Abstract
Background and Objective
As the spine is pivotal in the support and protection of human bodies, much attention is given to the understanding of spinal diseases. Quick, accurate, and automatic analysis of a spine image greatly enhances the efficiency with which spine conditions can be diagnosed. Deep learning (DL) is a representative artificial intelligence technology that has made encouraging progress in the last 6 years. However, it is still difficult for clinicians and technicians to fully understand this rapidly evolving field due to the diversity of applications, network structures, and evaluation criteria. This study aimed to provide clinicians and technicians with a comprehensive understanding of the development and prospects of DL spine image analysis by reviewing published literature.
Methods
A systematic literature search was conducted in the PubMed and Web of Science databases using the keywords “deep learning” and “spine”. Date ranges used to conduct the search were from 1 January, 2015 to 20 March, 2021. A total of 79 English articles were reviewed.
Key Content and Findings
The DL technology has been applied extensively to the segmentation, detection, diagnosis, and quantitative evaluation of spine images. It uses static or dynamic image information, as well as local or non-local information. The high accuracy of analysis is comparable to that achieved manually by doctors. However, further exploration is needed in terms of data sharing, functional information, and network interpretability.
Conclusions
The DL technique is a powerful method for spine image analysis. We believe that, with the joint efforts of researchers and clinicians, intelligent, interpretable, and reliable DL spine analysis methods will be widely applied in clinical practice in the future.
Keywords: Deep learning (DL), spine, image analysis, review
Introduction
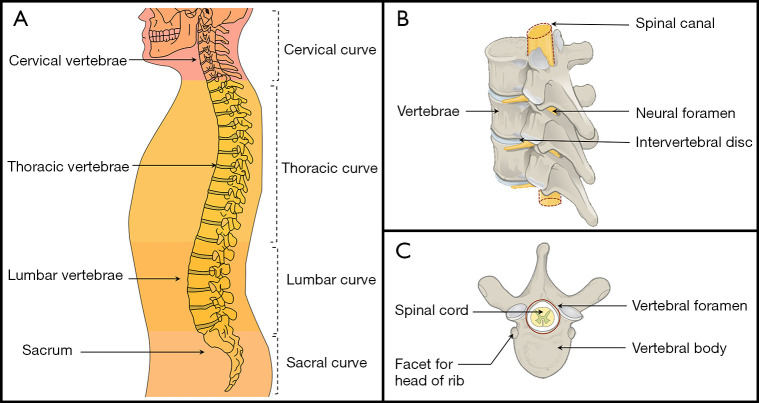
The spine is a critical supportive and protective structure in human bodies. The human spine and its regions consist mainly of cervical vertebrae, thoracic vertebrae, lumbar vertebrae, and the sacrum (1). The spine is composed of vertebrae, intervertebral discs (IVDs), neural foramina, and the spinal cord (Figure 1). Some spinal diseases have garnered more attention recently, including spinal deformities (2), back pain (3), and IVD disorder (4). For example, spine deformity affects 32–68% of individuals over 65 years old worldwide (5). Medical imaging plays a critical role in the diagnosis of spine diseases (6); however, the workload of radiologists has increased significantly due to the increasing number of people with spinal diseases (7,8). Further, even when following the same diagnostic standard, experienced radiologists can arrive at different evaluations (9). Therefore, a key challenge to spine image analysis is how to diagnose diseases quickly, automatically, and accurately.
Figure 1.
A schematic diagram of the main structure of the spine. On the left is a global diagram of the spine, while the right shows 2 local diagrams of the spine. (A) is derived from Illu vertebral column by Pixelsquid, used under Public Domain Mark 1.0. (B,C) Derived from Vertebra Superior View-en by Jmarchn, used under CC BY-SA 3.0. (B,C) Licensed under CC BY-SA 3.0 by Biao. Source: https://commons.wikimedia.org/wiki/File:Illu_vertebral_column.svg (A); https://commons.wikimedia.org/wiki/File:718_Vertebra-en.svg (B,C).
From the 1980s to the 1990s, researchers introduced pattern recognition methods to build task-specific spine image analysis methods (10), such as active contour models in the segmentation of vertebrae (11-13), watershed models in segmentation of IVDs and the spinal cord (14,15), and gradient-based methods in segmentation of the spine (16). These classic models rely on setting an edge detection strategy and manual thresholds to improve the segmentation performance (17), which may compromise the diagnostic performance. For example, the active contour model is sensitive to the manually selected initial contour position (18,19); the watershed model has a large number of minima in the embedded image or gradient, resulting in over-segmentation (20,21). These problems can be solved to some degree by gradient-based methods (18). For example, gradient vector flow was used as a new external force for the active contour model to reduce the sensitivity to the initial contour position (18). To eliminate the irrelevant local minima caused by noise and quantization errors in the gradient image, a watershed-based multi-scale gradient image segmentation algorithm was proposed to reduce over-segmentation and calculate time (22). Therefore, pattern recognition methods have pushed spine image analysis from manual setting to semi-automatic processing. However, the manual selection of initial features may still reduce the efficiency and accuracy of analysis (16).
The end of the 1990s saw machine learning replace semi-automatic work in spine image analysis, such as support vector machines (23,24) and random forests (25,26). The support vector machine finds the optimal hyperplane by learning samples and reducing the influence of outliers on the training data, thereby improving the robustness of the model in the reconstruction of three-dimensional scoliosis and diagnosis of IVD herniation (23,24). The random forest is a multivariate classifier that handles high-dimensional features and has been applied in spine classification with multiple objects (25,26). These machine learning methods have successfully transformed a human-designed system into a human training system (27). However, these machine learning systems still have hand-tailored characteristics since more complex feature engineering, such as the generation, extraction, and selection of features, needs to be completed by researchers (28,29).
The next step in spine image analysis is to determine how to enable computers to automatically learn data features. The powerful computing ability, such as the massive parallel computing with graphics processing units (GPUs) and big data in medical images, brings a great opportunity to the spinal imaging field. As a representative artificial intelligence technology, deep learning (DL) has revolutionized many fields (30), such as computer vision (31), natural language processing (32), biochemistry (33,34), signal processing (35,36), and medical image analysis (27,37-40). Exciting developments have been made with DL in spinal image analysis. For example, DL is used to generate a synthetic computed tomography (CT) images from radiation-free magnetic resonance imaging (MRI) and obtain measurement results equivalent to CT morphology, which provides new possibilities for radiation-free diagnosis (41). However, due to the diversity of applications, network structures, and evaluation criteria, it is still difficult to obtain an overall picture of this fast-developing field (Figure 2). Therefore, a systematic review is needed.
Figure 2.
A schematic diagram of the spine image analysis with DL: (A) segmentation, (B) detection, (C) diagnosis. [The spine image of (A,B) was reproduced with permission from the public dataset provided in (29)]. Source: https://www.spinesegmentation-challenge.com. DL, deep learning.
In this paper, we surveyed 79 papers on DL spine image analysis, to clarify the challenges, highlight the main contributions of published work, and for projection of future developments. This review attempts to provide clinicians and technicians with a comprehensive understanding of the current development and prospects of this active field.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-939/rc).
Methods
Search strategy
We conducted a comprehensive search of literature in the PubMed and Web of Science databases to review existing research about DL in spine image analysis. We searched for the following keywords in the title or abstract: “deep learning” AND “spine”. The search time range was from 1 January, 2015, to 20 March, 2021. The search strategy is shown in Table 1.
Table 1. The search strategy summary (Appendix 1).
| Items | Specification |
|---|---|
| Date of search | The search time range is from 1 January 2015 to 20 March 2021 |
| Databases and other sources searched | PubMed and Web of Science |
| Search terms used | Use “deep learning” + “spine” as keywords to search in the title or abstract of the literature |
| Timeframe | First search date: 15 October, 2020–5 November, 2020 |
| Second search date: 10 March, 2021–20 March, 2021 | |
| Literature selection: 21 March, 2021–15 April, 2021 | |
| Inclusion and exclusion criteria | Published English full-text journals and conference papers are selected, excluding reviews and non-English papers. Papers containing “deep learning” and descriptive words related to the spine are selected, otherwise they are excluded |
| Selection process | The literature selection was done independently by Biao Qu and Jianpeng Cao. Differences were resolved by consensus |
Inclusion criteria
The literature collection was conducted independently by 2 reviewers, and any differences were resolved by consensus. Papers were included if they were published in English full-text journals and conference papers, excluding reviews and non-English papers. Papers containing “deep learning” and descriptive words related to the spine were selected; Papers that do not contain both “deep learning” and descriptive words related to the spine will be excluded.
Data extraction
The data extraction was completed by 3 investigators (Biao Qu, Jianpeng Cao and Chen Qian). Author, publication date, target, disease, method, number of cases, modality, number of images, and metrics were extracted. These data were arranged into 4 tables which corresponded to different applications in DL spine image analysis.
Results
The review included 79 articles published in English. Figure 3 shows the number of papers published in the recent 6 years. The number of papers increased by a factor of 15 from 2015 to 2020. As an effective non-invasive examination method, MRI has become an important source of data, accounting for 40.5% of the 79 articles published papers. Figure 4 shows that the application of DL in spine image analysis mainly includes segmentation, detection, and diagnosis. Diagnosis is the most frequent application of DL in spinal imaging, accounting for 35.4% of included articles.
Figure 3.

Published papers on spine image analysis with DL. Source: PubMed and Web of Science. Search time range: 1 January 2015 to 20 March 2021. DL, deep learning; MRI, magnetic resonance imaging; CT, computed tomography.
Figure 4.
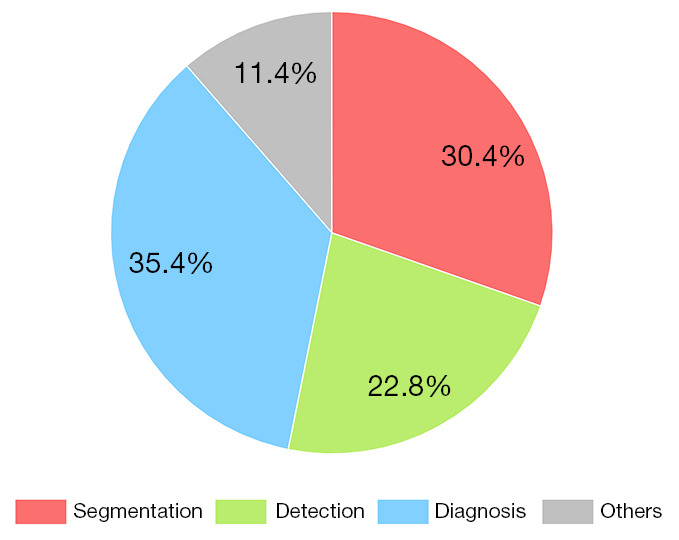
Application and distribution of DL in spine image analysis. DL, deep learning.
Discussion
After reviewing 79 papers, we found that different DL methods are suitable for different application scenarios of spine image analysis. In addition, the need for a large amount of training data in DL has also aroused patients’ concerns about privacy protection. Thus, this section discusses 3 aspects of DL spine image analysis: DL methods, main applications, and ethical and privacy issues.
DL methods
Here, we briefly discuss the common neural network structures (Figure 5) adopted in DL spine image analysis.
Figure 5.
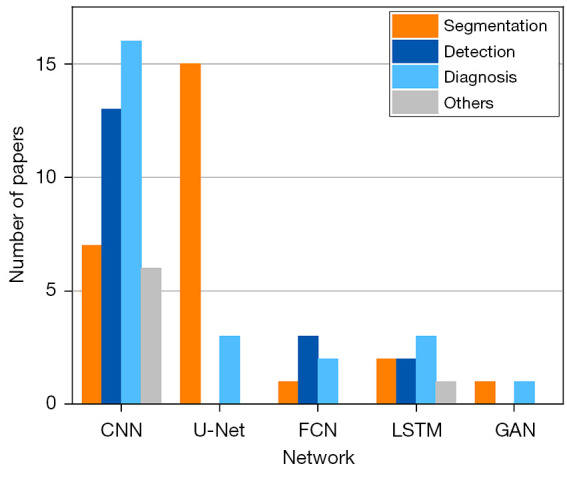
The number of commonly used network structures in DL spine image analysis. Source: PubMed and Web of Science. CNN. U-Net is a typical convolutional neural network (42), it has a contracting path for image downsampling and an expansive path for image upsampling. FCN; LSTM network; GAN. Search time range: 1 January, 2015 to 20 March, 2021. DL, deep learning; CNN, convolutional neural network; FCN, fully convolutional network; LSTM, long short-term memory; GAN, generative adversarial network.
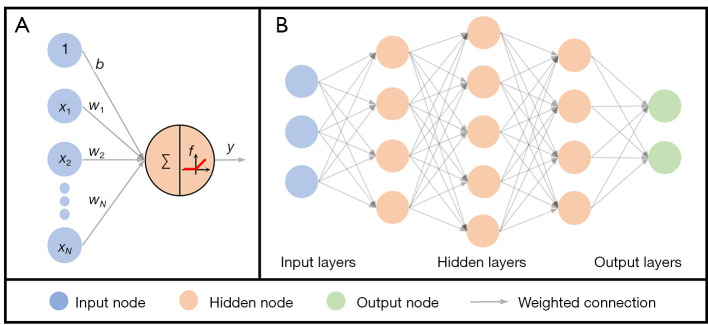
The artificial neuron is the basic unit of neural networks (Figure 6A). It comprises an input vector x, weights of a linear transform w, a bias b, and a nonlinear activation function f, as shown in Eq. [1]:
Figure 6.
