Abstract
Background:
Patients with breast cancer frequently report cognitive impairment both during and after completion of therapy. Evidence suggests that cancer-related cognitive impairments are related to widespread neural network dysfunction. The default mode network (DMN) is a large conserved network that plays a critical role in integrating the functions of various neural systems. Disruption of the network may play a key role in the development of cognitive impairment.
Methods:
We compared neuroimaging and neurocognitive data from 43 newly diagnosed primary breast cancer patients (mean age = 48, standard deviation [SD] = 8.9 years) and 50 frequency-matched healthy female controls (mean age = 50, SD = 10 years) before treatment and 1 year after treatment completion. Functional and effective connectivity measures of the DMN were obtained using graph theory and Bayesian network analysis methods, respectively.
Results:
Compared with healthy females, the breast cancer group displayed higher global efficiency and path length post-treatment (p < 0.03, corrected). Breast cancer survivors showed significantly lower performance on measures of verbal memory, attention, and verbal fluency (p < 0.05) at both time points. Within the DMN, local brain network organization, as measured by edge-betweenness centralities, was significantly altered in the breast cancer group compared with controls at both time points (p < 0.0001, corrected), with several connections showing a significant group-by-time effect (p < 0.003, corrected). Effective connectivity demonstrated significantly altered patterns of neuronal coupling in patients with breast cancer (p < 0.05). Significant correlations were seen between hormone blockade therapy, radiation therapy, chemotherapy cycles, memory, and verbal fluency test and edge-betweenness centralities.
Discussion:
This pattern of altered network organization in the default mode is believed to result in reduced network efficiency and disrupted communication. Subregions of the DMN, the orbital prefrontal cortex and posterior memory network, appear to be at the center of this disruption and this could inform future interventions.
Impact statement
This prospective study is the first to investigate how post-treatment changes in functional and effective connectivity in the regions of default mode network are related to cancer therapy and measures of memory and verbal learning in breast cancer patients. We demonstrate that the interactions between treatment, brain connectivity, and neurocognitive outcomes coalesce around a subgroup of brain structures in the orbital frontal and parietal lobe. This would suggest that interventions that target these regions may improve neurocognitive outcomes in breast cancer survivors.
Keywords: breast cancer, cognitive impairment, connectome, drug-related side effects and adverse reactions, survivorship
Introduction
Cancer-related cognitive impairment (CRCI) is a common adverse effect of cancer pathogenesis and its treatments. We have previously demonstrated that, even a year after completing chemotherapy treatment, 55% of patients with breast cancer have cognitive impairment (Henneghan et al., 2020b; Kesler et al., 2017b). Others have shown cognitive deficit in patients who completed chemotherapy decades earlier (Koppelmans et al., 2012), suggesting that CRCI is a persistent, long-term symptom. CRCI has been associated both acutely and chronically with widespread brain injury, including all cerebral lobes as well as subcortical areas (Conroy et al., 2013; Deprez et al., 2014; Lepage et al., 2014; Menning et al., 2017; Mo et al., 2017; Nudelman et al., 2014; Stouten-Kemperman et al., 2015), suggesting the involvement of large-scale, distributed networks.
The default mode network (DMN) is one of the most commonly observed functional brain networks and includes the precuneus, posterior cingulate, medial frontal, middle temporal, and lateral parietal regions, as well as the hippocampus (Damoiseaux et al., 2006). DMN is believed to support processes such as implicit learning, autobiographical memory, prospection, monitoring the external environment, creativity, and self-reflection (Abraham, 2013; Agnati et al., 2013; Qin and Northoff, 2011; Raichle, 2011; Takeuchi et al., 2012).
DMN has been shown to represent the brain's structural core, playing a critical role in integrating the functions of various neural systems (Hagmann et al., 2008). It requires a large percentage of cerebral metabolic resources, and, as a result, may be more vulnerable to the effects of aging, injury, and disease (Tomasi et al., 2013; Zhou et al., 2012). Accordingly, altered DMN connectivity is a consistent biomarker across neurologic conditions associated with cognitive dysfunction and decline (Santhanam et al., 2019; van Oort et al., 2020; Zhou and Seeley, 2014).
The DMN may be preferentially vulnerable to chemotherapy treatment. Specifically, Dumas and colleagues (2013) demonstrated that while connectivity of prefrontal-executive networks improved over time after chemotherapy, DMN impairment remained unchanged. Miao and colleagues (2016) observed that the DMN tends to become disconnected to other functional networks following breast cancer chemotherapy. Cheng and colleagues (2017) demonstrated abnormal connectivity between multiple brain regions within the DMN among chemotherapy-treated patients with breast cancer.
We previously demonstrated that DMN connectivity can be used to automatically distinguish chemotherapy-treated breast cancer survivors from chemotherapy-naive survivors and noncancer female controls (Kesler et al., 2013). We also conducted a meta-analysis of task-based functional magnetic resonance imaging (fMRI) studies and demonstrated that DMN regions were among the most commonly affected in breast cancer patients and survivors (Kesler, 2014). Certain DMN regions show subtle dysconnectivity before onset of breast cancer treatment (Kesler et al., 2017a), which tends to predict which patients are at highest risk for postchemotherapy cognitive impairment (Kesler et al., 2017b).
Resting-state fMRI (rsfMRI) is a noninvasive neuroimaging method for measuring the connectivity of functional brain networks. Existing studies, such as those mentioned above, have focused on functional connectivity, which is based on correlations among regional rsfMRI signals and therefore undirected. Functional connectivity measures are computationally simple and yield important information regarding brain network organization. However, brain networks are actually directed, in which there are causal interactions among brain regions known as effective connectivity (Friston, 2011). Directed networks provide novel mechanistic insights by revealing the trajectories of neural systems and information exchange (Stephan et al., 2017). Evaluating effective connectivity is much more complex, traditionally requiring a known or putative anatomical model.
However, Bayesian learning algorithms can infer directed networks in a data-driven manner (Rajapakse and Zhou, 2007; Zheng and Rajapakse, 2006). Previous studies have applied Bayesian network analysis to rsfMRI data to examine DMN effective connectivity in conditions related to CRCI, including age-related mild cognitive impairment (Li et al., 2013; Wu et al., 2016). No studies to date have evaluated effective connectivity associated with CRCI. We therefore aimed to measure changes in both functional and effective connectivity of the DMN in patients with breast cancer to provide novel insights regarding the mechanisms of post-treatment CRCI.
Materials and Methods
Participants
As part of our ongoing, prospective longitudinal study of breast cancer and cognition, we enrolled 43 newly diagnosed patients with primary breast cancer (stage I–III), age 34–65 years, and 50 frequency-matched healthy control females (Table 1). Exclusion criteria included death before long-term follow-up, stage IV disease or distant metastasis, secondary cancers or relapse requiring cranial radiation or central nervous system-directed chemotherapy, unrelated central nervous system injury/disease, or nonproficient in English. Patients were assessed before initiation of any treatment (including surgery with general anesthesia), 1 month after completing chemotherapy (not used in the current study), and again 1 year later. Controls were assessed at yoked intervals. For this study, we focused on the 1-year follow-up assessment to examine the neural mechanisms of chronic CRCI. The Stanford University Institutional Review Board approved this study and all procedures performed were in accordance with the ethical standards of the Declaration of Helsinki. Written informed consent was obtained from all participants included in the study.
Table 1.
