Abstract
BACKGROUND
Metabolic reprogramming has been identified as a core hallmark of cancer. Solute carrier family 2 is a major glucose carrier family. It consists of 14 members, and we mainly study solute carrier family 2 member 1 (SLC2A1) and solute carrier family 2 member 2 (SLC2A2) here. SLC2A1, mainly existing in human erythrocytes, brain endothelial cells, and normal placenta, was found to be increased in hepatocellular carcinoma (HCC), while SLC2A2, the major transporter of the normal liver, was decreased in HCC.
AIM
To identify if SLC2A1 and SLC2A2 were associated with immune infiltration in addition to participating in the metabolic reprogramming in HCC.
METHODS
The expression levels of SLC2A1 and SLC2A2 were tested in HepG2 cells, HepG215 cells, and multiple databases. The clinical characteristics and survival data of SLC2A1 and SLC2A2 were examined by multiple databases. The correlation between SLC2A1 and SLC2A2 was analyzed by multiple databases. The functions and pathways in which SLC2A1, SLC2A2, and frequently altered neighbor genes were involved were discussed in String. Immune infiltration levels and immune marker genes associated with SLC2A1 and SLC2A2 were discussed from multiple databases.
RESULTS
The expression level of SLC2A1 was up-regulated, but the expression level of SLC2A2 was down-regulated in HepG2 cells, HepG215 cells, and liver cancer patients. The expression levels of SLC2A1 and SLC2A2 were related to tumor volume, grade, and stage in HCC. Interestingly, the expression levels of SLC2A1 and SLC2A2 were negatively correlated. Further, high SLC2A1 expression and low SLC2A2 expression were linked to poor overall survival and relapse-free survival. SLC2A1, SLC2A2, and frequently altered neighbor genes played a major role in the occurrence and development of tumors. Notably, SLC2A1 was positively correlated with tumor immune infiltration, while SLC2A2 was negatively correlated with tumor immune infiltration. Particularly, SLC2A2 methylation was positively correlated with lymphocytes.
CONCLUSION
SLC2A1 and SLC2A2 are independent therapeutic targets for HCC, and they are quintessential marker molecules for predicting and regulating the number and status of immune cells in HCC.
Keywords: Hepatocellular carcinoma, Solute carrier family 2 member 1, Solute carrier family 2 member 2, Prognostic, Immune infiltration
Core Tip: We performed an integrated bioinformatics analysis to assess the influence of solute carrier family 2 member 1 (SLC2A1) and solute carrier family 2 member 2 (SLC2A2) in hepatocellular carcinoma (HCC). This study revealed that SLC2A1 and SLC2A2 are independent therapeutic targets in HCC and that they are correlated with immune infiltration, in addition to being involved in the metabolic reprogramming in HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the third most common cancer in the world[1], among which 55% of cases come from China and 80% of cases are caused by hepatitis B virus (HBV) infection[2]. HCC is an inflammation-induced tumor, and immune escape is one of the characteristics[3]. Immune cells and cytokines in the immune microenvironment play an important role in the occurrence and development of HCC[4]. Based on this, programmed cell death 1 (PD-1), CD274 molecule (PD-L1), and cytotoxic T-lymphocyte associated protein 4 (CTLA4) monoclonal antibodies prevent T cells from failing and thus activate the anti-cancer immunity[5]. However, clinical trials showed that PD-1 antibodies (nivolumab[6] and pembrolizumab[7]) and CTLA4 antibodies (tremelimumab[8]) improved clinical outcomes in a few patients or were not effective at all. Therefore, recognition molecules representing the patient's immune status will help identify subgroups sensitive to immunomodulatory drugs. Moreover, some independent target molecules associated with the tumor immune microenvironment are worth exploring in HCC.
Metabolic reprogramming was recognized as a core hallmark of cancer[9]. The solute carrier family 2 is the important carrier for glucose to enter target cells, and its ability to transport glucose is the first rate-determining step in tumor metabolic reprogramming[10]. Solute carrier family 2 member 1 (SLC2A1), one of the members of the solute carrier family 2, mainly exist in human erythrocytes, brain endothelial cells, and a normal placenta[11]. It is highly expressed in HCC[12]. Interestingly, solute carrier family 2 member 2 (SLC2A2), the major transporter of the normal liver[13], is low expressed when HCC occurs[14]. Lactic acid was produced and the pH changed by the Warburg effect, which was part of metabolic reprogramming affected immune cells in the tumor immune microenvironment[15]. A previous study showed that overexpression of SLC2A1 was correlated with suppressing CD8+ T cells and B cells in gastric cancer[16]. Nevertheless, the potential functions and mechanisms of SLC2A2 in HCC are still unclear.
In this study, multiple databases were used to estimate the expression levels of SLC2A1 and SLC2A2 in HCC. Then, expressions of SLC2A1 and SLC2A2 were correlated with the clinical characteristics and prognosis of patients. In addition, we also evaluated the pathways in which SLC2A1, SLC2A2, and frequently altered neighbor genes participated were involved in the occurrence and development of tumors. Notably, we analyzed the correlation between SLC2A1 and SLC2A2 and immune cells in the HCC microenvironment. Thus, this study clarified the crucial role of SLC2A1 and SLC2A2, and it was the first to propose that SLC2A1 and SLC2A2 are related to immune cells in HCC.
MATERIALS AND METHODS
National Center for Biotechnology Information's gene
National Center for Biotechnology Information's gene database (www.ncbi.nlm.nih.gov/gene/)[17] integrating gene-specific information from a wide range of species showed the expression of human genes SLC2A1 and SLC2A2 in normal liver tissues from 95 human individuals.
Gene expression series 121248 (GSE121248) in gene expression omnibus
Gene expression omnibus (www.ncbi.nlm.nih.gov/geo/)[18] is an international public repository of microarray chips, second-generation sequencing, and other high-throughput genetic data uploaded by researchers around the world. GSE121248[19], a cohort of HCC patients, demonstrated the expression levels of SLC2A1 and SLC2A2 compared with normal samples and the correlation between SLC2A1 and SLC2A2 expression in HCC.
The Cancer Genome Atlas
The Cancer Genome Atlas (TCGA) (portal.gdc.cancer.gov/)[20] was one of the most ambitious and successful cancer genomics programs to date. Raw counts of RNA sequencing data and corresponding clinical information from 371 HCC and normal tissue samples were available from TCGA. Clinical information of these patients is shown in Table 1.
Table 1.
Clinical information description of hepatocellular carcinoma patients in The Cancer Genome Atlas
|
Description
|
Samples, n = 371
|
Percentage
|
| Sex | ||
| Male | 249 | 67.11 |
| Female | 121 | 32.61 |
| Others | 1 | 0.27 |
| Race | ||
| White | 184 | 49.60 |
| Asian | 158 | 42.59 |
| Black or African American | 16 | 4.31 |
| Others | 13 | 3.50 |
| Vital status | ||
| Alive | 240 | 64.69 |
| Dead | 130 | 35.14 |
| Others | 1 | 0.27 |
| Age at diagnosis | ||
| ≥ 60-years-old | 197 | 53.10 |
| < 60-years-old | 169 | 45.55 |
| Others | 5 | 1.35 |
| AJCC pathologic T | ||
| T1 | 180 | 48.52 |
| T2 | 92 | 24.80 |
| T2a | 1 | 0.27 |
| T2b | 1 | 0.27 |
| T3 | 45 | 12.13 |
| T3a | 29 | 7.82 |
| T3b | 6 | 1.62 |
| T4 | 13 | 3.50 |
| TX | 1 | 0.27 |
| Others | 1 | 0.27 |
| AJCC system for tumor staging | ||
| N0 | 239 | 64.42 |
| N1 | 4 | 1.08 |
| Nx | 91 | 24.53 |
| Others | 2 | 0.54 |
| AJCC system for metastasis staging | ||
| M0 | 265 | 71.43 |
| M1 | 4 | 1.08 |
| Mx | 101 | 27.22 |
| Others | 1 | 0.27 |
| AJCC system for stage | ||
| Stage I | 170 | 45.82 |
| Stage II | 86 | 23.18 |
| Stage III | 3 | 0.81 |
| Stage IIIA | 65 | 17.52 |
| Stage IIIB | 8 | 2.16 |
| Stage IIIC | 9 | 2.43 |
| Stage IV | 2 | 0.54 |
| Stage IVA | 1 | 0.27 |
| Stage IVB | 2 | 0.54 |
| Others | 25 | 6.74 |
| Prior malignancy | ||
| Yes | 35 | 9.43 |
| No | 335 | 90.30 |
| Others | 1 | 0.27 |
| Treatment or therapy | ||
| Yes | 38 | 10.24 |
| No | 309 | 83.28 |
| Others | 24 | 6.47 |
AJCC: American Joint Committee on Cancer.
Human Protein Atlas
Human Protein Atlas (www.proteinatlas.org/)[21] provided information on the tissue and cellular distribution of 24000 human proteins. SLC2A1 and SLC2A2 immunofluorescence images of HepG2 cells were obtained. Next, immunohistochemistry images showed SLC2A1 and SLC2A2 in liver cancer patients and normal tissues.
Tumor immune estimation resource
Tumor immune estimation resource (TIMER) (cistrome.shinyapps.io/timer/)[22] is a comprehensive resource for the systematic analysis of immune infiltrates across diverse cancer types. It revealed the expression levels of SLC2A1 and SLC2A2 in different cancers. Furthermore, the expression of SLC2A1 and SLC2A2 and the level of immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells) were analyzed. Moreover, the correlation between SLC2A1 and SLC2A2 expression levels and immune cell markers [markers of CD8+ T cells, T cells (general), B cells, monocytes, tumor-associated macrophage (TAM), M1 macrophages, M2 macrophages, neutrophils, natural killer (NK) cells, dendritic cells, T helper (Th) 1 cells, Th2 cells, follicular helper T (Tfh) cells, Th17 cells, T cell regulatory (Tregs) and T cell exhaustion)] were analyzed in detail.
ONCOMINE
ONCOMINE (www.oncomine.org)[23] is an online cancer microarray database. It was used to analyze the expression levels of SLC2A1 and SLC2A2 in different cancers.
UALCAN
UALCAN (ualcan.path.uab.edu/index.html)[24] is a web resource for analyzing cancer omics data. Expressions of SLC2A1 and SLC2A2 in different tumor grades and stages of HCC were analyzed by UALCAN.
Gene expression profiling interactive analysis
Gene expression profiling interactive analysis (GEPIA) (gepia.cancer-pku.cn)[25] is a developed interactive website server for analyzing the RNA sequencing expression data from the TCGA and the Genotype-Tissue Expression projects. The correlation between SLC2A1 and SLC2A2 and immune marker genes of different immune cells was analyzed.
Kaplan-Meier plotter
The free online database Kaplan-Meier plotter (kmplot.com)[26], which includes gene expression data and survival information of 364 clinical HCC patients, was utilized to predict overall survival (OS) and relapse-free survival (RFS) of HCC patients. We assessed factors such as sex, race, disease stage, disease grade, American Joint Committee on Cancer system for tumor staging (AJCC-T), vascular infiltration, treatment with sorafenib, alcohol consumption, and HBV infection associated with survival.
String
Protein-protein interaction (PPI) networks of SLC2A1, SLC2A2, and their neighbors were constructed. Then, the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were obtained by String (string-db.org)[27]. Three aspects were contained in GO enrichment analysis: Biological process (BP), molecular function (MF), and cellular component (CC). GO and KEGG enrichment analysis results were selected by P values less than 0.05 as the critical criterion.
Cytoscape
PPI of SLC2A1, SLC2A2, and neighbor genes from the String database was reconstructed by Cytoscape (v.3.6.1, cytoscape.org/)[28].
Tumor and immune system interaction database
Tumor and immune system interaction database (cis.hku.hk/TISIDB/)[29] is a web portal for tumor and immune system interaction that integrates multiple heterogeneous data types. Immune infiltration associated with SLC2A2 methylation was searched to elucidate the tumor-immune system interaction.
R project
The RV4.0.3 project (www.r-project.org/) was used to visualize the top 20 GO and KEGG data in the String database. Meanwhile, data from TCGA were also processed and visualized.
Cell culture and drug treatment
Human hepatoma cell lines HepG2 cells and HepG2215 cells were acquired from the American Type Culture Collection (Manassas, VA, United States). They were maintained in Dulbecco’s Modified Eagle’s Medium (Gibco, Waltham, MA, United States) supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin. They were maintained in an atmosphere of 5% CO2 at 37°C.
RNA sequencing
Total RNA was extracted from the sample, and the quality was tested. Then, messenger RNA (mRNA) was extracted from total RNA by using poly-t oligo attached magnetic beads and disrupting to about 300 bp. The first strand of complementary DNA (cDNA) was synthesized with random primers and reverse transcriptase by using RNA as the template, and then the second strand of cDNA was synthesized by using the first strand of cDNA as the template. Polymerase chain reaction was used to amplify DNA, and 450 bp fragments were screened out to form the libraries. The total and effective concentrations were tested after libraries were inspected by using Agilent 2100 Bioanalyzer (Santa Clara, CA, United States). Libraries containing different index sequences were mixed proportionally according to the effective concentration and the amount of data required by libraries. Mixed libraries were uniformly diluted to 2 nmol/L, and single-chain libraries were formed by alkali denaturation. These libraries were paired-end (PE) sequenced by the Next-Generation Sequencing (NGS) based on the Illumina HiSeq platform (this work was entrusted to Nanjing Personal Gene Technology Co., LTD., Jiangsu, China)
Statistical analysis
Statistical analyses were performed by the Statistical Product and Service Solutions (SPSS) 23.0 statistics software (IBM Corp., Armonk, NY, United States), GraphPad Prism 8 software (San Diego, CA, United States), and Rv4.0.3 software. Each sample was repeated at least three times. If data were normally distributed, they are represented as mean ± standard deviation. When more than two groups were included, a one-way analysis of variance was utilized. Differences were considered statistically when P value was less than 0.05.
