ABSTRACT
Zika virus is a mosquito-borne flavivirus known to cause severe birth defects and neuroimmunological disorders. We have previously demonstrated that mosquito transmission of Zika virus decreases with temperature. While transmission was optimized at 29°C, it was limited at cool temperatures (<22°C) due to poor virus establishment in the mosquitoes. Temperature is one of the strongest drivers of vector-borne disease transmission due to its profound effect on ectothermic mosquito vectors, viruses, and their interaction. Although there is substantial evidence of temperature effects on arbovirus replication and dissemination inside mosquitoes, little is known about whether temperature affects virus replication directly or indirectly through mosquito physiology. In order to determine the mechanisms behind temperature-induced changes in Zika virus transmission potential, we investigated different steps of the virus replication cycle in mosquito cells (C6/36) at optimal (28°C) and cool (20°C) temperatures. We found that the cool temperature did not alter Zika virus entry or translation, but it affected genome replication and reduced the amount of double-stranded RNA replication intermediates. If replication complexes were first formed at 28°C and the cells were subsequently shifted to 20°C, the late steps in the virus replication cycle were efficiently completed. These data suggest that cool temperature decreases the efficiency of Zika virus genome replication in mosquito cells. This phenotype was observed in the Asian lineage of Zika virus, while the African lineage Zika virus was less restricted at 20°C.
IMPORTANCE With half of the human population at risk, arboviral diseases represent a substantial global health burden. Zika virus, previously known to cause sporadic infections in humans, emerged in the Americas in 2015 and quickly spread worldwide. There was an urgent need to better understand the disease pathogenesis and develop therapeutics and vaccines, as well as to understand, predict, and control virus transmission. In order to efficiently predict the seasonality and geography for Zika virus transmission, we need a deeper understanding of the host-pathogen interactions and how they can be altered by environmental factors such as temperature. Identifying the step in the virus replication cycle that is inhibited under cool conditions can have implications in modeling the temperature suitability for arbovirus transmission as global environmental patterns change. Understanding the link between pathogen replication and environmental conditions can potentially be exploited to develop new vector control strategies in the future.
KEYWORDS: RNA replication, Zika virus, temperature
INTRODUCTION
Diseases such as Zika, dengue, and chikungunya, once considered tropical and subtropical diseases, have spread explosively throughout the world due to climate change, globalization, and increased urbanization. With half of the human population at risk, arboviral diseases represent a substantial global health burden (1). Zika virus (ZIKV) is a mosquito-borne flavivirus known to cause sporadic and mild infections in humans. In 2015, ZIKV emerged in the Americas and within a year quickly spread to approximately 65 countries worldwide, resulting in over 360,000 suspected cases (https://www.paho.org/). Shortly after it was linked to birth defects (2) and neuroimmunological disorders (Guillain-Barré syndrome) (3), ZIKV was declared a “public health emergency of international concern” (4). With no therapeutics or vaccines to mitigate disease, ZIKV was quickly put at the forefront of research interest to fulfill the urgent need to better understand disease transmission, pathogenesis, and prevention.
Temperature is one of the strongest drivers of vector-borne disease transmission due to its profound impact on ectothermic mosquito vectors, pathogens, and their interactions (5). Numerous studies have investigated the effects of temperature on mosquito infection, the extrinsic incubation period (EIP), and overall transmission potential in a diversity of vector-borne disease systems (6–10). However, the exact mechanisms of how temperature shapes arboviral transmission are rarely elucidated. Temperature can affect virus replication directly or indirectly by altering mosquito physiology (11), immunity (12, 13), and cellular processes (14). Even though arboviruses evolved to replicate across a wide range of temperatures, from within invertebrate vectors to febrile mammalian and avian hosts, temperature can also modify virus replication. Studies have shown that temperature can alter virus structure (15, 16), induce the fluctuation of viral surface proteins required for entry in the host cell (17, 18), or affect genome replication (19, 20). Although a few studies have investigated ZIKV structure and thermal stability (21–23), no studies have investigated how temperature affects the ZIKV replication cycle.
In our previous work, we demonstrated that ZIKV transmission in Aedes aegypti mosquitoes was optimized at 29°C and had a thermal range of 22.7°C to 34.7°C. Although warm temperatures facilitated rapid virus replication, warm conditions (>36°C) also increased mosquito mortality and led to an overall decrease in transmission potential. However, cool conditions (<22°C) prevented the mosquitoes from becoming infectious due to poor midgut infection and escape (24). In order to understand the mechanisms of reduced ZIKV transmission potential at suboptimal temperatures, we investigated ZIKV replication in vitro using a mosquito cell line incubated across a range of temperatures. Here, we show that ZIKV replication in cell culture closely mirrors the findings in mosquitoes. ZIKV replication occurs faster as the temperature increases, until the temperature starts to induce cell death, while cool temperatures significantly decrease the replication kinetics. We formulated the following two, not mutually exclusive, hypotheses to address our observations. (i) The stress from cool temperatures alters the cellular environment, either limiting host factors necessary for viral replication or overproducing an inhibitory factor not produced at permissive temperatures. (ii) The suboptimal temperatures prevent a viral function required to complete the viral replication cycle and produce progeny virus. We found that reduced ZIKV replication at suboptimal temperature is not a result of acute cellular stress. In addition, maintaining cells at 20°C did not appear to affect early and late steps in the viral replication cycle; however, it did affect genome replication. While American isolates of ZIKV were restricted, African isolates were able to replicate more efficiently at 20°C.
RESULTS
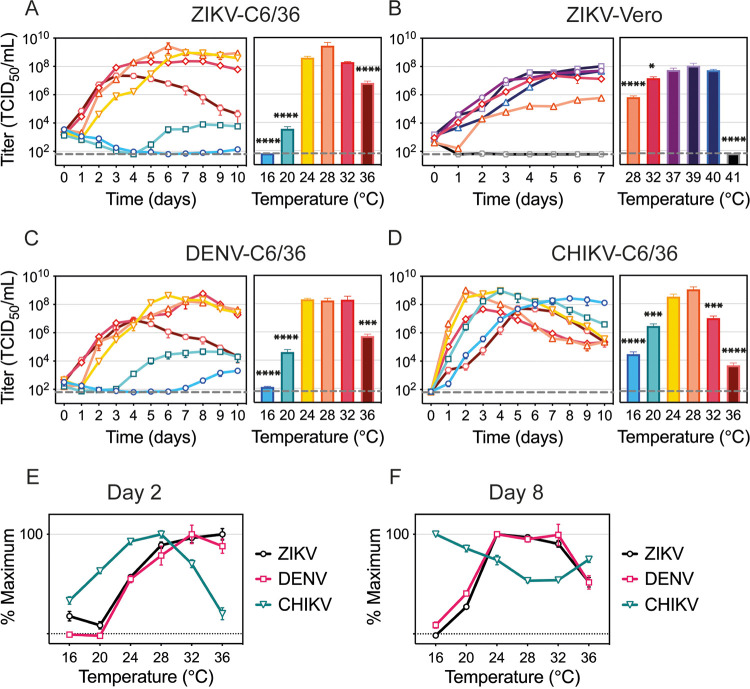
ZIKV, dengue virus (DENV), and chikungunya virus (CHIKV) replication curves at different temperatures.