Participant Characteristics at Baseline
| Breast cancer, N = 43, mean (SD) | Controls, N = 50, mean (SD) | T-score | p | |
|---|---|---|---|---|
| Age, years | 49 (8.9) | 50 (10) | 0.114 | 0.909 |
| Education, years | 16 (3) | 17 (2.2) | 1.58 | 0.119 |
| Chemotherapy cycles | 6.89 (3.7) |
| n (%) | ||||
|---|---|---|---|---|
| Anthracycline chemotherapy |
28 (65) |
|
|
|
| Radiation therapy |
30 (70) |
|
|
|
| Hormone blockade |
27 (63) |
|
|
|
| Selective estrogen receptor modulator |
25 (58) |
|
|
|
| Other hormone suppressors |
2 (5) |
|
|
|
| Aromatase inhibitors |
22 (51) |
|
|
|
| Combination therapy |
5 (12) |
|
|
|
| Stage at diagnosis | ||||
| Stage I |
6 (14) |
|
|
|
| Stage II |
29 (67) |
|
|
|
| Stage III |
8 (19) |
|
|
|
| Stage IV |
0 (0) |
|
|
|
| Surgery (including biopsy) | 43 (100) | |||
Data are shown as mean (SD) unless otherwise noted.
SD, standard deviation.
Neuroimaging acquisitions
RsMRI data were obtained while participants rested with eyes closed using a T2*-weighted (Glover and Lai, 1998) gradient echo spiral pulse sequence: repetition time (TR) = 2000 ms, echo time (TE) = 30 ms, flip angle = 80° and 1 interleave, field of view (FOV) = 22 cm, matrix = 64 × 64, in-plane resolution = 3.4375, and number of volumes = 216 at 3 Tesla. T1-weighted MRI: TR = 8.5, TE = minimum, flip = 15°, TI = 400 ms, BW = ±31.25 kHz, FOV = 22 cm, phase FOV: 0.75, slice thickness: 1.5 mm, 124 slices, 256 × 256 @ 1 NEX, and scan time: 4:33. Diffusion tensor imaging (DTI) was added later in the study protocol, and thus, data were collected for a subset of participants at Time 2 (16 breast cancer and 22 controls). DTI was acquired from 40 contiguous axial slices (thickness = 3 mm) using a single-shot echo planar imaging (EPI) sequence (TE = min, TR = 5000), FOV = 240 mm, and matrix size = 128 × 128 mm. Data included 2 images without diffusion weighting (b = 0 s/mm2) and diffusion-weighted images along 23 orthogonal directions (b = 850 s/mm2). The EPI sequence was repeated four times.
Identifying the DMN
RsfMRI data were preprocessed in CONN Toolbox v17 (Whitfield-Gabrieli and Nieto-Castanon, 2012) implemented in MATLAB v2019b (MathWorks, Inc., Natick, MA). Briefly, this involved realignment, coregistration with the segmented anatomic volume, spatial normalization, artifact detection, and smoothing (full-width-at-half-maximum = 8 mm) followed by bandpass filtering (0.008–0.09 Hz). The CompCor correction method was used to reduce physiological and other non-neuronal noise artifacts (Behzadi et al., 2007). Motion parameters from realignment were included as regressors and images identified as motion or signal outliers were excluded. Exclusion criteria included motion cutoffs of 1 mm translation, 0.05 radian rotation, and 3 SD (standard deviation) for global signal. We planned to discard data that had more than 10% volumes exceeding these thresholds in the time series. However, no participants in our study exceeded this criterion.
Temporal correlations between all possible pairs of 90 regions (Tzourio-Mazoyer et al., 2002) were computed and normalized resulting in a 90 × 90 functional connectivity matrix (network) for each participant and each time point (baseline = Time 1, 1-year follow-up = Time 2). We identified subnetworks by decomposing the functional network into nonoverlapping groups of regions (modules) that have maximal within-group connections and minimal between-group connections (Sporns and Betzel, 2016). We selected the DMN from among the modules as the one with the closest match to the known DMN configuration (Grayson and Fair, 2017; Meunier et al., 2009; Power et al., 2011). Being a data-driven process, DMN module configurations differed for each group at each time point. To facilitate comparisons, we selected the 11 regions that were common across groups and time points as the final DMN module (Supplementary Table S1).
Functional connectivity
We modeled the DMN for each participant as a system of nodes and edges representing regions and their normalized temporal correlations, respectively. Graphs were constructed with N = 11 nodes, network degree of E = number of edges, and a network density of D = E/[(N × (N − 1))/2] representing the fraction of present connections to all possible connections.
We measured the mean clustering coefficient, path length, transitivity, local and global efficiency of the DMN using Brain Connectivity Toolbox (Rubinov and Sporns, 2010) implemented in MATLAB v2019b (MathWorks, Inc.). These are commonly measured properties that provide insight regarding a network's specialization (clustering, transitivity, and local efficiency) and integration (path length and global efficiency) (Bullmore and Sporns, 2009; Schank and Wagner, 2005). We have previously shown that these properties are altered by chemotherapy in both humans and rodents (Bruno et al., 2012; Chiu et al., 2018; Kesler et al., 2017a, 2018b; Tang et al., 2019).
These properties were measured globally for the entire DMN. We also evaluated local DMN connectivity by measuring edge centrality (i.e., centrality), which indicates the importance of each edge for the network's integration (Kaiser, 2011). This resulted in an 11 × 11 matrix of edge centrality values for each participant.
As in prior studies by our group and others (Bassett et al., 2006, 2012; Kesler et al., 2018a), connectivity properties were measured across connection densities (thresholds) from minimum connection density to the maximum biologically plausible density of 50% (Kaiser and Hilgetag, 2006) at intervals of 1% connection density. The area under the curve across densities was computed and compared between groups at each time point and within groups across time using nonparametric permutation testing.
Specifically, we first calculated mean differences between the groups for each property, using the slope (Time 2 − Time 1) for longitudinal analyses. A permutation distribution of difference scores for each property was created by resampling participant data into two random groups of equal size to the original groups 5000 times. We then calculated the network properties for each random group. The differences between the randomized groups comprised the permutation distributions. The significance of the permutation test (p) was determined by calculating the mean number of instances that the permutation difference scores were greater than the original difference score, with false discovery rate (FDR) correction for multiple comparisons (Benjamini and Yekutieli, 2001). Permutation testing was conducted in MATLAB v2019b.
Effective connectivity
We performed a Bayesian network analysis, which learns the conditional dependencies between brain regions. This method characterizes the effective connectivity of the network without any prior assumptions (Rajapakse and Zhou, 2007). We implemented a hill-climbing approach (Gámez et al., 2011) in which the model that optimized the Bayesian information criterion score was considered the best fit network. Briefly, the conditional independence/dependence relationships among random variables were encoded with a directed acyclic graph: if a random node A directly depends on another random node B, even given all the other random nodes, then the dependence is encoded as an edge between nodes A and B (Li et al., 2008). The edges can be considered a set of linear regression models, and therefore, each edge is associated with a beta-coefficient. An edge was defined as having a significant coefficient (p < 0.05) after FDR correction.
Effective connectivity was compared graphically between groups at each time point and within groups across time points to indicate false-positive, false-negative, and true-positive edges. We used permutation testing as described above to determine if the number of false-negative/false-positive edges was significantly greater than that of random groups. We also compared the coefficients for true-positive edges (those that were common between groups at each time point or within groups across time points) using permutation testing. In this case, we calculated the original linear regression model for each edge for each of the 5000 random groups (Wu et al., 2011). The differences between the randomized groups in edge coefficients comprised the permutation distributions. Bayesian network analyses, including graphical comparisons and permutation testing, were conducted in the R Statistical Package v3.6.2 (R Foundation, Vienna, Austria), including the “bnlearn” library (Scutari, 2009).
Gray matter volumes
Gray matter maps were obtained using voxel-based morphometry (VBM) (Kurth et al., 2015). Images were first manually reoriented to the anterior and posterior commissures, then realigned, segmented into tissue compartments, spatially normalized to a template using diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL), modulated, and smoothed using the VBM8 Toolbox (Kurth et al., 2015). We then measured gray matter volume from each of the 11 DMN regions in mm3 using the “fslstats” function of the FMRIB Software Library (FSL) v6.0 (Jenkinson et al., 2012). We compared DMN gray matter volumes between groups at each time point and within groups across time using independent or paired t-tests, respectively, with FDR correction, in the R Statistical Package.
