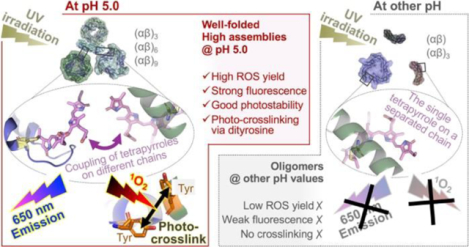
Abstract
Environment-triggered protein conformational changes have garnered wide interests in both fundamental research, for deciphering in vivo acclimatory responses, and practical applications, for designing stimuli-responsive probes. Here we propose a protein–chromophore regulatory mechanism that allows manipulation of C-phycocyanin (C-PC) from Spirulina platensis by environmental pH and UV irradiation. Using small-angle X-ray scattering, a pH-mediated C-PC assembly–disassembly pathway, from monomers to nonamers, was unraveled. Such flexible protein matrices impart tunability to the embedded tetrapyrroles, whose photochemical behaviors were found to be modulated by protein assembly states. UV irradiation on C-PC triggers pH-dependent singlet oxygen (1O2) generation and conformational changes. Intermolecular photo-crosslinking occurs at pH 5.0 via dityrosine species, which bridges solution-based C-PC oligomers into unprecedented dodecamers and 24-mers. These supramolecular assemblies impart C-PC at pH 5.0 significantly enhanced 1O2 yield, fluorescence, and photostability relative to those at other pH values, a finding that makes C-PC appealing for tumor-targeted photodynamic therapy.
Keywords: C-phycocyanin (C-PC), protein–chromophore complex, light-harvesting, photosensitization, pH-tunability, self-assembly, small-angle X-ray scattering (SAXS), electron paramagnetic resonance (EPR)
Graphical Abstract
INTRODUCTION
Organisms across all three domains of life have developed apparatus capable of capturing sunlight; however, intricate and complicated events of individual organisms demand their photosynthetic behaviors to be tailored to the individuals’ environments and needs. Phycobiliproteins are highly adaptable proteins in nature, a characteristic they achieve by exploiting their hierarchical protein structure as a scaffold for fine-tuning the light-harvesting performance of embedded tetrapyrrole chromophores.1–3 This tunability, in essence, relies on the tailor-made protein–chromophore architecture that can sense and respond to environmental changes.
Phycobiliproteins—comprising C-phycocyanin (C-PC), allophycocyanin, phycoerythrin, and phycoerythrocyanin—are a family of protein-chromophore complexes found in cynaobacteria and red algae.[1−2] The light absorption and energy transfer events during photosynthesis are enabled by open-chain tetrapyrrole chromophore cofactors attached covalently to the two phycobiliprotein subunits, the α- and β- subunits.[3] These subunits form (αβ) monomers that further assemble into ring-shaped (αβ)3 trimers, and the trimers can then stack to make rod-like (αβ)6 hexamers and higher assemblies, with other linker proteins acting as bridges at the core.[4–5] This modular and hierarchical structure tailors the energy states of the chromophores through diverse protein conformational changes, ensuring proper and efficient light-harvesting behaviors in fluctuating light environments.[6] The acclimation capability of phycobiliproteins has gained considerable scientific attention and may be harnessed for the design of biomimetic optical materials.
Using phycobiliproteins for in vitro purposes necessitates, firstly, uncovering structural mechanisms behind the acclimatory responses and the environmental conditions that regulate them. Whereas the crystalline structures of phycobiliproteins have been determined from a variety of species,[5, 7] little is known about their structures in solution and the changes under variable conditions. Solution-based structural characterizations, in particular for isolated phycobiliproteins, are complicated by the volatile protein–protein interactions. [8–10] These weak intermolecular interactions, mostly at the monomer–monomer and trimer–trimer interfaces of phycobiliproteins, can either form or break down in solution when exposed to environmental stressors, alternating the structures between mixed oligomers and supramolecular assemblies. The variability and the breadth of possible assembly states render characterization of the solution protein structures difficult.[11]
To fill in this knowledge gap, we offer here unique structural insights into phycobiliproteins’ assembly–disassembly pathways in solution state and the fundamental impact on photochemistry. We found that C-PC isolated from Spirulina platensis can be structurally tuned by changing solution pH and thus exhibit programmable photochemical responses to UV irradiation. The findings point to a protein–chromophore regulatory mechanism that dictates phycobiliproteins’ responses to light under different environments. Our work may inspire the design of new photo-responsive systems using naturally derived phycobiliproteins as templates.
EXPERIMENTAL SECTION
Extraction, purification, and sample preparation.
Protein extraction and purification were performed in the dark at 4 °C unless otherwise noted. The crude phycobiliproteins were extracted from spray-dried biomass of Spirulina platensis following our established protocol.4 The extract was then fractionated by precipitation with solid (NH4)2SO4 at 30% saturation to remove contaminating membrane proteins and linker proteins, followed by 55% saturation to precipitate the major phycocyanin fraction. The collected pellet was re-dissolved in a small volume of Na-phosphate buffer (5 mM, pH 7.0) and dialyzed overnight against the same buffer using a Slide-A-Lyzer G2 dialysis device (15 ml, 3.5K MWCO, Thermo Fisher). The dialyzed concentrates were centrifuged at 10,000g for 30 min and subjected to ion-exchange chromatography using DEAE-Sepharose® Fast Flow column (1.5 × 15 cm, GE Healthcare). A stepwise elution was carried out using a linear gradient of 0.15–0.22 M NaCl in K-phosphate buffer (50 mM, pH 7.4) at a flow rate of 1.0 ml/min. The effluent was collected in 2-mL fractions and the absorption of each fraction at 280 nm, 620 nm, 655 nm were recorded. All C-phycocyanin (C-PC) fractions having the purity ratio (A620/A280) > 4.5 and the separation factor (A620/A655) > 5.0 were pooled and then desalted on a Sephadex G-25 column. The final A620/A280 and A620/A655 of the collected C-PC sample were 5.03 and 6.79, respectively. The obtained pure C-PC was stored in dark at −20 °C as a lyophilized powder, which were pre-hydrated before use in Milli-Q water by stirring at 4 °C overnight and then filtrated through a 0.22 μm syringe filter (Celltreat). The C-PC concentration of the hydrated stock was adjusted to 5.0 mg/ml based on A280 (UV-2600 spectrophotometer, Shimadzu) using extinction coefficient of 29,590 M−1cm−1 or 0.8309 mg–1 ml cm−1 calculated from the amino acid composition.
Irradiation.
A 365-nm UV lamp (EL Series UVLS-28, 8 watt, Upland) with a measured fluence rate of 0.63 mW/cm2 was used as the light source. The prepared 0.25 mg/ml C-PC samples at pH 3.0, 5.0, 7.0, and 9.0, respectively, were bubbled with air for 20 min and packed into thin-walled borosilicate glass vials (VWR) with screw caps (sample volume: 5 ml). Then, the vials were laid flat and positioned parallel to the lamp at a set exposure distance of 2 cm to ensure the efficiency and uniformity of the irradiation.
SEC–SAXS.
SEC–SAXS experiments were conducted on beamline ID7A1 at the Macromolecular Diffraction Facility of the Cornell High Energy Synchrotron Source (MacCHESS)5, 6 using a dual Pilatus 100K-S SAXS/WAXS detector (Dectris, Switzerland). The sample-to-detector distance was 1.498 m, with the SAXS detector covering a collected q range of 0.007 < q < 0.27 Å–1 (q = (4π sin θ) /λ, where 2θ is the scattering angle). The photon energy of the X-ray beam was 9.808 keV (1.264 Å) at 8 × 1011 photons s–1, and the beam diameter was 250 × 250 μm. The beamline was outfitted with a ÄKTA purifier (GE Healthcare) equipped with a Superdex 200 Increase 10/300 GL column (GE Healthcare), which were preequilibrated at 4 °C with 50 mM citrate buffers (pH 3.0 and 5.0), HEPES buffer (pH 7.0), or Tris-base buffer (pH 9.0) containing 3% v/v glycerol for C-PC samples at corresponding pH. The non-irradiated and 12-h-irradiated C-PC samples at pH 3.0, 5.0, 7.0, and 9.0 were concentrated up to 5.0 mg/ml using an 10K MWCO Amicon Ultra-15 centrifugal filter (EMD Millipore), and centrifuged at 10,000g for 10 min before sample injection. Aliquots of 100 μl centrifuged sample was loaded at a flow rate of 0.12 ml/min and was directed continuously by the SEC system into the BioSAXS flow cell as the scattering data were collected (2 s exposure time per frame). Eluents of SEC system were monitored by absorption at 280 nm (A280). Approximately 1000 2-s exposures were collected per sample for further analysis of the structural parameters and modeling.
SAXS data analysis.
