Abstract
Vibrio cholerae identification based on molecular sequence data has been hampered by a lack of sequence variation from the closely related Vibrio mimicus. The two species share many genes coding for proteins, such as ctxAB, and show almost identical 16S DNA coding for rRNA (rDNA) sequences. Primers targeting conserved sequences flanking the 3′ end of the 16S and the 5′ end of the 23S rDNAs were used to amplify the 16S-23S rRNA intergenic spacer regions of V. cholerae and V. mimicus. Two major (ca. 580 and 500 bp) and one minor (ca. 750 bp) amplicons were consistently generated for both species, and their sequences were determined. The largest fragment contains three tRNA genes (tDNAs) coding for tRNAGlu, tRNALys, and tRNAVal, which has not previously been found in bacteria examined to date. The 580-bp amplicon contained tDNAIle and tDNAAla, whereas the 500-bp fragment had single tDNA coding either tRNAGlu or tRNAAla. Little variation, i.e., 0 to 0.4%, was found among V. cholerae O1 classical, O1 El Tor, and O139 epidemic strains. Slightly more variation was found against the non-O1/non-O139 serotypes (ca. 1% difference) and V. mimicus (2 to 3% difference). A pair of oligonucleotide primers were designed, based on the region differentiating all of V. cholerae strains from V. mimicus. The PCR system developed was subsequently evaluated by using representatives of V. cholerae from environmental and clinical sources, and of other taxa, including V. mimicus. This study provides the first molecular tool for identifying the species V. cholerae.
Vibrio cholerae is a noninvasive, gram-negative bacterium responsible for severe epidemics of cholera and endemic diarrhea in many parts of the world, especially developing countries (14, 33). On the basis of several genotypic and phenotypic characteristics, V. cholerae O1 strains can be subdivided into two biotypes, classical and El Tor. The current cholera pandemic, the seventh, which started in 1961, is caused by the El Tor biotype, whereas the classical O1 strains were responsible for previous pandemics (1881 to 1896 and 1899 to 1923). Non-O1 strains have not caused major cholera epidemics, until serotype O139, named Bengal, emerged in India in 1992 (1). Genetic and phenotypic evidence strongly suggests that the O139 strain arose from a V. cholerae O1 strain, probably an El Tor biotype, by horizontal gene transfer (3, 4, 15, 24, 49).
The species Vibrio mimicus is a biochemically atypical group of V. cholerae strains (17). V. mimicus produces a variety of toxins, including cholera toxin, and it causes sporadic diarrhea (8, 10, 37). It has been isolated from a number of environmental sources, including oysters, prawns, turtle eggs, rivers, and brackish waters, as well as clinical samples (8–10). On the basis of a full-length sequence comparison, V. mimicus has been determined to have genes coding for 16S rRNA (rDNA) nearly identical to those of V. cholerae, i.e., differing only in 6 of 1,456 nucleotides (41).
Genetic information derived from the rRNA (rrn) operon provides valuable taxonomic information. The rRNA-coding regions, notably 16S rDNA, have been used extensively to underpin phylogenetic structures at the species level or above (30, 51). Intergenic spacer regions (ISRs), especially those located between the 16S and 23S rDNAs, were thought to be under less evolutionary pressure and, therefore, to provide higher genetic variation than rRNA coding regions (20–23, 29, 31, 35, 44). In general, the ISR possesses a secondary structure and, frequently, tRNA genes (7). The number of rrn operons in bacteria varies from 1 to 11 and multiple operons often contain the same ISR. For example, Escherichia coli contains seven rrn operons, three of which comprise the ISR containing two tRNA genes for isoleucine and alanine; the remaining four have the ISR containing a single tRNA gene for glutamate (16). Therefore, genetic variations in ISR are not only interstrain but also intercistronic.
Nandi et al. (36) demonstrated that epidemic V. cholerae O1 and O139 strains have 9 rrn operons, whereas non-O1/non-O139 strains possess 10 operons. Using a pair of oligonucleotide primers flanking 16S and 23S rDNAs of E. coli, Coelho et al. (12) showed that ISR PCR amplification patterns from O1 classical, O1 El Tor, and O139 strains were different and, thereby, provide a potential tool for studying the epidemiology of V. cholerae. In the study reported here, the nucleotide sequences of ISR from V. cholerae O1 classical, O1 El Tor, O139, and non-O1/non-O139 strains, as well as those from V. mimicus strains, were obtained and analyzed to seek interspecies, interserotype, and intercistronic variations.
MATERIALS AND METHODS
Strains.
Strains of V. cholerae included in this study, listed in Table 1, were grown on Luria-Bertani agar (LB; Difco Laboratories, Detroit, Mich.) at 37°C and maintained on LB slants at room temperature or as suspensions in 25% glycerol at −70°C.
TABLE 1.
Test strains used in this study
| Strain no. | Species and serovar | Other designation(s) and information |
|---|---|---|
| RC2T | V. cholerae O1 classical | ATCC 14035 |
| RC25 | V. cholerae O1 El Tor | Clinical isolate; Mexico, 1991 |
| RC4 | V. cholerae O139 | Clinical isolate; Bangladesh, J. Johnson AI1877 |
| RC45 | V. cholerae O22 | J. Johnson Y334 |
| RC48 | V. cholerae O31 | J. Johnson Y1, NRT36S |
| RC42 | V. cholerae non-O1/non-O139 | ATCC 14547, V. albensis; fish from the Elbe River |
| RC44 | V. cholerae non-O1/non-O139 | ATCC 25874, clinical isolate |
| RC47 | V. cholerae non-O1/non-O139 | ATCC 25872, clinical isolate |
| RC5T | V. mimicus | ATCC 33653; isolated from human ear; North Carolina |
| RC55 | V. mimicus | Environmental isolate; Louisiana |
PCR primers for ISR amplification.
