Abstract
Background
Some people have characteristics of both asthma and COPD (asthma-COPD overlap), and evidence suggests they experience worse outcomes than those with either condition alone.
Research Question
What is the genetic architecture of asthma-COPD overlap, and do the determinants of risk for asthma-COPD overlap differ from those for COPD or asthma?
Study Design and Methods
We conducted a genome-wide association study in 8,068 asthma-COPD overlap case subjects and 40,360 control subjects without asthma or COPD of European ancestry in UK Biobank (stage 1). We followed up promising signals (P < 5 × 10–6) that remained associated in analyses comparing (1) asthma-COPD overlap vs asthma-only control subjects, and (2) asthma-COPD overlap vs COPD-only control subjects. These variants were analyzed in 12 independent cohorts (stage 2).
Results
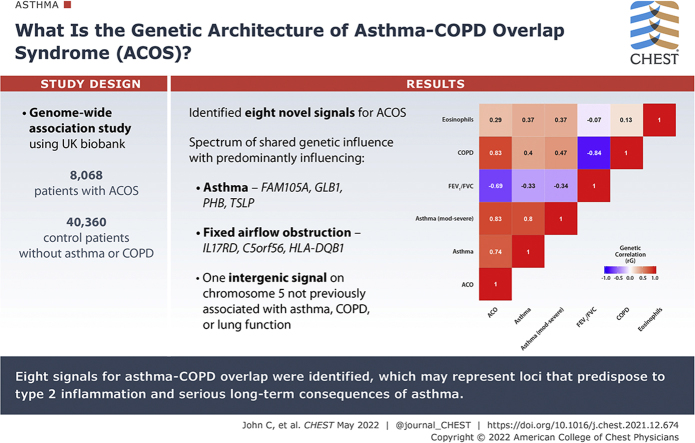
We selected 31 independent variants for further investigation in stage 2, and discovered eight novel signals (P < 5 × 10–8) for asthma-COPD overlap (meta-analysis of stage 1 and 2 studies). These signals suggest a spectrum of shared genetic influences, some predominantly influencing asthma (FAM105A, GLB1, PHB, TSLP), others predominantly influencing fixed airflow obstruction (IL17RD, C5orf56, HLA-DQB1). One intergenic signal on chromosome 5 had not been previously associated with asthma, COPD, or lung function. Subgroup analyses suggested that associations at these eight signals were not driven by smoking or age at asthma diagnosis, and in phenome-wide scans, eosinophil counts, atopy, and asthma traits were prominent.
Interpretation
We identified eight signals for asthma-COPD overlap, which may represent loci that predispose to type 2 inflammation, and serious long-term consequences of asthma.
Key Words: asthma, COPD, epidemiology, genome-wide association study, spirometry
Abbreviations: ACO, asthma-COPD overlap; EAF, effect allele frequency; eQTL, expression quantitative trait locus; GWAS, genome-wide association study; LDSC, linkage disequilibrium score regression; rg, genetic correlation; SNP, single-nucleotide polymorphism
Graphical Abstract
FOR EDITORIAL COMMENT, SEE PAGE 1125
Take-home Points.
Study Question: What are the genetic determinants of risk for asthma-COPD overlap, and how do these differ from those for COPD or asthma?
Results: We discovered eight novel signals for asthma-COPD overlap in a meta-analysis of 12,369 case subjects and 88,969 control subjects; most signals suggested a spectrum of shared genetic influences on asthma, COPD, or lung function, and in phenome-wide scans of these signals, eosinophil counts, atopy, and asthma traits were prominent.
Interpretation: We identified eight signals for asthma-COPD overlap, not driven by smoking or age at asthma diagnosis, which may represent loci that predispose to type 2 inflammation, and serious long-term consequences of asthma.
Asthma and COPD have substantial global impacts.1 They are heterogeneous conditions2, 3, 4 that share some common features, including airflow obstruction with differing degrees of reversibility. Inflammatory processes are important in both conditions, and cytokine profiles identify both distinct and overlapping groups of patients.5 People with characteristics of both conditions have until recently been referred to as having “asthma-COPD overlap” (ACO),4 and a number of studies have suggested that such patients have significantly worse outcomes than those with either condition alone.6, 7, 8, 9, 10, 11, 12, 13 Guidelines emphasize that asthma and COPD are different conditions, but may coexist in the same patient.14 People with features of both diseases risk being excluded from research that might provide evidence about the most effective treatment strategies.3
Environmental risk factors—notably smoking in COPD—are central, but genetics also plays an important role in both asthma and COPD,15, 16, 17 and it has long been hypothesized that there may be a shared, underlying genetic predisposition to both diseases.2,18 Genome-wide association studies (GWASs) examine variants across the genome in an unbiased manner, to identify variant-trait associations that inform our understanding of disease biology and potential treatment strategies. GWASs have identified many loci associated with asthma or COPD in European populations (e-Appendix 1). The genetic correlation (rg) between asthma and COPD is 0.38 (P = 6.2 × 10−5), suggesting a shared genetic etiology.19 A GWAS of ACO compared with COPD alone (n = 3,570) did not identify any variants associated at the conventional threshold,8 and a meta-analysis of an asthma and COPD GWAS found one association, driven by COPD.20 Eighteen loci outside the HLA (human leukocyte antigen) region have been identified as associated with both asthma and lung function/COPD at P < 5 × 10–8, but have not been specifically described as ACO loci.
Notwithstanding the controversies of changing terminology for people with both asthma and COPD, we refer to this case status as “ACO.” Improved knowledge of genetic variants associated with coexisting asthma and COPD would contribute to our understanding of underlying molecular pathways, and potentially inform diagnostic terminology and specific management strategies for those with coexisting asthma and COPD.
Accordingly, using spirometry, self-report, and electronic health care record data to define case subjects with both asthma and COPD (ACO) and suitable control subjects, we undertook the largest GWAS of coexisting asthma and COPD to date, including up to 12,369 case subjects and 88,969 control subjects, in a two-stage design incorporating 13 studies.
Study Design and Methods
Stage 1
The data source for this study was UK Biobank.21 Eligibility criteria, genotyping, and quality control are described in e-Appendix 1. A total of 321,057 people and 37 million single-nucleotide polymorphisms (SNPs) were included.
We defined cases of ACO if patients had self-reported asthma (see e-Appendix 1) and FEV1/FVC < 0.7 with Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2+ airflow limitation (FEV1 < 80% predicted). Case subjects who reported alpha-1 antitrypsin deficiency were excluded. Control subjects reported no asthma or COPD (e-Appendix 1), and had FEV1 ≥ 80% predicted and FEV1/FVC > 0.7. Five control subjects were randomly selected for each case. Case subjects and control subjects were unrelated (second degree or closer). Two additional control sets were defined for signal prioritization: people with asthma but without COPD, and people with COPD but without asthma. Asthma and COPD were defined as above.