After the images were collected, scattering profiles were generated for each sample with the software RAW.7 The middle portion of each SEC peak consisting of 10–20 frames with good signal to noise ratio was used for data analysis, and 50–100 buffer profiles preceding the elution peaks were averaged and used for the buffer subtraction. For C-PC at pH 3.0 (both non-irradiated and irradiated), elevated baselines resulting from radiation damage were noticed, thus baseline corrections using the integral baseline correction function in RAW were further applied for these two samples. The subtracted SAXS profiles were subjected to Guinier analysis performed using the automatically optimized q ranges, from which the radius of gyration (Rg), molecular weight (MW), and extrapolated zero-angle scattering (I(0) were determined. The pair-distance distribution functions (p(r)) were created by GNOM,8 which provided additional Rg and I(0) estimates and the maximum particle dimension, Dmax. Independent MW estimates were obtained by the adjusted Porod volume (Vp) and the volume-of-correlation (Vc) approaches. FoXS9, 10 was used to fit the experimental scattering profiles to the theoretical scattering curves calculated from the C-PC crystal model (PDB 1GH0)11 and to calculate the quality-of-fit (χ2). The input C-PC crystal structures in different oligomeric states, i.e., (αβ) monomers, (αβ)3 trimers, (αβ)6 hexamers, were derived from PDB 1GH0.11
Ab initio envelopes were reconstructed based on the whole range of q by GASBOR12 for all C-PC samples except for the irradiated fragment N at pH 5.0 (GASBOR is not used for this sample due to the MW < 700 kDa limitation), using symmetry and anisotropy assumption parameters in Table S1 for different C-PC samples. For structures with no known crystal models (irradiated fragment N at pH 5.0, irradiated and non-irradiated fragment 1 at pH 5.0) or partially unfolded (fragment 2 at pH 3.0 and irradiated C-PC at 9.0), ab initio were further performed by DAMMIN13 with no symmetry restraints to verify the computed shapes (see section 1.1.2 in SI for details).. To further optimize the arrangements of subdomains within the highly assembled structures (irradiated fragment N at pH 5.0, irradiated and non-irradiated fragment 1 at pH 5.0), rigid body refinements were performed by CORAL14 using (αβ)6 hexamer and (αβ)3 trimer models of C-PC (PDB 1GH0)11 as inputs. The reconstructed shapes by GASBOR, DAMMIN, and CORAL were visualized and superimposed on the corresponding C-PC crystal models using Pymol.15
Electron paramagnetic resonance (EPR) spectroscopy.
X-band continuous-wave (cw) EPR was carried out at room temperature on a Bruker ELEXSYS II EPR spectrometer at 9.8 GHz with 100 kHz modulation frequency, 2 G modulation amplitude, 2 mW power, and 20 dB attenuation. Aliquots of 150 μl samples were contained in WG-812 quartz flat cell (Wilmad) and the in situ irradiation during EPR experiments was provided by a ~8 watt UVA system placed in front of the optical window of the resonator. Each spectrum was acquired as a mean of 10 accumulated scans. The C-PC concentration for all EPR experiments were kept the same at 1 mg/ml. Spectral simulations were performed by EasySpin16 integrated into MATLAB using the “esfit” least-squares function and were used to assist with signal assignments and g-value determination.
For spin trapping experiments, the trapping agents, 2,2,6,6-tetramethylpiperidine (TEMP) or 5,5-dimethyl-1- pyrroline N-oxide (DMPO), were freshly prepared in DMSO at 10-fold concentration and mixed with C-PC samples at pH 3.0–9.0 before the measurements to a final concentration of 1.0 × 10−2 M. For quenching experiments, 8 mM sodium azide (NaN3) were added to the same C-PC samples prior to the addition of 1.0 × 10−2 M DMPO and the EPR measurements.
RESULTS AND DISCUSSION
pH-mediated assembly–disassembly
The relatively weak and dynamic intermolecular forces of C-PC enable it to rapidly respond to environmental stimuli and undergo a range of conformational changes. The C-PC primary sequence comprises abundant charged residues and residues with ionizable side chains (Figure S3A). Notably, most of these residues are dispersed across the monomer–monomer and trimer–trimer interfaces, allowing modular self-assemblies of C-PC through the key intermolecular salt bridges. For example, the salt bridges that form across the ends of adjacent (αβ) monomers can bridge three of them into an (αβ)3 trimer (Figure 1A). Likewise, two of the (αβ)3 trimers can be stacked face-to-face to form an (αβ)6 hexamer (Figure 1B). Extreme acidic or alkaline environments, characterized by pH values very different from the theoretical isoelectric point (pI) of C-PC (~5.23, see section 1.1.1 in SI), were found to trigger protonation or deprotonation of the residues and lead to a build-up of electrostatic repulsion across the chains (as indicated by the zeta potentials in Figure S3B). Consequently, the intermolecular salt bridges could be disrupted under the extreme pH conditions and induce protein disassembly.
Figure 1. pH-dependent salt bridges at C-PC intermonomer and intertrimer interfaces.
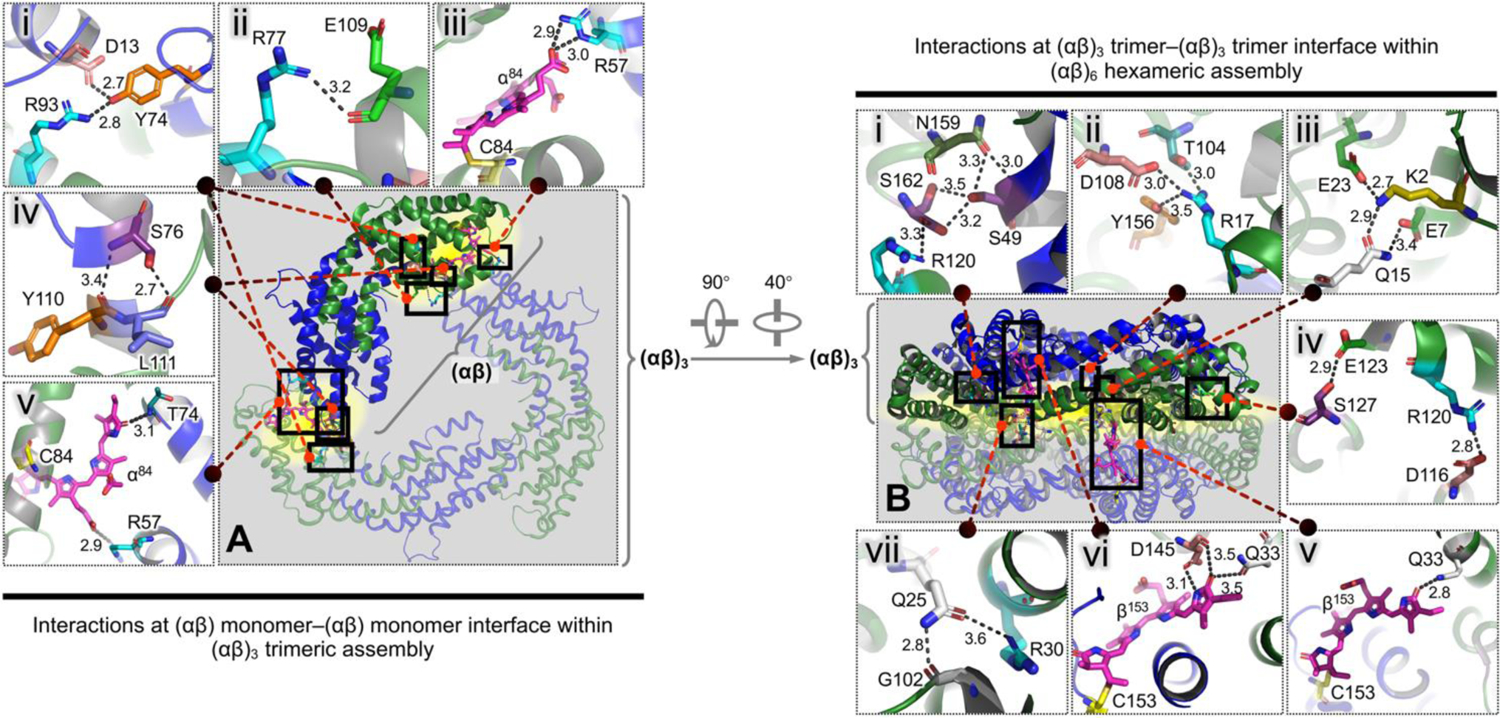
(A) Top view of the overall (αβ)3 trimeric assembly, with one (αβ) monomer highlighted in opaque thick representation. (B) Side view of the overall (αβ)6 hexameric assembly, with one (αβ)3 trimer highlighted in opaque thick representation. (i–vii) in both left and right panels, Close-ups of the detailed interactions across the described interfaces between protein residues or tetrapyrrole chromophores, which correspond to the black boxes in (A) and (B). The described interfaces are highlighted in yellow. The key salt bridges are denoted by the black dotted lines. The numbers beside the black dotted lines indicate interatomic distances (Å). All residues and chromophores involved in the interactions are labelled and shown as colored sticks (oxygen atoms, red; nitrogen atoms, blue). Helices of the α- and β- subunits are colored green and blue, respectively.
The assembly and disassembly of C-PC in response to pH changes in the 3.0–9.0 range were visualized by in-line size-exclusion chromatography with small-angle X-ray scattering (SEC–SAXS). All discussions in this section refer to the non-irradiated samples unless otherwise specified. Based on the SEC profiles (Figure 2A), C-PC at pH 9.0 (solid grey line) and pH 7.0 (solid blue line) were eluted as monodisperse peaks with gradually decreasing elution volumes. C-PC at pH 5.0 (solid green line in Figure 2A) gave rise to peaks characterized by high polydispersity, and the three well-separated fragments are hereafter referred to as Fragment 1, Fragment 2, and Fragment 3, respectively. Among these fragments, Fragment 2 was dominant, displaying a mass fraction of 42.69% (Table S1). All the fragments at pH 5.0–9.0 (non-irradiated) were homogeneous, displaying a consistent value of the radius of gyration, Rg, throughout each peak (Figure S4, B–D). Nevertheless, C-PC at pH 3.0 (solid red line in Figure 2A) was eluted as an aggregated species (Fragment 1) and another major fragment at larger elution volume (Fragment 2). Interpretable SAXS data was obtained only for the Fragment 2 (see section 1.2.1 in SI).
Figure 2. SEC–SAXS analysis reveals different assembly states of non-irradiated C-PC at pH 3.0–9.0.
