Abstract
The genetic diversity among a worldwide collection of 120 strains of Ralstonia solanacearum was assessed by restriction fragment length polymorphism (RFLP) analysis of amplified fragments from the hrp gene region. Five amplified fragments appeared to be specific to R. solanacearum. Fifteen different profiles were identified among the 120 bacterial strains, and a hierarchical cluster analysis distributed them into eight clusters. Each cluster included strains belonging to a single biovar, except for strains of biovars 3 and 4, which could not be separated. However, the biovar 1 strains showed rather extensive diversity since they were distributed into five clusters whereas the biovar 2 and the biovar 3 and 4 strains were gathered into one and two clusters, respectively. PCR-RFLP analysis of the hrp gene region confirmed the results of previous studies which split the species into an “Americanum” division including biovar 1 and 2 strains and an “Asiaticum” division including biovar 3 and 4 strains. However, the present study showed that most of the biovar 1 strains, originating from African countries (Reunion Island, Madagascar, Zimbabwe, and Angola) and being included in a separate cluster, belong to the “Asiaticum” rather than to the “Americanum” division. These African strains could thus have evolved separately from other biovar 1 strains originating from the Americas.
Ralstonia (formerly Pseudomonas) solanacearum (E. F. Smith) Yabuuchi et al. (47) is the causal agent of bacterial wilt, a severe and devastating plant disease in most tropical and subtropical and some warm temperate areas (22). Moreover, it can also occur in cool temperate areas (9, 33). Many economically important food crops such as potatoes, tomatoes, and bananas are affected. The disease was recorded on several hundred plant species distributed in more than 50 families (23). The species R. solanacearum is a complex taxonomic unit in which strains display an important diversity at different levels (physiological, serological, genetic characteristics, and host range). In order to describe this intraspecific variability, several systems of classification have been proposed. Thus, the species was subdivided into five races according to its host range (7, 25, 35) and into six biovars based on the utilization of three disaccharides and three hexose alcohols (21, 24, 25). Fatty acid analysis (26, 42) and protein profiling (15) were also performed but did not further clarify the relationships among R. solanacearum strains. Restriction fragment length polymorphism (RFLP) analysis (involving nine probes, seven of which encode information required for virulence and the hypersensitive response) (12–14) has provided a new classification scheme dividing the species into 46 groups in relation to geographic origin of strains and sometimes host range. The species was then separated into two major groups, the “Asiaticum” and the “Americanum” divisions, which regrouped strains from Asia and America, respectively. Further investigations comparing sequences of 16S rRNA (30, 40, 43) or using PCR amplification with tRNA consensus primers (39) supported the separation according to geographic origin.
Only a few strains originating from Africa, and only one from Reunion Island (21), were included in these previous studies. However, strains related to the three major biovars (1, 2, and 3) were isolated from various crops in Reunion (17). The aim of our study was to assess the genetic diversity within the local populations of R. solanacearum. Since we also wanted to develop molecular tools for the identification and detection of R. solanacearum biovars, we used the PCR-RFLP procedure to analyze the diversity. Recently, several authors have successfully performed PCR-RFLP analysis to assess genetic diversity among bacterial species (27, 28, 31, 44, 45). The hrp (hypersensitive reaction and pathogenicity) gene region, which is required by many phytopathogenic bacteria to produce symptoms on susceptible hosts and a hypersensitive reaction on resistant hosts or on nonhosts (1, 3, 4, 6, 18, 29), was explored for studying the variability within a collection of 120 strains isolated from different hosts over the five continents and belonging to biovars 1, 2, 3, and 4.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains studied (Table 1) included diverse strains of R. solanacearum with special attention to those isolated from Africa (51 strains including 28 from Reunion Island) and strains belonging to more or less closely related species (Ralstonia pickettii, Ralstonia eutropha, Burkholderia cepacia, Pseudomonas spp., Xanthomonas spp., Erwinia chrysanthemi, and Clavibacter michiganensis subsp. michiganensis). Identification of R. solanacearum strains at the biovar level was performed by using a modification of Hayward’s method (21). All strains were stored on beads in cryovials at −80°C (Microbank Pro-Lab Diagnostics). Nutrient broth cultures were grown for 24 h on a rotary shaker (150 rpm) at 28°C. Bacteria were cultivated either (R. solanacearum) on a modified Granada and Sequeira medium (19) (tryptone, 1 g/liter; peptone, 10 g/liter; agar, 18 g/liter; glycerine, 6.3 ml/liter; crystal violet, 0.002 g/liter; polymyxin sulfate, 0.01 g/liter; tyrothricin, 0.02 g/liter; chloramphenicol, 0.005 g/liter; triphenyltetrazolium chloride, 0.025 g/liter; propiconazole, 0.4 ml/liter; penicillin, 20 U/liter; pH 7.2) or (other species) on YPGA medium (yeast extract, 7 g/liter; peptone, 7 g/liter; glucose, 7 g/liter; agar, 15 g/liter; pH 7.2) and incubated for 3 days at 28°C.
TABLE 1.
Strains of R. solanacearum used in this study
| Strain | Other designation | Sourcea | Geographic origin | Host | Biovar | PCR-RFLP cluster and subclusterb |
|---|---|---|---|---|---|---|
| JS796 | CFBP1180 | B | Puerto Rico | Lycopersicon esculentum | 1 | Ia |
| JS927 | NCPPB1225 | C | Puerto Rico | Lycopersicon esculentum | 1 | |