To characterize the effect of temperature on ZIKV replication in mosquito cells, we infected C6/36 cells with ZIKV and incubated them at six temperatures ranging from 16°C to 36°C (Fig. 1A). To mirror our experiment in mosquitoes (24), six independent plates were infected at 28°C for 2 h, inoculum was removed, and the plates were then moved to the indicated temperatures, where they were maintained for the remainder of the experiment. Initial ZIKV replication and peak titers occurred more quickly as the temperature increased. Incubation at 16°C resulted in almost complete inhibition of virus production in C6/36 cells, while incubation at 20°C resulted in delayed particle release and more than a 5-log reduction in peak titers. While ZIKV replication started robustly at 36°C, virus replication was drastically reduced over time. This phenotype is a result of temperature-related cellular stress and was not observed in ZIKV-infected mammalian cells that optimally produce virus at 37°C (Fig. 1B). While virus replication peaked at 2.67 × 109 tissue culture infective doses (TCID50)/mL at 28°C in mosquito cells, the peak titer was 3.5-log lower at 28°C in Vero cells. In comparison, Vero cells infected at 37°C produced the highest titers of virus throughout the experiment, yet titers in C6/36 cells at 36°C peaked low and early and then fell over the course of the experiment. This suggests that peak virus replication is determined by both intracellular components and optimal environmental conditions.
FIG 1.
Temperature effects on ZIKV, DENV, and CHIKV replication. (A to D) Replication rates for ZIKV in C6/36 cells and titers on day 6 (A), ZIKV in Vero cells and titers on day 7 (B), DENV in C6/36 cells and titers on day 7 (C), and CHIKV in C6/36 cells and titers on day 2 (D). (E and F) Percent maximum titer on day 2 (E) and day 8 (F). C6/36 cells were incubated with ZIKV (MOI, 0.1), DENV (MOI, 0.1), or CHIKV (MOI, 0.005) for 2 h at 28°C, when infectious medium was removed and replaced with fresh medium. The cells were kept at 16°C, 20°C, 24°C, 28°C, 32°C, or 36°C for 10 days. Vero cells were infected with ZIKV (MOI, 0.1) for 1 h at 37°C. The cells were incubated at 28°C, 32°C, 37°C, 39°C, 40°C, or 41°C for 7 days. Supernatants were collected every 24 h and titrated on Vero cells. Titers are given in TCID50 units/mL. Dashed lines represent the limit of detection. Data shown are means ± SEM from three independent replicates for each virus. Bar graphs represent viral titers on the day peak titers were reached at optimal temperature (28°C for C6/36, 37°C for Vero). Statistical differences were determined with a one-way ANOVA on log-transformed data for each experiment. Dunnett’s test was used for multiple comparisons, where the optimal temperature (28°C or 37°C) was compared to all other temperatures. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Next, we compared ZIKV replication across six temperatures to another flavivirus (DENV) and to an alphavirus (CHIKV). DENV had replication dynamics similar to ZIKV, yet DENV was less restricted at cool temperatures, reaching 1.45 × 103 TCID50/mL at 16°C and 3.74 × 106 TCID50/mL at 20°C (Fig. 1C). CHIKV replication kinetics and its response to temperature greatly differed relative to both flaviviruses (Fig. 1D). In order to compare how the temperature differentially affects the three viruses, we compared how much virus was produced across the temperature range early (Fig. 1E) and late (Fig. 1F) during the infection. While CHIKV replication proceeded at a higher rate with both cool and warm temperatures inhibiting optimal viral yields early in the infection, ZIKV and DENV were observed to produce higher titers as the temperature increased (Fig. 1E). However, both ZIKV and DENV titers were low at 36°C late in infection, suggesting that prolonged high-temperature treatment may enhance early replication but eventually caused cell death (Fig. 1F). Interestingly, CHIKV peak TCID50 values were similar at 28°C and 20°C, simply delayed under cool conditions, suggesting that C6/36 cells are capable of producing virus particles at 20°C; however, low temperatures might affect ZIKV and CHIKV replication differently.
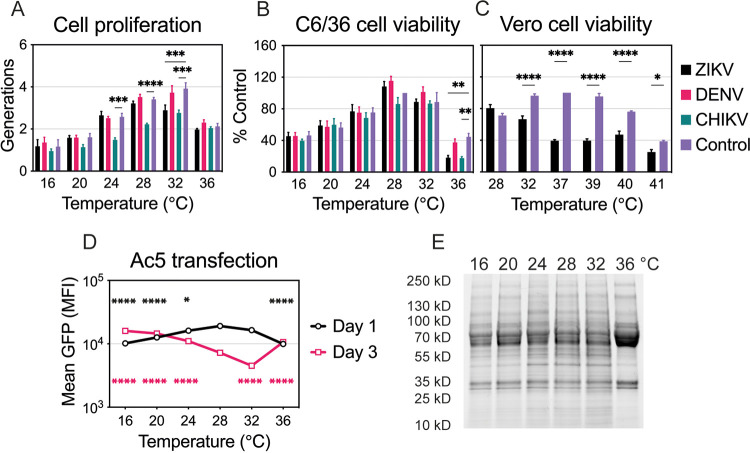
Temperature and virus effects on cell physiology.
Temperature profoundly affects cell physiology and metabolism; therefore, it can alter cell proliferation, viability, and protein production. We monitored uninfected as well as ZIKV-, DENV-, and CHIKV-infected C6/36 cell proliferation and viability at temperatures ranging from 16°C to 36°C. The number of cell generations over a 4-day period increased proportionally with temperature except for cells incubated at 36°C, where cell proliferation decreased (Fig. 2A). While DENV infection did not alter cell proliferation at any temperature, ZIKV infection reduced the number of generations at 32°C and CHIKV decreased cell proliferation at all temperatures. Low and high temperatures also affected cell viability, represented by lower ATP levels, after cells were maintained at the indicated temperature for 6 days (Fig. 2B). CHIKV was more cytopathic than ZIKV and DENV, while ZIKV affected cell viability only at warm temperatures. DENV-infected cells showed little to no reduction in ATP levels in comparison to uninfected cells. We also checked cell viability in uninfected and ZIKV-infected Vero cells at temperatures ranging from 28°C to 41°C (Fig. 2C). As expected, suboptimal and extreme temperatures (28°C, 40°C, 41°C) decreased cell viability. ZIKV infection was more cytopathic in mammalian than in mosquito cells, resulting in significantly reduced ATP levels at all temperatures except 28°C.
FIG 2.
Temperature and virus effects on cell proliferation, viability, and protein production. (A) Cell proliferation of uninfected or infected C6/36 cells at different temperatures (16°C to 36°C). Dyed cells were infected with ZIKV, DENV, or CHIKV as described in the legend to Fig. 1 and were incubated at one of the six temperatures for 4 days. Intracellular fluorescence was normalized to that of uninfected cells on day 0. Data are presented as the mean ± SEM from three independent experiments. (B) Cell viability of uninfected or infected C6/36 cells incubated at six temperatures (16°C to 36°C) for 6 days. Cell viability was normalized to that of uninfected cells maintained at 28°C, and data are presented as the mean ± SEM from three independent experiments. Statistical differences for cell proliferation and viability were assessed by a two-way ANOVA with Dunnett’s correction for multiple comparisons. For each temperature, uninfected cells were compared to those infected with each virus. (C) Cell viability of uninfected or ZIKV-infected Vero cells incubated at six temperatures (28°C to 41°C) for 4 days. Cell viability was normalized to that of uninfected cells maintained at 37°C, and data are presented as the mean ± SEM from three independent experiments. Statistical differences were determined by a two-way ANOVA, followed by Bonferroni’s test for multiple comparisons. (D) Protein production at each temperature (16°C to 36°C) on days 1 and 3 was assessed after transfection of C6/36 cells with Ac5-STABLE2-neo plasmid. The data shown, expressed as mean GFP fluorescence, are means ± SEM from three replicates. Statistically significant mean fluorescence was assessed by using a two-way ANOVA on log-transformed data, followed by Dunnett’s test for multiple comparisons. For days 1 and 3, 28°C was compared to all other temperatures. The color of the significance symbol corresponds to the color for each day. (E) Total protein lysate was prepared for uninfected C6/36 cells incubated at six temperatures (16°C to 36°C) for 4 days. Seventy-five micrograms of total protein was denatured and run on a TGX Stain-Free gel (Bio-Rad). The gel was activated with UV light and imaged with a ChemiDoc XRS digital imaging system (Bio-Rad). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In order to investigate the effects of temperature on protein production, we transfected C6/36 cells with an insect expression plasmid that encodes green fluorescent protein (GFP) (pAc5-GFP) at 28°C and 6 h later moved the cells to the indicated temperature (Fig. 2D). One day after transfection, cells produced less GFP at suboptimal (16°C, 20°C, 24°C) and hot (36°C) temperatures. Decreased protein production suggests that cells are slightly less metabolically active early after transfection at those temperatures. Three days after transfection, cells incubated at 28°C and 32°C were less fluorescent, likely due to faster cell proliferation. Slower proliferation at other temperatures resulted in higher mean GFP fluorescence as there were more GFP-positive cells. Therefore, while C6/36 cells do not proliferate as efficiently at low temperatures, they are still able to produce proteins. That was further verified by monitoring the endogenous protein levels across all six temperatures (Fig. 2E). Protein production was rather stable after 4 days under various temperature conditions with subtle decreases at cool temperatures (16°C and 20°C) and obvious differences when maintained at 36°C.
