Abstract
Twelve human and chicken isolates of Salmonella enterica serovar Enteritidis belonging to phage types 4, 8, 13a, and 23 were characterized for variability in lipopolysaccharide (LPS) composition. Isolates were differentiated into two groups, i.e., those that lacked immunoreactive O-chain, termed rough isolates, and those that had immunoreactive O-chain, termed smooth isolates. Isolates within these groups could be further differentiated by LPS compositional differences as detected by gel electrophoresis and gas liquid chromatography of samples extracted with water, which yielded significantly more LPS in comparison to phenol-chloroform extraction. The rough isolates were of two types, the O-antigen synthesis mutants and the O-antigen polymerization (wzy) mutants. Smooth isolates were also of two types, one producing low-molecular-weight (LMW) LPS and the other producing high-molecular-weight (HMW) LPS. To determine the genetic basis for the O-chain variability of the smooth isolates, we analyzed the effects of a null mutation in the O-chain length determinant gene, wzz (cld) of serovar Typhimurium. This mutation results in a loss of HMW LPS; however, the LMW LPS of this mutant was longer and more glucosylated than that from clinical isolates of serovar Enteritidis. Cluster analysis of these data and of those from two previously characterized isogenic strains of serovar Enteritidis that had different virulence attributes indicated that glucosylation of HMW LPS (via oafR function) is variable and results in two types of HMW structures, one that is highly glucosylated and one that is minimally glucosylated. These results strongly indicate that naturally occurring variability in wzy, wzz, and oafR function can be used to subtype isolates of serovar Enteritidis during epidemiological investigations.
Salmonella enterica serovar Enteritidis has been a major cause of a worldwide increase in the prevalence of human salmonellosis that has lasted for nearly two decades, due in part to its enhanced ability to contaminate hen eggs (1, 15, 17–19, 35). The structure of lipopolysaccharide (LPS) from serovar Enteritidis has been used to identify isolates that efficiently contaminate eggs (8, 10–12). Thus, understanding LPS structural variation is important, because isolates that can produce a large amount of high-molecular-weight (HMW) LPS contaminate eggs efficiently and increase chick mortality (8, 12). Specifically, these HMW LPS-producing isolates have a propensity to grow to high cell density (>1011 CFU/ml), hyperflagellate, and undergo swarming migration on solid agar (8, 10). Moreover, isolates with an orally invasive phenotype produce a dense cell surface matrix at 25°C that is composed primarily of glucosylated HMW LPS and bundled flagellar structures, although they do not swarm (11). Because both these phenotypes with specific roles in pathogenesis are differentiated from less-virulent smooth field isolates by production of HMW LPS, we believe that it is possible to monitor emerging virulence of serovar Enteritidis, and possibly other salmonella serovars, by analyzing LPS structural heterogeneity during epidemiological investigations (6, 7, 13, 14, 16, 27, 28, 30, 31). Thus, the objective of this study was to investigate what LPS phenotypes could be encountered during processing of field and clinical isolates.
By conducting compositional analysis of a dozen isolates and comparing results to those previously published and those obtained from a wzz mutant of serovar Typhimurium, we have gained new insight into the structure of LPS from serovar Enteritidis. Previous work had shown that HMW LPS has more than 11 O-chain repeat units and that 50% of these are glucosylated, whereas low-molecular-weight (LMW) LPS has an average O-chain length of 5 units, very few of them glucosylated (29). In this study, we demonstrate that variation in the composition of LPS from clinical isolates of serovar Enteritidis can be detected utilizing appropriate LPS extraction procedures. We also discuss possible genetic mechanisms for this LPS variation and describe how understanding these variations can be used to cluster data graphically to identify virulent isolates.
MATERIALS AND METHODS
Bacteria and media.
Isolates are identified by accession number in Table 1. Phage typing and identification of isolates as serovar Enteritidis were initially done at the contributing laboratory, either the Centers for Disease Control and Prevention, Atlanta, Ga., or the National Veterinary Services Laboratory, Ames, Iowa. Serovar classification was confirmed again at Southeast Poultry Research Laboratory by using O- and H-typing antisera (Difco) and a biochemical panel (Enterotube II; Fisher). Isolates were supplied on agar slants as low-passage isolates (fewer than five passages). Prior to inoculation of broth cultures, cells were streaked on Brilliant Green agar for isolation of colonies. Two liters of brain heart infusion (BHI) broth supplemented as described in the text below was inoculated with a single colony from Brilliant Green agar. Cultures were grown for 16 h without shaking at 42°C. Classification of isolates into rough (no O-chain) and smooth phenotypes was done by slide agglutination with antiserum specific for group D1 isolates producing tyvelose (factor 9) and glucosylated O-chain (factor 12) (Difco).
TABLE 1.
Sources of S. enterica serovar Enteritidis isolatesa
| SEPRL isolate | Supplier | PT | Accession no. |
|---|---|---|---|
| 7 | NVSL | PT13a | 93-09022.6839 |
| 19 | NVSL | PT8 | 92-44267-24697 |
| 25 | NVSL | PT23 | 93-02934-3184 |
| 27 | NVSL | PT13a | 93-03271-3682 |
| 30 | NVSL | PT8 | 92-44267-24699 |
| 37 | NVSL | PT23 | 93-00837-1174 |
| 38 | NVSL | PT23 | 92-37431-19510 |
| 39 | NVSL | PT13a | 92-37429-19575 |
| 40 | NVSL | PT13a | 93-03233-2573 |
| 41 | CDC | PT4 | 92-1250 |
| 46 | CDC | PT4 | 93-1123 |
| 58 | Scottish Reference Lab.; supplied by CDC | PT4 | 426 |
SEPRL = Southeast Poultry Research Laboratory; NVSL = National Veterinary Services Laboratory, Ames, Iowa; CDC = Centers for Disease Control and Prevention, Atlanta, Ga.