A deep feedforward network with 5 layers. (A) The input vector x=[x1,x2,…,xN] and the bias b, the weighted connection composed of weight vector w=[w1,w2,…,wN]T and the activation function f. (B) A deep feedforward network that is composed of 1 input layer, 3 hidden layers, and 1 output layer.
| [1] |
where y is an output that is received by 1neuron of the next layer. If the next layer has multiple neurons, an output vector y=[y1,…,yM]T is formed according to
| [2] |
which can be simplified as
| [3] |
where W=[w1,…,wM], b=[b1,…,bM]T and the Sigmoid or rectified linear unit (ReLU) function is commonly chosen as f. These nonlinear functions provide a new feature representation by nonlinear transformations (43).
Deep feedforward networks (DFN) (Figure 6B) approximate some complex target functions by many artificial neurons in a chain structure as shown in Eq. [4] (43),
| [4] |
where L is the length of the chain and gives the depth of the network, which is also called the total number of layers. The feedforward means that there are no feedback connections in which outputs of the model are fed back into itself. Aside from the output layer, training data does not give the desired output of other layers, which are therefore called hidden layers. Goodfellow et al. suggested viewing DFNs as function approximation machines that are designed for statistical generalization (43).
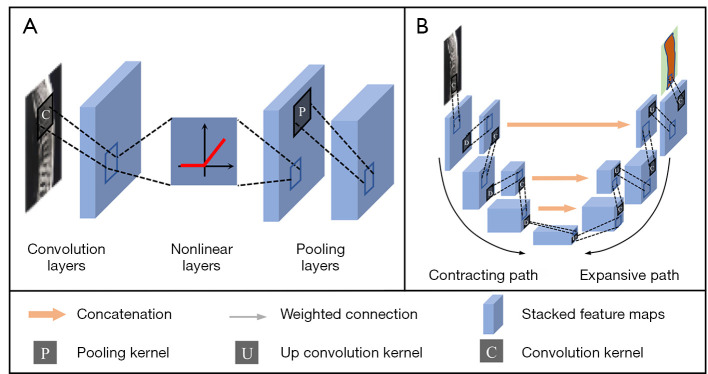
DFNs are fundamentals for many DL models (43). Convolutional neural network (CNN) is one of the most popular networks in spine image analysis. It consists of convolution layers, nonlinear layers, and pooling layers (Figure 7A). As the nonlinear layer is the same as DFNs, the following discussion focuses on the structures and functions of the other 2 layers.
Figure 7.
The network structure of CNN and U-Net (U-Net is a typical convolutional neural network (42), it has a contracting path for image downsampling and an expansive path for image upsampling). (A) CNN, (B) U-Net. Note: Stacked feature maps are a group of feature maps generated by the previous convolution, nonlinear or pooling layer. CNN, convolutional neural networks.
The convolution layers of CNN take the invariant translation statistical properties into account by sharing weight parameters across multiple image locations. The matrix multiplication WTx in Eq. [3] is replaced with the convolution in at least 1 layer. Given a convolution kernel k, Eq. [3] is rewritten as
| [5] |
where * denotes the convolution operation. A matrix form of Eq. [5] is
| [6] |
where k1,…kS−1,kS are the values of k. A smaller kernel size S means fewer input neurons are connected to the next layer and a sparser matrix W. Therefore, CNN enables neural networks with low memory to be computationally efficient. For the spine analysis, both the image x and convolution kernel k are in 2-dimensional (2D) form.
Pooling layers of CNN replace each output with a statistic of its nearby outputs. For example, max pooling and average pooling will output maximum and mean values within a rectangular neighborhood. Pooling operation brings invariance to local translation, which decreases the sensitivity of the networks to the feature location. With a huge amount of spine image 2D or 3D patches as the input and the labeled ground-truth as the output, CNN shows great ability to learn high-level features and improve the overall performance of spinal image segmentation and/or classification (28,44-46).
The U-Net is a typical CNN for image segmentation (42), which has a contracting path for image downsampling and an expansive path for image upsampling (Figure 7B). The contracting path consists of convolution and pooling layers where in spatial information of the image is reduced, and high-level features are extracted. In the expansive path, spatial information is concatenated with the feature from the contracting path. For spine segmented tasks, through the concatenation, multi-scale features are obtained to increase contrast and reduce blurred borders between vertebrae, IVDs, and the background, leading to more accurate spine segmentation (47-50).
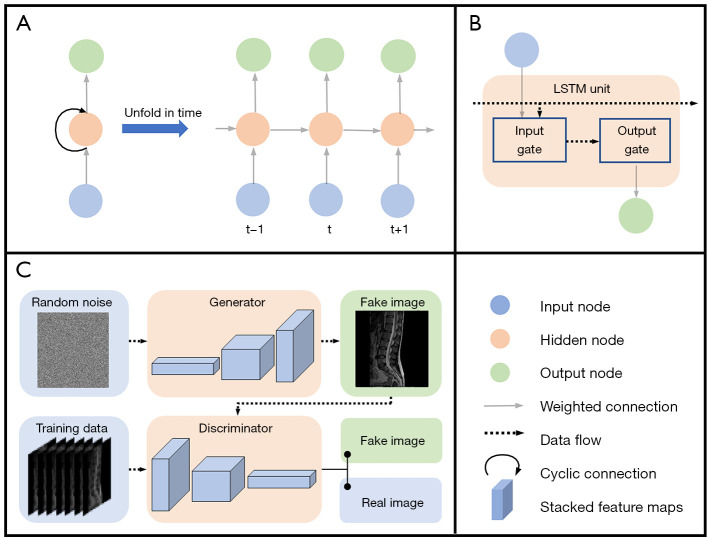
The recurrent neural network (RNN) is primarily designed for sequence data processing (51). The basic structure of RNNs consists of input nodes, output nodes, and hidden nodes with a cyclic connection. A computational graphic could be obtained by unrolling this recursive structure in time, leading to parameters sharing (Figure 8A). Usually, RNN records information from a previous input in a state vector, which is stored in hidden layers. The current output is determined by not only the current input, but also the state vector. However, gradient propagation over many stages tends to vanish or explode (43). Therefore, this structure fails to retain information for a long time, especially when the length of context increases (52). To solve this problem, the long short-term memory (LSTM) network employs different gates to control information flow into or out of the LSTM unit (53) (Figure 8B). Therefore, LSTM effectively mines the long-range contextual information that persists in the spinal images, and the large-scale memory information is conducive to improving the accuracy of detection (48).
Figure 8.
Network structures of a RNN, an LSTM unit and a GAN. (A) RNN has input nodes, output nodes, and hidden nodes with the cyclic connection. (B) The LSTM unit consists of input gate and output gate, which are built by neural network layers. Input gate adds significant information to data flow and ‘forgets’ unimportant parts. The output gate decides what LSTM unit is going to output. (C) The generator of the GAN produces fake images and the discriminator grades fake images and true images. [The spine image of (C) was reproduced with permission from the public dataset provided in Reference (29)]. Source: https://www.spinesegmentation-challenge.com. RNN, recurrent neural network; LSTM, long short-term memory; GAN, generative adversarial network.
Generative adversarial network (GAN) is another neural network that consists of 2 competitive components, 1 discriminator and 1 generator, which are playing in a 0-sum game (54) (Figure 8C). The generator produces samples, while the discriminator tries to distinguish between samples from the training data and the ‘fake samples’ from the generator. In practice, however, training a GAN is difficult because of the non-convex optimization of the generator and discriminator (43). Considerable efforts have been made to address this issue (55-57). In spine segmentation, the discriminator distinguishes predicted segmentation maps or ground-truth maps by scoring them (58). Then, scores direct the generator to narrow mismatches between predicted and ground-truth maps. This mechanism can correct prediction errors and break the limitations of small datasets, thereby increasing the continuity of data at the global level (55-58). Compared with using the ground-truth as labeled maps, GAN increases global-level accuracy and avoids over-fitting in spine image segmentation (58).
Freely accessible frameworks and increasingly powerful computing hardware are significant drivers of the popularity of DL. Frameworks such as Tensorflow (https://www.tensorflow.org/), Torch (https://pytorch.org/), and Keras (https://keras.io) make it easier to design or reproduce state-of-the-art networks. GPU and distributed computing promote the development of very large-scale networks with outstanding performance.
Applications
The neural network can explore the local and non-local information of spine images. Local information mainly includes edges and textures within each image patch, which can be extracted with the convolution kernel in DL. The non-local information commonly exists among multiple patches that are distant from each other. This non-local information could be the similarity and dissimilarity of adjacent patches (46), the connection and shape discrimination of adjacent vertebrae and IVDs (29), other information imbedded in inter-slices (11,59), and multi-modalities (60). From another view, neural networks can explore the static or dynamic spine image information. Static information commonly exists in a single image. Dynamic information is mainly represented by temporal changes in image sequences, such as the dynamic contrast enhancement in MRI and the motion mode of spine in video fluoroscopy (61,62).
This section discusses common applications of spine image analysis, including segmentation, detection, and diagnosis.
Segmentation
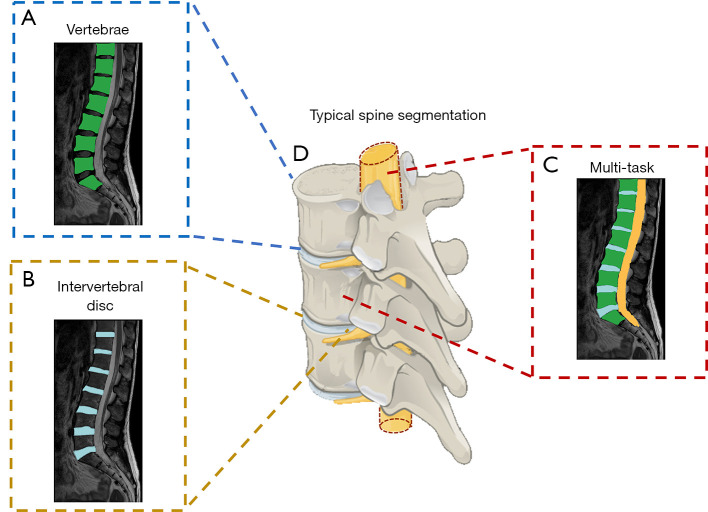
Object segmentation is an essential step for spine diagnosis and covers most DL methods in its image analysis. It is mainly used to distinguish the same type of objects from the background. This sub-section reviews the application of DL in spine image segmentation, including vertebrae segmentation (63-67), IVDs segmentation (13), and multi-task segmentation (68) (Figure 9; Table 2).
Figure 9.
Typical spine image segmentation with DL. (A) Vertebrae segmentation. (B) Intervertebral discs segmentation. (C) Multi-task segmentation. [The spine images of (A-C) were reproduced with permission from the public dataset provided in Reference (29)]. Source: https://www.spinesegmentation-challenge.com. (D) is derived from Vertebra Superior View-en by Jmarchn, used under Creative Commons Attribution Share Alike 3.0 Unported (CC BY-SA 3.0). (D) is licensed under CC BY-SA 3.0 by Biao. Source: https://commons.wikimedia.org/wiki/File:718_Vertebra-en.svg.) DL, deep learning.