| JS833 | UW30 | E | Trinidad and Tobago | Lycopersicon esculentum | 1 | |
| JR659 | CFBP2047c | B | United States | Lycopersicon esculentum | 1 | Ib |
| JS783 | CFBP1036 | B | United States | Lycopersicon esculentum | 1 | |
| JS967 | ICMP7963 | D | Kenya | Solanum tuberosum | 1 | |
| JS831 | UW26 | E | United States | Lycopersicon esculentum | 1 | Ic |
| JS768 | CFBP767 | B | Guadeloupe | Solanum tuberosum | 1 | II |
| JS794 | CFBP1162 | B | Guadeloupe | Nicotiana tabacum | 1 | |
| JS777 | CFBP770 | B | Guadeloupe | Lycopersicon esculentum | 1 | |
| JS733 | CFBP2478 | B | Guadeloupe | Lycopersicon esculentum | 1 | |
| JS784 | CFBP1036 | B | Martinique | Lycopersicon esculentum | 1 | |
| JS734 | CFBP2972 | B | Martinique | Solanum tuberosum | 1 | |
| JS716 | CFBP705 | B | Guyana | Lycopersicon esculentum | 1 | |
| JS837 | UW90 | E | Brazil | Nicotiana tabacum | 1 | |
| JS838 | UW275 | E | Costa Rica | Melampodium perfoliatum | 1 | |
| JS830 | UW256 | E | Costa Rica | Solanum tuberosum | 1 | |
| JS770 | CFBP712 | B | Burkina Faso | Solanum melongena | 1 | |
| JS779 | CFBP715 | B | Burkina Faso | Lycopersicon esculentum | 1 | |
| JS912 | CFBP3057 | B | Burkina Faso | Lycopersicon esculentum | 1 | |
| JS740 | CFBP1415 | B | Colombia | Solanum tuberosum | 1 | III |
| JS788 | CFBP1412 | B | Columbia | Musa sp. cv. plantain | 1 | |
| JS847 | CFBP1419 | B | Costa Rica | Musa sp. | 1 | |
| JT509 | A | Reunion Island | Lycopersicon esculentum | 2 | IV | |
| JT515 | A | Reunion Island | Lycopersicon esculentum | 2 | ||
| JT512 | A | Reunion Island | Lycopersicon esculentum | 2 | ||
| JT510 | A | Reunion Island | Solanum tuberosum | 2 | ||
| JT511 | A | Reunion Island | Solanum tuberosum | 2 | ||
| JT513 | A | Reunion Island | Solanum tuberosum | 2 | ||
| JT514 | A | Reunion Island | Solanum tuberosum | 2 | ||
| JT516 | A | Reunion Island | Solanum tuberosum | 2 | ||
| JS780 | CFBP2148 | B | Reunion Island | Solanum tuberosum | 2 | |
| JS931 | NCPPB1049 | C | Kenya | Lycopersicon esculentum | 2 | |
| JS948 | NCPPB2088 | C | Nigeria | Solanum tuberosum | 2 | |
| JS780 | CFBP2148 | B | Reunion Island | Solanum tuberosum | 2 | |
| JS931 | NCPPB1049 | C | Kenya | Lycopersicon esculentum | 2 | |
| JS948 | NCPPB2088 | C | Nigeria | Solanum tuberosum | 2 | |
| JS905 | CFBP3582 | B | Egypt | Solanum tuberosum | 2 | |
| JS939 | NCPPB1824 | C | Egypt | Solanum tuberosum | 2 | |
| JS908 | CFBP3525 | B | Morocco | Solanum tuberosum | 2 | |
| JS902 | CFBP3581 | B | France | Solanum tuberosum | 2 | |
| JS900 | CFBP3671 | B | France | Lycopersicon esculentum | 2 | |
| JS898 | CFBP3672 | B | France | Solanum tuberosum | 2 | |
| JS895 | CFBP3673 | B | France | Solanum tuberosum | 2 | |
| JS942 | NCPPB1019 | C | Portugal | Lycopersicon esculentum | 2 | |
| JS887 | CFBP3785 | B | Portugal | Unknown | 2 | |
| JS930 | NCPPB1489 | C | Madeira | Solanum tuberosum | 2 | |
| JS937 | NCPPB1789 | C | Greece | Solanum tuberosum | 2 | |
| JS935 | NCPPB339 | C | Israel | Unknown | 2 | |
| JS907 | CFBP3858 | B | The Netherlands | Solanum tuberosum | 2 | |
| JS928 | NCPPB2797 | C | Sweden | Solanum dulcamara | 2 | |
| JS929 | NCPPB2505 | C | Sweden | Solanum tuberosum | 2 | |
| JS792 | CFBP1810 | B | Haiti | Solanum tuberosum | 2 | |
| JS943 | NCPPB613 | C | Brazil | Solanum tuberosum | 2 | |
| JS758 | CFBP1420 | B | Colombia | Solanum phureja | 2 | |
| JS774 | CFBP1414 | B | Colombia | Solanum tuberosum | 2 | |
| JS897 | CFBP3103 | B | Peru | Solanum tuberosum | 2 | |
| JS926 | NCPPB1331 | C | India | Solanum tuberosum | 2 | |
| JS925 | NCPPB1323 | C | Sri Lanka | Solanum tuberosum | 2 | |
| JS932 | NCPPB1614 | C | Malaysia | Solanum tuberosum | 2 | |
| JS738 | CFBP1413 | B | Australia | Solanum tuberosum | 2 | |
| JS737 | CFBP1417 | B | Australia | Solanum tuberosum | 2 | |
| JS775 | CFBP1409 | B | Honduras | Musa sp. | 1 | V |
| JS730 | CFBP1482 | B | Panama | Musa sp. | 1 | |
| JS791 | CFBP1416 | B | Costa Rica | Musa sp. cv. plantain | 1 | |
| JS793 | CFBP1183 | B | Costa Rica | Heliconia sp. | 1 | |
| JS781 | CFBP1185 | B | Japan | Lycopersicon esculentum | 3 | VIa |
| JS945 | MAFF301860 | G | Japan | Capsicum annuum | 3 | |
| JS941 | NCPPB3190 | C | Malaysia | Lycopersicon esculentum | 3 | |
| JS836 | UW8 | E | Costa Rica | Eupatorium odoratum | 3 | |
| JS842 | UW119 | E | Costa Rica | Solanum tuberosum | 3 | |
| JS940 | NCPPB500 | C | Mauritius | Vicia faba | 3 | |
| JS944 | NCPPB501 | C | Mauritius | Brassica oleracea | 3 | |
| JS954 | NCPPB502 | C | Mauritius | Casuarina equisetifolia | 3 | |
| JS955 | NCPPB503 | C | Mauritius | Dahlia sp. | 3 | |
| JS834 | UW151 | E | Australia | Zingiber officinale | 4 | |
| JS835 | UW360 | E | China | Morus alba | 4 | |
| JS839 | UW369 | E | China | Arachis hypogaea | 4 | |
| JS832 | UW378 | E | China | Olea sp. | 4 | |
| JT517 | A | Reunion Island | Pelargonium asperum | 3 | VIb | |
| JT520 | A | Reunion Island | Pelargonium asperum | 3 | ||
| JT519 | A | Reunion Island | Pelargonium asperum | 3 | ||
| JS766 | CFBP726 | B | Reunion Island | Solanum melongena | 3 | |
| JT518 | A | Reunion Island | Solanum melongena | 3 | ||
| JT523 | A | Reunion Island | Solanum tuberosum | 3 | ||
| JS778 | CFBP2041 | B | Reunion Island | Solanum tuberosum | 3 | |
| JT521 | A | Reunion Island | Lycopersicon esculentum | 3 | ||
| JT522 | A | Reunion Island | Lycopersicon esculentum | 3 | ||
| JT524 | A | Reunion Island | Lycopersicon esculentum | 3 | ||
| JS841 | MAFF301418 | G | Japan | Lycopersicon esculentum | 4 | |
| JS933 | UW74 | E | Sri Lanka | Solanum tuberosum | 4 | |
| JS719 | CFBP2970 | B | Martinique | Capsicum annuum | 3 | VIc |
| JS715 | CFBP2976 | B | Martinique | Ensete ventricosum | 3 | |
| JS718 | CFBP2480 | B | Guadeloupe | Solanum melongena | 3 | |
| JS729 | CFBP2965 | B | Guadeloupe | Solanum melongena | 3 | |
| JS722 | CFBP1813 | B | Guyana | Solanum melongena | 3 | |
| JS753 | GMI1000 | F | Guyana | Lycopersicon esculentum | 3 | |