Association testing was undertaken in SNPTEST, version 2.5.2 (“score” option),22 under an additive model. Age, sex, smoking status (ever/never), genotyping array, and 10 principal components were included as covariates. Variants were filtered on the basis of minor allele frequency (MAF) > 0.01 and imputation quality (INFO) > 0.5. P values and SEs were adjusted for the linkage disequilibrium score regression (LDSC) intercept23 (e-Fig 1).
In stage 1, we defined distinct signals passing a P value threshold of P < 5 × 10–6. We defined regions of association around the most strongly associated variant (sentinel variant) ± 1 Mb. To identify distinct signals, and additional signals within the regions described above, conditional analyses were undertaken with GCTA-COJO24 (e-Appendix 1, e-Fig 2).
Two further “signal prioritization” analyses were undertaken to ascertain the extent to which signals were driven by association with COPD and/or asthma alone. These included the same cases as the primary analysis, plus the two additional control sets described above. Variants were selected for follow-up in stage 2 if they were associated at P < 5 × 10–6 in the main stage 1 analysis and at P < .01 in both signal prioritization analyses.
Stage 2 and Joint Analysis
SNPs identified in stage 1 signal prioritization analyses were tested for association in 12 independent studies of European ancestry (up to 4,301 case subjects and 48,609 control subjects, in CHS [Cardiovascular Health Study], COPDGene [Genetic Epidemiology of COPD], deCODE, ECLIPSE [Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints], EPIC-Norfolk [European Prospective Investigation of Cancer in Norfolk], FHS [Framingham Heart Study], Generation Scotland, GenKOLS [Genetics of Chronic Obstructive Lung Disease Study], Trøndelag Health Study [HUNT], Lovelace Smokers' Cohort, Rotterdam Study, and SPIROMICS [Subpopulations and Intermediate Outcome Measures in COPD Study]) and one cohort of participants of self-reported African American ethnicity (COPDGene; 297 case subjects, 1,335 control subjects) (e-Appendix 1, e-Tables 1, 2).
Case subjects had both asthma and COPD. Asthma was defined as any lifetime self-report of asthma, or asthma diagnosis in the health care record, including billing codes (see e-Appendix 1 for further details and validation).25 All case subjects had spirometry results indicating FEV1/FVC < 0.7, and FEV1 < 80% predicted. All control subjects had FEV1/FVC > 0.7, FEV1 ≥ 80% predicted, and no asthma diagnosis. When possible, studies excluded people with alpha-1 antitrypsin deficiency.
Details of statistical analysis in stage 2 studies are presented in e-Appendix 1 (e-Table 3). Results were combined across stage 2 studies, using fixed-effect meta-analysis. Heterogeneity was assessed using the I2 statistic. We combined these results with those from UK Biobank (stage 1).
We performed a sensitivity analysis to assess whether the way COPD was defined changed our stage 2 results (e-Appendix 1).
To assess whether associations with our stage 1 signals changed according to age at asthma diagnosis, we divided case subjects into those who self-reported their age at asthma diagnosis as < 12 years, and > 25 years.26 We then repeated the association tests in UK Biobank. In addition, we repeated association testing after stratifying our sample into ever/never smokers.
Definition of Top Signals for Bioinformatic Analyses
We undertook bioinformatic analyses on ACO signals reaching P < 5 × 10–8 in the joint analysis of stages 1 and 2, and which also had a lower P value in the joint analysis than in UK Biobank (stage 1) alone or had P < .05 in stage 2. For each of these, we identified the set of SNPs that was 99% likely to contain the causal variant (“99% credible set”), assuming that the causal variant was included in the data set (e-Appendix 1).27 For bioinformatic analysis methods, see e-Appendix 1.
Using LDSC,28 we computed genetic correlations between ACO (stage 1 results), asthma,29 moderate-severe asthma,30 COPD,31 eosinophil counts,32 and FEV1/FVC.33 We also computed genetic correlations between ACO and atopic, autoimmune, and smoking behavior traits.34
Approvals
The research was conducted using UK Biobank, under application 648. UK Biobank has ethical approval from the UK National Health Service National Research Ethics Service (11/NW/0382). All included studies were approved by the appropriate research ethics committee or institutional review board (e-Appendix 1). All participants gave informed consent.
Results
In stage 1, 8,068 ACO case subjects were selected from UK Biobank, and 40,360 as healthy control subjects free of asthma and COPD. For signal prioritization analyses, another 16,762 people were selected as control subjects with COPD alone (without asthma), and 26,815 as control subjects with asthma alone (without COPD). Descriptive statistics for case subjects and control subjects are presented in Table 1. ACO case subjects were slightly older than healthy control subjects, and included more men and ever smokers.
Table 1.
Descriptive Characteristics of Case Subjects and Control Subjects Included in Stage 1 (UK Biobank Primary and Signal Prioritization Analyses)
| Characteristic | ACO Case Subjects (n = 8,068) | Healthy Control Subjects (n = 40,360) | Control Subjects With COPD But No Asthma (n = 16,762) | Control Subjects With Asthma But No COPD (n = 26,815) |
|---|---|---|---|---|
| Age at recruitment, median (IQR), y | 60 (53-65) | 57 (49-63) | 62 (56-65) | 55 (48-61) |
| Sex, No. (%) | ||||
| Male | 4,179 (51.8%) | 17,598 (43.6%) | 9,147 (54.6%) | 9,703 (36.2%) |
| Female | 3,889 (48.2%) | 22,762 (56.4%) | 7,615 (45.4%) | 17,112 (63.8%) |
| Smoking status, No. (%) | ||||
| Ever smoked | 4,367 (54.1%) | 17,316 (42.9%) | 11,752 (70.1%) | 11,231 (41.9%) |
| Never smoked | 3,701 (45.9%) | 23,044 (57.1%) | 5,010 (29.9%) | 15,584 (58.1%) |
| Pack-years of smoking, median (IQR)a | 25.5 (13.5-39.5) | 15.8 (8.3-26.4) | 32.0 (19.0-45.5) | 16.5 (8.5-28.1) |
| Allergic rhinitis (including hay fever) or eczema, No. (%) | ||||
| Yes | 3,325 (41.2%) | 8,468 (21.0%) | 2,691 (16.1%) | 13,010 (48.5%) |
| No | 4,743 (58.8%) | 31,892 (79.0%) | 14,071 (83.9%) | 13,805 (51.5%) |
| Eosinophil count (median, IQR),b × 109 cells/L | 0.20 (0.13-0.32) | 0.13 (0.08-0.20) | 0.16 (0.10-0.24) | 0.21 (0.11-0.27) |
| Lung function, median (IQR) | ||||
| FEV1/FVC | 0.63 (0.58-0.67) | 0.78 (0.75-0.81) | 0.65 (0.61-0.68) | 0.77 (0.74-0.80) |
| % predicted FEV1 | 66.1% (56.5%-73.3%) | 97.3% (89.9%-105.6%) | 68.7% (60.0%-74.8%) | 90.8% (81.6%-100.0%) |
ACO = asthma-COPD overlap; IQR = interquartile range.