Isolates 19, 38, 39, 40, 41, and 46 were grown in BHI broth supplemented with 10 mM glucose, as recommended for recovery of LPS, while isolates 7, 25, 27, 30, 37, 58, and 46 were supplemented with 10 mM N-acetylglucosamine (GlcNAc) (32). By choosing a class of carbohydrate different from that usually used for supplementation, we introduced a source of variation that would help us assess more accurately the amount of sample variation that could be expected as different laboratories processed different isolates. GlcNAc was chosen as the alternative sugar to glucose, because it is an amino sugar that can be metabolized via two phosphotransferase systems (ptsG and ptsN systems), whereas only one system metabolizes the neutral sugar glucose (ptsG system) (23). These two pathways enable GlcNAc to be diverted directly to peptidoglycan and lipopolysaccharide biosynthesis in addition to being utilized, like glucose, as a carbon source (5). Experimental design would have been improved by directly comparing for each strain LPS yields after supplementation with glucose and after supplementation with GlcNAc, but this plan was followed to accommodate processing of the largest number of isolates possible by two different extraction methods, which preliminary data had suggested was a major source of sample variance. As a control for all variables, isolate 46 was supplemented with each metabolite in separate culture, and these two samples were then extracted by the two methods described in Materials and Methods, giving four samples in total for processing.
Water extraction of crude LPS.
LPS was prepared by a water-based extraction method used to obtain bacterial capsules (22). From each isolate, cells were pelleted at 10,000 × g, for 20 min at 4°C, and resuspended in 50 ml of water. The cell suspension, containing 5 × 1012 CFU, was vigorously stirred in a boiling water bath for 30 min and then cooled in an ice bath and stirred for another 90 min, after which the cell residue was removed by centrifugation (10,000 × g, for 30 min at 4°C). The supernatant was adjusted to 1% acetic acid, and crude polysaccharides were pelleted with 2.5 volumes of ethanol, for 24 h at −20°C. After centrifugation (10,000 × g, for 30 min at 4°C), the precipitate was dried, dissolved in 1.0 ml of nuclease buffer (0.01 M Tris [pH 7.8], 10 mM MgCl2), and incubated for 16 h at 37°C with DNase (2 μg/ml) and RNase I (10 μg/ml) (Boehringer Mannheim) (22). The sample was adjusted to include 0.5% sodium dodecyl sulfate (SDS) for incubation with proteinase K (50 μg/ml) (Amresco, Solon, Ohio) for 16 h at 42°C (11). An equal volume of phenol-chloroform (P:C) was used to remove hydrolytic enzymes, and LPS in the aqueous phase collected after centrifugation was precipitated with 2.5 volumes of ethanol for 16 h at −20°C. The precipitate was pelleted (10,000 × g, for 10 min at 4°C) and resuspended in 200 μl of water. Approximately 2 mg of LPS per sample was recovered from two liters of broth culture.
Organic solvent extraction of crude LPS.
A hot-water and P:C extraction method described elsewhere in detail was used to recover both smooth (O-antigen-positive) and rough (O-antigen-negative) LPSs (4, 12, 38, 40). Briefly, cells were pelleted from two liters of broth, suspended in 6 ml of TAE buffer (40 mM Tris acetate [pH 8.5], 2 mM EDTA) and 12 ml of lysis buffer (100 mM SDS, 50 mM Trizma base, 0.128 M NaOH), and then incubated at room temperature until the suspension cleared or for 10 min. An equal volume of P:C was added, and the mixture was vortexed vigorously for 1 min before heating of the sample at 65°C for 15 min. The aqueous phase was collected after centrifugation (10,000 × g, for 15 min at 4°C), and the organic phase was back-extracted with 2 ml of TAE. After another P:C extraction of the aqueous phase without heating, the sample was precipitated by addition of 6 ml of water, 1.5 ml of sodium acetate (NaOAc) (pH 5.2), and 2 volumes of ice-cold 100% ethanol. After overnight precipitation at −20°C, the pelleted sample was blown dry with air and resuspended in 1.0 ml of nuclease buffer for incubation with DNase, RNase, and proteinase K. Other steps are similar to those described above.
PAGE of LPS.
Polyacrylamide gel electrophoresis (PAGE) analysis was performed by using prerun 10- by 10-cm gels (Bio-Rad), prepared with deoxycholate (DOC), and a 5% stack (38, 40). Running buffer was 0.025 M Tris, 0.192 M glycine, and 0.1% SDS. DOC loading buffer was a 2× solution containing 0.02 g of bromphenol blue, 0.4 g of sucrose, and 0.04 g of DOC in 1 ml of water. Samples were boiled 3 min in loading buffer prior to electrophoresis. Bands were visualized by using a silver stain kit (Bio-Rad) and modifications that increase sensitivity (11, 38). Oxidation was attained by placing gels in a solution of 200 ml of water containing 1.4 g of periodic acid. Gels were then washed five times with water, and staining was achieved by immersion for 10 min in a solution containing 2 ml of concentrated NH4OH2, 28 ml of 0.1 N NaOH, 5 ml of 20% AgNO3, and 115 ml of water. Gels were washed three times in water and developed. To confirm that isolates were not producing a capsule, such as one containing colanic acid, that might skew results, samples were also electrophoresed in separate gels as already described but then stained with Alcian blue, which detects acidic sugars (21).
Glycosyl compositional analysis.
Composition analysis of LPS was by gas-liquid chromatography (GLC) of derivatized alditol acetates (42) with a Hewlett-Packard 5890A gas chromatographer equipped with a 15-m DB-1 column and a flame ionization detector. Briefly, samples were hydrolyzed in 2 M trifluoroacetic acid at 120°C for 3 h, reduced with sodium borohydride, and derivatized by acetylation to alditol acetates. Retention times and mass spectra were compared to those of standards. Inositol was included as an internal standard. Rhamnose was used as the sugar for describing O-chain composition (expressed as milligrams per 100 milligrams of crude LPS) as (i) it separates well from mannose and galactose by gas chromatography (GC) analysis, (ii) it is not present in the core of LPS or in other polysaccharide molecules produced by serovar Enteritidis, and (iii) it is a stable neutral hexose that occurs in 1:1:1 molar proportions with O-chain mannose and galactose (the immunodominant sugar tyvelose that determines the serovar designation for group D1 salmonellae is unstable upon derivatization and is thus not used to measure O-chain by GC).