A pair of oligonucleotides were designed to amplify the ISR of V. cholerae and related taxa. The forward primer, pr16S-1541F-PstI (5′-TTTCTGCAGYGGNTGGATCACCTCCTT-3′ (the PstI site is indicated by the underline), corresponding to 16S rDNA positions 1523 to 1541 of E. coli (5), was designed to match members of the domain bacteria, and the reverse primer, pr23S-25R-EcoRI (5′-ACGAATTCTGACTGCCMRGGCATCCA-3′ (the EcoRI site is indicated by the underline), corresponding to 23S rDNA positions 44 to 25 of E. coli (6), was designed based on sequences from E. coli (GenBank accession number V00331), Pseudomonas aeruginosa (Y00432), V. cholerae (U10956), and Vibrio vulnificus (U10951).
DNA isolation and PCR.
Chromosomal DNA was isolated by using guanidine thiocyanate according to the method of Chun and Goodfellow (11). Approximately 50 ng of DNA was subjected to PCR amplification, in a total volume of 50 μl containing primers (each at a concentration of 0.4 mM), a mixture of deoxynucleoside triphosphates (each at a concentration of 200 mM), Taq polymerase, and buffer (Promega, Madison, Wis.). A DNA thermal cycler (PTC-200; MJ Research) used for thermal amplification was programmed for the following: (i) an initial extensive denaturation step, consisting of treatment at 94°C for 2 min; (ii) 30 reaction cycles, with each cycle consisting of treatment at 94°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min; and (iii) a final extension step, consisting of treatment at 72°C for 10 min. The PCR products were separated by 1% agarose gel electrophoresis, stained with ethidium bromide, and visualized with UV light.
Cloning.
The ISR amplicons were purified with the Wizard PCR Mini-Prep Kit (Promega) according to the manufacturer’s instructions. The preparations were digested in tubes containing EcoRI, PstI, and buffer H (Promega) at 37°C for 1 h and then treated at 70°C for 15 min to inactivate the restriction enzymes. The digested ISR fragments were ligated into the predigested plasmids prepared as follows: pGEM-T Easy Vector (Promega) was recircularized by ligation, transformed into E. coli JM109, purified by using the Wizard Mini-Prep Kit, double-digested with EcoRI and PstI, and purified from 1% agarose gels by using the GeneClean II Kit (Bio 101, Vista, Calif.). The ligation was achieved by using T4 DNA ligase (Promega). The resultant mixture was transformed into highly competent E. coli JM109, and the recombinants were selected according to the standard blue-white cloning procedure (43). The selected clones were grown in LB broth containing ampicillin (100 μg ml−1), and the plasmids were purified with the Wizard Mini-Prep Kit. The size of the inserts was confirmed by 1% agarose gel electrophoresis after the EcoRI-PstI treatment.
Sequencing of ISR.
Nucleotide sequences of ISR inserts were determined by using the ABI PRISM Dye Terminator Cycle Sequencing Kit (Perkin-Elmer, Norwalk, Conn.) and an ABI 377 automated DNA sequencer. Two primers flanking the multiple cloning site of pGEM-T Easy Vector, prGTf (5′-TACGACTCACTATAGGGCGA-3′) and prGTr (5′-CTCAAGCTATGCATCCAACGC-3′), were synthesized and used to sequence both DNA strands.
Data analysis.
Nucleotide sequences were aligned by using the PILEUP program in the Genetics Computer Group package; the sequences were then adjusted manually. Evolutionary distances were calculated by using the model of Jukes and Cantor (25), and phylogenetic trees were inferred by the neighbor-joining method (42).
PCR for V. cholerae identification.
A pair of primers, namely, prVC-F (5′-TTAAGCSTTTTCRCTGAGAATG-3′; positions 227 to 248 of V. cholerae RC2 ISR-2) and prVCM-R (5′-AGTCACTTAACCATACAACCCG-3′; positions 501 to 12 of 23S rDNA of V. cholerae), were synthesized and used for V. cholerae-specific PCR experiments. PCR conditions were identical to those for ISR amplification, except that annealing was done at 60°C for 1 min and extension was done at 72°C for 1 min. The results were confirmed by 1.5% agarose gel electrophoresis. For routine identification, cells were scraped from LB plates, boiled in distilled water, and used as PCR template DNAs. False-negative results due to PCR inhibition and insufficient template DNA were checked by performing PCR targeting of the universal region of 16S rDNA.
Nucleotide sequence accession numbers.
Nucleotide sequences for ISRs determined in this study were deposited in GenBank under accession numbers AF114721 to AF114749.
RESULTS
PCR of ISR.
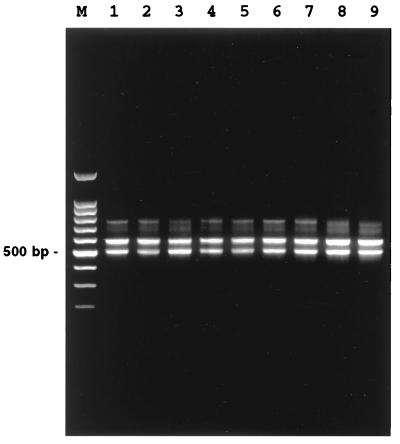
PCR with the two primers, p16S-1541F-PstI and p23S-25R-EcoRI flanking 16S-23S rDNA, yielded a nearly identical band pattern for the V. cholerae and V. mimicus strains containing the two major bands (ca. 580 and 500 bp) and one minor band (ca. 750 bp) (Fig. 1). In addition, a faint band of ca. 700 bp was visible for both species.
FIG. 1.
Electrophoresis with a 1.5% agarose gel of PCR-amplified 16S-23S rRNA intergenic spacer regions of V. cholerae and V. mimicus. Lanes: M, molecular weight marker (100-bp ladder); 1, V. cholerae O1 classical RC2; 2, V. cholerae O1 El Tor RC25; 3, V. cholerae O139 RC4; 4, V. cholerae O22 RC45; 5, V. cholerae O31 RC48; 6, V. cholerae non-O1/non-O139 RC42; 7, V. cholerae non-O1/non-O139 RC44; 8, V. mimicus RC5; and 9, V. mimicus RC55.
Sequence analysis of ISR.