| JS764 | GMI1336 | F | hrp mutant of GMI1000 | Lycopersicon esculentum | 3 | |
| JS843 | UW130 | E | Peru | Lycopersicon esculentum | 3 | |
| JS772 | CFBP707 | B | Tahiti | Lycopersicon esculentum | 3 | |
| JS773 | CFBP1960 | B | Algeria | Capsicum annuum | 3 | |
| JS759 | CFBP1168 | B | Trinidad and Tobago | Musa sp. | 3 | |
| JS840 | UW147 | E | Australia | Nicotiana tabacum | 3 | |
| JS947 | NCPPB1123 | C | Papua New Guinea | Lycopersicon esculentum | 4 | |
| JS953 | MAFF301552 | G | Japan | Lycopersicon esculentum | 3 | VId |
| JS936 | NCPPB3181 | C | Gambia | Solanum nicanum | 3 | VIe |
| JS950 | NCPPB1018 | C | Angola | Solanum tuberosum | 1 | VIIa |
| JT526 | A | Reunion Island | Pelargonium asperum | 1 | VIIb | |
| JT527 | A | Reunion Island | Pelargonium asperum | 1 | ||
| JT529 | A | Reunion Island | Pelargonium asperum | 1 | ||
| JT530 | A | Reunion Island | Pelargonium asperum | 1 | ||
| JT528 | A | Reunion Island | Solanum tuberosum | 1 | ||
| JT531 | A | Reunion Island | Solanum tuberosum | 1 | ||
| JS756 | CFBP2146 | B | Reunion Island | Pelargonium capitatum | 1 | |
| JT525 | A | Reunion Island | Pelargonium asperum | 1 | ||
| JT532 | A | Reunion Island | Unknown | 1 | ||
| JS767 | CFBP734 | B | Madagascar | Solanum tuberosum | 1 | |
| JS949 | NCPPB332 | C | Zimbabwe | Solanum tuberosum | 1 | |
| JS966 | ICMP748 | D | Zimbabwe | Solanum tuberosum | 1 | |
| JS946 | NCPPB283 | C | Zimbabwe | Solanum panduraforme | 1 | |
| JS951 | NCPPB505 | C | Zimbabwe | Symphytum sp. | 1 | |
| JS952 | NCPPB342 | C | Zimbabwe | Nicotiana tabacum | 1 | |
| JS934 | MAFF301558 | G | Japan | Solanum tuberosum | 3 | VIII |
Strains were contributed as follows: A, this study; B, Collection Française de Bactéries Phytopathogènes, Angers, France; C, National Collection of Plant Pathogenic Bacteria, Harpenden, United Kingdom; D, International Collection of Microorganisms from Plants, Auckland, New Zealand; E. D. Cook and L. Sequeira, Department of Plant Pathology, University of Wisconsin—Madison, Madison; F, M. Arlat and P. Barberis, CNRS-INRA, Auzeville, Castanet-Tolosan Cedex, France; G, K. Tsuchiya, Ministry of Agriculture, Forestry and Fisheries, National Institute of Agrobiological Resources, Tokyo, Japan.
PCR-RFLP clusters and subclusters were defined by this study.
Type strain.
DNA extraction.
DNA was extracted from R. solanacearum cells grown overnight at 28°C in 30 ml of YP (yeast extract, 7 g/liter; peptone, 7 g/liter) by the hexadecyltrimethylammonium bromide method (2). DNA concentration was estimated by fluorometry (TKO 100 minifluorometer; Hoefer Scientific Instruments, San Francisco, Calif.).
DNA amplification.
Pairs of primers from the nucleotide sequence of the hrp gene region of strain GMI1000 of R. solanacearum (accession no. Z14056 for EMBL-GenBank-DDBJ databases) were designed with Oligo 5.0 software (32). Eleven pairs were selected in order to explore the whole region. They delineated fragments with sizes ranging from 213 to 2,456 bp. Primers were synthesized by Genosys Biotechnologies, Cambridge, England.
PCRs were carried out in a total volume of 50 μl and performed in a thermocycler (GeneAmp PCR system 9600; Perkin-Elmer Corporation, Norwalk, Conn.). Two kinds of enzymes were used for PCR amplifications, either Taq DNA polymerase (GIBCO BRL Life Technologies, Cergy Pontoise, France) used with the 10× buffer (200 mM Tris-HCl, 500 mM KCl; pH 8.4) for an expected fragment length smaller than 1,000 bp, or a mix containing Taq and Pwo DNA polymerases used with buffer 3 (Expand Long Template PCR system; Boehringer Mannheim, Meylan, France) for a length over 1,000 bp. DNA, selected primers, MgCl2 (GIBCO BRL), dATP, dCTP, dGTP, dTTP (Boehringer Mannheim), and water (high-pressure liquid chromatography grade; Sigma-Aldrich, Steinheim, Germany) were added to the reaction mixture. Optimal conditions of amplification were determined by using the Taguchi methods as modified by Cobb and Clarkson (11).
PCR products were electrophoresed onto agarose gels and visualized with UV light after ethidium bromide staining (37).
Restriction fragment analysis.
The amplified DNA fragments considered to be specific to R. solanacearum were digested with restriction endonucleases according to the manufacturer’s directions (GIBCO BRL; Boehringer Mannheim). Enzymes were chosen on the basis of the nucleotide sequence of the hrp gene region of strain GMI1000 by using Oligo 5.0 software (32). Among all considered enzymes, only 13 available in the laboratory were retained: AvaI, BglII, BssHII, EclXI, EcoRI, HaeII, HindII, NotI, PstI, PvuI, PvuII, SacI, and SmaI. Restriction fragments were separated by electrophoresis and visualized as described previously (37).
Data analysis.
Data derived from the different RFLP patterns exhibited by the tested strains (presence or absence of bands) were used for a hierarchical cluster analysis (HCA). With Statlab software (41), clustering was based on the Euclidean distance between strains (Ward’s method [46]). The truncation level in the resulting dendrogram was thus determined to be that which provided the smallest number of clusters for which the variance within clusters was significantly (P = 0.05) different from the variance between clusters.
RESULTS
Specificity of primers to the hrp gene region of R. solanacearum.
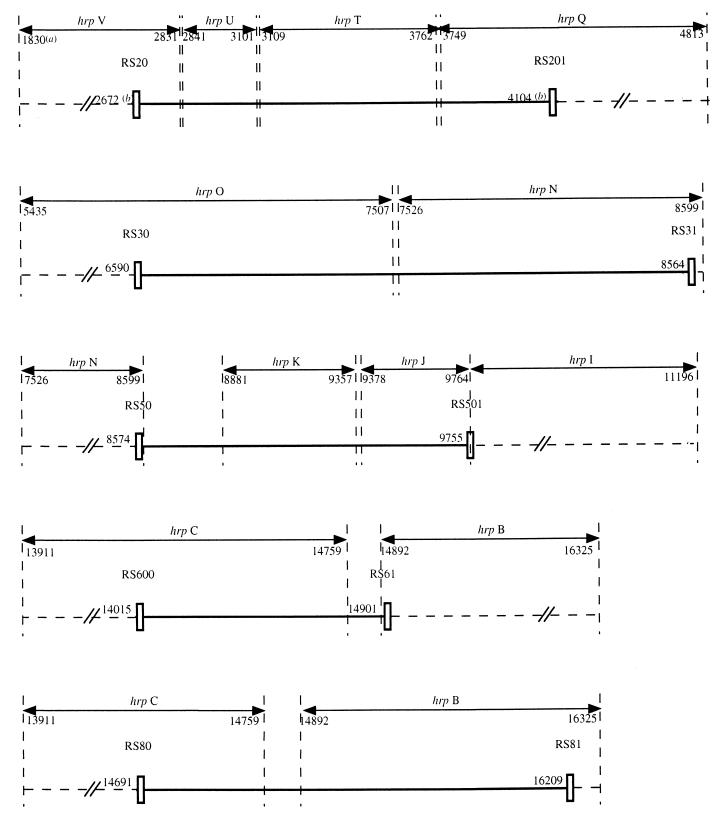
A suitable amplification pattern was obtained with only 6 of the 11 pairs of primers, giving fragments ranging from 213 to 1,993 bp (according to the sequence of the reference strain GMI1000). The amplified fragments were distributed along the hrp gene region and cover a variable number of genes (two to four) which have been previously defined: RS20-RS201, 1,452 bp over hrpV-hrpU-hrpT-hrpQ; RS30-RS31, 1,993 bp over hrpO-hrpN; RS50-RS501, 1,200 bp over hrpN-hrpK-hrpJ-hrpI; RS600-RS61, 905 bp, and RS80-RS81, 1,537 bp, both over hrpC-hrpB (Fig. 1); and RS90-RS91, 213 bp over hrpB-hrpA.