Fatty acid analysis.
Fatty acid analysis was by acid-catalyzed methanolysis (methanolic 1 M hydrochloric acid at 85°C for 20 min) and GLC-mass spectrometry of the resulting fatty acid methyl esters extracted with hexane (42). They were identified by their retention times compared to those of authentic standards and by their mass spectra. Heptadecanoic acid was included as an internal standard.
Construction of wzz (cld, rol) mutant.
A 1-kb fragment harboring the wzz gene was amplified from serovar Typhimurium by utilizing primer 1791 (5′-GGCTACACTGTCTCCAGC-3′) and primer 2848 (5′-ACGCGACCACCATCCGGC-3′) (GenBank accession no. M89933) (2). Next, we cloned the PCR-generated fragment into pGEM-T (Promega). To create an insertional mutation, this recombinant plasmid was opened at a unique BglII site within wzz, and a kan-containing BamHI fragment from pUC-4K was ligated into this site. The wzz::kan mutation was then introduced onto the chromosome by homologous recombination. First, the plasmid harboring the wzz::kan mutation was transformed into AA3007 (41), a polA strain, and kanamycin-resistant (Kanr) colonies were selected. Since pUC-based vectors do not replicate in polA mutants, Kanr transformants were the result of recombination between the chromosomal and plasmid wzz regions (7). The mutation was then introduced into SL1344 (16), a polA-carrying strain, by P22-mediated transduction selecting for Kanr transductants as described (25). The Kanr transductants were screened for ampicillin sensitivity, signifying the loss of the plasmid. PCR was used to verify the insertion on the chromosome (data not shown).
Statistical analysis.
Microsoft Excel software was used to perform Student’s t tests. P values of less than 0.05 and 0.01 were defined as indicating moderately and highly significant differences, respectively. Analysis was performed as a one-tailed test for paired samples or as a one-tailed test for unpaired samples as indicated in the text.
RESULTS
Compositional analysis of neutral sugars from isolate 46, a control for all experimental variables.
Experience with LPS structural analysis had suggested that sample variance could be decreased by combining an extraction method used to recover capsule from gram-negative bacteria with growth conditions recommended for recovering Salmonella LPS (see Materials and Methods). In order to establish what methodology was best for assessing LPS characteristics of Salmonella field isolates, we processed 12 isolates as paired samples using water and P:C for extraction. In addition, these 12 isolates were subdivided into two unpaired groups and supplemented with either glucose or GlcNAc to further analyze how much a seemingly small difference in growth conditions could contribute to final results. As a control for all variables, isolate 46 was processed under all conditions.
Recovery yields of O-chain neutral sugars from isolate 46 by water extraction were 17 and 14.8 mg of rhamnose per 100 mg of crude LPS from glucose- and GlcNAc-supplemented cultures, respectively, whereas yields after P:C extraction were 9.0 and 5.8 mg of rhamnose per 100 mg of crude LPS, respectively (Table 2). GlcNAc-supplemented samples formed a tenacious organic phase/aqueous phase interface during P:C extraction; thus, the lower yields of O-chain neutral sugars after GlcNAc supplementation could have been due to trapping of polysaccharides among denatured proteins (Table 2). This problem was minimized by careful separation of water and the aqueous layers. Overall, results suggested that water was better than P:C for recovery of HMW LPS.
TABLE 2.
Comparison of extraction methods for recovery of neutral sugars from serovar Enteritidis
| Isolatea | PT | Recovery (mg/100 mg of crude LPS extract) afterb:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water extraction
|
Extraction with organic solvent
|
||||||||||||
| Rha | Man | Gal | Glc | Hep | GlcNAc | Rha | Man | Gal | Glc | Hep | GlcNAc | ||
| 19 | 8 | 18.6 | 18.4 | 5.9 | 3.3 | 1.7 | 1.9 | 1.0 | 0.8 | 0.7 | 0.7 | 0.2 | 0.1 |
| 38 | 23 | 7.5 | 10.1 | 9.1 | 7.4 | 5.1 | 4.8 | 0.1 | 0.1 | 0.1 | 0.2 | 0.0 | 0.0 |
| 39 | 23 | 2.5 | 18.2 | 7.0 | 7.7 | 2.8 | 2.0 | 0.5 | 0.5 | 0.4 | 4.3 | 0.1 | 0.7 |
| 40 | 13a | 15.6 | 7.8 | 6.6 | 3.2 | 1.2 | 1.0 | 6.5 | 7.5 | 6.8 | 2.6 | 1.6 | 2.8 |
| 41 | 4 | 9.4 | 12.0 | 11.5 | 5.3 | 2.9 | 2.1 | 2.9 | 3.3 | 3.1 | 1.6 | 1.9 | 1.0 |
| 46 | 4 | 17.0 | 17.8 | 16.8 | 4.4 | 2.6 | 1.7 | 9.0 | 8.0 | 6.6 | 6.0 | 0.6 | 0.5 |
| Average | 11.77 | 14.05 | 9.48 | 5.22 | 2.72 | 2.25 | 2.20 | 2.43 | 2.22 | 1.88 | 0.76 | 0.92 | |
| 7 | 13a | 6.1 | 7.1 | 5.7 | 2.9 | 2.4 | 0.3 | 7.8 | 8.1 | 5.9 | 3.0 | 1.5 | 1.2 |
| 25 | 23 | 2.6 | 3.3 | 4.3 | 8.1 | 3.5 | 1.3 | 2.6 | 3.3 | 3.1 | 8.9 | 3.8 | 1.3 |
| 27 | 13a | 5.2 | 6.0 | 6.7 | 2.2 | 3.5 | 1.1 | 1.8 | 2.2 | 2.7 | 1.6 | 0.0 | 0.0 |
| 30 | 8 | 1.1 | 1.7 | 1.2 | 0.8 | 0.4 | 0.0 | 0.6 | 0.4 | 0.3 | 3.3 | 0.4 | 0.0 |
| 37 | 23 | 2.5 | 6.5 | 2.6 | 6.2 | 0.7 | 0.6 | 0.2 | 0.4 | 0.8 | 4.3 | 0.5 | 0.1 |
| 58 | 4 | 6.6 | 8.2 | 7.2 | 2.8 | 1.4 | 0.9 | 1.9 | 2.8 | 4.9 | 2.9 | 0.3 | 0.0 |
| 46 | 4 | 14.8 | 14.3 | 17.1 | 4.4 | 2.1 | 1.6 | 5.8 | 6.7 | 5.5 | 3.2 | 1.0 | 1.0 |
| Average | 5.56 | 6.73 | 6.40 | 3.91 | 2.00 | 0.83 | 2.48 | 2.87 | 2.95 | 4.00 | 1.08 | 0.43 | |
Isolates 19, 38 through 41, and 46 (top) were from cultures supplemented with glucose, and isolates 7, 25, 27, 30, 37, 58, and 46 (bottom) were from cultures supplemented with GlcNAc.