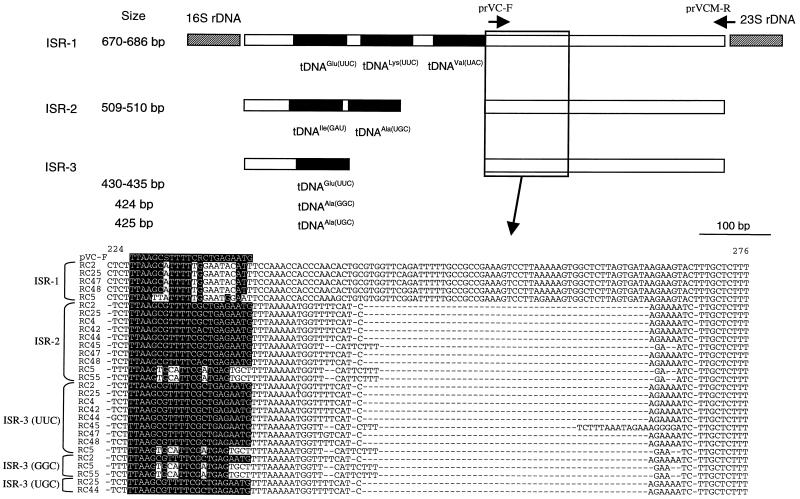
Recombinant plasmids containing different ISR amplicons were screened by simultaneously digesting with EcoRI and PstI, and those plasmids containing insert DNAs corresponding to three PCR amplicons of different sizes were identified and sequenced. The results of the sequence analyses are summarized in Fig. 2. In the type strain of V. cholerae (strain RC2T, O1 classical), the largest ISR (i.e., 750-bp amplicon; designated ISR-1) consisted of 686 nucleotides and contained three tRNA genes (tDNAs) coding for tRNAGlu(UUC), tRNALys(UUU), and RNAVal(UAC), respectively (anticodons are indicated in parentheses). Among the other V. cholerae strains, RC25 (O1 El Tor), RC47 (non-O1/non-O139), and RC48 (O31) showed an almost identical ISR-1 of the same length (Table 2) as that of RC2. In contrast, V. mimicus RC5T had a shorter version (i.e., 670 bp) and differed from the V. cholerae strains by 16 nucleotides. Such a length difference between two species is attributed to a 16-bp deletion in the V. mimicus strain, located between tDNAGlu and tDNALys, where V. cholerae and V. mimicus strains had 19- and 3-bp spacers, respectively.
FIG. 2.
Schematic representation of 16S-23S rRNA intergenic spacer regions of V. cholerae and V. mimicus. An alignment of ISR sequences, corresponding to positions 224 to 276 of V. cholerae RC2 ISR-2, is presented in detail, and identities are indicated by a solid box. The positions of PCR primers, namely, prVC-F and prVCM-R, are indicated by arrows. For details, see the text.
TABLE 2.
ISR-1 sequence similarity values among V. cholerae and V. mimicus strains
| Strain | Size (bp) | Sequence similarity valuesa
|
||||
|---|---|---|---|---|---|---|
| RC2 | RC25 | RC47 | RC48 | RC5 | ||
| V. cholerae O1 classical RC2T | 686 | 0/686 | 1/686 | 5/686 | 17/670 | |
| V. cholerae O1 El Tor RC25 | 686 | 100.0 | 1/686 | 5/686 | 17/670 | |
| V. cholerae NOb RC47 | 686 | 99.9 | 99.9 | 4/686 | 16/670 | |
| V. cholerae O31 RC48 | 686 | 99.3 | 99.3 | 99.4 | 16/670 | |
| V. mimicus RC5T | 670 | 97.5 | 97.5 | 97.6 | 97.6 | |
The values in the upper-right triangle indicate nucleotide differences/total number compared. The values in the lower-left triangle indicate the % similarity values.
NO, non-O1/non-O139.
In all of the V. cholerae and V. mimicus strains examined in this study, the 580-bp amplicon, designated ISR-2, invariably contained tDNAIle(GAU) and tDNAAla(UGC) and had a total size of either 509 or 510 bp. Intraspecies ISR-2 sequence similarities among the V. cholerae and V. mimicus strains ranged from 99.0 to 100%, whereas the corresponding values for the interspecies comparisons were lower, ranging from 97.0 to 98.0% (Table 3).
TABLE 3.
ISR-2 sequence similarity values among V. cholerae and V. mimicus strains
| Straina | Size (bp) | Sequence similarity valuesb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RC2 | RC25 | RC4 | RC42 | RC44 | RC45 | RC47 | RC48 | RC5 | RC55 | ||
| V. cholerae O1 classical RC2T | 510 | 2/510 | 1/510 | 4/510 | 1/510 | 4/505 | 2/510 | 5/510 | 13/505 | 12/505 | |
| V. cholerae O1 El Tor RC25 | 510 | 99.6 | 1/510 | 4/510 | 1/510 | 4/505 | 2/510 | 5/510 | 13/505 | 12/505 | |
| V. cholerae O139 RC4 | 510 | 99.8 | 99.8 | 3/510 | 0/510 | 3/505 | 1/510 | 4/510 | 12/505 | 11/505 | |
| V. cholerae NO RC42 | 510 | 99.2 | 99.2 | 99.4 | 3/510 | 4/505 | 4/510 | 3/510 | 15/505 | 14/505 | |
| V. cholerae NO RC44 | 510 | 99.8 | 99.8 | 100.0 | 99.4 | 3/505 | 1/510 | 4/510 | 12/505 | 11/505 | |
| V. cholerae O22 RC45 | 509 | 99.2 | 99.2 | 99.4 | 99.2 | 99.4 | 4/505 | 3/505 | 13/509 | 12/509 | |
| V. cholerae NO RC47 | 510 | 99.6 | 99.6 | 99.8 | 99.2 | 99.8 | 99.2 | 5/510 | 11/505 | 10/505 | |
| V. cholerae O31 RC48 | 510 | 99.0 | 99.0 | 99.2 | 99.4 | 99.2 | 99.4 | 99.0 | 14/505 | 13/505 | |
| V. mimicus RC5T | 509 | 97.4 | 97.4 | 97.6 | 97.0 | 97.6 | 97.4 | 97.8 | 97.2 | 3/509 | |
| V. mimicus RC55 | 510 | 97.6 | 97.6 | 97.8 | 97.2 | 97.8 | 97.6 | 98.0 | 97.4 | 99.4 | |
NO, non-O1/non-O139.
See Table 2, footnote a.