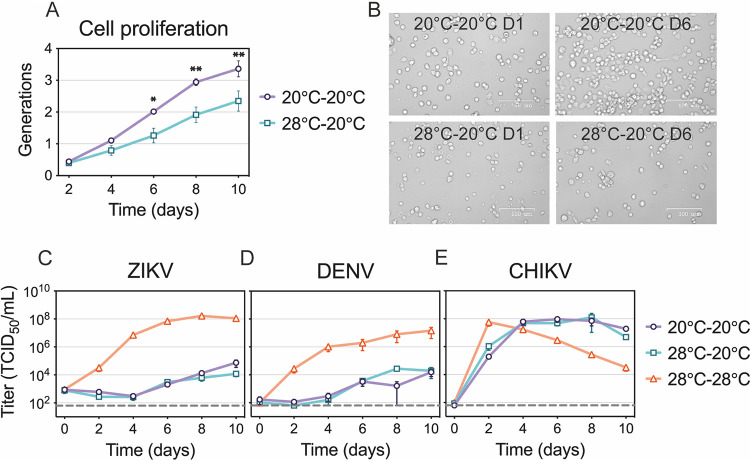
ZIKV, DENV, and CHIKV replication in 20°C-adapted C6/36 cells.
When the replication curves shown in Fig. 1 were determined, cells normally maintained at 28°C were infected and transferred to the different temperature treatments. Therefore, cells were subjected to temperature change during the initial infection time, which may have triggered acute cellular responses that would not be present if cells were maintained at the various temperature treatments prior to infection. To determine if virus replication was inhibited at 20°C due to acute cellular stress responses, we adapted C6/36 cells to grow at 20°C. After maintaining C6/36 cells at 20°C for several months, we compared the adapted cell morphology to the morphology of normal cells and observed no changes. We also demonstrated that 20°C-adapted cells proliferated faster at 20°C than the cells that were shifted from 28°C to 20°C (Fig. 3A and B). We then investigated ZIKV, DENV, and CHIKV replication in 20°C-adapted cells and found there was no difference in virus yields between adapted and nonadapted cells at 20°C (Fig. 3C to E). This suggests that the reduced ZIKV replication was not likely a result of acute cellular stress brought on by the sudden temperature shift.
FIG 3.
ZIKV, DENV, and CHIKV replication in 20°C-adapted C6/36 cells. (A and B) Cell proliferation of 20°C-adapted C6/36 cells in comparison to standard cells at 20°C. Both cell types were stained and maintained at 20°C for 10 days. Fluorescence was measured every 2 days and normalized to either 20°C-adapted or standard cells on day 0. Pictures of the cells were taken on days 0 and 6, and representative images are shown in panel B. The data shown are means ± SEM from three replicates. Statistical differences were determined by using a two-way ANOVA with Bonferroni’s correction, where adapted cells were compared to standard cells at each time point. (C to E) Replication rates of ZIKV (C), DENV (D), and CHIKV (E) in 20°C-adapted cells were compared to those of standard cells at 20°C and 28°C. 20°C-adapted cells were infected and kept at 20°C, while standard cells were infected at 28°C and kept at 20°C or 28°C. Supernatants were collected every 48 h and titrated. Dashed lines represent the limit of detection. Data shown are means ± SEM of results from three independent experiments for each virus. *, P < 0.05; **, P < 0.01.
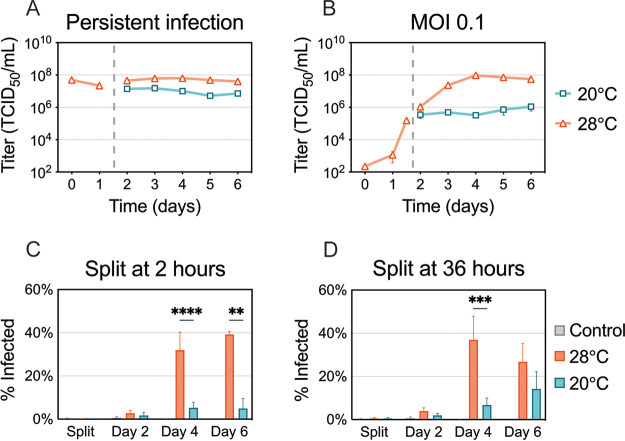
Effects of suboptimal temperature on ZIKV spread.
We next sought to determine which part of the virus replication cycle is affected by cool temperatures. We first examined if cells with established ZIKV infection are capable of producing infectious virus particles at 20°C (Fig. 4A). Persistently infected C6/36 cells were distributed into two dishes, one of which was transferred to 20°C, while the other was kept at 28°C. Interestingly, the amounts of infectious virus produced every 24 h for 5 days were similar at 20°C and 28°C. We observed a slight decrease in virus yields, which was likely due to decreased cellular metabolism at 20°C. Overall, this suggests that later steps in virus replication are not inhibited at cool temperatures.
FIG 4.
ZIKV production and spread at suboptimal temperature. (A) C6/36 cells with established ZIKV infection were split and incubated at 20°C or 28°C for 5 days. (B) C6/36 cells infected at an MOI of 0.1 were split at 36 h postinfection (p.i.) and incubated at 20°C and 28°C for 5 days. Supernatants were collected and replaced daily, and virus titers were determined. Data shown for persistent ZIKV infection are means ± SEM of results from two biological replicates, each consisting of three technical replicates (n = 6). Data shown for an MOI of 0.1 are means ± SEM of results from three independent experiments. Dashed lines represent when the cells were split. (C and D) The percentage of ZIKV-positive cells was assessed using flow cytometry. (C) Cells were infected with ZIKV (MOI 0.1) for 2 h at 28°C and then split and incubated at 20°C or 28°C. (D) Cells infected with ZIKV were kept at 28°C for 36 h and then split and incubated at 20°C or 28°C. ZIKV-positive cells were measured after splitting (2 or 36 h p.i.) and 2, 4, and 6 days p.i. Data shown are means ± SEM of results from seven independent experiments. Statistical differences were assessed by a two-way ANOVA with Bonferroni’s correction, where cells at 20°C were compared to cells at 28°C at every time point. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we infected C6/36 cells with a low multiplicity of infection (MOI, 0.1) and incubated them at 28°C for 36 h, during which ZIKV infection was established in a small proportion of cells (Fig. 4B). The cells were then split and incubated at 20°C or 28°C for five additional days. While virus titers increased up to 3 logs at 28°C, the titers plateaued in C6/36 cells maintained at 20°C. This plateau could be the result of inhibited viral spread from the previously infected cells to naive cells or an overall reduction in virus production in both cells with established infection and newly infected cells.