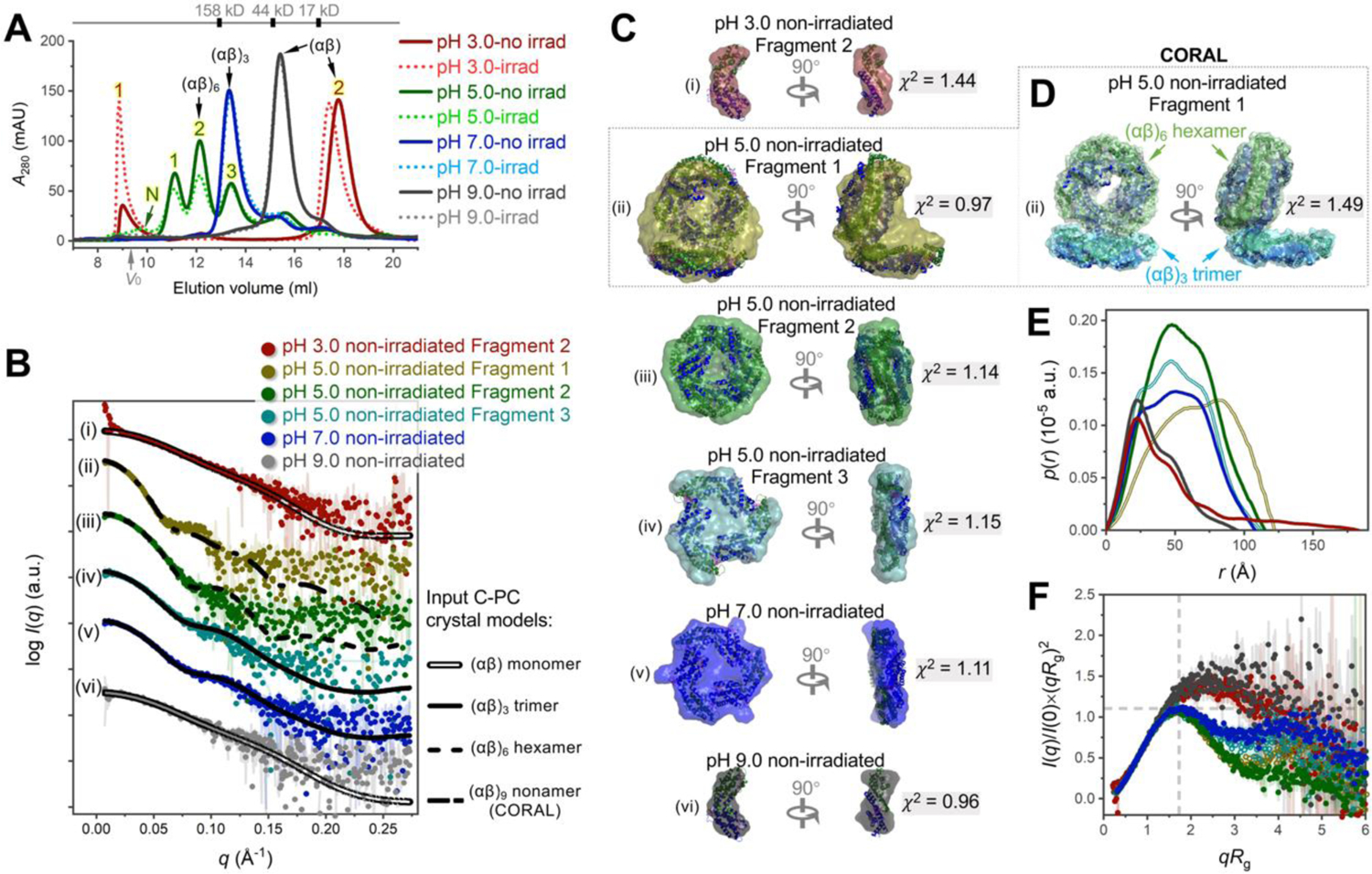
(A) Overlay of size-exclusion chromatography profiles. The solid and dotted lines are for non-irradiated and irradiated C-PC, respectively. The grey arrow indicates the void volume (V0). The approximate elution volumes of molecular weight standards are reported. Numbers 1, 2, 3, and N identify fragments used in subsequent SAXS analysis. (B) SAXS scattering profiles (colored dots) fitted with theoretical scattering curves (black lines) back-calculated from C-PC crystal models in different assembly states (PDB 1GH0).11 (i) Fragment 2 at pH 3.0 (red dots) fitted with (αβ) monomer model (double line). (ii) Fragment 1 at pH 5.0 (olive dots) fitted with a CORAL-derived (αβ)9 nonamer model shown in (D) (dash–dot line). (iii) Fragment 2 at pH 5.0 (green dots) fitted with (αβ)6 hexamer model (dashed line). (iv) Fragment 3 at pH 5.0 (teal dots) fitted with (αβ)3 trimer model (solid line). (v) Fragment at pH 7.0 (blue dots) fitted with (αβ)3 trimer model. (vi) Fragment at pH 9.0 (grey dots) fitted with (αβ) monomer model. Fittings were conducted by FoXS;9, 10 all χ2-values (Table S1) point to good fits. (C) GASBOR ab initio envelopes (surface representation) superimposed with the corresponding crystal models (cartoon). (D) The CORAL model of Fragment 1 at pH 5.0, consistent with the corresponding GASBOR envelope (ii in C). The indicated C-PC (αβ)3 trimer and (αβ)6 hexamer models were used as input rigid bodies. See Figure S6A for details. (E) Pair distance distribution functions. (F) Dimensionless Kratky plots. The cross-hair marks the Guinier–Kratky point (√3, 1.1), the peak position for an ideal globular structure. Panels (E–F) are colored as in (B).
For SAXS analysis, the Guinier plots (Figure S5) of all fragments obtained at pH 3.0–9.0, except for the inhomogeneous Fragment 1 at pH 3, displayed good linearity and no evidence of aggregation. The computed structural parameters for these fragments are summarized in Table S1. As the pH value decreased from 9.0 to 5.0, the key structural parameters derived from Guinier and pair distance distribution (p(r)) analyses (including Rg; extrapolated zero-angle scattering, I(0); and experimental maximum dimension, Dmax) gradually increased, suggesting that C-PC grew into higher assembly states. In Figure 2B, the experimental SAXS scattering profiles were compared to the theoretical data back-calculated from C-PC crystal models in different assembly states (PDB 1GH0,11 several basic structures are given in Figure S3C). Good fits were obtained by superimposing the scattering profiles of an (αβ)6 hexamer model—a face-to-face assembly of two (αβ)3 trimers—with Fragment 2 at pH 5 (curves iii, χ2 = 1.56); a threefold symmetric (αβ)3 trimer model with Fragment 3 at pH 5 (curves iv, χ2 = 1.23) and the fragment at pH 7.0 (curves v, χ2 = 0.93); and an (αβ) monomer model with the fragment at pH 9.0 (curves vi, χ2 = 0.91). The ab initio envelopes reconstructed by GASBOR13 for the above fragments also resulted in the expected shapes, which were closely superimposable with the crystal models in the corresponding assembly states (iii–v in Figure 2C); only one exception existed: the envelope at pH 9.0 (vi in Figure 2C) had slight protrusions on its sides, reflecting a further extension of the actual structure at this pH than was suggested by the crystal model.
The highly assembled Fragment 1 at pH 5 was determined as an (αβ)9 nonameric assembly comprising an (αβ)3 trimer and an (αβ)6 hexamer (see section 1.2.2 in SI for details). These two subdomains were offset and organized in columns (Figure 2D; Figure S6, A and G) rather than in the common side-by-side arrangements of intact phycobiliprotein antennas.17, 18 The computed conformation, however, is in good agreement with previously reported structures of the isolated phycobiliproteins,19–21 whereby the subdomains were offset due to the absence of the linker proteins that bridge the rods. Notably, our study revealed that C-PC was further offset in solution, that is, an even wider angle was present between the upper and lower subdomains than was the case in the reported crystal structures.
For the Fragment 2 at pH 3, interpretable data was only obtained at a higher q range of 0.023–0.045 Å−1 due to the slight radiation damage (see section 1.2.1 in SI). The SAXS-derived parameters (Table S1) and ab initio reconstructions (i in Figure 2C) suggested that Fragment 2 at pH 3 was an (αβ) monomer with an even more unfolded and flexible conformation than the above-described monomeric C-PC at pH 9.0. Such a high conformational flexibility would explain why C-PC at pH 3.0 was aggregation-prone and susceptible to radiation damage.
The structures at different pH values were further compared by p(r) and dimensionless Kratky plots (Figure 2, E and F; the curves are colored as in Figure 2B). The extended shapes of C-PC at pH 3.0 and pH 9.0 were evidenced by the early peaks in the p(r) plots with roughly linear fall-offs and long tails. By contrast, C-PC at pH 5.0 and pH 7.0 yielded prominent and nearly symmetrical bell-shaped peaks with multiple shoulders that correlated well with the numbers of subdomain structures. Likewise, the dimensionless Kratky plots of C-PC at pH 5.0 and 7.0 exhibited well-defined peaks at Guinier–Kratky point (marked by cross-hair), which is typical of fully folded globular proteins; by contrast, the peaks of C-PC at pH 3.0 and pH 9.0 were broadened and remarkably shift away from the Guinier–Kratky point, suggesting that these structures tend to extend flexibly rather than collapse in a globular fashion. Folding and unfolding of the fragments could also be monitored by circular dichroism (CD) and intrinsic protein fluorescence (Figure S7 and the supplemental notes; however, smaller structures, which were successfully captured by SEC–SAXS, were deemed hard to identify by CD and fluorescence assays alone owing to the coexisting larger aggregates.