RESULTS
SLC2A1 expression was increased whereas SLC2A2 expression was decreased in liver cancer
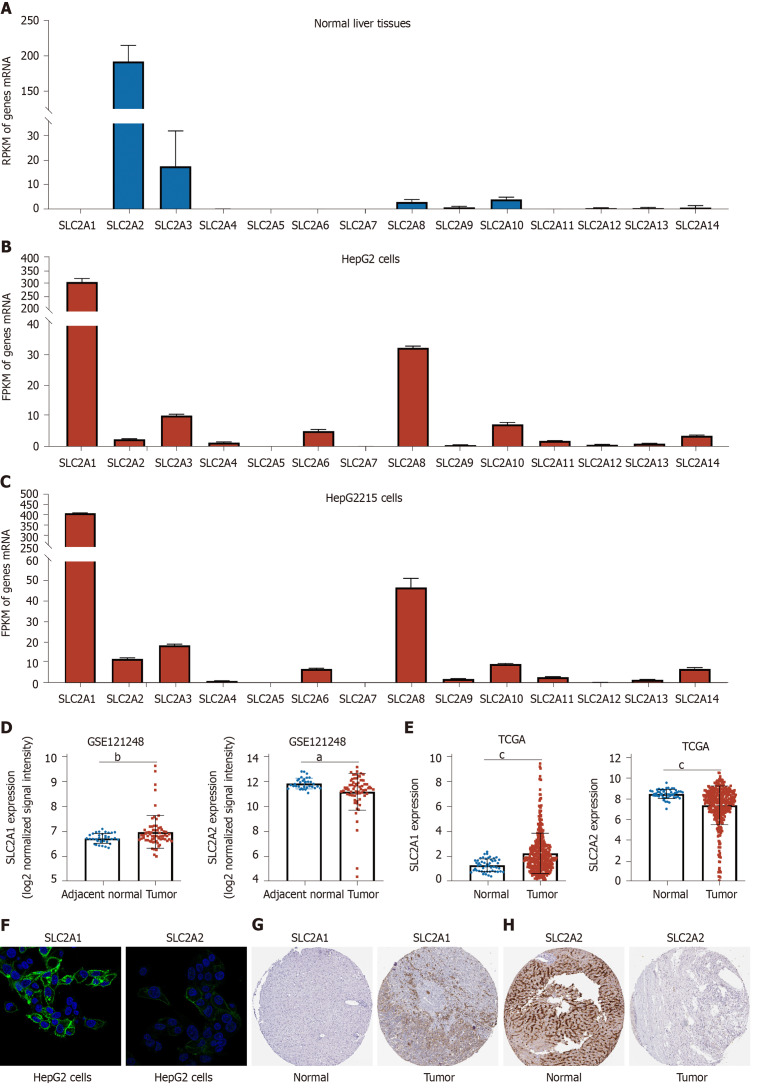
We investigated the expression of solute carrier family 2 in normal liver tissues by the National Center for Biotechnology Information's gene. We found that SLC2A2, as the normal liver transporter, was highly expressed compared with other members of the family (Figure 1A). Next, we compared the mRNA level of solute carrier family 2 in HepG2 and HepG2215 cells. Interestingly, we found that SLC2A2 expression was less but SLC2A1 was elevated in these two cell lines (Figure 1B and C). Similarly, the cohort (GSE121248) of HCC patients showed that SLC2A1 expression was elevated while SLC2A2 expression was reduced compared with the normal samples (Figure 1D). Consistently, TCGA data also indicated that SLC2A1 was up-regulated and SLC2A2 was down-regulated in HCC patients (Figure 1E). Notably, the Human Protein Atlas database revealed that SLC2A1 protein expression was expressed and SLC2A2 protein expression was weakly expressed in HepG2 cells (Figure 1F). As expected, SLC2A1 protein expression (Figure 1G) was increased and SLC2A2 protein expression (Figure 1H) was decreased in liver cancer in the Human Protein Atlas database. Taken together, SLC2A1 was up-regulated while SLC2A2 was down-regulated in liver cancer patients.
Figure 1.
Solute carrier family 2 member 1 expression was increased whereas solute carrier family 2 member 2 expression was decreased in liver cancer. A: The reads per kilobase million (RPKM) of solute carrier family 2 in normal liver tissues from 95 human individuals by National Center for Biotechnology Information's gene database. The RPKM of solute carrier family 2 member 2 (SLC2A2) was 193.619, while solute carrier family 2 member 1 (SLC2A1) was almost undetectable; B: The fragments per kilobase million (FPKM) of solute carrier family 2 in HepG2 cells by RNA sequencing. The FPKM of SLC2A1 was 314.336 while SLC2A2 was 2.68857 in HepG2 cells; C: The FPKM of solute carrier family 2 in HepG2215 cells by RNA sequencing. The FPKM of SLC2A1 was 412.048 while SLC2A2 was 12.0905 in HepG2215 cells; D: The messenger RNA expression level of SLC2A1 was increased while that of SLC2A2 was decreased in hepatocellular carcinoma (HCC) in the cohort [gene expression series 121248 (GSE121248)]; E: The messenger RNA expression level of SLC2A1 was increased while that of SLC2A2 was decreased in HCC compared with normal tissues in The Cancer Genome Atlas (TCGA); F: Immunofluorescence analysis revealed that SLC2A1 protein expression was expressed and SLC2A2 protein expression was weakly expressed in HepG2 cells in the Human Protein Atlas; G: Immunohistochemical analysis showed the expression level of SLC2A1 was increased while SLC2A2 was decreased in liver cancer compared with normal tissues in the Human Protein Atlas. aP < 0.05; bP < 0.01; cP < 0.001.
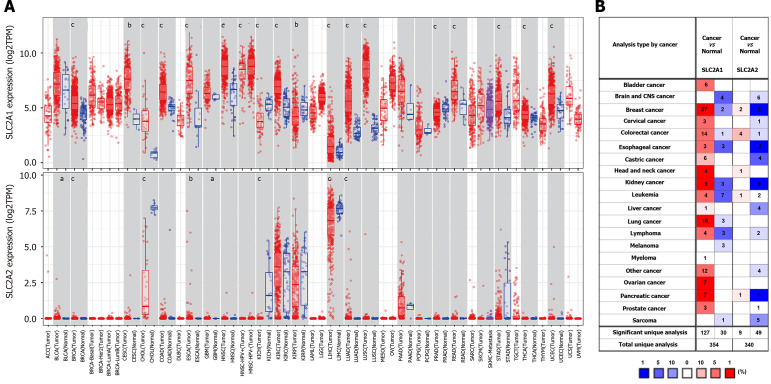
SLC2A1 and SLC2A2 mRNA expression levels in different human cancers were changed by using TIMER and ONCOMINE databases
To see if this phenomenon was universal, the mRNA expressions of SLC2A1 and SLC2A2 in different human cancers were investigated by using the TIMER database. This analysis revealed that the SLC2A1 was higher in breast invasive carcinoma, cervical squamous cell carcinoma and endometrial adenocarcinoma, cholangiocarcinoma, colon adenocarcinoma, esophageal carcinoma, head and neck cancer, kidney renal clear cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, rectum adenocarcinoma, stomach adenocarcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma (Figure 2A). It was, however, lower in kidney chromophobe, kidney renal papillary cell carcinoma, and prostate adenocarcinoma (Figure 2A). Similarly, SLC2A2 was lower in cholangitis carcinoma, kidney chromophobe, and liver hepatocellular carcinoma (Figure 2A), but it was higher in lung adenocarcinoma, bladder urothelial carcinoma, breast invasive carcinoma, esophageal carcinoma, and glioblastoma multiforme (Figure 2A). Although SLC2A2 was increased in these cancers, it was expressed at low or absent levels in these cancers and the corresponding normal samples.
Figure 2.
Solute carrier family 2 member 1 and solute carrier family 2 member 2 messenger RNA expression levels in different human cancers were changed by using tumor immune estimation resource and ONCOMINE databases. A: The messenger RNA expression levels of solute carrier family 2 member 1 (SLC2A1) and solute carrier family 2 member 2 (SLC2A2) in different types of human cancers by the Tumor Immune Estimation Resource database; B: The messenger RNA expression levels of SLC2A1 and SLC2A2 in different human cancers by ONCOMINE database. aP < 0.05, bP < 0.01, cP < 0.001. TPM: Transcripts per million; ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endometrial adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and neck cancer; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute Myeloid Leukemia; LGG: Brain Lower Grade Glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MESO: Mesothelioma; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin Cutaneous Melanoma; STAD stomach adenocarcinoma; TGCT: Testicular Germ Cell Tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma.
To verify further expressions of SLC2A2 and SLC2A1 in cancer, we examined their expressions in the ONCOMINE database. The result showed that SLC2A1 was up-regulated in most cancers and SLC2A2 was down-regulated in most cancers in most datasets. SLC2A1 has been observed to be up-regulated in bladder cancer, breast cancer, cervical cancer, colorectal cancer, esophageal cancer, gastric cancer, head and neck cancer, kidney cancer, leukemia, liver cancer, lung cancer, lymphoma, myeloma, ovarian cancer, pancreatic cancer, and prostate cancer. However, some datasets showed that SLC2A1 was down-regulated in brain and central nervous system cancer, breast cancer, colorectal cancer, esophageal cancer, kidney cancer, leukemia, lung cancer, lymphoma, melanoma, and sarcoma (Figure 2B). Similarly, the result showed the expression of SLC2A2 was reduced in brain and central nervous system cancer, breast cancer, cervical cancer, colorectal cancer, esophageal cancer, gastric cancer, kidney cancer, leukemia, liver cancer, lymphoma, pancreatic cancer, prostate cancer, and sarcoma. In other datasets, SLC2A2 was up-regulated in breast cancer, colorectal cancer, head and neck cancer, leukemia, and pancreatic cancer (Figure 2B). In addition to HCC, SLC2A1 elevation and SLC2A2 reduction have been observed in several other cancers. SLC2A2, as a key transporter in normal liver tissue, was decreased in the occurrence of HCC, while SLC2A1 expression was increased. Thus, we still discuss relevant issues of SLC2A1 and SLC2A2 in HCC.
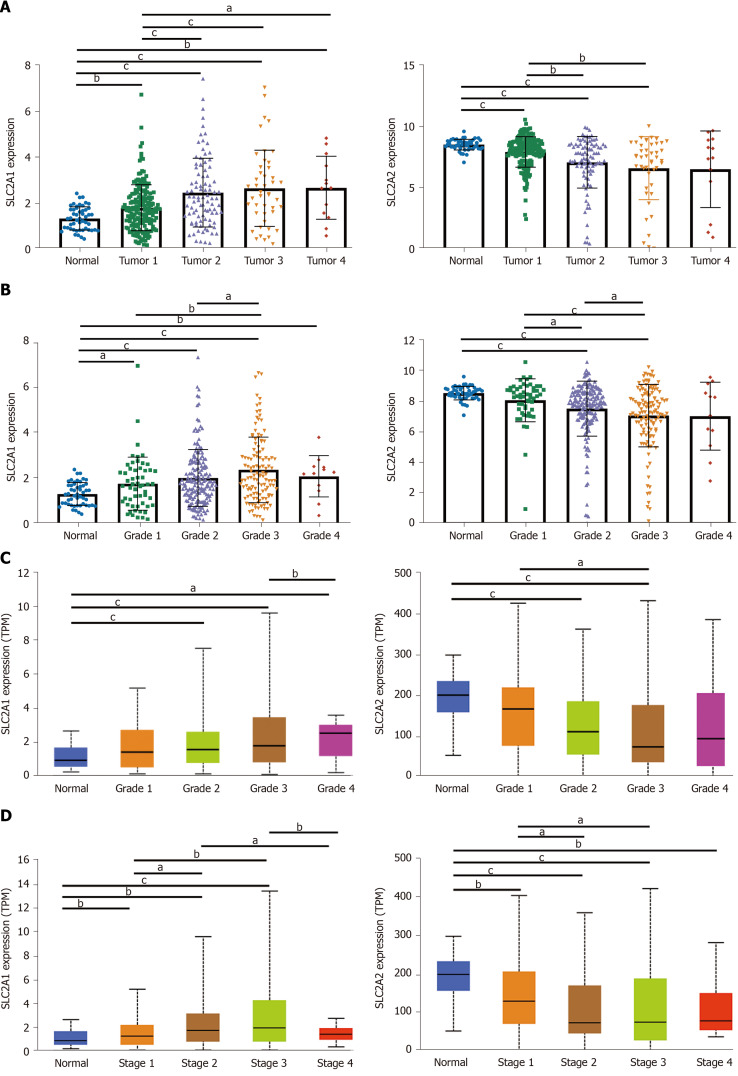
SLC2A1 and SLC2A2 transcription levels in HCC patients were correlated with tumor volume, grade, and stage
Next, we investigated the correlation between SLC2A1 and SLC2A2 expression levels and tumor volume, grade, and stage in HCC patients in TCGA data and UALCAN database. The results showed that the SLC2A1 level gradually increased while the SLC2A2 level gradually decreased with the process of primary tumor volume enlargement in TCGA data (Figure 3A). Consistently, highly differentiated HCC indicated lower SLC2A1 and higher SLC2A2 compared with poorly differentiated HCC in TCGA data and UALCAN database (Figure 3B and C). Notably, the expression of SLC2A1 was higher while the expression of SLC2A2 was lower with the progression of HCC grade (Figure 3D) in UALCAN database. Collectively, with the development of HCC, the expression of SLC2A1 was increased, while the expression of SLC2A2 was decreased.
Figure 3.
Solute carrier family 2 member 1 and solute carrier family 2 member 2 transcription levels in hepatocellular carcinoma patients were correlated with tumor volume, grade, and stage. A: Solute carrier family 2 member 1 (SLC2A1) level gradually increased whereas solute carrier family 2 member 2 (SLC2A2) level gradually decreased with the process of primary tumor volume enlargement in The Cancer Genome Atlas (TCGA) data; B: SLC2A1 level gradually increased, whereas SLC2A2 level gradually decreased with the process of hepatocellular carcinoma (HCC) grade in TCGA data; C: SLC2A1 level gradually increased whereas SLC2A2 level gradually decreased with the process of HCC grade in the UALCAN database; D: SLC2A1 level gradually increased, whereas SLC2A2 level gradually decreased with the process of HCC stage in UALCAN database. aP < 0.05; bP < 0.01; cP < 0.001.
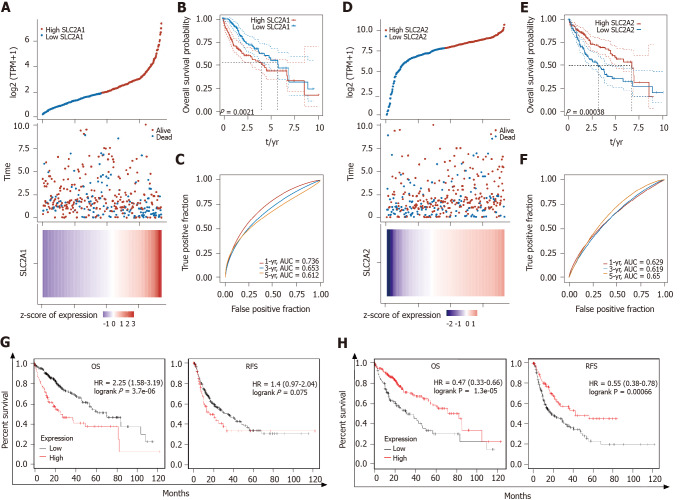
SLC2A1 and SLC2A2 mRNA levels were correlated with survival rates of HCC patients in TCGA data and the Kaplan-Meier Plotter database
To clarify the impact of the expression levels of SLC2A1 and SLC2A2 on patient prognosis, survival analysis was utilized to analyze the correlation between these expression levels and the survival rates of HCC patients.