To determine if the plateau in virus titers was due to inhibition of virus spread, we quantified the number of infected cells over time. We infected cells with a low MOI and incubated them at 28°C for either 2 h, to mimic the original replication curves (Fig. 4C), or 36 h, to mimic the later experiment (Fig. 4D). The cells were then maintained at 20°C or 28°C, stained for ZIKV antigen, and analyzed using flow cytometry. We saw that cultures maintained at 20°C displayed low levels of ZIKV-positive cells, suggesting that the virus was unable to spread and replicate in naive cells (Fig. 4C). When the cells were maintained at 28°C for 36 h before shifting, more ZIKV-positive cells were detected at 20°C; however, 28°C enhanced the production of ZIKV antigens. Taken together, these data suggest that the cells with an established infection are capable of producing infectious particles but that the virus produced is not able to efficiently spread and establish infection in uninfected cells at 20°C.
ZIKV entry kinetics.
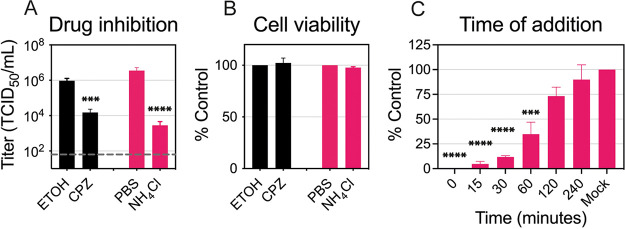
In order to elucidate the kinetics of ZIKV internalization and fusion in C6/36 cells, infected cells were treated with chlorpromazine (CPZ) and ammonium chloride (NH4Cl). CPZ inhibits clathrin-mediated endocytosis, an entry pathway used by ZIKV in mammalian cells (25, 26). NH4Cl is a weak base that increases the endosomal pH and prevents the low-pH-dependent conformational changes in the E protein required for fusion (27–29). To ensure that these compounds inhibit ZIKV entry in C6/36 cells, we first confirmed that both drugs reduced viral titers while remaining noncytotoxic (Fig. 5A and B). We then performed a time-of-addition assay with NH4Cl to establish the dynamics of internalization and fusion steps in C6/36 cells (Fig. 5C). We treated the cells with NH4Cl at the indicated time points to determine when ZIKV completes fusion and entry in cells maintained at 28°C. If NH4Cl was added 1 h after infection, it blocked 65% of the virus produced in the mock control, while only 25% of the virus was inhibited if added at 2 h. This suggests that most ZIKV particles enter C6/36 cells within the first 2 h of infection at 28°C.
FIG 5.
ZIKV entry dynamics in C6/36 cells. (A) Drug inhibition assay. CPZ (50 μM) and NH4Cl (25 mM) were added 2 h prior to ZIKV infection (MOI, 0.1) and were kept during infection. After 2 h of incubation at 28°C, the infectious medium with the drug or vehicle was removed and replaced with fresh medium. Supernatants were collected at 48 h p.i. and titrated. The dashed line represents the limit of detection. (B) Cell viability. The cells were treated with the same concentrations of the drug or the same amount of vehicle for 4 h, and ATP levels were measured after 2 days. Data are presented as the mean ± SEM from three independent experiments for each drug. Statistical differences were assessed by using a two-way ANOVA, followed by Bonferroni’s test for multiple comparisons. (C) Time of addition. NH4Cl (25 mM) was added to the ZIKV-infected cells at 0, 15, 30, 60, 120, and 240 min p.i. and kept for 4 h. Supernatants were collected at 48 h p.i. and titrated. Cell titers were normalized to mock infection, and data are presented as the mean ± SEM from three independent experiments. Statistical significance was determined by using a one-way ANOVA on log-transformed data with Dunnett’s correction, where mock infection was compared to the time of drug addition. *, P < 0.05.
Effects of suboptimal temperature on ZIKV entry.
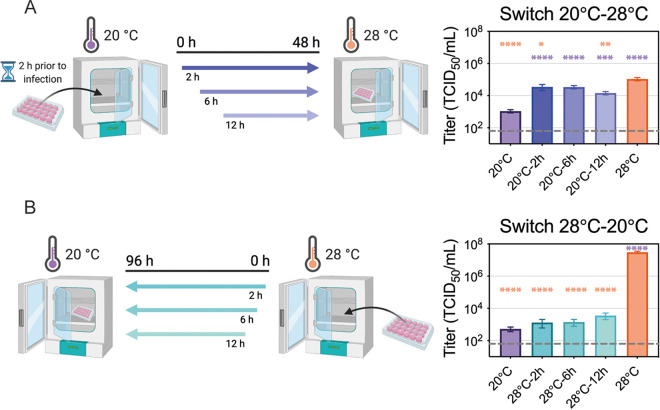
Next, we wanted to determine when the cool temperature most affects ZIKV replication. We evaluated the effect of transferring infected cells between suboptimal (20°C) and optimal (28°C) environments on viral titers. If the cells were infected and maintained at 20°C for 2 or 6 h before being shifted to 28°C, the virus titers were similar to that of cells maintained at 28°C for the entire time period (Fig. 6A). Based on the previously established entry dynamics, this suggests that ZIKV entry was not inhibited at the suboptimal temperature. However, if cells were infected at 20°C and maintained at the cool temperature for 12 h before being shifted to 28°C, fewer particles were produced. When infected cells were shifted from warm (28°C) to cool (20°C) environments, we found that considerably fewer particles were produced if the shift occurred at any point within the first 12 h after infection (Fig. 6B). This suggests that ZIKV requires more than 12 h at 28°C to establish a productive infection. In combination with our previous findings (Fig. 4 and 5), these data suggest that cool temperatures block efficient replication after fusion, but late stages of the virus replication cycle in cells with established infection can proceed at cool temperatures.
FIG 6.
ZIKV entry at suboptimal temperature. (A) Cool-to-warm temperature switch. C6/36 cells (6 × 105 cells/mL) were plated and incubated at 20°C 2 h prior to infection. Cells were then inoculated with ZIKV (MOI, 0.1) for 2 h and moved to 28°C at 2, 6, or 12 h p.i. As a control, one set of cells was maintained at 20°C while another was maintained at 28°C throughout the experiment. Supernatants were collected at 48 h p.i. and titrated. (B) Warm-to-cool temperature switch. C6/36 cells were plated at the same density as described for panel A and infected with ZIKV (MOI, 0.1) for 2 h. The cells were transferred from 28°C to 20°C at 2, 6, or 12 h p.i. As a control, one set of cells was maintained at 28°C while another was maintained at 20°C throughout the experiment. Supernatants were collected at 96 h p.i. and titrated. Dashed lines represent the limit of detection. Experimental designs were created with BioRender.com. Data are presented as the mean ± SEM from three replicates for each experiment. Statistical differences were determined by a one-way ANOVA on log-transformed data, followed by Dunnett’s test for multiple comparisons, where each condition was compared to both 20°C (purple significance symbol) and 28°C (orange significance symbol) controls. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Effects of suboptimal temperature on virus recovery, translation, replication, and double-stranded RNA (dsRNA) production.
Our data indicate that cooler temperature primarily inhibits ZIKV replication after the initial 2-h viral entry period. To further confirm the phenotype, we bypassed binding, internalization, fusion, and nucleocapsid disassembly steps by transfecting full-length genomes into the cells and quantifying the amount of infectious viral particles produced (Fig. 7A). If the cells were transfected and maintained at 28°C, infectious ZIKV particles were readily detected after 2 days. However, if the cells were transfected at 28°C and transferred to 20°C 1 h following transfection, no infectious virus was produced. If 20°C-adapted cells were transfected and kept at 20°C, no virus was produced, but if transferred to 28°C 1 h following transfection, virus was produced at levels similar to those of the cells maintained at 28°C.