White matter integrity
DTI data were processed as in our prior studies (Kesler et al., 2015, 2016, 2018b). Briefly, eddy current correction was performed in FSL. A binary brain mask of the B0 image was created using FSL Brain Extraction Tool and applied to corrected DTI volumes to constrain tensor fitting and tractography to brain tissue. Tensor reconstruction and deterministic tractography were performed in Diffusion Toolkit v0.6.4.1 and TrackVis v0.6.1 (Feigl et al., 2014; Wang et al., 2007), respectively. Tractography used the fiber assignment by continuous tracking algorithm (Mori and van Zijl, 2002). We used a curvature threshold of 60° and streamlines were smoothed using a spline filter.
Virtual fibers were mapped between each pair of 11 DMN regions. Mean fractional anisotropy (FA), apparent diffusion coefficient (ADC), and length of virtual fibers (edges) were measured using the UCLA Multimodal Connectivity Package (Brown, 2014) resulting in an 11 × 11 matrix for each metric for each participant. These were compared at Time 2 (the only available time point) using nonparametric permutation testing as described above for functional connectivity, with FDR correction.
Cognitive outcomes and imaging correlates
Verbal learning, interference, and retention were measured using the Rey Auditory Verbal Learning Test (RAVLT) Immediate, Interference, and Delayed Recall tests, respectively (Schmidt, 2012). Attention and processing speed were measured using Trail 1 of the Comprehensive Trail Making Test (CTMT), and attention, processing speed, and cognitive flexibility using Trail 5 of the CTMT (Moses, 2004). Verbal fluency was assessed using the Controlled Oral Word Association (COWA) (Ruff et al., 1996). In addition, we measured depression, anxiety, and fatigue using the total score from the clinical assessment of depression (CAD) (Aghakhani and Chan, 2007).
Test scores were compared between groups at each time point using independent t-tests and across time using linear mixed modeling. Two-tailed Pearson or spearman correlations, as appropriate, were conducted to explore relationships between DMN measures and test scores that were significantly different between groups. We also examined relationships between DMN, cognitive performance, and clinical variables in the breast cancer group. Given that longitudinal analyses were not significant, correlations were conducted at Time 2 within each group to focus on associations of chronic CRCI and to reduce the number of comparisons.
For each comparison for a given hypothesis (i.e., edge centrality in survivor's vs. controls or global efficiency in survivor's vs. controls), the FDR method was used to correct for the multiple comparison problem. However, analyses of cognitive tests were not corrected for multiple comparisons given the known limited sensitivity to CRCI of these tests compared with neuroimaging metrics (Horowitz et al., 2018; Kesler et al., 2020). Statistical tests were performed in the R Statistical Package including the “lmerTest” library for linear mixed modeling (Kuznetsova et al., 2017).
Results
Functional connectivity
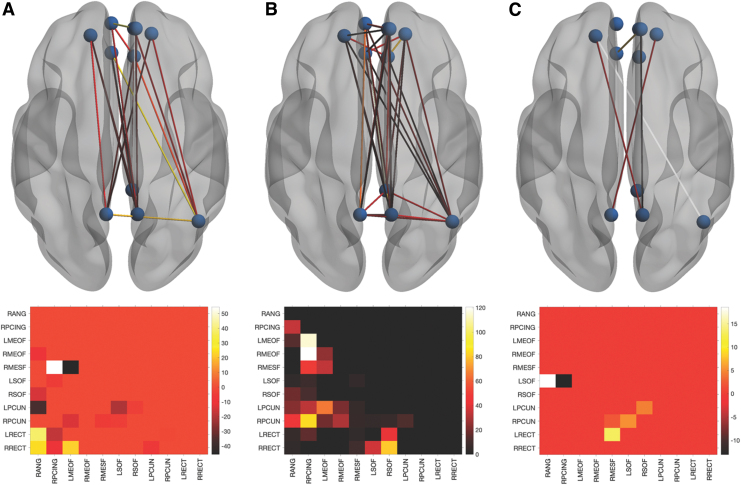
At Time 1, there were no between-group differences in global DMN properties (Table 2). However, there were multiple local differences in which the breast cancer group showed significantly (p < 0.013, FDR corrected) higher or lower edge centrality compared with controls (Fig. 1). At Time 2, the breast cancer group showed significantly higher global efficiency and path length compared with controls (p < 0.03, FDR corrected, Table 2). At Time 2, centrality was significantly lower for multiple edges in the breast cancer group compared with controls (p < 0.0001, FDR corrected), with several edges showing a significant group-by-time effect (p < 0.003, FDR corrected, Fig. 1).
Table 2.
Global Default Mode Network Functional Connectivity Properties
| Breast cancer |
Control |
Permutation test |
|||||
|---|---|---|---|---|---|---|---|
| Time 1, N = 43 | Time 2, N = 31 | Time 1, N = 50 | Time 2, N = 43 | Mean slope differencea | 95% CI | p Value (FDR corrected) | |
| Mean clustering coefficient | 0.285 (0.034) | 0.275 (0.032) | 0.282 (0.029) | 0.278 (0.033) | 0.0051 | −0.0116 to 0.0119 | 0.173 |
| Transitivity | 0.329 (0.033) | 0.312 (0.045) | 0.319 (0.039) | 0.325 (0.045) | 0.0137 | −0.0144 to 0.0145 | 0.078 |
| Global efficiencyb | 0.249 (0.018) | 0.258 (0.018) | 0.252 (0.016) | 0.247 (0.018) | −0.0080 | −0.0065 to 0.0066 | 0.496 |
| Mean local efficiency | 0.313 (0.032) | 0.304 (0.029) | 0.312 (0.028) | 0.307 (0.032) | 0.0046 | −0.0112 to 0.0114 | 0.173 |
| Path lengthb | 0.808 (0.073) | 0.853 (0.080) | 0.824 (0.076) | 0.806 (0.081) | −0.0347 | −0.0289 to 0.0296 | 0.496 |
Data are shown as mean (SD).
Control minus breast cancer.
Significantly higher in breast cancer compared with controls at Time 2 (p < 0.03, FDR corrected).
CI, confidence interval; FDR, false discovery rate.
FIG. 1.
Local DMN functional connectivity. Differences in edge centrality represented in the upper row by ball and stick models with all 11 nodes as spheres and significant edges as lines. The bottom row presents the corresponding lower triangle heatmap representing the mean difference (control minus breast cancer). At Time 1 (A), the breast cancer group showed several edges with significantly higher (negative value, darker color) or lower (positive value, lighter color) centrality compared with controls (p < 0.013, FDR corrected). At Time 2 (B), there were even more group differences characterized by exclusively lower edge centrality in the breast cancer group (p < 0.0001, FDR corrected). There was a significant group-by-time effect (C) for several edges (p < 0.003, FDR corrected). DMN, default mode network; FDR, false discovery rate; L/RMEOF, left/right medial orbital frontal; L/RPCUN, left/right precuneus; L/RRECT, left/right rectus gyrus; L/RSOF, left/right superior orbital frontal; RANG, right angular gyrus; RMESF, right medial superior frontal; RPCING, right posterior cingulate. Color images are available online.