Patients with high expression of SLC2A1 had poor OS, which suggested that high SLC2A1 expression posed a risk factor for HCC patients (Figure 4A and B). Consistently, higher expression of SLC2A2 patients had a better OS, which suggested high SLC2A2 expression was a protective factor in HCC (Figure 4D and E). One-year, three-year, and five-year areas under the curve of SLC2A1 and SLC2A2 were all greater than 0.6, which suggested the prediction result was reliable (Figure 4C and F). In addition, low expression SLC2A1 and high expression SLC2A2 have better OS and RFS in the Kaplan-Meier Plotter database (Figure 4G and H).
Figure 4.
Solute carrier family 2 member 1 and solute carrier family 2 member 2 messenger RNA levels were correlated with survival rates of hepatocellular carcinoma patients in The Cancer Genome Atlas data and the Kaplan-Meier Plotter database. A: In The Cancer Genome Atlas (TCGA) database, the top graph represents the scatter graph of solute carrier family 2 member 1 (SLC2A1) expression from low to high, where the blue represented the low expression group and the red represented the high expression group. The middle graph represents the scatter graph of the survival time and survival state corresponding to the SLC2A1 expression level of different samples. The bottom graph is a heat map of the expression of SLC2A1 in diverse samples; B: According to the expression level of SLC2A1, the patients were divided into the high expression group and the low expression group, and the results suggested that the low expression group had better overall survival (OS); C: Area under the curve (AUC) of SLC2A1 in 1, 3, and 5 years. The higher the value was, the stronger the predictive ability of the gene was; D: In TCGA database, the top graph represents the scatter graph of solute carrier family 2 member 2 (SLC2A2) expression from low to high, where the blue represents the low expression group and the red represents the high expression group. The middle graph represents the scatter graph of the survival time and survival state corresponding to the SLC2A2 expression level of different samples. The bottom graph is a heat map of the expression of SLC2A2 in diverse samples; E: According to the expression level of SLC2A2, the patients were divided into the high expression group and the low expression group and the results suggested that the low expression group had poor OS; F: AUC of SLC2A2 in 1, 3, and 5 years. The higher the value was, the stronger the prognostic ability of the gene; G: Hepatocellular carcinoma (HCC) patients with high expression of SLC2A1 had poorer OS [hazard ratio (HR): 2.25, P = 3.7e-06] and relapse-free survival (RFS) (HR: 1.4, P = 0.075); H: HCC patients with high expression of SLC2A2 had better OS (HR: 2.25, P = 3.7e-06) and RFS (HR: 0.55, P = 6.6e-04).
Next, we analyzed the correlation between SLC2A1 and SLC2A2 expression and clinical characteristics. These clinical characteristics included sex, race, stage, grade, and AJCC-T and whether the patients received sorafenib treatment, were infected with HBV, consumed alcohol, and had vascular invasion (Table 2). These results suggested that high SLC2A1 and low SLC2A2 were independent risk factors for HCC, independent of these factors. Notably, prominent expression of SLC2A1 was more dangerous for males than females, Asians than whites, and those who were infected with HBV than uninfected. Confusingly, high expression of SLC2A1 was more dangerous in non-alcoholics than in alcoholics. Notably, with the progression of the stage, the grade, and AJCC-T of HCC, the high expression of SLC2A1 and low expression of SLC2A2 became worse for patients.
Table 2.
Correlation of solute carrier family 2 member 1 and solute carrier family 2 expression and clinical prognosis in hepatocellular carcinoma with different clinicopathological factors in Kaplan-Meier plotter
| Clinicopathological characteristics |
Solute carrier family 2 member 1
|
Solute carrier family 2 member 2
|
||||||||||
|
Overall survival
|
Relapse-free survival
|
Overall survival
|
Relapse-free survival
|
|||||||||
|
n
|
Hazard ratio
|
P
value
|
n
|
Hazard ratio
|
P
value
|
n
|
Hazard ratio
|
P
value
|
n
|
Hazard ratio
|
P
value
|
|
| Sex | ||||||||||||
| Male | 246 | 2.71 (1.73-4.24) | 5.40E-06 | 210 | 1.36 (0.89-2.08) | 0.149 | 246 | 0.3 (0.0.19-0.48) | 5.80E-08 | 210 | 0.5 (0.34-0.75) | 0.00068 |
| Female | 118 | 1.89 (1.06-3.38) | 0.0287 | 106 | 1.61 (0.89-2.9) | 0.1109 | 118 | 1.48 (0.75-2.93) | 0.26 | 106 | 0.47 (0.26-0.85) | 0.0108 |
| Race | ||||||||||||
| Asian | 155 | 3.14 (1.71-5.77) | 1.00E-04 | 143 | 1.54 (0.93-2.56) | 0.092 | 155 | 0.2 (0.11-0.38) | 3.80E-08 | 143 | 0.41 (0.23-0.73) | 0.0017 |
| White | 181 | 1.98 (1.24-3.16) | 0.0034 | 147 | 1.41 (0.88-2.27) | 0.1527 | 181 | 0.61 (0.35-1.05) | 0.072 | 147 | 0.53 (0.33-0.84) | 0.0065 |
| Black or African American | 17 | NA | NA | 13 | NA | NA | 17 | NA | NA | 13 | NA | NA |
| Sorafenib treatment | ||||||||||||
| Yes | 29 | 2.19 (0.61-7.89) | 0.219 | 22 | 1.43 (0.59-3.47) | 0.4312 | 29 | 0.23 (0.06-0.88) | 0.0205 | 22 | 0.61 (0.24-1.53) | 0.2867 |
| None | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Risk factors | ||||||||||||
| Alcohol consumption | ||||||||||||
| Yes | 115 | 1.94 (1-3.78) | 0.0464 | 99 | 1.61 (0.86-3.03) | 0.1365 | 115 | 0.42 (0.22-0.8) | 0.0063 | 99 | 0.37 (0.2-0.67) | 0.00061 |
| None | 202 | 2.34 (1.46-3.73) | 0.00025 | 183 | 1.42 (0.9-2.23) | 0.1284 | 202 | 0.41 (0.26-0.65) | 9.70E-05 | 183 | 0.57 (0.34-0.96) | 0.0337 |
| Hepatitis virus | ||||||||||||
| Yes | 150 | 2.99 (1.5-5.95) | 0.0011 | 139 | 1.45 (0.87-2.39) | 0.149 | 150 | 0.32 (0.16-0.62) | 0.0004 | 139 | 0.63 (0.37-1.05) | 0.0749 |
| None | 167 | 2.39 (1.45-3.96) | 0.00046 | 143 | 1.92 (1.12-3.32) | 0.0168 | 167 | 0.47 (0.27-0.83) | 0.0083 | 143 | 0.41 (0.25-0.68) | 0.00038 |
| Pathology | ||||||||||||
| Stage | ||||||||||||
| 1 | 170 | 2.49 (1.33-4.64) | 0.0031 | 153 | 1.67 (0.97-2.89) | 0.0617 | 170 | 0.42 (0.22-0.77) | 0.0044 | 153 | 0.41 (0.22-0.76) | 0.0035 |
| 1+2 | 253 | 2.09 (1.27-3.44) | 0.003 | 228 | 1.22 (0.8-1.86) | 0.35 | 253 | 0.54 (0.33-0.87) | 0.0107 | 228 | 0.4 (0.23-0.7) | 0.00091 |
| 2 | 83 | 2.12 (0.79-5.64) | 0.1258 | 75 | 0.5 (0.25-0.99) | 0.0424 | 83 | 0.34 (0.1-1.14) | 0.0665 | 75 | 0.56 (0.27-1.14) | 0.1046 |
| 2+3 | 166 | 2.27 (1.39-3.7) | 0.00072 | 145 | 1.44 (0.89-2.35) | 0.1389 | 166 | 0.47 (0.26-0.83) | 0.0077 | 145 | 0.65 (0.41-1.03) | 0.064 |
| 3 | 83 | 2.78 (1.52-5.06) | 0.00052 | 70 | 2.29 (1.21-4.3) | 0.0086 | 83 | 0.31 (0.16-0.57) | 0.00011 | 70 | 0.54 (0.29-0.99) | 0.0428 |
| 3+4 | 87 | 2.67 (1.49-4.78) | 6.00E-01 | 70 | 2.29 (1.21-4.3) | 0.0086 | 87 | 0.34 (0.19-0.62) | 0.0003 | 70 | 0.54 (0.29-0.99) | 0.0428 |
| 4 | 5 | NA | NA | 0 | NA | NA | 4 | NA | NA | 0 | NA | NA |
| Grade | ||||||||||||
| 1 | 55 | 0.67 (0.26-1.7) | 0.3967 | 45 | 3.65 (0.83-16.1) | 0.067 | 55 | 0.42 (0.14-1.27) | 0.1114 | 45 | 0.45 (0.17-1.17) | 0.0919 |
| 2 | 174 | 2.16 (1.29-3.61) | 0.0025 | 149 | 1.49 (0.89-2.5) | 0.1246 | 174 | 0.48 (0.28-0.81) | 0.0051 | 149 | 0.48 (0.29-0.78) | 0.0029 |
| 3 | 118 | 3.11 (1.69-5.7) | 0.00012 | 107 | 2.16 (1.2-3.86) | 0.0082 | 118 | 0.3 (0.14-0.66) | 0.0014 | 107 | 0.62 (0.34-1,.14) | 0.1221 |
| 4 | 12 | NA | NA | 11 | NA | NA | 12 | NA | NA | 11 | NA | NA |
| American Joint Committee on Cancer system for tumor staging | ||||||||||||
| 1 | 180 | 2.31 (1.27-4.21) | 0.0047 | 160 | 1.51 (0.89-2.58 | 0.1245 | 180 | 0.39 (0.22-0.71) | 0.0014 | 160 | 0.41 (0.22-0.75) | 0.0027 |
| 2 | 90 | 2.07 (0.79-5.44) | 0.1303 | 80 | 0.48 (0.25-0.92) | 0.023 | 90 | 0.45 (0.16-1.29) | 0.1272 | 80 | 2.02 (0.9-4.56) | 0.0836 |
| 3 | 78 | 3.04 (1.64-5.64) | 0.00022 | 67 | 2.23 (1.11-4.48) | 0.0209 | 78 | 0.29 (0.15-0.57) | 0.00011 | 67 | 0.43 (0.22-0.83) | 0.01 |
| 4 | 13 | NA | NA | 6 | NA | NA | 13 | NA | NA | 6 | NA | NA |
| Vascular invasion | ||||||||||||
| None | 203 | 2.18 (91.26-3.78) | 0.0043 | 175 | 1.32 (0.77-2.28) | 0.3144 | 203 | 0.39 (0.23-0.66) | 0.00027 | 175 | 0.45 (0.25-0.83) | 0.0089 |
| Micro | 90 | 2.16 (1.01-4.62) | 0.042 | 82 | 0.46 (0.24-0.88) | 0.0163 | 90 | 0.32 (0.1-1.08) | 0.0531 | 82 | 0.73 (0.37-1.44) | 0.3593 |
| Macro | 16 | NA | NA | 14 | NA | NA | 16 | NA | NA | 14 | NA | NA |
NA: Not applicable.
Together, both the high expression of SLC2A1 and the low expression of SLC2A2 were independent risk factors for HCC patients and led to poor prognosis.
SLC2A1 and SLC2A2 showed negative correlations in HCC patients in GSE121248, TCGA, TIMER database, and GEPIA database
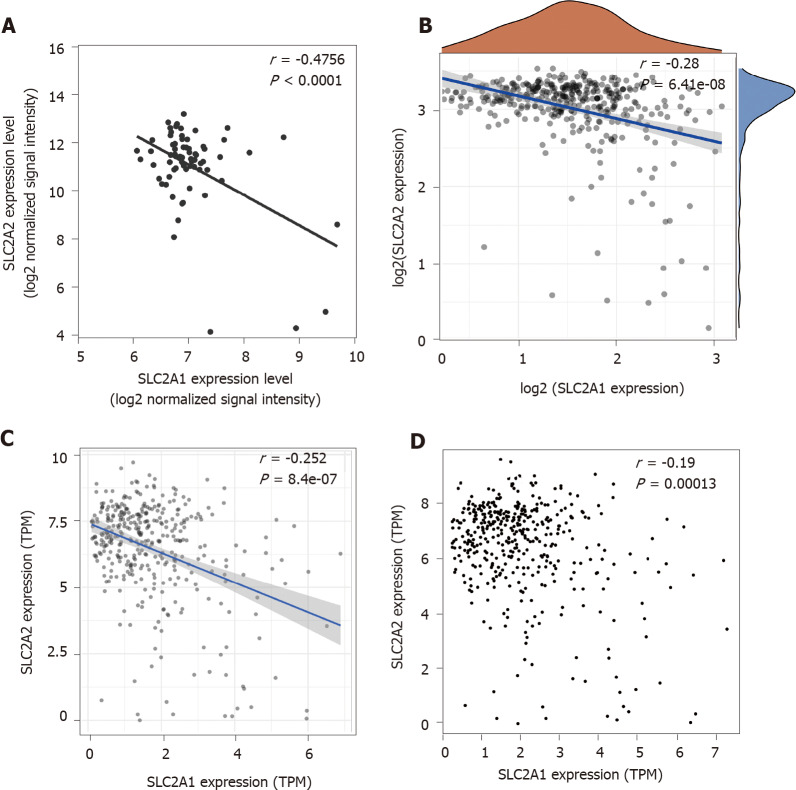
To investigate whether the opposite trend of SLC2A1 and SLC2A2 expression was accidental, we proceeded to correlation analysis of the expression levels. The results showed that there were always negative correlations between SLC2A1 and SLC2A2 expression levels by the cohort GSE121248 (r = -0.4756) (Figure 5A), TCGA data (r = -0.28) (Figure 5B), TIMER database (r = -0.252) (Figure 5C), and GEPIA (r = -0.19) (Figure 5D) database. It was inevitable that SLC2A1 and SLC2A2 had an opposite expression tendency in HCC, because they may be involved in some biological processes besides serving as glucose transporters together.
Figure 5.
Solute carrier family 2 member 1 and solute carrier family 2 member 2 showed negative correlations in hepatocellular carcinoma patients in multiple databases. A: solute carrier family 2 member 1 (SLC2A1) was negatively correlated with solute carrier family 2 member 2 (SLC2A2) in the cohort gene expression series 121248 (r = -0.4756, P < 0.0001); B: SLC2A1 was negatively correlated with SLC2A2 in The Cancer Genome Atlas data (r = -0.28, P = 6.41e-08); C: SLC2A1 was negatively correlated with SLC2A2 in the Tumor Immune Estimation Resource database (r = -0.252, P = 8.4e-07); D: SLC2A1 was negatively correlated with SLC2A2 in the gene expression profiling interactive analysis database (r = -0.19, P = 1.3e-04); TPM: Transcripts per million.