In ever smokers with nonmissing data, 3,270 of 4,367 case subjects, 11,196 of 17,316 main control subjects, 9,672 of 11,752 COPD-not asthma control subjects, 7,443 of 11,231 asthma-not COPD control subjects
In those with nonmissing data after cleaning as per Astle et al,32 n = 7,666 case subjects, n = 38,259 main control subjects, n = 15,845 COPD-not asthma control subjects, n = 25,292 asthma-not COPD control subjects.
After filtering on MAF and INFO, 7,693,381 variants were analyzed. The LDSC intercept was 1.018, suggesting that results were not strongly inflated due to population structure (e-Fig 1).28
ACO Association Signals
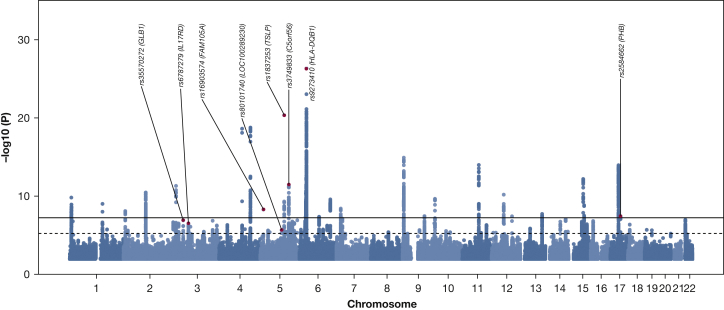
In stage 1, there were 83 distinct signals at P < 5 × 10–6 (Fig 1,32 e-Appendix 1, e-Fig 2 for the signal selection; e-Table 4 for results). Of these, 31 retained significance (P < .01) in signal prioritization analyses comparing ACO case subjects separately with either COPD case subjects or asthma case subjects, to determine whether signals were driven by asthma or COPD alone (e-Table 4). In stage 2, comprising 12 independent cohorts (4,301 case subjects, 48,609 control subjects) (e-Tables 1, 2), 26 of 31 signals had a direction of effect concordant with stage 1 (e-Table 5), and the median value for heterogeneity (I2) across these signals was 15%. Although the sample size of participants of African American ethnicity was small (297 case subjects, 1,335 control subjects) and CIs were broad, 22 of 31 signals had a direction of effect consistent with the European ancestry studies (e-Table 5).
Figure 1.
Manhattan plot of association results for asthma-COPD overlap in stage 1 (UK Biobank). The x axis shows genomic location by chromosome, the y axis shows the –log10P value, corrected for the intercept of linkage disequilibrium score regression (1.018). The eight top signals (from joint analysis) are highlighted in red, and labeled with rsIDs (reference SNP [single-nucleotide polymorphism] ID numbers). The black line indicates P = 5 × 10–8 (commonly known as genome-wide significance), and the dotted line corresponds to P = 5 × 10–6 (genome-wide suggestive threshold). A quantile-quantile plot is shown in e-Figure 1. For further details on the eight SNPs shown here, see also Table 2.
Results for the stage 2 sensitivity analysis (9,638 case subjects and 128,273 control subjects from 15 studies), in which COPD was defined either by available spirometry or, alternatively, by electronic health care record diagnoses (e-Appendix 1), are in e-Table 6.
Subgroup Analyses
Effect sizes for the 31 signals among case subjects with childhood-onset asthma were highly correlated with those among people with adult-onset asthma (R = 0.883) (e-Table 7, e-Fig 3). Effect sizes in ever and never smokers were also closely correlated (R = 0.911) (e-Table 7, e-Fig 4).
Eight Top Signals for ACO Defined From Joint Analysis
After meta-analysis combining stage 1 and stage 2, 13 signals were genome-wide significant (P < 5 × 10–8) (e-Table 4; e-Fig 2 for flow diagram). Of these, eight either had a lower P value in the joint analysis than in stage 1 alone, or P < .05 in stage 2 studies alone (Table 2, e-Figs 5, 6). None of these eight signals were previously reported as associated specifically with ACO.8
Table 2.
Eight Genome-Wide Signals for Asthma-COPD Overlap
| rsID | Chr:Pos (Effect/Noneffect Allele) | Nearest Gene | Location | EAF | Stage 1 (UK Biobank; Case Subjects, 8,068; Control Subjects, 40,360) |
Stage 2 (12 Independent Studies; Case Subjects, 4,301; Control Subjects, 48,609)a |
Joint Analysis of Stage 1 and Stage 2 |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |||||
| rs80101740 | 5:98471135 (C/A) | LOC100289230 | Intergenic | 0.015 | 1.44 (1.24-1.68)a | 1.87 × 10–6a | 1.37 (1.10-1.71) | 5.49 × 10–3 | 1.42 (1.25-1.61) | 3.72 × 10–8 |
| rs35570272 | 3:33047662 (T/G) | GLB1 | Intronic | 0.398 | 1.11 (1.07-1.15) | 1.06 × 10–7 | 1.08 (1.02-1.14) | 4.67 × 10–3 | 1.10 (1.06-1.13) | 2.44 × 10–9 |
| rs16903574 | 5:14610309 (G/C) | FAM105A | Exonic | 0.077 | 1.23 (1.15-1.32) | 4.47 × 10–9 | 1.13 (1.03-1.25) | 9.96 × 10–3 | 1.20 (1.13-1.27) | 3.8 × 10–10 |
| rs2584662 | 17:47470487 (C/A) | PHB | Intergenic | 0.42 | 0.90 (0.86-0.94) | 3.20 × 10–8 | 0.95 (0.90-1.00) | 5.89 × 10–2 | 0.92 (0.89-0.95) | 2.21 × 10–8 |
| rs1837253 | 5:110401872 (C/T) | TSLP | Intergenic | 0.739 | 1.22 (1.17-1.27) | 4.22 × 10–21 | 1.06 (1.00-1.12) | 4.44 × 10–2 | 1.16 (1.12-1.20) | 1.53 × 10–18 |
| rs6787279 | 3:57163751 (C/T) | IL17RD | Intronic | 0.169 | 0.88 (0.84-0.92) | 2.69 × 10–7 | 0.91 (0.85-0.97) | 6.51 × 10–3 | 0.89 (0.86-0.93) | 7.87 × 10–9 |
| rs9273410 | 6:32627250 (A/C) | HLA-DQB1 | UTR3 | 0.445 | 1.24 (1.19-1.29) | 4.37 × 10–27 | 1.11 (1.05-1.18) | 6.42 × 10–4 | 1.20 (1.16-1.24) | 9.19 × 10–28 |
| rs3749833 | 5:131799626 (C/T) | C5orf56 | ncRNA intronic | 0.263 | 1.16 (1.11-1.21) | 3.10 × 10–12 | 1.06 (1.00-1.12) | 4.21 × 10–2 | 1.12 (1.09-1.16) | 9.37 × 10–12 |
Variants were annotated with nearest gene and type of region, using ANNOVAR software (and genome build hg19). OR, 95% CI, and P value were all calculated by score testing. The Firth test for rs80101740 gave OR, 1.40 (95% CI, 1.22-1.60) and P = 1.56 × 10–6. Chr:Pos = chromosome:position; EAF = effect allele frequency; ncRNA = noncoding RNA; rsID = reference SNP (single-nucleotide polymorphism) ID number; UTR3 = 3' (three prime) untranslated region.