Rha, rhamnose; Man, mannose; Gal, galactose; Glc, glucose; Hep, heptose.
Compositional analysis of neutral sugars from all other isolates.
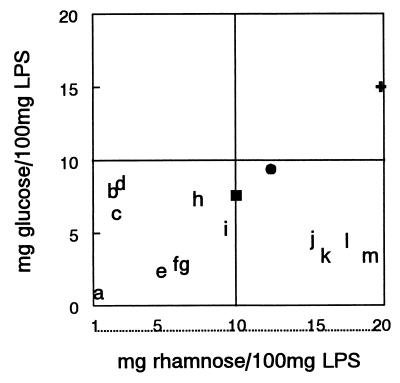
Water extraction of the 12 isolates (13 samples) yielded significantly more O-chain neutral sugar and core hexoses than did extraction with P:C (p of less than 0.005 that data sets are the same) (Table 2). This result is important because P:C extraction failed to detect O-chain neutral sugars that indicated that isolates 19, 40, and 46 were producing HMW LPS (Table 2). These isolates formed a cluster that had on average 16.5 ± 1.3 mg of rhamnose per 100 mg of crude LPS (Fig. 1). Analysis also indicated that all other isolates, whether rough or smooth, clustered together and yielded 1.1 to 9.4 mg of rhamnose per 100 mg of LPS (Fig. 1). Recovery from isolate 30 of all LPS constituents (O-chain, core, and fatty acids) was very low, which perhaps indicated that this sample was inadvertently degraded during derivatization. Two other prominent surface-associated polysaccharides produced by the salmonellae, colanic acid and enterobacterial common antigen, might have yielded other distinguishing neutral sugars such as fucose or excessive amounts of GlcNAc, but neither was detected. Furthermore, Alcian blue staining of gels for acidic polysaccharide confirmed that none of these isolates produced colanic acid. All three of the smooth phage types (PTs) analyzed here, namely, PT4, PT8, and PT13a, were represented in both HMW and LMW LPS clusters, which indicated that PT does not predict or confirm differences in O-chain composition for those isolates with group D1 immunoreactivity (Fig. 1).
FIG. 1.
Cluster analysis of S. enterica serovar Enteritidis rhamnose and glucose yields. Letters on the cluster analysis graph show relative position of each veterinary and clinical isolate listed in Table 1 (PTs are given in parentheses), as follows: a, 30 (PT8); b, 39 (PT23); c, 37 (PT23); d, 25 (PT23); e, 27 (PT13a); f, 7 (PT13a); g, 58 (PT4); h, 38 (PT23); i, 41 (PT4); j, 46 (PT4) from GlcNAc-supplemented culture; k, 40 (PT13a); l, 46 (PT4) from glucose-supplemented culture; m, 19 (PT8). Other strains that were characterized either here or previously and are included in the graph to aid interpretation are wzz Typhimurium (●), avirulent Enteritidis (■), and virulent Enteritidis (+).
Overall, glucose supplementation aided recovery of O-chain from the other isolates, as it did with isolate 46, to a greater extent than GlcNAc supplementation. The differences in yields were significant, but not highly so (Tables 2 and 3; P = 0.0320, one-tailed test for unpaired samples). As occurred with strain 46, all of the organic and aqueous layers from cultures grown with GlcNAc were difficult to separate. Thus, we conclude that glucose is the better growth supplement for recovery of LPS. Exactly why it improved the lubricity of the organic phase/aqueous phase interface is not known, although we suggest a possibility in the portion of the text describing results from fatty acid analysis.
TABLE 3.
Comparison of extraction methods for recovery of fatty acids from serovar Enteritidis
| Isolatea | PT | Recovery (μg/mg of crude LPS extract) after:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Water extraction
|
Extraction with organic solvent
|
||||||||
| C12:O | C14:O | C16:O | 3-OH C14:O | C12:O | C14:O | C16:O | 3-OH C14:O | ||
| 19 | 8 | 0.2 | 0.3 | 0.2 | 0.6 | 1.1 | 1.1 | 2.7b | 0.7 |
| 38 | 23 | 0.3 | 0.5 | 10.3b | 0.9 | 0.8 | 0.9 | 1.0 | 2.0 |
| 39 | 23 | 0.3 | 0.6 | 0.2 | 0.8 | 0.8 | 0.8 | 1.0 | 1.8 |
| 40 | 13a | 1.7 | 1.7 | 1.8 | 3.0 | 0.2 | 0.4 | 1.5b | 0.3 |
| 41 | 4 | 2.3 | 3.3 | 3.2 | 5.4 | 8.5b | 9.5b | 9.0b | 5.1 |
| 46 | 4 | 0.2 | 0.3 | 0.2 | 0.6 | 0.1 | 0.2 | 0.2b | 0.1 |
| Average | 0.83 | 1.12 | 2.65 | 1.88 | 1.92 | 2.15 | 2.57 | 1.67 | |
| 7 | 13a | 3.0 | 4.0b | 4.0b | 3.7 | 0.2 | 0.3 | 1.5 | 0.3 |
| 25 | 23 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.1 |
| 27 | 13a | 1.2 | 0.7 | 0.8 | 3.0 | 0.6 | 1.0 | 1.0 | 2.3 |
| 30 | 8 | 0.2 | 0.6 | 1.0 | 1.0 | 0.0 | 0.1 | 0.1 | 0.1 |
| 37 | 23 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 |
| 58 | 4 | 0.2 | 0.4 | 0.7 | 1.0 | 0.1 | 0.2 | 0.2 | 0.2 |
| 46 | 4 | 0.2 | 0.3 | 0.2 | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 |
| Average | 0.87 | 0.34 | 0.97 | 1.34 | 0.15 | 0.24 | 0.42 | 0.47 | |
Isolates 19, 38 through 41, and 46 (top) were from cultures supplemented with glucose, and isolates 7, 25, 27, 30, 37, 58, and 46 (bottom) were from cultures supplemented with GlcNAc.