The smallest ISR amplicon, designated ISR-3, corresponding to the ca. 500-bp PCR product, was 430 to 445 bp long and contained one tDNA coding for tRNAGlu(UUC). In contrast to ISR-1 and ISR-2, ISR-3 from some strains contained single tDNAAla, in addition to ISR-3 containing tRNAGlu (Table 4). The additional ISR-3 containing one tRNAAla(GGC) was recovered from V. cholerae RC2 and V. mimicus RC5 and RC55. Similarly, ISR-3 with tDNAAla(UGC) was found in V. cholerae RC25 and RC44.
TABLE 4.
ISR-3 sequence similarity values among V. cholerae and V. mimicus strains
| Straina | Size (bp) | Sequence similarity valuesb with tDNA found in ISR-3:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tDNAGlu(UUC)
|
tDNAAla(GGC)
|
tDNAAla(UGC)
|
|||||||||||||
| RC2 | RC25 | RC4 | RC42 | RC44 | RC45 | RC47 | RC48 | RC5 | RC2 | RC5 | RC55 | RC25 | RC44 | ||
| tDNAGlu(UUC) | |||||||||||||||
| V. cholerae O1 classical RC2T | 431 | 0/431 | 0/431 | 0/431 | 1/431 | 6/429 | 1/431 | 2/431 | 14/426 | 46/424 | 56/419 | 57/419 | 42/424 | 42/424 | |
| V. cholerae O1 El Tor RC25 | 431 | 100.0 | 0/431 | 0/431 | 1/431 | 6/429 | 1/431 | 2/431 | 14/426 | 46/424 | 56/419 | 57/419 | 42/424 | 42/424 | |
| V. cholerae O139 RC4 | 431 | 100.0 | 100.0 | 0/431 | 1/431 | 6/429 | 1/431 | 2/431 | 14/426 | 46/424 | 56/419 | 57/419 | 42/424 | 42/424 | |
| V. cholerae NO RC42 | 431 | 100.0 | 100.0 | 100.0 | 1/431 | 6/429 | 1/431 | 2/431 | 14/426 | 46/424 | 56/419 | 57/419 | 42/424 | 42/424 | |
| V. cholerae NO RC44 | 431 | 99.8 | 99.8 | 99.8 | 99.8 | 7/429 | 2/431 | 3/431 | 15/426 | 47/424 | 57/419 | 58/419 | 43/424 | 43/424 | |
| V. cholerae O22 RC45 | 445 | 98.6 | 98.6 | 98.6 | 98.6 | 98.4 | 6/429 | 6/429 | 15/429 | 52/422 | 57/422 | 58/422 | 46/422 | 48/422 | |
| V. cholerae NO RC47 | 431 | 99.8 | 99.8 | 99.8 | 99.8 | 99.5 | 98.6 | 3/431 | 14/426 | 47/424 | 56/419 | 57/419 | 43/424 | 43/424 | |
| V. cholerae O31 RC48 | 431 | 99.5 | 99.5 | 99.5 | 99.5 | 99.3 | 98.6 | 99.3 | 12/426 | 48/424 | 56/419 | 57/419 | 42/424 | 44/424 | |
| V. mimicus RC5T | 430 | 96.7 | 96.7 | 96.7 | 96.7 | 96.5 | 96.5 | 96.7 | 97.2 | 58/419 | 44/423 | 49/423 | 52/419 | 54/419 | |
| tDNAAla(GGC) | |||||||||||||||
| V. cholerae O1 classical RC2T | 424 | 89.2 | 89.2 | 89.2 | 89.2 | 88.9 | 87.7 | 88.9 | 88.7 | 86.2 | 14/420 | 15/420 | 8/425 | 4/425 | |
| V. mimicus RC5T | 424 | 86.6 | 86.6 | 86.6 | 86.6 | 86.4 | 86.5 | 86.6 | 86.6 | 89.6 | 96.7 | 5/424 | 18/420 | 16/420 | |
| V. mimicus RC55 | 424 | 86.4 | 86.4 | 86.4 | 86.4 | 86.2 | 86.3 | 86.4 | 86.4 | 88.4 | 96.4 | 98.8 | 19/420 | 17/420 | |
| tDNAAla(UGC) | |||||||||||||||
| V. cholerae O1 El Tor RC25 | 425 | 90.1 | 90.1 | 90.1 | 90.1 | 89.9 | 89.1 | 89.9 | 90.1 | 87.6 | 98.1 | 95.7 | 95.5 | 4/425 | |
| V. cholerae NO RC44 | 425 | 90.1 | 90.1 | 90.1 | 90.1 | 89.9 | 88.6 | 89.9 | 89.6 | 87.1 | 99.1 | 96.2 | 96.0 | 99.1 | |
NO, non-O1/non-O139.
See Table 2, footnote a.
The corresponding clone containing a faint band of ca. 700 bp was not recovered. Therefore, it is not clear whether it resulted from nonspecific PCR or from amplification of authentic ISR.
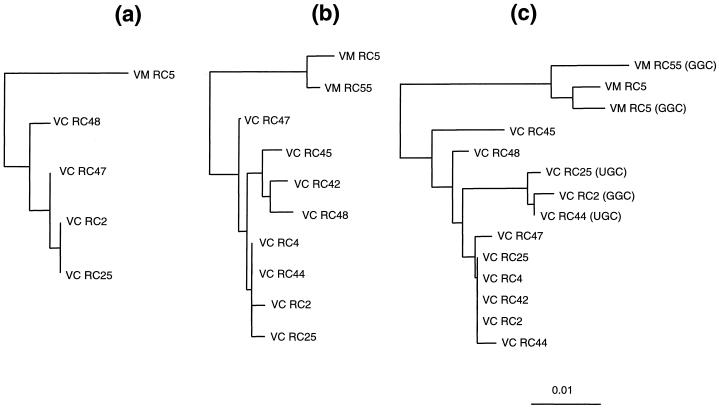
Phylogenetic analysis based on ISR.