Stage 2 studies: CHS (Cardiovascular Health Study), COPDGene (Genetic Epidemiology of COPD), deCODE, ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints), EPIC-Norfolk (European Prospective Investigation of Cancer in Norfolk), FHS (Framingham Heart Study), Generation Scotland, GenKOLS (Genetics of Chronic Obstructive Lung Disease Study), Trøndelag Health Study (HUNT), Lovelace Smokers' Cohort, Rotterdam Study, SPIROMICS (Subpopulations and Intermediate Outcome Measures in COPD Study).
For the novel intergenic ACO signal on chromosome 5 (rs80101740, effect allele frequency [EAF], 0.015; OR, 1.42; P = 3.72 × 10–8) (e-Table 5), which has not been previously associated with asthma, lung function, or COPD, the sentinel SNP had the largest posterior probability (0.77) of being the true causal variant, assuming the causal variant was genotyped/imputed (e-Table 8). There was no evidence of colocalization with expression quantitative trait locus (eQTL) signals at this locus (e-Tables 9, 10), and no chromatin interactions were identified.
Four of our novel signals for ACO were previously reported for asthma but not COPD/lung function.35, 36, 37 For rs35570272 in GLB1 (OR, 1.10; EAF, 0.398; P = 2.44 × 10–9), there were 11 SNPs in the credible set, and the intronic sentinel SNP had the highest posterior probability (0.655). There were significant chromatin interactions nearby in GLB1 in fetal lung fibroblasts. GLB1 encodes the β-galactosidase enzyme involved in lysosomal function, and an elastin-binding protein involved in extracellular elastic fiber formation. Two SNPs (both with a posterior probability of ∼0.13) in the 99% credible set, rs7646283 and rs34064757, were eQTLs for the gene encoding cartilage-associated protein (CRTAP) in lung (e-Table 9), implicated in bone development and osteogenesis imperfecta.
Another signal (previously reported for asthma)26,35 was rs16903574 (EAF, 0.077; OR, 1.20; P = 3.8 × 10–10), a missense variant in FAM105A, deleterious according to its combined annotation-dependent depletion (CADD) score (22.6).38 FAM105A encodes a pseudoenzyme, possibly involved in protein-protein interactions.39 This sentinel had a posterior probability of 0.99. A previous study in asthma predicted FAM105A as the target based on chromatin interactions and correlation between enhancer epigenetic marks and gene expression, although we did not identify any eQTL evidence in lung or whole blood.35 We also identified a highly significant chromatin interaction in fetal lung fibroblasts overlapping FAM105A and another nearby gene (TRIO), but not in adult lung.
An intergenic signal between PHB and ZNF652 (rs2584662) (EAF, 0.42; OR, 0.92; P = 2.21 × 10–8) was previously associated with asthma and reported as a blood eQTL for GNGT2 (implicated in NF-κB activation),29,35 although we did not identify this in our eQTL analysis. In our analysis, eight SNPs were in the credible set (posterior probabilities all ≤ 0.2). Hi-C data suggested a significant chromatin interaction in ZNF652, with another, less significant peak close to GNGT2. Nearby loci in ZNF652 have previously been associated with asthma/allergic disease and moderate-to-severe asthma.35
We also identified rs1837253, an intergenic signal near TSLP (EAF, 0.739; OR, 1.16; P = 1.53 × 10–18), with a posterior probability of 1, that is, the only variant in the credible set. No eQTL evidence was identified. There were highly significant chromatin interactions with SLC25A46 in fetal lung fibroblasts and in adult lung tissue, and with a region between TSLP and SLC25A46 in fetal lung fibroblasts only. The gene encoding thymic stromal lymphopoietin (TSLP) was implicated in asthma and allergic disease before the GWAS era,40 and an anti-TSLP antibody has been trialled in allergic asthma.41
Another signal, rs6787279 in IL17RD (EAF, 0.169; OR, 0.89; P = 7.87 × 10–9), has been previously reported for lung function and COPD.31,42 There were 55 variants in the credible set, all with posterior probability ≤ 0.12, meaning it is not yet possible to fine-map this signal confidently. One SNP in the credible set was exonic and possibly damaging (rs17057718), but the posterior probability was only 0.012. Multiple SNPs at this locus were eQTLs for IL17RD in lung, with the ACO risk allele corresponding to decreased IL17RD expression. IL17RD is in the IL-17 signaling pathway, which is implicated in asthma,43 and in COPD pathogenesis,44,45 potentially by mediating effects of cigarette smoke.