Taken together, these results reveal that pH changes in the 3.0–9.0 range mediated large-scale assembly and disassembly of C-PC in solution state. Specifically, the partially unfolded monomers found in both acidic (pH 3.0) and alkaline (pH 9.0) conditions can be converted to the well-folded trimers at pH 7.0 and high assemblies—a mixture of trimers, hexamers, and nonamers—at pH 5.0.
pH-tunable photochemical behaviors
The conformational variability of C-PC protein matrix led us to investigate whether the photochemical behavior of the attached tetrapyrrole chromophores could also be tuned by pH via the assembly–disassembly pathways. Data and detailed discussions are provided in Figure S8A and the supplemental notes. Briefly, red-shifted fluorescence emissions and blue-shifted Q-band absorptions were observed for higher C-PC assemblies, and the shifts were accompanied by apparent color and intensity changes. Such spectroscopic behaviors reflected a protein–chromophore regulatory mechanism, whereby the protein assembly state of C-PC does not control only the positions and conformations of single chromophores but also interchromophoric couplings. Based on this regulatory mechanism, the embedded chromophores gained tunability owing to the attached protein matrix that assembles and disassembles according to the environmental pH. The found pH-tunable photochemical behaviors of C-PC, with respect to fluorescence, Q-band absorption, and color, may be exploited in the future for the development of photochromic pigments22, 23 and smart materials for imaging,24 photosensing,25 and biolabeling.26
pH-dependent spectral and conformational responses to UV irradiation
In vivo, phycobiliproteins serve as essential photosynthetic apparatuses because of their light-harvesting abilities and flexible structures that allow the organisms to cope with diverse environments.27 Our results suggested that isolated C-PC in vitro can also sense and respond to light while exhibiting large-scale spectral and conformational changes in a pH-dependent manner. An important evidence of C-PC’s light sensitivity was the photobleaching phenomenon, whereby the blue color of C-PC progressively bleached under constant 365-nm UV irradiation (Figure 3, A and B). Intriguingly, the kinetic results (Figure 3C) indicated that the photobleaching rate was remarkably decreased in the following order: pH 3.0 > pH 9.0 > pH 7.0 > pH 5.0. Given the above SEC–SAXS-derived structural information, it was evident that the photobleaching was inhibited as the protein matrix folds and assembles into higher-order structures.
Figure 3. Spectroscopic analyses for irradiation-induced photobleaching and conformational change at pH 3.0–9.0.
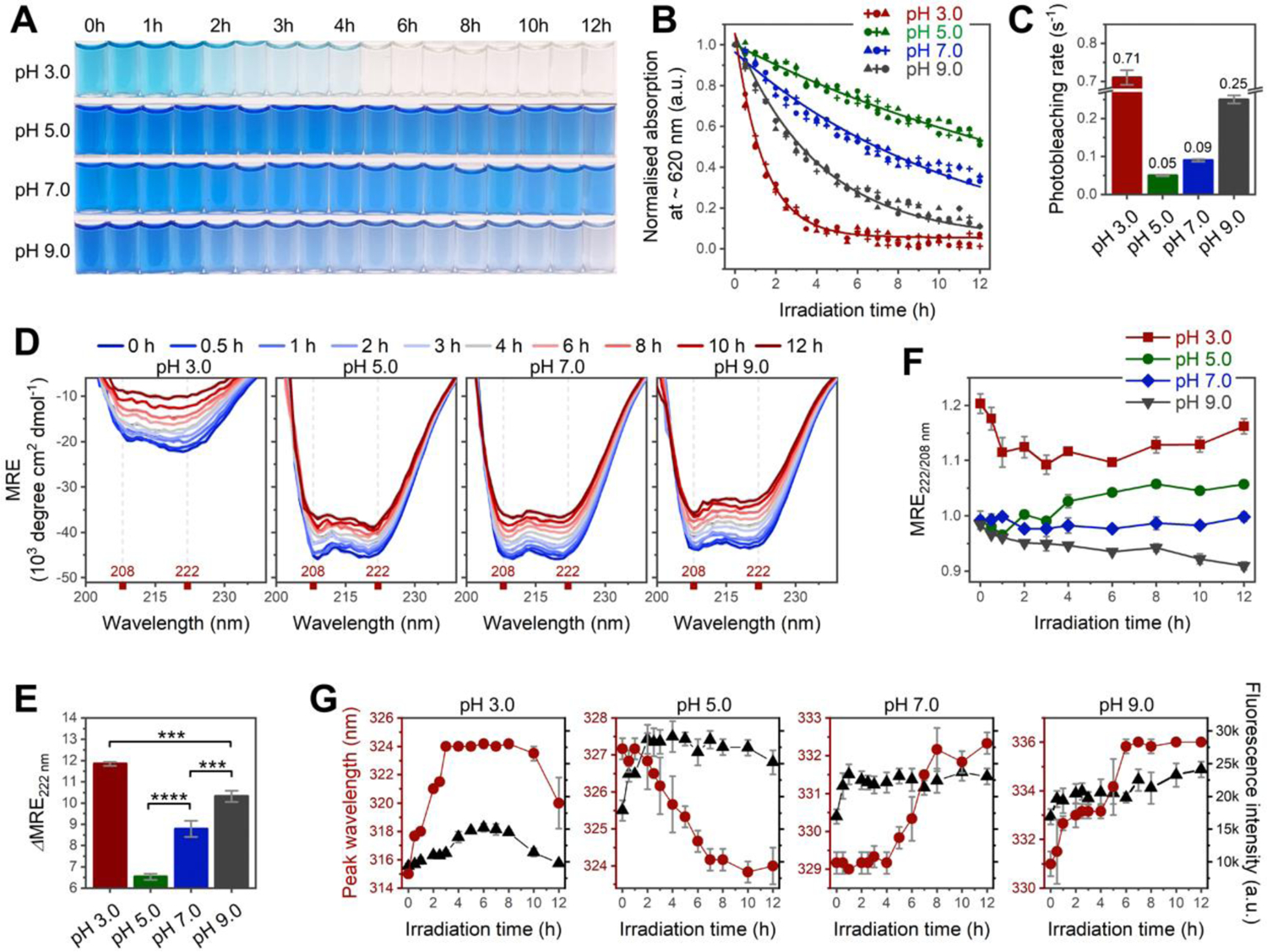
(A–B) Changes in color and Q-band absorption of C-PC as a function of irradiation time. Absorptions in (B) are recorded at peak maxima indicated in Figure S8C. n = 3 independent samples; each individual point represents the mean of three technical replicates. (C) Photobleaching rate constant calculated from data points in (B) (see section 1.1.6 in SI for details). (D) Far-UV CD spectra of C-PC at pH 3.0–9.0 after 0−12 h of irradiation. (E) Irradiation-induced secondary structural changes calculated by the difference in mean residue ellipticity (MRE) minima at 222 nm before and after 12 h of irradiation (ΔMRE222). (F) Irradiation-induced changes in interhelical contacts traced by ratios of MRE minima at 222 and 208 nm (MRE222/208). (G) Intensity (black) and peak wavelength (red) of protein intrinsic fluorescence plotted against irradiation time. All irradiations were performed under a 365-nm UV lamp; n = 3 independent samples, mean ± s.d. Statistical significance in (E) was calculated using one-way ANOVA with Tukey’s honest significant difference test. ***P<0.001; ****P<0.0001. Panels (F–G) reveal irradiation-induced aggregation at pH 5.0 versus irradiation-induced unfolding at pH 3.0, 7.0, and 9.0.
Monitoring C-PC conformational changes over a 12-h irradiation period by CD and protein intrinsic fluorescence (Figure 3D–G, details are given in section 1.3 in SI) revealed that highly assembled C-PC at pH 5.0 tended to further aggregate under irradiation, and this increased aggregation is in opposition to the tendency of irradiation to cause protein unfolding at other pH values (pH 3.0, 7.0, and 9.0).
In order to gain further insight into how the conformation of differently assembled C-PC fragments changes as a result of irradiation, 12-h-irradiated C-PC samples were also subjected to SEC–SAXS analysis, and the results thus obtained were compared with those of the corresponding non-irradiated fragments. It was found that all the interpretable fragments at pH 3.0, 7.0, and 9.0 displayed increased conformational flexibility after irradiation but no discernible aggregation, cleavage or disorder (see SI section 1.2.3 for details).
In the case of C-PC at pH 5.0, the irradiated Fragment 2 and Fragment 3 (Table S2; Figure 4, panels ii and iii in A–E) also displayed nearly identical structural parameters and ab initio envelopes to their non-irradiated counterparts. However, a highly assembled Fragment 1 and a new fragment characterized by a large Rg value (hereafter Fragment N) were observed to have formed following irradiation; SEC–SAXS data of the two fragmetns are given in Figures 2A (green dotted line) and S4F (yellow and red boxes), and detailed SAXS analysis are given in section 1.2.4 in SI. In brief, the irradiated Fragment 1 at pH 5.0 was determined to be an (αβ)12 dodecamer consisting of two offset (αβ)6 hexamer subdomains (Figure S6B), which were arranged in a similar offset and columnar conformation to its non-irradiated counterpart (Figure S6A). Concurrently, the new Fragment N was a rod-like (αβ)24 24-mer composed of four offset and tightly packed (αβ)6 hexamers (Figure S6C). These identified supramolecular assemblies further corroborated the CD and protein intrinsic fluorescence results that irradiation induced protein aggregation at pH 5.0.
Figure 4. SAXS analysis reveals irradiation-induced conformational changes of C-PC at pH 3.0–9.0.