Phylogenetic trees based on ISR-1, ISR-2, and ISR-3 (Fig. 3) showed consistent genealogical relationships among the V. cholerae and V. mimicus strains. In the case of ISR-3, the tDNA region was excluded from the neighbor-joining analysis, as it was not considered homologous. It was clear from the phylogenetic analyses and sequence comparisons that the ISRs from epidemic V. cholerae O1 classical, O1 El Tor, and O139 strains are very similar, if not identical. V. cholerae RC44 (non-O1/non-O139) was also found to be closely related to the epidemic V. cholerae strains. Other serotypes, including V. cholerae RC42, RC45, RC47, and RC48, showed significant sequence variation in the ISR. However, ISR-3 from RC42 was identical to the epidemic V. cholerae strains.
FIG. 3.
Phylogenies of V. cholerae and V. mimicus strains based on ISR-1 (a), ISR-2 (b), and ISR-3 (c) sequences. A region coding for tRNA was excluded from analysis for ISR-3. Anticodons of tDNAs in ISR-3, other than tDNAGlu, are indicated in parentheses. The unrooted evolutionary trees were inferred by using the neighbor-joining method. The scale bar represents 0.01-nucleotide substitutes per position. Abbreviations: VC, V. cholerae; VM, V. mimicus.
V. mimicus strains formed a separate monophyletic clade. The intraspecies ISR similarity values were 99.5 ± 0.4 and 99.1 ± 0.3 for V. cholerae and V. mimicus, respectively, whereas the corresponding value for interspecies comparisons was 97.3 ± 0.4. ISR-3 containing tDNAAla(UGC) or tDNAAla(GGC) formed a subclade within the V. cholerae clade. However, such a division was not observed for the V. mimicus strains (Fig. 3c).
Sequence analysis of tRNA genes found in ISRs.
The primary structures of five tDNAs found in ISR-1 and ISR-2 were identical among the V. cholerae and V. mimicus strains. tDNAGlu(UUC) in ISR-3 was identical to tDNAGlu(UUC) in ISR-1. The results of a BLAST search of tDNAs are summarized in Table 5. None of the tDNAs from V. cholerae and V. mimicus was identical to published or deposited sequences in GenBank to date. tDNAGlu(UUC), which was found in both ISR-1 and ISR-3, showed the greatest similarity to Aeromonas tDNAGlu(UUC) found in ISR-3 (19). Similarly, two tDNAs coding for tRNAIle and tRNAAla located in ISR-2 showed the greatest similarity to those found in ISR-2 of Aeromonas hydrophila. tDNALys and tDNAVal found in ISR-1 were most similar to those found in E. coli and Haemophilus influenzae, respectively; the latter did not originate from ISR but from tRNA gene clusters. tDNAAla(GGC) obtained from ISR-3 of strains RC2, RC5, and RC6 was nearly identical to the tDNAAla(GGC) of E. coli, which was found in one of tRNA operons (26).
TABLE 5.
Sequence similarity of tDNAs found in 16S-23S rRNA intergenic spacer regions of V. cholerae and V. mimicus
| ISR and tDNA | Matching organism(s) | % Similaritya |
|---|---|---|
| ISR-1 | ||
| tDNAGlu(UUC) | Aeromonas salmonicida | 93.4 (5/76) |
| Aeromonas hydrophila | 93.3 (5/75) | |
| Plesiomonas shigelloides, Escherichia coli, Klebsiella pneumoniae | 92.1 (6/76) | |
| tDNALys(UUU) | Escherichia colib | 90.7 (7/75) |
| Synechocystis sp.b | 77.3 (14/75) | |
| Mycoplasma sp.b | 76.3 (16/75) | |
| tDNAVal(UAC) | Haemophilus influenzaeb | 94.7 (4/76) |
| Acholeplasma laidlawii,bEscherichia coli,bPseudomonas aeruginosab | 85.5 (11/76) | |
| ISR-2 | ||
| tDNAIle(GAU) | Aeromonas hydrophila | 94.8 (4/77) |
| Bacillus subtilis, Haemophilus influenzae, Actinobacillus pleuropneumoniae | 93.5 (5/77) | |
| tDNAAla(UGC) | Aeromonas hydrophila, Escherichia coli | 96.1 (3/76) |
| Haemophilus influenzae | 94.7 (4/76) | |
| ISR-3 | ||
| tDNAGlu(UUC) | Identical to tDNAGlu(UUC) found in ISR-1 | |
| tDNAAla(GGC)c | Escherichia colib | 97.4 (2/76) |
| Aeromonas hydrophila,eEscherichia colie | 93.4 (5/76) | |
| tDNAAla(UGC)d | Vibrio cholerae, Vibrio mimicuse | 98.7 (1/76) |
| Escherichia coli, Aeromonas hydrophila | 97.4 (2/76) | |
| tDNAAla(UGC)f | Escherichia coli, Salmonella enterica | 96.1 (3/76) |
| Vibrio choleraee | 94.7 (4/76) | |
| Haemophilus influenzae, Pseudomonas syringae | 92.1 (6/76) |
Numbers of different nucleotides/total nucleotides compared are given in parentheses.
tDNAs recovered from non-ISR.
Obtained from strains RC2, RC5, and RC55.
Obtained from strain RC25.
tDNAAla(UGC) found in ISR-2.
Obtained from strain RC44.
PCR specific for V. cholerae.
Even though the ISR sequences for V. cholerae and V. mimicus were very similar, a region containing substantial genetic variation was identified next to the last tRNA coding genes, starting at position 224 of V. cholerae RC2T ISR-2 (Fig. 2). A stretch of 17 nucleotides in ISR-2 and ISR-3 was consistently conserved within the species and different between species, from which the V. cholerae specific primer, named prVC-F, was designed (Fig. 2). The primer contains two degenerate nucleotides and five mismatches compared to V. mimicus. The reverse primer, prVCM-R, was derived from the sequence encompassing the 3′ end of ISR and the 5′ end of 23S rDNA and was complementary to all of the ISRs for V. cholerae and V. mimicus.