Table 2. Summary of papers on deep learning spine segmentation.
| Author | Target | Method | Cases (source) | Modality | Images | Metric value |
|---|---|---|---|---|---|---|
| Kervadec et al. (63) (2019) | Vertebrae | 2D CNN | 23 (Z) | MRI | NA | DC 86.04% |
| Lavdas et al. (10) (2017) | Vertebrae | 3D CNN | 51 | MRI | NA | DC 81%±13% |
| Rak et al. (46) (2019) | Vertebrae | 3D CNN | 64+23 (Z) | MRI | NA | DC 93.8%±2.6% |
| Bae et al. (45) (2020) | Vertebrae | 2D U-Net | 41 | CT | 4,589 | DC 88.67%±5.82% |
| Chuang et al. (50) (2019) | Vertebrae | 3D U-Net | 32 | CT | NA | DC 92.6% |
| Fan et al. (64) (2019) | Vertebrae | 3D U-Net | 50 (S) | CT | NA | DC 94.5% |
| Fang et al. (69) (2021) | Vertebrae | 2D U-Net | 1449 | CT | NA | DC 82.3% |
| Kim et al. (65) (2020) | Vertebrae | 2D U-Net | 100 | CT | 344 | DC 90.4% |
| Kolarik et al. (66) (2019) | Vertebrae | 3D U-Net | 10 (S) | CT | NA | DC 97.08% |
| Lessmann et al. (11) (2019) | Vertebrae | 3D U-Net | 15 | MRI/CT | NA | DC 96.3%±1.3% |
| Rehman et al. (49) (2020) | Vertebrae | 2D U-Net | 45 | CT | NA | DC 96.4%±0.8% |
| Zhang et al. (47) (2021) | Vertebrae | 2D U-Net | 240 | MRI | NA | DC 92.6% |
| Zhou et al. (67) (2020) | Vertebrae | 2D U-Net | 57 | MRI | 1,140 | DC 84.9%±9.1% |
| Al Arif et al. (70) (2018) | Vertebrae | 2D FCN/U-Net | NA | X-ray | 296 | DC 84% |
| Xia et al. (71) (2020) | Vertebrae | 3D FCN/CNN | 10 (S) | CT | 10,991 | DC 94.84% |
| Iriondo et al. (13) (2020) | IVDs | 2D CNN | 31 | MRI | NA | DC 91.5% |
| Kim et al. (28) (2018) | IVDs | 2D U-Net | 20 (S) | MRI | NA | DC 89.44% |
| Li et al. (12) (2018) | IVDs | 3D FCN | 12 | MRI | NA | DC 91.34%±2.16% |
| Tam et al. (72) (2020) | IVDs; Vertebrae | 2D CNN | 222 (C) | MRI/CT | 1,413 | DC 93% |
| Pang et al. (29) (2021) | IVDs; Vertebrae | 3D CNN | 215 | MRI | NA | DC 87.49%±3.81% |
| Rehman et al. (73) (2019) | IVDs; Vertebrae | 2D U-Net | 20 (S)+173 | MRI/CT | NA | DC 90.37% ± 0.9% |
| Huang et al. (48) (2020) | IVDs; SC; Vertebrae | 2D U-Net | 100 | MRI | 300 | IoU 94.7% |
| Han et al. (58) (2018) | IVDs; NF; Vertebrae | 2D GAN | 253 | MRI | 3,564 | DC 87.1% |
| Hong et al. (68) (2020) | IVDs; NF; Vertebrae | 2D CNN | 200 | MRI | 3,400 | DC 90.6% |
C, http://csi-workshop.weebly.com/challenges.html; S, Spineweb (http://spineweb.digitalimaginggroup.ca); Z, http://dx.doi.org/10.5281/zenodo.22304. IVDs, intervertebral discs; NF, neural foramen; SC, spinal canal; CNN, convolutional neural network; DC, dice coefficient; FCN, fully convolutional network; GAN, generative adversarial network; MRI, magnetic resonance imaging; CT, computed tomography; N, not available; IoU, intersection over union.
Vertebrae segmentation
Vertebrae segmentation is the most common topic in spine segmentation papers and faces difficulties such as the complexity of structure, similarity of adjacent bones, and the low contrast of images (45,46,49) (Figure 9A; Table 2).
Initially, CNN was adopted to address these difficulties (10). A CNN cuts the spine image into many patches, performs pixel-level discrimination with learnable filters, and classifies patches through a sliding window. However, the patch-based perception area can only extract local features, which ignores the contextual information of the vertebrae and limits the segmentation performance.
Compared with CNN, U-Net obtains complete contextual information of the vertebrae by connecting up/down-sampling image features with skip connection operations (27). Thus, various advanced spine image segmentation methods adopt U-Net as the basic network structure.
The fundamental U-Net in spine segmentation is in 2D form. Each slice of the spine image is independently input to the network (45,69). This operation significantly saves the computation cost and increases the number of training samples by one order. By labelling the upper and lower vertebrae as the output of the network, the segmentation and separation of 3D vertebrae are resolved through 2D network training (45). However, the segmentation accuracy may be compromised by the irregular shape, artifacts, and large variability between slices (59). To adapt to the highly variable topological shape of vertebrae, Rehman et al. (73) integrated the parameter level set method into U-Net and improved the robustness of the network (74). To make use of the contextual information of adjacent vertebrae, Al Arif et al. (70) combined fully convolutional network (FCN) and 2D U-Net to enable global localization, center localization, and vertebrae segmentation in a single thread. The inter-slice information was also explored by Zhang et al. (59) with an inter-slice attention module, which substantially improved the segmentation accuracy compared to 2D U-Net and achieved comparable or slightly better performance than 3D U-Net.
A 3D U-Net treats 3D image patches of the multi-slice image as the network input. Lessmann et al. (11) proposed to use large size 3D image patches (128×128×128) to cover a whole vertebra, thus avoiding missing information of each vertebra in the network input. Chuang et al. (50) improved this method by changing the skip connection in the network to reduce computing with memory. Rak et al. (46) used a similar 3D U-Net network to combine the constraint of star convex cuts between adjacent patches, which solved the problem of blurred segmentation among adjacent cones.
IVD segmentation
An IVD lies between adjacent vertebrae and acts as a ligament to hold the vertebrae together and absorb shocks for the spine (Figure 9B).
Unlike the traditional U-Net for medical image segmentation (42), the boundary of IVDs has been treated carefully to realize the fine segmentation and improve the dice similarity coefficient by 3% (28). Different modal MRI images further improve the segmentation accuracy with multi-scale and modal dropout learning (12). To exploit the complementary information of different modalities, the “modal dropout learning” strategy randomly zeros out the voxel portion of the randomly selected modal image (49). The random dropout of voxels will allow the network to avoid redundant features, which can reduce co-adaption issues and improve the discrimination ability of the network (49). By combing the atlas-based registration and statistical parameter mapping, Iriondo et al. (13) quantitatively analyzed IVD degeneration.
Multi-task segmentation
The traditional single-task segmentation network focuses only on the segmentation of 1 class of objects. Multi-task segmentation simultaneously segments multiple classes of objects and combines segmentation with other tasks such as classification. For instance, a simultaneous segmentation and classification of spine images is displayed in Figure 9C. Due to the similarity of the edges of vertebrae and IVDs, the generalization and segmentation capabilities can be improved, and lower memory usage and faster inference calculation can be achieved by sharing the network parameters among multiple tasks (75-77).
To complete the semantic segmentation of the IVD, vertebral body, and neural foramen in 1 network, Han et al. (58) proposed a spine-GAN network to address the high diversity and variability of complex spine structure and used the LSTM to find the spatial pathological correlation between multiple objects. By modelling the dependency between adjacent vertebrae and IVDs with a probability map, Pang et al. (29) combined a 3D graph convolutional network to extract low-resolution features through the dependency and a 2D residual U-Net to refine segmentation with high-resolution image slices. To address the nonlinear relationship between different organs, Tam et al. (72) proposed the holistic multi-task regression method using multi-scale and fused image features and achieved outstanding portability and adaptability. To realize the segmentation and classification of multiple vertebral bodies simultaneously, Xia et al. (71) designed a 3D FCN with coarse segmentation and a cascaded CNN with fine segmentation.
Although many DL spine segmentation methods have been explored, there are still many problems that are worthy of continued research efforts. For example, it is hard to accurately segment small and complex organs, such as the spinal canal and intervertebral foramen. The number of cases in publications is still small (smaller than 100 in many publications, and the paper with the most contained 1,449 cases) and this may affect the applicability of these methods (Table 2).
Detection
Spine detection mainly includes localization and identification of vertebrae and IVDs (12,78-85) (Table 3). As it is manually performed by clinicians, traditional detection is time-consuming (87,89); there is a need for fast and automatic detection. Automatic detection will also resolve other difficulties, such as similar vertebrae structure (86), different field of view or resolution (90), different appearances caused by pathological variation, and image artefacts (90).
Table 3. Summary of papers on deep learning spine detection.
| Author | Target | Method | Cases (source) | Modality | Images | Metric value |
|---|---|---|---|---|---|---|
| Li et al. (12) (2018) | IVDs | 3D FCN | 12 (M16) | MRI/CT | NA | DC 91.2% |
| Amin et al. (78) (2015) | LV | 3D CNN | 32 | MRI | NA | IR 91% |
| Zhang et al. (86) (2020) | LV | 2D CNN/LSTM | 407 | MRI | NA | AC 93.55% |
| Zhou et al. (79) (2019) | LV | 2D CNN | 1,318 | MRI | 2,739 | AC 98.6% |
| Cai et al. (60) (2016) | Vertebrae | 2D CNN | 150 (S) | MRI/CT | NA | AC 95% |
| Chen et al. (87) (2020) | Vertebrae | 3D FCN | 302 (M14) | CT | NA | IR 94.67% |
| Forsberg et al. (80) (2017) | Vertebrae | 2D CNN | NA | CT | 720 | AC 99.1% |
| Zhang et al. (47) (2021) | Vertebrae | 2D SCRL | 240 | MRI | NA | AC 96.4% |
| Roggen et al. (81) (2020) | LV; TV | 2D CNN | 13 | X-ray | 952 | MV 0.23 mm |
| Chen et al. (88) (2015) | LV; TV; Sacrum | 2D CNN | 302 (M14) | CT | 67,145 | ME 8.82±13.04 mm |
| Liao et al. (89) (2018) | LV; TV; Sacrum | 3D CNN | 302 (S) | CT | NA | IR 88.3% |
| Wang et al. (82) (2019) | LV; TV; Sacrum | 2D CNN | 98 (S) | CT | 1,078 | IR 82.19% |
| Zhao et al. (90) (2021) | LV; TV; Sacrum | 2D CNN | NA | MRI | 450 | AC 95.5% |
| Hetherington et al. (83) (2017) | IVDs; LV; Sacrum | 2D CNN | 20 | US | 8,850 | AC 91% |
| Netherton et al. (84) (2020) | SC; Vertebrae | 2D CNN | 897 (M14) | CT | 2,296 | IR 86.8% |
| Jakubicek et al. (85) (2020) | IVDs; SC; Vertebrae | 2D CNN | 421 | CT | NA | IR 87.1% |
| Wimmer et al. (91) (2018) | IVDs; LV; TV; Sacrum | 2D CNN | NA (S) | MRI/CT | 1,659 | AC 92.5% |
IVDs, intervertebral discs; SC, spinal canal; LV, lumbar vertebrae; TV, thoracic vertebrae; CNN, convolutional neural network; SNN, siamese neural network; FCN, fully convolutional network; LSTM, long short-term memory; SCRL, sequential conditional reinforcement learning network; US, ultrasound; MRI, magnetic resonance imaging; CT, computed tomography; NA, not available, AC, accuracy; ALE, average localization error; DC, dice coefficient; IR, identification rate; ME, mean error; MV, median value; M14, MICCAI 2014; M16, MICCAI 2016; S, Spineweb (http://spineweb.digitalimaginggroup.ca/).
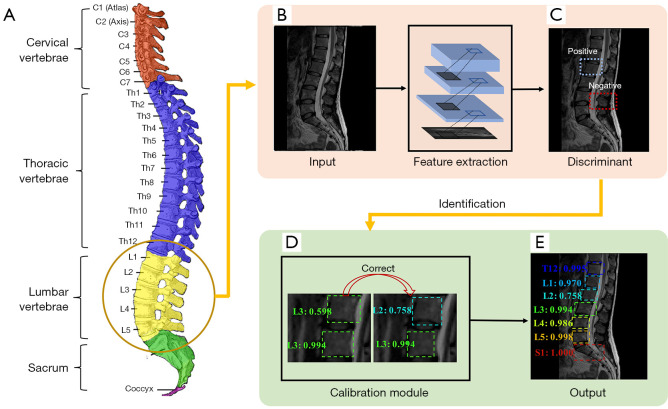
Figure 10 shows a typical process of automatic vertebrae detection. First, the vertebrae features are extracted through the neural network. Second, vertebrae are marked with a square box and only the positive boxes, identified through comparing with the manually labelled ones, are preserved to show the vertebrae location. Third, the wrong labels are corrected (90). Finally, the network generates the label and the recognition confidence score.
Figure 10.
A schematic diagram of the vertebrae detection process. (A) The anatomy of the human spine [(A) is derived from Gray 111 - Vertebral column-colored by Henry, used under Public Domain Mark 1.0, https://commons.wikimedia.org/w/index.php?curid=1282158]. (B) The spine image as an input. (C) Vertebrae discrimination. (D) Correction. (E) The identification as an output. (C-E) Adapted from Figure 3 in Ref. (90). Note: The square box is used to locate the vertebrae and the number after the label represents the recognition confidence score. MRI image was reproduced with permission from the public dataset provided in Reference (29). Source: https://www.spinesegmentation-challenge.com. MRI, magnetic resonance imaging.
In the first step of Figure 10, the shapes of vertebrae are first trained from the labelled data and then used to detect every possible vertebra in the target spine image. This approach is efficient since it usually divides the image into small regions or patches and requires a relatively small amount of training data. However, neighboring or global information of multiple vertebrae is not explored, and false positive detection may thus occur. To address this problem, the second and third steps of Figure 10 are adopted (sometimes the 2 steps may be integrated). For example, Chen et al. (88) combined the CNN and random forest classifier to slide extract vertebrae candidates, which increased the identification rate from 77.13% to 84.16% in typical CNN-based detection. By mining the long-range contextual information that existed in the fixed spatial order of vertebrae, Liao et al. (89) developed a bidirectional multi-task learning RNNs to jointly learn this contextual information from 2 directions (from cervical vertebrae to sacral vertebrae and the other way around) and further increased the identification rate to 88.3%. Zhang et al. (47) proposed to model the sequential correlations of vertebrae from top to bottom as dynamic-interaction processes and introduce deep reinforcement learning to segment and detect vertebrae concurrently. Since the sequential correlation was introduced, this method has effectively handled the complex background and pathological or anatomic variations (47). Zhang et al. (86) further applied a multi-task relational learning to locate, identify, and segment the vertebrae simultaneously, which avoided the overfitting of a single task, corrected each other, and pushed the identification rate to 93.55%.