Samples with obvious phospholipid contamination. Values are included in the average yield.
Identification of two types of rough isolates.
Three PT23 isolates, 39, 25, and 37, which should lack O-chain, yielded 2.53 ± 0.044 mg of rhamnose per 100 mg of LPS when water extraction was used (Table 2). The nearly identical values for rhamnose yield from these three rough isolates indicated that sample variance was indeed small for water-extracted samples. Results also indicated that each rough strain produced a small amount of O-chain, an amount that was below a threshold needed for a positive serological reaction. However, strain 38, which lacked O-chain immunoreactivity and was also a PT23 strain, yielded 7.5 mg of rhamnose/100 mg of LPS. This was a surprising result, because this was as much O-chain as or more O-chain than that produced by several of the smooth strains (Table 2). Additional analysis with gel electrophoresis revealed that this unique PT23 belonged to a different chemotype, as is discussed in a portion of the text that follows.
Cluster analysis of LPS structures from serovar Enteritidis and identification of two types of HMW LPS.
Glucosylation of O-chain is a nonstoichiometric modification to Salmonella LPS that is likely to differ between isolates. It is important to address glucosylation as a separate topic in regards to evaluating LPS structure, because efficient glucosylation correlates with enhanced oral invasiveness in chicks (11). To evaluate glucosylation, glucose yields were compared to those for rhamnose, which is a stoichiometric component of LPS and required in every O-chain repeating unit. For these data, cluster analysis comparing LPS glucose and rhamnose yields revealed that isolates 19, 40, and 46, which produced HMW LPS, were poorly glucosylated and did not produce a structure like that of Salmonella typhi but were still distinct from strains producing LMW LPS (Fig. 1). In contrast, the virulent serovar Enteritidis, which killed chicks and efficiently contaminated eggs, produced glucosylated HMW LPS with a glucose/rhamnose ratio that placed it well into the upper right quadrant of the cluster analysis graph (Fig. 1). An attenuated isogenic variant, which failed to kill chicks or produce contaminated eggs of the virulent serovar Enteritidis yielded primarily LMW LPS and a trace of HMW LPS, as is usual for most field isolates, and its glucose/rhamnose ratio placed it in the midst of the 12 isolates examined here (Fig. 1). Rough isolates yielded the highest ratios of LPS-associated glucose to LPS-associated rhamnose, which is an expected result due to there being a high percentage of glucose in the core region (Table 1). The average yield of glucose from rough isolates 25, 37, 38, and 39 was 7.35 ± 0.819 mg/100 mg of LPS, in contrast to an average yield of 3.13 ± 1.367 mg/100 mg of LPS for smooth isolates. This difference was highly significant (P = 0.0001). Isolates 19, 40, and 46, which produced HMW LPS, yielded no more glucose on average than did isolates producing LMW LPS (3.67 ± 0.723 and 3.125 ± 1.629 mg/100 mg of LPS, respectively, for water-extracted samples).
Cluster analysis of LPS from a wzz mutant of serovar Typhimurium.
Graphing of the glucose/rhamnose ratio of a wzz mutant of serovar Typhimurium helped to reveal the relationship between LPS glucose and LPS rhamnose further. The wzz Typhimurium made no HMW LPS, but instead it incorporated its excess of O-units into LMW LPS, as is discussed in the portion of the text describing gel electrophoresis patterns. The wzz mutant produced 9.4 mg of glucose and 12 mg of rhamnose per 100 mg of LPS, and cluster analysis placed its glucose/rhamnose ratio in a position intermediate between the ratios for the veterinary clinical isolates that produced HMW LPS and those that produced LMW LPS (Fig. 1). Therefore, the wzz mutant fell into an unusual category, because it produced an elongated LMW structure that could be more efficiently glucosylated.
Thus, glucosylation of HMW LPS O-chain is a complex topic, which can be summarized based on all available data as follows. First, immunogenic O-chain is inefficiently glucosylated when recovered in low yields, which is especially evident when an adjustment is made for the relative contribution that the core makes to total glucose yields (Fig. 1, points e, f, and g). In this case, previous analysis had indicated that one of eight O-chain units from LMW LPS was glucosylated (29). The frequency of glucosylation then increases concurrently with chain length and forms a linear relationship with rhamnose as the contribution of glucose from the core to the total yield of neutral sugars becomes insignificant (Fig. 1, points ■, ●, and +). In this case, previous analysis indicated that at least one of every two O-chain units was glucosylated. However, results from this study indicated that glucosylation of HMW LPS can be specifically inhibited (Fig. 1, points j, k, l, and m).
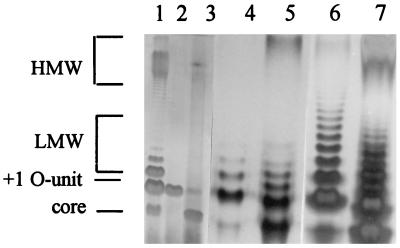
Comparison of compositional results to PAGE patterns.
