Abstract
This study used phylogenetic probes in hybridization analysis to (i) determine in situ microbial community structures in regions of a shallow sand aquifer that were oxygen depleted and fuel contaminated (FC) or aerobic and noncontaminated (NC) and (ii) examine alterations in microbial community structures resulting from exposure to toluene and/or electron acceptor supplementation (nitrate). The latter objective was addressed by using the NC and FC aquifer materials for anaerobic microcosm studies in which phylogenetic probe analysis was complemented by microbial activity assays. Domain probe analysis of the aquifer samples showed that the communities were predominantly Bacteria; Eucarya and Archaea were not detectable. At the phylum and subclass levels, the FC and NC aquifer material had similar relative abundance distributions of 43 to 65% β- and γ-Proteobacteria (B+G), 31 to 35% α-Proteobacteria (ALF), 15 to 18% sulfate-reducing bacteria, and 5 to 10% high G+C gram positive bacteria. Compared to that of the NC region, the community structure of the FC material differed mainly in an increased abundance of B+G relative to that of ALF. The microcosm communities were like those of the field samples in that they were predominantly Bacteria (83 to 101%) and lacked detectable Archaea but differed in that a small fraction (2 to 8%) of Eucarya was detected regardless of the treatment applied. The latter result was hypothesized to reflect enrichment of anaerobic protozoa. Addition of nitrate and/or toluene stimulated microbial activity in the microcosms, but only supplementation of toluene alone significantly altered community structures. For the NC material, the dominant subclass shifted from B+G to ALF, while in the FC microcosms 55 to 65% of the Bacteria community was no longer identifiable by the phylum or subclass probes used. The latter result suggested that toluene exposure fostered the proliferation of phylotype(s) that were otherwise minor constituents of the FC aquifer community. These studies demonstrated that alterations in aquifer microbial communities resulting from specific anthropogenic perturbances can be inferred from microcosm studies integrating chemical and phylogenetic probe analysis and in the case of hydrocarbon contamination may facilitate the identification of organisms important for in situ biodegradation processes. Further work integrating and coordinating microcosm and field experiments is needed to explore how differences in scale, substrate complexity, and other hydrogeological conditions may affect patterns observed in these systems.
Aromatic hydrocarbons are pervasive contaminants of terrestrial environments, particularly shallow aquifers. There are a variety of anthropogenic sources for these chemicals, but the most environmentally significant are hydrocarbon fuel mixtures leaking from buried storage tanks. In situ bioremediation is often considered for cleanup or containment of hydrocarbon-contaminated groundwater, and these applications have generated increasing interest in anaerobic processes. Electron acceptor demands typically created in aquifers following fuel contamination greatly exceed dissolved oxygen supplies (9, 28). Thus, the systems become anaerobic, and biodegradation processes operative under these conditions are important for supporting intrinsic bioremediation (9, 28). Anaerobic processes have also been considered for potential use in enhanced bioremediation to obviate difficulties associated with introducing, dispersing, and maintaining oxygen at levels adequate to sustain aerobic biodegradation (33). These applications for electron acceptor supplementation with nitrate have attracted attention because of its high water solubility and general lack of noxious by-products (23, 24).
Elucidation of the microbiological and hydrogeochemical influences on anaerobic biodegradation of hydrocarbons in aquifers is needed to gain an improved understanding of both intrinsic and enhanced bioremediation processes (33). Microbiological investigations focusing on laboratory microcosm and selective enrichment experiments have demonstrated anaerobic degradation of aromatic hydrocarbons under redox conditions ranging from nitrate-reducing to methanogenic (18, 24, 29) and allowed identification of axenic cultures (14, 15, 26, 27, 37) or defined consortia (7) of anaerobic, aromatic hydrocarbon degraders. Hydrogeochemical studies have demonstrated the impacts of hydrocarbon contamination on altering temporal and spatial zonation of dominant terminal electron-accepting processes (7, 25). Collectively, these findings indicate that in contaminated aquifers, anaerobic degradation of hydrocarbons is mediated by a diversity of organisms, the nature and variety of which change in time and space.
While community successions appear to be a fundamental aspect of the microbial ecology underlying anaerobic hydrocarbon biodegradation, there is relatively little information on this process, partly because the diversity of the microbial trophic groups that comprise these communities has rendered culture-based techniques alone inadequate to comprehensively evaluate community structure. However, the recent development of phylogenetic probes, which may be used to detect and quantify specific taxonomically defined groups of microorganisms without culturing, offers microbial ecologists the possibility of gaining new insights into microbial population dynamics. For analysis of microbial community structure in complex environments, a suite of phylogenetically nested probes may be used to conduct analysis of the population in hybridization assays from the top down (5). In these experiments, amounts of broad-spectrum (e.g., domain-level) probes hybridizing to nucleic acids extracted from the environment are quantified, and the community structure is further defined in subsequent hybridization assays by using probes with progressively greater specificity. However, because phylogenetic probes do not directly assay function, their utility for deciphering potential activities of the targeted organisms is strengthened when they are integrated with physical-chemical analysis of the environment that facilitates the relation of phylotype to ecotype.
Phylogenetic probe analysis has been applied to study microbial communities in a variety of environments (12, 35, 39, 42), but there have been relatively few examinations of aquifers. Fry et al. (16) used this approach to analyze groundwater from two deep (>300 m below ground) aquifers and showed that the populations were similar in that they were dominated by Bacteria, with Eucarya comprising a smaller but significant fraction (5 to 14%). Archaea were detected at low levels (<2%) and were more abundant in groundwater that was methanogenic. Hess et al. (21) applied in situ hybridization in a laboratory study of fuel-contaminated (FC) aquifer material incubated under denitrifying conditions. These experiments demonstrated that the microbial community structure varied with electron acceptor gradients and that the β-Proteobacteria appeared to predominate under denitrifying conditions. Detection of Azoarcus spp. with specific probes perhaps strengthened this hypothesis, as this organism is classified under β-Proteobacteria and has members that are known aromatic hydrocarbon-degrading denitrifiers (15). Dojka et al. (13) conducted a phylogenetic survey of an anaerobic contaminated aquifer. They examined partial 16S rRNA gene sequences from 104 clones and identified representatives of established phylogenetic groups as well as several that were apparently of unknown lineage. In addition, some gene sequences were encountered more frequently in areas with a particular redox condition. This study provided evidence of the vast diversity of microorganisms inhabiting contaminated aquifers but did not develop correlations between changes in groundwater chemistry and changes in microbial populations, which might help to identify organisms involved in biodegradation processes.