Effective connectivity
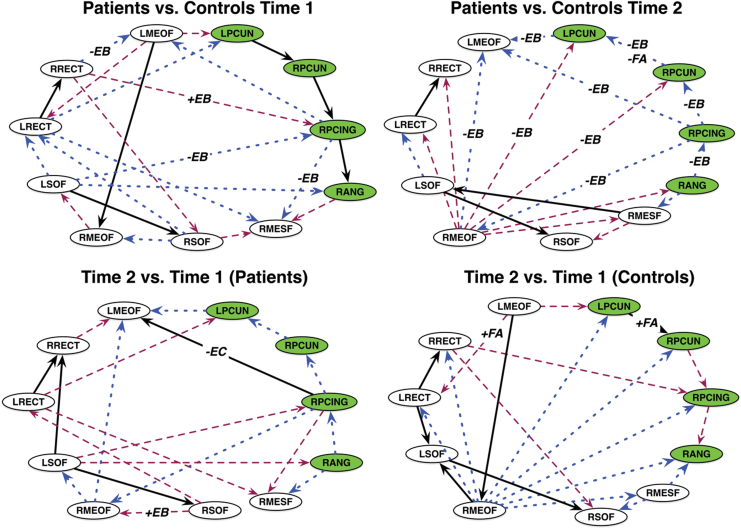
Patterns of effective connectivity among DMN regions for each group are shown in Supplementary Figure S1. In this analysis, false positive is a connection between two regions, which, either connects where there is no true connection, or connects two regions where there is a true connection but takes a pathway that is not true to the underlying anatomy. A false negative is the failure to generate a connection between regions for which there is a true connection (Drakesmith et al., 2015).
As shown in Figure 2, at Time 1, the breast cancer group, compared with controls, showed 6 true-positive-, 10 false-positive-, and 7 false-negative-directed edges. At Time 2, there were nine false positives (two common with Time 1), seven false negatives (one common with Time 1 but reversed direction), and three true positives (two common with Time 1). The number of false-positive (p = 0.05)- and false-negative (p = 0.02)-directed edges in the breast cancer group were significantly greater than that of random groups at T2, but not at T1 (p > 0.10). The true-positive edges did not differ significantly in terms of edge coefficients at Time 1 (p > 0.235, FDR corrected) or at Time 2 (p > 0.53, FDR corrected).
FIG. 2.
Graphical comparisons of DMN effective connectivity. Between-group (top row) and within-group (bottom row) comparisons based on graphical comparison. Black lines denote true positives, blue are false positives, and maroon are false negatives for patients compared with controls, or Time 2 compared with Time 1. Edge labels indicate overlap with functional connectivity and/or white matter integrity comparisons: +/−EB = higher/lower edge centrality +/−FA = higher/lower fractional anisotropy. The +/−EC edge label indicates a significant difference in edge coefficient (p = 0.03, FDR corrected). Nodes in the PMN are colored green. PMN, posterior memory network. Color images are available online.
For the breast cancer group, there were eight false-positive, eight false-negative, and four true-positive edges at Time 2 compared with Time 1 (Fig. 2). The true-positive edge from the right posterior cingulate to left medial orbital frontal gyrus showed decreased connectivity strength over time as indicated by the change in edge coefficient (p = 0.03, FDR corrected, Fig. 2). In controls, there were nine false-positive and six false-negative and six true-positive edges at Time 2 compared with Time 1 (Fig. 2). None of the true-positive edges changed significantly across time in terms of connectivity strength for controls.
Gray matter volumes
There were no significant cross-sectional or longitudinal differences in gray matter volume for any DMN nodes (p > 0.28, FDR corrected, Supplementary Table S2).
White matter integrity
The mean FA of several DMN edges was significantly lower in patients with breast cancer compared with controls (p < 0.008, FDR corrected, Supplementary Fig. S2) at Time 2. ADC and length were not significant (p > 0.126, FDR corrected).
Cognitive outcomes and imaging correlates
At Time 1, Immediate Recall, Trail 1, and COWA scores were significantly lower in the breast cancer group compared with controls (p < 0.02, Table 3). CAD score was significantly higher in patients with breast cancer (p < 0.001). At Time 2, the breast cancer group demonstrated significantly lower Interference Recall, Trail 1, and COWA performance (p < 0.048), and the CAD score was significantly higher (p = 0.011). Both groups tended to improve in cognitive performance over time, but there were no group-by-time effects (Table 3). CAD score also remained unchanged over time (Table 3).
Table 3.
Cognitive-Behavioral Assessment Scores
| Breast cancer |
Controls |
LMM (group-by-time) |
||||
|---|---|---|---|---|---|---|
| Time 1, N = 43 | Time 2, N = 31 | Time 1, N = 50 | Time 2, N = 43 | F | p (uncorrected) | |
| Immediate recalla | 53 (10) | 58 (10) | 58 (10) | 59 (11) | 1.35 | 0.249 |
| Interference recallb | 52 (9) | 53 (14) | 56 (11) | 60 (17) | 0.949 | 0.333 |
| Delayed recall | 52 (11) | 53 (11) | 56 (9) | 55 (9) | 0.078 | 0.930 |
| Trail 1a,b | 52 (8) | 53 (9) | 56 (10) | 58 (10) | 0.092 | 0.762 |
| Trail 5 | 51 (9) | 54 (9) | 54 (10) | 57 (9) | 0.032 | 0.860 |
| COWAa,b | 44 (12) | 45 (10) | 50 (13) | 52 (13) | 0.419 | 0.519 |
| CAD totala,b | 52 (10) | 51 (9) | 43 (9) | 44 (11) | 1.10 | 0.297 |
Data are shown as mean (SD).
Significantly different between groups at Time 1 (p < 0.02, uncorrected).
Significantly different between groups at Time 2 (p < 0.048, uncorrected).
CAD, clinical assessment of depression; COWA, Controlled Oral Word Association; LMM, linear mixed model.
As shown in Table 4, in general, cognitive performance showed both negative and positive correlations with edge centrality and FA in both groups. In the breast cancer group, hormone blockade therapy (yes/no) was negatively correlated with centrality of three edges. Radiation therapy (yes/no) was positively correlated with centrality for one edge and negatively correlated with two others, as well as negatively correlated with the FA of one edge. The number of chemotherapy cycles was positively correlated with centrality for four different edges. CAD was not significantly correlated with DMN measures or cognitive testing scores in either group.
Table 4.
Time 2 Correlations Between Default Mode Network Connectivity, Cognitive Function, and Clinical Variables
| Behavioral/Clinical variable | DMN variable | Connectivity variable | r | p (uncorrected) |
|---|---|---|---|---|
| Breast cancer | ||||
| Interference recall | LSOF-RANG | Centrality | −0.361 | 0.029 |
| LMEOF-LRECT | FA | −0.545 | 0.015 | |
| COWA | RSOF-LRECT | FA | −0.785 | 0.0002 |
| Number of chemotherapy cycles | RMESF-LMEOF | Centrality | 0.667 | <0.0001 |
| RMESF-LRECT | Centrality | 0.361 | 0.030 | |
| RMESF-LSOF | Centrality | 0.333 | 0.042 | |
| RMESF-RRECT | Centrality | 0.357 | 0.031 | |
| Radiation therapy | RSOF-RANG | Centrality | 0.376 | 0.024 |
| RPCING-RPCUN | Centrality | −0.393 | 0.019 | |
| RMESF-LRECT | Centrality | −0.325 | 0.046 | |
| LSOF-LMEOF | FA | −0.533 | 0.017 | |
| Hormone blockade therapy | RMESF-RPCING | Centrality | −0.348 | 0.035 |
| LPCUN-RPCUN | Centrality | −0.364 | 0.028 | |
| RANG-LRECT | Centrality | −0.319 | 0.049 | |
| Controls | ||||
| Interference recall | LRECT-RMESF | Centrality | −0.255 | 0.049 |
| LSOF-RPCUN | Centrality | 0.276 | 0.036 | |
| Trail 1 | LSOF-RPCUN | Centrality | 0.285 | 0.032 |
| COWA | LSOF-LMEOF | FA | −0.435 | 0.023 |
Only significant (p < 0.05) correlations are shown.
