Abstract
A purified bacteriocin produced by Enterococcus faecium BFE 900 isolated from black olives was shown by Edman degradation and mass spectrometric analyses to be identical to enterocin B produced by E. faecium T136 from meat (P. Casaus, T. Nilsen, L. M. Cintas, I. F. Nes, P. E. Hernández, and H. Holo, Microbiology 143:2287–2294, 1997). The structural gene was located on a 2.2-kb HindIII fragment and a 12.0-kb EcoRI chromosomal fragment. The genetic characteristics and production of EntB by E. faecium BFE 900 differed from that described so far by the presence of a conserved sequence like a regulatory box upstream of the EntB gene, and its production was constitutive and not regulated. The 2.2-kb chromosomal fragment contained the hitherto undetected immunity gene for EntB in an atypical orientation that is the reverse of that of the structural gene. Typical transport and other genes associated with bacteriocin production were not detected on the 12.0-kb chromosomal fragment containing the EntB structural gene. This makes the EntB genetic system different from most other bacteriocin systems, where transport and possible regulatory genes are clustered. EntB was subcloned and expressed by the dedicated secretion machinery of Carnobacterium piscicola LV17A. The structural gene was amplified by PCR, fused to the divergicin A signal peptide, and expressed by the general secretory pathway in Enterococcus faecalis ATCC 19433.
Members of the genus Enterococcus are among the lactic acid bacteria (LAB) that are of importance in foods (18, 39). Phylogenetically, they are more closely related to the genus Carnobacterium than to the streptococci or lactococci to which they were historically linked (14). Enterococci are used as starter cultures in some cheeses and as animal and human probiotic cultures; however, when they are present in foods adventitiously, they may be used as indicators of unsanitary handling (24) and they may cause spoilage of heat-treated foods (2, 23). Many enterococci produce bacteriocins. The best-characterized Enterococcus bacteriocins are enterocins A, B, and P, which are produced by strains of Enterococcus faecium isolated from fermented meats (3, 8, 12).
Most of the well-characterized bacteriocins that have been isolated from LAB, with the notable exception of the class I lantibiotic nisin, are class II bacteriocins (25, 29). The class II bacteriocins are ribosomally synthesized, small (4 to 6 kDa), heat-stable peptides that, unlike the lantibiotics, do not undergo extensive posttranslational modification. Considerable emphasis has been placed on the pediocin-like structure and the anti-Listeria activity of the class IIa bacteriocins that contain a YGNGVXC amino acid motif near the N terminus of the active peptide. Enterocin B (EntB) does not contain this amino acid motif; nonetheless, it has sequence similarity to bacteriocins of the pediocin family (8). The class II bacteriocins have distinct similarities in their genetic organizations, consisting of the structural gene, followed immediately by a gene encoding the immunity protein and genes for dedicated transport and accessory proteins (29). Some class IIa bacteriocins are regulated and may have an induction factor (IF) as well as regulatory genes, as demonstrated for carnobacteriocin (Cbn) B2 (32) and sakacin P (6).
EntB production has been described for E. faecium T136 and CTC 492 isolated from fermented sausage in Spain (8, 30). The structural gene coding for the bacteriocin was cloned from the chromosome of E. faecium T136. Apart from this gene, no other genes involved with EntB production were described previously (8). The regulation of enterocin A and B production by an induction factor has also been reported (30). In this study we characterized EntB production by E. faecium BFE 900 isolated from black olives (17) and analyzed the genetic composition of both a 2.2-kb fragment and a 12.0-kb fragment of chromosomal DNA. The objective of this study was to clone the genetic determinants for EntB production and immunity for future incorporation into a multiple bacteriocin cassette that is contained in a food-grade vector. The study also aimed to localize and express the hitherto unidentified immunity gene for EntB and to show that EntB production in E. faecium BFE 900 is constitutive and thus different from the regulated production of EntB that occurs in E. faecium CTC 492 (30).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The bacteriocin producer E. faecium BFE 900 was previously isolated from black olives and unequivocally identified by biochemical and physiological tests, as well as DNA-DNA hybridization and sodium dodecyl sulfate-polyacrylamide gel electrophoresis of total cell protein studies (17). This strain and other LAB were grown in Lactobacilli MRS broth (Difco Laboratories Inc., Detroit, Mich.) or APT (All Purpose Tween) broth (Difco) at 30°C. Carnobacterium strains were grown in APT broth at 25°C. For bacteriocin purification, E. faecium BFE 900 was grown in APT broth at 30°C. Escherichia coli strains were grown on a rotary shaker at 250 rpm in Luria-Bertani broth (Becton Dickinson, Cockeysville, Md.) at 37°C. All cultures were incubated aerobically. Antibiotics were added as selective agents when appropriate, as follows: erythromycin (200 μg/ml) and ampicillin (150 μg/ml) for E. coli; erythromycin at 5 μg/ml for carnobacteria and Lactococcus lactis ATCC 19435 or at 50 μg/ml for Enterococcus faecalis ATCC 19433. Stock cultures were made in the same medium used for culturing the bacterial strain, with 15% (vol/vol) glycerol, and were stored at −80°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. faecalis ATCC 19433 | EntBr | |
| L. sake DSM 20017 | EntBs | |
| L. lactis ATCC 19435 | EntBs CbnAr | |
| E. faecium | ||
| BFE 900 | entB+ eniB+, plasmidless | 17 |
| CTC 492 | entB+ eniB+ entA+ eniA+ | 3 |
| C. piscicola | ||
| UAL26 | Bac+ EntBr | 1 |
| LV17C | EntBs CbnAs, plasmidless | 1 |
| LV17A | cbnA+ cbiA+, containing pCP49 | 1, 44 |
| E. coli | ||
| DH5α | F−endA1 hsdR17 (rk− mk+) supE44 thi-1 λ−recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 φ80dlacZΔM15 | BRL Life Technologies Inc. |
| MH1 | MC1061 derivative; araD139 lacX74 galU galK hsr hsm+ strA | 7 |
| Plasmids | ||
| pUC118 | lacZ′ Ampr, 3.2 kb | 42 |
| pMG36e | Expression vector, Emr, 3.6 kb | 41 |
| pRW19e | pMG36e derivative containing divergicin A structural and immunity genes | 27 |
| pCMAP01 | pUC118 containing 12.0-kb EcoRI fragment, entB+ eniB+ | This study |
| pCMAP02 | pUC118 containing 2.2-kb HindIII fragment, entB+ eniB+ | This study |
| pCMAP03 | pRW19e containing 200-bp HindIII-KpnI PCR product insert that creates the divergicin A signal peptide::enterocin B gene fusion | This study |
| pCMAP04 | pMG36e containing 2.1-kb XbaI-PstI fragment, entB+ eniB+ | This study |
| pCMAP05 | pMG36e containing 197-bp XbaI-HindIII PCR product of eniB | This study |
| pCMAP06 | pUC118 containing 138-bp XbaI-HindIII PCR product of a part of the gene coding for mature enterocin A | This study |
Bacteriocin activity assays and induction.