String database was used to predict the effects of SLC2A1, SLC2A2, and frequently altered adjacent genes expression changes on functions and pathways
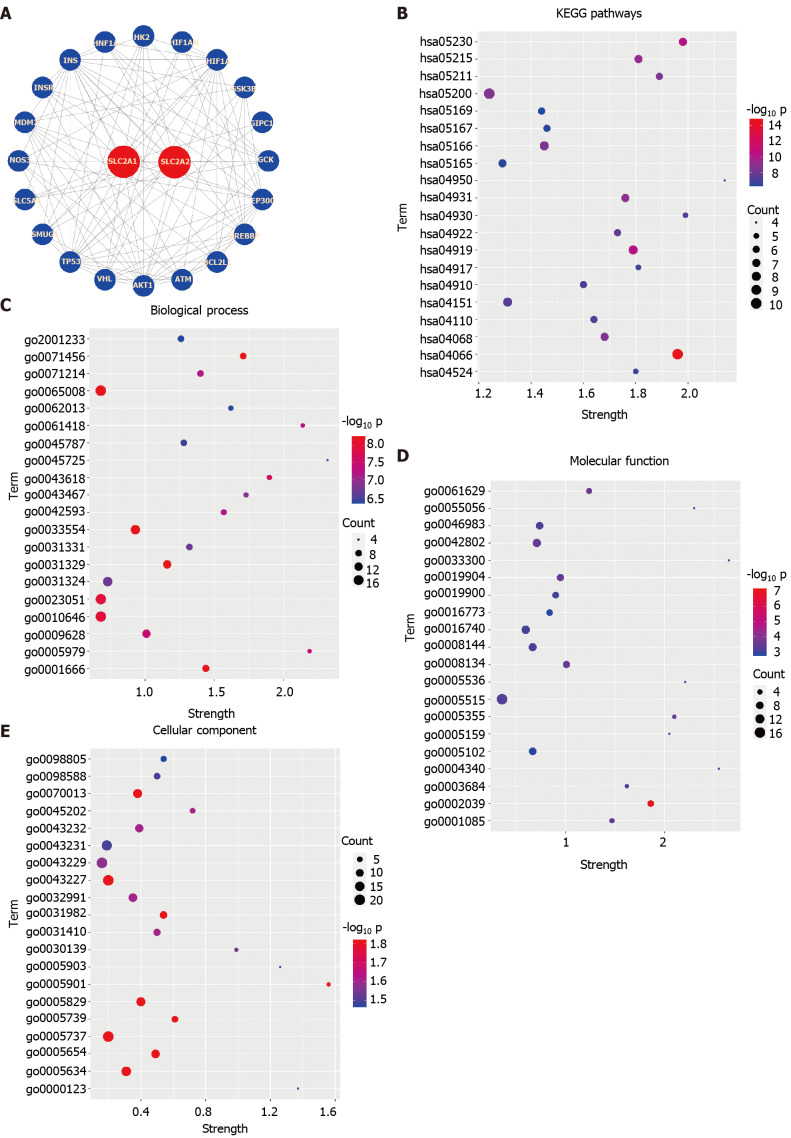
The expression levels of SLC2A1 and SLC2A2 were modified in HCC, and their expressions were negatively correlated. Therefore, we explored the influence of changes of them and their neighbor genes on the functions and pathways in the String database. We structured the PPI network of SLC2A1, SLC2A2, and their neighbor genes in the String database, which contained 22 nodes and 107 edges (Figure 6A). In this network diagram, genes tumor protein p53 (TP53), ATM serine/threonine kinase (ATM), MDM2 proto-oncogene (MDM2), protein kinase-B (AKT1), hypoxia inducible factor 1 subunit alpha (HIF1A), binding protein p300 (EP300), insulin (INS), glucokinase (GCK), and hexokinase 2 (HK2) were included.
Figure 6.
The String database was used to predict the effects of solute carrier family 2 member 1, solute carrier family 2 member 2, and frequently altered adjacent genes expression changes on functions and pathways. A: The protein-protein interaction networks of solute carrier family 2 member 1 (SLC2A1), solute carrier family 2 member 2 (SLC2A2), and their neighbor genes in the String database were constructed, which contained 22 nodes and 107 edges. In this networks diagram, genes tumor protein p53, ATM serine/threonine kinase, MDM2 proto-oncogene, protein kinase-B, hypoxia inducible factor 1 subunit alpha, binding protein p300, insulin, glucokinase, and hexokinase 2 were included; B: SLC2A1, SLC2A2, and their neighbors were analyzed by Kyoto Encyclopedia of Genes and Genomes, and the top 20 genes were ranked by P value from small to large in the String database; C: SLC2A1, SLC2A2, and their neighbors were analyzed by biological process (BP), and the top 20 genes were ranked by P value from small to large in the String database; D: SLC2A1, SLC2A2, and their neighbors were analyzed by biological process (MF), and the top 20 genes were ranked by P value from small to large in the String database; E: SLC2A1, SLC2A2, and their neighbors were analyzed by cellular component (CC), and the top 20 genes were ranked by P value from small to large in the String database. HIF1AN: Hypoxia inducible factor 1 subunit alpha inhibitor; GSK3B: Glycogen synthase kinase 3 beta; GIPC1: GIPC PDZ domain containing family member 1; CREBBP: CREB binding protein; BCL2L1: BCL2 Like 1; VHL: von Hippel-Lindau tumor suppressor; SMUG1: Single-strand-selective monofunctional uracil-DNA glycosylase 1; SLC5A1: Solute carrier family 5 member 1; NOS3: Nitric oxide synthase 3; INSR: Insulin receptor; HNF1A: HNF1 homeobox A.
Next, we carried out GO analysis on these genes in the database, and GO analysis results included three different levels of BP, CC, and MF. In Figures 6C, D, and E, we selected and showed the first 20 P values from small to large. The results indicated that cellular response to hypoxia (GO:0001666), cellular response to stress (GO:0033554), regulation of cell communication (GO:0010646), regulation of transcription from RNA polymerase II promoter in response to stress (GO:0043618), glucose homeostasis (GO:0042593), negative regulation of cellular metabolic process (GO:0031324), negative regulation of cellular metabolic process (GO:0031324), positive regulation of cellular catabolic process (GO:0045787), regulation of apoptotic signaling pathway (GO:2001233), and others were involved.
Then, to explore which pathways these genes influenced together, we performed KEGG analysis and showed the top 20 from small to large according to the P value. KEGG pathway analysis of these genes presented hypoxia-inducible factor 1 signaling pathway (hsa04066), central carbon metabolism in cancer (hsa05230), pathways in cancer (hsa05200), Forkhead box O (FOXO) signaling pathway (hsa04068), phosphatidylinositol 3-kinase/AKT signaling pathway (hsa04151), cell cycle pathway (hsa04110), insulin resistance pathway (hsa04931), glucagon signaling pathway (hsa04922), insulin signaling pathway (hsa04910), and others were involved (Figure 6B).
These complex regulatory relationships indicated that SLC2A1 and SLC2A2 were not only directly involved in the metabolism of glucose but also participated in many important pathways together with other neighbor genes. Although GO and KEGG have proved that SLC2A1, SLC2A2, and their neighbor genes are involved in the occurrence and development of tumors, they have not proved the influence on the immune microenvironment. These results were based on previously discovered interrelationships but did not include what has not been discovered.
Landscape of infiltrating immune cells in HCC was different from that in normal liver tissues and expressions of SLC2A1 and SLC2A2 correlated with immune infiltration levels in HCC
It has been noted that the lactic acid microenvironment associated with the Warburg effect always affected the number and function of some immune cells[30]. Some studies have also found that SLC2A1 expression levels were correlated with immune cells in some tumors[16,31]. However, the relationship between glucose transporters SLC2A1 and SLC2A2 and the immune microenvironment was unclear in HCC.
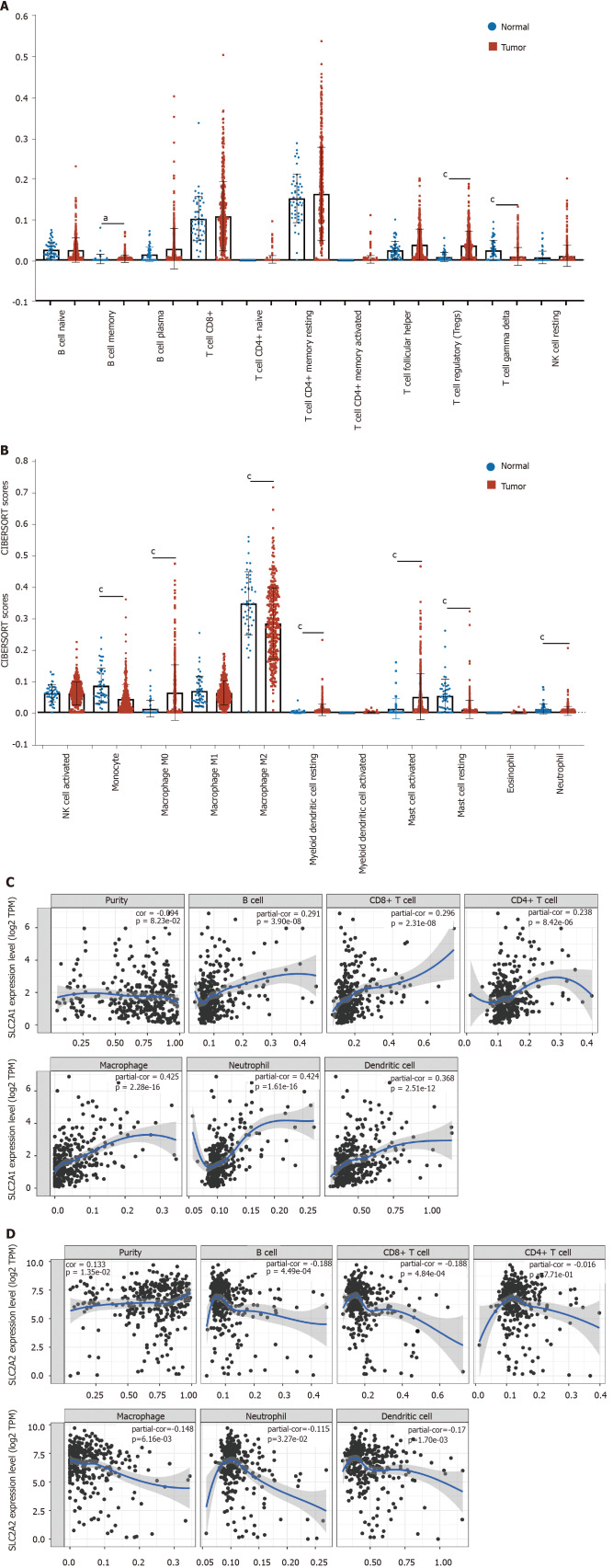
The Cell Type Identification by Estimating Relative RNA Transcript subsets was performed firstly. The results showed that HCC had different levels of immune invasion compared to normal tissues. In comparison to normal tissues, the proportions of B cell memory, Tregs, T cell gamma delta, monocyte, macrophage M0, myeloid dendritic cell resting, mast cell activated, mast cell resting, and neutrophil had changed in HCC (Figure 7A and B).
Figure 7.
The landscape of infiltrating immune cells in hepatocellular carcinoma was different from that in normal liver tissues and expressions of solute carrier family 2 member 1 and solute carrier family 2 member 2 correlated with immune infiltration level in hepatocellular carcinoma. A and B: The pattern of immune cells by the signature gene expression profile in hepatocellular carcinoma compared with normal samples with the Cell Type Identification by Estimating Relative RNA Transcript subsets method. In comparison to normal tissues, the proportions of B cell memory, regulatory T, T cell gamma delta, monocyte, macrophage M0, macrophage M2, myeloid dendritic cell resting, mast cell activated, mast cell resting, and neutrophil had changed; C: Correlation analysis between solute carrier family 2 member 1 (SLC2A1) transcription level and immune cell infiltration level. The immune cells included B cells (partial.cor (r) = 0.291), CD8+ T cells (r = 0.296), CD4+ T cells (r = 0.238), macrophages (r = 0.425), neutrophils (r = 0.424), and dendritic cells (r = 0.368); D: Correlation analysis between solute carrier family 2 member 1 (SLC2A2) transcription level and immune cell infiltration level. The immune cells included B cells (r = -0.188), CD8+ T cells (r = -0.188), macrophages (r = -0.148), neutrophils (r = -0.115) and dendritic cells (r = -0.17). aP < 0.05; cP < 0.001.
Next, SLC2A1 and SLC2A2 were investigated to determine whether their expressions were correlated to the level of immune cells with infiltration by TIMER database. The result showed that SLC2A1 expression had a weak negative correlation with tumor purity, but the SLC2A1 expression level had a positive correlation with infiltrating levels of B cells (r = 0.291), CD8+ T cells (r = 0.296), CD4+ T cells (r = 0.238), macrophages (r = 0.425), neutrophils (r = 0.424), and dendritic cells (r = 0.368) (Figure 7C). Conversely, SLC2A2 expression showed a weak positive correlation with tumor purity, but the SLC2A2 expression level had a negative correlation with infiltrating levels of B cells (r = -0.188), CD8+ T cells (r = -0.188), macrophages (r = -0.148), neutrophils (r = -0.115) and dendritic cells (r = -0.17) (Figure 7D).
SLC2A1 and SLC2A2 expressions correlated with immune marker genes in HCC by using TIMER and GEPIA databases
To understand further the relationship between SLC2A1 and SLC2A2 and various immune cells in HCC, we studied the correlation between SLC2A1 and SLC2A2 and immune marker genes of different immune cells in TIMER and GEPIA databases. These immune marker genes included CD8+ T cells, T cells (general), B cells, monocytes, TAM, M1 macrophages, M2 macrophages, neutrophils, NK cells, and dendritic cells. We also analyzed the different functional T cells, such as Tregs, Th1 cells, Th2 cells, Tfh cells, Th17 cells, and T cells exhaustion. After the correlation adjustment by purity, the result showed that the SLC2A1 expression was positively correlated with most immune marker genes. On the contrary, SLC2A2 was negatively correlated with these immune cells markers.
Notably, these results showed that SLC2A1 had a strong positive correlation with the immune markers of CD8+ T cells, T cells (general), and B cells, while SLC2A2 had a weak negative correlation with these markers in TIMER (Table 3) and GEPIA (Table 4) databases. These markers included CD8A, CD8B, CD3D, CD3E, CD2, CD19, and CD79A.
Table 3.