Two ACO signals have previously been reported separately for both asthma and lung function or COPD: rs9273410 in HLA-DQB1 (EAF, 0.445; OR, 1.20; P = 9.19 × 10–28) and rs3749833 in C5orf56 (EAF, 0.263; OR, 1.12; P = 9.37 × 10–12). HLA-DQB1 encodes a major histocompatibility complex type II molecule involved in antigen presentation. HLA-DQB1 alleles are associated with numerous inflammatory and autoimmune diseases. In our analysis, the sentinel was the only SNP in the credible set. For lung function, an amino acid change in the gene product HLA-DQβ1 has been identified as the main driver of signals in the major histocompatibility complex region.33 Analyses in asthma have identified HLA-DQA1 as the likely driver gene.35
C5orf56 is located on a cytokine gene cluster on chromosome 5, including IL3, IL4, and IL5. Several interleukins in this region have been considered as therapeutic targets in asthma. In severe eosinophilic asthma, the anti-IL-5 monoclonal antibodies mepolizumab and reslizumab reduce exacerbations and improve quality of life.46, 47, 48 SNPs in the credible set were eQTLs in lung and/or blood for SLC22A5, AC116366.6, RAD50, and a noncoding Y RNA. SLC22A4 has been identified as the most likely candidate gene for the lung function association.33 The gene products of SLC22A4 and SLC22A5 are involved in bronchial uptake of bronchodilators and anticholinergic drugs.49 An analysis in asthma predicted C5orf56 (which encodes the interferon regulatory factor 1 antisense RNA, IRF1-AS1) as the causal gene.35
In our phenome-wide scan, all ACO loci previously associated with asthma showed association with blood cell counts, particularly eosinophils and neutrophils, and atopic traits (e-Table 11). The HLA locus was associated with a wide range of autoimmune/inflammatory traits. Another locus (rs2584662, near PHB and ZNF652) was associated with anthropometric traits, cardiovascular phenotypes, and chronic diseases/multimorbidity, whereas rs3749833 (near C5orf56) was associated with anthropometric traits and inflammatory bowel disease. SNPs in the credible set for the intergenic chromosome 5 signal (rs80101740) were associated with cardiovascular and a range of other traits.
ACO Shares Genetic Architecture With Other Traits
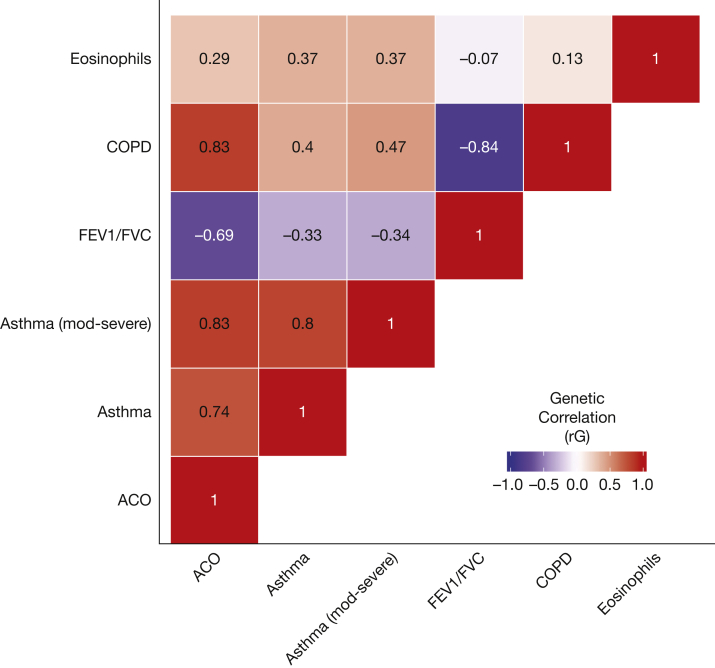
We observed genetic correlations (rg) of broadly similar magnitude between ACO and COPD (rg = 0.828; P = 3.19 × 10–299), ACO and asthma (rg = 0.743; P = 6.18 × 10−44), and ACO and FEV1/FVC (rg = –0.692; P = 7.48 × 10−33) (Fig 2, e-Table 12). The genetic correlation (rg) between asthma and FEV1/FVC was –0.333 (P = 8.71 × 10−7) (ie, increased asthma risk was correlated with lower FEV1/FVC). Blood eosinophil counts were moderately correlated with ACO (rg = 0.292; P = 4.87 × 10−11), similar in magnitude to the correlation of eosinophils with asthma (rg = 0.371; P = 3.15 × 10−7), whereas the correlations of eosinophils with FEV1/FVC (rg = –0.070; P = .002) and COPD (rg = 0.130; P = 4.83 × 10–6) were smaller. We additionally computed genetic correlations between ACO and 16 autoimmune traits, and between ACO and smoking behavior (rg = 0.046, P = .417) (e-Table 12). After asthma, the next strongest correlation was with eczema (rg = 0.255, P = .004), then multiple sclerosis (rg = 0.323, P = .011).
Figure 2.
Genetic correlations between asthma-COPD overlap (ACO) and asthma, moderate-severe asthma, COPD, FEV1/FVC, and blood eosinophil counts. Genetic correlations were computed by linkage disequilibrium score regression. The annotation in each tile represents the magnitude of the genetic correlation estimate (rG), and intensity is proportional to the magnitude of effect. Note that for FEV1/FVC, a negative correlation shows that the other trait is associated with reduced FEV1/FVC (reduced FEV1/FVC implies worse lung function). Data sets used: ACO = current discovery results from UK Biobank; Asthma = GWAS results from Demenais et al29; Asthma (moderate-severe) = genome-wide association study (GWAS) of asthma by Shrine et al30; COPD = GWAS of COPD by Sakornsakolpat et al31; FEV1/FVC = GWAS of FEV1/FVC (UK Biobank and SpiroMeta) by Shrine et al33; Eosinophils = blood eosinophil counts published by Astle et al.32
Discussion
We conducted the largest GWAS of ACO to date, and identified 83 independent signals associated at P < 5 × 10–6 in stage 1. After excluding variants associated with asthma only or COPD only, we studied 31 variants in stage 2, with eight distinct signals for ACO showing genome-wide significance (P < 5 × 10–8) in a stage 1 and stage 2 meta-analysis.
Our study contributes to understanding of the genetic architecture of ACO. We showed strong genetic correlation between ACO and COPD/lung function, and between ACO and asthma, especially moderate-severe asthma. Furthermore, we showed that the genetic correlation of blood eosinophil counts with ACO was more similar in magnitude to the genetic correlation of eosinophils with asthma than of eosinophils with FEV1/FVC and COPD. Increased eosinophils are associated with asthma and COPD exacerbations,50, 51, 52 and with lung function decline in subjects with and without asthma.53 Eosinophil counts, atopy, and asthma traits were prominent in phenome-wide scans of our top eight signals, consistent with an important role for type 2 inflammation in ACO.54,55
One intergenic signal on chromosome 5 (rs80101740) is not previously reported as associated with asthma, COPD, or lung function. Although near to a putative signal for lung function without replication support (rs377731, r2 = 0.02 with rs80101740),33 the ACO sentinel was independent of this lung function signal in conditional analyses. Evidence from eQTL studies suggests that the nearby lung function signal is associated with RGMB and LINC02062 expression.