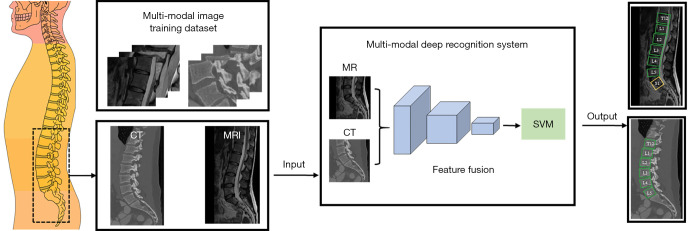
Unlike the previous practice of training a dedicated network from a single modality or contrast, multi-modality or multi-contrast image information can improve the spine detection ability of DL. Cai et al. (60) fused the MRI and CT image features with a transformed deep CNN (Figure 11). Single-modal features are initially detected and then fused in the intermediate layers of neural network, enhancing the invariance of vertebra patterns under different contrasts and modalities. An entropy-optimized texture model was introduced by Wimmer et al. (91) for seed point localization and iterative labelling, which enabled the use of a single learning-based pipeline without parameterizing it to different imaging modalities. Combing this model with CNN, automatic cross-modality sacral region detection of IVDs and vertebrae is achieved (91). If the multi-contrast images are not aligned, image registration is necessary since objects may move during the time gap between 1 to 9 min (93). Then, a multi-channel image, emulating the red/green/blue channel images in computer vision, is created for these multi-contrast images (93).
Figure 11.
The multi-modal recognition for spine images. Pictures are reproduced based on the idea in Fig. 1 of Reference (60). The CT image source is VerSe2019 (92) (https://osf.io/nqjyw/files/) and the MRI was reproduced with permission from the public dataset provided in Reference (29). Source: https://www.spinesegmentation-challenge.com. CT, computed tomography; MR, magnetic resonance; MRI, magnetic resonance imaging; SVM, support vector machine.
In summary, CNN is the most frequently adopted neural network and enables automatic spine localization and identification. In the future, multi-modal and multi-contrast information would be valuable research areas to improve the accuracy of location and identification rates.
Diagnosis
Diagnosis from spinal imaging includes the diagnosis of scoliosis, foraminal stenosis, metastatic spinal lesions, and spinal stenosis (94-111) (Table 4; Figure 12). However, conventional approaches may be time-consuming (121,122), cannot handle low-contrast spine images very well, and are prone to high inter- and intra-observer variability (112,115).
Table 4. Overview of papers using deep learning in the application of spinal diagnosis.
| Author | Disease | Method | Subjects (Source) | Modality | Images | Metric Value |
|---|---|---|---|---|---|---|
| Chen et al. (112) (2019) | Scoliosis | 2D CNN | 581 | X-ray | NA | RRMSE 11.88% |
| Galbusera et al. (95) (2019) | Scoliosis | 2D FCN | 493 | X-ray | NA | Standard angle errors 2.7°–11.5° |
| He et al. (96) (2021) | Scoliosis | 2D CNN | 525 | X-ray | NA | AC 80.36% |
| Horng et al. (111) (2019) | Scoliosis | 2D U-Net | NA | X-ray | 595 | DC 95.1%±2.7% |
| Kim et al. (113) (2020) | Scoliosis | 2D U-Net | NA | X-ray | 609 | CMAE 3.51° |
| Kokabu et al. (97) (2021) | Scoliosis | 2D CNN | 160 | X-ray | NA | AC 94% |
| Mandel et al. (98) (2021) | Scoliosis | 2D CNN | 139 | MRI | 695 | Errors 1.8±0.8 mm |
| Pan et al. (99) (2019) | Scoliosis | 2D CNN | 248 | X-ray | NA | SE 89.59% |
| Ito et al. (100) (2021) | Scoliosis | 2D CNN | 50 | X-ray | NA | Mean angle error 0.5° |
| Wu et al. (114) (2018) | Scoliosis | 2D CNN | 154 | X-ray | 526 | CMAE 4.04° |
| Yang et al. (115) (2019) | Scoliosis | 2D CNN | 3,240(B) | X-ray | NA | AC 94.6% |
| Jamaludin et al. (101) (2017) | Spinal stenosis | 3D CNN | 2,009 | MRI | 12,018 | AC 95.6% |
| Won et al. (116) (2020) | Spinal stenosis | 2D CNN | NA | MRI | 542 | AC 83% |
| Al-kafri et al. (93) (2019) | Spinal stenosis | 2D CNN | 515 | MRI | 48,345 | IoU 92% |
| Fan et al. (102) (2020) | Spinal stenosis | 3D U-Net | 31 | CT | 1,681 | DC 94% |
| Gaonkar et al. (117) (2019) | Spinal stenosis | 2D U-Net | 1,755 | MRI | NA | DC 88% |
| Han et al. (103) (2018) | Foraminal stenosis | 2D GAN | 253 | MRI | NA | AC 96.2%±0.3% |
| Han et al. (118) (2018) | Foraminal stenosis | 2D FCN | 200 | MRI | NA | PR 84.5% |
| LewandrowskI et al. (119) (2020) | Foraminal stenosis | 2D CNN | 3,560 | MRI | 17,800 | AC 86.2% |
| Chmelik et al. (120) (2018) | MSL | 2D CNN | 31 | CT | 626 | SE 92.0% |
| Lang et al. (61) (2019) | MSL | 2D LSTM | 61 | MRI | NA | AC 81%±3.4% |
| Wang et al. (104) (2017) | MSL | 2D SNN | 26 | MRI | NA | FP 0.40 |
| Löffler et al. (105) (2021) | Osteoporosis | 2D CNN | 192 | CT | NA | SE 84% |
| Zhang et al. (106) (2020) | Osteoporosis | 2D CNN | 910 | X-ray | 1,820 | AC 76.7% |
| Li et al. (107) (2021) | Fractures | 2D CNN | 941 | CT/MRI | NA | AC 93% |
| Maki et al. (108) (2020) | SSM | 2D CNN | 84 | MRI | 534 | AC 87.6% |
| Kim et al. (109) (2018) | Spondylitis | 2D CNN | 161 | MRI | 3,489 | AC 80.2% |
| Ma et al. (110) (2020) | Spinal cord injury | 2D CNN | 1,500 | MRI | 5,000 | PR 88.6% |
MSL, metastatic spinal lesions; SSM, spinal schwannoma and meningioma; LSTM, long short term memory network; CNN, convolutional neural network; FCN, fully convolutional network; MRI, magnetic resonance imaging; CT, computed tomography; NA, not available; CMAE, circular mean absolute error; IoU, intersection over union; DC, dice coefficient; FP, false positive; AC, accuracy; PR, precision; RRMSE, relative root mean squared error; SE, sensitivity; B, https://pan.baidu.com/s/1z9ipKpy0H09ceZtBDaJ09Q#list/path=%2F.
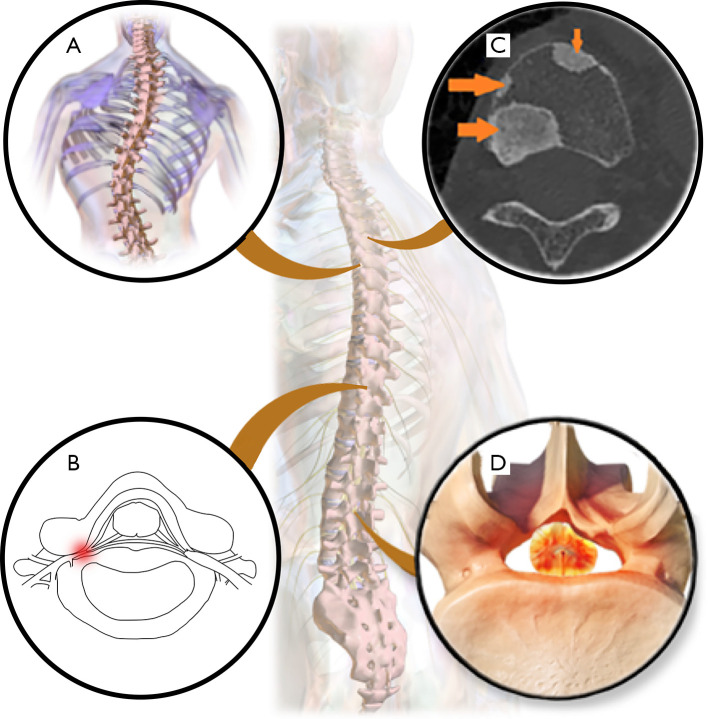
Figure 12.
Diagnosed diseases of spine images by DL. (A) Scoliosis. (B) Foraminal stenosis. (C) Metastatic spinal lesions, arrows indicate areas of metastatic spinal lesions. [Reproduced with permission in Figure 1 in Ref. (120)]. (D) Spinal stenosis. [(A) is derived from Blausen 0785 Scoliosis 01by BruceBlaus, used under Creative Commons Attribution Share Alike 3.0 Unported (CC BY-SA 3.0). (A) is licensed under CC BY-SA 3.0 by Biao. Source: https://commons.wikimedia.org/wiki/File:Blausen_0785_Scoliosis_01.png. (D) is derived from Spinal Stenosisby BruceBlaus, used under CC BY-SA 3.0. (D) is licensed under CC BY-SA 3.0 by Biao]. Source: https://commons.wikimedia.org/wiki/File:Spinal_Stenosis.png. DL, deep learning.
The X-ray is the most used imaging technique to diagnose scoliosis. Measurement of the Cobb angle is the gold standard for this disease (114) (Figure 13). The accuracy of manual measurement is susceptible to observer bias (124), and automatic measurements are highly anticipated. However, the anatomical variability and low tissue contrast of spine X-ray images make automated measurement of Cobb angles difficult (112). Recently, DL methods have been applied to automatically measure the Cobb angle. Yang et al. (115) proposed to automatically screen scoliosis on unclothed back images, avoiding X-ray radiation. Their work achieved superior accuracy over human specialists in detecting scoliosis cases with a curve ≥20° and severity grading. Chen et al. (112) enhanced the Cobb angle estimation with an alternative error correction net and integrated a high-precision calculation into the network, which reduced the error of the estimated angle by 50%. Wu et al. (114) combined anterior-posterior and lateral views through a convolutional layer and mitigated the problem of occlusion (e.g., vertebral occlusion in lateral view caused by the ribcage). The circular mean absolute error of Cobb angles in anterior-posterior and lateral views was significantly reduced to 4.04° and 4.07°, respectively. Kim et al. (113) built a scoliosis diagnosis system that provided not only automatic Cobb angle assessment but also localized and identified all vertebrae, which achieved a circular mean absolute error of 3.51°. Bernstein et al. (124) proposed a DL method to use a spline constructed from vertebra centroids to resemble spinal curve characteristics more closely and further enhance precision by automatically detecting the centroids. Their results showed that, even in the case of poor X-ray image quality, the error of measuring the Cobb angle was only one-tenth of manual measurement error, which greatly improved the measurement accuracy.
Figure 13.

A schematic diagram of measuring Cobb angle method (123). Cobb angle α involves measuring the angle between the endplate tangents of the upper and lower vertebrae. 0°<α≤10°, 10°<α≤20°, 20°<α≤40°, α>40° are defined as spine curve, mild scoliosis, moderate scoliosis, and severe scoliosis (111), respectively. [Note: The figure is adapted from Fig. 3 in Ref. (111)].
The diagnosis of spinal degenerative diseases, such as intervertebral foramen stenosis, spinal stenosis, and IVD herniation, are also handled well with DL. (I) Spinal stenosis: Al-Kafri et al. (93) studied lumbar spinal stenosis through a CNN semantic segmentation followed by delineating four regions, including IVDs, posterior element, thecal sac, and the area between anterior and posterior vertebrae elements in MRI images. Gaonkar et al. (117) adopted U-Net to segment discs on 100 sagittal MRI images and then analyzed changes in the lumbar spinal canal in terms of age, gender, and height. To evaluate the diagnostic agreement between DL and 2 experts, Won et al. (116) adopted CNN as the classifier in the grading of spinal stenosis on labelled axial MRI images, reporting an agreement accuracy of 75% and no statistical significance between the automatic the classifier and the human analyzer was observed. (II) Intervertebral foramen stenosis: For realizing early diagnosis and comprehensive evaluation, Han et al. (118) studied the pathology of lumbar foraminal stenosis through the location and grading of a neural foramen in terms of multiple goals, multiple scales, and multiple tasks. (III) IVD herniation: Staartjes et al. (125) used postoperative pain scores to train a preoperative clinical prediction model and then predict postoperative back pain in patients with lumbar disc herniation. Their model was not limited to the image features; class definitions and pathologies that existed in the written reports of radiologists were also chosen to train the DL model and generate the automatic diagnosis report for the central canal and neural foramina (119).
Few DL methods have been developed for the diagnosis of spinal cancer. Metastatic cancer is the most common malignant tumor in the spine, but the primary tumor location is unknown for approximately 30% of patients (61,126). To address this problem, Lang et al. (61) proposed to learn the temporal changes in dynamic contrast-enhanced MRI with convolutional LSTM network, which achieved slightly better accuracy (0.81) of metastases origination than that of the compared best radiomics method (0.79). Chmelik et al. (120) adopted the deep CNN and proposed medial axis transform based on the random forest to simplify the shape of the segmented lesion area, which solved the problem of distinguishing soluble and sclerosing metastatic lesions.