Examination of LPS samples by DOC-PAGE confirmed GC results indicating that rough isolates of serovar Enteritidis could be divided into two different types of PT23 (Fig. 2). One pattern was identical to the classic Ra chemotype that produces at most a trace of O-chain either due to mutation in linking enzymes, for example, WaaL, or due to loss of O-chain biosynthesis resulting from some change in the wba operon. This pattern was associated with a mean yield of 2.53 ± 0.044 mg of rhamnose/100 mg of crude LPS (Fig. 2, lane 3). A second pattern was that of a Wzy phenotype, which failed to polymerize O-chain but was linkage proficient and added a single unit to the core (Fig. 2, lane 2). Compositional analysis had shown that the Wzy phenotype (isolate 38) produced 7.5 mg of rhamnose/100 mg of LPS (Fig. 2, lane 1). Gel patterns suggested that free O-chain repeat units produced by the Wzy phenotype drove linkage to the core, because no bands migrating faster than that for core plus one O-chain unit were detected for this strain (Fig. 2, lane 2). In contrast, polymerization diverted O-chain units into a few HMW molecules produced by most field isolates, which left some core unlinked to any O-chain (Fig. 2, lanes 1, 5, and 7).
FIG. 2.
Gel electrophoresis of LPS from Salmonella. Gels are 12% DOC as described in Materials and Methods. Lane 1, LPS from a smooth serovar Enteritidis isolate, i.e., isolate 46; lane 2, LPS with core plus one O-unit from a rough serovar Enteritidis isolate exhibiting the Wzy phenotype, i.e., isolate 38; lane 3, LPS from a rough serovar Enteritidis isolate exhibiting the Ra chemotype, i.e., isolate 37; lane 4, LPS from a wzz mutant of serovar Typhimurium; lane 5, LPS from wild-type serovar Typhimurium; lane 6, threefold more LPS from wzz mutant than included in lane 4; lane 7, threefold more LPS from wild-type serovar Typhimurium than included in lane 5. LMW LPS is core plus 2 to 6 O-units for lanes 1, 4, and 5, core plus 2 to 12 O-units for lane 6, and core plus 2 to 8 O-units for lane 7.
Thus, the Wzy null phenotype yielded a core molecule that was uniformly linked to a single O-unit, which is perhaps unusual to encounter in field isolates but is probably not rare. In contrast, the Ra core (bottom band) of the wzz mutant in gels was noticeably diminished as compared to typical serovar Typhimurium LPS (Fig. 2, lanes 4 and 6), whereas wild-type serovar Typhimurium produced the usual salmonella bimodal structure with LMW and HMW regions (Fig. 2, lanes 5 and 7). This indicated that the wzz mutant had considerable wzy character as seen by detection of a prominent band for core plus one O-unit. In addition, the wzz mutant produced more-elongated LMW LPS than did the wild type, and LMW chain lengths of 12 repeating units were observed for the wzz mutant, in contrast to 8 for the wild type (Fig. 2, lanes 6 and 7, respectively).
Relative yields of β-hydroxymyristic acid, an LPS-associated fatty acid.
Different yields of fatty acids were obtained by the two extraction methods, as indicated by yields of β-hydroxymyristic acid (3-OH C14:O), which is a prominent LPS-associated fatty acid component of lipid A (Table 3). In order of decreasing β-hydroxymyristic acid yields, the following ranks were obtained for the four different conditions used to examine LPS: (i) water extraction with glucose supplementation (1.88 μg/mg of LPS), (ii) P:C extraction with glucose supplementation (1.67 μg/mg of LPS), (iii) water extraction with GlcNAc supplementation (1.34 μg/mg of LPS), and (iv) P:C extraction with GlcNAc supplementation (0.47 μg/mg). P:C extraction with GlcNAc supplementation was a combination that was especially likely to yield nondetectable amounts of fatty acids, and thus we conclude that this is indeed a poor combination to use for recovery of LPS (Table 3).
Average recovery of lipid A from the rough PT23 isolates by using P:C extraction was near that of smooth isolates (all other PTs), with rough isolates and smooth isolates yielding 1.05 and 1.14 μg of β-hydroxymyristic acid/mg of LPS, respectively. Conversely, water extraction was better for recovering fatty acids from smooth strains and on average yielded 3.17 μg of β-hydroxymyristic acid/mg of LPS, in contrast to an 0.45-μg/mg yield for rough strains. These are important results for two reasons. First, yields of fatty acids obtained by using organic solvent extraction confirm previous observations that P:C extraction recovers both rough and smooth LPS structures from serovar Enteritidis equally (4). However, these results also indicate that there are hydrophilic and hydrophobic LPS molecules and that hydrophilic structures are better recovered by using water-based extraction methods designed for recovering bacterial capsules than by using P:C extraction. Water extraction does recover LPS from rough strains, but it might lose a population of core molecules that are totally devoid of any O-chain.
Yields of other fatty acids that can be part of either LPS or phospholipid.
Palmitic acid (C16:O) in LPS samples has two sources. It can be a secondary acyl group in the structure of lipid A via a transacylation reaction, or it can be recovered as contaminating phospholipid in crude LPS preparations. Thus, it is not unusual to recover some palmitic acid in LPS samples, but yields higher than those of β-hydroxymyristic acid should be interpreted as contamination. P:C extraction of glucose-supplemented smooth PTs (isolates 19, 40, 41, and 46) was especially likely to result in gross phospholipid contamination as measured by yields of palmitic acid (C16:O) (Table 3). One water-extracted sample from a culture supplemented with glucose was grossly contaminated (isolate 38), whereas one water-extracted sample from a culture supplemented with GlcNAc was minimally contaminated (isolate 7) (Table 3). These results perhaps indicate that a change in outer membrane fatty acid composition or in the interaction of acyl groups with LPS and proteins explains why glucose supplementation aided extractions to a greater extent than GlcNAc supplementation. Further investigation is required to ascertain exactly why water extraction in combination with glucose supplementation is the best combination for recovery of HMW LPS.