FIG 7.
Virus recovery, translation, and replication at suboptimal temperature. (A) Virus recovery. Standard and 20°C-adapted cells were transfected with ZIKV RNA at their respective temperatures for 1 h. After transfection, one plate with standard cells was kept at 28°C and one was transferred to 20°C, and one plate with 20°C-adapted cells was kept at 20°C and one was transferred to 28°C. Supernatants were collected and titrated at the indicated time points. The dashed line represents the limit of detection. Data are presented as the mean ± SEM from at least three independent experiments. (B) Translation. Standard and 20°C-adapted C6/36 cells were transfected with a translation reporter construct at their respective temperatures. One plate with standard cells was kept at 28°C and one was switched to 20°C 1 h after transfection, while 20°C-adapted cells were kept at 20°C. At the indicated time points, cells were lysed and luminescence was measured. Luminescence was normalized to the 28°C control at 1 h after transfection. Data shown are means ± SEM from three independent experiments, each performed in duplicate wells. (C) Reverse transcription (RT)-qPCR. Standard and 20°C-adapted cells were infected with ZIKV (MOI, 0.1) at their respective temperatures for 2 h. After the virus inoculum was removed, one plate with standard cells was kept at 28°C and one was transferred to 20°C, and one plate with 20°C-adapted cells was kept at 20°C and one was transferred to 28°C. At the indicated time points, cell lysates were collected, RNA was isolated and reversed transcribed to cDNA, and genome copies were measured by using a qPCR. ZIKV copy numbers were compared to RPL32 transcripts (2−ΔΔCT) and normalized to 2 h after infection. Data shown are means ± SEM from three independent experiments, and each qPCR was performed in duplicate wells. Statistical significance was assessed by using a two-way ANOVA with Dunnett’s correction (data were first log transformed for panels A and C). For each time point, standard cells kept at 28°C (28°C-28°C) were compared to all other conditions. (D and E) ZIKV replicon. Standard and 20°C-adapted C6/36 cells were transfected with a CMV-driven ZIKV replicon or a control replicon containing a GDD-AAA mutation in NS5. Two plates were kept at their respective temperatures, and two were placed in the opposite-temperature incubators after transfection for 6 days. Luminescence was measured each day from duplicate wells. Data shown are means ± SEM from three or six independent experiments. Statistical significance was determined by using a two-way ANOVA on log-transformed data, followed by Bonferroni’s test for multiple comparisons. For each time point, wt replicon was compared to its control replicon. The color of the significance symbol corresponds to the color for each condition. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Efficient translation is required for virus to be produced from the transfected genomes. To directly measure translation efficiency, we designed a translation reporter construct containing a luciferase coding region flanked by the 5′ and 3′ ZIKV untranslated (UTR) regions and transfected the RNA into the cells (Fig. 7B). There was no difference in luciferase signal between the cells that were transfected and maintained at 28°C and the 20°C-adapted cells that were transfected and maintained at 20°C. Interestingly, there was a slight increase in luciferase production in the cells that were transfected at 28°C and incubated at 20°C for 8 h.
Since translation was not inhibited by the cool temperature, we wanted to investigate the effect of the cool temperature on genome replication. First, we infected the cells with ZIKV and measured ZIKV genome copies relative to Aedes albopictus RPL32 transcripts by using the ΔΔCT method (Fig. 7C). Cells maintained at 28°C rapidly produced ZIKV RNA; however, cells maintained at 20°C did not produce RNA at levels above the incoming particles 2 h after infection. We then utilized a cytomegalovirus (CMV)-driven ZIKV replicon containing nanoluciferase and a control replicon with an NS5 mutation (GDD-AAA) that prevents ZIKV amplification (Fig. 7D and E). While plasmid transcription will induce nanoluciferase production, amplification of the replicon by ZIKV replication machinery will be evident by an increase in signal by the replication-competent replicon compared to the replication-incompetent version. There was no difference in nanoluciferase signal between the cells transfected with replication-competent (wild-type [wt] replicon) and -incompetent (GDD-AAA replicon) plasmids at 20°C (Fig. 7D). Furthermore, there was no difference in the signals between the 20°C-adapted cells transfected and maintained at 20°C and the standard cells transfected at 28°C and transferred to 20°C, indicating that there is no significant difference in transfection efficiency at 20°C versus 28°C and no difference between the cells adapted to 20°C and standard cells moved to 20°C. In contrast, we detected an amplification of the luciferase signal in cells transfected with replication-competent (wt replicon) plasmid if maintained at 28°C after transfection (Fig. 7E). In cells with replication-incompetent (GDD-AAA) replicons, we detected only the baseline signal from the plasmid.
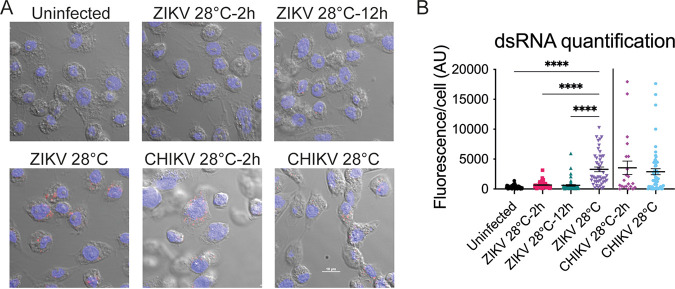
In addition, we investigated genome replication by staining cells with an antibody that detects double-stranded RNA (dsRNA), an intermediate produced during genome replication (Fig. 8A). When ZIKV-infected cells were transferred to 20°C 2 or 12 h after transfection, we were unable to detect dsRNA intermediates at the cool temperature after 48 h. In contrast, there was no significant difference in dsRNA intermediates between CHIKV-infected cells that were transferred to 20°C 2 h after transfection or those that were maintained at 28°C (Fig. 8B). Taken together, our data suggest that ZIKV genome replication is inefficient at cool temperatures in mosquito cells.
FIG 8.
dsRNA production at suboptimal temperature. (A) Accumulation of dsRNA intermediates. C6/36 cells were infected with a high MOI of ZIKV (MOI >10) and with CHIKV (MOI 1) at 28°C and were shifted to 20°C at 2 or 12 h p.i. Cells were processed for immunofluorescence using a monoclonal antibody to detect dsRNA at 48 h p.i. Uninfected cells were used as a control for specificity of the antibody against dsRNA. Nuclei are shown in blue, and dsRNA intermediates are shown in red. (B) Quantification of dsRNA fluorescence signal in ZIKV- and CHIKV-infected cells as described for panel A. Points represent individual cells analyzed. Statistical significance for ZIKV was assessed by using a one-way ANOVA with Dunnett’s correction, where cells infected and kept at 28°C were compared to each condition. Statistical significance for CHIKV was determined by using an unpaired t test with Welch’s correction. AU, arbitrary units. ****, P < 0.0001.
Replication curves of different ZIKV strains at suboptimal temperature.
Lastly, we wanted to explore if limited virus replication in C6/36 cells at 20°C was common among different ZIKV isolates or lineages. We compared replication curves of ZIKV Mex 1-44 to another Asian lineage, SPH (Fig. 9A), and to two African lineages, MR-766 and IbH (Fig. 9B and C). As previously observed with Mex 1-44 (Fig. 1A), SPH replication was reduced at 20°C; low levels of virus were detected on day 6 and only slightly increased through day 10. In contrast, both African lineage strains replicated faster, with detectable replication occurring earlier at day 4 and viral production at both 20°C and 28°C reaching similar values over the course of the experiment.