In summary, the accuracy of diagnosis can be improved with DL, and studies have shown that the accuracy of DL is similar to that of human analysis. Although the diagnosis of scoliosis and spinal degenerative diseases has been studied well, the use of DL for diagnosing spinal cancer DL remains under-explored.
Other applications
In addition to applications mentioned earlier, DL has been extended to other spine image analyses (62,127-129) (Table 5).
Table 5. Overview of papers using deep learning in other applications that are related to spine image analysis.
| Author | Task | Method | Subjects | Modality | Images | Metric Value |
|---|---|---|---|---|---|---|
| Liu et al. (62) (2019) | Spine tracking | 2D SNN | 47 | DVFI | 14,000 | ME 0.5° |
| Han et al. (127) (2021) | Report generation | 2D GAN | 253 | MRI | NA | AC 96.5% |
| Pang et al. (128) (2019) | Measurement | 2D CARN | 235 | MRI | NA | ME 1.22±1.04 mm |
| Shen et al. (129) (2021) | Measurement | 2D CNN | 120 | MRI | NA | IoU 92.2% |
| Schwartz et al. (130) (2021) | Measurement | 2D CNN | 816 | X-ray | NA | DC 95.1% |
| Korez et al. (131) (2020) | Measurement | 2D CNN | 55 | X-ray | NA | ME 1.2° |
| Esfandiari et al. (132) (2018) | Pose estimation | 2D CNN | 100 | X-ray | NA | ME 1.93°±0.64° |
| Lee et al. (133) (2020) | Prediction of BMD | 2D CNN | 334 | X-ray | NA | AUC 0.74 |
| Yasaka et al. (134) (2020) | Prediction of BMD | 2D CNN | 183 | CT | 1665 | AUC 0.97 |
| Pesteie et al. (135) (2019) | Data augmentation | 2D Auto-Encoders | 20 | US | 16,850 | AC 92% |
BMD, bone mineral density; CARN, cascade amplifier regression network; CNN, convolutional neural network; GAN, generative adversarial network; DVFI, digitalized video fluoroscopic imaging; US, ultrasound; MRI, magnetic resonance imaging; CT, computed tomography; DC, dice coefficient; NA, not available; AC, accuracy; AUC, area under curve; IoU, intersection over union; ME, mean error; SNN, siamese neural network.
In the automated quantitative measurement of the spine, Pang et al. (128) adopted the cascade amplifier regression network to measure the vertebral body height and IVD height, where the body height was valuable for assessing the risk of vertebral fractures and disc height decreased with the IVD degeneration. Other quantitative measurements, including pelvic incidence, pelvic tilt, and sacral slope, were evaluated with CNN by Korez et al. (131) and Schwartz et al. (130).
People with high-risk osteoporosis may be alerted as early as possible if bone mineral density can be accurately predicted (133). Yasaka et al. (134) exploited CNN to predict bone mineral density of lumbar vertebrae. The predicted density values from unenhanced abdominal CT images were significantly correlated with those values obtained with dual-energy X-ray absorptiometry (134). To compare the performance of different prediction models, Lee et al. (133) used 4 feature extraction models (AlexNet, VGGnet, Inception-V3, and ResNet-50) and 3 classification algorithms (Support vector machine, K-nearest neighbor, and random forest) to predict bone mineral density from spine X-ray images, and concluded that the combination of VGGnet for feature extraction and random forest for classification yielded the best overall performance.
Effects of different modalities in DL methods
Image modality plays an important role when devising a solution strategy for medical images. In this section, we discuss the effects of different modalities on various DL methods in image analysis applications for the spine.
Ultrasound is one of the most used imaging modalities due to its non-ionizing radiation and cost-effectiveness (136). However, ultrasound images are of low imaging quality due to speckle noise and reverberation artifacts (137). This problem can be alleviated by employing multi-scale and multi-directional image features (137). For example, multi-scale Hadamard features can be combined with convolutional feature maps for automatic localization of epidural needle insertion (137,138).
In the clinical examination of scoliosis, X-ray is regarded as the gold standard (139) and Cobb angle has been widely employed in the diagnosis decisions (112). Therefore, X-ray images are often used to automatically measure the Cobb angle (95,113).
Low spatial resolution is a common problem of CT. Before training of the network, CT images need to be pre-processed, such as improving image contrasts, by setting the appropriate window width and level (45,69,140) or utilizing contrast adaptive histogram equalization (71,107).
For MRI, the image intensity gradually varies within the single target, resulting in the poor performance of many pixel intensity-based methods. For example, intensities in magnetic resonance (MR) images are non-uniform due to the non-uniformity of the radio-frequency coil. Pre-processing for non-uniformity correction is considered necessary when the image is fed into the network, including N3 bias field correction and DL-based nonuniformity field correction (141,142). In addition, MRI has a variety of imaging sequences, such as T1-weighted, T2-weighted, and functional MRI (143). Since the contrast and information of the sequences are different, cross-modal convolution can effectively pool this complementary information and improve the accuracy of DL methods (143-145).
Ethical issues and privacy
The DL spine image analysis methods are not only employed in medical research but have also been approved by the Food and Drug Administration (FDA) of the United States and commercialized by companies such as Aidoc (146).
Commercialization raises ethical and privacy concerns. The first issue is decision-making accountability. If the DL algorithm makes a mistake, who is to blame: the radiologist operating the commercial product or the manufacturer of the product? Another issue is patient privacy protection. In general, leakage of patients’ privacy will not only damage the reputation of patients and cause mental stress to individuals, but also may cause economic losses (147). For example, after private medical data is leaked, the patient may be distressed by the potential of others knowing or even misusing their medical information. Patient long-term care insurance premiums may subsequently rise, and insurance companies may directly refuse to provide life insurance (148). In addition, employment discrimination may also occur (147,149). Therefore, it is necessary to protect patient privacy in the process of sharing and using patients’ data (147,150,151).
Conclusions and outlooks
In spine image analysis, DL has been extensively applied to the segmentation, detection, diagnosis, and quantitative evaluation. It can utilize static or dynamic image information and local or non-local information. The accuracy rates of DL analysis are almost as high as those of doctors and discrepancy between DL and radiologists has been shown to be lower than that between different radiologists. If these automatic methods can be integrated systematically, fully automatic processing could be possible.
However, these DL methods still face challenges, such as deficiency of data and interpretability. Privacy concerns make sharing medical data difficult (152). In addition, high-quality labelled data is scarce because collection and labelling by clinicians is time-consuming (153,154). This requires patients’ support and trust in sharing data and clinicians’ efforts to collect and label data. In addition, DL is often described as “black box medicine” (155), with little explanation for why certain features are chosen over others during training (156), which hinders the recognition of algorithms by clinicians and patients. Therefore, machine learning researchers are encouraged to increase the interpretability when designing DL models.
To solve these problems, we offer the following 4 suggestions:
Shared huge datasets are needed. First, protecting patient privacy during data sharing is essential so that patients are less worried and more willing to share their data. Personally identifiable information, such as names and email addresses, should be removed when sharing data (148). Another option is to form an authoritative steering board with representative patients to decide which data requests are allowed, and under which circumstances (148). Second, practitioners in this field are encouraged to jointly maintain a public dataset sharing platform and integrate datasets of all spine image analysis comparisons onto 1 platform. Furthermore, labelling these images is onerous. Therefore, it is necessary to develop an intelligent labelling system on this platform. Authorized automatic transmission from the picture archiving and communication systems (PACS) of the hospital to the labelling system, and real-time desensitization can save a lot of time to transfer the data. The optimal system would have both smart segmentation and good visualization functions to improve the efficiency of labelling by radiologists.
Reduce the need for training data. Transfer learning can address the issue of small datasets (157). It attempts to transfer knowledge from previous tasks (source domain) to a target task (target domain), and it is hoped that this knowledge can be adapted to new target tasks, thereby reducing the dependence on the amount of target task data (158). To achieve transformation capabilities, the network that is pre-trained in the source domain is used as the feature extractor in the target domain and fine-tuned with fixed network weights (159). Transfer learning is divided into cross-domain transfer learning and cross-modal transfer learning. Cross-domain transfer learning focuses on knowledge transfer across domains, and it is required that the training samples have sufficient generalization ability to adapt to the target domain, such as, models trained on natural images may not enough adapt to medical images. Cross-modal transfer learning is needed because spine imaging modalities are diverse, such as the multi-modalities (ultrasound, X-ray, CT, MRI), multi-sequences (T1-weighted imaging, T2-weighted imaging, and functional MRI), and multi-hospital data collection.
Learning functional information. Most current DL spine image analysis methods focus on structural features. The functional information may further expand the scope of intelligent analysis and fit for the clinical practice in spine diagnosis. For example, magnetic resonance hydrography can diagnose whether the nerve damage caused by foraminal stenosis is compressed or displaced (160). How to use neuroimaging and genetic information to intelligently analyze the innervation area to track the origin of spinal cord tumors is worth exploring. For MRI, perfusion (161), water-fat separation (162), and metabolic concentration (163) may improve the accuracy of diagnosing and predicting diseases such as tumors and IVD degeneration (164).
Improve the interpretability of networks. For machine learning researchers, combining the logical reasoning capabilities of other models with DL may be a solution. For example, the Bayesian DL model can use the perception ability of neural networks and the logical inference ability from the probabilistic graphical model (156), which can improve model interpretability (165). Another promising technique, called algorithm unrolling, was developed and can connect traditional iterative algorithms (such as those used for sparse coding) to neural network architectures (166). The unrolled network naturally inherits prior knowledge from iterative algorithms, and is thus no longer purely data-driven.
Summary
Reliable, intelligent, and interpretable DL spine image analysis requires the long-term efforts of machine learning researchers and clinicians and also the trust and support of patients. Perseverance from all parties will help to make DL available and widely accepted in clinical practice.
Acknowledgments
Funding: This work was financially supported by the Science and Technology Planning Project of Fujian Province (No. 2020H6003), the Xiamen Municipal Science and Technology Project (No. 3502Z20193015), the National Key R&D Program of China (No. 2017YFC0108703), the National Natural Science Foundation of China (Nos. 61971361, 61871341, and 62122064), the Health Education Joint Project of Fujian Province (No. 2019-WJ-31), and the Xiamen University Nanqiang Outstanding Talents Program.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-939/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-939/coif). China Mobile Group funded the computational resources of the Biomedical Intelligent Cloud R&D Center. QH is a representative of China Mobile Group and acts as vice director to co-supervise the running of the center. The other authors have no conflicts of interest to declare.