Bacteriocin activity was quantified and expressed as activity units (AU) per milliliter by the critical dilution method (17), with the neutralized culture supernatant of the producer culture grown overnight at 30°C. Indicator bacteria were inoculated into the appropriate soft (0.75%) agar medium to give a concentration of 106 CFU/ml. Unless otherwise stated, Lactobacillus sake DSM 20017 was used as the indicator strain for assays of bacteriocin activity. For induction experiments, E. faecium BFE 900 and CTC 492 (M. Hugas, Centre de Technologia de la Carn, Monells, Spain) were grown overnight in APT broth at 30°C. Each culture was serially diluted to extinction in APT broth by using 12 10-fold dilutions. The diluted cultures were incubated at 30°C and allowed to grow until turbid. The neutralized culture supernatant was prepared from all dilutions that grew, and bacteriocin activity was determined by critical dilution assay. In addition, the presence of the EntB compound in the supernatant of all cultures that grew after dilution was confirmed by matrix-adsorbed laser desorption–ionization–time-of-flight (MALDI-TOF) mass spectrometry (described below). All induction tests were done in duplicate.
Enterocin B purification.
E. faecium BFE 900 was grown aerobically with gentle stirring in 3 liters of APT broth supplemented with 1% glucose at 30°C for 18 h. Cells were removed by centrifugation, and enterocin was purified by hydrophobic interaction chromatography with Amberlite XAD8 (BDH Chemicals Ltd., Poole, United Kingdom), concentrated to 100 ml by rotary evaporation, and cation-exchange chromatography with SP Sepharose Fast Flow (Pharmacia Biotech, Baie D’Urfé, Quebec, Canada) in a way similar to the method of Casaus et al. (8). Following cation-exchange chromatography the bacteriocin fraction was desalted with a Sep Pak C18 reverse-phase column (Waters Ltd., Mississauga, Ontario, Canada) and freeze-dried. The freeze-dried protein was resuspended in 1.5 ml of 0.1% trifluoroacetic acid (TFA) and purified by high-pressure liquid chromatography (HPLC) by injecting 200-μl aliquots on a C18 reverse-phase column (Waters Delta-Pak; 8 by 100 mm, 15-μm particle size, 300-Å pore size, a flow rate of 1.0 ml/min, a mobile phase of 0.1% TFA [A] and 95% ethanol [B]). Bacteriocin was eluted by a gradient method, consisting of first 35 to 60% solvent B in 7 min and then 60 to 70% solvent B in 10 min. Fractions were monitored for activity, and the A218 was determined. Bacteriocin activity of all fractions against L. sake DSM 20017 was determined by the critical dilution method. The protein concentrations of these fractions were determined by a modified Lowry method (20).
N-terminal amino acid sequencing and mass spectrometry.
Purified EntB was subjected to Edman degradation analysis by methods described earlier (44). The mass spectra of purified EntB were obtained by the direct injection of a solution (50% aqueous acetonitrile, 0.1% TFA) onto a VG ZabSpec sector instrument (Fisons, Manchester, United Kingdom) with an electrospray ionization source. For MALDI-TOF mass spectrometry the culture supernatant was analyzed on a linear Bruker Proflex III MALDI-TOF instrument equipped with delayed-extraction technology and a 125-cm flight tube (Bruker Analytical Systems, Billerica, Mass.) by methods described earlier (34). For this process, 0.5 μl of supernatant was spotted onto a probe. The sample was air dried, washed for 30 s by immersion into sterile distilled water, and allowed to air dry. A 0.5-μl volume of matrix consisting of 2 parts 0.1% TFA and 1 part acetonitrile, saturated with sinapinic acid (Sigma, St. Louis, Mo.), was spotted onto the sample and air dried before analysis on the MALDI-TOF mass spectrometer. All spectra were acquired in positive-ion linear mode with a nitrogen laser (λ = 337 nm) for desorption-ionization of the samples and an acceleration voltage of 20 kV. The spectra are representative of 60 consecutive laser shots and were smoothed. External mass calibration was performed with two points that bracket the mass range of the analyte. Angiotensin (MH+ = 1046.542) and bovine insulin (MH+ = 5734.557) were used as calibrants (34).
DNA isolation, manipulation, and hybridization.
Large-scale plasmid DNA preparations (44), small-scale plasmid isolations from E. coli (35) and LAB (40), and chromosomal DNA isolation (33) were done by established techniques. DNA fragments were recovered from agarose gels with either Geneclean II (Bio 101 Inc., La Jolla, Calif.) or QIAEX II (Qiagen, Chatsworth, Calif.). Restriction enzymes, T4 DNA ligase, T4 polynucleotide kinase (New England Biolabs, Mississauga, Ontario, Canada), and Klenow enzyme (Promega, Madison, Wis.) were used as recommended by the suppliers. DNA manipulations, cloning, and hybridizations were done as described by Sambrook et al. (35). Competent cells of E. coli were prepared and transformed according to the one-step method of Chung et al. (11). Recombinant pMG36e plasmids were first transformed into E. coli MH1 before being transformed into LAB. Carnobacteria and L. lactis ATCC 19435 cells were transformed by electroporation (45), while E. faecalis ATCC 19433 and L. sake DSM 20017 cells were electroporated according to the methods of Cruz-Rodz and Gilmore (13) and Berthier et al. (5), respectively.
Southern and colony blot hybridizations with Hybond N (Amersham Canada, Oakville, Ontario, Canada) nylon membranes were done by standard methods (35). A 32-mer degenerate probe, CFR-01 (5′-GAA AAT GAT CAT [C/A]G[T/A] ATG CC[T/A] AAT GAA CT[T/A] AA-3′), corresponding to the N terminus of the EntB structural gene (entB), was used to identify entB in Southern and colony blot hybridizations. Oligonucleotides were synthesized on an Applied Biosystems 391 PCR MATE synthesizer (Applied Biosystems, Foster City, Calif.) and used without further purification. Oligonucleotides were end labeled with [γ-32P]ATP (Amersham Canada). Hybridizations were done in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) hybridization solution (35) at 44°C with 5% formamide and probe CFR-01.