FIG 9.
Replication of different ZIKV strains at suboptimal temperatures. Replication rates for SPH (A), MR-766 (B), and IbH (C) at cool temperature. The cells were infected at an MOI of 0.1 for 2 h at 28°C and incubated at either 20°C or 28°C for 10 days. Supernatants were collected every 48 h and titrated. Dashed lines represent the limit of detection. Data shown are means ± SEM of results from five replicates for each virus.
DISCUSSION
In order to be successfully transmitted, arboviruses need to adapt to distinct hosts and persist within a wide temperature range. Environmental temperature plays a critical role in arboviral transmission since mosquito vectors are susceptible to changes in temperature (8, 30–34). We have previously demonstrated that ZIKV transmission in A. aegypti mosquitoes is optimized at 29°C with a thermal range of 22.7°C to 34.7°C (24). Low ambient temperatures that are well within the range for survival of Aedes species are known to inhibit arboviral replication and transmission (11, 35). However, the molecular mechanisms underlying the effects of temperature on virus replication efficiency and vector competence remain unclear. Here, we present data that focus on how temperature alters ZIKV replication in vitro in C6/36 cells to characterize this relationship in a minimal system that excludes systemic host responses. We hypothesized that suboptimal temperatures alter the intracellular environment, which consequently inhibits virus replication, and/or that temperature directly inhibits virus replication. First, we showed that ZIKV replication is inhibited at suboptimal temperatures even when cells are adapted to those temperatures, suggesting that the reduced replication is not due to changes caused by acute temperature stress. We further clarified the steps of the virus replication cycle that are most affected by cool temperatures, examining viral entry, translation, and genome replication. We found that cool temperatures did not inhibit ZIKV entry or translation but affected genome replication and significantly reduced the production of dsRNA replication intermediates (Fig. 10A and B). However, if cells with an established infection were shifted to 20°C, all the subsequent steps were efficiently completed, suggesting that cool temperatures decrease the efficiency of early stages in genome replication (Fig. 10C).
FIG 10.
Model of ZIKV replication cycle at 20°C and 28°C. (A) Replication of ZIKV efficiently occurs at 28°C, with all steps completed. (B) When ZIKV infection is initiated at 20°C, infection is stalled after capsid disassembly but before dsRNA intermediates can be detected. (C) Late stages in the ZIKV replication cycle can occur at 20°C if RNA replication steps occur at 28°C (C). Created in Biorender.com.
Temperature is known to induce molecular changes that impact structures and functions of proteins, lipids, and nucleic acids, which in turn can affect intracellular processes, enzymatic activity, membrane fluctuations, cell metabolism, and viability and induce temperature-shock responses. Therefore, temperature modifies virions and their interactions with cellular components and is one of the most important factors impacting infection dynamics during viral replication (36). Although arboviruses can tolerate drastic temperature changes, replicating efficiently in both mammalian (~37°C) and mosquito (~28°C) hosts, we found that exposing ZIKV-infected mosquito cells to a range of temperatures affected virus yields over time. During early infection, we observed slower replication kinetics at lower temperatures and faster replication at higher temperatures (Fig. 1E). These higher replication rates of ZIKV at high temperatures could be caused by increased cellular metabolism, enzymatic activity, and fluidity of cellular membranes. Moreover, there is evidence that the heat shock response can facilitate entry and replication of Zika, dengue, West Nile, and Japanese encephalitis viruses (37–39). It has also been suggested that high temperatures have a direct influence on viral RNA structure and long-range RNA-RNA interactions (20, 40) as well as on the conformation of RNA-dependent RNA polymerase (RdRp) (19), resulting in enhanced replication and higher titers. Later in infection, we observed decreased viral titers at cool and high temperatures and increased titers around intermediate temperatures (Fig. 1F). The drop in virus yields observed under high-temperature conditions was caused by a prolonged heat shock response that affected cell viability. Cool temperatures, on the other hand, resulted in low virus yields at all time points. Although cold temperatures decrease cellular metabolism, enzymatic activity, and membrane fluidity (41, 42), none of these potential alterations in the intracellular environment inhibited DENV or CHIKV replication in C6/36 cells at 20°C to the same extent that ZIKV replication was inhibited, suggesting that ZIKV is more sensitive to temperature than the other viruses.
We evaluated ZIKV entry, which includes virus-cell attachment, internalization, and envelope-cell membrane fusion, at 20°C. ZIKV enters the cells through clathrin-mediated endocytosis and requires low pH for fusion (25). By adding ammonium chloride at different times after ZIKV infection, we established that most ZIKV virions enter C6/36 cells within 2 h at 28°C. Since cells were infected at 28°C for 2 h before being shifted to other temperatures in the majority of the experiments, we suggest that the temperature inhibition was preventing a step in the replication cycle following membrane fusion. Moreover, transfecting the cells with viral RNA, thereby bypassing entry and nucleocapsid disassembly steps, resulted in an inhibition of virus production similar to infecting the cells at 20°C (Fig. 7A). However, while transfecting viral RNA into cells maintained at 20°C prevented virus production, transfecting a translation reporter construct (with untypical type-0 cap) or a nanoluciferase replicon (with typical type-1 cap) demonstrated that translation is not dramatically affected by cool temperature (Fig. 7B and D). When we specifically measured and investigated genome replication by quantitative PCR (RT-qPCR) and a ZIKV replicon, we could not detect any amplification of the ZIKV genome or the nanoluciferase signal by ZIKV replication machinery at 20°C, respectively. Furthermore, we could not readily observe ZIKV dsRNA replication intermediates at cool temperature. Late steps in the virus replication cycle, such as budding, maturation, and viral egress, are not likely disrupted by cool temperature, since the cells with an established infection can produce similar numbers of infectious particles at 20°C and 28°C (Fig. 4A). While our study used C6/36 cells, these findings should be validated in other cell lines and in mosquitoes.
There are several steps viruses undergo after entry and before genome replication that could be sensitive to cooler temperatures. After translation and polyprotein processing, ZIKV must induce extensive ultrastructural changes and rearrangement of the intracellular membranes to provide the platforms required for the formation of replication complexes (43, 44). Replication inside membrane compartments concentrates the molecules necessary for replication and particle assembly but also helps evade host antiviral defense mechanisms (45, 46). The endoplasmic reticulum (ER) is the primary site for flavivirus replication, and DENV is known to perturb lipid homeostasis in mosquito cells (47) and to reprogram the ER protein synthesis and processing environment to promote viral replication (48). DENV utilizes fatty acid biosynthesis resulting in lipid enrichment that has the capacity to destabilize and change the curvature and permeabilization of membranes (47), and the NS3 protein has been identified as redistributing fatty acid synthase (FAS) to the site of viral replication (49). It is well known that cool temperatures can affect membrane fluidity, but it remains unclear how temperature could alter the ability of viral nonstructural proteins to interact with cellular factors and disturb membrane rearrangement.
RNA cyclization, another posttranslational step in the virus replication cycle, is known to be temperature dependent, and it is a prerequisite for the initiation of flavivirus replication. Studies have shown that cultivation temperature decisively determines the replication efficiency of West Nile virus (WNV) and ZIKV RNA in mammalian and mosquito cells (20, 40). WNV long-range RNA-RNA interactions in vitro are enhanced at high temperatures (37°C), while at low temperatures (28°C), genome cyclization is less efficient and therefore replication starts at later time points. It has been suggested that this “RNA thermometer” activity could be involved in modulating the replication efficiency during host switching (20). A study by Li et al. has shown that a single-nucleotide mutation in the DAR (downstream of AUG region) is sufficient to abolish long-range RNA-RNA interactions and de novo RNA synthesis of ZIKV at 28°C but not at 37°C. A more restricted DAR complementarity for viral replication at lower temperatures is additional evidence that temperature is a critical determinant of replication kinetics (40). Another study suggested that the active site of the DENV RdRp exists in two conformational states, a more rigid conformation at lower temperatures and a more mobile conformation at higher temperatures, and that temperature determines how the enzyme recognizes the conformation of the template RNA and affects RNA synthesis (19).