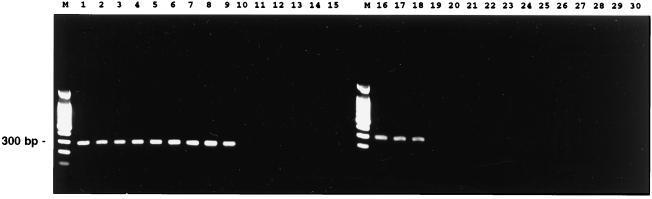
The results of PCR, based on the ISR sequence data, are shown in Fig. 4. The amplicon, the size of which was expected to be 295 to 310 bp, was apparent among more than 100 V. cholerae strains, including fresh clinical and environmental isolates from Bangladesh and the Chesapeake Bay, but not in V. mimicus strains. In addition, we confirmed negative results for Listonella anguillarum ATCC 19264T, Listonella pelagia ATCC 25916T, Salinivibrio costicola ATCC 33508T, Photobacterium damselae subsp. damselae ATCC 33539T, V. aestuarianus ATCC 35048T, V. alginolyticus ATCC 17749T, V. campbellii ATCC 25920T, V. carchariae ATCC 35084T, V. diazotrophicus ATCC 33466T, V. fischeri ATCC 7744T, V. fluvialis ATCC 33809T, V. furnissii ATCC 35016T, V. hollisae ATCC 33564T, V. natriegens ATCC 14048T, V. nigripulchritudo ATCC 27043T, V. orientalis ATCC 33934T, V. proteolyticus ATCC 15338T, V. salmonicida ATCC 43839T, V. splendidus ATCC 33125T, V. tubiashii ATCC 19109T, and V. vulnificus ATCC 27562T.
FIG. 4.
Identification of V. cholerae by using PCR based on the 16S-23S rRNA ISR. Lanes: M, molecular weight marker (100-bp ladder): 1, V. cholerae O1 classical RC2; 2 to 5, V. cholerae O1 El Tor clinical isolates; 6 to 9, V. cholerae O139 clinical isolates; 10, V. mimicus RC5; 11 to 15, V. mimicus isolates; 16 to 18, V. cholerae non-O1/non-O139 isolates; 19, V. aestuarianus ATCC 35048T; 20, V. alginolyticus ATCC 17749T; 21, V. campbellii ATCC 25920T; 22, V. carchariae ATCC 35084T; 23, V. diazotrophicus ATCC 33466T; 24, V. fischeri ATCC 7744T; 25, V. fluvialis ATCC 33809T; 26, V. furnissii ATCC 35016T; 27, V. hollisae ATCC 33564T; 28, V. natriegens ATCC 14048T; 29, V. salmonicida ATCC 43839T; and 30, V. vulnificus ATCC 27562T.
DISCUSSION
Genetic information derived from the 16S-23S rRNA ISR can be used to differentiate closely related organisms (20, 22, 29, 39, 40, 44, 47, 52). One of the goals of this study was to underpin genealogical variation among V. cholerae species, especially those responsible for large-scale cholera epidemics. It was, therefore, disappointing to find so very little variation between the V. cholerae O1 and O139 strains. It has been pointed out by many investigators (3, 4, 15, 24, 48, 49) that serotype O139 may have arisen when a V. cholerae strain, probably an O1 El Tor, picked up genes responsible for its O antigen synthesis from other bacteria by lateral gene transfer. The close relationship between O1 and O139 strains, based on ISR sequence data, supports this hypothesis. Non-O1/non-O139 strains showed very little, albeit significant, variation from the O1 and O139 strains, except for the V. cholerae non-O1/non-O139 strain RC44.
The ISR PCR band patterns generated from V. cholerae and V. mimicus were almost identical (Fig. 1), which is in disagreement with the previous study of Coelho et al. (12), who reported that V. cholerae O1 classical, O1 El Tor, and O139 showed different ISR PCR patterns with low-stringency PCR conditions and primers based on E. coli. However, when their primer (NB-2) was compared with the recently available 23S rDNA sequence (GenBank accession number U10956) for V. cholerae, three mismatches, including two nucleotides at the 5′ end, were noted. It is therefore not clear that the PCR amplicons generated in the study of Coelho et al. originated from 16S-23S rRNA ISR. In contrast, our study was based on available 23S rDNA of V. cholerae and highly stringent PCR conditions.
The tRNA genes found in the bacterial 16S-23S rRNA ISR varied in number. A. hydrophila and E. coli contained two ISR types, one containing tDNAGlu(UUC) and the other tDNAIle(GAU)-tDNAAla(UGC) (16, 19). In contrast, Staphylococcus aureus has three types, i.e., no tDNA, one tDNAIle(GAU), and tDNAIle(GAU)-tDNAAla(UGC) (21). The ISRs from mycobacteria have no tDNA (39, 47). It is interesting that the ISR containing tDNAIle(GAU)-tDNAAla(UGC), which is equivalent to the ISR-2 found in V. cholerae and V. mimicus, was also present in a variety of bacterial taxa, including proteobacteria (16, 19, 23, 27, 29, 31, 34, 38), cytophaga (2), gram-positive bacteria with a low G+C DNA (20, 28, 35, 45), and cyanobacteria and chloroplasts (46, 50). The recent finding of the same ISR type in Aquifex aeolicus (18), thought to represent the deepest evolutionary branch of bacteria, strongly suggests that this ISR type may be widespread among bacteria and present in the common ancestor of the domain Bacteria. Even though it is not possible to align ISR sequences between distantly related bacteria, two tDNAs found in ISR-2 can be compared that are likely homologous, i.e., originate from the same gene in an ancestral organism. The phylogenetic relationship based on tDNAIle(GAU)-tDNAAla(UGC) sequences found in ISR-2 was readily comparable to current bacterial taxonomy based on 16S rDNA sequence data (data not shown). However, this type of ISR was not found among the archaea, actinomycetes, and mitochondria.
To date, the number of tDNAs found in the 16S-23S rRNA ISRs ranges from zero to two. It is, therefore, unexpected and surprising that the largest ISR amplicon of V. cholerae and V. mimicus, i.e., ISR-1, contained three tDNAs. In addition, the presence of tDNALys or tDNAVal in 16S-23S rRNA ISRs has not been reported before to occur in prokaryotes. We hypothesize that ISR-1 was generated from homologous recombination events between ISR-3 containing tDNAGlu(UUC) and other tRNA gene clusters containing tDNALys and tDNAVal, since the region consisting of 144 bp of the 5′ end, including 76 bp coding for tRNAGlu(UUC), and of 235 bp of the 3′ end of ISR-1 was almost identical to ISR-3 among V. cholerae and V. mimicus. This event might have occurred in a common ancestor of V. cholerae and V. mimicus. At this stage of analysis, how far this event will date back is not certain, though additional 16S-23S rRNA ISR analyses with other vibrios and related taxa may provide an answer.