DNA and amino acid sequence analysis.
DNA was sequenced bidirectionally and analyzed in an Applied Biosystems 373A sequencer with fluorescent dideoxy chain terminators. The recombinant plasmids pCMAP01 and pCMAP02 (Table 1) were used as templates. Primers used for sequencing were forward and reverse universal primers for the pUC plasmid series. In addition, specific oligonucleotides were synthesized on an Applied Biosystems 391 PCR MATE synthesizer and used for sequencing in a primer-walking strategy. The nucleotide sequence was analyzed with the DNAStrider program (version 1.2).
PCR amplification. (i) Reaction 1.
Primer CFR-02 (5′-TAT ATC TAG AAA ATA TTA TGG AAA TGG AGT GTA T-3′) was complementary to the YGNGVXC consensus motif at the 5′ end of the gene encoding mature EntA, while CFR-03 (5′-TAT ACT GCA GGC ACT TCC CTG GAA TTG CTC C-3′) was complementary to the 3′ end of the EntA gene (3). Primers CFR-02 and CFR-03 contained XbaI and PstI restriction sites, respectively (underlined in each sequence).
(ii) Reaction 2.
The open reading frame (ORF) eniB located adjacent to the enterocin structural gene was amplified by PCR with the primers CFR-04 (5′-TTA AGC TTT TAC GAG TTT TTT CTC TTC T-3′) and CFR-05 (5′-AAT CTA GAA AAG AGA GGA TGT TTA TAT T-3′), complementary to the 5′ and 3′ ends of eniB, respectively. Primer CFR-04 contains a HindIII restriction site, while CFR-05 contains an XbaI restriction site (underlined in each sequence above).
(iii) Reaction 3.
The primers CFR-06 (5′ CCC AAG CTT CTG CTG AAA ATG ATC ACA GAA TGC CTA A-3′) and CFR-07 (5′ CCC CTG CAG CAT GCT TAG TTG CAT TTA GAG TAT AC-3′) were used to create a divergicin A signal peptide::EntB fusion construct, similar to that used by McCormick et al. (27) for a divergicin A signal peptide::CbnB2 fusion construct. Primer CFR-07 contained adjacent PstI and SphI restriction sites (underlined), in which one base is common to both restriction enzyme sites, and it is complementary to the 3′ end of the EntB structural gene on pCMAP02. CFR-06 contained a HindIII restriction site (underlined), which together with the next five nucleotides (CTGCT) encodes the carboxy terminus of the divergicin A signal peptide (27, 45). The nucleotides which follow CTGCT encode the amino terminus of mature EntB. Primer CFR-06 created an in-frame fusion of the 3′ end of the divergicin A signal peptide and the EntB structural gene.
For all PCR, DNA was amplified in a 100-μl volume with a temperature cycler (Omnigene; InterSciences Inc., Markham, Ontario, Canada). PCR mixtures contained 1.0 μM concentrations of each of the respective primers, 200 μM concentrations of deoxynucleoside triphosphates, 3 mM MgCl2, 2.5 U of Taq DNA polymerase (Perkin-Elmer), and a 1× reaction buffer (Perkin-Elmer). E. faecium BFE 900 chromosomal DNA was used as template DNA for reaction 1, and pCMAP02 was used for reactions 2 and 3. DNA was amplified in 32 cycles (denaturation, 94°C for 1 min; annealing, 56°C for reaction 1, 48°C for reaction 2, and 54°C for reaction 3 for 1 min; extension, 72°C for 1 min). The PCR products were cloned into pUC118 for sequencing and to confirm the fidelity of the reactions.
Expression of enterocin B structural and immunity genes in heterologous hosts.
Plasmid pRW19e is a derivative of pMG36e and contains the divergicin A structural gene bearing a signal peptide as well as the divergicin immunity gene. Digestion with HindIII and KpnI removes the 3′ end of the divergicin A signal peptide and the divergicin A structural and immunity genes (27). The PCR product of reaction 3 above was cloned into pUC118. A divergicin A signal peptide::EntB structural gene fusion was created by excising the PCR product from pUC118 with HindIII and KpnI and inserting it into the restriction sites of pRW19e, resulting in plasmid pCMAP03 (Table 1).
E. faecalis ATCC 19433 was transformed with pCMAP03 and tested for bacteriocin production, by using L. sake DSM 20017 containing pMG36e as the bacteriocin-sensitive indicator. E. faecalis ATCC 19433 and E. faecium BFE 900 both containing pMG36e were used as negative and positive controls, respectively. Carnobacterium piscicola LV17A containing pCP49 (CbnA+ Imm+) was transformed with pCMAP04 and tested for bacteriocin production by the deferred antagonism assay (1). C. piscicola LV17A containing pMG36e and C. piscicola LV17C (CbnAs EntBs) containing pCMAP04 were used as negative controls. L. lactis ATCC 19435 containing pMG36e was used as an EntB-sensitive, CbnA-resistant indicator.
The immunity of L. sake DSM 20017 containing either pCMAP04 or pCMAP05 (Table 1) was confirmed with the transformants as indicators in deferred inhibition assays with E. faecalis ATCC 19433 containing pCMAP03 as the producer strain. L. sake DSM 20017 containing pMG36e was used as an EntB-sensitive control. The presence of recombinant pCMAP plasmids in transformed strains was confirmed by small-scale plasmid isolation and electrophoresis on 0.7% agarose gels in Tris-acetate/EDTA buffer (35).
Tests for multiple bacteriocin production.
To investigate the possibility that E. faecium BFE 900 produced more than one bacteriocin, E. faecium BFE 900 transformed with pMG36e and E. faecalis ATCC 19433 transformed with pCMAP03 (EntB+) were used in the deferred inhibition assay against the indicator strains L. sake DSM 20017, C. piscicola LV17C, and C. piscicola UAL26, each containing pMG36e (Table 1). For deferred inhibition assays the E. faecium and E. faecalis strains were spotted onto the same APT agar plate and grown overnight at 30°C before being overlaid with the indicator. In addition, the production of EntA by E. faecium BFE 900 in culture supernatants was confirmed by detecting this compound with MALDI-TOF mass spectrometry by the methods described above.
Nucleotide sequence accession number.