The objectives of this study were to evaluate the use of phylogenetic probes to identify alterations in aquifer microbial community structure elicited by anthropogenic perturbances. Analyses were done on FC and noncontaminated (NC) aquifer material taken directly from the field and on aquifer materials used to establish anaerobic microcosms supplemented with toluene and/or nitrate. The latter experiments were done to gain information on the effects of specific, isolated variables relevant to the bioremediation of anaerobic aquifers and to facilitate the establishment of phylotype-ecotype linkages.
MATERIALS AND METHODS
Aquifer characterization and sampling.
Aquifer samples were obtained from a military installation (Fort McCoy; Sparta, Wis.) where fuel hydrocarbons leaking from buried tanks and transfer lines had contaminated a shallow (1.4 m below ground surface) sand aquifer. Groundwater samples were collected from miniature multilevel sampling devices (37) installed to sample the aquifer near the water table surface at 0.6-m intervals to a depth of 11 m. A peristaltic pump was attached to the sampling lines, and after approximately 3 well volumes were withdrawn, water samples were collected for analysis of redox indicators (dissolved oxygen, NO3−, Fe2+, SO42−, HS−) and volatile organic compounds (VOC). All redox indicators except HS− were analyzed immediately in the field by using colorimetric techniques with CHEMets kits (CHEMetrics, Inc., Claverton, Va.). Sulfide analysis was done by ion chromatography at the University of Wisconsin-Madison, Soil and Plant Analysis Laboratory, and VOC determinations were done by the U.S. Army Analytical Laboratory (Sauk City, Wis.).
To obtain aquifer material samples, a Geoprobe (Geoprobe Systems, Salina, Kans.) was used to core to a depth of 1.4 m. The coring tube was retrieved, a sterile acrylic sleeve was inserted, and then the device was driven an additional 0.6 m into the aquifer. The acrylic sleeve was then removed, capped, sealed with duct tape, and stored on ice for transport to the laboratory. The same procedure was used to obtain aquifer samples from a parallel (80-m horizontal separation), noncontaminated portion of the aquifer. Cores were used immediately for microcosm establishment and/or DNA extraction. All of these procedures were completed by the end of the day following sample acquisition. Additional core samples were taken for physical-chemical characterization, including organic matter content, which was done at the University of Wisconsin-Madison, Soil and Plant Analysis Laboratory, by dry combustion. Direct counts of microbial population densities were done by staining with 4,6-diamidino-2-phenylindole (DAPI) and epifluorescence microscopy as described by Bottomley (8).
Microcosm experiments.
Microcosms were established in 100-ml (nominal volume) serum bottles under an N2 atmosphere in an anaerobic glovebox (model HE-493; Vacuums Atmosphere Co., Hawthorn, Calif.). Each microcosm contained 50 g of homogenized, sediment material from FC and NC regions of the aquifer and 50 ml of base medium. The base medium was composed of 330 μM KH2PO4 and 70 μM NH4Cl and was adjusted to pH 7 with NaOH (24). Duplicate microcosms were then amended to a final concentration of 2 mM KNO3 and/or 250 μM toluene. Duplicate microcosms containing the sediment and base medium alone were used as nonamended controls. The serum bottles were sealed with Teflon-coated butyl rubber stoppers affixed with aluminum crimp caps and then incubated inverted in the dark with continuous, gentle agitation on an orbital shaker (150 rpm) at 24°C. Agitation was applied to facilitate diffusion and did not generate a turbulent, water sediment suspension or disturb the sediment in any way.
During incubation, headspace and/or aqueous samples were taken and analyzed to track substrate consumption (toluene, NO3−) and product formation (NO2−, N2O, CO2). To monitor NO2− and NO3− levels, aqueous samples (0.5 ml) were periodically taken with a 1-ml tuberculin syringe and assayed by colorimetric methods (19, 43). Additional 0.5-ml aliquots were taken for toluene analysis. These samples were immediately injected into sealed gas chromatograph (GC) vials (2-ml nominal volume) containing 0.5 ml of n-pentane. The vials were mixed by vortexing (5 s), and 0.3-ml aliquots of the organic layer were transferred to sealed 2-ml GC vials. Toluene was determined by using a Hewlett-Packard (Palo Alto, Calif.) 6890A GC equipped with a Hewlett-Packard 5972A mass-selective detector, Rtx-Wax capillary column (30 m by 250 μm; film thickness, 0.25 μm; Alltech Associates, Deerfield, Ill.), a Hewlett-Packard 61513A autosampler, a split-splitless capillary column injection port (held at 30°C), and a constant helium carrier gas flow of 1.0 ml min−1. After injection (1 μl), the oven was held at 30°C for 5 min and then heated at 5°C min−1 to 80°C (1 min hold). The detector was operated in electron ionization mode (70 eV) scanning atomic mass units ranging from 50 to 550 at 1.53 s decade−1. Carbon dioxide and N2O were analyzed in 1-ml headspace samples on a Hewlett-Packard 5890A GC fitted with a thermal conductivity detector (TCD). Isothermal (45°C) separation was achieved with a Haysep R packed column (6 ft by 1/8 in.; Alltech) and a helium carrier gas flow rate of 25 ml min−1. The injector and detector were held at 80 and 100°C, respectively. The packed column-TCD method also allowed separation and quantification of CH4, but this gas was never detected in headspace samples from the microcosms. The detection limits were (analyte) 13 nmol (CH4), 30 nmol (CO2), and 40 nmol (N2O).
For mass balance calculations, growth equation 1 was as follows: 1.1272 C7H8 + NH4+ + 0.08 H2PO4− + 0.03 SO42− + 4.6 NO3− + 3.74 H+ = C4H7.3O1.8NP0.08S0.03 (bacterial cells) + 3.8906 CO2 + 2.3 N2 + 4.7218 H2O. Including trace elements, the formula weight for a nitrogen (N) mole of bacterial cells was 103.5, which was assumed to be 4% (wt/wt) DNA (20).
Statistical analysis.
Data were analyzed with repeated-measures analysis of variance. Means were compared with pairwise comparisons of the estimated population marginal means, by using the statistical procedure PROC MIXED (SAS Institute Inc., Cary, N.C.).