FIG. 1.
Location of the five selected pairs of primers (RS20-RS201, RS30-RS31, RS50-RS501, RS600-RS61, and RS80-RS81) within the hrp genes of R. solanacearum (strain GMI1000). (a), number of the base on the 5′-3′ DNA sequence; (b), number of the base at the 5′ end of the primer.
For each of the six pairs of primers which led to a suitable amplification and for all of the tested strains which belonged to R. solanacearum, a single band with the expected size was observed. However, the density of the band appeared to be variable depending upon the pair of primers and the bacterial DNA. In contrast, no amplification could be obtained for strains belonging to another bacterial species even for such closely related species as R. eutropha or R. pickettii (data not shown).
Restriction endonuclease analysis of specific amplified hrp sequences.
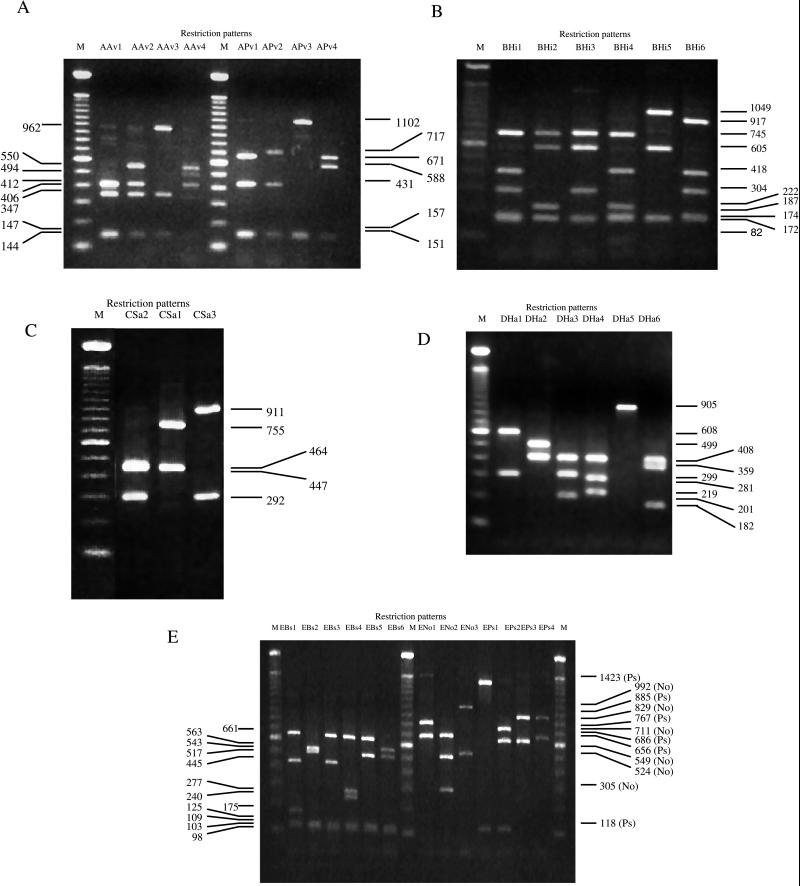
The six amplified DNA fragments, which were considered to be specific to R. solanacearum, were digested with the 13 selected restriction endonucleases. Different restriction patterns among the 120 strains of R. solanacearum were observed with different restriction endonucleases: AvaI (four patterns) and PvuII (four patterns) for the RS20-RS201 sequence; HindII (six patterns) for the RS30-RS31 sequence; SacI (three patterns) for the RS50-RS501 sequence; HaeII (six patterns) for the RS600-RS61 sequence; BssHII (six patterns), NotI (three patterns), and PstI (four patterns) for the RS80-RS81 sequence (Fig. 2).
FIG. 2.
Restriction patterns (see explanation in Table 2) of the five amplified fragments of the hrp gene region of R. solanacearum when digested by the designated enzymes. (A) RS20-RS201 AvaI (left side) and PvuII (right side); (B) RS30-RS31 HindII; (C) RS50-RS501 SacI; (D) RS600-RS61 HaeII; (E) RS80-RS81 BssHII (left side), NotI (right side), and PstI (right side). M, molecular size markers (100-bp ladder; GIBCO BRL). The size (in base pairs) of the bands was estimated from the sequence of the hrp gene region of the GMI1000 strain.
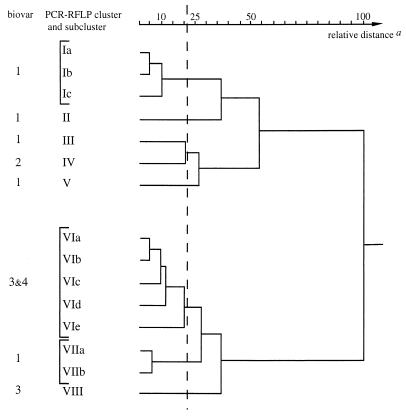
Clustering of the PCR-RFLP profiles.
Among the 120 strains of R. solanacearum, 15 different profiles could be distinguished (Table 2). A profile was the result of the combination of the RFLP patterns given by the eight restriction endonucleases which generated polymorphism and which were selected for the data analysis. The HCA resulted in a dendrogram showing the genetic relatedness between strains (Fig. 3). The truncation level allowed separation of eight PCR-RFLP clusters designated clusters I to VIII. While five clusters (II, III, IV, V, and VIII) contained a unique profile, the three remaining contained two (cluster VII), three (cluster I), and five (cluster VI) different profiles. The number of strains in each cluster ranged from 1 to 40, but each cluster contained strains belonging to the same biovar, with the exception of cluster VI, which included strains of biovars 3 and 4. Biovar 1 strains were distributed over five clusters (I, II, III, V, and VII), biovar 3 was grouped into both cluster VI and cluster VIII (only one strain within the latter), and the 36 strains belonging to biovar 2 were grouped together in cluster IV.
TABLE 2.