DISCUSSION
In spite of its genetic homogeneity, serovar Enteritidis is phenotypically pleomorphic and isolates vary in their ability to contaminate eggs and to contribute to human disease (14, 20, 26, 34, 36, 37, 39). Results presented here give a first indication that Wzy activity is a central source of variation among natural field isolates of smooth serovar Enteritidis and that its loss defines a subset of PT23 isolates. In contrast, we could find little evidence that Wzz function was variable among field isolates, because the unusual phenotype of the wzz mutant was not detected here or during previous analyses of LPS structure that characterized egg-contaminating serovar Enteritidis. The finding that the wzz mutant increases the length of LMW LPS O-chain without yielding any HMW LPS suggests that for the pathogenic salmonellae, wzz is required to produce HMW LPS. It is also possible that the variations are due to changes in the ratio of Wzy and Wzz, similar to what has been described for Shigella (3).
When information from other studies is considered with these results, it appears that there are two types of HMW LPS structures, i.e., those that are highly glucosylated and those that are not. O-chain glucosylation is the function of the gene oafR, which is located outside the LPS biosynthetic waa (rfa) and wba (rfb) operons (13, 24, 33), and previous analyses had shown that HMW O-chain is the preferred substrate for glucosylation (29). This means that oafR activity is dependent upon functional wzy and wzz genes. There is some indication that temperature and other environmental conditions alter the preference of the enzyme for O-chain, as growth at ambient temperatures enhances glucosylation of LMW LPS compared to that at 37°C (9, 11). It will be interesting to determine what biological effect is associated with variable glucosylation of HMW and LMW structures, which we can now do because of the unique phenotype of the wzz mutant.
Previous attempts to characterize LPS microstructural heterogeneity were hampered by extraction techniques that limited the ability to process many isolates or yielded samples too crude for derivatization. Water extraction still produces a crude preparation, but it is clean enough of extraneous phospholipid and other cell membrane components for GLC analysis of (i) the O-chain sugars rhamnose, mannose, galactose, and glucose, (ii) the core sugars heptose and GlcNAc, and (iii) lipid A fatty acids. Also, the amount of organic compounds used during water extraction is minimal, which makes it easier and safer to perform extractions. Water extraction in combination with glucose supplementation aided recovery of LPS, possibly by altering the phospholipid composition of the outer membrane and increasing the lubricity of denatured proteins. Overall, these findings indicate that specific growth conditions should be used when LPS is characterized as part of an epidemiological investigation and that HMW LPS is best treated as a hydrophilic structure similar to some capsules.
Thus, the broad range of LPS microstructural heterogeneity seen here strongly suggests that regulation of LPS-modifying enzymes located outside of the main waa and wba biosynthetic operons of the salmonellae results in differences between isolates that can alter epidemiological patterns in humans and animals. As proposed by others, the importance of O-chain serotype in general is to determine what problems will be caused by any one serotype (30). So far, only the group D broad-host-range serovar Enteritidis routinely has contaminated hen eggs and caused disease in people, which is indeed a very narrow problem within the spectrum of food-borne disease. We thus extend the concept that serotype is important to the epidemiology of the salmonellae to include intraserovar differences that do not always produce a gross immunological change, for example, as seen when smooth strains convert to rough. Thus, variable activity of LPS-modifying enzymes could lead to noticeable and prolonged changes in outbreak incidence as exemplified by the current problem with serovar Enteritidis. These analytical methods are being used to investigate genetic differences between isolates of serovar Enteritidis.
ACKNOWLEDGMENTS
This research was supported by USDA CRIS 6612-32000-017 to the Southeast Poultry Research Laboratory and a grant from the USDA/CSREES NRICGP, Food Safety Division (98-35201-6281), to J.G.-P. The collaboration of R.W.C. was made possible by a grant from the D.O.E. (DE-FG05-93ER20097) to the CCRC.
We thank Murry Stein, University of Vermont, for providing materials for the construction of the wzz mutant, and T. J. Humphrey, PHLS, Exeter, United Kingdom, for thoughts and advice on the limitations of phage typing.
REFERENCES
- 1.Bean N H, Griffin P M. Foodborne disease outbreaks in the USA 1973–1987. Pathogens, vehicles and trends. J Food Prot. 1990;53:804–817. doi: 10.4315/0362-028X-53.9.804. [DOI] [PubMed] [Google Scholar]
- 2.Collins L V, Hackett J. Molecular cloning, characterization, and nucleotide sequence of the rfc gene, which encodes an O-antigen polymerase of Salmonella typhimurium. J Bacteriol. 1991;173:2521–2529. doi: 10.1128/jb.173.8.2521-2529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels C, Vindurampulle C, Morona R. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy) Mol Microbiol. 1998;28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 4.Darveau R P, Hancock R E W. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium isolates. J Bacteriol. 1983;155:831–838. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrogosz W J. N-Acetylglucosamine assimilation in Escherichia coli and its relation to catabolite repression. J Bacteriol. 1968;95:585–591. doi: 10.1128/jb.95.2.585-591.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman R C, Leive L. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem. 1980;107:145–153. doi: 10.1111/j.1432-1033.1980.tb04635.x. [DOI] [PubMed] [Google Scholar]
- 7.Groisman E A, Sturmoski M A, Solomon F R, Lin R, Ochman H. Molecular, functional, and evolutionary analysis of sequences specific to Salmonella. Proc Natl Acad Sci USA. 1993;90:1033–1037. doi: 10.1073/pnas.90.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guard-Petter J. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl Environ Microbiol. 1998;64:2166–2172. doi: 10.1128/aem.64.6.2166-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guard-Petter, J. Unpublished data.