References
- 1.Rupp TK, Ehlers W, Karajan N, Günther M, Schmitt S. A forward dynamics simulation of human lumbar spine flexion predicting the load sharing of intervertebral discs, ligaments, and muscles. Biomech Model Mechanobiol 2015;14:1081-105. 10.1007/s10237-015-0656-2 [DOI] [PubMed] [Google Scholar]
- 2.Diebo BG, Shah NV, Boachie-Adjei O, Zhu F, Rothenfluh DA, Paulino CB, Schwab FJ, Lafage V. Adult spinal deformity. Lancet 2019;394:160-72. 10.1016/S0140-6736(19)31125-0 [DOI] [PubMed] [Google Scholar]
- 3.Vlaeyen JWS, Maher CG, Wiech K, Van Zundert J, Meloto CB, Diatchenko L, Battié MC, Goossens M, Koes B, Linton SJ. Low back pain. Nat Rev Dis Primers 2018;4:52. 10.1038/s41572-018-0052-1 [DOI] [PubMed] [Google Scholar]
- 4.Yang S, Zhang F, Ma J, Ding W. Intervertebral disc ageing and degeneration: The antiapoptotic effect of oestrogen. Ageing Res Rev 2020;57:100978. 10.1016/j.arr.2019.100978 [DOI] [PubMed] [Google Scholar]
- 5.Hoy D, Bain C, Williams G, March L, Brooks P, Blyth F, Woolf A, Vos T, Buchbinder R. A systematic review of the global prevalence of low back pain. Arthritis Rheum 2012;64:2028-37. 10.1002/art.34347 [DOI] [PubMed] [Google Scholar]
- 6.Mandl P, Navarro-Compán V, Terslev L, Aegerter P, van der Heijde D, D'Agostino MA, et al. EULAR recommendations for the use of imaging in the diagnosis and management of spondyloarthritis in clinical practice. Ann Rheum Dis 2015;74:1327-39. 10.1136/annrheumdis-2014-206971 [DOI] [PubMed] [Google Scholar]
- 7.Saffari A, Kölker S, Hoffmann GF, Weiler M, Ziegler A. Novel challenges in spinal muscular atrophy - How to screen and whom to treat? Ann Clin Transl Neurol 2018;6:197-205. 10.1002/acn3.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Guo R, Lv X, Lai Q, Xie B, Jiang X, Dai M, Zhang B. Challenges in diagnosis of spinal epidural abscess: A case report. Medicine (Baltimore) 2019;98:e14196. 10.1097/MD.0000000000014196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady AP. Error and discrepancy in radiology: inevitable or avoidable? Insights Imaging 2017;8:171-82. 10.1007/s13244-016-0534-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavdas I, Glocker B, Kamnitsas K, Rueckert D, Mair H, Sandhu A, Taylor SA, Aboagye EO, Rockall AG. Fully automatic, multiorgan segmentation in normal whole body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs), and a multi-atlas (MA) approach. Med Phys 2017;44:5210-20. 10.1002/mp.12492 [DOI] [PubMed] [Google Scholar]
- 11.Lessmann N, van Ginneken B, de Jong PA, Išgum I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal 2019;53:142-55. 10.1016/j.media.2019.02.005 [DOI] [PubMed] [Google Scholar]
- 12.Li X, Dou Q, Chen H, Fu CW, Qi X, Belavý DL, Armbrecht G, Felsenberg D, Zheng G, Heng PA. 3D multi-scale FCN with random modality voxel dropout learning for intervertebral disc localization and segmentation from multi-modality MR Images. Med Image Anal 2018;45:41-54. 10.1016/j.media.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 13.Iriondo C, Pedoia V, Majumdar S. Lumbar intervertebral disc characterization through quantitative MRI analysis: An automatic voxel-based relaxometry approach. Magn Reson Med 2020;84:1376-90. 10.1002/mrm.28210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieniewski M, Serneels R. Segmentation of spinal cord images by means of watershed and region merging together with inhomogeneity correction. Mach Graph Vis 2002;11:101-22. [Google Scholar]
- 15.Najman L, Schmitt M. Watershed of a continuous function. Signal Process 1994;38:99-112. 10.1016/0165-1684(94)90059-0 [DOI] [Google Scholar]
- 16.Koh J, Kim T, Chaudhary V, Dhillon G. Automatic segmentation of the spinal cord and the dural sac in lumbar MR images using gradient vector flow field. Annu Int Conf IEEE Eng Med Biol Soc 2010;2010:3117-20. [DOI] [PubMed] [Google Scholar]
- 17.Ke R, Bugeau A, Papadakis N, Kirkland M, Schuetz P, Schonlieb CB. Multi-task deep learning for image segmentation using recursive approximation tasks. IEEE Trans Image Process 2021;30:3555-67. 10.1109/TIP.2021.3062726 [DOI] [PubMed] [Google Scholar]
- 18.Xu C, Prince JL. Generalized gradient vector flow external forces for active contours. Signal Process 1998;71:131-9. 10.1016/S0165-1684(98)00140-6 [DOI] [Google Scholar]
- 19.Beck A, Teboulle M. A fast iterative shrinkage-thresholding algorithm for linear inverse problems. SIAM J Imaging Sci 2009;2:183-202. 10.1137/080716542 [DOI] [Google Scholar]
- 20.Beucher S. The watershed transformation applied to image segmentation. Scanning Microsc 1992;6:28. [Google Scholar]
- 21.Beucher S. Watershed, hierarchical segmentation and waterfall algorithm. Conference: Mathematical morphology and its applications to image processing 1994;69-76.
- 22.Wang D. A multiscale gradient algorithm for image segmentation using watershelds. Pattern Recognit 1997;30:2043-52. 10.1016/S0031-3203(97)00015-0 [DOI] [Google Scholar]
- 23.Oktay AB, Albayrak NB, Akgul YS. Computer aided diagnosis of degenerative intervertebral disc diseases from lumbar MR images. Comput Med Imaging Graph 2014;38:613-9. 10.1016/j.compmedimag.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 24.Lecron F, Boisvert J, Mahmoudi S, Labelle H, Benjelloun M. Three-dimensional spine model reconstruction using one-class SVM regularization. IEEE Trans Biomed Eng 2013;60:3256-64. 10.1109/TBME.2013.2272657 [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S, Chaudhary V. Supervised methods for detection and segmentation of tissues in clinical lumbar MRI. Comput Med Imaging Graph 2014;38:639-49. 10.1016/j.compmedimag.2014.03.005 [DOI] [PubMed] [Google Scholar]
- 26.Chu C, Belavý DL, Armbrecht G, Bansmann M, Felsenberg D, Zheng G. Fully automatic localization and segmentation of 3D vertebral bodies from CT/MR images via a learning-based method. PLoS One 2015;10:e0143327. 10.1371/journal.pone.0143327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Bae WC, Masuda K, Chung CB, Hwang D. Fine-grain segmentation of the intervertebral discs from MR spine images using deep convolutional neural networks: BSU-Net. Appl Sci (Basel) 2018;8:1656. 10.3390/app8091656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang S, Pang C, Zhao L, Chen Y, Su Z, Zhou Y, Huang M, Yang W, Lu H, Feng Q. SpineParseNet: spine parsing for volumetric MR image by a two-stage segmentation framework with semantic image representation. IEEE Trans Med Imaging 2021;40:262-73. 10.1109/TMI.2020.3025087 [DOI] [PubMed] [Google Scholar]
- 30.Litjens G, Ciompi F, Wolterink JM, de Vos BD, Leiner T, Teuwen J, Išgum I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc Imaging 2019;12:1549-65. 10.1016/j.jcmg.2019.06.009 [DOI] [PubMed] [Google Scholar]
- 31.Murphy RR. Computer vision and machine learning in science fiction. Sci Robot 2019;4:eaax7421. 10.1126/scirobotics.aax7421 [DOI] [PubMed] [Google Scholar]
- 32.Young T, Hazarika D, Poria S, Cambria E. Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag 2018;13:55-75. 10.1109/MCI.2018.2840738 [DOI] [Google Scholar]
- 33.Chen D, Wang Z, Guo D, Orekhov V, Qu X. Review and prospect: deep learning in nuclear magnetic resonance spectroscopy. Chemistry 2020;26:10391-401. 10.1002/chem.202000246 [DOI] [PubMed] [Google Scholar]
- 34.Qu X, Huang Y, Lu H, Qiu T, Guo D, Agback T, Orekhov V, Chen Z. Accelerated nuclear magnetic resonance spectroscopy with deep learning. Angew Chem Int Ed Engl 2020;59:10297-300. 10.1002/anie.201908162 [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Guo D, Tu Z, Huang Y, Zhou Y, Wang J, Feng L, Lin D, You Y, Agback T, Orekhov V, Qu X. A sparse model-inspired deep thresholding network for exponential signal reconstruction--application in fast biological spectroscopy. IEEE Trans Neural Netw Learn Syst 2022. [Epub ahead of print]. doi: . 10.1109/TNNLS.2022.3144580 [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, Zhao J, Wang Z, Orekhov V, Guo D, Qu X. Exponential signal reconstruction with deep hankel matrix factorization. IEEE Trans Neural Netw Learn Syst 2021. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 37.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44-56. 10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 38.Lin W, Tong T, Gao Q, Guo D, Du X, Yang Y, Guo G, Xiao M, Du M, Qu X, Alzheimer’s Disease Neuroimaging Initiative . Convolutional neural networks-based MRI image analysis for the alzheimer’s disease prediction from mild cognitive impairment. Front Neurosci 2018;12:777. 10.3389/fnins.2018.00777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu T, Zhang X, Huang Y, Guo D, Huang F, Xu Q, Hu Y, Ou-Yang L, Lin J, Yan Z, Qu X. pFISTA-SENSE-ResNet for parallel MRI reconstruction. J Magn Reson 2020;318:106790. 10.1016/j.jmr.2020.106790 [DOI] [PubMed] [Google Scholar]
- 40.Zeng G, Guo Y, Zhan J, Wang Z, Lai Z, Du X, Qu X, Guo D. A review on deep learning MRI reconstruction without fully sampled k-space. BMC Med Imaging 2021;21:195. 10.1186/s12880-021-00727-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morbée L, Chen M, Herregods N, Pullens P, Jans LBO. MRI-based synthetic CT of the lumbar spine: Geometric measurements for surgery planning in comparison with CT. Eur J Radiol 2021;144:109999. 10.1016/j.ejrad.2021.109999 [DOI] [PubMed] [Google Scholar]
- 42.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-assisted Intervention; 2015:234-41. [Google Scholar]
- 43.Goodfellow I, Bengio Y, Courville A, Bengio Y. Deep learning. MIT press; 2016. [Google Scholar]
- 44.Jamaludin A, Kadir T, Zisserman A. SpineNet: Automated classification and evidence visualization in spinal MRIs. Med Image Anal 2017;41:63-73. 10.1016/j.media.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 45.Bae HJ, Hyun H, Byeon Y, Shin K, Cho Y, Song YJ, Yi S, Kuh SU, Yeom JS, Kim N. Fully automated 3D segmentation and separation of multiple cervical vertebrae in CT images using a 2D convolutional neural network. Comput Methods Programs Biomed 2020;184:105119. 10.1016/j.cmpb.2019.105119 [DOI] [PubMed] [Google Scholar]
- 46.Rak M, Steffen J, Meyer A, Hansen C, Tönnies KD. Combining convolutional neural networks and star convex cuts for fast whole spine vertebra segmentation in MRI. Comput Methods Programs Biomed 2019;177:47-56. 10.1016/j.cmpb.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Chen B, Li S. Sequential conditional reinforcement learning for simultaneous vertebral body detection and segmentation with modeling the spine anatomy. Med Image Anal 2021;67:101861. 10.1016/j.media.2020.101861 [DOI] [PubMed] [Google Scholar]
- 48.Huang J, Shen H, Wu J, Hu X, Zhu Z, Lv X, Liu Y, Wang Y. Spine Explorer: a deep learning based fully automated program for efficient and reliable quantifications of the vertebrae and discs on sagittal lumbar spine MR images. Spine J 2020;20:590-9. 10.1016/j.spinee.2019.11.010 [DOI] [PubMed] [Google Scholar]
- 49.Rehman F, Ali Shah SI, Riaz MN, Gilani SO, R F. A region-based deep level set formulation for vertebral bone segmentation of osteoporotic fractures. J Digit Imaging 2020;33:191-203. 10.1007/s10278-019-00216-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang CH, Lin CY, Tsai YY, Lian ZY, Xie HX, Hsu CC, Huang CL. Efficient triple output network for vertebral segmentation and identification. IEEE Access 2019;7:117978-85.
- 51.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986;323:533-6. 10.1038/323533a0 [DOI] [Google Scholar]
- 52.Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw 1994;5:157-66. 10.1109/72.279181 [DOI] [PubMed] [Google Scholar]
- 53.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735-80. 10.1162/neco.1997.9.8.1735 [DOI] [PubMed] [Google Scholar]
- 54.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. Adv Neural Inform Process Syst 2014;27:2672-80. [Google Scholar]
- 55.Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv:1511.06434v2. 2016 Jan 7. Available online: https://arxiv.org/abs/1511.06434
- 56.Lai WS, Huang JB, Ahuja N, Yang MH. Deep laplacian pyramid networks for fast and accurate super-resolution. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017:624-32. [Google Scholar]
- 57.Yu Y, Gong Z, Zhong P, Shan J. Unsupervised representation learning with deep convolutional neural network for remote sensing images. International Conference on Image and Graphics; 2017:97-108. [Google Scholar]
- 58.Han Z, Wei B, Mercado A, Leung S, Li S. Spine-GAN: Semantic segmentation of multiple spinal structures. Med Image Anal 2018;50:23-35. 10.1016/j.media.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Yuan L, Wang Y, Zhang J. SAU-Net: efficient 3D spine MRI segmentation using inter-slice attention. Proceedings of the Third Conference on Medical Imaging with Deep Learning; 2020:903-13. [Google Scholar]
- 60.Cai Y, Landis M, Laidley DT, Kornecki A, Lum A, Li S. Multi-modal vertebrae recognition using transformed deep convolution network. Comput Med Imaging Graph 2016;51:11-9. 10.1016/j.compmedimag.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 61.Lang N, Zhang Y, Zhang E, Zhang J, Chow D, Chang P, Yu HJ, Yuan H, Su MY. Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn Reson Imaging 2019;64:4-12. 10.1016/j.mri.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Sui X, Liu C, Kuang X, Hu Y. Automatic lumbar spine tracking based on siamese convolutional network. J Digit Imaging 2020;33:423-30. 10.1007/s10278-019-00273-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kervadec H, Dolz J, Tang M, Granger E, Boykov Y, Ben Ayed I. Constrained-CNN losses for weakly supervised segmentation. Med Image Anal 2019;54:88-99. 10.1016/j.media.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 64.Fan G, Liu H, Wu Z, Li Y, Feng C, Wang D, Luo J, Wells WM, 3rd, He S. Deep learning-based automatic segmentation of lumbosacral nerves on CT for spinal intervention: A translational study. AJNR Am J Neuroradiol 2019;40:1074-81. 10.3174/ajnr.A6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim YJ, Ganbold B, Kim KG. Web-based spine segmentation using deep learning in computed tomography images. Healthc Inform Res 2020;26:61-7. 10.4258/hir.2020.26.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolarik M, Burget R, Uher V, Riha K, Dutta MK. Optimized high resolution 3D Dense-U-Net network for brain and spine segmentation. Appl Sci-Basel 2019;9:17. 10.3390/app9030404 [DOI] [Google Scholar]
- 67.Zhou J, Damasceno PF, Chachad R, Cheung JR, Ballatori A, Lotz JC, Lazar AA, Link TM, Fields AJ, Krug R. Automatic vertebral body segmentation based on deep learning of dixon images for bone marrow fat fraction quantification. Front Endocrinol (Lausanne) 2020;11:612. 10.3389/fendo.2020.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong YF, Wei BZ, Han ZY, Li X, Zheng YJ, Li S. MMCL-Net: spinal disease diagnosis in global mode using progressive multi-task joint learning. Neurocomputing 2020;399:307-16. 10.1016/j.neucom.2020.01.112 [DOI] [Google Scholar]
- 69.Fang Y, Li W, Chen X, Chen K, Kang H, Yu P, Zhang R, Liao J, Hong G, Li S. Opportunistic osteoporosis screening in multi-detector CT images using deep convolutional neural networks. Eur Radiol 2021;31:1831-42. 10.1007/s00330-020-07312-8 [DOI] [PubMed] [Google Scholar]
- 70.Al Arif SMMR, Knapp K, Slabaugh G. Fully automatic cervical vertebrae segmentation framework for X-ray images. Comput Methods Programs Biomed 2018;157:95-111. 10.1016/j.cmpb.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 71.Xia L, Xiao L, Quan G, Bo W. 3D cascaded convolutional networks for multi-vertebrae segmentation. Curr Med Imaging 2020;16:231-40. 10.2174/1573405615666181204151943 [DOI] [PubMed] [Google Scholar]
- 72.Tam CM, Zhang D, Chen B, Peters T, Li S. Holistic multitask regression network for multiapplication shape regression segmentation. Med Image Anal 2020;65:101783. 10.1016/j.media.2020.101783 [DOI] [PubMed] [Google Scholar]
- 73.Rehman F, Shah SIA, Riaz N, Gilani SO. A robust scheme of vertebrae segmentation for medical diagnosis. IEEE Access 2019;7:120387-98.