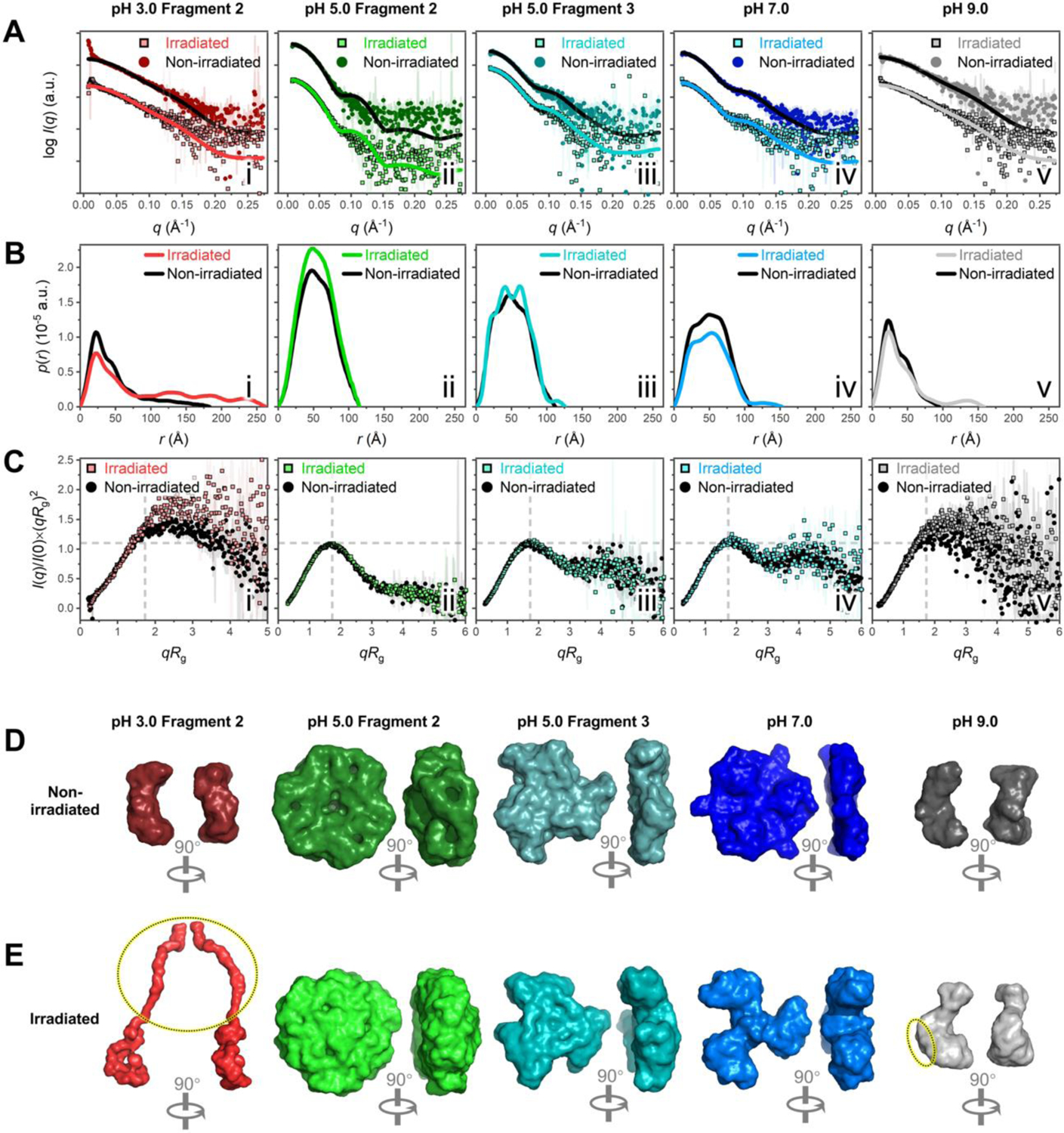
(A) SAXS scattering profiles (dots) fitted with theoretical scattering curves (lines) back-calculated from corresponding C-PC crystal models (PDB 1GH0).11 Non-irradiated fragments are colored lines on square dots; irradiated fragments are black lines on circular dots. The following C-PC crystal models in different assembly states were used for fitting: (αβ) monomer in i, v; (αβ)3 trimer in iii, iv; and (αβ)6 hexamer in ii. (B) Dimensionless Kratky plots. (C) Pair distance distribution functions. (D–E) Surface representations of GASBOR ab initio envelopes for non-irradiated (D) and irradiated (E) C-PC fragments. For Fragment 2 at pH 3.0 (red) and the fragment at pH 9.0 (grey), the dotted ovals in (E) highlight the pronounced protrusions that arised in the structures after irradiation, which were absent from the corresponding non-irradiated structures in (D). The corresponding DAMMIN envelopes in Figure S6, E and F, reveal similar protrusions in the structures.
For the first time we constructed and visualized stable, highly assembled solution structures of isolated C-PC as offset (αβ)12 dodecamers and rod-like (αβ)24 24-mers, both of which were formed at pH 5.0 after irradiation. Normally, the weak intermolecular interactions of phycobiliproteins determine that large assemblies in solution are easily disrupted by environmental changes and do not further associate into rods, in the absence of linker proteins or stabilizing agents (e.g., 0.5–0.9 M phosphate21, 28).29, 30 Our finding suggested that additional intermolecular forces that bridge and strengthen the assemblies may have arisen at pH 5.0 after irradiation; evidences will be provided later to support this hypothesis.
pH-controlled photogeneration of singlet oxygen (1O2)
To elucidate the photochemical mechanisms behind the structural changes, we monitored irradiation-induced radical formation by C-PC at pH 3.0–9.0. Generation of 1O2, one of the main products of photosensitized reactions,31, 32 was determined by electron paramagnetic resonance (EPR) spectroscopy using 2,2,6,6-tetramethylpiperidine (TEMP) as the trapping agent (Figure 5A). In the absence of irradiation, no signals due to 1O2 were detected at any pH; upon in situ irradiation, however, 1O2 production was immediately detected via appearance of the 1:1:1 triplet lines (aN ≅ 16.2 G, g = 2.006) characteristic of the paramagnetic nitroxide radical, 2,2,6,6-tetramethylpiperidine-N-oxyl (TEMPO). The intensity of the triplet signal at each pH increased to reach a maximum within ~10 min; it then slowly decayed over ~15 min, indicating the dependency of 1O2 formation on irradiation time and the probable existence of other reactions competing for the generated TEMPO radicals. Notably, the 1O2 generation displayed a pronounced pH dependency and the yield decreased as follows: pH 5.0 > pH 3.0 > pH 7.0 > pH 9.0, whereby the 1O2 yield at pH 5.0 was 2–5 times higher than that at other pH values. In the present context, we envisage the following situation: if the C-PC chromophores serve as photosensitizers to trigger 1O2 generation, then the highly assembled C-PC at pH 5.0, which assisted hierarchical chromophore couplings at intermolecular interfaces (discussed in Figure S8), may also facilitate photosensitized interactions and boost 1O2 yield.
Figure 5. X-Band EPR detection of photogenerated radicals from C-PC at pH 3.0–9.0.
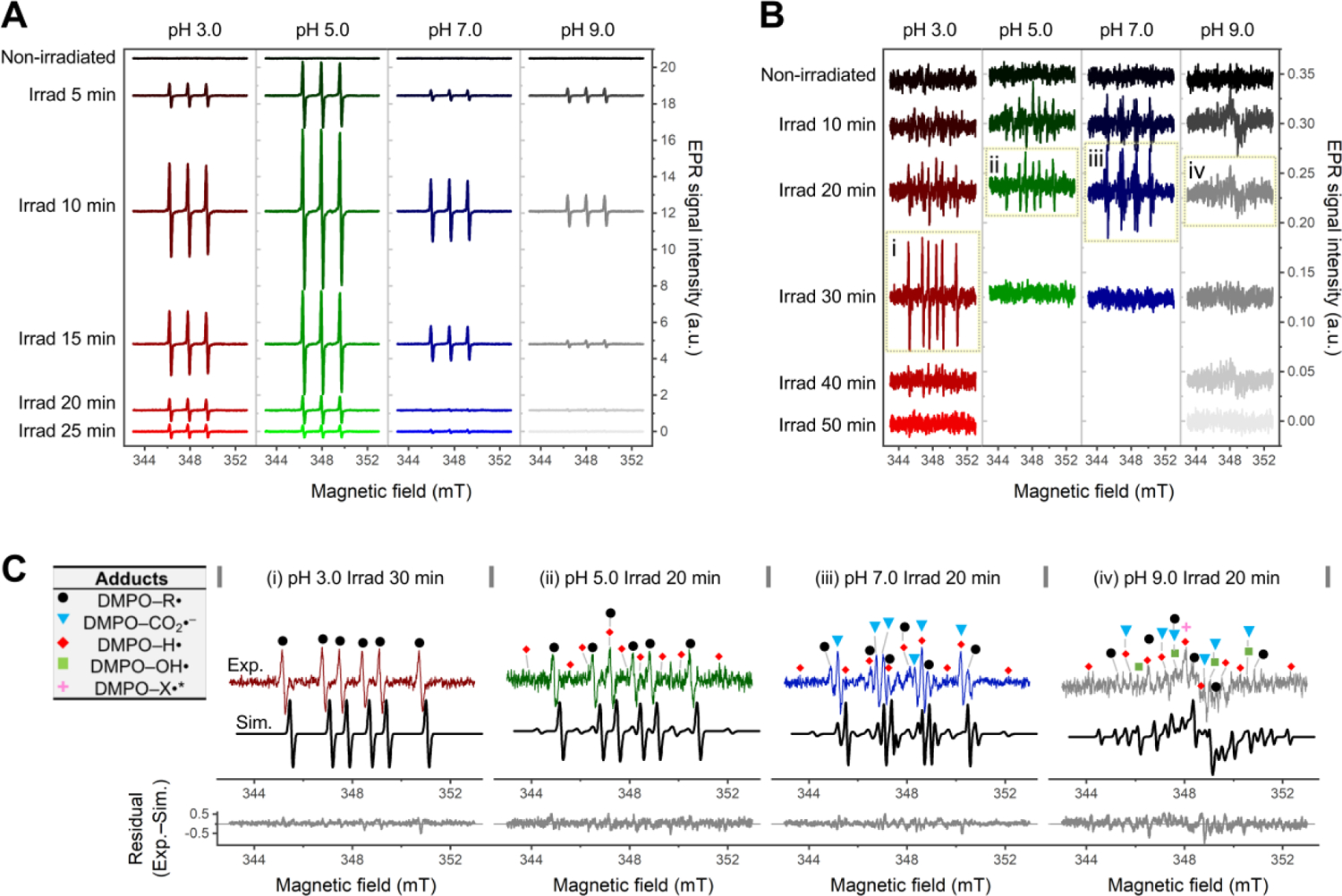
(A–B) Time-dependent EPR spectra of 2,2,6,6-tetramethylpiperidine (TEMP)/1O2 (A) and 5,5-dimethyl-1-pyrroline N-oxide (DMPO) adducts (B) collected under in situ UVA irradiation for the indicated times. The spectra are shifted by an arbitrary offset for clarity. Panels i–iv in (B) are used for the spectral simulations in (C). (C) Experimental (colored lines) and simulated (black lines) EPR spectra of photogenerated DMPO adducts with residuals of the fits reported below. The input spectra are C-PC at pH 3.0 after 30 min irradiation (i); and C-PC at pH 5.0 (ii), 7.0 (iii) and 9.0 (iv) after 20 min irradiation. The symbols mark line components belonging to DMPO–R• (black dots), DMPO–CO2•– (light-blue triangles), DMPO–H• (red diamonds), DMPO–OH• (light-green squares), and an unidentified DMPO–X• (magenta asterisk); see Table S3 for details of spectral simulation. In all cases, the C-PC concentration was 1 mg/ml.