As in the case of the 16S rRNA sequence data, the 16S-23S rRNA ISR also provides a limited, albeit much higher, range of genetic variation. At the higher taxonomical level, it was not possible to compare the ISR with that of other bacteria examined to date except for the tDNAs. At the lower level, differentiation between the epidemic V. cholerae strains could not be achieved. However, information coded in the ISR was useful in separating V. cholerae from the closely related V. mimicus, a species category previously created for strains biochemically resembling V. cholerae. V. mimicus strains, like V. cholerae, are capable of producing many toxins, including cholera toxin, and share ecological niches with V. cholerae. It is therefore encouraging that V. cholerae can be identified by using the species-specific PCR method presented here. Currently available methods for identifying V. cholerae rely mainly on conventional biochemical tests that are time-consuming, are not always accurate, and require culturing. Techniques based on serology are critical, but they are useful only for detecting specific serogroups. In contrast, the PCR-based identification techniques are accurate, sensitive, and permit a large throughput. Furthermore, they allow direct detection without the necessity of culture. The latter is important, since it is now well documented that V. cholerae is present in the environment in the viable-but-nonculturable state. Monitoring for V. cholerae, at the species level, has significance because of the potential for conversion between V. cholerae serogroups; notably, conversion from non-O1 to O1 can occur in vitro and, probably, in vivo (13, 32). In conclusion, it is fair to say that the PCR-mediated identification system developed in the present study will provide an ecological and epidemiological tool for detecting V. cholerae in both the natural and clinical environments.
ACKNOWLEDGMENTS
This study was supported by NIH grant number R01 AI39129-01A1 and EPA grant number R824995-01-0.
We thank Judy Johnson for providing some of the strains used in this study.
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Andresson O S, Fridjonsson O H. The sequence of the single 16S rRNA gene of the thermophilic eubacterium Rhodothermus marinus reveals a distant relationship to the group containing Flexibacter, Bacteroides, and Cytophaga species. J Bacteriol. 1994;176:6165–6169. doi: 10.1128/jb.176.20.6165-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik E M, Gouw R D, Mooi F R. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius J, Dull T J, Noller H F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 8.Campos E, Bolános H, Acúna M T, Díaz G, Matamoros M C, Raventós H, Sánchez L M, Sánchez O, Barquero C. Vibrio mimicus diarrhea following ingestion of raw turtle eggs. Appl Environ Microbiol. 1996;62:1141–1144. doi: 10.1128/aem.62.4.1141-1144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury M A, Aziz K M, Kay B A, Rahim Z. Toxin production by Vibrio mimicus strains isolated from human and environmental sources in Bangladesh. J Clin Microbiol. 1987;25:2200–2203. doi: 10.1128/jcm.25.11.2200-2203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury M A, Yamanaka H, Miyoshi S, Aziz K M, Shinoda S. Ecology of Vibrio mimicus in aquatic environments. Appl Environ Microbiol. 1989;55:2073–2078. doi: 10.1128/aem.55.8.2073-2078.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun J, Goodfellow M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol. 1995;45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- 12.Coelho A, Vicente A C, Baptista M A, Momen H, Santos F A, Salles C A. The distinction of pathogenic Vibrio cholerae groups using arbitrarily primed PCR fingerprints. Res Microbiol. 1995;146:671–683. doi: 10.1016/0923-2508(96)81064-3. [DOI] [PubMed] [Google Scholar]
- 13.Colwell R R, Huq A, Chowdhury M A R, Brayton P R, Xu B. Serogroup conversion of vibrio cholerae. J Can Microbiol. 1995;41:946–950. doi: 10.1139/m95-131. [DOI] [PubMed] [Google Scholar]
- 14.Colwell R R. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 15.Comstock L E, Johnson J A, Michalski J M, Morris J G, Jr, Kaper J B. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 16.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis B R, Fanning G R, Madden J M, Steigerwalt A G, Bradford H B, Jr, Smith H L, Jr, Brenner D J. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus. J Clin Microbiol. 1981;14:631–639. doi: 10.1128/jcm.14.6.631-639.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 19.East A K, Collins M D. Molecular characterization of DNA encoding 23S rRNA and 16S-23S rRNA intergenic spacer regions of Aeromonas hydrophila. FEMS Microbiol Lett. 1993;106:129–133. doi: 10.1111/j.1574-6968.1993.tb05947.x. [DOI] [PubMed] [Google Scholar]
- 20.Graham T A, Golsteyn-Thomas E J, Thomas J E, Gannon V P. Inter- and intraspecies comparison of the 16S-23S rRNA operon intergenic spacer regions of six Listeria spp. Int J Syst Bacteriol. 1997;47:863–869. doi: 10.1099/00207713-47-3-863. [DOI] [PubMed] [Google Scholar]
- 21.Gurtler V, Barrie H D. Typing of Staphylococcus aureus strains by PCR-amplification of variable-length 16S-23S rDNA spacer regions: characterization of spacer sequences. Microbiology. 1995;141:1255–1265. doi: 10.1099/13500872-141-5-1255. [DOI] [PubMed] [Google Scholar]
- 22.Hain T, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E, Rainey F A. Discrimination of Streptomyces albidoflavus strains based on the size and number of 16S-23S ribosomal DNA intergenic spacers. Int J Syst Bacteriol. 1997;47:202–206. doi: 10.1099/00207713-47-1-202. [DOI] [PubMed] [Google Scholar]