The nucleotide sequences of both the 2.2-kb and 12.0-kb cloned chromosomal DNA fragments from E. faecium BFE 900 were submitted to GenBank (Los Alamos, N.Mex.) and were given accession no. AF076604 for the 2.2-kb HindIII fragment and AF121254 for the 12.0-kb EcoRI fragment, respectively.
RESULTS
Regulation of bacteriocin production.
To examine whether bacteriocin production by E. faecium BFE 900 is regulated, similar to the case of enterocins A and B produced by E. faecium CTC 492 (30), or constitutive, these two strains were used in induction experiments. Bacteriocin activity (3,200 AU/ml) against L. sake DSM 20017 was detected in cell-free culture supernatants from all tubes of E. faecium BFE 900 that grew after serial dilution to extinction. Because an induction factor is not involved in constitutive bacteriocin production, the dilution of a culture in growth medium to extinction will not result in a loss of bacteriocin production. E. faecium BFE 900 produced bacteriocin in all cultures that grew after dilution to extinction; hence, production is considered to be constitutive. In contrast, with a serial dilution of E. faecium CTC 492, bacteriocin activity (3,200 AU ml−1) was detected only in culture supernatants from tubes that were diluted to 10−4. In tubes at dilutions of >10−5 in which the bacteria grew (dilutions of 10−5 to 10−10) bacteriocin activity was not detected in the supernatant, indicating the absence of an induction factor and, therefore, regulated bacteriocin production. Supernatants of EntB-producing E. faecalis ATCC 19433 containing plasmid pCMAP03 failed to induce bacteriocin production by bacteriocin-negative E. faecium CTC 492, indicating that the bacteriocin EntB itself was not the induction factor.
Enterocin B purification and mass spectral analysis.
EntB was purified from the culture supernatant with a 1,205-fold purification (Table 2). Approximately 1 mg of pure EntB was recovered. The overall recovery was 1.1% of the activity detected in the culture supernatant. EntB was eluted as a single peak on a C18 reverse-phase column. The purity of the HPLC fraction was confirmed by Tricine–sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A single band of approximately 5 kDa showed antimicrobial activity (result not shown) when the gel was overlayered with L. sake DSM 20017. N-terminal amino acid analysis of the HPLC-purified EntB revealed the following 53-amino-acid sequence: Glu-Asn-Asp-His - Arg - Met - Pro - Asn - Glu - Leu - Asn - Arg - Pro - Asn - Asn - Leu - Ser - Lys - Gly - Gly - Ala - Lys - Xaa - Gly - Ala - Ala - Ile - Ala - Gly - Gly - Leu - Phe - Gly - Ile - Pro - Lys - Gly - Pro - Leu - Ala - Trp - Ala - Ala - Gly -Leu-Ala-Asn-Val-Tyr-Ser-(Leu/Lys)-Xaa-(Leu/Asn).
TABLE 2.
Purification of enterocin B produced by E. faecium BFE 900 in 3:1 APT broth culture at 30°C
| Method | Fraction | Volume (ml) | Total activity (AU) | Activity recovery (%) | Total protein (mg) | Sp act (AU/mg) | Purification (fold) |
|---|---|---|---|---|---|---|---|
| Culturing and harvesting of supernatant | I | 3,000 | 1.92 × 107 | 100 | 1.32 × 105 | 1.46 × 102 | 1 |
| Hydrophobic interaction chromatography, rotary evaporation | II | 100 | 1.02 × 107 | 53 | 3.61 × 102 | 2.82 × 104 | 193 |
| Cation exchange, Sep Pak desalting, and freeze-drying | III | 1.5 | 3.07 × 105 | 1.6 | 2.33 | 1.32 × 105 | 904 |
| Reverse-phase HPLC of purified enterocin B | IV | 1 | 2.04 × 105 | 1 | 1.16 | 1.76 × 105 | 1,205 |
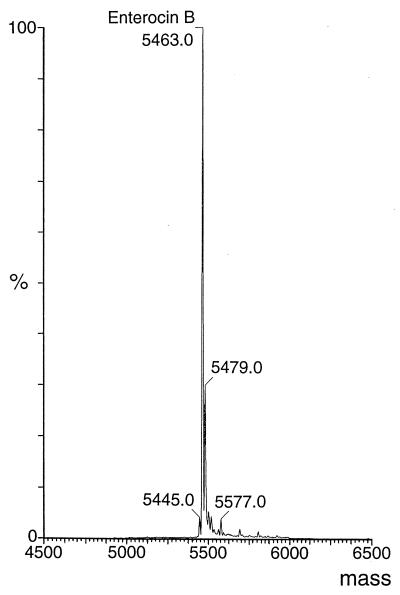
The interpretation of the nucleotide sequence indicated that the amino acids at positions 23 and 52 were cysteines. The nucleotide sequence also showed that amino acids 51 and 53 were incorrectly identified as leucine and that they are lysine and asparagine, as indicated above in parentheses. The average molecular mass of EntB determined by mass spectral analysis was 5,463.0 ± 1 Da (Fig. 1).
FIG. 1.
Electrospray mass spectrum of enterocin B. Arbitrary intensity (percent). Mass, average molecular mass (Daltons).
Our study of the regulation of bacteriocin production showed that EntB production by E. faecium BFE 900 was constitutive. However, both EntA and EntB are produced by E. faecium BFE 900, so it was unclear whether EntB, EntA, or both were constitutively produced. To determine this, the supernatants of diluted cultures of E. faecium BFE 900 and CTC 492 were analyzed by MALDI-TOF mass spectrometry. A compound with a molecular mass of 5,480 ± 5 Da was detected in the supernatants of all E. faecium BFE 900 cultures that grew after dilution to extinction. This approximates the mass of EntB (see above), assuming that the oxidation of a methionine residue had occurred. In addition, a compound with a molecular mass of 4,836 ± 5 Da was detected in the supernatants. This approximates the mass reported for EntA (3), also assuming that the oxidation of a methionine residue had occurred. The mass spectra performed on supernatants of E. faecium BFE 900 cultures that grew after dilution to 10−1, 10−5, and 10−9 are shown in Fig. 2. The presence of enterocins A and B in the supernatants of all E. faecium BFE 900 cultures that grew indicates that both enterocins A and B are constitutively produced.
FIG. 2.
MALDI-TOF mass spectrum of supernatants from E. faecium BFE 900 (A) and CTC 492 (B) cultures that were grown after dilution to 10−1 (graphs A), 10−5 (graphs B), and 10−10 (graphs C) in APT broth. Mass spectra show the masses of the ionized enterocin B (EntB) and enterocin A (EntA) compounds. a.i., arbitrary intensity; m/z, mass-to-charge ratio (Daltons).