Nucleic acid extraction techniques.
A variety of nucleic acid extraction procedures were evaluated to determine the optimal technique for extracting the most DNA from the aquifer materials used in this study. The nucleic acid extraction protocol ultimately adopted for routine use was a lysozyme–freeze-thaw cell lysis technique for DNA extraction modified from that reported by Tsai and Olsen (41). Briefly, aquifer material (50 g) was suspended in 50 ml of TE buffer containing 0.3% Tween 20 and mixed for 2 h on a rotary shaker (150 rpm, 26°C). The supernatant was transferred to a 35-ml centrifuge tube and spun at 8,200 × g (20 min, 4°C). The pellets were resuspended in 5 ml of lysis solution (15 mg of lysozyme in TE ml−1) and incubated at 37°C for 1.5 h. After incubation, sodium dodecyl sulfate was added to a final concentration of 2% (wt/vol) and the sample was subjected to three freeze-thaw cycles (−70°C, 65°C). The solution was then extracted twice (1:1 [vol/vol]) with TE-buffered phenol and then twice (1:1 [vol/vol]) with chloroform. DNA was precipitated with ethanol and sodium acetate as described above with 2 volumes of 100% ethanol and 0.1 volume of 3 M sodium acetate overnight at −20°C. The solutions were then centrifuged (8,200 × g, 20 min), and the resulting DNA pellets were washed once with 75% ethanol and then dissolved in double-distilled water (ddH2O).
Nucleic acid purification and quantification.
A two-step chromatographic purification system was adopted for routine application to the extracts, consisting of sequential filtration over Sepharose 2B (Sigma, St. Louis, Mo.) and Wizard Plus Minipreps (Promega, Madison, Wis.). For the first step, Sepharose gels were hydrated in 40 ml of TE buffer overnight and loaded into 5-ml syringes plugged with glasswool, which were then spun at 1,200 × g (10 min). Next, crude DNA extracts (200 to 400 μl) were loaded, and the tubes were spun at 1,200 × g (10 min). The effluents were collected, and the columns were washed twice with 100 μl of TE buffer. The effluents were then combined, and DNA was precipitated as described above and dissolved in ddH2O. In the second step, the samples were further purified by using Wizard spin-columns according to the manufacturers’ instructions.
To determine DNA yields and recoveries, aliquots of crude or purified extracts were electrophoresed in 1% agarose gels, stained with ethidium bromide, and photographed. The photos were then scanned into an image analysis program (IPLab gel; Scanalytics Inc., Vienna, Va.), and the DNA content of the samples was determined by interpolation from standard response curves developed with bacteriophage lambda DNA. Based on these DNA yields (140 to 740 ng g−1), and assuming an average cellular DNA content of 2 fg (6), the calculated microbial population density was 2 × 107 to 4 × 108 cells g−1; this was in good agreement with population densities determined by epifluorescence microscopy direct counts.
Hybridization analysis.
Nucleic acid samples were transferred to Hybond-N+ nylon membranes (Amersham, Arlington Heights, Ill.), by using a minifold II 72-well slot-blot manifold (Schleicher and Schuell, Keene, N.H.), and cross-linked to the membranes by exposure to 120 mJ of UV light energy with a UV Stratalinker 1800 (Stratagene, Torrey Pines, Calif.). All oligonucleotides used as probes (Table 1) were conjugated at the 5′ ends to digoxigenin by the supplier (NBI, Plymouth, Minn.). The probes were tested and calibrated with reference genomic DNAs prepared from 19 organisms representing the α-, β-, and γ-Proteobacteria, sulfate-reducing bacteria, high-G+C gram positives, low-G+C gram positives, Eucarya, and Archaea (Table 2). Hybridization and wash conditions were empirically optimized for each probe. Probe-target hybrids were detected following a ≤45-min exposure of the membranes to X-ray film by using the Genius chemiluminescence system (Boehringer Mannheim, Indianapolis, Ind.). Rapid-Hyb buffer (Amersham) was used instead of the Easi-Hyb buffer supplied with the Genius system, because in comparative tests the former yielded much stronger hybridization signals.
TABLE 1.
Summary of sequences, target groups, and hybridization conditions for oligonucleotides used as phylogenetic probes
| Probea | Sequence (5′ to 3′) | Target | Temp (°C)b | Reference |
|---|---|---|---|---|
| S-*-Univ-1390-a-A-18 | GAC GGG CGG TGT GTA CAA | All organisms (universal) | 54 | 44 |
| S-D-Bact-0338-a-A-18 | GCT GCC TCC CGT AGG AGT | Bacteria | 42 | 4 |
| S-D-Euca-1379-a-A-16 | TAC AAA GGG CAG GGA C | Eucarya | 40 | 22 |
| S-D-Arch-0915-a-A-20 | GTG CTC CCC CGC CAA TTC CT | Archaea | 50 | 39 |
| S-Sc-aProt-0019-a-A-17 | CGT TCG (C/T)TC TGA GCC AG | α-Proteobacteria | 47 | 31 |
| L-Sc-bProt-1027-a-A-17 | GCC TTC CCA CTT CGT TT | β-Proteobacteria | 50 | 31 |
| L-Sc-gProt-1027-a-A-17 | GCC TTC CCA CAT CGT TT | γ-Proteobacteria | 47 | 31 |
| S-*-Srb-0385-a-A-18 | CGG CGT CGC TGC GTC AGG | Sulfate-reducing bacteria | 55 | 3 |
| L-P-Grps-1901-a-A-18 | TAT AGT TAC CAC CGC CGT | High-G+C gram positives | 47 | 36 |
| L-S-E. coli-1531-a-A-21 | CAC CGT AGT GCC TCG TCA TCA | E. coli | 50 | 34 |
Naming convention proposed by Alm et al. (1).
Empirically optimized for hybridization and membrane washes.
TABLE 2.