- 23.Jagoueix S, Bove J M, Garnier M. Comparison of the 16S/23S ribosomal intergenic regions of “Candidatus Liberobacter asiaticum” and “Candidatus Liberobacter africanum,” the two species associated with citrus huanglongbing (greening) disease. Int J Syst Bacteriol. 1997;47:224–227. doi: 10.1099/00207713-47-1-224. [DOI] [PubMed] [Google Scholar]
- 24.Johnson J A, Salles C A, Panigrahi P, Albert M J, Wright A C, Johnson R J, Morris J G., Jr Vibrio cholerae O139 synonym Bengal is closely related to Vibrio cholerae El Tor but has important differences. Infect Immun. 1994;62:2108–2110. doi: 10.1128/iai.62.5.2108-2110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 26.Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 27.La Fontaine S, Rood J I. Organization of ribosomal RNA genes from the footrot pathogen Dichelobacter nodosus. Microbiology. 1996;142:889–899. doi: 10.1099/00221287-142-4-889. [DOI] [PubMed] [Google Scholar]
- 28.Loughney K, Lund E, Dahlberg J E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982;10:1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luz S P, Rodriguez-Valera F, Lan R, Reeves P S. Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J Bacteriol. 1998;180:2144–2151. doi: 10.1128/jb.180.8.2144-2151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montilla R, Chowdhury M A, Huq A, Xu B, Colwell R R. Serogroup conversion of Vibrio cholerae non-O1 to Vibrio cholerae O1: effect of growth state of cells, temperature, and salinity. Can J Microbiol. 1996;42:87–93. doi: 10.1139/m96-014. [DOI] [PubMed] [Google Scholar]
- 33.Mooi F R, Bik E M. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 1997;5:161–165. doi: 10.1016/S0966-842X(96)10086-X. [DOI] [PubMed] [Google Scholar]
- 34.Moreira D, Amils R. PCR-mediated detection of the chemolithotrophic bacterium Thiobacillus cuprinus using 23S rDNA- and 16S/23S intergenic spacer region-targeted oligonucleotide primers. FEMS Microbiol Lett. 1996;142:289–293. doi: 10.1111/j.1574-6968.1996.tb08445.x. [DOI] [PubMed] [Google Scholar]
- 35.Naimi A, Beck G, Branlant C. Primary and secondary structures of rRNA spacer regions in enterococci. Microbiology. 1997;143:823–834. doi: 10.1099/00221287-143-3-823. [DOI] [PubMed] [Google Scholar]
- 36.Nandi S, Khetawat G, Sengupta S, Majumder R, Kar S, Bhadra R K, Roychoudhury S, Das J. Rearrangements in the genomes of Vibrio cholerae strains belonging to different serovars and biovars. Int J Syst Bacteriol. 1997;47:858–862. doi: 10.1099/00207713-47-3-858. [DOI] [PubMed] [Google Scholar]
- 37.Ramamurthy T, Albert M J, Huq A, Colwell R R, Takeda Y, Takeda T, Shimada T, Mandal B K, Nair G B. Vibrio mimicus with multiple toxin isolated from human and environmental sources. J Med Microbiol. 1994;40:194–196. doi: 10.1099/00222615-40-3-194. [DOI] [PubMed] [Google Scholar]
- 38.Rijpens N P, Jannes G, Van Asbroeck M, Rossau R, Herman L M. Direct detection of Brucella spp. in raw milk by PCR and reverse hybridization with 16S-23S rRNA spacer probes. Appl Environ Microbiol. 1996;62:1683–1688. doi: 10.1128/aem.62.5.1683-1688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth A, Fischer M, Hamid M E, Michalke S, Ludwig W, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roux V, Raoult D. The 16S-23S rRNA intergenic spacer region of Bartonella (Rochalimaea) species is longer than usually described in other bacteria. Gene. 1995;156:107–111. doi: 10.1016/0378-1119(94)00919-j. [DOI] [PubMed] [Google Scholar]
- 41.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christen R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 42.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sawada H, Takeuchi T, Matsuda I. Comparative analysis of Pseudomonas syringae pv. actinidiae and pv. phaseolicola based on phaseolotoxin-resistant ornithine carbamoyltransferase gene (argK) and 16S-23S rRNA intergenic spacer sequences. Appl Environ Microbiol. 1997;63:282–288. doi: 10.1128/aem.63.1.282-288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smart C D, Schneider B, Blomquist C L, Guerra L J, Harrison N A, Ahrens U, Lorenz K H, Seemuller E, Kirkpatrick B C. Phytoplasma-specific PCR primers based on sequences of the 16S-23S rRNA spacer region. Appl Environ Microbiol. 1996;62:2988–2993. doi: 10.1128/aem.62.8.2988-2993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takaiwa F, Sugiura M. Nucleotide sequence of the 16S-23S spacer region in an rRNA gene cluster from tobacco chloroplast DNA. Nucleic Acids Res. 1982;10:2665–2676. doi: 10.1093/nar/10.8.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Giessen J W, Haring R M, van der Zeijst B A. Comparison of the 23S ribosomal RNA genes and the spacer region between the 16S and 23S rRNA genes of the closely related Mycobacterium avium and Mycobacterium paratuberculosis and the fast-growing Mycobacterium phlei. Microbiology. 1994;140:1103–1108. doi: 10.1099/13500872-140-5-1103. [DOI] [PubMed] [Google Scholar]
- 48.Vimont S, Dumontier S, Escuyer V, Berche P. The rfaD locus: a region of rearrangement in Vibrio cholerae O139. Gene. 1997;185:43–47. doi: 10.1016/s0378-1119(96)00625-7. [DOI] [PubMed] [Google Scholar]
- 49.Waldor M K, Mekalanos J J. Vibrio cholerae O139 specific gene sequences. Lancet. 1994;343:1366. doi: 10.1016/s0140-6736(94)92504-6. [DOI] [PubMed] [Google Scholar]
- 50.Wilmotte A, Neefs J M, De Wachter R. Evolutionary affiliation of the marine nitrogen-fixing cyanobacterium Trichodesmium sp. strain NIBB 1067, derived by 16S ribosomal RNA sequence analysis. Microbiology. 1994;140:2159–2164. doi: 10.1099/13500872-140-8-2159. [DOI] [PubMed] [Google Scholar]
- 51.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon J H, Lee S T, Kim S B, Goodfellow M, Park Y H. Inter- and intraspecific genetic analysis of the genus Saccharomonospora with 16S to 23S ribosomal DNA (rDNA) and 23S to 5S rDNA internally transcribed spacer sequences. Int J Syst Bacteriol. 1997;47:661–669. doi: 10.1099/00207713-47-3-661. [DOI] [PubMed] [Google Scholar]