DMN, default mode network; FA, fractional anisotropy; L/RMEOF, left/right medial orbital frontal; L/RPCUN, left/right precuneus; L/RRECT, left/right rectus gyrus; L/RSOF, left/right superior orbital frontal; RANG, right angular gyrus; RMESF, right medial superior frontal; RPCING, right posterior cingulate.
Discussion
Global efficiency and path length, indicating the overall functional connectivity of the DMN, were significantly higher post-treatment in the breast cancer group compared with controls. These are measures of network integration and suggest that regions have higher direct information exchange (Latora and Marchiori, 2001). However, greater network integration comes at an increased wiring and metabolic cost (Achard and Bullmore, 2007). The brain network shows a small-world organization characterized by high local connectivity and economical long-range connectivity to balance the opposing demands of specialization and integration (Bassett and Bullmore, 2017). Accordingly, we have previously demonstrated that too much, as well as too little, connectivity is associated with CRCI (Kesler et al., 2016). Above normal integration has also been noted previously in other cognitive disorders, including Alzheimer's disease (Kesler et al., 2018a; Wang et al., 2013).
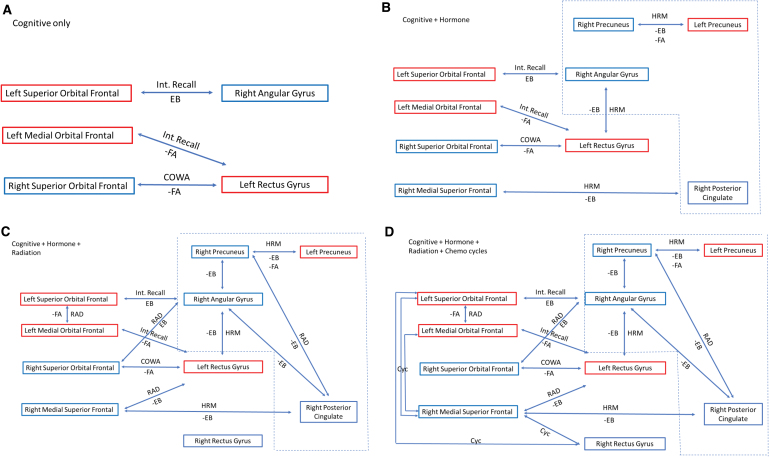
Locally, DMN functional connectivity was altered in patients with breast cancer at baseline, characterized by some DMN edges showing higher connectivity, while others showed lower connectivity compared with controls. This is consistent with previous studies (Kaiser et al., 2019; Patel et al., 2015) and suggests potential effects of tumor pathogenesis, inflammation, and/or patient-related factors on CRCI. Post-treatment, alterations in local connectivity among patients with breast cancer were more widespread throughout the DMN. A summary of correlations between functional connectivity and cognitive performance as it relates to therapy is presented in Figure 3. Local connectivity was measured using edge-betweenness centrality, which provides insights regarding the importance, or centrality, of a particular edge to the network (Girvan and Newman, 2002; Newman and Girvan, 2004). Even though the centrality of certain edges actually increased over time in patients with breast cancer, all remained significantly lower compared with controls at Time 2. This suggests that several DMN connections may become damaged after breast cancer treatments and show insufficient recovery over time such that they play a diminished role in information exchange.
FIG. 3.
Summary of correlations between cognitive testing, treatment, structural, functional, and effective connectivity at Time 2. Correlations for the breast cancer group for FA and EB, therapy, and cognitive deficits are linked to form a network that may inform future interventions. Negative correlations are denoted by the minus (−) sign. Bilateral superior and left medial orbital frontal are part of the OPFC. Red box denotes left hemisphere and blue denotes right hemisphere structures. Dash line box demotes structures associated with the PMN. (A) Denotes correlations between brain regions associated with changes in cognitive testing. (B) Additional connections associated with HRM form circuits with structures linked to the OPFC and PMN. (C) Associations with RAD further consolidate the interactions between treatment, connectivity, and the OPFC and PMN. (D) The number of chemotherapy cycles (Cyc) has a positive association with the OPFC and may represent an unidentified association between anthracyclines and strengthening of maladaptive networks. COWA, Controlled Oral Word Association; HRM, hormone blockade therapy; OPFC, orbital prefrontal cortex; RAD, radiation dose. Color images are available online.
Our results indicated significant differences in effective connectivity for patients with breast cancer post-treatment, but not pretreatment. Post-treatment differences included several false-positive- and false-negative-directed edges compared with controls, indicating significantly altered patterns of efferent and afferent projections. Lower edge-betweenness centralities overlapped with 29% of the false-negative- and 78% of the false-positive-directed edges (Fig. 2), suggesting that the direction of information exchange may alter or be altered by edge importance.
Interestingly, many of the effective connectivity differences involved a purported subnetwork called the posterior memory network (PMN), which sits adjacent to and may be separate from the DMN (Gilmore et al., 2015). This incidental finding is important for a number of reasons. At Time 1, the PMN was seen in both controls and patients. However, at Time 2, this network in patients shifted to a false positive (i.e., connections were missing or had different directions than those in the control network). The PMN has been associated with memory encoding and retrieval, which patients showed difficulties at both time points.
Both groups demonstrated longitudinal changes in effective DMN (and PMN) connectivity. The stability of the PMN over time is unknown, although there is evidence of longitudinal change in DMN functional connectivity (Staffaroni et al., 2018). Since the stability of PMN is unknown, future studies should be conducted to determine if it is a good target for interventions in this cohort. Brain connectivity in general is very dynamic and appears to be modulated by the DMN (Betzel et al., 2016; Fukushima et al., 2018).
Greater number of chemotherapy cycles was correlated with higher betweenness centralities, especially for edges involving the right medial superior frontal (RMESF) cortex. We previously noted higher functional connectivity in this region among long-term breast cancer survivors who had received anthracycline chemotherapies compared with chemotherapy-naive survivors (Kesler and Blayney, 2016), and most patients in the present study received anthracycline chemotherapies. Increased connectivity of RMESF has also been associated with increased cerebrospinal fluid amyloid-beta levels (Canuet et al., 2015). Increased RMESF connectivity is also present in cognitively normal adults with an apolipoprotein e4 genotype, which is associated with increased amyloid-beta aggregation (Yang et al., 2014). Interestingly, anthracyclines are used as a treatment for amyloidosis and are believed to work by binding to and dissolving amyloid (Gianni et al., 1995). If RMESF tends to have a relatively higher amyloid concentration normally, anthracycline chemotherapies may be preferentially drawn to it. We have shown in vitro that even very small amounts of doxorubicin can cause significant, long-term neuron death in cortical tissue, and importantly, this damage was reduced by levetiracetam (Manchon et al., 2016).
Radiation therapy was also associated with altered local functional connectivity as well as lower white matter integrity. The relationships between radiation therapy and edge-betweenness centralities were both positive and negative. The clinical significance of the positive correlations between breast cancer treatments and edge centralities is difficult to interpret given that some edges significantly increased in centrality over time. Since edge centralities in the patient group post-treatment were significantly lower than controls, this might suggest that chemotherapy and/or radiation have a positive effect on certain aspects of local functional connectivity.
However, the only correlation between local connectivity and cognitive function was inverse; lower interference memory was associated with higher centrality for the edge between left superior orbital frontal and right angular gyri. The brain network is a very complex system, and thus, alterations in connectivity by treatments in certain regions may propagate changes in other areas that result in cognitive impairment. Previous studies have suggested that pathology spreads through the brain networks from targeted epicenters (Crossley et al., 2014; Zhou et al., 2012). We previously demonstrated that physical exercise can protect large-scale white matter organization from radiation damage, including damage resulting in higher path length (Sahnoune et al., 2018).