Two compounds with similar molecular masses were observed in the mass spectra of culture supernatants from E. faecium CTC 492 that had been diluted to 10−1 (Fig. 2), while the EntB compound was not detected in supernatants of cultures that grew after dilution to 10−4. The mass spectra done on supernatants of E. faecium CTC 492 cultures that grew after dilution to 10−5 and 10−10 are also shown in Fig. 2. The absence of the EntB compound in the supernatants of E. faecium CTC 492 cultures diluted to 10−5 or lower confirmed the results of Nilsen et al. (30) showing that EntB production in this strain is regulated.
Nucleotide sequence and identification of the enterocin B structural gene.
Plasmid DNA was not isolated from E. faecium BFE 900 with small-scale (17) and large-scale (this study) plasmid isolation methods. Probe CFR-01 hybridized to both 2.2-kb HindIII and 12.0-kb EcoRI chromosomal DNA fragments. These fragments were cloned separately into pUC118 and completely sequenced in both directions. Analysis of the nucleotide sequence of the 2.2-kb fragment revealed eight possible ORFs, two oriented in the 5′ to 3′ direction (orf1 and entB) and six in the opposite direction (orf2 to orf6 and eniB [Fig. 3]). The translation of the entB ORF matched the amino acid sequence determined by Edman degradation analysis for EntB. The first amino acid of the N-terminal sequence (Glu) matches the 19th amino acid deduced from the nucleotide sequence. The enterocin prepeptide consists of an 18-amino-acid N-terminal extension ending in a double-glycine cleavage site and a 53-amino-acid bacteriocin identical to the prepeptide reported for EntB from E. faecium T136 (8). A probable ribosome binding site (RBS) for the EntB structural gene (AAAGGAG) was located 11 bases upstream of the initiation codon. Possible −10 and −35 promoter sequences were detected upstream of the structural gene. A conserved sequence like a regulatory box, consisting of the direct repeat sequences TTCAGGAAT (left) and TTCAGGAAG (right) separated by 13 nucleotides, was located upstream of the −35 site. An imperfect inverted repeat starting with the third base of the last amino acid codon (Asn) of the enterocin structural gene had the characteristics of a possible Rho-independent terminator. A second conserved sequence like a regulatory box, consisting of the sequences TTCAGGATA (right) and TTCAGGAAG (left) separated by 13 nucleotides, was located upstream of orf6.
FIG. 3.
Organization of genes on the cloned 12.0- and 2.2-kb chromosomal fragments from E. faecium BFE 900 associated with enterocin B production. Constructs used for enterocin B immunity testing are shown. A terminator structure is indicated by a stem-loop vertical line, and the P32 promoter is indicated by a box and arrow. Chromosomal fragments and constructs are contained in plasmids pCMAP01, pCMAP02 (b), pCMAP04 (c) and pCMAP05 (d).
Amino acid homology.
EntB from E. faecium BFE 900 is identical to that produced by E. faecium T136 and E. faecium CTC 492 (8, 30). A search of the protein data banks revealed that none of the ORFs on the cloned 2.2-kb fragment or the 12.0-kb fragment showed homology to reported bacteriocin immunity proteins, but immunity proteins in general have little similarity (3, 29). The product of orf6 has homology to the N-terminal extensions of the sakacin A prepeptide and the precarnobacteriocins A, B2, and BM1. The first 18 amino acids of this protein are characteristic of bacteriocin leader peptides of the double-glycine type; however, the mature peptide following the N-terminal extension is unlikely to encode a class II bacteriocin, because it contains only 21 amino acids. A double-glycine-type leader peptide followed by a small mature peptide is characteristic of an induction factor (30). The putative products of orf2 to orf4 all have low-level similarity (7.2, 7.2, and 6.8% identity, respectively) to a UV resistance protein produced by E. faecalis that is transcribed from a single ORF in that strain (31). The putative products of orf1 and orf5 show no clear homology to reported amino acid sequences.
The nucleotide sequence from the 12.0-kb EcoRI chromosomal fragment consists of approximately 3 kb of DNA sequenced downstream and 9 kb sequenced upstream of the EntB structural gene. Analysis of the DNA sequence downstream of entB revealed the presence of four additional ORFs (Fig. 3). The putative products of these ORFs did not have homology to reported amino acid sequences in the data banks. Analysis of the DNA sequence upstream of entB revealed the presence of 11 additional ORFs, six of which were in an orientation opposite to that of the entB structural gene (Fig. 3). The first (bglR) gene, located immediately upstream of orf1, encoded a protein of 281 amino acids, which showed homology to transcription antiterminator proteins of the BglG family, such as LicT of Bacillus subtilis (37.5% identity), BglG of E. coli K-12 (34.5% identity), ArbG of Erwinia chrysanthemi (31.8% identity), and BglR of L. lactis (28.3% identity) (4, 16, 36, 37, 47). The ORF immediately following it (bglS) showed homology to the β-glucoside-specific transport proteins (enzyme IIBgl) such as BglS of E. coli K-12 (31.7% identity) and ArbF of E. chrysanthemi (33.0% identity) (16, 37). The next ORF (bglB) has homology to phospho-β-glucosidase hydrolyzing enzymes such as BglH of B. subtilis (45.8% identity), ArbB of E. chrysanthemi (43.2% identity), and BglB of E. coli K-12 (41.5% identity) (16, 26, 37). The putative products of the seven ORFs (orf1 to orf7) located downstream of bglB did not show clear homology to reported amino acid sequences. The 5′ end of the ORF pepC was detected at the proximal end of the cloned fragment, but the 3′ end of this ORF was not located within the cloned fragment. This truncated ORF showed homology to cysteine aminopeptidase proteins such as PepC of Streptococcus thermophilus (30.3% identity) and L. lactis subsp. cremoris (28.7% identity) (9, 10). These are based on the amino acid sequence derived from the truncated E. faecium BFE 900 PepC gene, present on the cloned fragment in this study, and can be expected to be greater for the entire E. faecium BFE 900 PepC protein.
Heterologous expression of enterocin B.
E. faecalis ATCC 19433 was electrotransformed with plasmid pCMAP03 (Table 1) for heterologous bacteriocin expression by the sec pathway, and L. sake DSM 20017 containing pMG36e was used as the indicator strain. Transformants containing pCMAP03 exhibited activity against the indicator, whereas E. faecalis ATCC 19433 containing pMG36e did not. L. lactis ATCC 19435 was used as the indicator for heterologous bacteriocin expression by the dedicated secretion machinery, because this strain is sensitive to EntB but not to CbnA. Transformants of C. piscicola LV17A with plasmid pCMAP04 exhibited antimicrobial activity against this indicator, whereas transformants of C. piscicola LV17C containing plasmid pCMAP04 did not.