- 74.Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vis 1988;1:321-31. 10.1007/BF00133570 [DOI] [Google Scholar]
- 75.Liang J, Liu Z, Zhou J, Jiang X, Zhang C, Wang F. Model-protected multi-task learning. IEEE Trans Pattern Anal Mach Intell 2022;44:1002-19. 10.1109/TPAMI.2020.3015859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vandenhende S, Georgoulis S, De Brabandere B, Van Gool L. Branched multi-task networks: deciding what layers to share. arXiv:1904.02920v5. 2020 Aug 13. Available online: https://arxiv.org/abs/1904.02920
- 77.Lu Y, Kumar A, Zhai S, Cheng Y, Javidi T, Feris R. Fully-adaptive feature sharing in multi-task networks with applications in person attribute classification. Proceedings of the IEEE conference on computer vision and pattern recognition 2017;5334-43. [Google Scholar]
- 78.Amin S, Abtin R, Alexander S, Sidney F, Robert NR, Purang A. Deep learning for automatic localization, identification, and segmentation of vertebral bodies in volumetric MR images. Proc SPIE 2015;9415:14-21. [Google Scholar]
- 79.Zhou Y, Liu Y, Chen Q, Gu G, Sui X. Automatic lumbar MRI detection and identification based on deep learning. J Digit Imaging 2019;32:513-20. 10.1007/s10278-018-0130-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forsberg D, Sjöblom E, Sunshine JL. Detection and labeling of vertebrae in MR images using deep learning with clinical annotations as training data. J Digit Imaging 2017;30:406-12. 10.1007/s10278-017-9945-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roggen T, Bobic M, Givehchi N, Scheib SG. Deep Learning model for markerless tracking in spinal SBRT. Phys Med 2020;74:66-73. 10.1016/j.ejmp.2020.04.029 [DOI] [PubMed] [Google Scholar]
- 82.Wang X, Zhai S, Niu Y. Automatic vertebrae localization and identification by combining deep SSAE contextual features and structured regression forest. J Digit Imaging 2019;32:336-48. 10.1007/s10278-018-0140-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hetherington J, Lessoway V, Gunka V, Abolmaesumi P, Rohling R. SLIDE: automatic spine level identification system using a deep convolutional neural network. Int J Comput Assist Radiol Surg 2017;12:1189-98. 10.1007/s11548-017-1575-8 [DOI] [PubMed] [Google Scholar]
- 84.Netherton TJ, Rhee DJ, Cardenas CE, Chung C, Klopp AH, Peterson CB, Howell RM, Balter PA, Court LE. Evaluation of a multiview architecture for automatic vertebral labeling of palliative radiotherapy simulation CT images. Med Phys 2020;47:5592-608. 10.1002/mp.14415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jakubicek R, Chmelik J, Jan J, Ourednicek P, Lambert L, Gavelli G. Learning-based vertebra localization and labeling in 3D CT data of possibly incomplete and pathological spines. Comput Methods Programs Biomed 2020;183:105081. 10.1016/j.cmpb.2019.105081 [DOI] [PubMed] [Google Scholar]
- 86.Zhang R, Xiao X, Liu Z, Li Y, Li S. MRLN: Multitask relational learning network for MRI vertebral localization, identification, and segmentation. IEEE J Biomed Health Inform 2020;24:2902-11. 10.1109/JBHI.2020.2969084 [DOI] [PubMed] [Google Scholar]
- 87.Chen Y, Gao Y, Li K, Zhao L, Zhao J. Vertebrae identification and localization utilizing fully convolutional networks and a hidden markov model. IEEE Trans Med Imaging 2020;39:387-99. 10.1109/TMI.2019.2927289 [DOI] [PubMed] [Google Scholar]
- 88.Chen H, Shen C, Qin J, Ni D, Shi L, Cheng JCY, Heng PA. Automatic localization and identification of vertebrae in spine CT via a joint learning model with deep neural networks. International Conference on Medical Image Computing and Computer-assisted Intervention; 2015:515-22. [Google Scholar]
- 89.Liao H, Mesfin A, Luo J. Joint vertebrae identification and localization in spinal CT images by combining short- and long-Rrange contextual information. IEEE Trans Med Imaging 2018;37:1266-75. 10.1109/TMI.2018.2798293 [DOI] [PubMed] [Google Scholar]
- 90.Zhao S, Wu X, Chen B, Li S. Automatic vertebrae recognition from arbitrary spine MRI images by a category-Consistent self-calibration detection framework. Med Image Anal 2021;67:101826. 10.1016/j.media.2020.101826 [DOI] [PubMed] [Google Scholar]
- 91.Wimmer M, Major D, Novikov AA, Bühler K. Fully automatic cross-modality localization and labeling of vertebral bodies and intervertebral discs in 3D spinal images. Int J Comput Assist Radiol Surg 2018;13:1591-603. 10.1007/s11548-018-1818-3 [DOI] [PubMed] [Google Scholar]
- 92.Löffler MT, Sekuboyina A, Jacob A, Grau AL, Scharr A, El Husseini M, Kallweit M, Zimmer C, Baum T, Kirschke JS. A vertebral segmentation dataset with fracture grading. Radiol Artif Intell 2020;2:e190138. 10.1148/ryai.2020190138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al-Kafri AS, Sudirman S, Hussain A, Al-Jumeily D, Natalia F, Meidia H, Afriliana N, Al-Rashdan W, Bashtawi M, Al-Jumaily M. Boundary delineation of MRI images for lumbar spinal stenosis detection lough semantic segmentation using deep neural networks. IEEE Access 2019;7:43487-501.
- 94.Binder DK, Schmidt MH, Weinstein PR. Lumbar spinal stenosis. Semin Neurol 2002;22:157-66. 10.1055/s-2002-36539 [DOI] [PubMed] [Google Scholar]
- 95.Galbusera F, Niemeyer F, Wilke HJ, Bassani T, Casaroli G, Anania C, Costa F, Brayda-Bruno M, Sconfienza LM. Fully automated radiological analysis of spinal disorders and deformities: a deep learning approach. Eur Spine J 2019;28:951-60. 10.1007/s00586-019-05944-z [DOI] [PubMed] [Google Scholar]
- 96.He Z, Wang Y, Qin X, Yin R, Qiu Y, He K, Zhu Z. Classification of neurofibromatosis-related dystrophic or nondystrophic scoliosis based on image features using Bilateral CNN. Med Phys 2021;48:1571-83. 10.1002/mp.14719 [DOI] [PubMed] [Google Scholar]
- 97.Kokabu T, Kanai S, Kawakami N, Uno K, Kotani T, Suzuki T, Tachi H, Abe Y, Iwasaki N, Sudo H. An algorithm for using deep learning convolutional neural networks with three dimensional depth sensor imaging in scoliosis detection. Spine J 2021;21:980-7. 10.1016/j.spinee.2021.01.022 [DOI] [PubMed] [Google Scholar]
- 98.Mandel W, Oulbacha R, Roy-Beaudry M, Parent S, Kadoury S. Image-guided tethering spine surgery with outcome prediction using spatio-temporal dynamic networks. IEEE Trans Med Imaging 2021;40:491-502. 10.1109/TMI.2020.3030741 [DOI] [PubMed] [Google Scholar]
- 99.Pan Y, Chen Q, Chen T, Wang H, Zhu X, Fang Z, Lu Y. Evaluation of a computer-aided method for measuring the Cobb angle on chest X-rays. Eur Spine J 2019;28:3035-43. 10.1007/s00586-019-06115-w [DOI] [PubMed] [Google Scholar]
- 100.Ito S, Ando K, Kobayashi K, Nakashima H, Oda M, Machino M, Kanbara S, Inoue T, Yamaguchi H, Koshimizu H, Mori K, Ishiguro N, Imagama S. Automated detection of spinal schwannomas utilizing deep learning based on object detection from magnetic resonance imaging. Spine (Phila Pa 1976) 2021;46:95-100. 10.1097/BRS.0000000000003749 [DOI] [PubMed] [Google Scholar]
- 101.Jamaludin A, Lootus M, Kadir T, Zisserman A, Urban J, Battié MC, Fairbank J, McCall I; Genodisc Consortium. ISSLS PRIZE IN BIOENGINEERING SCIENCE 2017: Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J 2017;26:1374-83. 10.1007/s00586-017-4956-3 [DOI] [PubMed] [Google Scholar]
- 102.Fan G, Liu H, Wang D, Feng C, Li Y, Yin B, Zhou Z, Gu X, Zhang H, Lu Y, He S. Deep learning-based lumbosacral reconstruction for difficulty prediction of percutaneous endoscopic transforaminal discectomy at L5/S1 level: A retrospective cohort study. Int J Surg 2020;82:162-9. 10.1016/j.ijsu.2020.08.036 [DOI] [PubMed] [Google Scholar]
- 103.Han Z, Wei B, Leung S, Chung J, Li S. Towards automatic report generation in spine radiology using weakly supervised framework. International Conference on Medical Image Computing and Computer-assisted Intervention; 2018:185-93. [Google Scholar]
- 104.Wang J, Fang Z, Lang N, Yuan H, Su MY, Baldi P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput Biol Med 2017;84:137-46. 10.1016/j.compbiomed.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Löffler MT, Jacob A, Scharr A, Sollmann N, Burian E, El Husseini M, Sekuboyina A, Tetteh G, Zimmer C, Gempt J, Baum T, Kirschke JS. Automatic opportunistic osteoporosis screening in routine CT: improved prediction of patients with prevalent vertebral fractures compared to DXA. Eur Radiol 2021;31:6069-77. 10.1007/s00330-020-07655-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang B, Yu K, Ning Z, Wang K, Dong Y, Liu X, et al. Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: A multicenter retrospective cohort study. Bone 2020;140:115561. 10.1016/j.bone.2020.115561 [DOI] [PubMed] [Google Scholar]
- 107.Li YC, Chen HH, Horng-Shing Lu H, Hondar Wu HT, Chang MC, Chou PH. Can a deep-learning model for the automated detection of vertebral fractures approach the performance level of human subspecialists? Clin Orthop Relat Res 2021;479:1598-612. 10.1097/CORR.0000000000001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maki S, Furuya T, Horikoshi T, Yokota H, Mori Y, Ota J, Kawasaki Y, Miyamoto T, Norimoto M, Okimatsu S, Shiga Y, Inage K, Orita S, Takahashi H, Suyari H, Uno T, Ohtori S. A deep convolutional neural network with performance comparable to radiologists for differentiating between spinal schwannoma and meningioma. Spine (Phila Pa 1976) 2020;45:694-700. 10.1097/BRS.0000000000003353 [DOI] [PubMed] [Google Scholar]
- 109.Kim K, Kim S, Lee YH, Lee SH, Lee HS, Kim S. Performance of the deep convolutional neural network based magnetic resonance image scoring algorithm for differentiating between tuberculous and pyogenic spondylitis. Sci Rep 2018;8:13124. 10.1038/s41598-018-31486-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma S, Huang Y, Che X, Gu R. Faster RCNN-based detection of cervical spinal cord injury and disc degeneration. J Appl Clin Med Phys 2020;21:235-43. 10.1002/acm2.13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Horng MH, Kuok CP, Fu MJ, Lin CJ, Sun YN. Cobb angle measurement of spine from X-ray images using convolutional neural network. Comput Math Methods Med 2019;2019:6357171. 10.1155/2019/6357171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen B, Xu QH, Wang LS, Leung S, Chung J, Li S. An automated and accurate spine curve analysis system. IEEE Access 2019;7:124596-605.
- 113.Kim KC, Yun HS, Kim S, Seo JK. Automation of spine curve assessment in frontal radiographs using deep learning of vertebral-tilt vector. IEEE Access 2020;8:84618-30.