Correlation analysis between solute carrier family 2 member 1 and solute carrier family 2 and genes markers of immune cells in tumor immune estimation resource
| Description | Gene markers |
Solute carrier family 2 member 1
|
Solute carrier family 2 member 2
|
||||||
|
None
|
Purity
|
None
|
Purity
|
||||||
|
cor
|
P
value
|
cor
|
P
value
|
cor
|
P
value
|
cor
|
P
value
|
||
| CD8+ T cell | |||||||||
| CD8A | 0.216 | 2.88E-05 | 0.173 | 1.27E-03 | -0.103 | 4.85E-02 | -0.027 | 6.15E-01 | |
| CD8B | 0.229 | 8.60E-06 | 0.195 | 2.72E-04 | -0.199 | 1.14E-04 | -0.138 | 1.04E-02 | |
| T cell (general) | |||||||||
| CD3D | 0.241 | 2.75E-06 | 0.22 | 3.73E-05 | -0.327 | 1.33E-10 | -0.276 | 1.88E-07 | |
| CD3E | 0.201 | 1.01E-04 | 0.162 | 2.59E-03 | -0.165 | 1.44E-03 | -0.087 | 1.08E-01 | |
| CD2 | 0.178 | 5.64E-04 | 0.138 | 1.04E-02 | -0.194 | 1.75E-04 | -0.121 | 2.48E-02 | |
| B cell | |||||||||
| CD19 | 0.208 | 5.50E-05 | 0.176 | 1.04E-03 | -0.137 | 8.06E-03 | -0.089 | 9.95E-02 | |
| CD79A | 0.099 | 5.72E-02 | 0.045 | 4.01E-01 | -0.138 | 7.84-03 | -0.07 | 1.92E-01 | |
| Monocyte | |||||||||
| CD86 | 0.413 | 0.00E+00 | 0.415 | 8.08E-16 | -0.196 | 1.50E-04 | -0.15 | 5.21E-03 | |
| CD115 (CSF1R) | 0.41 | 0.00E+00 | 0.408 | 3.06E-15 | -0.175 | 7.02E-04 | -0.126 | 1.96E-02 | |
| Tumor-associated macrophage | |||||||||
| CCL2 | 0.206 | 6.86E-05 | 0.162 | 2.53E-03 | -0.117 | 2.47E-02 | -0.042 | 4.32E-01 | |
| CD68 | 0.38 | 4.24E-14 | 0.381 | 2.40E-13 | -0.235 | 5.28E-06 | -0.192 | 3.26E-04 | |
| IL10 | 0.326 | 1.26E-10 | 0.309 | 4.43E-09 | -0.131 | 1.15E-02 | -0.078 | 1.48E-01 | |
| M1 macrophage | |||||||||
| INOS (NOS2) | 0.068 | 1.91E-01 | 0.052 | 3.39E-01 | 0.175 | 7.12E-04 | 0.18 | 7.95E-04 | |
| IRF5 | 0.142 | 6.33E-03 | 0.138 | 1.05E-02 | 0.011 | 8.39E-01 | 0.003 | 9.63E-01 | |
| COX2 (PTGS2) | 0.32 | 2.91E-10 | 0.313 | 2.80E-09 | -0.06 | 2.45E-01 | -0.007 | 9.03E-01 | |
| M2 macrophage | |||||||||
| CD163 | 0.28 | 3.94E-08 | 0.253 | 1.91E-06 | 0.046 | 3.72E-01 | 0.125 | 2.00E-02 | |
| VSIG4 | 0.353 | 3.65E-12 | 0.336 | 1.56E-10 | -0.06 | 2.46E-01 | -0.007 | 8.96E-01 | |
| MS4A4A | 0.337 | 3.49E-11 | 0.33 | 3.15E-10 | -0.057 | 2.69E-01 | 0.01 | 9.85E-01 | |
| CD66b (CEACAM8) | 0.086 | 9.68E-02 | 0.084 | 1.20E-01 | -0.036 | 4.89E-01 | -0.027 | 6.12E-01 | |
| CD11b (ITGAM) | 0.428 | 0.00E+00 | 0.405 | 4.71E-15 | -0.18 | 5.06E-04 | -0.156 | 3.75E-03 | |
| CCR7 | 0.088 | 8.89E-02 | 0.016 | 7.70E-01 | -0.032 | 5.35E-01 | -0.045 | 4.00E-01 | |
| KIR2DL1 | 0.083 | 1.09E-01 | 0.063 | 2.46E-01 | -0.003 | 9.51E-01 | 0.019 | 7.23E-01 | |
| KIR2DL3 | 0.179 | 5.16E-04 | 0.167 | 1.82E-03 | -0.006 | 9.15E-01 | -0.038 | 4.79E-01 | |
| KIR2DL4 | 0.234 | 5.38E-06 | 0.212 | 7.12E-05 | -0.133 | 1.06E-02 | -0.107 | 4.78E-02 | |
| KIR3DL1 | 0.146 | 4.97E-03 | 0.12 | 2.64E-02 | 0.079 | 1.30E-01 | 0.116 | 3.15E-02 | |
| KIR3DL2 | 0.098 | 6.04E-02 | 0.08 | 1.38E-01 | -0.027 | 6.02E-01 | 0.003 | 9.60E-01 | |
| KIR3DL3 | 0.06 | 2.53E-01 | 0.046 | 3.91E-01 | -0.021 | 6.91E-01 | -0.015 | 7.87E-01 | |
| KIR2DS4 | 0.117 | 2.39E-02 | 0.13 | 1.57E-02 | 0.04 | 4.37E-01 | 0.052 | 3.37E-01 | |
| HLA-DPB1 | 0.319 | 4.21E-10 | 0.296 | 2.07E-08 | -0.191 | 2.25E-04 | -0.138 | 1.03E-02 | |
| HLA-DQB1 | 0.221 | 1.77E-05 | 0.192 | 3.35E-04 | -0.219 | 2.09E-05 | -0.171 | 1.43E-03 | |
| HLA-DRA | 0.35 | 5.25E-12 | 0.326 | 5.24E-10 | -0.117 | 2.37E-02 | -0.055 | 3.09E-01 | |
| HLA-DPA1 | 0.338 | 3.01E-11 | 0.316 | 1.92E-09 | -0.114 | 2.88E-02 | -0.052 | 3.34E-01 | |
| BDCA-1 (CD1C) | 0.061 | 2.45E-01 | 0.024 | 6.52E-01 | 0.004 | 9.40E-01 | 0.073 | 1.77E-01 | |
| BDCA-4 (NRP1) | 0.357 | 1.99E-12 | 0.352 | 1.70E-11 | 0.043 | 4.13E-01 | 0.087 | 1.08E-01 | |
| CD11c (ITGAX) | 0.382 | 2.93E-14 | 0.38 | 2.50E-13 | -0.154 | 2.93E-03 | -0.118 | 2.86E-02 | |
| Th1 | |||||||||
| T-bet (TBX21) | 0.095 | 6.88E-02 | 0.042 | 4.32E-01 | 0.025 | 6.38E-01 | 0.116 | 3.07E-02 | |
| STAT4 | 0.082 | 1.13E-01 | 0.045 | 4.04E-01 | -0.106 | 4.22E-02 | -0.058 | 2.84E-01 | |
| STAT1 | 0.314 | 7.29E-10 | 0.29 | 4.18E-08 | 0.023 | 6.52E-01 | 0.058 | 2.84E-01 | |
| IFN-γ (IFNG) | 0.237 | 3.81E-06 | 0.212 | 7.39E-05 | -0.169 | 1.09E-03 | -0.122 | 2.31E-02 | |
| TNF-α (TNF) | 0.277 | 6.15E-08 | 0.257 | 1.34E-06 | -0.147 | 4.67E-03 | -0.092 | 8.91E-02 | |
| Th2 | |||||||||
| GATA3 | 0.248 | 1.34E-06 | 0.235 | 1.03E-05 | -0.176 | 6.76E-04 | -0.108 | 4.52E-02 | |
| STAT6 | 0.27 | 1.35E-07 | 0.255 | 1.63E-06 | 0.178 | 5.68E-04 | 0.19 | 3.82E-04 | |
| STAT5A | 0.37 | 1.83E-13 | 0.338 | 1.18E-10 | -0.097 | 6.27E-02 | -0.055 | 3.12E-01 | |
| IL13 | -0.081 | 1.19E-01 | -0.113 | 3.56E-02 | 0.077 | 1.38E-01 | 0.09 | 9.40E-02 | |
| Tfh | |||||||||
| BCL6 | 0.267 | 2.04E-07 | 0.281 | 1.16E-07 | 0.227 | 1.08E-05 | 0.228 | 1.94E-05 | |
| IL21 | 0.124 | 1.65E-02 | 0.13 | 1.55E-02 | -0.065 | 2.15E-01 | -0.06 | 2.68E-01 | |
| Th17 | |||||||||
| IL17A | -0.047 | 3.68E-01 | -0.033 | 5.46E-01 | 0.04 | 4.47E-01 | -0.051 | 3.41E-01 | |
| T cell regulatory | |||||||||
| FOXP3 | 0.068 | 1.89E-01 | -0.039 | 4.68E-01 | 0.169 | 1.07E-03 | 0.189 | 4.24E-04 | |
| CCR8 | 0.286 | 1.95E-08 | 0.264 | 6.44E-07 | -0.056 | 2.79E-01 | -0.008 | 8.86E-01 | |
| STAT5B | 0.156 | 2.63E-03 | 0.173 | 1.29E-03 | 0.402 | 0.00E+00 | 0.397 | 1.83E-14 | |
| TGFβ (TGFB1) | 0.314 | 7.92E-10 | 0.303 | 8.83E-09 | -0.281 | 4.30E-08 | -0.242 | 5.53E-06 | |
| T cell exhaustion | |||||||||
| PD-1 (PDCD1) | 0.256 | 5.91E-07 | 0.232 | 1.31E-05 | -0.281 | 3.50E-08 | -0.238 | 7.71E-06 | |
| CTLA4 | 0.284 | 2.57E-08 | 0.266 | 5.31E-07 | -0.327 | 1.15E-10 | -0.283 | 8.93E-08 | |
| LAG3 | 0.156 | 2.67E-02 | 0.127 | 1.83E-02 | -0.195 | 1.66E-04 | -0.165 | 2.15E-03 | |
| TIM-3 (HAVCR2) | 0.434 | 0.00E+00 | 0.437 | 1.52E-17 | -0.255 | 6.91E-07 | -0.228 | 1.89E-05 | |
| GZMB | 0.176 | 6.91E-04 | 0.131 | 1.51E-02 | -0.124 | 1.70E-02 | -0.068 | 2.10E-01 | |
cor: Correlation coefficient; L10: Interleukin 10; INOS: Nitric oxide synthase 2; IRF5: Interferon regulatory factor 5; COX2: Cytochrome c oxidase subunit II; VSIG4: V-set and immunoglobulin domain containing 4; MS4A4A: Membrane spanning 4-domains A4A; CCR7: C-C motif chemokine receptor 7; KIR2DL1: Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 1; KIR2DL3: Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 3; KIR2DL4: Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4; KIR3DL1: Killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 1; KIR3DL2: Killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 2; KIR3DL3: Killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 3; KIR2DS4: Killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 4; HLA-DPB1: Major histocompatibility complex, class II, DP beta 1; HLA-DQB1: Major histocompatibility complex, class II, DQ beta 1; HLA-DRA: Major histocompatibility complex, class II, DR alpha; HLA-DPA1: Major histocompatibility complex, class II, DP alpha 1; BDCA-4: Neuropilin 1; T-bet: T-box transcription factor 21; STAT4: Signal transducer and activator of transcription 4; STAT1: Signal transducer and activator of transcription 1; IFN-γ: Interferon gamma; TNF-α: Tumor necrosis factor; GATA3: GATA binding protein 3; STAT6: Signal transducer and activator of transcription 6; STAT5A: Signal transducer and activator of transcription 5A; IL13: Interleukin 13; BCL6: BCL6 transcription repressor; IL21: Interleukin 21; IL17A: Interleukin 17A; FOXP3: Forkhead box P3; CCR8: C-C motif chemokine receptor 8; STAT5B: Signal transducer and activator of transcription 5B; TGFβ: Transforming growth factor-beta; PD-1: Programmed cell death 1; CTLA4: Cytotoxic T-lymphocyte associated protein 4; LAG3: Lymphocyte activating 3; TIM-3: Hepatitis A virus cellular receptor 2; GZMB: Granzyme B; Tfh: Follicular helper T.
Table 4.