Multiple bacteriocin production.
The nucleotide sequence from the PCR amplicon in pCMAP06 (Table 1) obtained from PCR 1 encoded amino acids identical to the last 42 amino acids of the EntA gene, as determined previously by Aymerich et al. (3). The codon usage in the sequence derived from E. faecium BFE 900 chromosomal DNA was identical to that reported for E. faecium CTC 492 by Aymerich et al. (3) (results not shown). The use of MALDI-TOF mass spectrometry on supernatants of E. faecium BFE 900 revealed two peaks, which corresponded to the experimentally determined masses of enterocins A and B (Fig. 2). The production of additional bacteriocin was also indicated by deferred-inhibition tests with E. faecium BFE 900 and an EntB-producing clone of E. faecalis. These tests showed that a zone of activity was obtained with both producers when L. sake DSM 20017 was used as the indicator. Although inhibition zones were obtained with E. faecium BFE 900, zones were not observed with the E. faecalis clone when C. piscicola LV17C and C. piscicola UAL26 were used as indicators.
Immunity gene.
The possibility that one of the putative ORFs on the 2.2-kb cloned fragment encoded the immunity protein for EntB was investigated. When L. sake DSM 20017 containing pCMAP04 was used as an indicator in a deferred-inhibition test with E. faecalis containing pCMAP03 as a producer, a smaller zone of inhibition resulted than that of the L. sake control containing pMG36e (Fig. 4). This indicated that one or more ORFs on the 2.2-kb fragment were involved in immunity. The ORF eniB was considered a likely candidate, because it is located adjacent to the enterocin structural gene albeit in an opposite orientation (Fig. 3) and it has a relatively strong RBS. This ORF was amplified by PCR and cloned into pMG36e (pCMAP05) in correct orientation in relation to the P32 promoter for expression (Fig. 3) and transformed into L. sake DSM 20017 for use as an indicator in the deferred-inhibition assay with E. faecalis ATCC 19433 containing pCMAP03. The absence of a zone of inhibition in this assay (Fig. 4) indicated that eniB is the immunity gene for EntB. This gene encodes a putative product of 58 amino acids that has charged amino acids at both its ends. By using the PepTool protein structure prediction software (version 1.0.0B1; BioTools Ltd., Edmonton, Alberta, Canada) the central region (residues 9 to 49) was predicted to form an α-helix that could insert itself into the membrane. The polar ends were assumed to be in a random coil.
FIG. 4.
Deferred-inhibition test with E. faecalis ATCC 19433 containing pCMAP03 (EntB+) against L. sake DSM 20017 containing plasmids pMG36e (A), pCMAP04 (B), and pCMAP05 (C).
DISCUSSION
E. faecium BFE 900 was originally isolated from olives (17), and in our study it was shown that it produces a bacteriocin identical to EntB from E. faecium T136 that was isolated from meat (8). The EntB prepeptides in E. faecium T136 and BFE 900 also have identical leader peptides. As mentioned by Casaus et al. (8) EntB has strong sequence similarity to CbnA produced by C. piscicola LV17A (44). Both consist of 53 amino acids, and they have 47% identical amino acids. Their leader peptides both contain 18 amino acids and have 72% similarity (8). The comparison of the mature EntB and CbnA peptides by Lipman-Pearson protein alignment indicates 58.5% similarity. Although the N-terminal extension of EntB has homology to leader sequences of other bacteriocins, the mature bacteriocin has homology only with CbnA (8).
The experimentally determined molecular mass of EntB was 2 Da less than the calculated mass, indicating that a disulfide bridge is probably formed between the two cysteine residues at positions 23 and 52. The molecular mass of EntB determined in this study is identical to that reported by Casaus et al. (8). A second peak with a molecular mass of 5,479.0 Da was detected in mass spectral analysis, which was similar to the mass of EntB determined by MALDI-TOF mass spectrometry. This peak was 16 Da more than EntB and probably represents EntB with an oxidized methionine residue. The peaks obtained for enterocins A and B by MALDI-TOF mass spectrometry differed from the molecular masses reported for these bacteriocins by up to 5 Da (3, 8). This could be explained by the fact that the accuracy of MALDI-TOF mass spectrometry is in the range of 0.01 to 0.1% (38). Furthermore, in complex analytes, moderate concentrations of salt and detergent detract significantly from the desorption-ionization efficiency of peptides, reducing the mass range and resolution of mass spectra (46). Internal calibrants were not used together with the culture supernatants in MALDI-TOF mass spectrometry. Theoretically the use of such internal calibrants should result in more accurate mass determinations; however, they may interfere with the mass spectrometry of compounds that are present in small amounts in the culture supernatant by suppressing analyte ion formation, leading to the detection of only the calibrants.
Three base pairs upstream of the −35 putative promoter sequence for entB is a conserved sequence like a regulatory box. Diep et al. (15) suggested that such sequences are conserved in regulated bacteriocins. The conserved sequence like a regulatory box upstream of the EntB promoter region suggests that EntB production is regulated and that it could rely on the gene for an induction factor (see below) as well as an unidentified response regulator and a histidine kinase. A conserved regulatory sequence was not observed upstream of entB in E. faecium T136 (8). This is surprising because the nucleotide sequence reported for entB by Casaus et al. (8) is identical to the sequence determined in our study; however, the nucleotide sequence upstream of the RBS of entB in this study differs from that reported by Casaus et al. (8).