The impact of hormone modulation therapy on targeted brain regions demonstrated in our study could be driven by estrogen receptor (ER) blockade or aromatase-regulated estradiol 2 (E2) production. A previous study demonstrated that postmenopausal women taking tamoxifen showed lower metabolic rates of glucose metabolism in the orbital frontal and dorsolateral prefrontal cortices compared with woman taking estrogen (Eberling et al., 2004). Interestingly, the nucleus accumbens is ER-beta (ER-β) rich and protrudes into the posterior rectus gyrus, which lies adjacent to the medial orbital gyrus. This could indicate that ER activity in this region may play some role in connectivity changes since tamoxifen is known to bind with equal affinity to the ER-β receptor (Kuiper et al., 1997). Both E2 and tamoxifen have been reported to negatively affect glutamate binding to N-methyl-d-aspartate (NMDA) receptors (Cyr et al., 2001). Thus, the effect of estrogen modulation therapy on brain neural networks may be additionally or alternatively mediated through NMDA receptor activity.
Interestingly, all the interlinkages between connectivity, treatment, and neurocognitive outcomes center around the orbital prefrontal cortex (OPFC) and PMN. This would suggest that interventions that target the OPFC or PMN may be beneficial. Animal studies have shown that oxytocin receptors are in higher concentrations in the nucleus accumbens and OPFC (Insel and Shapiro, 1992; Jurek and Neumann, 2018). Studies in noncancer female participants have shown that oxytocin improves memory performance when tied to an emotional stimulus (Brambilla et al., 2016). The OPFC processes the physical, chemical, and emotional properties of sensory information and has been associated with learning and memory and evaluation of positive and negative enforcers (Kringelbach, 2005). This aligns with neuroimaging studies that demonstrated, when synced with an emotional cue, oxytocin significantly enhanced connectivity between the orbital frontal cortex, precuneus, and angular gyrus (Riem et al., 2012).
These results have been reproduced in multiple studies and, in females, suggest that brain network changes may be context dependent (Bethlehem et al., 2013). In addition, oxytocin has been shown to mediate the beneficial effects of exercise training on improved memory outcomes in breast cancer survivors who engage in a formal exercise program during chemotherapy (Alizadeh et al., 2018; Salerno et al., 2019). Finally, no study has investigated the use of intranasal delivered oxytocin during therapy, although studies in noncancer populations have shown mixed effects on long-term memory (Brambilla et al., 2016).
Limitations of this study include missing data for Time 2 and no DTI for Time 1. Our cognitive testing battery was very limited to reduce participant burden. It also relied on neuropsychological tests that are known to have limited sensitivity and specificity for CRCI (Horowitz et al., 2018). These factors likely increased our false-negative rate regarding cognitive effects and correlations. Previous studies have demonstrated evidence of microbleed and altered cerebral perfusion post-treatment in breast cancer survivors, which may impact MRI signal detection or neurovascular coupling (Koppelmans et al., 2015; Nudelman et al., 2014). However, a growing body of evidence has demonstrated the reliability and reproducibility of detecting DMN abnormalities in breast cancer survivors (Conroy et al., 2013; de Ruiter et al., 2011; Kesler et al., 2009, 2011). Measures of estrogen levels, and amyloid or NMDA activity were not acquired during this study, and so, our potential intervention targets require further study. Finally, there are many different options for conducting functional and effective connectivity analyses that may yield different findings.
However, this is a comprehensive, multimodal, longitudinal study demonstrating novel neural mechanisms underlying CRCI that point to potential intervention targets. This is the first investigation of effective connectivity associated with CRCI and our findings provided unique insights regarding the PMN, a specific subnetwork within the DMN that may be affected by breast cancer and its treatments. We have initiated much larger studies that will include cognitive neuroscience-based tasks to provide greater insight into component cognitive processes. We will also compare alternate methods for functional and effective connectivity measurement. We have implemented an innovative assay for measuring amyloid-beta and tau (Henneghan et al., 2020a), and we will collect data pertaining to estrogen levels in future studies.
Conclusion
In this prospective cohort study, we show differences between survivors and healthy community controls. We demonstrate associations between treatment and survivors' effective and functional brain connectivity, and neurocognitive outcomes. The novelty of this study includes the identification of subregions of the DMN whose disruption is associated with cognitive impairment in breast cancer patients. Our findings highlight that the orbital frontal cortex is a suitable target for an intervention study that may prevent or manage CRCI in female breast cancer patients.
Supplementary Material
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authors' Contributions
This study was developed by the principal investigator (S.R.K.). Data preparation was conducted by V.R., L.K., S.M., R.V., and S.R.K. Neuroimaging analysis was conducted by V.R. and S.R.K. Statistical analysis was conducted by V.R., L.K., and S.R.K. The article was prepared and edited by N.S.P., K.K., and S.R.K. All authors approved the article for submission. V.R., L.K., R.V., S.M., and S.R.K. had access to the raw data.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study was funded by the National Institutes of Health (R01CA172145 and R01CA226080 to Shelli R. Kesler, and T32CA225590 to Kevin Krull).
Supplementary Material
References
- Abraham A. 2013. The world according to me: personal relevance and the medial prefrontal cortex. Front Hum Neurosci 7:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard S, Bullmore E. 2007. Efficiency and cost of economical brain functional networks. PLoS Comput Biol 3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghakhani A, Chan EK. 2007. Test reviews: Bracken, B.A., & Howell, K. (2004). Clinical assessment of depression. J Psychoeduc Assess 25:416–422. [Google Scholar]
- Agnati LF, Guidolin D, Battistin L, et al. . 2013. The neurobiology of imagination: possible role of interaction-dominant dynamics and default mode network. Front Psychol 4:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AM, Heydari Z, Rahimi M, et al. . 2018. Oxytocin mediates the beneficial effects of the exercise training on breast cancer. Exp Physiol 103:222–235. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET. 2017. Small-world brain networks revisited. Neuroscientist 23:499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Meyer-Lindenberg A, Achard S, et al. . 2006. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc Natl Acad Sci U S A 103:19518–19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Nelson BG, Mueller BA, et al. . 2012. Altered resting state complexity in schizophrenia. Neuroimage 59:2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, et al. . 2007. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage 37:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188. [Google Scholar]
- Bethlehem RA. I., van Honk J, Auyeung B, et al. . 2013. Oxytocin, brain physiology, and functional connectivity: A review of intranasal oxytocin fMRI studies. Psychoneuroendocrinology 38:962–974. [DOI] [PubMed] [Google Scholar]
- Betzel RF, Fukushima M, He Y, et al. . 2016. Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage 127:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla M, Manenti R, de Girolamo G, et al. . 2016. Effects of intranasal oxytocin on long-term memory in healthy humans: a systematic review. Drug Dev Res 77:479–488. [DOI] [PubMed] [Google Scholar]
- Brown J. 2014. UCLA Multimodal Connectivity Package (UMCP). https://www.ccn.ucla.edu/wiki/index.php/UCLA_Multimodal_Connectivity_Package. Last accessed March 6, 2020.