Correlation analysis between solute carrier family 2 member 1 and solute carrier family 2 and genes markers of immune cells in gene expression profiling interactive analysis
| Description | Gene markers |
Solute carrier family 2 member 1
|
Solute carrier family 2 member 2
|
||
|
cor
|
P
value
|
cor
|
P
value
|
||
| CD8+ T cell | |||||
| CD8A | 0.24 | 3.10E-06 | -0.084 | 1.10E-01 | |
| CD8B | 0.26 | 3.20E-07 | -0.13 | 1.40E-02 | |
| T cell (general) | |||||
| CD3D | 0.18 | 5.10E-04 | -0.23 | 1.10E-05 | |
| CD3E | 0.15 | 4.00E-03 | -0.11 | 3.90E-02 | |
| CD2 | 0.17 | 1.50E-03 | -0.12 | 2.60E-02 | |
| B cell | |||||
| CD19 | 0.22 | 1.60E-05 | -0.073 | 1.60E-01 | |
| CD79A | 0.036 | 4.90E-01 | -0.59 | 2.60E-01 | |
| Monocyte | |||||
| CD86 | 0.39 | 1.50E-14 | -0.16 | 1.70E-03 | |
| CD115 (CSF1R) | 0.36 | 1.40E-12 | -0.14 | 5.40E-03 | |
| Tumor-associated macrophage | |||||
| CCL2 | 0.16 | 2.30E-03 | -0.13 | 1.50E-02 | |
| CD68 | 0.27 | 9.20E-08 | -0.14 | 6.50E-03 | |
| IL10 | 0.32 | 2.50E-10 | -0.095 | 6.70E-02 | |
| M1 macrophage | |||||
| INOS (NOS2) | -0.031 | 5.50E-01 | 0.2 | 1.20E-04 | |
| IRF5 | 0.14 | 7.40E-03 | 0.044 | 4.00E-01 | |
| COX2 (PTGS2) | 0.16 | 1.50E-03 | -0.028 | 5.90E-01 | |
| M2 macrophage | |||||
| CD163 | 0.31 | 7.00E-10 | -0.13 | 1.40E-02 | |
| VSIG4 | 0.36 | 1.10E-12 | -0.12 | 1.80E-02 | |
| MS4A4A | 0.29 | 1.50E-08 | -0.11 | 2.80E-02 | |
| CD66b (CEACAM8) | 0.074 | 1.60E-01 | -0.018 | 7.30E-01 | |
| Neutrophil | |||||
| CD11b (ITGAM) | 0.28 | 5.80E-08 | -0.074 | 1.60E-01 | |
| CCR7 | 0.039 | 4.60E-01 | -0.028 | 5.90E-01 | |
| Natural killer cell | |||||
| KIR2DL1 | 0.087 | 9.40E-02 | -0.059 | 2.60E-01 | |
| KIR2DL3 | 0.13 | 1.40E-02 | -0.05 | 3.40E-01 | |
| KIR2DL4 | 0.18 | 7.10E-04 | -0.12 | 2.00E-02 | |
| KIR3DL1 | 0.086 | 9.70E-02 | -0.058 | 2.70E-01 | |
| KIR3DL2 | 0.069 | 1.90E-01 | -0.0081 | 8.80E-01 | |
| KIR3DL3 | 0.08 | 1.30E-01 | -0.068 | 1.90E-01 | |
| KIR2DS4 | 0.044 | 4.00E-01 | 0.02 | 7.10E-01 | |
| Dendritic cell | |||||
| HLA-DPB1 | 0.25 | 1.40E-06 | -0.16 | 2.20E-03 | |
| HLA-DQB1 | 0.14 | 8.10E-03 | -0.19 | 2.60E-04 | |
| HLA-DRA | 0.26 | 6.00E-07 | -0.15 | 5.00E-03 | |
| HLA-DPA1 | 0.2 | 8.00E-05 | -0.12 | 2.30E-02 | |
| BDCA-1 (CD1C) | 0.12 | 2.40E-02 | -0.044 | 4.00E-01 | |
| BDCA-4 (NRP1) | 0.23 | 6.80E-06 | -0.037 | 4.80E-01 | |
| CD11c (ITGAX) | 0.24 | 2.00E-06 | -0.13 | 1.40E-02 | |
| Th1 | |||||
| T-bet (TBX21) | 0.088 | 9.00E-02 | -0.058 | 2.70E-01 | |
| STAT4 | 0.0027 | 9.60E-01 | -0.041 | 4.40E-01 | |
| STAT1 | 0.13 | 1.10E-02 | 0.017 | 7.50E-01 | |
| IFN-γ (IFNG) | 0.19 | 3.30E-04 | -0.1 | 4.70E-02 | |
| TNF-α (TNF) | 0.12 | 2.00E-02 | -0.1 | 5.40E-02 | |
| Th2 | |||||
| GATA3 | 0.31 | 2.10E-09 | -0.13 | 1.20E-02 | |
| STAT6 | 0.15 | 4.80E-03 | 0.19 | 2.40E-04 | |
| STAT5A | 0.2 | 8.20E-05 | -0.07 | 1.80E-01 | |
| IL13 | -0.025 | 6.30E-01 | -0.01 | 8.40E-01 | |
| Tfh | |||||
| BCL6 | 0.1 | 4.70E-02 | 0.24 | 2.70E-06 | |
| IL21 | 0.072 | 1.70E-01 | -0.044 | 3.90E-01 | |
| Th17 | |||||
| IL17A | -0.022 | 6.70E-01 | 0.071 | 1.80E-01 | |
| T cell regulatory | |||||
| FOXP3 | -0.036 | 4.90E-01 | 0.17 | 1.40E-03 | |
| CCR8 | 0.16 | 1.90E-03 | -0.082 | 1.20E-01 | |
| STAT5B | 0.073 | 1.60E-01 | 0.43 | 0.00E+00 | |
| TGFβ (TGFB1) | 0.24 | 2.10E-06 | -0.25 | 1.40E-06 | |
| T cell exhaustion | |||||
| PD-1 (PDCD1) | 0.08 | 1.20E-01 | -0.12 | 2.00E-02 | |
| CTLA4 | 0.17 | 1.10E-03 | -0.18 | 7.20E-04 | |
| LAG3 | 0.23 | 7.60E-06 | -0.19 | 3.30E-04 | |
| TIM-3 (HAVCR2) | 0.22 | 1.30E-05 | -0.17 | 1.30E-03 | |
| GZMB | 0.17 | 8.10E-04 | -0.16 | 2.20E-03 | |
cor: Correlation coefficient; L10: Interleukin 10; INOS: Nitric oxide synthase 2; IRF5: Interferon regulatory factor 5; COX2: Cytochrome c oxidase subunit II; VSIG4: V-set and immunoglobulin domain containing 4; MS4A4A: Membrane spanning 4-domains A4A; CCR7: C-C motif chemokine receptor 7; KIR2DL1: Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 1; KIR2DL3: Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 3; KIR2DL4: Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4; KIR3DL1: Killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 1; KIR3DL2: Killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 2; KIR3DL3: Killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 3; KIR2DS4: Killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 4; HLA-DPB1: Major histocompatibility complex, class II, DP beta 1; HLA-DQB1: Major histocompatibility complex, class II, DQ beta 1; HLA-DRA: Major histocompatibility complex, class II, DR alpha; HLA-DPA1: Major histocompatibility complex, class II, DP alpha 1; BDCA-4: Neuropilin 1; T-bet: T-box transcription factor 21; STAT4: Signal transducer and activator of transcription 4; STAT1: Signal transducer and activator of transcription 1; IFN-γ: Interferon gamma; TNF-α: Tumor necrosis factor; GATA3: GATA Binding protein 3; STAT6: Signal transducer and activator of transcription 6; STAT5A: Signal transducer and activator of transcription 5A; IL13: Interleukin 13; BCL6: BCL6 transcription repressor; IL21: Interleukin 21; IL17A: Interleukin 17A; FOXP3: Forkhead box P3; CCR8: C-C motif chemokine receptor 8; STAT5B: Signal transducer and activator of transcription 5B; TGFβ: Transforming growth factor-beta; PD-1: Programmed cell death 1; CTLA4: Cytotoxic T-lymphocyte associated protein 4; LAG3: Lymphocyte activating 3; TIM-3: Hepatitis A virus cellular receptor 2; GZMB: Granzyme B; Th: T helper; Tfh: Follicular helper T.
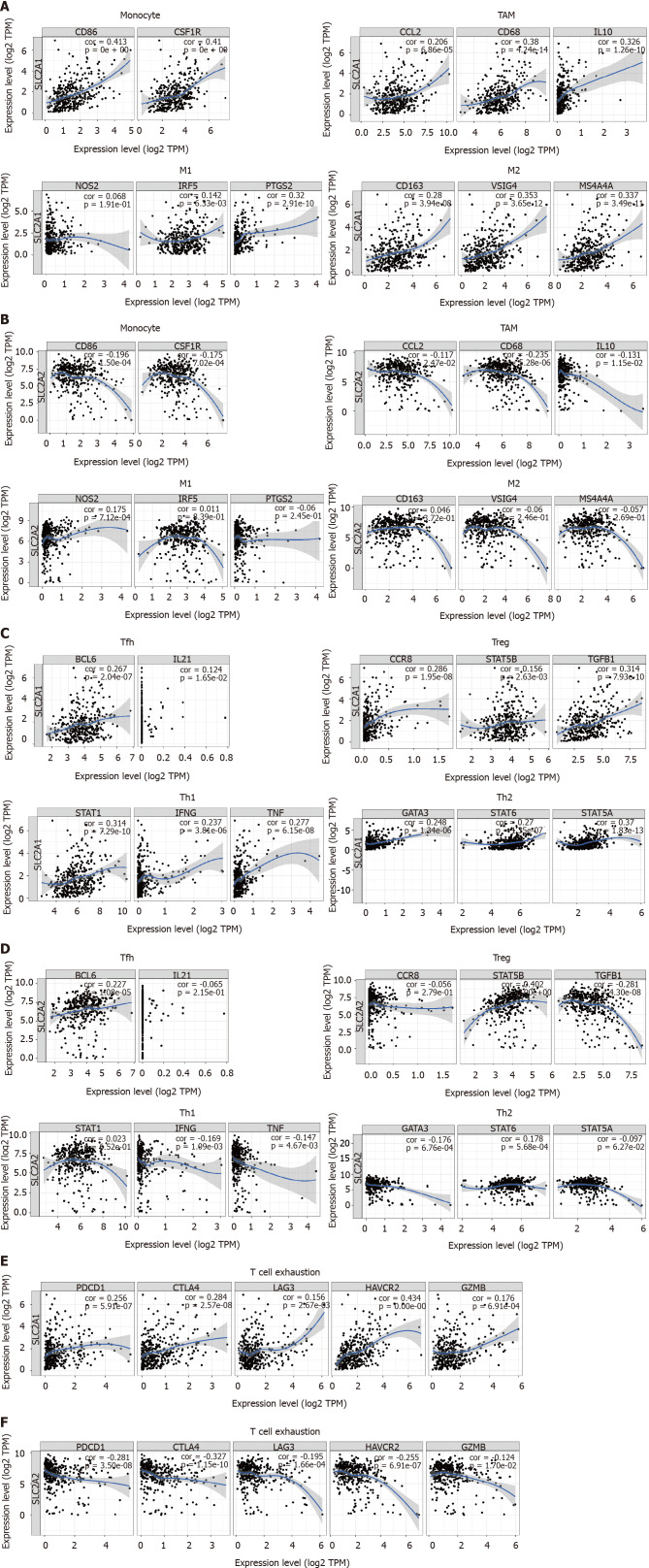
Next, SLC2A1 had a strong positive correlation with monocytes, TAM, M1, and M2 macrophages, and the correlation with M2 macrophage makers was stronger than that of M1 macrophage. In contrast, SLC2A2 presented a negative correlation with CD86 and CD115 of monocytes and C-C motif chemokine ligand 2 (CCL2), CD68, and interleukin 10 (IL10) of TAM and only presented a positive correlation with nitric oxide synthase 2 of M1 macrophage but had no obvious or negative correlation with M2 macrophage. (Figure 8A, B, Table 3 and Table 4)
Figure 8.
Solute carrier family 2 member 1 and solute carrier family 2 member 2 expressions correlated with immune marker genes in hepatocellular carcinoma by using tumor immune estimation resource and gene expression profiling interactive analysis databases. A: Solute carrier family 2 member 1 (SLC2A1) expression correlated with macrophage polarization in hepatocellular carcinoma (HCC). Markers included CD86 and CD115 of monocytes; C-C motif chemokine ligand 2, CD68, and interleukin (IL) 10 of tumor-associated macrophage; nitric oxide synthase 2, interferon regulatory factor 5, and COX2 cytochrome c oxidase subunit II of M1 macrophages; and CD163, V-set and immunoglobulin domain containing 4, and membrane spanning 4-domains A4A of M2 macrophages; B: Solute carrier family 2 member 2 (SLC2A2) expression correlated with macrophage polarization in HCC. Markers included CD86 and CD115 of monocytes; C-C motif chemokine ligand 2, CD68, and IL10 of tumor-associated macrophage; nitric oxide synthase 2, interferon regulatory factor 5, and COX2 cytochrome c oxidase subunit II of M1 macrophages; and CD163, V-set and immunoglobulin domain containing 4, and membrane spanning 4-domains A4A of M2 macrophages; C: SLC2A1 expression was correlated with functional T cells in HCC. Markers included BCL6 transcription repressor and IL21 of follicular helper T; C-C motif chemokine receptor 8, signal transducer and activator of transcription (STAT) 5B, and transforming growth factor-beta markers of T cell regulatory; STAT1, interferon gamma, and tumor necrosis factor of T helper (Th)1; and GATA binding protein 3, STAT6, and STAT5A of Th2; D: SLC2A2 expression was correlated with functional T cells in HCC. Markers included BCL6 transcription repressor and IL21 of follicular helper T; C-C motif chemokine receptor 8,, STAT5B, and transforming growth factor-beta markers of T cell regulatory; STAT1, interferon gamma, and tumor necrosis factor alpha of Th1 and GATA binding protein 3, STAT6 and STAT5A of Th2; E: SLC2A1 expression correlated with T cell exhaustion in HCC. Markers included programmed cell death 1, cytotoxic T-lymphocyte associated protein 4, lymphocyte activating 3, hepatitis A virus cellular receptor 2, and granzyme B; F: SLC2A2 expression correlated with T cell exhaustion in HCC. Markers included programmed cell death 1, cytotoxic T-lymphocyte associated protein 4, lymphocyte activating 3, hepatitis A virus cellular receptor 2, and granzyme B. TAM: Tumor-associated macrophage; CCL2: C-C motif chemokine ligand 2; IL10: interleukin 10; NOS2: nitric oxide synthase 2; IRF5: interferon regulatory factor 5; PTGS2: COX2 cytochrome c oxidase subunit II;VSIG4: V-set and immunoglobulin domain containing 4; MS4A4A: membrane spanning 4-domains A4A; Tfh: Follicular helper T; BCL6: BCL6 transcription repressor; IL21: Interleukin 21; Treg: T cell regulatory; CCR8: C-C motif chemokine receptor 8; STAT5B: signal transducer and activator of transcription 5B; TGFB1: transforming growth factor-beta; Th: T helper; STAT1: signal transducer and activator of transcription 1; IFNG: interferon gamma; TNF: tumor necrosis factor; GATA3: GATA binding protein 3; STAT6: signal transducer and activator of transcription 6; STAT5A: signal transducer and activator of transcription 5A; PDCD1: programmed cell death 1; CTLA4: cytotoxic T-lymphocyte associated protein 4; LAG3: lymphocyte activating 3; HAVCR2: hepatitis A virus cellular receptor 2; GZMB: granzyme B.
Furthermore, high expression of SLC2A1 was always accompanied by high dendritic cell [major histocompatibility complex, class II, DP beta 1 (HLA-DPB1), major histocompatibility complex, class II, DQ beta 1 (HLA-DQB1), major histocompatibility complex, class II, DR alpha (HLA-DRA), major histocompatibility complex, class II, DP alpha 1 (HLA-DPA1), neuropilin 1 (BDCA-4) and integrin subunit alpha X (CD11c)], Tfh [BCL6 transcription repressor (BCL6) and interleukin 21 (IL21)], Th1 [signal transducer and activator of transcription 1 (STAT1), interferon gamma (IFN-γ) and tumor necrosis factor (TNF-α)] and Th2 [GATA binding protein 3 (GATA3), signal transducer and activator of transcription 6 (STAT6) and signal transducer and activator of transcription 5A (STAT5A)] infiltration. However, high expression SLC2A2 was always accompanied by low dendritic cell (HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, BDCA-4, and CD11c), Th1 (IFN-γ and TNF-α), and Th2 (GATA3) infiltration. (Figure 8C, D, Table 3 and Table 4)
Moreover, we also found that SLC2A1 showed a strong positive correlation with many markers of T cells exhaustion, while SLC2A2 showed a strong negative correlation with these markers. These biomarkers included PD-1, CTLA4, lymphocyte activating 3 (LAG3), hepatitis A virus cellular receptor 2 (TIM-3), and granzyme B (GZMB). Notably, SLC2A1 was always positively correlated with C-C motif chemokine receptor 8 (CCR8), signal transducer and activator of transcription 5B (STAT5B), and transforming growth factor beta 1 (TGF-β1) markers of Tregs, while SLC2A2 was positively correlated with forkhead box P3 (FOXP3) and signal transducer and activator of transcription 5B (STAT5B) markers of Tregs but negatively correlated with TGF-β (Figure 8E, F, Table 3 and Table 4)
SLC2A2 methylation correlated with immune cells in HCC by using the Tumor and immune system interaction database database
Previous studies have revealed that the reduced expression of SLC2A2 in HCC was due to the methylation of SLC2A2[32]. Therefore, we assessed the correlation between SLC2A2 methylation and immune cells.