1O2 reacts with nearby molecules and initiates the cascade generation of other reactive species.33–35 The secondary radicals generated upon irradiation were detected by using 5,5-dimethyl-1-pyrroline N-oxide (DMPO) as the trapping agent (Figure 5B). Although the signals due to DMPO adducts at different pH values were 2–3 orders of magnitude weaker than those due to 1O2, their increases and decays followed a similar trend. Notably, the identified carbon-centred radicals at all pH values (black dots in Figure 5C, see Table S3 for details) corroborated that C-PC’s protein matrix or chromophores were involved in the radical formation.
We also observed that formation of the DMPO adducts at all pH values could be significantly suppressed by the addition of sodium azide (NaN3), a selective 1O2 quencher (Figure S9); this further confirmed the role of 1O2 in the generation of secondary radicals trapped by DMPO. Moreover, as radicals were being directly and indirectly scavenged by NaN3, C-PC photobleaching was inhibited by 30.5–37.1% (Figure S10) with respect to solutions lacking NaN3; this indicated that, in addition to C-PC’s protein matrix, the embedded tetrapyrrole chromophores are also a main target of radical attacks.
Intriguingly, our results suggested that high 1O2 yields do not necessarily signify high photosensitizer consumption. At pH 5.0, whereby the highest 1O2 yield was observed, the chromophores displayed the most photostable blue color (Figure 3A–C, described above). This phenomenon can be interpreted as an indication that the C-PC photosensitization proceeded via different mechanisms. Theoretically, photosensitizers participate in photosensitized reactions via either Type I photochemistry, to generate radials/radical ions (e.g., sensitiser radical anions, OH• and O2•–) and get oxidized, or Type II photochemistry, to generate 1O2 species that react with other substrates instead of the photosensitizer (leading to almost no photosensitizer loss).36 Multiple radical products were detected in our case, indicating that the photosensitization of C-PC may proceed via both Type I and II mechanisms.35 However, the radical composition differed substantially for C-PC at different pH, which suggested that the protein assembly state determined the predominant type of photosensitized reaction in a given solution condition, and thus determined to what extent the embedded tetrapyrrole chromophores, as photosensitizers, are consumed and photobleached during the reaction.
Intermolecular photo-crosslinking via Tyr residues
SDS-PAGE analysis suggested that C-PC was also covalently modified by UV irradiation in a pH-dependent manner (see Figure S11A for details). Generally, photo-crosslinking in proteins often occurs between several photoactive residues, such as Cys, His, Trp, and Tyr, forming disulphide, histidine–lysine, or dityrosine linkages.37–39 In our case, the photo-crosslinking—occurred intermolecularly via nonreducible covalent bonds—most likely involves Tyr residues, given their high abundance in C-PC relative to the other photoactive residues (Figure S3A). Moreover, it was noticed that most of the Tyr residues in C-PC locate around the chromophores40 and at the interfaces across α- and β- subunits (Figure 1), making it possible for the photo-crosslinking to happen intermolecularly across separate chains as was suggested by SDS-PAGE gels. Indeed, the involvement of Tyr residues in photo-crosslinking was experimentally corroborated by the Tyr-derived radicals (direct EPR, Figure S11B) and dityrosine species (dityrosine-specific fluorescence, Figure S11C) extensively formed in irradiated C-PC at pH 5.0.
Collectively, irradiation at pH 5.0 induced intermolecular photo-crosslinking via dityrosine linkages, which was not detected or barely detected for C-PC at other pH values. Thus, it is reasonable that these intermolecular covalent bonds provide additional bridging forces between the assemblies and prompted the formation of the described supramolecular fragments at pH following irradiation. The pronounced pH-dependency of the photo-crosslinking could explain why C-PC exhibited opposite conformational changes—the aggregation at pH 5.0 versus the unfolding at pH 3.0, 7.0, and 9.0—under irradiation. In brief, we propose that the intermolecular photo-crosslinking detected at pH 5.0 more than compensates for the intrinsic instability of isolated phycobiliproteins in solution and the irradiation-induced unfolding tendency predominant at other pH values.
CONCLUSIONS
The in vivo light-adaptability of C-PC highlights this protein’s conformational flexibility and controllability, which set the stage for its in vitro utilization as tunable photoactive systems. However, manipulating C-PC in vitro requires an in-depth understanding of its behavior in solution, especially the structural and photochemical responsiveness to pre-specified changes in environments. Among the common environmental stimuli proteins can respond to (e.g., pH, ionic strength, concentration, and heat), we paid particular attention to pH changes, which drive large-scale but reversible conformational changes in C-PC. By controlling the solution pH, the tendency of isolated C-PC to disintegrate became suppressible, thus we were able to construct and characterize the diverse C-PC structures in solution using SEC–SAXS. These differently assembled structures were used as starting materials to investigate the assembly–disassembly pathways and the protein–chromophore regulatory mechanisms related to phycobiliproteins’ light acclimatory responses.
Our results unveiled that C-PC protein matrix’s mode of assembly governs the photochemical functions of the embedded tetrapyrrole chromophores, which is achieved via multiple mechanisms. The spectroscopic behavior of the chromophores varied according to the protein assembly state, mirroring the regulatory role of the protein matrix on chromophores’ conformations and pairwise interactions. With the chromophores functioning as photosensitizers, UV irradiation of C-PC induced pH-dependent 1O2 generation, which in turn initiated radical chain reactions with the chromophores and nearby amino acid residues. These events were made evident by the observations of residue-derived radicals, photo-crosslinking via dityrosine linkages, as well as the NaN3-inhibitable radical formation and photobleaching. However, although the chromophores were shown to act as both the source and target of the photogenerated radicals, it was the protein matrix that actually dictated C-PC’s light responsiveness. Indeed, the protein matrix’s mode of assembly determined not only the predominant photosensitized reactions, but also the distinct conformational changes under irradiation, that is, the irradiation-induced unfolding at pH 3.0, 7.0 and 9.0 versus the photo-crosslinking and aggregation at pH 5.0.
Additionally, of all the pH conditions investigated, it was the highly assembled C-PC at pH 5.0 that unexpectedly displayed both the highest 1O2 yield and the highest color stability under irradiation. These observations highlighted another potential protein–chromophore regulatory mechanism, that is, the protein matrix can impart photoprotection on the embedded chromophores via the assembly–disassembly pathways. Cyanobacteria or other light-harvesting organisms are known to have evolved photoprotective mechanisms to mitigate the detrimental effects of sunlight on their photosystems.41, 42 Such mechanisms, however, usually require synergy between phycobiliproteins and other in vivo substances, such as carotenoids.43 Our results demonstrated a self-photoprotective mechanism on chromophores imparted solely by phycobiliproteins’ protein matrix, which has not been previously reported. In fact, high-order protein assemblies are abundant in cells that are frequently exposed to stressed environments.44 In these cells, protein assembly or disassembly can be triggered by many in vivo signaling pathways that alters cellular conditions, such as ion concentration, osmotic pressure, pH, and heat, etc. Transitions of protein assembly states enable rapid and reversable changes in protein function and thus fast adaptation of the cells to external fluctuations. In terms of phycobiliproteins, the weak protein intermolecular interactions dictate that the high assemblies can rapidly disintegrate under undesirable conditions, like nutrient stress45 or excess light,46, 47 in order to avoid unnecessary photosynthesis and energy loss. In this view, high phycobiliprotein assemblies can promote cellular survival under extreme environments via flexible assembly–disassembly transitions.