- 10.Guard-Petter J, Henzler D, Rahman M M, Carlson R W. On-farm monitoring of invasive Salmonella enteritidis from the spleens of mice captured in the hen house environment. Appl Environ Microbiol. 1997;63:1588–1593. doi: 10.1128/aem.63.4.1588-1593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guard-Petter J, Keller L H, Rahman M M, Carlson R W, Silvers S. O-antigen variation, matrix formation, and virulence of Salmonella enteritidis. Epidemiol Infect. 1996;117:219–231. doi: 10.1017/s0950268800001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guard-Petter J, Lakshmi B, Carlson R, Ingram K. Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method. Appl Environ Microbiol. 1995;61:2845–2851. doi: 10.1128/aem.61.8.2845-2851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helander I M, Moran A P, Makela P H. Separation of two lipopolysaccharide populations with different contents of O-antigen 122 in Salmonella enterica serovar Typhimurium. Mol Microbiol. 1992;6:2857–2862. doi: 10.1111/j.1365-2958.1992.tb01465.x. [DOI] [PubMed] [Google Scholar]
- 14.Helmuth R, Schroeter A. Molecular typing methods for serovar Enteritidis. Int J Food Microbiol. 1994;21:69–77. doi: 10.1016/0168-1605(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 15.Hogue A, White P, Guard-Petter J, Schlosser W, Gast R, Ebel E, Farrer J, Gomez T, Madden J, Madison M, McNamara A M, Morales R, Parham D, Sparling P, Sutherlin W, Swerdlow D. Epidemiology and control of egg-associated Salmonella Enteritidis in the United States of America. Rev Sci Tech Off Int Epizoot. 1997;16:542–553. doi: 10.20506/rst.16.2.1045. [DOI] [PubMed] [Google Scholar]
- 16.Hoiseth S K, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey T J. Contamination of egg shell and contents with Salmonella enteritidis: a review. Int J Food Microbiol. 1994;21:31–40. doi: 10.1016/0168-1605(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey T J, Baskerville A, Mawer S, Rowe B, Hoppers B. Salmonella enteritidis phage type 4 from the contents of intact eggs: a study involving naturally infected hens. Epidemiol Infect. 1989;103:415–424. doi: 10.1017/s0950268800030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey T J, Hinton M H. Salmonella enteritidis PT 4 super bug. J Appl Bacteriol. 1989;67:XXIV. [Google Scholar]
- 20.Kantama L, Jayanetra P. Salmonella enteritidis outbreak in Thailand: study by random amplified polymorphic DNA (RAPD) analysis. Southeast Asian J Trop Med Public Health. 1996;27:119–125. [PubMed] [Google Scholar]
- 21.Kim J S, Reuhs B L, Rahman M M, Ridley B, Carlson R W. Separation of bacterial capsular and lipopolysaccharides by preparative electrophoresis. Glycobiology. 1996;6:433–437. doi: 10.1093/glycob/6.4.433. [DOI] [PubMed] [Google Scholar]
- 22.Lee L, Cherniak B. Capsular polysaccharide of Clostridium perfringens Hobbs 10. Infect Immun. 1974;9:318–322. doi: 10.1128/iai.9.2.318-322.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin E C C. Dissimilatory pathways for sugars, polyols and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 307–433. [Google Scholar]
- 24.Makela P H, Stocker B A D. Genetics of lipopolysaccharide. In: Rietschel E T, editor. Handbook of endotoxin. 1. Chemistry of endotoxin. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1984. pp. 59–137. [Google Scholar]
- 25.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 26.Millemann Y, Lesage-Descauses M-C, Lafont J P, Chaslus-Dancla E. Comparison of random amplified polymorphic DNA analysis and enterobacterial repetitive intergenic consensus-PCR for epidemiological studies of Salmonella. FEMS Immunol Med Microbiol. 1996;14:129–134. doi: 10.1111/j.1574-695X.1996.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 27.Munford R S, Hall C L, Rick P D. Size heterogeneity of Salmonella typhimurium lipopolysaccharides in outer membranes and culture supernatant membrane fragments. J Bacteriol. 1980;144:630–640. doi: 10.1128/jb.144.2.630-640.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson A A, Haug A, McGroarty E J. Physical properties of short- and long-O-antigen-containing fractions of lipopolysaccharide from Escherichia coli 0111:B4. J Bacteriol. 1986;165:116–122. doi: 10.1128/jb.165.1.116-122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman M M, Guard-Petter J, Carlson R W. A virulent isolate of Salmonella enteritidis produces a Salmonella typhi-like lipopolysaccharide. J Bacteriol. 1997;179:2126–2131. doi: 10.1128/jb.179.7.2126-2131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves P. Evolution of Salmonella O antigen variation involved in interspecies gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 31.Reeves P. Biosynthesis and assembly of lipopolysaccharide. New Compr Biochem. 1994;27:281–317. [Google Scholar]
- 32.Schlecht S, Galanos C. Influence of the glucose concentration on the yield of biomass and lipopolysaccharides in Salmonella cultures. Int J Med Microbiol Virol Parasitol Infect Dis. 1994;281:30–37. doi: 10.1016/s0934-8840(11)80634-1. [DOI] [PubMed] [Google Scholar]
- 33.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley J, Saunders N. DNA insertion sequences and the molecular epidemiology of Salmonella and Mycobacterium. J Med Microbiol. 1996;45:236–251. doi: 10.1099/00222615-45-4-236. [DOI] [PubMed] [Google Scholar]
- 35.St. Louis M E, Morse D L, Potter M E, DeMelfi T M, Guzewuch J J, Tauxe V, Blake P A. The emergence of grade A eggs as a major source of Salmonella enteritidis infections: new implications for the control of salmonellosis. JAMA. 1988;259:2103–2109. [PubMed] [Google Scholar]
- 36.Stubbs A D, Hickman-Brenner F W, Cameron D N, Farmer J J., III Differentiation of Salmonella enteritidis phage type 8 strains: evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns, and biotyping. J Clin Microbiol. 1994;32:199–201. doi: 10.1128/jcm.32.1.199-201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thong K-L, Ngeow Y-F, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai C M, Frasch C E. A sensitive silver strain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 39.Usera M A, Popovic T, Bopp C A, Strockbine N A. Molecular subtyping of Salmonella enteritidis phage type 8 strains from the United States. J Clin Microbiol. 1994;32:194–198. doi: 10.1128/jcm.32.1.194-198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of this procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 41.Whitfield H J, Levine G. Isolation and characterization of a mutant of Salmonella typhimurium deficient in a major deoxyribonucleic acid polymerase activity. J Bacteriol. 1973;116:54–58. doi: 10.1128/jb.116.1.54-58.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York W S, Darvill A G, McNeil M, Stevenson T T, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]