The putative protein product of orf6 had characteristics of an IF. Similar to other IF genes, a conserved sequence like a regulatory box (a 9-bp repeat spaced by 13 nucleotides) is located upstream of orf6. This putative IF differs from EntF, the IF previously described to be responsible for the induction of enterocins A and B (30). EntF consists of a 41-amino-acid prepeptide with a 16-amino-acid double-glycine-type leader peptide (30). Induction experiments in parallel with MALDI-TOF mass spectrometry showed conclusively that the production of EntB in E. faecium BFE 900 was constitutive, whereas the production of EntB in E. faecium CTC 492 was induced. The constitutive production of EntB in strain BFE 900 suggests that the product of orf6, which has the characteristics of an IF, is probably not associated with bacteriocin production. It is possible that EntB production in strain BFE 900 was regulated at an earlier time and that during the course of evolution the bacteriocin may have become constitutively produced as a result of a mutational event. This could explain the presence of a conserved sequence like a regulatory box upstream of the EntB structural gene. This is the first study to describe the constitutive and regulated production of an identical bacteriocin by different bacteria. Although the production of identical bacteriocins by different bacterial strains, species, or genera has been reported before for pediocin PA-1/AcH (17, 22, 28) and leucocin A (19, 20, 21), a difference in the regulation of production of these has not yet been described. Based on our MALDI-TOF mass spectrometry data, the production of EntA in E. faecium BFE 900 and CTC 492 also appears to be constitutive, because this compound was detected in supernatants of cultures of E. faecium BFE 900 and CTC 492 that grew after dilution to 10−10 in induction experiments.
EntB differs from other class IIa bacteriocins because it does not contain a YGNGVXC consensus motif near the N terminus (25), but similar to class IIa bacteriocins it has a double-glycine-type leader peptide that is usually associated with a dedicated bacteriocin transport system (29). Heterologous expression could not be attempted with EntB transporter proteins, because the genes for EntB-dedicated ATP-binding cassette transporter and accessory proteins were not found on either the 2.2- or 12.0-kb cloned chromosomal fragments. This could mean that the secretion of EntB depends on the bacteriocin-dedicated transport proteins of a different bacteriocin, such as EntA. Deferred-inhibition tests with E. faecium BFE 900 and an EntB+ E. faecalis clone against various indicators suggested that the wild-type strain produced multiple bacteriocins. We obtained a PCR amplicon that included part of the mature EntA gene identical to that produced by E. faecium T136 (8) and E. faecium CTC 492 (30). In addition, results from MALDI-TOF mass spectrometry indicated that enterocins A and B are both produced by E. faecium BFE 900. The results of this study, therefore, contribute to our understanding of the transport of multiple bacteriocins from a cell. From our study we expect that the two different bacteriocins may be secreted by one set of transport proteins. C. piscicola LV17 produces three bacteriocins, carnobacteriocins A, BM1, and B2 (33, 44). The secretion of CbnBM1, for which the structural and immunity genes are located on the chromosome, is dependent on bacteriocin-dedicated transport proteins associated with CbnB2 that are located on a plasmid (33). In contrast, the genetic loci for CbnA and CbnB2 include genes encoding their own dedicated transport proteins which differ in amino acid sequence (33, 43). Thus, multiple bacteriocin production can occur by either of these two transport possibilities.
For most class II bacteriocins described to date, the structural gene is in an operon with an immunity gene located adjacent to and downstream of the structural gene (25, 29). This was not the case for EntB; it resembles CbnA, in which an immunity gene was not detected in the same operon as the bacteriocin structural gene (44). However, the immunity gene for EntB was located in the opposite orientation, immediately downstream of the EntB structural gene, which is different from CbnA. This study is the first to report the EntB immunity gene and the first to report an immunity gene for class IIa bacteriocins that do not contain the N-terminal YGNGVXC motif but have sequence homology at the C terminus.
Apart from the EntB structural and immunity genes, no other genes associated with bacteriocin production could be unequivocally identified from the sequenced 12.0-kb EcoRI chromosomal DNA fragment of E. faecium BFE 900. This is in contrast to the presence and arrangement of genes in bacteriocin loci of other bacteriocin-producing bacteria, including the similar bacteriocin CbnA (6, 25, 29, 32, 40, 43). The genetic locus of EntB production is therefore atypical. The instance of isolated structural and immunity genes is rare and is paralleled only by the case of CbnBM1, in which the similarly isolated structural and immunity genes are located on the chromosome of C. piscicola LV17 (33). In contrast, the EntB structural and immunity genes also are not arranged in an operon.
The leader peptide of EntB is similar to that of CbnA, with 13 of 18 amino acids identical, which prompted the investigation of the heterologous expression of EntB with the CbnA transport proteins. The cloning of the EntB structural gene into C. piscicola LV17A resulted in the activity of this strain against L. lactis ATCC 19435, an EntB-sensitive, CbnA-resistant indicator. This suggested that the enterocin leader peptide was recognized by the ATP-binding cassette transporter for CbnA. The heterologous expression of EntB was also achieved by the sec pathway.
As the EntB structural and immunity genes have been identified and cloned they are now available for incorporation in a multiple-bacteriocin cassette, the construction of which is one of the ultimate goals of our research. As shown in this study, the secretion of EntB in a heterologous host can be achieved either by utilizing the CbnA dedicated bacteriocin transport system or by accessing the sec pathway by fusing the EntB mature bacteriocin gene to the divergicin A signal peptide. This multiple-bacteriocin cassette is envisaged for use in a food-grade starter strain for the biopreservation of foods.
ACKNOWLEDGMENTS
We thank M. Hugas for supplying E. faecium CTC 492. We also thank Ken Roy, Marco van Belkum, Liang Yan, John McCormick, Adam Szpacenko, Lynn Elmes, and Natisha Rose for technical assistance or helpful advice.
This research was funded by a strategic grant from the Natural Sciences and Engineering Research Council of Canada. C.M.A.P.F. gratefully acknowledges the financial assistance of a University of Alberta dissertation fellowship.