Although radical generation during photosynthesis is an unwanted side reaction in vivo, which leads to photo-oxidative stress and cell death, it enables C-PC to be photoactivated for potential chemical and biomedical applications. Toward this end, this work dissected the intriguing UV-activable and pH-tunable photoactivity of C-PC, featuring remarkably enhanced 1O2 yields and fluorescence emissions at pH 5.0 with respect to C-PC at other pH values. From an application point of view, the enhanced activation of C-PC in weakly acidic environments (pH 5.0) is of particular biomedical significance, since tumor microenvironments are characterized by similar acidity, for example, the pH of lysosome is 4.5−5.0,48 relative to healthy tissues, a trait typically exploited in cancer treatment.49 In this sense, C-PC could be utilized in the design of novel pH-responsive fluorescent imaging probes50, 51 or 1O2 generators52 to boost photodynamic activities at acidic tumor sites while minimizing phototoxicity to non-target healthy tissues.53 Previously, tetrapyrrole-containing proteins, naturally derived or synthetic, have been used to fabricate photosensitizers51, 54, 55 and fluorescent probes.56–58 In most of these studies, however, external carriers, chemotherapeutic agents, or fluorophores had to be attached59 in order to gain ROS formation and tumor selectivity—that is, the tetrapyrroles and phycobiliproteins merely functioned as photosensory pigments (light absorbers and emitters). While the referenced cases demonstrate the potential photodynamic applications, the intrinsic photoactivity and tunability of phycobiliproteins, offered by their modular and flexible protein architecture, are overlooked and underutilized. Therefore, it is envisaged that with further development, the remarkable pH-responsiveness, photoactivity, and conformational flexibility of C-PC demonstrated in this work, along with this protein’s inherent biocompatibility and wide availability, will open new routes for its applications in cancer diagnosis, photodynamic therapy (PDT), drug delivery, and oxidation catalysis60–62. Our future work will be directed toward a series of application-centered studies based on the identified tunability and photoactivity of C-PC. First, in vivo and in vitro tests will be conducted to assess the intracellular uptake, therapeutic efficacy, and tumor cell selectivity of C-PC under pH controls. Second, we seek to further explore how C-PC respond to other physical or chemical stimuli; priorities will be given to the investigations of protein concentration, ionic strength, surfactants, and redox agents—in light of the identified sensitivity of C-PC to electrostatic/redox signals and solution conditions. For example, changes in protein concentration63 may be used in conjunction with the pH effect to further populate the high C-PC assemblies at pH 5.0 and thus boost the photosensitized reactions. Similar extension of the current work by applying multiple physical or chemical stimuli may reveal phycobiliprotein systems with more versatile responsiveness, thus, propelling the applications in exciting new directions.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by USDA NIFA award 2020-67017-31274 to A.A. We thank the Cornell High Energy Synchrotron Source (CHESS) and National Biomedical Center for Advanced ESR Technologies (ACERT) for access to data collection facilities. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-1829070, and the Macromolecular Diffraction at CHESS (MacCHESS) resource is supported by NIH/NIGMS award GM-124166. ACERT is supported by the NIH/NIGMS award P41 GM103521. We thank Dr. Boris Dzikovski for assistance running the EPR experiments.
Footnotes
SUPPORTING INFORMATION
Supporting Information includes Supporting Experimental Section, Supporting Text, 11 figures, and 3 tables.
REFERENCES
- 1.Wiltbank LB; Kehoe DM, Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol 2019, 17 (1), 37–50. [DOI] [PubMed] [Google Scholar]
- 2.Scholes GD; Fleming GR; Olaya-Castro A; van Grondelle R, Lessons from nature about solar light harvesting. Nat. Chem 2011, 3 (10), 763–774. [DOI] [PubMed] [Google Scholar]
- 3.Tamara S; Hoek M; Scheltema RA; Leney AC; Heck AJR, A Colorful Pallet of B-Phycoerythrin Proteoforms Exposed by a Multimodal Mass Spectrometry Approach. Chem 2019, 5 (5), 1302–1317. [Google Scholar]
- 4.Li Y; Zhang Z; Paciulli M; Abbaspourrad A, Extraction of phycocyanin—A natural blue colorant from dried spirulina biomass: Influence of processing parameters and extraction techniques. J. Food Sci 2020, 85 (3), 727–735. [DOI] [PubMed] [Google Scholar]
- 5.Acerbo AS; Cook MJ; Gillilan RE, Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J. Synchrotron Radiat 2015, 22 (1), 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skou S; Gillilan RE; Ando N, Synchrotron-based small-angle X-ray scattering of proteins in solution. Nat. Protoc 2014, 9 (7), 1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen SS; Toft KN; Snakenborg D; Jeppesen MG; Jacobsen JK; Vestergaard B; Kutter JP; Arleth L, BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Crystallogr 2009, 42 (5), 959–964. [Google Scholar]
- 8.Svergun DI, Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr 1992, 25 (4), 495–503. [Google Scholar]
- 9.Schneidman-Duhovny D; Hammel M; Sali A, FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010, 38 (suppl_2), W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneidman-Duhovny D; Hammel M; Tainer John A.; Sali A, Accurate SAXS Profile Computation and its Assessment by Contrast Variation Experiments. Biophys. J 2013, 105 (4), 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X-Q; Li L-N; Chang W-R; Zhang J-P; Gui L-L; Guo B-J; Liang D-C, Structure of C-phycocyanin from Spirulina platensis at 2.2 A resolution: a novel monoclinic crystal form for phycobiliproteins in phycobilisomes. Acta Crystallogr. Sect. D. Biol. Crystallogr 2001, 57 (6), 784–792. [DOI] [PubMed] [Google Scholar]
- 12.Svergun DI; Petoukhov MV; Koch MHJ, Determination of Domain Structure of Proteins from X-Ray Solution Scattering. Biophys. J 2001, 80 (6), 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svergun DI, Restoring Low Resolution Structure of Biological Macromolecules from Solution Scattering Using Simulated Annealing. Biophys. J 1999, 76 (6), 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petoukhov MV; Franke D; Shkumatov AV; Tria G; Kikhney AG; Gajda M; Gorba C; Mertens HDT; Konarev PV; Svergun DI, New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr 2012, 45 (2), 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrodinger LLC, The PyMOL Molecular Graphics System, Version 1.8. 2015.
- 16.Stoll S; Schweiger A, EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson 2006, 178 (1), 42–55. [DOI] [PubMed] [Google Scholar]
- 17.Ma J; You X; Sun S; Wang X; Qin S; Sui S-F, Structural basis of energy transfer in Porphyridium purpureum phycobilisome. Nature 2020, 579 (7797), 146–151. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J; Ma J; Liu D; Qin S; Sun S; Zhao J; Sui S-F, Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 2017, 551 (7678), 57–63. [DOI] [PubMed] [Google Scholar]
- 19.Padyana AK; Bhat VB; Madyastha KM; Rajashankar KR; Ramakumar S, Crystal Structure of a Light-Harvesting Protein C-Phycocyanin from Spirulina platensis. Biochem. Biophys. Res. Commun 2001, 282 (4), 893–898. [DOI] [PubMed] [Google Scholar]
- 20.Stec B; Troxler RF; Teeter MM, Crystal Structure of C-Phycocyanin from Cyanidium caldarium Provides a New Perspective on Phycobilisome Assembly. Biophys. J 1999, 76 (6), 2912–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marx A; Adir N, Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly. Biochim. Biophys. Acta 2013, 1827 (3), 311–318. [DOI] [PubMed] [Google Scholar]
- 22.Lan H; Lv G; Wen Y; Mao Y; Huang C; Yi T, The synergic control of photocyclization and fluorescence in a pH-gated photochromic system. Dyes Pigm. 2016, 131, 18–23. [Google Scholar]
- 23.Zhu J-L; Xu L; Ren Y-Y; Zhang Y; Liu X; Yin G-Q; Sun B; Cao X; Chen Z; Zhao X-L; Tan H; Chen J; Li X; Yang H-B, Switchable organoplatinum metallacycles with high quantum yields and tunable fluorescence wavelengths. Nat. Commun 2019, 10 (1), 4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P; Sun M; Xu Z; Liu X; Zhao W; Gao W, Site-Selective in Situ Growth-Induced Self-Assembly of Protein–Polymer Conjugates into pH-Responsive Micelles for Tumor Microenvironment Triggered Fluorescence Imaging. Biomacromolecules 2018, 19 (11), 4472–4479. [DOI] [PubMed] [Google Scholar]
- 25.Li Z; Wang Y; Li M; Chen H; Xie Y; Li P; Guo H; Ya H, Solvent-dependent and visible light-activated NIR photochromic dithienylethene modified by difluoroboron β-diketonates as fluorescent turn-on pH sensor. Dyes Pigm. 2019, 162, 339–347. [Google Scholar]