- Bruno J, Hosseini SM, Kesler S. 2012. Altered resting state functional brain network topology in chemotherapy-treated breast cancer survivors. Neurobiol Dis 48:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10:186–198. [DOI] [PubMed] [Google Scholar]
- Canuet L, Pusil S, Lopez ME, et al. . 2015. Network disruption and cerebrospinal fluid amyloid-beta and phospho-tau levels in mild cognitive impairment. J Neurosci 35:10325–10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Li W, Gong L, et al. . 2017. Altered resting-state hippocampal functional networks associated with chemotherapy-induced prospective memory impairment in breast cancer survivors. Sci Rep 7:45135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu GS, Boukelmoune N, Chiang ACA, et al. . 2018. Nasal administration of mesenchymal stem cells restores cisplatin-induced cognitive impairment and brain damage in mice. Oncotarget 9:35581–35597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy SK, McDonald BC, Smith DJ, et al. . 2013. Alterations in brain structure and function in breast cancer survivors: effect of post-chemotherapy interval and relation to oxidative DNA damage. Breast Cancer Res Treat 137:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Scott J, et al. . 2014. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137(Pt 8):2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr M, Thibault C, Morissette M, et al. . 2001. Estrogen-like activity of tamoxifen and raloxifene on NMDA receptor binding and expression of its subunits in rat brain. Neuropsychopharmacology 25:242–257. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, et al. . 2006. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruiter MB, Reneman L, Boogerd W, et al. . 2011. Cerebral hyporesponsiveness and cognitive impairment 10 years after chemotherapy for breast cancer. Hum Brain Mapp 32:1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez S, Vandenbulcke M, Peeters R, et al. . 2014. Longitudinal assessment of chemotherapy-induced alterations in brain activation during multitasking and its relation with cognitive complaints. J Clin Oncol 32:2031–2038. [DOI] [PubMed] [Google Scholar]
- Drakesmith M, Caeyenberghs K, Dutt A, et al. . 2015. Overcoming the effects of false positives and threshold bias in graph theoretical analyses of neuroimaging data. Neuroimage 118:313–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas JA, Makarewicz J, Schaubhut GJ, et al. . 2013. Chemotherapy altered brain functional connectivity in women with breast cancer: a pilot study. Brain Imaging Behav 7:524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberling JL, Wu C, Tong-Turnbeaugh R, et al. . 2004. Estrogen- and tamoxifen-associated effects on brain structure and function. Neuroimage 21:364–371. [DOI] [PubMed] [Google Scholar]
- Feigl GC, Hiergeist W, Fellner C, et al. . 2014. Magnetic resonance imaging diffusion tensor tractography: evaluation of anatomic accuracy of different fiber tracking software packages. World Neurosurg 81:144–150. [DOI] [PubMed] [Google Scholar]
- Friston KJ. 2011. Functional and effective connectivity: a review. Brain Connect 1:13–36. [DOI] [PubMed] [Google Scholar]
- Fukushima M, Betzel RF, He Y, et al. . 2018. Fluctuations between high- and low-modularity topology in time-resolved functional connectivity. Neuroimage 180(Pt B):406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámez JA, Mateo JL, Puerta JM. 2011. Learning Bayesian networks by hill climbing: efficient methods based on progressive restriction of the neighborhood. Data Min Knowl Discov 22:106–148. [Google Scholar]
- Gianni L, Bellotti V, Gianni AM, et al. . 1995. New drug therapy of amyloidoses: resorption of AL-type deposits with 4'-iodo-4'-deoxydoxorubicin. Blood 86:855–861. [PubMed] [Google Scholar]
- Gilmore AW, Nelson SM, McDermott KB. 2015. A parietal memory network revealed by multiple MRI methods. Trends Cogn Sci 19:534–543. [DOI] [PubMed] [Google Scholar]
- Girvan M, Newman ME. 2002. Community structure in social and biological networks. Proc Natl Acad Sci U S A 99:7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Lai S. 1998. Self-navigated spiral fMRI: interleaved versus single-shot. Magn Reson Med 39:361–368. [DOI] [PubMed] [Google Scholar]
- Grayson DS, Fair DA. 2017. Development of large-scale functional networks from birth to adulthood: A guide to the neuroimaging literature. Neuroimage 160:15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, et al. . 2008. Mapping the structural core of human cerebral cortex. PLoS Biol 6:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneghan A, Haley AP, Kesler S. 2020a. Exploring relationships among peripheral amyloid beta, tau, cytokines, cognitive function, and psychosomatic symptoms in breast cancer survivors. Biol Res Nurs 22:126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneghan AM, Gibbons C, Harrison RA, et al. . 2020b. Predicting patient reported outcomes of cognitive function using connectome-based predictive modeling in breast cancer. Brain Topogr 33:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz TS, Suls J, Treviño M. 2018. A call for a neuroscience approach to cancer-related cognitive impairment. Trends Neurosci 41:493–496. [DOI] [PubMed] [Google Scholar]
- Insel TR, Shapiro LE. 1992. Oxytocin receptor distribution reflects social-organization in monogamous and polygamous voles. Proc Natl Acad Sci U S A 89:5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, et al. . 2012. Fsl. Neuroimage 62:782–790. [DOI] [PubMed] [Google Scholar]
- Jurek B, Neumann ID. 2018. The oxytocin receptor: from intracellular signaling to behavior. Physiol Rev 98:1805–1908. [DOI] [PubMed] [Google Scholar]
- Kaiser J, Dietrich J, Amiri M, et al. . 2019. Cognitive performance and psychological distress in breast cancer patients at disease onset. Front Psychol 10:2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M. 2011. A tutorial in connectome analysis: topological and spatial features of brain networks. Neuroimage 57:892–907. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. 2006. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput Biol 2:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR. 2014. Default mode network as a potential biomarker of chemotherapy-related brain injury. Neurobiol Aging 35(Suppl 2):S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Acton P, Rao V, et al. . 2018a. Functional and structural connectome properties in the 5XFAD transgenic mouse model of Alzheimer's disease. Netw Neurosci 2:241–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Adams M, Packer M, et al. . 2017a. Disrupted brain network functional dynamics and hyper-correlation of structural and functional connectome topology in patients with breast cancer prior to treatment. Brain Behav 7:e00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Bennett FC, Mahaffey ML, et al. . 2009. Regional brain activation during verbal declarative memory in metastatic breast cancer. Clin Cancer Res 15:6665–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Blayney DW. 2016. Neurotoxic effects of anthracycline- vs nonanthracycline-based chemotherapy on cognition in breast cancer survivors. JAMA Oncol 2:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Gugel M, Huston-Warren E, et al. . 2016. Atypical structural connectome organization and cognitive impairment in young survivors of acute lymphoblastic leukemia. Brain Connect 6:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Kent JS, O'Hara R. 2011. Prefrontal cortex and executive function impairments in primary breast cancer. Arch Neurol 68:1447–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Ogg R, Reddick WE, et al. . 2018b. Brain network connectivity and executive function in long-term survivors of childhood acute lymphoblastic leukemia. Brain Connect 8:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Petersen ML, Rao V, et al. . 2020. Functional connectome biotypes of chemotherapy-related cognitive impairment. J Cancer Surviv 14:483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Rao A, Blayney DW, et al. . 2017b. Predicting long-term cognitive outcome following breast cancer with pre-treatment resting state fMRI and random forest machine learning. Front Hum Neurosci 11:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Watson CL, Blayney DW. 2015. Brain network alterations and vulnerability to simulated neurodegeneration in breast cancer. Neurobiol Aging 36:2429–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Wayfull JS, Hosseini SM, et al. . 2013. Default mode network connectivity distinguishes chemotherapy-treated breast cancer survivors from controls. Proc Natl Acad Sci U S A 110:11600–11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppelmans V, Breteler MM, Boogerd W, et al. . 2012. Neuropsychological performance in survivors of breast cancer more than 20 years after adjuvant chemotherapy. J Clin Oncol 30:1080–1086. [DOI] [PubMed] [Google Scholar]