Characterization of the 15 PCR-RFLP profiles identified among the 120 strains of R. solanacearum
| PCR-RFLP profilea | Cluster or subcluster |
|---|---|
| AAv2; APv1; BHi4; CSa2; DHa4; EBs4; ENo2; EPs3 | Ia |
| AAv3; APv1; BHi4; CSa2; DHa4; EBs4; ENo2; EPs3 | Ib |
| AAv2; APv3; BHi4; CSa2; DHa4; EBs4; ENo2; EPs3 | Ic |
| AAv2; APv1; BHi2; CSa1; DHa5; EBs5; ENo2; EPs3 | II |
| AAv4; APv2; BHi2; CSa2; DHa5; EBs2; ENo2; EPs2 | III |
| AAv2; APv2; BHi2; CSa2; DHa2; EBs2; ENo2; EPs2 | IV |
| AAv2; APv2; BHi2; CSa2; DHa6; EBs6; ENo3; EPs4 | V |
| AAv2; APv1; BHi3; CSa1; DHa3; EBs3; ENo1; EPs1 | VIa |
| AAv3; APv1; BHi3; CSa1; DHa3; EBs3; ENo1; EPs1 | VIb |
| AAv2; APv3; BHi3; CSa1; DHa3; EBs3; ENo1; EPs1 | VIc |
| AAv3; APv4; BHi3; CSa1; DHa3; EBs3; ENo1; EPs1 | VId |
| AAv2; APv1; BHi6; CSa1; DHa3; EBs3; ENo1; EPs1 | VIe |
| AAv1; APv4; BHi1; CSa1; DHa1; EBs1; ENo1; EPs1 | VIIa |
| AAv1; APv1; BHi1; CSa1; DHa1; EBs1; ENo1; EPs1 | VIIb |
| AAv4; APv2; BHi5; CSa3; DHa1; EBs3; ENo1; EPs1 | VIII |
A profile was the combination of eight restriction patterns generated through the digestion of the five amplified fragments by the designated enzymes. Each pattern was given a code containing three letters and one number: the first letter refers to the amplified fragment (A as delineated by primers RS20-RS201, B by RS30-RS31, C by RS50-RS501, D by RS600-R61, and E by RS80-RS81), the following letters indicate the enzyme (AvaI, PvuII, HindII, SacI, HaeII, BssHII, NotI, and PstI), and the number refers to the pattern generated by the enzyme.
FIG. 3.
Dendrogram resulting from an HCA based on the restriction patterns of the five amplified fragments within the hrp gene region of 120 strains of R. solanacearum. a, the relative distance between the farthest clusters was assumed to be 100.
Most restriction patterns were common to different clusters. However, the restriction patterns AAv1, BHi1, DHa1 (one exception), and EBs1 appeared to be specific to cluster VII whereas BHi4 and DHa4 characterized cluster I (pattern designations are explained in Table 2). In addition, BHi5 and CSa3 were characteristic of the unique strain within cluster VIII, and DHa6, EBs6, ENo3, and EPs4 seemed to be specific to cluster V. Similarly, when the distribution of the restriction patterns within the biovars was analyzed DHa2 was found only in biovar 2 strains and BHi3, DHa3, and EBs3 were found only in biovars 3 and 4.
The restriction patterns generated by HaeII (DHa1 to DHa6) and BssHII (EBs1 to EBs6) appeared to be the most useful for separating the eight clusters and distinguishing the three biovars (Table 3). Strains of biovars 2 and 3 or 4 had a unique distinctive profile, DHa2-EBs2 and DHa3-EBs3, respectively, and were classified either in cluster IV or in clusters VI and VIII, while the biovar 1 strains displayed five different profiles which characterized the five remaining clusters.
TABLE 3.
Distribution of the restriction patterns generated by HaeII on the RS600-RS61-amplified fragment and by BssHII on the RS80-RS81-amplified fragment of the hrp gene region of R. solanacearum according to the PCR-RFLP cluster and to the biovar
| Biovar | PCR-RFLP cluster | Restriction pattern with:
|
|
|---|---|---|---|
| HaeII | BssHII | ||
| 1 | I | DHa4 | EBs4 |
| 1 | II | DHa5 | EBs5 |
| 1 | III | DHa5 | EBs2 |
| 1 | V | DHa6 | EBs6 |
| 1 | VII | DHa1 | EBs1 |
| 2 | IV | DHa2 | EBs2 |
| 3/4 | VI | DHa3 | EBs3 |
| 3 | VIII | DHa1 | EBs3 |
In addition, the dendrogram obtained suggests that these R. solanacearum strains can be separated into two distinct groups, namely, clusters I to V (all biovar 2 strains and approximately 64% of biovar 1 strains) and clusters VI to VIII (all biovar 3 and 4 strains and about 36% of biovar 1 strains). The diversity within biovar 1 appeared to be correlated with geographic origin since all strains belonging to cluster VII were isolated from Africa, mainly from the southern part (Angola, Zimbabwe, Madagascar, and Reunion Island), while most (85%) of those included in clusters I, II, III, and V originated from the Americas. Some strains isolated from northern Africa (Burkina Faso and Kenya) belonged, however, to clusters I and II. The 36 biovar 2 strains fell into cluster IV regardless of geographic origin: Africa, Americas, Asia, Europe, or Oceania. There were no differences in profile between biovar 3 and biovar 4 strains, and all (one exception) were gathered in one cluster (VI), but five profiles were identified, which separated five subclusters. The subcluster VIa included many Asiatic strains (46%), VIb included most African strains (83%), and 69% of the American strains were included in subcluster VIc.
Cluster V contained four strains which were isolated from hosts of the Musaceae family and was characterized by a profile which included the specific restriction patterns DHa6, EBs6, ENo3, and EPs4. It must be noted that the sum of sizes of the restriction fragments included in DHa6 and EPs4 appeared to be higher (41 and 30 bp, respectively) than the total size of the corresponding amplified fragment, suggesting that an inserted sequence may be present (Fig. 4). This was characteristic only of the strains belonging to cluster V. Three other strains isolated from Musa sp. were distributed either into cluster III or into cluster VI. There was no obvious correlation between the host origin of strains and their distribution into clusters.
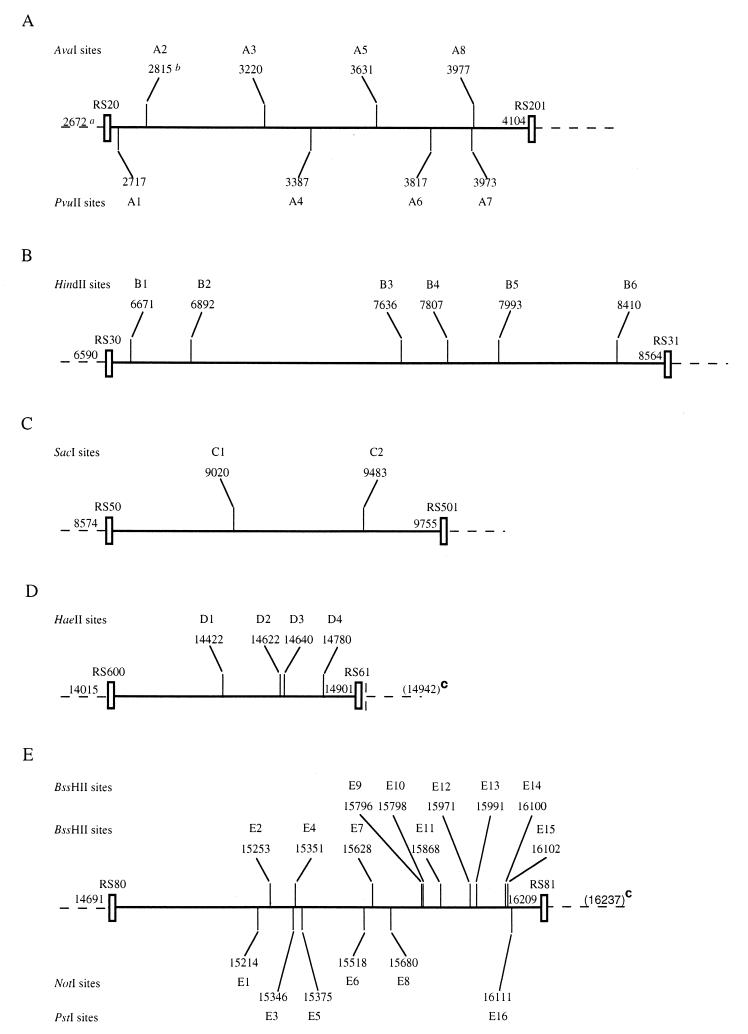
FIG. 4.