- 114.Wu H, Bailey C, Rasoulinejad P, Li S. Automated comprehensive adolescent idiopathic scoliosis assessment using MVC-Net. Med Image Anal 2018;48:1-11. 10.1016/j.media.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 115.Yang J, Zhang K, Fan H, Huang Z, Xiang Y, Yang J, et al. Development and validation of deep learning algorithms for scoliosis screening using back images. Commun Biol 2019;2:390. 10.1038/s42003-019-0635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Won D, Lee HJ, Lee SJ, Park SH. Spinal stenosis grading in magnetic resonance imaging using deep convolutional neural networks. Spine (Phila Pa 1976) 2020;45:804-12. 10.1097/BRS.0000000000003377 [DOI] [PubMed] [Google Scholar]
- 117.Gaonkar B, Villaroman D, Beckett J, Ahn C, Attiah M, Babayan D, Villablanca JP, Salamon N, Bui A, Macyszyn L. Quantitative analysis of spinal canal areas in the lumbar spine: an imaging informatics and machine learning study. AJNR Am J Neuroradiol 2019;40:1586-91. 10.3174/ajnr.A6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Han Z, Wei B, Leung S, Nachum IB, Laidley D, Li S. Automated pathogenesis-based diagnosis of lumbar neural foraminal stenosis via deep multiscale multitask learning. Neuroinformatics 2018;16:325-37. 10.1007/s12021-018-9365-1 [DOI] [PubMed] [Google Scholar]
- 119.LewandrowskI KU, Muraleedharan N, Eddy SA, Sobti V, Reece BD, Ramírez León JF, Shah S. Feasibility of deep learning algorithms for reporting in routine spine magnetic resonance imaging. Int J Spine Surg 2020;14:S86-97. 10.14444/7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chmelik J, Jakubicek R, Walek P, Jan J, Ourednicek P, Lambert L, Amadori E, Gavelli G. Deep convolutional neural network-based segmentation and classification of difficult to define metastatic spinal lesions in 3D CT data. Med Image Anal 2018;49:76-88. 10.1016/j.media.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 121.H A , Prabhu GK. Automatic quantification of spinal curvature in scoliotic radiograph using image processing. J Med Syst 2012;36:1943-51. 10.1007/s10916-011-9654-9 [DOI] [PubMed] [Google Scholar]
- 122.Anitha H, Karunakar A, Dinesh K. Automatic extraction of vertebral endplates from scoliotic radiographs using customized filter. Biomed Eng Lett 2014;4:158-65. 10.1007/s13534-014-0129-z [DOI] [Google Scholar]
- 123.Greiner KA. Adolescent idiopathic scoliosis: radiologic decision-making. Am Fam Physician 2002;65:1817-22. [PubMed] [Google Scholar]
- 124.Bernstein P, Metzler J, Weinzierl M, Seifert C, Kisel W, Wacker M. Radiographic scoliosis angle estimation: spline-based measurement reveals superior reliability compared to traditional COBB method. Eur Spine J 2021;30:676-85. 10.1007/s00586-020-06577-3 [DOI] [PubMed] [Google Scholar]
- 125.Staartjes VE, de Wispelaere MP, Vandertop WP, Schröder ML. Deep learning-based preoperative predictive analytics for patient-reported outcomes following lumbar discectomy: feasibility of center-specific modeling. Spine J 2019;19:853-61. 10.1016/j.spinee.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 126.Lemay A, Gros C, Zhuo Z, Zhang J, Duan Y, Cohen-Adad J, Liu Y. Automatic multiclass intramedullary spinal cord tumor segmentation on MRI with deep learning. Neuroimage Clin 2021;31:102766. 10.1016/j.nicl.2021.102766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Han Z, Wei B, Xi X, Chen B, Yin Y, Li S. Unifying neural learning and symbolic reasoning for spinal medical report generation. Med Image Anal 2021;67:101872. 10.1016/j.media.2020.101872 [DOI] [PubMed] [Google Scholar]
- 128.Pang S, Su Z, Leung S, Nachum IB, Chen B, Feng Q, Li S. Direct automated quantitative measurement of spine by cascade amplifier regression network with manifold regularization. Med Image Anal 2019;55:103-15. 10.1016/j.media.2019.04.012 [DOI] [PubMed] [Google Scholar]
- 129.Shen H, Huang J, Zheng Q, Zhu Z, Lv X, Liu Y, Wang Y. A deep-learning-based, fully automated program to segment and quantify major spinal components on axial lumbar spine magnetic resonance images. Phys Ther 2021;101:pzab041. [DOI] [PubMed]
- 130.Schwartz JT, Cho BH, Tang P, Schefflein J, Arvind V, Kim JS, Doshi AH, Cho SK. Deep learning automates measurement of spinopelvic parameters on lateral lumbar radiographs. Spine (Phila Pa 1976) 2021;46:E671-8. [DOI] [PubMed] [Google Scholar]
- 131.Korez R, Putzier M, Vrtovec T. A deep learning tool for fully automated measurements of sagittal spinopelvic balance from X-ray images: performance evaluation. Eur Spine J 2020;29:2295-305. 10.1007/s00586-020-06406-7 [DOI] [PubMed] [Google Scholar]
- 132.Esfandiari H, Newell R, Anglin C, Street J, Hodgson AJ. A deep learning framework for segmentation and pose estimation of pedicle screw implants based on C-arm fluoroscopy. Int J Comput Assist Radiol Surg 2018;13:1269-82. 10.1007/s11548-018-1776-9 [DOI] [PubMed] [Google Scholar]
- 133.Lee S, Choe EK, Kang HY, Yoon JW, Kim HS. The exploration of feature extraction and machine learning for predicting bone density from simple spine X-ray images in a Korean population. Skeletal Radiol 2020;49:613-8. 10.1007/s00256-019-03342-6 [DOI] [PubMed] [Google Scholar]
- 134.Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Prediction of bone mineral density from computed tomography: application of deep learning with a convolutional neural network. Eur Radiol 2020;30:3549-57. 10.1007/s00330-020-06677-0 [DOI] [PubMed] [Google Scholar]
- 135.Pesteie M, Abolmaesumi P, Rohling RN. Adaptive augmentation of medical data using independently conditional variational auto-encoders. IEEE Trans Med Imaging 2019;38:2807-20. 10.1109/TMI.2019.2914656 [DOI] [PubMed] [Google Scholar]
- 136.Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, Ni D, Wang T. Deep learning in medical ultrasound analysis: a review. Engineering 2019;5:261-75. 10.1016/j.eng.2018.11.020 [DOI] [Google Scholar]
- 137.Pesteie M, Lessoway V, Abolmaesumi P, Rohling RN. Automatic localization of the needle target for ultrasound-guided epidural injections. IEEE Trans Med Imaging 2018;37:81-92. 10.1109/TMI.2017.2739110 [DOI] [PubMed] [Google Scholar]
- 138.Pesteie M, Lessoway V, Abolmaesumi P, Rohling R. Automatic midline identification in transverse 2-D ultrasound images of the spine. Ultrasound Med Biol 2020;46:2846-54. 10.1016/j.ultrasmedbio.2020.04.018 [DOI] [PubMed] [Google Scholar]
- 139.Knott P, Pappo E, Cameron M, Demauroy J, Rivard C, Kotwicki T, Zaina F, Wynne J, Stikeleather L, Bettany-Saltikov J, Grivas TB, Durmala J, Maruyama T, Negrini S, O'Brien JP, Rigo M. SOSORT 2012 consensus paper: reducing x-ray exposure in pediatric patients with scoliosis. Scoliosis 2014;9:4. 10.1186/1748-7161-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Koehler PR, Anderson RE, Baxter B. The effect of computed tomography viewer controls on anatomical measurements. Radiology 1979;130:189-94. 10.1148/130.1.189 [DOI] [PubMed] [Google Scholar]
- 141.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29:1310-20. 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen L, Wu Z, Hu D, Wang F, Smith JK, Lin W, Wang L, Shen D, Li G, Consortium FUBCP. ABCnet: Adversarial bias correction network for infant brain MR images. Med Image Anal 2021;72:102133. 10.1016/j.media.2021.102133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Valindria VV, Pawlowski N, Rajchl M, Lavdas I, Aboagye EO, Rockall AG, Rueckert D, Glocker B. Multi-modal learning from unpaired images: application to multi-organ segmentation in CT and MRI. IEEE Winter Conference on Applications of Computer Vision; 2018:12-5. [Google Scholar]
- 144.Zhou T, Ruan S, Canu S. A review: deep learning for medical image segmentation using multi-modality fusion. Array 2019;3-4:100004. 10.1016/j.array.2019.100004 [DOI] [Google Scholar]
- 145.Tseng KL, Lin YL, Hsu W, Huang CY. Joint sequence learning and cross-modality convolution for 3d biomedical segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017:6393-6400. [Google Scholar]
- 146.Voter AF, Larson ME, Garrett JW, Yu JJ. Diagnostic accuracy and failure mode analysis of a deep learning algorithm for the detection of cervical spine fractures. AJNR Am J Neuroradiol 2021;42:1550-6. 10.3174/ajnr.A7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jia L, Fan W. Medical sports data privacy protection method based on legal risk control. J Healthc Eng 2021;2021:6630429. 10.1155/2021/6630429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Price WN, 2nd, Cohen IG. Privacy in the age of medical big data. Nat Med 2019;25:37-43. 10.1038/s41591-018-0272-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun J, Zhu X, Zhang C, Fang Y. HCPP: cryptography based secure EHR system for patient privacy and emergency healthcare. 31st International Conference on Distributed Computing Systems 2011;373-82. [Google Scholar]
- 150.Flanagin A, Bauchner H, Fontanarosa PB. Patient and study participant rights to privacy in journal publication. JAMA 2020;323:2147-50. 10.1001/jama.2020.3590 [DOI] [PubMed] [Google Scholar]
- 151.Chalmers J, Muir R. Patient privacy and confidentiality. BMJ 2003;326:725-6. 10.1136/bmj.326.7392.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 2016;35:1153-9. 10.1109/TMI.2016.2553401 [DOI] [Google Scholar]
- 153.Ker J, Wang L, Rao J, Lim T. Deep learning applications in medical image analysis. IEEE Access 2017;6:9375-89.
- 154.Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018;321:321-31. 10.1016/j.neucom.2018.09.013 [DOI] [Google Scholar]
- 155.Ford RA, Price W, Nicholson I. Privacy and accountability in black-box medicine. Mich Telecomm Tech L Rev 2016;23:1. [Google Scholar]
- 156.Chakraborty S, Tomsett R, Raghavendra R, Harborne D, Alzantot M, Cerutti F, Srivastava M, Preece A, Julier S, Rao RM. Interpretability of deep learning models: a survey of results. Available online: https://ieeexplore.ieee.org/document/8397411
- 157.Zhuang F, Qi Z, Duan K, Xi D, Zhu Y, Zhu H, Xiong H, He Q. A comprehensive survey on transfer learning. Proc IEEE 2020;109:43-76. 10.1109/JPROC.2020.3004555 [DOI] [Google Scholar]
- 158.Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng 2010;22:1345-59. 10.1109/TKDE.2009.191 [DOI] [Google Scholar]
- 159.Azizi S, Mousavi P, Yan P, Tahmasebi A, Kwak JT, Xu S, Turkbey B, Choyke P, Pinto P, Wood B, Abolmaesumi P. Transfer learning from RF to B-mode temporal enhanced ultrasound features for prostate cancer detection. Int J Comput Assist Radiol Surg 2017;12:1111-21. 10.1007/s11548-017-1573-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang B, Pu F, Wu Q, Zhang Z, Shao Z. Presacral tarlov cyst as an unusual cause of abdominal pain: new case and literature review. World Neurosurg 2018;110:79-84. 10.1016/j.wneu.2017.10.135 [DOI] [PubMed] [Google Scholar]
- 161.Järnum H, Steffensen EG, Knutsson L, Fründ ET, Simonsen CW, Lundbye-Christensen S, Shankaranarayanan A, Alsop DC, Jensen FT, Larsson EM. Perfusion MRI of brain tumours: a comparative study of pseudo-continuous arterial spin labelling and dynamic susceptibility contrast imaging. Neuroradiology 2010;52:307-17. 10.1007/s00234-009-0616-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bley TA, Wieben O, François CJ, Brittain JH, Reeder SB. Fat and water magnetic resonance imaging. J Magn Reson Imaging 2010;31:4-18. 10.1002/jmri.21895 [DOI] [PubMed] [Google Scholar]
- 163.Majumdar S. Magnetic resonance imaging and spectroscopy of the intervertebral disc. NMR Biomed 2006;19:894-903. 10.1002/nbm.1106 [DOI] [PubMed] [Google Scholar]
- 164.Wang YX, Griffith JF. Effect of menopause on lumbar disk degeneration: potential etiology. Radiology 2010;257:318-20. 10.1148/radiol.10100775 [DOI] [PubMed] [Google Scholar]
- 165.Wang H, Yeung DY. Towards Bayesian deep learning: a framework and some existing methods. IEEE Trans Knowl Data Eng 2016;28:3395-408. 10.1109/TKDE.2016.2606428 [DOI] [Google Scholar]
- 166.Gregor K, LeCun Y. Learning fast approximations of sparse coding. Proceedings of the 27th International Conference on Machine Learning; 2010:399-406. [Google Scholar]