REFERENCES
- 1.Ahn C, Stiles M E. Plasmid-associated bacteriocin production by a strain of Carnobacterium piscicola from meat. Appl Environ Microbiol. 1990;56:2503–2510. doi: 10.1128/aem.56.8.2503-2510.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André Gordon C L, Ahmad M H. Thermal susceptibility of Streptococcus faecium strains isolated from frankfurters. Can J Microbiol. 1991;37:609–612. doi: 10.1139/m91-103. [DOI] [PubMed] [Google Scholar]
- 3.Aymerich T, Holo H, Håvarstein L S, Hugas M, Garriga M, Nes I F. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol. 1996;62:1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardowski J, Ehrlich S D, Chopin A. BglR protein, which belongs to the BglG family of transcriptional antiterminators, is involved in β-glucoside utilization in Lactococcus lactis. J Bacteriol. 1994;176:5681–5685. doi: 10.1128/jb.176.18.5681-5685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthier F, Zagorec M, Champomier-Vergès M, Ehrlich S D, Morel-Deville F. Efficient transformation of Lactobacillus sake by electroporation. Microbiology. 1996;142:1273–1279. doi: 10.1099/13500872-142-5-1273. [DOI] [PubMed] [Google Scholar]
- 6.Brurberg M B, Nes I F, Eijsink V G H. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban M C, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Casaus P, Nilsen T, Cintas L M, Nes I F, Hernández P E, Holo H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology. 1997;143:2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- 9.Chapot-Chartier M-P, Nardi M, Chopin M-C, Chopin A, Gripon J-C. Cloning and sequencing of pepC, a cysteine aminopeptidase gene from Lactococcus lactis subsp. cremoris AM2. Appl Environ Microbiol. 1993;59:330–333. doi: 10.1128/aem.59.1.330-333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapot-Chartier M-P, Rul F, Nardi M, Gripon J-C. Gene cloning and characterization of PepC, a cysteine aminopeptidase from Streptococcus thermophilus, with sequence similarity to the eucaryotic bleomycin hydrolase. Eur J Biochem. 1994;224:497–506. doi: 10.1111/j.1432-1033.1994.00497.x. [DOI] [PubMed] [Google Scholar]
- 11.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cintas L M, Casaus P, Håvarstein L S, Hernández P E, Nes I F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol. 1997;63:4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruz-Rodz A L, Gilmore M S. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990;224:152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- 14.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 15.Diep D B, Håvarstein L S, Nes I F. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178:4472–4483. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Hassouni M, Henrissat B, Chippaux M, Barras F. Nucleotide sequences of the arb genes, which control β-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new β-glycohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J Bacteriol. 1992;174:765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franz C M A P, Schillinger U, Holzapfel W H. Production and characterization of enterocin 900, a bacteriocin produced by Enterococcus faecium BFE 900 from black olives. Int J Food Microbiol. 1996;29:255–270. doi: 10.1016/0168-1605(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 18.Giraffa G, Carminati D, Neviani E. Enterococci isolated from dairy products: a review of risks and potential technological use. J Food Prot. 1997;60:732–738. doi: 10.4315/0362-028X-60.6.732. [DOI] [PubMed] [Google Scholar]
- 19.Hastings J W, Gibson P T, Chauhan R, Dykes G A, von Holy A. Similarity of bacteriocins from spoiled meat lactic acid bacteria. S Afr J Sci. 1996;92:376–380. [Google Scholar]
- 20.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastings J W, Stiles M E, von Holy A. Bacteriocins of leuconostocs isolated from meat. Int J Food Microbiol. 1994;24:75–81. doi: 10.1016/0168-1605(94)90107-4. [DOI] [PubMed] [Google Scholar]
- 22.Henderson J T, Chopko A L, van Wasseman P D. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 23.Houben J H. Heat resistance of Streptococcus faecium in pasteurized ham. Fleischwirtschaft. 1982;62:490–493. [Google Scholar]
- 24.Jay J M. Modern food microbiology. 5th ed. New York, N.Y: Chapman and Hall; 1996. pp. 395–400. [Google Scholar]
- 25.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 26.Le Coq D, Lindner C, Krüger S, Steinmetz M, Stülke J. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick J K, Worobo R W, Stiles M E. Expression of the antimicrobial peptide carnobacteriocin B2 by a signal peptide-dependent general secretory pathway. Appl Environ Microbiol. 1996;62:4095–4099. doi: 10.1128/aem.62.11.4095-4099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motlagh A M, Bhunia A K, Szostek F, Hansen T R, Johnson M C, Ray B. Nucleotide and amino acid sequence of pap gene (pediocin AcH) produced in Pediococcus acidilactici H. Lett Appl Microbiol. 1992;15:45–48. doi: 10.1111/j.1472-765x.1992.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 29.Nes I F, Diep D B, Håvarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 30.Nilsen T, Nes I F, Holo H. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J Bacteriol. 1998;180:1848–1854. doi: 10.1128/jb.180.7.1848-1854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozawa Y, Tanimoto K, Fujimoto S, Tomita H, Ike Y. Cloning and genetic analysis of the UV resistance determinant (uvr) encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pAD1. J Bacteriol. 1997;179:7468–7475. doi: 10.1128/jb.179.23.7468-7475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quadri L E N, Kleerebezem M, Kuipers O P, de Vos W M, Roy K L, Vederas J C, Stiles M E. Characterization of a locus from Carnobacterium piscicola LV17B involved in bacteriocin production and immunity: evidence for global inducer-mediated transcriptional regulation. J Bacteriol. 1997;179:6163–6171. doi: 10.1128/jb.179.19.6163-6171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 34.Rose N L, Sporns P, McMullen L M. Detection of bacteriocins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Appl Environ Microbiol. 1999;65:2238–2242. doi: 10.1128/aem.65.5.2238-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schnetz K, Stülke J, Gertz S, Krüger S, Krieg M, Hecker M, Rak B. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J Bacteriol. 1996;178:1971–1979. doi: 10.1128/jb.178.7.1971-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnetz K, Toloczyki C, Rak B. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siuzdak G. The emergence of mass spectrometry in biochemical research. Proc Natl Acad Sci USA. 1994;91:11290–11297. doi: 10.1073/pnas.91.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stiles M E, Holzapfel W H. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 40.van Belkum M J, Stiles M E. Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl Environ Microbiol. 1995;61:3573–3579. doi: 10.1128/aem.61.10.3573-3579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Guchte M, van der Vossen J M B M, Kok J, Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:224–228. doi: 10.1128/aem.55.1.224-228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 43.Worobo R W. Characterization of two bacteriocins and a food grade plasmid from carnobacteria. Ph.D. thesis. Edmonton, Alberta, Canada: University of Alberta; 1996. [Google Scholar]
- 44.Worobo R W, Henkel T, Sailer M, Roy K L, Vederas J C, Stiles M E. Characteristics and genetic determinant of a hydrophobic peptide bacteriocin, carnobacteriocin A, produced by Carnobacterium piscicola LV17A. Microbiology. 1994;140:517–526. doi: 10.1099/00221287-140-3-517. [DOI] [PubMed] [Google Scholar]
- 45.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worrall T A, Cotter R J, Woods A S. Purification of contaminated peptides and proteins on synthetic membrane surfaces for matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 1998;70:750–756. doi: 10.1021/ac970969e. [DOI] [PubMed] [Google Scholar]
- 47.Zukowski M M, Miller L, Cogswell P, Chen K, Aymerich S, Steinmetz M. Nucleotide sequence of the sacS locus of Bacillus subtilis reveals the presence of two regulatory genes. Gene. 1990;90:153–155. doi: 10.1016/0378-1119(90)90453-x. [DOI] [PubMed] [Google Scholar]