- Koppelmans V, Vernooij MW, Boogerd W, et al. . 2015. Prevalence of cerebral small-vessel disease in long-term breast cancer survivors exposed to both adjuvant radiotherapy and chemotherapy. J Clin Oncol 33:588–593. [DOI] [PubMed] [Google Scholar]
- Kringelbach ML. 2005. The human orbitofrontal cortex: linking reward to hedonic experience. Nat Rev Neurosci 6:691–702. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Carlsson B, Grandien K, et al. . 1997. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology 138:863–870. [DOI] [PubMed] [Google Scholar]
- Kurth F, Gaser C, Luders E. 2015. A 12-step user guide for analyzing voxel-wise gray matter asymmetries in statistical parametric mapping (SPM). Nat Protoc 10:293–304. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmerTest package: tests in linear mixed effects models. J Stat Softw 82:26. [Google Scholar]
- Latora V, Marchiori M. 2001. Efficient behavior of small-world networks. Phys Rev Lett 87:198701. [DOI] [PubMed] [Google Scholar]
- Lepage C, Smith AM, Moreau J, et al. . 2014. A prospective study of grey matter and cognitive function alterations in chemotherapy-treated breast cancer patients. Springerplus 3:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang ZJ, Palmer SJ, et al. . 2008. Dynamic Bayesian network modeling of fMRI: a comparison of group-analysis methods. Neuroimage 41:398–407. [DOI] [PubMed] [Google Scholar]
- Li R, Yu J, Zhang S, et al. . 2013. Bayesian network analysis reveals alterations to default mode network connectivity in individuals at risk for Alzheimer's disease. PLoS One 8:e82104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchon JF, Dabaghian Y, Uzor NE, et al. . 2016. Levetiracetam mitigates doxorubicin-induced DNA and synaptic damage in neurons. Sci Rep 6:25705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menning S, de Ruiter MB, Veltman DJ, et al. . 2017. Changes in brain activation in breast cancer patients depend on cognitive domain and treatment type. PLoS One 12:e0171724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Fornito A, et al. . 2009. Hierarchical modularity in human brain functional networks. Front Neuroinform 3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Chen X, Yan Y, et al. . 2016. Functional connectivity change of brain default mode network in breast cancer patients after chemotherapy. Neuroradiology 58:921–928. [DOI] [PubMed] [Google Scholar]
- Mo C, Lin H, Fu F, et al. . 2017. Chemotherapy-induced changes of cerebral activity in resting-state functional magnetic resonance imaging and cerebral white matter in diffusion tensor imaging. Oncotarget 8:81273–81284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. 2002. Fiber tracking: principles and strategies—a technical review. NMR Biomed 15:468–480. [DOI] [PubMed] [Google Scholar]
- Moses J. 2004. Comprehensive trail making test (CTMT) by Cecil R. Reynolds. Austin, Texas: PRO-ED, Inc., 2002. Arch Clin Neuropsychol 19:703–708. [DOI] [PubMed] [Google Scholar]
- Newman ME, Girvan M. 2004. Finding and evaluating community structure in networks. Phys Rev E 69:026113. [DOI] [PubMed] [Google Scholar]
- Nudelman KN, Wang Y, McDonald BC, et al. . 2014. Altered cerebral blood flow one month after systemic chemotherapy for breast cancer: a prospective study using pulsed arterial spin labeling MRI perfusion. PLoS One 9:e96713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SK, Wong AL, Wong FL, et al. . 2015. Inflammatory biomarkers, comorbidity, and neurocognition in women with newly diagnosed breast cancer. J Natl Cancer Inst 107:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, et al. . 2011. Functional network organization of the human brain. Neuron 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Northoff G. 2011. How is our self related to midline regions and the default-mode network? Neuroimage 57:1221–1233. [DOI] [PubMed] [Google Scholar]
- Raichle ME. 2011. The restless brain. Brain Connect 1:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse JC, Zhou J. 2007. Learning effective brain connectivity with dynamic Bayesian networks. Neuroimage 37:749–760. [DOI] [PubMed] [Google Scholar]
- Riem MME, van IJzendoorn MH, Tops M, et al. . 2012. No laughing matter: intranasal oxytocin administration changes functional brain connectivity during exposure to infant laughter. Neuropsychopharmacology 37:2174–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Ruff RM, Light RH, Parker SB, et al. . 1996. Benton Controlled Oral Word Association Test: reliability and updated norms. Arch Clin Neuropsychol 11:329–338. [PubMed] [Google Scholar]
- Sahnoune I, Inoue T, Kesler SR, et al. . 2018. Exercise ameliorates neurocognitive impairments in a translational model of pediatric radiotherapy. Neuro Oncol 20:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno EA, Rowland K, Kramer AF, et al. . 2019. Acute aerobic exercise effects on cognitive function in breast cancer survivors: a randomized crossover trial. BMC Cancer 19:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam P, Wilson SH, Oakes TR, et al. . 2019. Effects of mild traumatic brain injury and post-traumatic stress disorder on resting-state default mode network connectivity. Brain Res 1711:77–82. [DOI] [PubMed] [Google Scholar]
- Schank T, Wagner D. 2005. Approximating clustering coefficient and transitivity. J Graph Algorithms Appl 9:265–275. [Google Scholar]
- Schmidt M. 2012. Rey Auditory Verbal Learning Test (RAVLT): A Handbook. Lutz, FL: Psychological Assessment Resources. [Google Scholar]
- Scutari M. 2009. Learning Bayesian networks with the bnlearn R package. arXiv Preprint arXiv:0908.3817. [Google Scholar]
- Sporns O, Betzel RF. 2016. Modular brain networks. Annu Rev Psychol 67:613–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffaroni AM, Brown JA, Casaletto KB, et al. . 2018. The longitudinal trajectory of default mode network connectivity in healthy older adults varies as a function of age and is associated with changes in episodic memory and processing speed. J Neurosci 38:2809–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Schlagenhauf F, Huys QJM, et al. . 2017. Computational neuroimaging strategies for single patient predictions. Neuroimage 145(Pt B):180–199. [DOI] [PubMed] [Google Scholar]
- Stouten-Kemperman MM, de Ruiter MB, Koppelmans V, et al. . 2015. Neurotoxicity in breast cancer survivors >/ = 10 years post-treatment is dependent on treatment type. Brain Imaging Behav 9:275–284. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Taki Y, Hashizume H, et al. . 2012. The association between resting functional connectivity and creativity. Cereb Cortex 22:2921–2929. [DOI] [PubMed] [Google Scholar]
- Tang TT, Zawaski JA, Kesler SR, et al. . 2019. A comprehensive preclinical assessment of late-term imaging markers of radiation-induced brain injury. Neurooncol Adv 1:vdz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Wang GJ, Volkow ND. 2013. Energetic cost of brain functional connectivity. Proc Natl Acad Sci U S A 110:13642–13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. . 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289. [DOI] [PubMed] [Google Scholar]
- van Oort J, Kohn N, Vrijsen JN, et al. . 2020. Absence of default mode downregulation in response to a mild psychological stressor marks stress-vulnerability across diverse psychiatric disorders. Neuroimage Clin 25:102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zuo X, Dai Z, et al. . 2013. Disrupted functional brain connectome in individuals at risk for Alzheimer's disease. Biol Psychiatry 73:472–481. [DOI] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen AG, et al. . 2007. Diffusion Toolkit: a software package for diffusion imaging data processing and tractography. Proc Int Soc Mag Reson Med 15:3720. [Google Scholar]
- Whitfield-Gabrieli S, Nieto-Castanon A. 2012. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect 2:125–141. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Q, Yu X, et al. . 2016. A triple network connectivity study of large-scale brain systems in cognitively normal APOE4 carriers. Front Aging Neurosci 8, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li R, Fleisher AS, et al. . 2011. Altered default mode network connectivity in Alzheimer's disease—a resting functional MRI and Bayesian network study. Hum Brain Mapp 32:1868–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AC, Huang CC, Liu ME, et al. . 2014. The APOE varepsilon4 allele affects complexity and functional connectivity of resting brain activity in healthy adults. Hum Brain Mapp 35:3238–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Rajapakse JC. 2006. Learning functional structure from fMR images. Neuroimage 31:1601–1613. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, et al. . 2012. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 73:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Seeley WW. 2014. Network dysfunction in Alzheimer's disease and frontotemporal dementia: implications for psychiatry. Biol Psychiatry 75:565–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.