Location of the 36 restriction sites identified in the five amplified fragments of the hrp gene region of R. solanacearum when digested by AvaI and PvuII for RS20-RS201 (A); by HindII for RS30-RS31 (B); by SacI for RS50-RS501 (C); by HaeII for RS600-RS61 (D); and by BssHII, NotI, and PstI for RS80-RS81 (E), as estimated from the DNA sequence of strain GMI1000 and the size of the bands of the restriction patterns. a, number of the base at the 5′ end of the primer; b, number of the base at left of the restriction site; c, the size of the amplified fragment was higher for the four strains of cluster V.
Variability in restriction sites in the hrp gene region of R. solanacearum.
Thirty-six restriction sites were identified within the five amplified fragments when the eight selected enzymes were used (Fig. 4). Eight sites appeared to be common to all 120 strains (A5, A7, B4, B5, E12, E13, E14, and E15). The occurrence of the 28 remaining sites was variable according to the RFLP clustering of the strains. However, 14 among them appeared to be particularly remarkable since they could separate the biovars and/or the geographic or botanical origins (Table 4).
TABLE 4.
Occurrence of 14 discriminating restriction sites identified within the five amplified fragments of the hrp gene region of R. solanacearum according to biovar typing, geographic or host origin, and PCR-RFLP clustering
| Biovar | Origin | PCR-RFLP cluster | Result for restriction site:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1bi | A2ai | B1cj | B2cj | C1dk | D2el | D3el | D4el | E2fm | E5gm | E6hm | E7fm | E8hm | E11fm | |||
| 1 | Americasn | I | + | − | + | + | + | − | + | − | − | − | + | + | − | + |
| 1 | Americaso | II | + | − | + | + | + | − | − | − | − | − | + | − | − | + |
| 1 | Africa | VII | + | + | − | + | + | + | − | − | − | − | + | − | − | − |
| 1 | Musaceaep | III | − | − | + | + | + | − | − | − | + | − | + | − | − | + |
| 1 | Musaceae | V | − | − | + | + | + | − | − | + | + | + | − | − | + | + |
| 2 | Worldwide | IV | − | − | + | + | + | − | − | − | + | − | + | − | − | + |
| 3 | Worldwide | VI | + | − | − | + | + | + | − | − | − | − | + | − | − | + |
| 3 | Asia | VIII | − | − | − | − | − | + | − | − | − | − | + | − | − | + |
| 4 | Asia, Oceania | VI | + | − | − | + | + | + | − | − | − | − | + | − | − | + |
AvaI site.
PvuII site.
HindII site.
SacI site.
HaeII site.
BssHII site.
NotI site.
PstI site.
Located between the RS20 and RS201 pair of primers.
Located between RS30 and RS31.
Located between RS50 and RS501.
Located between RS600 and RS61.
Located between RS80 and RS81.
All strains originated from the Americas except for one from Africa.
All strains originated from the Americas except for three from Africa.
All strains were isolated from musaceous plants except for one from potato.
A1 was identified within all but one biovar 3 or biovar 4 strain and within all biovar 1 strains except those isolated from musaceous plants. A2 characterized the African biovar 1 strains (cluster VII). In contrast, E11 was absent only from these African biovar 1 strains. B1 was present in all biovar 2 strains and also in all biovar 1 strains except those originating from Africa (cluster VII). D2 was found in biovar 3 (and biovar 4) strains and in the African biovar 1 strains (cluster VII). B2 and C1 were present in all strains except one strain (cluster VIII). D3 and E7 were characteristic of American biovar 1 strains (cluster I). E2 characterized all biovar 2 strains and also biovar 1 strains isolated from musaceous plants (clusters III and V). These musaceous clusters could be separated by four sites characterizing strains grouped in cluster V: D4, E5, E6, and E8.
An additional HCA, based on the presence or absence of discriminating restriction sites, gave a slightly different cluster distribution. The truncation then separated six groups, one joining the PCR-RFLP clusters I and II and the other joining clusters III and IV.
DISCUSSION
The exploration of the hrp gene region with 11 selected pairs of primers gave six amplicons which were confirmed to be specific to R. solanacearum. Indeed, no amplification was observed with DNA from strains belonging to other bacterial species and even from such closely related species as R. pickettii, R. eutropha, and B. cepacia. Consequently, the hrp region seems to be useful for the identification and specific detection of strains of R. solanacearum. However, since we did not succeed in getting strains of Pseudomonas celebense and Pseudomonas syzygii from laboratory collections, which are also species close to R. solanacearum, the amplification within their hrp region when the selected primers were used was not checked. Nevertheless, since our main objective was to develop molecular tools for the detection of populations of R. solanacearum on Reunion Island and since these particular pathogenic species (P. celebense and P. syzygii) were recorded only in Indonesia on bananas and cloves, respectively, a lack of specificity in that case would be of no consequence.
Although we concentrated mainly on the strains originating from Reunion Island (28 strains), the remaining 92 isolates were chosen to represent the broad host range, wide geographic distribution, and metabolic diversity (biovars) of R. solanacearum. Although our analysis gave a lower resolution level than that seen after genomewide RFLP analysis (12–14), we found that it gave reliable estimates of phylogenetic relationships among strains of R. solanacearum. Whereas the 46 described RFLP profiles were correlated with geographic origin, biochemical typing, and host origin, we identified 15 PCR-RFLP profiles distributed into eight clusters, these clusters being correlated with biochemical typing and to a lesser degree with geographic origin. Since some PCR-RFLP patterns correlated well with biovar typing, the PCR-RFLP procedure provides a complementary or alternative method for biovar determination.
The PCR-RFLP analysis confirmed the great variability within R. solanacearum. Biovar 1 strains showed the greatest diversity since they were distributed into five of the eight clusters. Six biovar 1 strains originating from the Musaceae family were distributed into two specific clusters, one (cluster V) including four of these strains and the other (cluster III) comprising the two remaining strains together with one strain isolated from potato. Among the 36 restriction sites identified on the five amplified fragments, only 25 were common to the musaceous strains, whereas there were 28 sites common to biovar 2 and biovar 3 strains. Moreover, among the 12 discriminating restriction sites located on the fragment delineated by the RS80-RS81 primer pair, only three were common to both clusters III and V while all were shared by clusters III and IV. These features suggest that there are important differences between the musaceous strains distributed in two separate clusters. All of these strains came from Central America or northern South America, but nothing was known of their pathogenicity, and no clear indication of the race to which they belonged was reported. The strains of cluster V could belong to race 2, whereas those of cluster III might be associated with race 1, which could explain the presence of the Colombian strain isolated from potato within the cluster. One particular strain isolated from Musa sp. and characterized as belonging to biovar 3 (36) fell into cluster VI, as most of the strains were related to the same biovar. Further studies incorporating more strains isolated from Musaceae and belonging either to race 1 or to race 2 are required.