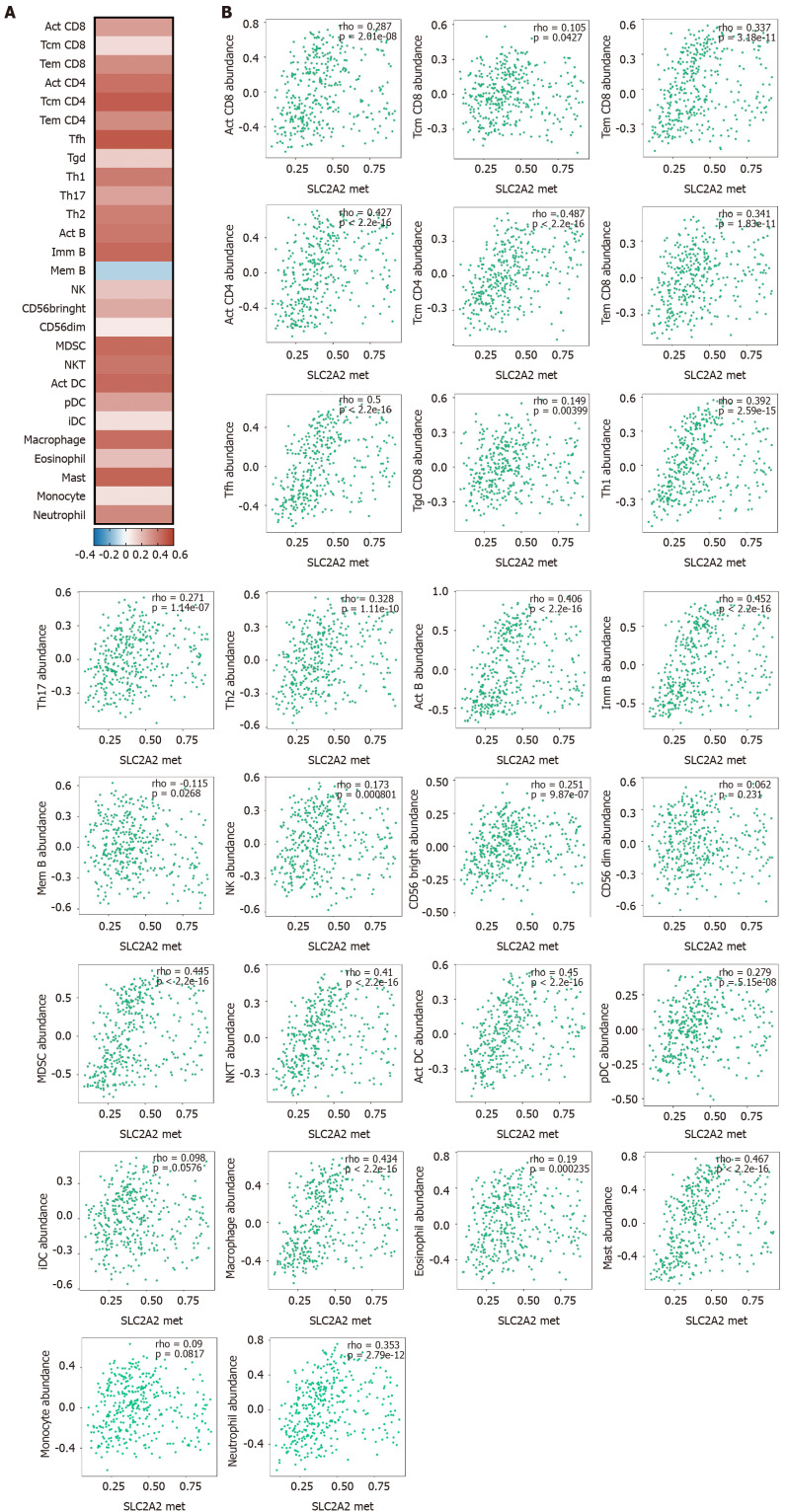
These results suggested that SLC2A2 methylation was positively correlated with lymphocytes (activated (Act) CD8, central memory CD8, effector memory CD8, Act CD4, central memory CD4, effector memory CD4, Tfh, gamma-delta T cells, Th1, Th17, Th2, Act B, immune B, memory B, NK, CD56bringht, CD56dim, myeloid-derived suppressor, NKT, Act DC, plasmacytoid DC, interstitial DC, macrophage, eosinophil, mast, monocyte, and neutrophil) (Figure 9A and 9B). These results were consistent with the correlation between lower SLC2A2 expression and positive immune infiltration in HCC. In conclusion, both elevated SLC2A1 and decreased SLC2A2 indicated high immune infiltration in HCC patients.
Figure 9.
Solute carrier family 2 member 2 methylation correlated with immune cells in hepatocellular carcinoma by using the tumor and immune system interaction database. A: Heat map of the correlation between solute carrier family 2 member 2 (SLC2A2) transcription level and the level of immune cell infiltration; B: Scatter plot of the correlation between SLC2A2 transcription level and the level of immune cell infiltration.Act: activated; Tcm: central memory T; Tem: effector memory T; Tfh: Follicular helper T; Tgd: gamma-delta T cells; Th: T helper; Imm: immune; Mem: memory; NK: natural killer; MDSC: myeloid-derived suppressor; NKT: natural killer T; pDC: plasmacytoid dendritic cells; iDC: interstitial dendritic cells.
DISCUSSION
Several immune checkpoint inhibitors have been approved for clinical trials in HCC. However, they were effective for only a small percentage of patients or were not effective at all[6-8]. Therefore, it is important to identify sensitive populations with immunotherapeutic drugs or search for more effective immunotherapeutic drugs to improve the survival time in HCC. In other words, knowing the immune status of patients in advance and selecting appropriate immunotherapeutic drugs are methods to improve the survival rate of patients. A previous study found that lactic acid and other related substances that were produced by the Warburg effect affected the number and function of immune cells in the tumor microenvironment[15]. SLC2A1 and SLC2A2 have been confirmed to be involved in the Warburg effect of tumors as glucose transporters[33], but whether they are related to immune cells has not been reported in HCC in detail. In our study, we identified that SLC2A1 and SLC2A2 were independent target molecules for HCC, but their expressions were negatively correlated. Secondly, both high expression SLC2A1 and low expression SLC2A2 were correlated with higher levels of immune cells. Thirdly, SLC2A2 methylation was positively correlated with the levels of immune infiltration. This study clarified that SLC2A1 and SLC2A2 were important target molecules, and it was proposed that SLC2A1 and SLC2A2 were related to immune cells in HCC.
In our study, SLC2A1 was overexpressed while SLC2A2 was low expressed in HepG2, HepG2215 cells, and liver cancer patients. A previous study had shown that SLC2A1 was highly expressed in HepG2, Hep3B, and SK-HEP1 cells compared with normal hepatocytes[34]. Another study also confirmed that SLC2A1 protein expression increased while SLC2A2 protein expression decreased with the progression of HCC[35]. In addition, the trend was becoming worse and worse with the progress of tumor volume, grade, and stage of HCC. We also analyzed the expression of SLC2A1 and SLC2A2 in other cancers, and the results showed that SLC2A1 and SLC2A2 were increased in some cancers but decreased in others compared with normal samples. This may be related to tumor heterogeneity or the different collection criteria and calculation methods in different data sets.
Interestingly, our study found that the expression levels of SLC2A1 and SLC2A2 were negatively correlated in HCC. Another study had also found that their expressions were negatively correlated at the protein level[35]. One possible reason was that the SLC2A2 glucose transporter was highly reversible whereas the SLC2A1 glucose transporter was not. They had this mode of expression ensured for higher glucose flux in cancer cells[9]. It has been reported that cancer cells expressing oncogene KRAS proto-oncogene, GTPase (KRAS), or oncogene B-Raf proto-oncogene, serine/threonine kinase (BFAF) required SLC2A1[36], which may be one of the reasons for the SLC2A1 overexpression in HCC. Other studies have found that activation of pregnane X receptor dysregulated SLC2A2 expression and subcellular localization in the liver[37]. However, other reasons for increased SLC2A1 expression and decreased SLC2A2 expression in HCC are still worth exploring.
Meanwhile, we showed that high SLC2A1 expression and low SLC2A2 expression were positively correlated with the poor OS and RFS in HCC. Notably, high expression of SLC2A1 and low expression of SLC2A2 were more dangerous for males, Asians, non-alcoholics, infected by HBV, high stage, high grade, and high AJCC-T than that in the control group. When primary rat hepatocytes were infected with an adenoviral vector expressing portions of the HBV genome, up-regulation of SLC2A1 and down-regulation of SLC2A2 were observed[38]. Teng et al[39] have demonstrated that HBV Pre-S2 mutant up-regulated SLC2A1 expression through the mammalian target of rapamycin/YY1 transcription factor/MYC proto-oncogene signaling. Besides, down-regulation of SLC2A2 has also been revealed in HBV replicon transfected Huh-7 cells[40]. Other studies have also found that high SLC2A1 expression level was associated with advanced tumor stage, high tumor grade, depth of the invasion, and poor differentiation in various cancers[41-43]. The study showed that SLC2A2 protein expression was decreased following the malignant progression of HCC[35]. Therefore, we suggested that SLC2A1 and SLC2A2 were independent prognostic factors for HCC.
Next, we discussed that SLC2A1 and SLC2A2 and their neighbor genes were involved in pathways in the String database. Notably, genes TP53, ATM, MDM2, AKT1, HIF1A, EP300, INS, GCK, and HK2 were included. Studies have demonstrated that tumor-associated mutation P53 stimulated the Warburg effect by promoting SLC2A1 translocation to the plasma membrane[44]. HIF1A promoted the Warburg effect of various cancers by increasing the expression of multiple glycolysis genes (such as SLC2A1 and carbonic anhydrase 9)[45]. When these oncogenes or tumor suppressor genes functioned, they all seemed to be associated with the SLC2A1-induced Warburg effect. The study indicated that SLC2A1, SLC2A2, and their neighbors were involved in the central carbon metabolism in cancer (hsa05230), pathways in cancer (hsa 05200), FOXO signaling pathway (hsa04068), phosphatidylinositol 3-kinase-Akt signaling pathway (hsa 04151), and so on. FOXOs were involved in cellular differentiation, apoptosis, cell proliferation, DNA damage and repair, and as mediators of oxidative stress[46]. Studies reported that Akt activation, caused by disturbances of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha or phosphatase and tensin homolog, mediated increased glucose uptake and solute carrier family 2 overexpression[33]. In conclusion, SLC2A1, SLC2A2, and their neighbor genes played an important role in the occurrence and development of tumors. Regrettably, we had not enriched some pathways associated with immunity in the String database. This may be attributed to the lack of database algorithms or the lack of studies on the relationship between glucose transporters and immune cells.
A previous study found that lactic acid and other related substances produced from the Warburg effect affected the number and function of immune cells in the tumor microenvironment[15]. High expression of SLC2A1 in gastric cancer cells was associated with suppressing CD8+ T cells and B cells[16], and an inverse correlation between SLC2A1 expression and the number of CD8+ T cells in renal cell carcinoma[31] was showed. Other studies reported that T cells, CD8+ T cells, and B cells reduced in the SLC2A1 expressive group of human papillomavirus type 16-positive cervical cancer[47]. However, the relationship between SLC2A2 and immunity in tumors has rarely been reported, which may be due to its low or no expression in various cancers. These results suggested that SLC2A1 and SLC2A2 affected the number and function of immune cells in the tumor microenvironment.
In this study, the results suggested that HCC had different levels of immune invasion compared to normal tissues. Of course, immune dysregulation in HCC has long been recognized[48]. Notably, it was presented for the first time that expressions of SLC2A1 and SLC2A2 were correlated with the infiltration levels of various immune cells and immune marker genes in HCC. Furthermore, both high SLC2A1 and low SLC2A2 expressions, as independent risk factors for HCC, pointed to a common outcome of high levels of immune cell infiltration. These results suggested that the expression of SLC2A1 and SLC2A2 played an important role in the number and function of immune cells in HCC.
Firstly, high expression of SLC2A1 and low expression of SLC2A2 had positive correlations with infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, and neutrophils in HCC.
Secondly, the expression of SLC2A1 was correlated with the expressions of monocytes markers (CD86 and CD115), TAM markers (CCL2, CD68, and IL10), M1 macrophage (interferon regulatory factor 5, and cytochrome c oxidase subunit II), and M2 macrophage (CD163, V-set and immunoglobulin domain containing 4, and membrane spanning 4-domains A4A). Monocytes and macrophages were involved in tumor initiation, growth, migration, vascularization, invasion, and metastasis[49]. Studies have found that lactic acid produced by the Warburg effect could promote Tcell apoptosis by activating M2-TAM and regulating PD-1/PD-L1 signals[50]. These results indicated that SLC2A1 had potential regulatory effects on tumor-associated macrophages.
Thirdly, SLC2A1 and SLC2A2 were correlated with several dendritic cell markers, indicating that they had an important relationship with the normal function of dendritic cells. Tumor-derived lactic acid regulated the activation and antigen expression of dendritic cells[51]. Therefore, one of the reasons why SLC2A1 and SLC2A2 affected dendritic cell function was that they could affect cell function through the lactic acid produced by the mediated Warburg effect.
Fourthly, the expression level of SLC2A1 was also correlated with the expressions of several markers of T helper cells. They included Th1 cell makers (STAT1, IFN-γ, and TNF-α), Th2 cell markers (GATA3, STAT6, and STAT5A), and Tfh cell makers (BCL6 and IL21). These results indicated that SLC2A1 expression level played an important role in the function of Th1cells, Th2 cells, and Tfh cells.
Notably, the results revealed that high expression of SLC2A1 and low expression of SLC2A2 had positive correlations with Treg marker TGFβ and T cell exhaustion markers (PD-1, CTLA4, LAG3, TIM-3, and GZMB). Furthermore, high expression of SLC2A1 and low expression of SLC2A2 had the potential to induce T cell exhaustion. The PD-1/PD-L1 axis inhibited T cell activation, proliferation, survival, and cytotoxic secretion in tumors[52]. CTLA4 interacted with its ligand to inhibit T cell activation, but the precise mechanism was not fully understood[53]. Increased TGFβ caused T cell exhaustion by up-regulation of PD-1, and inhibiting TGFβ might directly enhance antitumor immunity in HCC[54]. High expression of TIM-3 indicated T cell exhaustion[55], which together with Tregs supported exhausted CD8+ T cell development and limiting the expansion of CD4+ and CD8+ T cells[56]. TIM-3 expression inhibited glucose uptake, glucose consumption, and lactic acid release. Concomitantly, TIM-3 expression inhibited SLC2A1 expression but not SLC2A2 expression in Jurkat T cells[57].
Notably, previous studies have found that the reduced expression of SLC2A2 in HCC was due to the methylation of SLC2A2[32], and we analyzed the correlation between SLC2A2 methylation and immune cells. These results suggested that SLC2A2 methylation or low expression SLC2A2 was positively correlated with immune cells. However, larger studies are required to elucidate further the relationship between expressions of SLC2A1 and SLC2A2 and the immune microenvironment.
CONCLUSION
Collectively, SLC2A1 and SLC2A2 are independent therapeutic targets for HCC. Both high SLC2A1 and low SLC2A2 expression pointed to a common outcome of high levels of immune cell infiltration. However, larger studies are required to clarify the preliminary findings.