Microbial cultures used to prepare reference DNAs for initial optimization of probe hybridization conditions and routine probe calibration
| Taxonomic group and organism | Culture sourceb |
|---|---|
| Eucarya | |
| Penicillium crustosuma | 1 |
| Archaea | |
| Methanobacterium formicicuma | 2 |
| Bacteria | |
| Proteobacteria | |
| α | |
| Azospirillum brasilensea | 3 |
| Rhodospirillum rubrum | 4 |
| Sinorhizobium meliloti | 5 |
| Rhizobium loti | 5 |
| β | |
| Burkholderia cepacia | 6 |
| Alcaligenes sp.a | 7 |
| γ | |
| Escherichia colia | 8 |
| Pseudomonas fluorescens | 9 |
| Pseudomonas aureofaciens | 9 |
| Serratia plymuthicaa | 9 |
| Xanthomonas campestris | 9 |
| Enterobacter cloacae | 9 |
| Azotobacter vinelandii | 4 |
| Klebsiella pneumoniae | 10 |
| δ | |
| Desulfovibrio desulfuricansa | 11 |
| Gram positive | |
| Low G+C | |
| Bacillus cereus | 9 |
| High G+C | |
| Arthrobacter oxydans | 12 |
| Arthrobacter globiformisa | 12 |
| Streptomyces bikiniensis | 12 |
| Streptomyces coelicolor | 12 |
| Streptomyces griseus | 12 |
DNA from this organism was used for routine probe calibration.
Sources were as follows: 1, J. Parke (University of Wisconsin-Madison); 2, P. Weimer (University of Wisconsin-Madison); 3, American Type Culture Collection (ATCC; Rockville, Md.) strain 29145; 4, G. Roberts (University of Wisconsin-Madison); 5, J. Lindquist (University of Wisconsin-Madison); 6, S. Fetzner (University of Hohenheim); 7, R. C. Wyndham (Carleton University); 8, authors’ lab collection; 9, R. Zablotowicz (U.S. Department of Agriculture-ARS, Stoneville, Miss.); 10, B. Thede (Ciba-Giegy Corp., Greensboro, N.C.); 11, ATCC strain 27774; and 12, J. Ensign (University of Wisconsin-Madison).
Under the optimized hybridization conditions, the probes effectively discriminated between target and nontarget groups. However, we did observe the β-Proteobacteria probe hybridized to γ-Proteobacteria DNA, consistent with the findings of Manz et al. (31), who developed the probe. There is currently no comprehensive probe for the δ-Proteobacteria, but several oligonucleotides have been described that vary in coverage of this subclass (3, 11). From these, we selected the SRB385 probe designed by Amann et al. (3), because comparison of this oligonucleotide to the Ribosomal Database Project (30) gave the greatest number of exact matches to sulfate-reducing bacteria (SRB). In addition to SRBs, this probe may detect other δ-Proteobacteria, some α-Proteobacteria, and some gram positives (2). Thus, references herein to SRB detected by this probe are made with the realization that the scope of this probe may extend beyond sulfate reducers.
Hybridization signals for each probe were routinely calibrated by hybridizing to genomic DNA extracted from representative organisms (Table 2). Blots with DNAs from the indicated groups were used to establish conditions that gave maximum signal to the intended targets and minimal cross-reaction with nontargets. Under the optimized conditions, cross-reactions were negligible to nondetectable. The relative abundance of domains or subgroups was determined by normalizing hybridization signals to the signal generated from hybridization to a universal probe (16, 44). Aliquots of ddH2O containing the denaturing solution were blotted as negative controls and used for background correction. To quantify hybridization signals, the membranes were scanned and analyzed by image analysis as described above.
RESULTS AND DISCUSSION
Chemical and molecular characterization of field samples.
In the FC region of the aquifer, total BTEX (benzene, toluene, ethylbenzene, and xylene) levels ranged from 4,000 to >5,000 μg liter−1, reflecting significant spatial and temporal variation. Natural organic matter content ranged from 18 to 53 g kg−1, reflecting vertical heterogeneity of the aquifer material resulting from shifting patterns of fluvial deposition and development of peat in a riparian wetland. Samples collected from the NC portion of the aquifer were lower in natural organic matter (2 to 31 g kg−1), and groundwater from this location contained no detectable fuel hydrocarbons. Although peat layers also existed in the NC portion of the aquifer, these were not recovered in the samples examined in these experiments. The NC and FC areas also differed in that the groundwater chemistry of the former indicated predominantly aerobic conditions (pH 7.0 to 7.5) with 1 to 5 mg of dissolved oxygen (DO) liter−1 and no detectable Fe2+. In the FC region, DO concentrations across the 0.6-m sampling interval ranged from 0 (nondetectable) to 0.1 mg liter−1, while Fe2+ levels were 10 to 100 mg liter−1 (pH 5.6 to 6.2). Additional evidence for the occurrence of anaerobic processes in the FC area was a depletion of other dissolved electron acceptors compared to the concentrations measured in the NC section. Specifically, in the NC area NO3− ranged from 6 to 10 mg liter−1 and SO4−2 ranged from 10 to 20 mg liter−1, while in the FC region NO3− was nondetectable and SO4−2 concentrations varied from 0 to 10 mg liter−1, respectively.
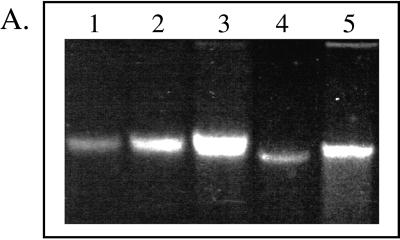
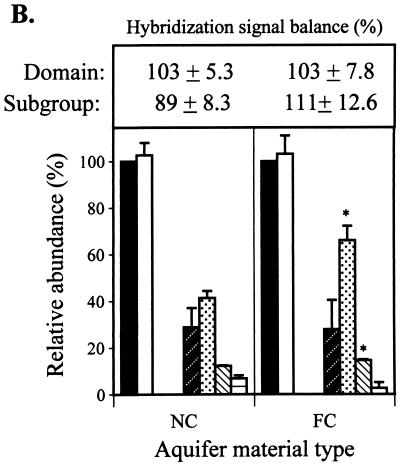
High-molecular-weight DNA was recovered from aquifer material samples (Fig. 1A). DNA yields from the NC and FC aquifer material were 7 and 37 μg, respectively. The greater DNA yield from the latter material may have reflected a higher microbial population density established in response to the higher native organic matter levels and/or the introduction of fuel hydrocarbons. Domain probe analysis of the aquifer samples showed that the DNA extracts were essentially all Bacteria (Fig. 1B); Eucarya and Archaea were nondetectable (≤0.5 ng of DNA or 0.25% of the community DNA analyzed in a typical blot). At the phylum and subclass levels, the FC and NC aquifer material showed similar relative abundance patterns: 43 to 65% β- and γ-Proteobacteria (B+G) > 31 to 35% α-Proteobacteria (ALF) > 15 to 18% SRB > 5 to 10% high-G+C gram positives (HGC). Compared to the NC zone, the FC aquifer community structure had a significantly greater abundance of B+G (P ≤ 0.05). The suite of probes applied gave sufficiently comprehensive coverage of the community, as reflected in the sums of normalized domain signals and subgroup probes (Fig. 1).