Four of the eight signals identified as novel (GLB1, FAM105A, PHB, TSLP) are known signals for asthma or allergic disease, but not COPD. Our results suggest that these loci also have a role in COPD. All four have been associated with child- and adult-onset asthma, and could represent an opportunity to intervene in early life to prevent serious long-term sequelae.26 One ACO signal (IL17RD) is a known lung function and COPD locus; our findings demonstrate its relevance in reversible airflow obstruction. Together, these loci could represent targets for intervention, potentially to prevent development of fixed airflow obstruction.
Two signals are known to be associated with asthma and COPD/lung function, including the HLA-DQB1 locus (the first signal identified as associated with both asthma and COPD), and a signal at C5orf56, encoding IRF1-AS1, on chromosome 5, near a cytokine gene cluster.
In subgroup analyses, there was a strong positive correlation between stage 1 effect sizes for ACO in ever and never smokers, suggesting that ACO is not due solely to smoking in people with asthma, although childhood asthma in smokers increases COPD risk compared with nonasthmatics, possibly via early lung development.56 Similarly, when stratifying by child- vs adult-onset asthma, there was a strong correlation between effect sizes in both groups. Nevertheless, for some of the eight top signals, we found evidence of chromatin interactions in fetal but not adult lung. Although this may implicate developmental processes in ACO, inference is difficult, due to differences in experimental conditions, sample sizes, and reporting practices. Clearer conclusions may become possible as functional genomic assays advance.
Our study has some potential limitations. The stage 2 sample size (4,301 case subjects) was substantial, although relatively underpowered compared with stage 1 (8,068 case subjects). All signals reported met commonly adopted criteria for genome-wide significance, but stricter criteria are starting to be used for genome sequencing studies57; future work using sequence data would provide an opportunity to reevaluate the genomic regions we highlight. Misclassification of asthma and COPD diagnoses is possible: asthma in older patients may mimic COPD, and physicians may be less likely to suspect COPD in nonsmokers. To mitigate this, we used GOLD 2+ spirometric criteria to define COPD wherever possible, and note that self-reported asthma has been shown to accurately identify subjects with clinical and genetic characteristics of asthma.56 We hypothesize that any remaining misclassification would attenuate effect estimates toward the null, that is, reduce power to detect true genetic associations with ACO. Our main analysis was undertaken in European ancestry populations only; although for many loci there was good concordance in a small sample of participants of African American ethnicity, it is essential to study this trait further in diverse populations.
Interpretation
In the largest genome-wide association study to date, we identified eight signals associated with ACO. Our findings suggest a spectrum of shared genetic influences, from variants predominantly influencing asthma to those predominantly influencing fixed airflow obstruction. We focus on variants that tend toward an intermediate phenotype with features of both asthma and fixed airflow obstruction, with pathways implicating innate and adaptive immunity and potentially bone development, and signals for which the biology remains unclear. Further biological understanding is likely to be important for therapeutics to prevent the development of fixed airflow obstruction among people with asthma.
Acknowledgments
Author contributions: C. J., A. L. G., and M. D. T. act as guarantors for the content of the manuscript. C. J., A. L. G., L. V. W., and M. D. T. contributed to the conception and design of the study. C. J., A. L. G., N. S., T. A. O., J. Liu, L. P. H., S. H. C., J. T. K., J. Luan, X. L., N. T., H. X., T. M. B., H. P., S. L., A. C., A. I. H. C., and M. O. undertook data analysis. C. J., A. L. G., N. S., R. P., S. A. B., M. O., G. T., D. A. M., E. R. B., L. C. S., C. I., Y. T., S. A. G., J. D., G. B., L. L., V. E. O., I. J., J. K. Q., D. D. S., Y. B., M. v. d. B., D. N., I. S., I. P. H., C. L., S. R., T. L., A. C. W., J. L.-S., P. B., A. G., C. P. H., C. H., A. L., B. B., K. S., M. H. C., L. V. W., and M. D. T. contributed to data acquisition and/or interpretation. C. J., A. L. G., L. V. W., and M. D. T. drafted the manuscript. All authors critically reviewed the manuscript before submission.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: C. J. held a Medical Research Council Clinical Research Training Fellowship (MR/P00167X/1). A. L. G. was funded by internal fellowships at the University of Leicester from the Wellcome Trust Institutional Strategic Support Fund (204801/Z/16/Z) and the BHF Accelerator Award (AA/18/3/34220). L. P. H. is funded by the NIH/NHLBI [5K23HL136851]. S. H. C. is funded by the NIH/NHLBI [1K01HL153941-01]. A. I. H. C. is supported by Mitacs Accelerate. C. H. was supported by a Medical Research Council Human Genetics Unit programme grant (U.MC_UU_00007/10). M. H. C. is supported by R01 HL137927, R01 HL089856, and R01 HL147148. L. V. W. holds a GSK/British Lung Foundation Chair in Respiratory Research. M. D. T. is supported by a Wellcome Trust Investigator Award (WT202849/Z/16/Z). M. D. T. and L. V. W. have been supported by the MRC (MR/N011317/1). M. D. T. and I. P. H. hold NIHR Senior Investigator Awards. This research was partially supported by the NIHR Leicester Biomedical Research Centre and the NIHR Nottingham Biomedical Research Centre; the views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. C. J. and M. D. T. were involved in all stages of study development and delivery, and M. D. T. had full access to all data in the study and final responsibility for the decision to submit for publication. This research was funded in whole, or in part, by the Wellcome Trust. For the purpose of open access, the authors have applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. Please see e-Appendix 1 for additional study acknowledgments and funding statements. T. A. O., G. T., I. J., and K. S. are employees of deCODE Genetics/Amgen Inc. E. R. B. has undertaken clinical trials through his employer, Wake Forest School of Medicine and University of Arizona, for AstraZeneca, MedImmune, Boehringer Ingelheim, Genentech, Novartis, Regeneron, and Sanofi Genzyme. E. R. B. has also served as a paid consultant for ALK-Abelló, AstraZeneca, GlaxoSmithKline (GSK), MedImmune, Novartis, Regeneron, Sanofi Genzyme, and TEVA. M. v. d. B. reports grants paid to the University from AstraZeneca, Teva, GSK, and Chiesi outside the submitted work. D. N. has been a Merck & Co. employee during this study and is now an employee at Biogen Inc. I. S. has received support from GSK and Boehringer Ingelheim. I. P. H. has funded research collaborations with GSK, Boehringer Ingelheim, and Orion. M. H. C. has received grant support from GSK and Bayer, and consulting and speaking fees from Genentech, Illumina, and AstraZeneca. L. V. W. receives funding from GSK for a collaborative research project outside of the submitted work. M. D. T. receives funding from GSK and Orion for collaborative research projects outside of the submitted work.