Clusters I and II included all biovar 1 strains originating from the Americas, more specifically, either from North America for cluster I or from Central America for cluster II. The fact that four African isolates fell into these clusters suggests that they could have been introduced from the Americas. Most biovar 1 strains isolated from African countries, however, were included in cluster VII. All of these strains originated from southern Africa, including Reunion Island, Madagascar, Zimbabwe, and Angola, whereas the African isolates from clusters I and II came from the northern part of Africa (Burkina Faso and Kenya). Thus, Africa may have two different biovar 1 populations, either endemic and commonly isolated in southern countries or introduced from the Americas through direct or indirect commercial exchanges. Although both populations belonged to the same biovar, there was no indication that they have similar host ranges (and/or similar virulence).
The 36 strains of biovar 2 displayed a similar profile which was characterized by the specific restriction pattern DHa2 and were included in cluster IV, close to those encompassing the biovar 1 strains of American origin. The consistent homogeneity of biovar 2 strains, although they were collected from 20 countries distributed worldwide, could be attributed to their narrow host range, including only potato and tomato plants. The result agrees with the commonly accepted hypothesis of a common origin for all the biovar 2 strains. South America is the presumed origin, and the wide distribution of these strains is probably due to the dissemination of latently infected plant material (particularly potato tubers) by humans (8, 10, 22). The biovar 2 strains (cluster IV) are closely related to cluster III strains, since 34 of the 36 restriction enzyme sites were common to both clusters, underlining the proximity of some musaceous isolates to race 3 strains.
Compared to biovar 1 strains, biovar 3 strains showed rather modest genetic diversity since they could be assigned to one major cluster. An additional cluster with a unique strain originating from Japan was also described. The few biovar 4 strains fell into the same cluster as most biovar 3 strains, indicating that there were only slight differences between these biovars. However, six different profiles more or less correlated with geographic origin (Asia, cluster VIa; Reunion Island, cluster VIb; America, cluster VIc) were identified.
The dendrogram resulting from an HCA revealed the separation of R. solanacearum into two major divisions. This result confirmed the conclusion of many previous studies on DNA homologies and physiological characterization of strains (20, 34) and more recently of RFLP analysis (12–14), of 16S rRNA sequencing (30, 40, 43), or of PCR amplification with tRNA consensus primers (39). The first division, Americanum sensu Cook et al. (12), contains biovar 1 and 2 strains, and the second division, Asiaticum sensu Cook et al. (12), includes biovar 3 and 4 strains. Thus, compared to other genomic regions (16S rRNA and tRNA), the hrp gene region, which is involved in host-pathogen interactions, revealed the same major trend of diversity, suggesting that hrp genes have evolved in parallel with 16S rRNA and tRNA.
The amplified fragment delineated by the RS80-RS81 pair of primers provided much more polymorphism than all the others: 12 discriminating restriction sites were identified and permitted separation of certain groups of strains. All of these polymorphisms were located within the hrpB regulatory gene (16). This observation confirms that regulatory systems of bacteria seem to be less conserved than those genes whose function they govern (5). Furthermore, this result suggests that the hrp regulatory gene may have other metabolic functions besides its role in pathogenic diversity of R. solanacearum. More precise analysis of the hrpB gene in different strains in the future might provide a useful way of relating pathogenicity gene function to genetic diversity.
Although the dendrogram confirms the separation of R. solanacearum into two groups, the distribution of biovar 1 strains, which displayed a rather wide variability, did not agree completely with the scheme proposed by Cook et al. (12). Clusters I, II, III, and V (American biovar 1 strains) were close to cluster IV, which included biovar 2 strains, and would thus belong to the Americanum division, whereas cluster VII (African biovar 1 strains) near cluster VI (biovar 3 and 4 strains) would be separated and connected rather to the Asiaticum division. The African strains included in cluster VII could have evolved separately as a result of geographic isolation and thereby have contributed to increasing the diversity of the species. Clearly, further analysis with other techniques such as DNA probes for RFLP analysis (12–14) and/or 16S rRNA sequencing (30, 40, 43) to confirm other characteristic features of these strains would be of interest. Preliminary results obtained with the R. solanacearum-specific primer pair PS96-H and PS96-I (38) support the hypothesis of separate evolution of these strains, since these primers never led to amplification of any biovar 1 strain originating from Reunion Island, Madagascar, Zimbabwe, or Angola (data not shown). Whatever the explanation, these African biovar 1 strains shared more sites with biovar 3 strains (23 sites) than with the American biovar 1 strains (13 to 19 sites according to the cluster), and the conclusion is that American and African biovar 1 strains are phylogenetically distinct, the latter being more closely related to Asiatic (biovar 3 and 4) strains.
Our study of the genetic diversity of the hrp gene region of R. solanacearum thus provides discriminating tools which besides being useful for fundamental research can also be used for diagnostic purposes. For example, biovars 1 and 2 and the combination of biovars 3 and 4 can easily be distinguished from each other by the restriction pattern generated after amplification with the RS600 and RS61 primers when digested by HaeII: DHa3 for biovars 3 and 4; DHa2 for biovar 2; and DHa1, DHa4, DHa5, or DHa6 for biovar 1. Moreover, the restriction pattern could give useful information about the geographic origin of the biovar 1 strain. As PCR amplification is known to be a very sensitive technique, such primers could be used to detect the populations of R. solanacearum in plant, irrigation water, or soil extracts. They could also be employed to clarify some aspects of the epidemiology of bacterial wilt regarding, for instance, seed as a vehicle of disease spread or some weeds or resistant plants as possible carriers of low levels of infectious populations.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Ministère de l’Enseignement Supérieur et de la Recherche.
We are grateful to A. Couteau and J. J. Chéron for technical assistance. We thank N. Grimsley, P. Prior, C. Boucher, and O. Pruvost for critical reading of the manuscript and L. Gardan, A. Aspin, J. Young, C. Allen, C. Boucher, and K. Tsuchiya for providing strains.
REFERENCES
- 1.Arlat M, Gough C L, Zischek C, Barberis P A, Trigalet A, Boucher C A. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1992;5:187–193. doi: 10.1094/mpmi-5-187. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates-Wiley Interscience; 1991. [Google Scholar]
- 3.Beer S V, Bauer D W, Jiang X H, Laby R J, Sneath B J, Wei Z M, Wilcox D A, Zumoff C H. The hrp cluster of Erwinia amylovora. In: Henneke H, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 53–60. [Google Scholar]
- 4.Bonas U, Schulte R, Fenselau S, Minsavage G V, Staskawicz B J, Stall R E. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol Plant-Microbe Interact. 1991;4:81–88. [Google Scholar]
- 5.Boucher, C. A. Personal communication.