FIG. 1.
(A) Agarose gel analysis of DNA extracted from aquifer
field samples and standards. Lanes 1 to 3, Pseudomonas
genomic DNA loaded at 0.59, 1.18, and 2.47 μg; lanes 4 and 5,
aliquots (10 μl) of purified DNAs extracted from the NC or FC
sediment (total volumes of the DNA extracts were 100 and 600 μl for
the NC and FC extracts, respectively). (B) Relative abundance
(universal probe-normalized signals) of domains and subgroups (phylum
and subclass levels) in the NC and FC aquifer samples. The sums of
domain and subgroup hybridization signals (± standard deviations) are
given in the box above the graph. Legend for probe target groups: ■,
universal; □, Bacteria;
 ,
Eucarya;
,
Eucarya;
 , ALF;
, B+G;
, ALF;
, B+G;
 , SRB;
, SRB;
 , HGC. Data are
means of duplicate measurements (± standard deviations). An asterisk
indicates that the hybridization signal from a given probe differed
significantly (P ≤ 0.05) from that for the nonamended
control.
, HGC. Data are
means of duplicate measurements (± standard deviations). An asterisk
indicates that the hybridization signal from a given probe differed
significantly (P ≤ 0.05) from that for the nonamended
control.
The differences in levels of oxidized and reduced electron acceptors between the FC and NC zones were consistent with the expected effect of the increased organic electron donor level imparted by fuel contaminants. Conditions in the NC area were predominantly aerobic, while those in the FC region were microaerophilic to anaerobic, with the terminal electron-accepting processes (TEAPs) that were potentially operative ranging from aerobic respiration to sulfate reduction. Thus, the FC region was perhaps best characterized as a heterogeneous environment in terms of TEAP use.
The dominance of Bacteria, and the Proteobacteria specifically, might have been anticipated, given that organisms comprising the latter group are collectively capable of using all of the TEAPs indicated by the chemical analyses of groundwater as potentially important in the FC and NC zones. The only electron acceptor excluded from use by the Proteobacteria is carbon dioxide (methanogenesis), which the lack of detectable Archaea suggested was not a major process in the aquifer. The most obvious difference in community structure between the NC and FC regions was the greater abundance of B+G relative to ALF in the FC sample. This shift could be interpreted to indicate that ALF and B+G were codominant in the pristine aquifer but that positive- and/or negative-selective pressures imposed by fuel contamination, as well as shifting electron acceptor availability, resulted in proliferation of B+G at the expense of ALF.
A more subtle difference between the aquifer samples was the increased abundance of SRB (P ≤ 0.05) in the FC zone. Since the SRB probe is expected to detect many types of sulfate reducers, these results could be regarded as corroborating the chemical evidence for sulfate reduction as a functional TEAP in the FC region. However, because the SRB probe range extends beyond the sulfate reducers, it is also possible that the increased SRB abundance reflects the proliferation of phylotypes with TEAPs other than (or in addition to) sulfate reduction. The SRB also constituted a significant fraction of the microbial community in the aerobic, NC region. The establishment of this SRB population did not appear to be attributable to the transient development of anaerobic conditions because more than 2 years’ monitoring of groundwater conditions in this region has consistently shown moderate to high DO levels (38). Thus, SRB probe hybridization to the NC region community probably indicates detection of organisms other than sulfate reducers.
Chemical and molecular analyses of NC sediment microcosms.
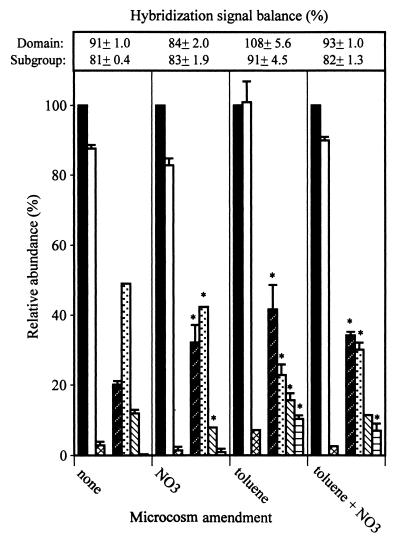
Toluene supplementation stimulated microbial growth as indicated by levels of CO2 production and DNA yields compared to those of nonamended controls (Table 3). Toluene degradation was detectable between days 10 and 12 and coincided with the period of enhanced CO2 production. The total amount of CO2 was not significantly different from the 32-μmol amount expected based on equation 1. The DNA extracted from the nonamended and toluene-amended microcosms totaled 6 and 9 μg, respectively. The amount of toluene added should have supported the production of ca. 88 μg of bacterial biomass containing 3.5 μg of DNA. Thus, the 3 μg of additional DNA produced in the toluene-amended treatment agreed well with the theoretical yield and indicated effective recovery of DNA from the microcosms. Domain probe analysis showed that in both the toluene-amended and nonamended flasks, Bacteria accounted for 85 to 95% of the community DNA, Eucarya accounted for 2 to 8%, and Archaea were nondetectable (Fig. 2). Shifts in microbial community structure were detectable at the phylum and subclass levels. In the nonamended microcosm, the relative abundance was 50% B+G, 20% ALF, 15% SRB, and 2% HGC (Fig. 2). In the toluene-supplemented microcosms there was reversal of ALF versus B+G as the dominant subclass and significant increases in the SRB and HGC groups (Fig. 2).
TABLE 3.