Role of sponsors: The funders had no role in the design of the analyses or conduct of the study.
Data sharing statement: Individual-level participant data are from UK Biobank (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access; accessed February 18, 2022). Summary-level genome-wide association statistics will be made publicly available via the EBI GWAS catalog on publication (https://www.ebi.ac.uk/gwas/; accessed February 18, 2022).
Additional information: The e-Appendix, e-Figures, and e-Tables are available online under “Supplementary Data.”
Footnotes
C. John and A. L. Guyatt contributed equally to this manuscript.
FUNDING/SUPPORT: This work is supported by BREATHE, the Health Data Research Hub for Respiratory Health [MC_PC_19004] in partnership with SAIL Databank. BREATHE is funded through the UK Research and Innovation Industrial Strategy Challenge Fund and delivered through Health Data Research UK.
Supplementary Data
References
- 1.GBD Chronic Respiratory Disease Collaborators Prevalence and attributable health burden of chronic respiratory diseases, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir Med. 2020;8(6):585–596. doi: 10.1016/S2213-2600(20)30105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman E.D., Reddel H.K., van Zyl-Smit R.N., Agusti A. The asthma-COPD overlap syndrome: towards a revised taxonomy of chronic airways diseases? Lancet Respir Med. 2015;3(9):719–728. doi: 10.1016/S2213-2600(15)00254-4. [DOI] [PubMed] [Google Scholar]
- 3.Postma D.S., Rabe K.F. The asthma-COPD overlap syndrome. N Engl J Med. 2015;373(13):1241–1249. doi: 10.1056/NEJMra1411863. [DOI] [PubMed] [Google Scholar]
- 4.Global Initiative for Asthma; Global Initiative for Chronic Obstructive Lung Disease Diagnosis of Diseases of Chronic Airflow Limitation: Asthma, COPD, and Asthma-COPD Overlap Syndrome (ACOS). 2015. https://goldcopd.org/wp-content/uploads/2016/04/GOLD_ACOS_2015.pdf
- 5.Ghebre M.A., Bafadhel M., Desai D., et al. Biological clustering supports both “Dutch” and “British” hypotheses of asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2015;135(1):63–72. doi: 10.1016/j.jaci.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Marco R., Marcon A., Rossi A., et al. Asthma, COPD and overlap syndrome: a longitudinal study in young European adults. Eur Respir J. 2015;46(3):671–679. doi: 10.1183/09031936.00008615. [DOI] [PubMed] [Google Scholar]
- 7.Miravitlles M., Soriano J.B., Ancochea J., et al. Characterisation of the overlap COPD-asthma phenotype: focus on physical activity and health status. Respir Med. 2013;107(7):1053–1060. doi: 10.1016/j.rmed.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Hardin M., Cho M., McDonald M.L., et al. The clinical and genetic features of COPD-asthma overlap syndrome. Eur Respir J. 2014;44(2):341–350. doi: 10.1183/09031936.00216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menezes A.M.B., Montes de Oca M., Perez-Padilla R., et al. Increased risk of exacerbation and hospitalization in subjects with an overlap phenotype: COPD-asthma. Chest. 2014;145(2):297–304. doi: 10.1378/chest.13-0622. [DOI] [PubMed] [Google Scholar]
- 10.Lange P., Colak Y., Ingebrigtsen T.S., Vestbo J., Marott J.L. Long-term prognosis of asthma, chronic obstructive pulmonary disease, and asthma-chronic obstructive pulmonary disease overlap in the Copenhagen City Heart Study: a prospective population-based analysis. Lancet Respir Med. 2016;4(6):454–462. doi: 10.1016/S2213-2600(16)00098-9. [DOI] [PubMed] [Google Scholar]
- 11.Kauppi P., Kupiainen H., Lindqvist A., et al. Overlap syndrome of asthma and COPD predicts low quality of life. J Asthma. 2011;48(3):279–285. doi: 10.3109/02770903.2011.555576. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Guzman E., Khosravi M., Mannino D.M. Asthma, chronic obstructive pulmonary disease, and mortality in the U.S. population. COPD. 2011;8(6):400–407. doi: 10.3109/15412555.2011.611200. [DOI] [PubMed] [Google Scholar]
- 13.Henriksen A.H., Langhammer A., Steinshamn S., Mai X.M., Brumpton B.M. The prevalence and symptom profile of asthma-COPD overlap: the HUNT Study. COPD. 2018;15(1):27–35. doi: 10.1080/15412555.2017.1408580. [DOI] [PubMed] [Google Scholar]
- 14.Global Initiative for Chronic Obstructive Lung Disease Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease: 2020 Report. https://goldcopd.org/wp-content/uploads/2019/12/GOLD-2020-FINAL-ver1.2-03Dec19_WMV.pdf
- 15.Zhou J.J., Cho M.H., Castaldi P.J., Hersh C.P., Silverman E.K., Laird N.M. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am. J Respir Crit Care Med. 2013;188(8):941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Los H., Koppelman G.H., Postma D.S. The importance of genetic influences in asthma. Eur Respir J. 1999;14(5):1210–1227. doi: 10.1183/09031936.99.14512109. [DOI] [PubMed] [Google Scholar]
- 17.Duffy D.L., Martin N.G., Battistutta D., Hopper J.L., Mathews J.D. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142(6 Pt 1):1351–1358. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 18.Sluiter H.J., Koëter G.H., de Monchy J.G., Postma D.S., de Vries K., Orie N.G. The Dutch hypothesis (chronic non-specific lung disease) revisited. Eur Respir J. 1991;4(4):479–489. [PubMed] [Google Scholar]
- 19.Hobbs B.D., de Jong K., Lamontagne M., et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49(3):426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smolonska J., Koppelman G.H., Wijmenga C., et al. Common genes underlying asthma and COPD? Genome-wide analysis on the Dutch hypothesis. Eur Respir J. 2014;44(4):860–872. doi: 10.1183/09031936.00001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bycroft C., Freeman C., Petkova D., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchini J. Band G. SNPTEST. https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
- 23.Bulik-Sullivan B. Finucane H. LDSC (LD Score) https://github.com/bulik/ldsc
- 24.Yang J. GCTA-COJO: multi-SNP-based conditional & joint association analysis using GWAS summary data. https://yanglab.westlake.edu.cn/software/gcta/#COJO