- 6.Boucher C A, Gough C L, Arlat M. Molecular genetics of pathogenicity determinants of Pseudomonas solanacearum with special emphasis on hrp genes. Annu Rev Phytopathol. 1992;30:443–461. [Google Scholar]
- 7.Buddenhagen I W, Sequeira L, Kelman A. Designation of races of Pseudomonas solanacearum. Phytopathology. 1962;52:726. [Google Scholar]
- 8.Buddenhagen I W. Bacterial wilt revisited. ACIAR Proc. 1986;13:126–143. [Google Scholar]
- 9.Ciampi L, Sequeira L. Multiplication of Pseudomonas solanacearum in resistant potato plants and the establishment of latent infections. Am Potato J. 1980;57:307–316. [Google Scholar]
- 10.Ciampi L, Sequeira L, French E R. Latent infection of potato tubers by Pseudomonas solanacearum. Am Potato J. 1980;57:377–386. [Google Scholar]
- 11.Cobb B D, Clarkson J M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 1994;22:3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook D, Barlow E, Sequeira L. Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Mol Plant-Microbe Interact. 1989;2:113–121. [Google Scholar]
- 13.Cook D, Barlow E, Sequeira L. DNA probes as tools for the study of host-pathogen evolution: the example of Pseudomonas solanacearum. In: Henneke H, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 103–108. [Google Scholar]
- 14.Cook D, Sequeira L. Strain differentiation of Pseudomonas solanacearum by molecular genetic methods. In: Hayward A C, Hartman G L, editors. Bacterial wilt, the disease and its causative agent, Pseudomonas solanacearum. Wallingford, United Kingdom: CAB International; 1994. pp. 77–93. [Google Scholar]
- 15.Dianese J C, Dristig M C G. Strain characterization of Pseudomonas solanacearum based on membrane protein patterns. In: Hayward A C, Hartman G L, editors. Bacterial wilt, the disease and its causative agent, Pseudomonas solanacearum. Wallingford, United Kingdom: CAB International; 1994. pp. 113–121. [Google Scholar]
- 16.Genin S, Gough C L, Zischek C, Boucher C A. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol Microbiol. 1992;6:3065–3076. doi: 10.1111/j.1365-2958.1992.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 17.Girard J C, Nicole J F, Cheron J J, Gaubiac A M, Huvier O, Oudard B, Suzor H. Bacterial wilt due to Pseudomonas solanacearum in Reunion: general situation and current research. ACIAR Proc. 1993;45:343–347. [Google Scholar]
- 18.Gopalan S, He S Y. Bacterial genes involved in the elicitation of hypersensitive response and pathogenesis. Plant Dis. 1996;80:604–610. [Google Scholar]
- 19.Granada G A, Sequeira L. A new selective medium for Pseudomonas solanacearum. Plant Dis. 1983;67:1084–1088. [Google Scholar]
- 20.Harris D C. Proceedings of the 3rd International Conference on Plant Pathogenic Bacteria, Wageningen, The Netherlands. 1972. Intraspecific variations in Pseudomonas solanacearum; pp. 289–292. [Google Scholar]
- 21.Hayward A C. Characteristics of Pseudomonas solanacearum. J Appl Bacteriol. 1964;27:265–277. [Google Scholar]
- 22.Hayward A C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–89. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 23.Hayward A C. The hosts of Pseudomonas solanacearum. In: Hayward A C, Hartman G L, editors. Bacterial wilt, the disease and its causative agent, Pseudomonas solanacearum. Wallingford, United Kingdom: CAB International; 1994. pp. 9–25. [Google Scholar]
- 24.Hayward A C, El-Nashaar H M, Nydegger U, De Lindo L. Variation in nitrate metabolism in biovars of Pseudomonas solanacearum. J Appl Bacteriol. 1990;69:269–280. [Google Scholar]
- 25.He L Y, Sequeira L, Kelman A. Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 1983;67:1357–1361. [Google Scholar]
- 26.Janse J D. Infra and intraspecific classification of Pseudomonas solanacearum strains using whole cell fatty acid analysis. Syst Appl Microbiol. 1991;14:335–345. [Google Scholar]
- 27.Laguerre G, Allard M R, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leite R P, Jr, Minsavage G V, Bonas U, Stall R E. Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol. 1994;60:1068–1077. doi: 10.1128/aem.60.4.1068-1077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindgren P B, Peet R C, Panopoulos N J. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Dorsch M, Del Dot T, Sly L I, Stackebrandt E, Hayward A C. Phylogeny of biovars of Pseudomonas solanacearum based on sequencing of 16S rRNA. ACIAR Proc. 1993;45:93–95. [Google Scholar]
- 31.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Biosciences, Inc. Oligo, primer analysis software, version 5.0. Plymouth, Minn: National Biosciences, Inc.; 1996. [Google Scholar]
- 33.Olsson K. Experience of brown rot caused by Pseudomonas solanacearum (Smith) in Sweden. EPPO Bull. 1976;6:199–207. [Google Scholar]
- 34.Palleroni N J, Doudoroff M. Phenotypic characterization and deoxyribonucleic acid homologies of Pseudomonas solanacearum. J Bacteriol. 1971;107:690–696. doi: 10.1128/jb.107.3.690-696.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pegg K, Moffett M. Host range of the ginger strain of Pseudomonas solanacearum in Queensland. Aust J Exp Agric Anim Husb. 1971;11:696–698. [Google Scholar]
- 36.Prior P, Steva H. Characteristics of strains of Pseudomonas solanacearum from the French West Indies. Plant Dis. 1990;74:13–17. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Seal S E, Jackson L A, Daniels M J. Isolation of a Pseudomonas solanacearum-specific DNA probe by subtraction hybridization and construction of species-specific oligonucleotide primers for sensitive detection by the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3751–3758. doi: 10.1128/aem.58.11.3751-3758.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seal S E, Jackson L A, Daniels M J. Use of tRNA consensus primers to indicate subgroups of Pseudomonas solanacearum by polymerase chain reaction amplification. Appl Environ Microbiol. 1992;58:3759–3761. doi: 10.1128/aem.58.11.3759-3761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seal S E, Jackson L A, Young J P W, Daniels M J. Differentiation of Pseudomonas solanacearum, Pseudomonas syzygii and the blood disease bacterium by partial 16S rRNA sequencing: construction of oligonucleotide primers for sensitive detection by polymerase chain reaction. J Gen Microbiol. 1993;139:1587–1594. doi: 10.1099/00221287-139-7-1587. [DOI] [PubMed] [Google Scholar]
- 41.SLP Statistiques. Statlab software, version 2.0. Monterey, Calif: SLP Statistiques; 1994. [Google Scholar]
- 42.Stead D E. Classification and identification of Pseudomonas solanacearum and other pseudomonads by fatty acid profiling. ACIAR Proc. 1993;45:49–53. [Google Scholar]
- 43.Taghavi M, Hayward C, Sly L I, Fegan M. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1996;46:10–15. doi: 10.1099/00207713-46-1-10. [DOI] [PubMed] [Google Scholar]
- 44.Urakawa H, Kita-Tsukamoto K, Ohwada K. 16S rRNA genotyping using PCR/RFLP (restriction fragment length polymorphism) analysis among the family Vibrionaceae. FEMS Microbiol Lett. 1997;152:125–132. doi: 10.1111/j.1574-6968.1997.tb10418.x. [DOI] [PubMed] [Google Scholar]
- 45.Vallaeys T, Persello-Cartieaux F, Rouard N, Lors C, Laguerre G, Soulas G. PCR-RFLP analysis of 16S rRNA, tfdA and tfdB genes reveals a diversity of 2,4-D degraders in soil aggregates. FEMS Microbiol Lett. 1997;24:269–278. [Google Scholar]
- 46.Ward J H. Hierarchical grouping to optimize an objective function. Am Stat Assoc J. 1963;58:236–244. [Google Scholar]
- 47.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal for Ralstonia pickettii, Ralstonia solanacearum and Ralstonia eutropha. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]