Chemical indicators of microbial activity measured in the NC microcosmsa
| Indicator | Microbial activity for microcosm with
indicated amendment (μmol)
|
|||
|---|---|---|---|---|
| None | Toluene | NO3− | Toluene + NO3− | |
| CO2 | 13.22 ± 4.91 | 30.71 ± 3.99b | 39.46 ± 7.82b | 33.10 ± 4.84b |
| NO3− | 0.77 ± 1.11 | 0.49 ± 0.03 | 84.00 ± 6.98 | 15.01 ± 3.72 |
| NO2− | NDc | ND | 1.79 ± 2.19 | 1.83 ± 0.06 |
| N2O | ND | ND | 9.12 ± 1.03b | 1.34 ± 0.32 |
Determined at the conclusion of the 18-day incubation. Data are means ± standard deviations.
This value is significantly different (P < 0.05) from that for the nonamended control (CO2) or those for the nitrate-amended treatments (NO3−, NO2−, and N2O).
ND, not detected.
FIG. 2.
Molecular analysis of DNAs extracted from microcosms established with the NC aquifer material. Relative abundance of the domain or subgroups was determined by hybridization to the indicated probe (see the legend to Fig. 1 for the key to probe abbreviations). Bars marked with an asterisk were significantly different (P ≤ 0.05) from those for the nonamended microcosms. Probes for which data are not plotted gave hybridization signals that were below background.
Nitrate amendment also stimulated CO2 production and enhanced DNA yields (Table 3). The total DNA yield from nitrate-amended microcosms was 17 μg. The electron donor source in this case was presumed to be native organic material. Nitrate consumption paralleled CO2 formation and production of NO2− and N2O. Nitrite accumulations were greatest within the first week of incubation (4 to 12 μmol) and subsequently steadily decreased. Production of N2O was also detectable by the first week, and after 8 to 10 days reached a consistent level of 12 μmol. Nitrite and N2O were not detected in microcosms that did not receive NO3− supplementation. Formation of these nitrate reduction products provided evidence that populations of denitrifying organisms were stimulated. However, molecular analysis of the NO3−-amended aquifer material showed no significant differences in community structure at the domain or subgroup level compared to that of the nonamended control.
Combining the amendments did not significantly increase CO2 production or DNA yields (13 μg total) above levels obtained with the individually applied supplements (Table 3). Nitrate addition supported more rapid toluene degradation compared to that of the microcosms spiked with toluene alone, and toluene supplementation decreased levels of NO2− and N2O accumulation. These results suggested linkage of toluene degradation to nitrate reduction. The community structure at all the levels examined was similar to that determined for the microcosms amended with toluene alone. The suite of probes applied gave sufficiently comprehensive coverage of the microbial population as reflected in the sums of normalized hybridization signals (Fig. 2). For the NC sediment, the sum of domain level signals (normalized to the universal probe) ranged from 84 to 108% and from 81 to 91% for the phylum and subclass categories combined (Fig. 2).
Two lines of evidence allowed us to rule out methanogenesis as a significant TEAP. First, molar ratios of CO2 to CH4 produced during chemoorganotrophic growth of methanogens might range from 1:1 to 1:3; chemolithotrophic growth results in carbon dioxide depletion and methane enrichment (17). Given that the measured CO2 levels were on the order of 20 to 40 μmol (Tables 3 and 4), methane should have been easily detectable (i.e., >1,000-fold excess of the detection limit) even if methanogenesis accounted for only a fraction of the total CO2 production. Second, methanogenesis could be dismissed as occurring in the nitrate-amended microcosms according to the first principle of thermodynamics, namely, that in an anaerobic environment dissimilatory nitrate reduction will be the dominant TEAP as long as nitrate is present at appreciable levels. The chemical analysis of the microcosms (Tables 3 and 4) showed that this was clearly the case throughout the course of the study.
TABLE 4.
Chemical indicators of microbial activity measured in the FC microcosmsa
| Indicator | Microbial activity for microcosm with
indicated amendment (μmol)
|
|||
|---|---|---|---|---|
| None | Toluene | NO3− | Toluene + NO3− | |
| CO2 | 27.18 ± 3.36 | 41.16 ± 7.97b | 27.66 ± 6.18 | 42.84 ± 4.40b |
| NO3− | NDc | 0.01 ± 0.01 | 17.52 ± 4.69 | 15.55 ± 3.16 |
| NO2− | ND | ND | 1.14 ± 0.87 | 0.39 ± 0.07 |
| N2O | ND | ND | 11.62 ± 4.14b | 0.23 ± 0.34 |
Determined at the conclusion of the 18-day incubation. Data are means ± standard deviations.
This value is significantly different (P < 0.05) from that for the nonamended control (CO2) or those for the nitrate-amended treatments (NO3−, NO2−, and N2O).
ND, not detected.
Chemical and molecular analyses of FC aquifer microcosms.
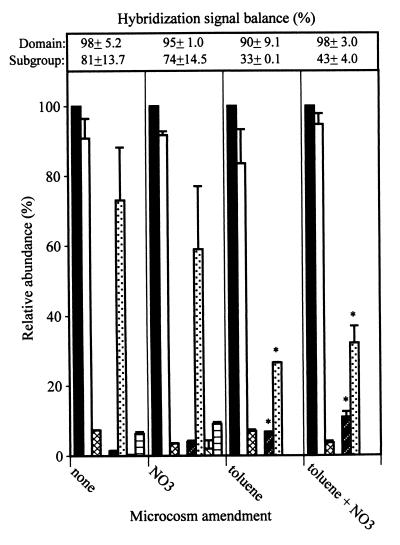
Toluene supplementation increased total CO2 production to ca. 40 μmol in the nonamended treatment compared to ca. 25 μmol in the nitrate-amended treatments (Table 4). Since the theoretical CO2 production should be about 32 μmol, this result may have indicated that microbial activity was supported in part by oxidation of native organic materials. While CO2 analysis indicated that toluene supplementation stimulated microbial activity, there was no significant effect on DNA yield. This result may have been attributed to the high organic matter content of the FC sediment interfering with DNA recovery from these materials. Toluene degradation was detectable between days 6 and 10 and coincided with the period of enhanced CO2 production. Toluene degradation was more rapid than that observed in the FC sediment and may have reflected the effect of prior fuel exposure on acclimation of the microbial population. Results of domain probe analysis were similar to those of the NC sediment in abundance patterns and the lack of significant treatment effects on domain abundance (Fig. 3). However, significant effects of toluene supplementation on community structure were detected at the lower taxonomic levels. The FC sediment showed a 40% decrease in B+G and an 8% increase in the ALF. Most striking, however, was the fact that the majority of the DNA (67%) was not accounted for by the phylum or subclass probe (Fig. 3).