- 26.Cai W; Gentle IR; Lu GQ; Zhu J-J; Yu A, Mesoporous Silica Templated Biolabels with Releasable Fluorophores for Immunoassays. Anal. Chem 2008, 80 (14), 5401–5406. [DOI] [PubMed] [Google Scholar]
- 27.Pi X; Zhao S; Wang W; Liu D; Xu C; Han G; Kuang T; Sui S-F; Shen J-R, The pigment-protein network of a diatom photosystem II–light-harvesting antenna supercomplex. Science 2019, 365 (6452), aax4406. [DOI] [PubMed] [Google Scholar]
- 28.Adir N, Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynthesis Res. 2005, 85 (1), 15–32. [DOI] [PubMed] [Google Scholar]
- 29.Collier JL; Grossman AR, A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994, 13 (5), 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx A; Adir N, Structural characteristics that stabilize or destabilize different assembly levels of phycocyanin by urea. Photosynthesis Res. 2014, 121 (1), 87–93. [DOI] [PubMed] [Google Scholar]
- 31.Henke P; Dolanský J; Kubát P; Mosinger J, Multifunctional Photosensitizing and Biotinylated Polystyrene Nanofiber Membranes/Composites for Binding of Biologically Active Compounds. ACS Appl. Mater. Interfaces 2020, 12 (16), 18792–18802. [DOI] [PubMed] [Google Scholar]
- 32.Hirakawa K; Umemoto H; Kikuchi R; Yamaguchi H; Nishimura Y; Arai T; Okazaki S; Segawa H, Determination of Singlet Oxygen and Electron Transfer Mediated Mechanisms of Photosensitized Protein Damage by Phosphorus(V)porphyrins. Chem. Res. Toxicol 2015, 28 (2), 262–267. [DOI] [PubMed] [Google Scholar]
- 33.He J; Wang Y; Missinato MA; Onuoha E; Perkins LA; Watkins SC; St Croix CM; Tsang M; Bruchez MP, A genetically targetable near-infrared photosensitizer. Nat. Methods 2016, 13 (3), 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Mascio P; Martinez GR; Miyamoto S; Ronsein GE; Medeiros MHG; Cadet J, Singlet Molecular Oxygen Reactions with Nucleic Acids, Lipids, and Proteins. Chem. Rev 2019, 119 (3), 2043–2086. [DOI] [PubMed] [Google Scholar]
- 35.Rinalducci S; Pedersen JZ; Zolla L, Generation of reactive oxygen species upon strong visible light irradiation of isolated phycobilisomes from Synechocystis PCC 6803. Biochim. Biophys. Acta 2008, 1777 (5), 417–424. [DOI] [PubMed] [Google Scholar]
- 36.Spikes JD, Photosensitization. In The Science of Photobiology, Smith KC, Ed. Springer, Boston, MA: Boston, MA, 1989; pp 79–110. [Google Scholar]
- 37.Müller F; Graziadei A; Rappsilber J, Quantitative Photo-crosslinking Mass Spectrometry Revealing Protein Structure Response to Environmental Changes. Anal. Chem 2019, 91 (14), 9041–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kampf CJ; Liu F; Reinmuth-Selzle K; Berkemeier T; Meusel H; Shiraiwa M; Pöschl U, Protein Cross-Linking and Oligomerization through Dityrosine Formation upon Exposure to Ozone. Environ. Sci. Technol 2015, 49 (18), 10859–10866. [DOI] [PubMed] [Google Scholar]
- 39.Tan R; Shin J; Heo J; Cole BD; Hong J; Jang Y, Tuning the Structural Integrity and Mechanical Properties of Globular Protein Vesicles by Blending Crosslinkable and NonCrosslinkable Building Blocks. Biomacromolecules 2020, 21 (10), 4336–4344. [DOI] [PubMed] [Google Scholar]
- 40.Kannaujiya VK; Rahman A; Adinath; Sundaram S; Sinha RP, Structural and functional dynamics of tyrosine amino acid in phycocyanin of hot-spring cyanobacteria: A possible pathway for internal energy transfer. Gene Rep. 2016, 5, 83–91. [Google Scholar]
- 41.Demmig-Adams B; Adams WW, Photoprotection and Other Responses of Plants to High Light Stress. Annu. Rev. Plant Physiol. Plant Mol. Biol 1992, 43 (1), 599–626. [Google Scholar]
- 42.Ruban AV; Johnson MP; Duffy CDP, The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta 2012, 1817 (1), 167–181. [DOI] [PubMed] [Google Scholar]
- 43.Bao H; Melnicki MR; Pawlowski EG; Sutter M; Agostoni M; Lechno-Yossef S; Cai F; Montgomery BL; Kerfeld CA, Additional families of orange carotenoid proteins in the photoprotective system of cyanobacteria. Nat. Plants 2017, 3 (8), 17089. [DOI] [PubMed] [Google Scholar]
- 44.Rabouille C; Alberti S, Cell adaptation upon stress: the emerging role of membrane-less compartments. Curr. Opin. Cell Biol 2017, 47, 34–42. [DOI] [PubMed] [Google Scholar]
- 45.Nagarajan A; Zhou M; Nguyen AY; Liberton M; Kedia K; Shi T; Piehowski P; Shukla A; Fillmore TL; Nicora C, Proteomic Insights into Phycobilisome Degradation, A Selective and Tightly Controlled Process in The Fast-Growing Cyanobacterium Synechococcus elongatus UTEX 2973. Biomolecules 2019, 9 (8), 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamary E; Kiss V; Nevo R; Adam Z; Bernát G; Rexroth S; Rögner M; Reich Z, Structural and functional alterations of cyanobacterial phycobilisomes induced by high-light stress. Biochim. Biophys. Acta 2012, 1817 (2), 319–327. [DOI] [PubMed] [Google Scholar]
- 47.Lao K; Glazer AN, Ultraviolet-B photodestruction of a light-harvesting complex. Proc. Natl. Acad. Sci. USA 1996, 93 (11), 5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng T; Lim S, Trimer-Based Design of pH-Responsive Protein Cage Results in Soluble Disassembled Structures. Biomacromolecules 2011, 12 (9), 3131–3138. [DOI] [PubMed] [Google Scholar]
- 49.Tian J; Ding L; Xu H-J; Shen Z; Ju H; Jia L; Bao L; Yu J-S, Cell-Specific and pH-Activatable Rubyrin-Loaded Nanoparticles for Highly Selective Near-Infrared Photodynamic Therapy against Cancer. J. Am. Chem. Soc 2013, 135 (50), 18850–18858. [DOI] [PubMed] [Google Scholar]
- 50.Cheng H-B; Zhang Y-M; Liu Y; Yoon J, Turn-On Supramolecular Host-Guest Nanosystems as Theranostics for Cancer. Chem 2019, 5 (3), 553–574. [Google Scholar]
- 51.Du S-W; Zhang L-K; Han K; Chen S; Hu Z; Chen W; Hu K; Yin L; Wu B; Guan Y-Q, Combined Phycocyanin and Hematoporphyrin Monomethyl Ether for Breast Cancer Treatment via Photosensitizers Modified Fe3O4 Nanoparticles Inhibiting the Proliferation and Migration of MCF-7 Cells. Biomacromolecules 2018, 19 (1), 31–41. [DOI] [PubMed] [Google Scholar]
- 52.Lu C; Zhang C; Wang P; Zhao Y; Yang Y; Wang Y; Yuan H; Qu S; Zhang X; Song G; Pu K, Light-free Generation of Singlet Oxygen through Manganese-Thiophene Nanosystems for pH-Responsive Chemiluminescence Imaging and Tumor Therapy. Chem 2020, 6 (9), 2314–2334. [Google Scholar]
- 53.Luby BM; Walsh CD; Zheng G, Advanced Photosensitizer Activation Strategies for Smarter Photodynamic Therapy Beacons. Angew. Chem 2019, 58 (9), 2558–2569. [DOI] [PubMed] [Google Scholar]
- 54.Wan D-H; Ma X-Y; Lin C; Zhu D-H; Li X; Zheng B-Y; Li J; Ke M-R; Huang J-D, Noncovalent Indocyanine Green Conjugate of C-Phycocyanin: Preparation and Tumor-Associated Macrophages-Targeted Photothermal Therapeutics. Bioconjug. Chem 2020, 31 (5), 1438–1448. [DOI] [PubMed] [Google Scholar]
- 55.Pu Y; Wei M; Witkowski A; Krzywda M; Wang Y; Li W, A hybrid biomaterial of biosilica and C-phycocyanin for enhanced photodynamic effect towards tumor cells. Biochem. Biophys. Res. Commun 2020, 533 (3), 573–579. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J; Campbell RE; Ting AY; Tsien RY, Creating new fluorescent probes for cell biology. Nature Reviews Molecular Cell Biology 2002, 3 (12), 906–918. [DOI] [PubMed] [Google Scholar]
- 57.Fuenzalida Werner JP; Mishra K; Huang Y; Vetschera P; Glasl S; Chmyrov A; Richter K; Ntziachristos V; Stiel AC, Structure-Based Mutagenesis of Phycobiliprotein smURFP for Optoacoustic Imaging. ACS Chem. Biol 2019, 14 (9), 1896–1903. [DOI] [PubMed] [Google Scholar]
- 58.Ghosh T; Mondal A; Vyas A; Mishra S, A ‘one–tube’ synthesis of a selective fluorescence ‘turn off/on’ DNA probe based on a C-phycocyanin-graphene oxide (CPC-GO) bio composite. Int. J. Biol. Macromol 2020, 163, 977–984. [DOI] [PubMed] [Google Scholar]
- 59.Dognini P; Coxon CR; Alves WA; Giuntini F, Peptide-Tetrapyrrole Supramolecular Self-Assemblies: State of the Art. Molecules 2021, 26 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du B; Jia S; Wang Q; Ding X; Liu Y; Yao H; Zhou J, A Self-Targeting, Dual ROS/pH-Responsive Apoferritin Nanocage for Spatiotemporally Controlled Drug Delivery to Breast Cancer. Biomacromolecules 2018, 19 (3), 1026–1036. [DOI] [PubMed] [Google Scholar]
- 61.Chandna S; Thakur NS; Kaur R; Bhaumik J, Lignin–Bimetallic Nanoconjugate Doped pH-Responsive Hydrogels for Laser-Assisted Antimicrobial Photodynamic Therapy. Biomacromolecules 2020, 21 (8), 3216–3230. [DOI] [PubMed] [Google Scholar]
- 62.Dag A; Cakilkaya E; Omurtag Ozgen PS; Atasoy S; Yigit Erdem G; Cetin B; Çavuş Kokuroǧlu A; Gürek AG, Phthalocyanine-Conjugated Glyconanoparticles for Chemo-photodynamic Combination Therapy. Biomacromolecules 2021, 22 (4), 1555–1567. [DOI] [PubMed] [Google Scholar]
- 63.Eisenberg I; Harris D; Levi-Kalisman Y; Yochelis S; Shemesh A; Ben-Nissan G; Sharon M; Raviv U; Adir N; Keren N; Paltiel Y, Concentration-based self-assembly of phycocyanin. Photosynthesis Res. 2017, 134 (1), 39–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


