0140-6736(93)90416-e. [DOI] [PubMed] [Google Scholar]
- Ely E.W., Ramanan A.V., Kartman C.E., de Bono S., Liao R., Piruzeli M.L.B., Goldman J.D., Saraiva J.F.K., Chakladar S., Marconi V.C., et al. Efficacy and safety of baricitinib plus standard of care for the treatment of critically ill hospitalised adults with COVID-19 on invasive mechanical ventilation or extracorporeal membrane oxygenation: an exploratory, randomised, placebo-controlled trial. Lancet Respir. Med. 2022;10:327–336. doi: 10.1016/S2213-2600(22)00006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroshenko N., Gill T., Keaveney M.K., Church G.M., Trevejo J.M., Rajaniemi H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat. Biotechnol. 2020;38:789–791. doi: 10.1038/s41587-020-0577-1. [DOI] [PubMed] [Google Scholar]
- Falasca L., Nardacci R., Colombo D., Lalle E., Di Caro A., Nicastri E., Antinori A., Petrosillo N., Marchioni L., Biava G., et al. Postmortem findings in Italian patients with COVID-19: a descriptive full autopsy study of cases with and without comorbidities. J. Infect. Dis. 2020;222:1807–1815. doi: 10.1093/infdis/jiaa578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford B.D., Moncada Giraldo D., Margaroli C., Giacalone V.D., Brown M.R., Peng L., Tirouvanziam R. Functional and transcriptional adaptations of blood monocytes recruited to the cystic fibrosis airway microenvironment in vitro. Int. J. Mol. Sci. 2021;22:2530. doi: 10.3390/ijms22052530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest O.A., Ingersoll S.A., Preininger M.K., Laval J., Limoli D.H., Brown M.R., Lee F.E., Bedi B., Sadikot R.T., Goldberg J.B., et al. Frontline Science: pathological conditioning of human neutrophils recruited to the airway milieu in cystic fibrosis. J. Leukoc. Biol. 2018;104:665–675. doi: 10.1002/JLB.5HI1117-454RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwell J.R., Giacalone V.D., Stephenson S., Margaroli C., Dobosh B.S., Brown M.R., Fitzpatrick A.M., Tirouvanziam R. Neutrophil dysfunction in the airways of children with acute respiratory failure due to lower respiratory tract viral and bacterial coinfections. Sci. Rep. 2019;9:2874. doi: 10.1038/s41598-019-39726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L.A., Canna S.W., Schulert G.S., Volpi S., Lee P.Y., Kernan K.F., Caricchio R., Mahmud S., Hazen M.M., Halyabar O., et al. On the alert for cytokine storm: immunopathology in COVID-19. Arthritis Rheumatol. 2020;72:1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T.N., Pino M., Boddapati A.K., Viox E.G., Starke C.E., Upadhyay A.A., Gumber S., Nekorchuk M., Busman-Sahay K., Strongin Z., et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell. 2021;184:460–475.e21. doi: 10.1016/j.cell.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira C., Crespo A., Ranjbar S., Ingber J., Parry B., Ravid S., de Lacerda L.B., Lewandrowski M., Clark S., Ho F., et al. SARS-CoV-2 infects blood monocytes to activate NLRP3 and AIM2 inflammasomes, pyroptosis and cytokine release. medRxiv. 2021 doi: 10.1101/2021.03.06.21252796. Preprint at. [DOI] [Google Scholar]
- Kalil A.C., Patterson T.F., Mehta A.K., Tomashek K.M., Wolfe C.R., Ghazaryan V., Marconi V.C., Ruiz-Palacios G.M., Hsieh L., Kline S., et al. Baricitinib plus remdesivir for hospitalized adults with covid-19. N. Engl. J. Med. 2021;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S., Lockey R.F., Kolliputi N. Soluble ACE2 as a potential therapy for COVID-19. Am. J. Physiol. Cell Physiol. 2021;320:C279–C281. doi: 10.1152/ajpcell.00478.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucirica D.R., Schofield C.J., McLean S.A., Margaroli C., Agudelo-Romero P., Stick S.M., Tirouvanziam R., Kicic A., Garratt L.W., Western Australian Epithelial Research Program (WAERP) Australian Respiratory Early Surveillance Team for CF (AREST CF) Pseudomonas aeruginosa modulates neutrophil granule exocytosis in an in vitro model of airway infection. Immunol. Cell Biol. 2022;100:352–370. doi: 10.1111/imcb.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang Y., Lu J., Li L., Gao H., Ma C., Dai E., Wei L. Asymptomatic COVID-19 individuals tend to establish relatively balanced innate and adaptive immune responses. Pathogens. 2021;10:1105. doi: 10.3390/pathogens10091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55:2000607. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B.K., Francisco E.B., Yogendra R., Long E., Pise A., Rodrigues H., Hall E., Herrera M., Parikh P., Guevara-Coto J., Triche T.J., Scott P., Hekmati S., Maglinte D., Chang X., Mora-Rodriguez R.A., Mora J. Persistence of SARS CoV-2 S1 protein in CD16+ monocytes in post-acute sequelae of COVID-19 (PASC) up to 15 Months post-infection. Front. Immunol. 2021;12:746021. doi: 10.3389/fimmu.2021.746021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Alba E., Nuzzolo-Shihadeh L., Aguirre-Garcia G.M., Espinosa-Mora J., Lecona-Garcia J.D., Flores-Perez R.O., Mendoza-Garza M., Camacho-Ortiz A. Baricitinib plus dexamethasone compared to dexamethasone for the treatment of severe COVID-19 pneumonia: a retrospective analysis. J. Microbiol. Immunol. Infect. 2021;54:787–793. doi: 10.1016/j.jmii.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontelli M.C., Castro I.A., Martins R.B., Veras F.P., Serra L.L., Nascimento D.C., Cardoso R.S., Rosales R., Lima T.M., Souza J.P., et al. Infection of human lymphomononuclear cells by SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.07.28.225912. Preprint at. [DOI] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V.U.S., Arakeri G., Subash A., Rao J., Jadhav S., Suhail Sayeed M., Rao G., Brennan P.A. COVID-19: loss of bridging between innate and adaptive immunity? Med. Hypotheses. 2020;144:109861. doi: 10.1016/j.mehy.2020.109861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra N.G., Alfajaro M.M., Gasque V., Huston N.C., Wan H., Szigeti-Buck K., Yasumoto Y., Greaney A.M., Habet V., Chow R.D., et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021;19:e3001143. doi: 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Kramer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E., Qu R., Junqueira C., Kaffe E., Mirza H., Zhao J., Brewer J.R., Han A., Steach H.R., Israelow B., et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. bioRxiv. 2022 doi: 10.1101/2021.09.27.461948. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siren J., Valimaki N., Makinen V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans Comput. Biol. Bioinform. 2014;11:375–388. doi: 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- Stebbing J., Krishnan V., de Bono S., Ottaviani S., Casalini G., Richardson P.J., Monteil V., Lauschke V.M., Mirazimi A., Youhanna S., et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 2020;12:e12697. doi: 10.15252/emmm.202012697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., Shi Y. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16:e1008536. doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., et al. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94 doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Yang M.L., Duan Z.L., Liu F.L., Jin L., Long C.B., Zhang M., Tang X.P., Xu L., Li Y.C., et al. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S., Frieman M.B. COVID-19: knowns, unknowns, and questions. mSphere. 2020;5:e00203–e00220. doi: 10.1128/mSphere.00203-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhou X., Zhu C., Song Y., Feng F., Qiu Y., Feng J., Jia Q., Song Q., Zhu B., Wang J. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front. Mol. Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host. Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E.I., Macal M., Lewis G.M., Harker J.A. Innate and adaptive immune regulation during chronic viral infections. Annu. Rev. Virol. 2015;2:573–597. doi: 10.1146/annurev-virology-100114-055226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Row 9 contains the list of all genes related to the cellular process GO term relevant to Figure 1C. Please refer to Table S1 for expression values.
All sequences are written from 5’->3’.
All sequences are written as the top strand 5’->3’.
Data Availability Statement
-
•
RNA-seq data has been deposited at Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. It can be accessed under accession: GEO: GSE186460 with full link here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186460.
This paper analyzes existing, publicly available scRNA-seq data (Liao et al., 2020) and can be accessed from the GEO database under accession code GEO: GSE145926 with full link here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145926.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.