FIG. 3.
Molecular analysis of DNA extracted from microcosms established with the FC aquifer material. Relative abundance of the domain or subgroups was determined by hybridization to the indicated probe (see the legend to Fig. 1 for the key to probe abbreviations). Bars marked with an asterisk were significantly different (P ≤ 0.05) from those for the nonamended microcosms. Probes for which data are not plotted gave hybridization signals that were below background.
In contrast to results with the NC aquifer material, NO3− amendment alone did not enhance CO2 production relative to that of the nonamended control (Table 4). The FC microcosms were similar to the NC microcosms in patterns of NO3− consumption and NO2− and N2O production. Nitrite and N2O were not detected in microcosms that did not receive NO3− supplementation. As in the NC microcosms, there were no significant differences in community structure between the NO3−-supplemented and nonamended aquifer materials (Fig. 3).
The combined amendment supported the highest level of CO2 production through the first 16 days of incubation. By day 18, cumulative CO2 production in these flasks was still greater than that for either the nonamended controls or the NO3− amended treatments but was the same as that in the microcosms amended with toluene alone. As with the NC microcosms, there were interactions between amendments. NO3− addition supported more rapid toluene degradation than toluene addition alone, and toluene supplementation decreased accumulations of NO3− reduction products, particularly N2O (Table 4). The community structure was similar to that of the FC microcosms amended with toluene alone (Fig. 3). Normalized domain signals equaled 98%, while those of subgroups equaled only 43% (Fig. 3).
A consistent result from phylogenetic probe analysis of the FC microcosms was the low balance of hybridization signals from the toluene-amended aquifer materials. This could have been explained by lack of homology between probes and the dominant phylotypes or perhaps by unknown matrix effects that interfered with hybridization. To examine the latter possibility, the FC microcosm extracts were spiked with genomic DNA from Escherichia coli as an internal standard and then hybridized with an E. coli-specific probe. All E. coli DNA-spiked extracts hybridized to the E. coli probe without any significant differences in signals, and there was no hybridization to the extracts not spiked with E. coli DNA (data not shown). Thus, the significant decrease in subgroup probe signal balances in toluene-amended microcosms was not attributable to sample matrix effects but, rather, indicated a major community structure shift; specifically, the dominant phylotype(s) established following exposure to toluene constituted a minor fraction of the community in the absence of this chemical.
Identifying the phylotypes established in the toluene-amended FC sediment could provide insights into a segment of the microbial community that is important for fuel degradation. Since domain-level phylotype abundance was not affected by toluene exposure, and good signal balances were obtained with the domain probes, it is reasonable to infer that these organisms were Bacteria. Beyond this point we can hypothesize only as to the organisms’ taxonomic affiliation(s), but we believe the δ-Proteobacteria are likely candidates, because organisms within this subclass are mainly anaerobes capable of using a variety of TEAPs, and many of these might not be detected by the SRB probe. An example is Geobacter, which can grow anaerobically with monoaromatic compounds as its sole carbon and energy sources, possesses a membrane-bound nitrate reductase and is widely distributed in sedimentary environments like aquifers (10, 26, 27, 32).
Comparison of field sample analysis and microcosm experiments: insights into effects of hydrocarbon contaminants and/or nitrate supplementation on microbial community structure.
At the domain level, the microbial community structure showed no significant differences regardless of the aquifer material type or microcosm amendment. The detection of Eucarya in the microcosms was in agreement with the hybridization analysis of Fry et al. (16), who reported that Eucarya comprised 6 to 14% of the microbial community in anaerobic groundwater from deep aquifers. While the nature and function of Eucarya observed in the microcosms are unknown, the detection of a consistent fraction suggests that these organisms were not directly affected by the treatments applied. These Eucarya could be anaerobic protozoa grazing on the microbial populations that were stimulated in the microcosms.
A comparison of the phylogenetic probe and chemical analyses showed that both toluene and NO3− stimulated microbial activity, but only the former had a significant effect on community structure at the phylum or subclass level. The lack of community structure shifts associated with NO3− supplementation may have reflected the widespread distribution of denitrification abilities and/or phylogenetic overlap with organisms mediating other anaerobic processes. These results were in contrast to those of Telang et al. (40) who, based on reverse sample genome probing, concluded that NO3− injection significantly altered a sulfidogenic oil field microbial community by resulting in the proliferation of a single organism. Further studies are needed to establish whether or not differing impacts of NO3− supplementation on community structure may be anticipated based on intrinsic characteristics of the environment and/or microbial community.
The consistent effect of toluene on altering microbial community structure in both the FC and NC sediments indicated that organisms able to grow anaerobically on this compound were present in multiple phylogenetic groups. Furthermore, the fact that community structure alterations induced by exposure to toluene were the same with or without the addition of nitrate suggested that the phylotypes stimulated in either sediment type were able to couple toluene degradation to energy-conserving processes other than respiratory denitrification. Collectively, these results may reflect diversity in toluene degradation pathways within the phylotypes stimulated and/or the potential for these pathways to be coupled with multiple TEAPs. While the microbial community structures in the NC and FC microcosms were similar in that they were significantly affected by toluene exposure, the patterns of these shifts were clearly distinct. Presumably, these differences were at least partly attributable to the impacts of prior exposure to hydrocarbon contamination and/or anaerobic conditions. It was also clear, however, that community structures in the toluene-amended NC and FC microcosms did not resemble those of field samples from the NC and FC regions, which were rather similar to each other. Incongruities such as these might be expected, given that the microcosms were designed to accentuate community responses to individual environmental variables (e.g., toluene exposure), whereas the field samples represented the summation of a more complex series of environmental interactions acting over vastly different temporal and spatial scales.
Ideally, all questions about the behavior of microbes in aquifers could be addressed by direct analysis of field samples, but because of the myriad factors affecting microbial populations in aquifers, questions about the mechanisms controlling the activities or composition of microbial communities cannot be unambiguously resolved by the analysis of field samples alone. Analysis of microcosms, established and maintained under controlled conditions, can be used to establish causal relationships between environmental variables and microbial activities. However, these systems are undeniably altered in some way(s). Thus, if the understanding of microbial populations is to progress, both field samples and microcosms should be analyzed together in a coordinated, iterative manner.