- 25.John C., Reeve N.F., Free R.C., et al. Cohort profile: Extended Cohort for E-health, Environment and DNA (EXCEED) Int J Epidemiol. 2019;48(3):678–679j. doi: 10.1093/ije/dyz073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pividori M., Schoettler N., Nicolae D.L., Ober C., Im H.K. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med. 2019;7(6):509–522. doi: 10.1016/S2213-2600(19)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakefield J. Reporting and interpretation in genome-wide association studies. Int J Epidemiol. 2008;37(3):641–653. doi: 10.1093/ije/dym257. [DOI] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan B., Finucane H.K., Anttila V., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demenais F., Margaritte-Jeannin P., Barnes K.C., et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50(1):42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shrine N., Portelli M.A., John C., et al. Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study. Lancet Respir Med. 2019;7(1):20–34. doi: 10.1016/S2213-2600(18)30389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakornsakolpat P., Prokopenko D., Lamontagne M., et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet. 2019;51(3):494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Astle W.J., Elding H., Jiang T., et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167(5):1415–1429.e1419. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrine N., Guyatt A.L., Erzurumluoglu A.M., et al. Understanding Society Scientific Group New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51(3):481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng J., Erzurumluoglu A.M., Elsworth B.L., et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2016;33(2):272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira M.A., Vonk J.M., Baurecht H., et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49(12):1752–1757. doi: 10.1038/ng.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirota T., Takahashi A., Kubo M., et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43(9):893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira M.A.R., Matheson M.C., Tang C.S., et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol. 2014;133(6):1564–1571. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ceccarelli D.F., Ivantsiv S., Mullin A.A., et al. FAM105A/OTULINL Is a pseudodeubiquitinase of the OTU-class that localizes to the ER membrane. Structure. 2019;27(6):1000–1012.e6. doi: 10.1016/j.str.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vicente C.T., Revez J.A., Ferreira M.A.R. Lessons from ten years of genome-wide association studies of asthma. Clin Transl Immunology. 2017;6(12):e165. doi: 10.1038/cti.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauvreau G.M., O’Byrne P.M., Boulet L.P., et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370(22):2102–2110. doi: 10.1056/NEJMoa1402895. [DOI] [PubMed] [Google Scholar]
- 42.Wyss A.B., Sofer T., Lee M.K., et al. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nat Commun. 2018;9(1):2976. doi: 10.1038/s41467-018-05369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hynes G.M., Hinks T.S.C. The role of interleukin-17 in asthma: a protective response? ERJ Open Res. 2020;6(2):00364–2019. doi: 10.1183/23120541.00364-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miossec P., Kolls J.K. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11(10):763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 45.Mellett M., Atzei P., Horgan A., et al. Orphan receptor IL-17RD tunes IL-17A signalling and is required for neutrophilia. Nat Commun. 2012;3:1119. doi: 10.1038/ncomms2127. [DOI] [PubMed] [Google Scholar]
- 46.Ortega H.G., Liu M.C., Pavord I.D., et al. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med. 2014;371(13):1198–1207. doi: 10.1056/NEJMoa1403290. [DOI] [PubMed] [Google Scholar]
- 47.Chupp G.L., Bradford E.S., Albers F.C., et al. Efficacy of mepolizumab add-on therapy on health-related quality of life and markers of asthma control in severe eosinophilic asthma (MUSCA): a randomised, double-blind, placebo-controlled, parallel-group, multicentre, phase 3b trial. Lancet Respir Med. 2017;5(5):390–400. doi: 10.1016/S2213-2600(17)30125-X. [DOI] [PubMed] [Google Scholar]
- 48.Castro M., Zangrilli J., Wechsler M.E., et al. Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: results from two multicentre, parallel, double-blind, randomised, placebo-controlled, phase 3 trials. Lancet Respir Med. 2015;3(5):355–366. doi: 10.1016/S2213-2600(15)00042-9. [DOI] [PubMed] [Google Scholar]
- 49.Mukherjee M., Cingolani E., Pritchard D.I., Bosquillon C. Enhanced expression of organic cation transporters in bronchial epithelial cell layers following insults associated with asthma: impact on salbutamol transport. Eur J Pharm Sci. 2017;106:62–70. doi: 10.1016/j.ejps.2017.05.052. [DOI] [PubMed] [Google Scholar]
- 50.Zeiger R.S., Schatz M., Li Q., et al. High blood eosinophil count is a risk factor for future asthma exacerbations in adult persistent asthma. J Allergy Clin Immunol Pract. 2014;2(6):741–750. doi: 10.1016/j.jaip.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Price D., Wilson A.M., Chisholm A., et al. Predicting frequent asthma exacerbations using blood eosinophil count and other patient data routinely available in clinical practice. J Asthma Allergy. 2016;9:1–12. doi: 10.2147/JAA.S97973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siva R., Green R.H., Brightling C.E., et al. Eosinophilic airway inflammation and exacerbations of COPD: a randomised controlled trial. Eur Respir J. 2007;29(5):906–913. doi: 10.1183/09031936.00146306. [DOI] [PubMed] [Google Scholar]
- 53.Hancox R.J., Pavord I.D., Sears M.R. Associations between blood eosinophils and decline in lung function among adults with and without asthma. Eur Respir J. 2018;51(4):1702536. doi: 10.1183/13993003.02536-2017. [DOI] [PubMed] [Google Scholar]
- 54.Miravitlles M. Diagnosis of asthma-COPD overlap: the five commandments. Eur Respir J. 2017;49(5):1700506. doi: 10.1183/13993003.00506-2017. [DOI] [PubMed] [Google Scholar]
- 55.Christenson S.A., Steiling K., van den Berge M., et al. Asthma-COPD overlap: clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191(7):758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayden L.P., Cho M.H., Raby B.A., Beaty T.H., Silverman E.K., Hersh C.P. Childhood asthma is associated with COPD and known asthma variants in COPDGene: a genome-wide association study. Respir Res. 2018;19(1):209. doi: 10.1186/s12931-018-0890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulit S.L., de With S.A., de Bakker P.I. Resetting the bar: statistical significance in whole-genome sequencing-based association studies of global populations. Genet Epidemiol. 2017;41(2):145–151. doi: 10.1002/gepi.22032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.