Interestingly, we found that low temperature affected Asian lineage ZIKV isolates (Mex 1-44 and SPH) and African lineage ZIKV isolates (MR-766 and IbH) differently (Fig. 9). While Mex 1-44 and SPH replicated at a lower rate and had lower virus titers at 20°C, MR-766 and IbH replicated faster, and titers peaked at similar levels at 20°C and 28°C. African and Asian lineages were previously shown to have different phenotypes in cell culture and A. aegypti mosquitoes (50). While African isolates have higher replication rates in C6/36 cells than Asian lineage isolates, they seem to be less infectious to A. aegypti mosquitoes (50). Moreover, ZIKV isolates from Asian and African lineages displayed different replication kinetics, cytopathic effects, and impacts on human neural progenitor cell function (51, 52), as well as the ability to infect human placental trophoblast (53), cause severe brain damage (54), and cause disease progression in mice (55). Comparative analysis of viral entry showed that divergent ZIKV strains enter cells in a highly conserved manner (29), suggesting that genetic variation between ZIKV isolates affects some other aspect of ZIKV replication cycle with consequences for infection kinetics and pathogenesis.
We have shown that temperature affects flavivirus and alphavirus replication kinetics differently. ZIKV and DENV responded similarly to temperatures; however, the extent to which temperature affected virus yields differed. DENV replication was more efficient and DENV reached higher titers at cool temperatures than ZIKV, which is consistent with our in vivo study showing that the predicted thermal minimum for DENV is 5°C lower than that of ZIKV in A. aegypti mosquitoes (24) and even lower in A. albopictus mosquitoes (10). While flavivirus titers increased with the increase in temperature early during infection, CHIKV titers were the highest at intermediate temperatures and lower at both low and high temperatures. Previous work suggests that CHIKV replication kinetics, rather than high titers, are more important for midgut escape and transmission (56), since slowly replicating variants are less able to overcome the midgut escape barrier. This could also explain the block in transmission of ZIKV at 20°C (24).
After exposing A. albopictus-derived C6/36 cells infected with ZIKV to the same temperatures that we used previously for ZIKV-infected A. aegypti mosquitoes (24), we observed similar qualitative trends. High temperatures caused a higher replication rate in vitro, as well as a higher proportion of infected mosquitoes and a shorter EIP in vivo, while affecting cell viability and mosquito survival. Low temperatures caused a significant reduction in viral titers in vitro and a low proportion of infected mosquitoes, longer EIP, and impaired transmission in vivo. We have also investigated the transcriptome profile of ZIKV-infected mosquitoes at low and high temperatures and showed that temperature dramatically shapes mosquito gene expression (57). Although temperature causes systemic changes in mosquitoes that can alter virus replication, the direct effect of temperature on ZIKV replication was still visible at the cellular level. While the effects of temperature on ZIKV transmission and vector competence have been well documented (6, 24, 58–60), Murrieta et al. recently demonstrated that temperature also impacts virus evolution. They showed that temperature significantly modifies the selective environment within mosquitoes and that fluctuating temperatures cause strong purifying selection mostly in ZIKV NS5 (61). Future experiments should thus characterize the phenotypes at suboptimal temperatures and investigate potential evolutionary patterns associated with ZIKV replication at cool temperatures.
Understanding transmission dynamics and epidemiology of infectious diseases is crucial for successfully controlling current outbreaks and preventing future ones. Characterizing the link between pathogen replication and environmental conditions is important to better understand how environmental factors shape transmission.
MATERIALS AND METHODS
Cell lines.
C6/36 mosquito cells (ATCC CRL-1660) from A. albopictus were maintained in Leibovitz’s L-15 medium supplemented with 10% fetal bovine serum (FBS) at 28°C. Some C6/36 cells were adapted to grow at 20°C. To adapt the cells, they were initially maintained in L-15 medium with a higher FBS content (20%). After the cells were maintained at 20°C for several weeks, the FBS content was gradually reduced to 10% over several months. Vero (African green monkey kidney) cells were cultured in Dulbecco’s modified Eagle medium (DMEM) with 5% FBS at 37°C, 5% CO2.
Virus isolates and viral titer determination.
All viral stocks were generated in Vero cells and tested negative for Mycoplasma contamination by use of a MycoSensor PCR assay kit (Agilent). Detailed passage histories of the ZIKV isolates used in this study were previously described (50). ZIKV Mex 1-44 was isolated from a field-caught A. aegypti mosquito in Chiapas, Mexico, in 2016 and was obtained from the University of Texas Medical Branch (62). ZIKV SPH originated from a Brazilian clinical sample in 2015 and was obtained from the Oswaldo Cruz Foundation (63). Two African lineage isolates, ZIKV MR-766 and IbH, were purchased from the American Type Culture Collection (catalog no. VR-1838 and VR-1829, respectively) (64). The DENV-2 isolate (PRS 225488) originated from human serum collected in Thailand in 1974 and was obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. An attenuated CHIKV strain (181/c25) was generated as previously described (65). Viral RNA was produced by in vitro transcription of pSinRep5-181/25ic, a gift from Terence Dermody (Addgene plasmid no. 60078) (66). Capped RNA was transfected into 293T cells, and virus-containing supernatants were collected when cells showed signs of cytopathic effect. All viral titers were determined by endpoint dilutions on Vero cells. The 50% tissue culture infectious dose (TCID50) was determined using the Spearman-Karber method (67).
Virus replication curves.
C6/36 cells were seeded in 12-well plates at a density of 4 × 105 cells/mL several hours prior to infection. Cells were inoculated with ZIKV using a multiplicity of infection (MOI) of 0.1, with DENV at an MOI of 0.1, or with CHIKV at an MOI of 0.005 for 2 h. The cells and medium were kept at the indicated constant temperatures for 10 days. Every 24 or 48 h, half of the supernatant was collected and replaced with fresh medium equilibrated to each temperature. Similarly, Vero cells were plated at a density of 2.5 × 105 cells/mL and inoculated with ZIKV (MOI, 0.1) for 1 h at 37°C. The cells were incubated at different constant temperatures, and half of the cell supernatant was collected every 24 h for 7 days.
Cell proliferation.
C6/36 cells were stained using the CellTrace violet cell proliferation kit (Invitrogen) per the manufacturer’s instructions and plated at a density of 4 × 105 cells/mL in 24-well plates. The cells were maintained at different constant temperatures, and intracellular fluorescence, which decreases with every generation, was determined using flow cytometry at the indicated time points. The mean fluorescence intensities (MFI) were normalized to the MFI value determined at the time of staining to calculate the number of divisions that occurred.
Cell viability.
Cell viability was monitored using the CellTiter-Glo luminescent cell viability assay (Promega) and a Glomax Explorer (Promega) plate reader per the manufacturer’s instructions. C6/36 cells were plated at a density of 4 × 105 cells/mL in 24-well plates and treated with the virus, temperature, or drug. Vero cells were plated at a density of 2.5 × 105 cells/mL in 24-well plates and infected with ZIKV or left uninfected. At the indicated time points, cell viability was determined and compared to that of the indicated controls.
pAc5-STABLE2-neo transfection.