ACKNOWLEDGMENTS
We thank Kurt Brownell for permitting access to the study site, John DeWilde for operation of the Geoprobe, Paul Ludden for use of the anaerobic glovebox, and the individuals identified herein who provided microbial cultures used for probe calibration.
These studies were supported by U.S. EPA cooperative agreement number CR-824670 to W.J.H. and by the UW-system groundwater research program (project A349454 to J.M.B. and W.J.H.).
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. Personal communication.
- 3.Amann R I, Binder B J, Olsen R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R, Ludwig W, Schleifer K-H. Identification of uncultured bacteria: a challenging task for molecular taxonomists. ASM News. 1994;60:360–365. [Google Scholar]
- 6.Bakken L R, Olsen R A. DNA content of soil bacteria of different cell size. Soil Biol Biochem. 1989;21:789–793. [Google Scholar]
- 7.Bossert I D, Rivera M D, Young L Y. p-Cresol biodegradation under denitrifying conditions: isolation of a bacterial co-culture. FEMS Microbiol Ecol. 1986;38:313–319. [Google Scholar]
- 8.Bottomley P J. Weaver et al. (ed.), Methods of soil analysis. Part 2, Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1994. Light microscopic methods for studying soil microorganisms; pp. 81–106. [Google Scholar]
- 9.Chapelle F H, McMahon P B, Dubrovsky N M, Fujii R F, Oaksford E T, Vroblesky D A. Deducing the distribution of terminal electron-accepting processes in hydrologically diverse groundwater systems. Water Resour Res. 1995;31:359–371. [Google Scholar]
- 10.Coates J D, Phillips E J P, Lonergan D J, Jenter H, Lovely D. Isolation of Geobacterspecies from diverse sedimentary environments. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.5.1531-1536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 12.DiChristina T J, DeLong E F. Design and application of rRNA-targeted oligonucleotide probes for the dissimilatory iron- and manganese-reducing bacterium Shewanella putrefaciens. Appl Environ Microbiol. 1993;59:4152–4160. doi: 10.1128/aem.59.12.4152-4160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans P J, Mang D T, Kim K S, Young L Y. Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol. 1991;57:1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry N K, Fredrickson J K, Fishbain S, Wagner M, Stahl D A. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl Environ Microbiol. 1997;63:1498–1504. doi: 10.1128/aem.63.4.1498-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschalk G. Bacterial metabolism. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 18.Grbic-Galic D, Vogel T M. Transformation of toluene and benzene by mixed methanogenic cultures. Appl Environ Microbiol. 1987;53:254–260. doi: 10.1128/aem.53.2.254-260.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson R S, Phillips J A. Chemical composition. In: Gerhardt P, et al., editors. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. pp. 328–364. [Google Scholar]
- 20.Harris R F, Arnold S M. Redox and energy aspects of soil bioremediation. In: Skipper H D, Turco R F, editors. Bioremediation: science and applications. Madison, Wis: Soil Science Society of America; 1995. pp. 55–85. [Google Scholar]
- 21.Hess A, Zarda B, Hahn D, Haner A, Stax A, Hohener P, Zeyer J. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl Environ Microbiol. 1997;63:2136–2141. doi: 10.1128/aem.63.6.2136-2141.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks R, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′-6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchins S R, Downs W C, Wilson J T, Smith G B, Kovacs D A, Fine D D, Douglass R H, Hendrix D J. Effect of nitrate addition on biorestoration of fuel contaminated aquifer: field demonstration. Ground Water. 1991a;29:571–580. [Google Scholar]
- 24.Hutchins S R, Sewell G W, Kovacs D A, Smith G A. Biodegradation of aromatic hydrocarbons by aquifer microorganisms under denitrifying conditions. Environ Sci Technol. 1991;25:68–76. [Google Scholar]
- 25.Jackson C R, Harper J P, Willoughby D, Roden E C, Churchhill P F. A simple, efficient method for separation of humic substances and DNA from environmental samples. Appl Environ Microbiol. 1997;63:4993–4995. doi: 10.1128/aem.63.12.4993-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovley D R, Lonergan D J. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism GS-15. Appl Environ Microbiol. 1990;61:1858–1964. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovley D R, Giovannoni S J, White D C, Chamoine J E, Phillips E J, Goby Y A, Goodwin S. Geobacter metallireducensgen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 28.Lovley D R, Chapelle F H, Woodward J C. Use of dissolved H2concentrations to determine distribution of microbially catalyzed redox reactions in anoxic groundwater. Environ Sci Technol. 1994;28:1205–1210. doi: 10.1021/es00056a005. [DOI] [PubMed] [Google Scholar]
- 29.Lovley D R, Coates J D, Woodward J C, Phillips E J P. Benzene oxidation coupled to sulfate reduction. Appl Environ Microbiol. 1995;61:953–958. doi: 10.1128/aem.61.3.953-958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 32.Naik R R, Murillo F M, Stolz J F. Evidence for a novel nitrate reductase in the dissimilatory iron-reducing bacterium Geobacter metallireducens. FEMS Microbiol Lett. 1993;106:53–58. [Google Scholar]
- 33.National Research Council. In situ bioremediation, when does it work? Washington, D.C: National Academy Press; 1994. [Google Scholar]
- 34.Poulsen L K, Lan F, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situhybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raskin L, Poulsen L K, Noguera D R, Rittman B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of gram-positive bacteria with high G+C content using 23S rRNA-targeted oligonucleotides. Appl Environ Microbiol. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 37.Schocher R J, Seyfried B, Vazquez F, Zeyer J. Anaerobic degradation of toluene by pure cultures of denitrifying bacteria. Arch Microbiol. 1991;157:7–12. doi: 10.1007/BF00245327. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber, M. E. Unpublished data.
- 39.Stahl D A, Amann R. Development an application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 205–248. [Google Scholar]
- 40.Telang A J, Ebert S, Foght J M, Westlake D W S, Jenneman G E, Gervertz D, Voordouw G. Effect of nitrate injection on the microbial community in an oil field as monitored by reverse sample genome probing. Appl Environ Microbiol. 1997;63:1785–1793. doi: 10.1128/aem.63.5.1785-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai Y-L, Olsen B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for Proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weatherburn M W. Phenol-hypochlorite reaction for the determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- 44.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]