ARTICLE HIGHLIGHTS
Research background
Metabolic reprogramming has been identified as a core hallmark of cancer. Solute carrier family 2 is a major glucose carrier family. The solute carrier family 2 is an important carrier for glucose to enter target cells, and its ability to transport glucose is the first rate-determining step in tumor metabolic reprogramming. It consists of 14 members, and we mainly study solute carrier family 2 member 1 (SLC2A1) and solute carrier family 2 member 2 (SLC2A2) here.
Research motivation
Hepatocellular carcinoma (HCC) is still characterized by late diagnosis and limited effective treatment options. Immune checkpoint inhibitors are also less effective than expected. The discovery of new biomarkers indicating the patient's immune status is essential for the treatment of HCC with immune checkpoint inhibitors.
Research objectives
To identify if SLC2A1 and SLC2A2 were associated with immune infiltration in addition to participating in the metabolic reprogramming in HCC.
Research methods
SLC2A1 and SLC2A2 expression were tested in HepG2 cells, HepG215 cells, and multiple databases. The clinical characteristics of SLC2A1 and SLC2A2 were examined by multiple databases. The correlation between SLC2A1 and SLC2A2 was analyzed by multiple databases. The functions and pathways in which SLC2A1, SLC2A2, and neighbor genes were involved were discussed. Immune infiltration levels and immune marker genes associated with SLC2A1 and SLC2A2 were discussed by multiple databases.
Research results
SLC2A1 was increased but the expression level SLC2A2 was decreased in HepG2 cells, HepG215 cells, and liver cancer patients. The expression levels of SLC2A1 and SLC2A2 were related to HCC progression. Interestingly, the expression levels of SLC2A1 and SLC2A2 were negatively correlated. Further, high SLC2A1 expression and low SLC2A2 expression were related to poor overall survival and relapse-free survival. Moreover, SLC2A1, SLC2A2, and neighbor genes played a major role in the occurrence and development of tumors. Notably, SLC2A1 was positively correlated with tumor immune infiltration, while SLC2A2 was negatively correlated with tumor immune infiltration. Particularly, SLC2A2 methylation was positively correlated with lymphocytes.
Research conclusions
SLC2A1 and SLC2A2 are independent therapeutic targets for HCC, and they are quintessential marker molecules for predicting and regulating the number and status of immune cells in HCC.
Research perspectives
Clinical trials showed that programmed cell death 1 and cytotoxic T-lymphocyte associated protein 4 antibodies improved clinical outcomes in a few patients or were not effective at all in HCC. Therefore, recognition molecules representing the patient's immune status will help identify subgroups sensitive to immunomodulatory drugs. Moreover, some independent target molecules associated with the tumor immune microenvironment are worth exploring in HCC.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Yu-Qiang Xue for the statistical methods of this study.
Footnotes
Conflict-of-interest statement: The authors declare no competing interest.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: April 21, 2021
First decision: August 8, 2021
Article in press: February 25, 2022
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Farid K, Egypt; Florentino RM, United States S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Wu RR
Contributor Information
Qing Peng, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Li-Yuan Hao, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Ying-Lin Guo, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Zhi-Qin Zhang, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Jing-Min Ji, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Yu Xue, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Yi-Wei Liu, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Jun-Lan Lu, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Cai-Ge Li, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China.
Xin-Li Shi, Department of Pathobiology and Immunology, Hebei University of Chinese Medicine, Shijiazhuang 050200, Hebei Province, China. sxlsunshine@sina.com.
Data sharing statement
No additional data are available.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin . 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, Tateishi R, Han KH, Chawla YK, Shiina S, Jafri W, Payawal DA, Ohki T, Ogasawara S, Chen PJ, Lesmana CRA, Lesmana LA, Gani RA, Obi S, Dokmeci AK, Sarin SK. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int . 2017;11:317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shlomai A, de Jong YP, Rice CM. Virus associated malignancies: the role of viral hepatitis in hepatocellular carcinoma. Semin Cancer Biol . 2014;26:78–88. doi: 10.1016/j.semcancer.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruf B, Heinrich B, Greten TF. Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol . 2021;18:112–127. doi: 10.1038/s41423-020-00572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donisi C, Puzzoni M, Ziranu P, Lai E, Mariani S, Saba G, Impera V, Dubois M, Persano M, Migliari M, Pretta A, Liscia N, Astara G, Scartozzi M. Immune Checkpoint Inhibitors in the Treatment of HCC. Front Oncol . 2020;10:601240. doi: 10.3389/fonc.2020.601240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, Meyer T, Kang YK, Yeo W, Chopra A, Anderson J, Dela Cruz C, Lang L, Neely J, Tang H, Dastani HB, Melero I. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet . 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finn RS, Ryoo BY, Merle P, Kudo M, Bouattour M, Lim HY, Breder V, Edeline J, Chao Y, Ogasawara S, Yau T, Garrido M, Chan SL, Knox J, Daniele B, Ebbinghaus SW, Chen E, Siegel AB, Zhu AX, Cheng AL KEYNOTE-240 investigators. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J Clin Oncol . 2020;38:193–202. doi: 10.1200/JCO.19.01307. [DOI] [PubMed] [Google Scholar]
- 8.Duffy AG, Ulahannan SV, Makorova-Rusher O, Rahma O, Wedemeyer H, Pratt D, Davis JL, Hughes MS, Heller T, ElGindi M, Uppala A, Korangy F, Kleiner DE, Figg WD, Venzon D, Steinberg SM, Venkatesan AM, Krishnasamy V, Abi-Jaoudeh N, Levy E, Wood BJ, Greten TF. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol . 2017;66:545–551. doi: 10.1016/j.jhep.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer . 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatanaka M. Transport of sugars in tumor cell membranes. Biochim Biophys Acta . 1974;355:77–104. doi: 10.1016/0304-419x(74)90008-0. [DOI] [PubMed] [Google Scholar]
- 11.Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med . 2013;34:121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann T, Maegdefrau U, Hartmann A, Agaimy A, Marienhagen J, Weiss TS, Stoeltzing O, Warnecke C, Schölmerich J, Oefner PJ, Kreutz M, Bosserhoff AK, Hellerbrand C. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol . 2009;174:1544–1552. doi: 10.2353/ajpath.2009.080596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorens B, Cheng ZQ, Brown D, Lodish HF. Liver glucose transporter: a basolateral protein in hepatocytes and intestine and kidney cells. Am J Physiol . 1990;259:C279–C285. doi: 10.1152/ajpcell.1990.259.2.C279. [DOI] [PubMed] [Google Scholar]
- 14.Kim YH, Jeong DC, Pak K, Han ME, Kim JY, Liangwen L, Kim HJ, Kim TW, Kim TH, Hyun DW, Oh SO. SLC2A2 (GLUT2) as a novel prognostic factor for hepatocellular carcinoma. Oncotarget . 2017;8:68381–68392. doi: 10.18632/oncotarget.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon C, O'Farrelly C, Robinson MW. The Immune Consequences of Lactate in the Tumor Microenvironment. Adv Exp Med Biol . 2020;1259:113–124. doi: 10.1007/978-3-030-43093-1_7. [DOI] [PubMed] [Google Scholar]
- 16.Min KW, Kim DH, Son BK, Moon KM, Kim SM, Intazur Rahaman M, Kim SW, Kim EK, Kwon MJ, Koh YW, Oh IH. High SLC2A1 expression associated with suppressing CD8 T cells and B cells promoted cancer survival in gastric cancer. PLoS One . 2021;16:e0245075. doi: 10.1371/journal.pone.0245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown GR, Hem V, Katz KS, Ovetsky M, Wallin C, Ermolaeva O, Tolstoy I, Tatusova T, Pruitt KD, Maglott DR, Murphy TD. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res . 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res . 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang SM, Ooi LL, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res . 2007;13:6275–6283. doi: 10.1158/1078-0432.CCR-06-2236. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnström H, Glimelius B, Sjöblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science . 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res . 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia . 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia . 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res . 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci . 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res . 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res . 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics . 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 30.Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat . 2018;38:1–11. doi: 10.1016/j.drup.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Singer K, Kastenberger M, Gottfried E, Hammerschmied CG, Büttner M, Aigner M, Seliger B, Walter B, Schlösser H, Hartmann A, Andreesen R, Mackensen A, Kreutz M. Warburg phenotype in renal cell carcinoma: high expression of glucose-transporter 1 (GLUT-1) correlates with low CD8(+) T-cell infiltration in the tumor. Int J Cancer . 2011;128:2085–2095. doi: 10.1002/ijc.25543. [DOI] [PubMed] [Google Scholar]
- 32.Jin B, Seong JK, Ryu DY. Tissue-specific and de novo promoter methylation of the mouse glucose transporter 2. Biol Pharm Bull . 2005;28:2054–2057. doi: 10.1248/bpb.28.2054. [DOI] [PubMed] [Google Scholar]
- 33.Barron CC, Bilan PJ, Tsakiridis T, Tsiani E. Facilitative glucose transporters: Implications for cancer detection, prognosis and treatment. Metabolism . 2016;65:124–139. doi: 10.1016/j.metabol.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J, Zhang Z, Nie D, Ma H, Yuan G, Su S, Liu S, Tang G. PET Imaging of Hepatocellular Carcinomas: 18F-Fluoropropionic Acid as a Complementary Radiotracer for 18F-Fluorodeoxyglucose. Mol Imaging . 2019;18:1536012118821032. doi: 10.1177/1536012118821032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei Y, Hu Q, Gu J. Expressions of Carbohydrate Response Element Binding Protein and Glucose Transporters in Liver Cancer and Clinical Significance. Pathol Oncol Res . 2020;26:1331–1340. doi: 10.1007/s12253-019-00708-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, Diaz LA Jr, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science . 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassani-Nezhad-Gashti F, Rysä J, Kummu O, Näpänkangas J, Buler M, Karpale M, Hukkanen J, Hakkola J. Activation of nuclear receptor PXR impairs glucose tolerance and dysregulates GLUT2 expression and subcellular localization in liver. Biochem Pharmacol . 2018;148:253–264. doi: 10.1016/j.bcp.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Lamontagne J, Mell JC, Bouchard MJ. Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression. PLoS Pathog . 2016;12:e1005438. doi: 10.1371/journal.ppat.1005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng CF, Hsieh WC, Wu HC, Lin YJ, Tsai HW, Huang W, Su IJ. Hepatitis B Virus Pre-S2 Mutant Induces Aerobic Glycolysis through Mammalian Target of Rapamycin Signal Cascade. PLoS One . 2015;10:e0122373. doi: 10.1371/journal.pone.0122373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jagya N, Varma SP, Thakral D, Joshi P, Durgapal H, Panda SK. RNA-seq based transcriptome analysis of hepatitis E virus (HEV) and hepatitis B virus (HBV) replicon transfected Huh-7 cells. PLoS One . 2014;9:e87835. doi: 10.1371/journal.pone.0087835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamura T, Kusakabe T, Sugino T, Watanabe K, Fukuda T, Nashimoto A, Honma K, Suzuki T. Expression of glucose transporter-1 in human gastric carcinoma: association with tumor aggressiveness, metastasis, and patient survival. Cancer . 2001;92:634–641. doi: 10.1002/1097-0142(20010801)92:3<634::aid-cncr1364>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 42.Mori Y, Tsukinoki K, Yasuda M, Miyazawa M, Kaneko A, Watanabe Y. Glucose transporter type 1 expression are associated with poor prognosis in patients with salivary gland tumors. Oral Oncol . 2007;43:563–569. doi: 10.1016/j.oraloncology.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Kim E, Wu HG, Keam B, Kim TM, Kim DW, Paeng JC, Kim HJ, Chang JH. Significance of 18F-FDG PET Parameters According to Histologic Subtype in the Treatment Outcome of Stage III Non-small-cell Lung Cancer Undergoing Definitive Concurrent Chemoradiotherapy. Clin Lung Cancer . 2019;20:e9–e23. doi: 10.1016/j.cllc.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, Lin M, Yu H, Liu L, Levine AJ, Hu W, Feng Z. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun . 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schito L, Semenza GL. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer . 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int J Biol Sci . 2017;13:815–827. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim BH, Chang JH. Differential effect of GLUT1 overexpression on survival and tumor immune microenvironment of human papilloma virus type 16-positive and -negative cervical cancer. Sci Rep . 2019;9:13301. doi: 10.1038/s41598-019-49928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J Hepatol . 2015;62:1420–1429. doi: 10.1016/j.jhep.2015.02.038. [DOI] [PubMed] [Google Scholar]
- 49.Gouveia-Fernandes S. Monocytes and Macrophages in Cancer: Unsuspected Roles. Adv Exp Med Biol . 2020;1219:161–185. doi: 10.1007/978-3-030-34025-4_9. [DOI] [PubMed] [Google Scholar]
- 50.Shan T, Chen S, Chen X, Wu T, Yang Y, Li S, Ma J, Zhao J, Lin W, Li W, Cui X, Kang Y. M2TAM subsets altered by lactic acid promote Tcell apoptosis through the PDL1/PD1 pathway. Oncol Rep . 2020;44:1885–1894. doi: 10.3892/or.2020.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gottfried E, Kunz-Schughart LA, Ebner S, Mueller-Klieser W, Hoves S, Andreesen R, Mackensen A, Kreutz M. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood . 2006;107:2013–2021. doi: 10.1182/blood-2005-05-1795. [DOI] [PubMed] [Google Scholar]
- 52.Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res . 2020;10:727–742. [PMC free article] [PubMed] [Google Scholar]
- 53.Rowshanravan B, Halliday N, Sansom DM. CTLA-4: a moving target in immunotherapy. Blood . 2018;131:58–67. doi: 10.1182/blood-2017-06-741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Gingold JA, Su X. Immunomodulatory TGF-β Signaling in Hepatocellular Carcinoma. Trends Mol Med . 2019;25:1010–1023. doi: 10.1016/j.molmed.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 55.Huang YH, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, Dougan SK, Petersen BS, Melum E, Pertel T, Clayton KL, Raab M, Chen Q, Beauchemin N, Yazaki PJ, Pyzik M, Ostrowski MA, Glickman JN, Rudd CE, Ploegh HL, Franke A, Petsko GA, Kuchroo VK, Blumberg RS. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature . 2015;517:386–390. doi: 10.1038/nature13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakuishi K, Ngiow SF, Sullivan JM, Teng MW, Kuchroo VK, Smyth MJ, Anderson AC. TIM3+FOXP3+ regulatory T cells are tissue-specific promoters of T-cell dysfunction in cancer. Oncoimmunology . 2013;2:e23849. doi: 10.4161/onci.23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee MJ, Yun SJ, Lee B, Jeong E, Yoon G, Kim K, Park S. Association of TIM-3 expression with glucose metabolism in Jurkat T cells. BMC Immunol . 2020;21:48. doi: 10.1186/s12865-020-00377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.