C6/36 cells were seeded in two T-25 flasks at a density of 6 × 106 cells per flask the night before transfection. One flask was transfected with Ac5-STABLE2-neo plasmid, a gift from Rosa Barrio and James Sutherland (Addgene plasmid no. 32426) (68), using Lipofectamine 3000 transfection reagent (Invitrogen) per the manufacturer’s instructions. The plasmid encodes Flag-mCherry, GFP, and Neor under the control of the insect Actin5C promoter. Six hours after transfection, both transfected and control cells were lifted, plated in six 24-well plates, and transferred to the indicated constant-temperature incubators. Cells were resuspended in phosphate-buffered saline (PBS), and GFP production was measured using a flow cytometer (BD-LSRII) 24 and 72 h after temperature exposure.
Monitoring of endogenous protein levels.
C6/36 cells were plated at a density of 8 × 105 cells per well in 6-well plates and incubated at 28°C for 4 h to allow the cells time to adhere. The cells were then distributed to incubators at the indicated temperatures and incubated for 4 days. Cells were gently washed in PBS, scraped in a small volume, and lysed (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100). Aliquots of each sample were diluted in PBS and quantified using a Bradford assay. Equivalent amounts of protein were denatured/reduced (200 mM Tris [pH 6.8], 8 M urea, 5% SDS, 0.1 mM EDTA, 5 mM dithiothreitol, 0.03% bromophenol blue) and run on a 10% TGX Stain-Free gel. The protein stain was activated with UV light and imaged per the manufacturer’s instructions (Bio-Rad).
ZIKV production during temperature shift experiments.
Persistently infected ZIKV C6/36 cells were produced by maintaining an infected culture in a T-75 flask at 28°C for several weeks. To monitor viral production, 1 mL of cell supernatant was collected on two consecutive days. The cells were then seeded in two 6-well plates at a density of 6 × 105 cells/mL. One plate was maintained at 28°C, and the second was placed at 20°C. Every 24 h, the entire cell supernatant was collected and replaced to measure the daily virus production.
For the experiment with actively spreading infection, C6/36 cells were seeded in a T-25 flask at a density of 6 × 105 cells/mL. The following day, the cells were infected with ZIKV (MOI, 0.1) for 2 h at 28°C. After incubation, infectious medium was removed, and the cells were washed once before fresh medium was added. Supernatant was collected at the indicated time points. After the 36-h time point sample was collected, the cells were washed and seeded in two 6-well plates. Each plate was incubated at either 20°C or 28°C for 5 days. Every 24 h, the entire cell supernatant was collected and replaced to measure the daily virus production.
Detection of ZIKV-positive cells at optimal and suboptimal temperatures using flow cytometry.
C6/36 cells were plated in three T-25 flasks at a density of 2 × 106 cells/mL. Two flasks were infected with ZIKV (MOI, 0.1) for 2 h at 28°C. Cells from one flask were split into two 6-well plates and transferred to 20°C or 28°C at 2 h following infection. Infected and uninfected control cells from the other two flasks were kept at 28°C and were split and transferred 36 h following infection. The cells were stained for intracellular ZIKV protein NS1, and the percentage of ZIKV-positive cells was determined using a flow cytometer at the indicated time points. Briefly, the cells were resuspended in fresh medium, fixed with 4% paraformaldehyde for 15 min, and permeabilized with 0.1% Triton X-100 in 1× PBS for 15 min. The cells were centrifuged at 300 × g for 10 min and resuspended in the primary antibody (anti-ZIKV NS1 mouse MAb; BioFront Technologies) diluted in permeabilization buffer at a concentration of 1:500. After 45 min of incubation at 37°C, the cells were washed three times in permeabilization buffer by centrifugation for 5 min at 300 × g. After the last spin, the cells were resuspended in the secondary antibody (Alexa Fluor 647-conjugated goat anti-mouse Ab; Invitrogen) at a concentration of 1:1,000 and incubated for 30 min at 37°C. The cells were washed three times and resuspended in 1× PBS. The percentage of infected cells for each treatment was determined for 20,000 single cells.
Drug inhibition and time of addition.
Chlorpromazine hydrochloride (Millipore Sigma) was dissolved in ethanol and ammonium chloride (Millipore Sigma) in 1× PBS (Millipore Sigma). C6/36 cells were seeded at a density of 6 × 105 cells/mL in a 24-well plate and were treated in parallel with the inhibitor or the equivalent amount of vehicle. Cells were infected with ZIKV (MOI, 0.1), and inhibitors were added at the indicated time points. Each treatment with CPZ or NH4Cl was for 4 h, after which time the inhibitors were removed and fresh medium was added. The cells were kept at 28°C for 48 h, at which time the supernatant was collected and titrated.
ZIKV RNA transfection.
Infectious viral RNA was isolated from viral particles. Briefly, five 15-cm dishes of Vero cells were inoculated with ZIKV (MOI, 0.01) for 6 days. Supernatant was clarified (1,000 × g, 3 min) to remove the cell debris, and the virus was then pelleted through a 20% sucrose cushion at 37,000 rpm in an F37L-8×100 rotor in a Sorvall WX ultracentrifuge. Viral particle pellets were lysed in 500 μL TRIzol reagent (Thermo Fisher Scientific), and the RNA was extracted per the manufacturer’s instructions. Viral RNA was quantified using a Take3 microvolume plate and BioTek microplate reader. Standard and 20°C-adapted C6/36 cells were plated at a density of 7 × 105 cells/mL in 48-well plates the day before transfection. Infectious RNA (3 μg) was diluted in 100 μL of serum-free L-15 medium and mixed with Lipofectamine 3000 (Invitrogen) transfection reagents in accordance with the manufacturer’s instructions. The transfection mixture was equally distributed over three wells on three plates. Each plate was incubated for 2 h at its respective temperatures. The medium was replaced, and some plates were switched to different temperatures. Supernatant was collected at the indicated time points, and viral titers were determined.
Translation quantification.
A translation reporter construct, synthesized by Genewiz, that encodes NanoLuc luciferase with the 5′ and 3′ UTRs of ZIKV was amplified based on a previously published protocol (69). The DNA was in vitro transcribed by using the HiScribe T7 ARCA mRNA kit (NEB), and capped RNA was isolated by using an RNA Clean & Concentrator (Zymo Research) in accordance with the manufacturer’s instructions. Standard and 20°C-adapted C6/36 cells were plated at a density of 1 × 106 cells/mL in 96-well plates and incubated at their respective temperatures for 3 h. Capped RNA (3 μg) was mixed with Lipofectamine 3000 (Invitrogen) transfection reagents, in accordance with the manufacturer’s instructions, and used to transfect 12 wells on each of three 96-well plates. After 1 h of incubation at the respective temperatures, one plate was shifted from 28°C to 20°C. At the indicated time points, the medium was removed from duplicate wells, and the cells were lysed in lysis buffer and stored at 4°C. Once cells from all time points were collected, the Nano-Glo substrate (Promega) was added, and the luminescence was measured in a Glomax Explorer (Promega) plate reader.
qPCR.
Standard and 20°C-adapted C6/36 cells were plated at a density of 1 × 106 cells/mL in 12-well plates and were infected with ZIKV (MOI, 0.1) for 2 h at their respective temperatures. The virus was then removed, cells were washed with medium, fresh medium was replaced, and the cells were moved to the indicated temperatures. At the indicated time points, cells were lysed, and RNA was isolated by using a Quick-RNA miniprep kit (Zymo Research) per the manufacturer’s instructions. One hundred nanograms of RNA was reverse transcribed into cDNA by using a High-Capacity RNA-to-cDNA kit (Thermo Scientific). qPCR was performed using the iQ SYBR green supermix with ZIKV NS5 primers (F, GACTGGGTTCCAACTGGGAG; R, CCACACTCTGTTCCACACCA) (70) and A. albopictus RPL32 control gene (F, TATGACAAGCTTGCCCCCAA; R, AGGAACTTCTTGAATCCGTTGG) (71). The qPCR cycling protocol consisted of a denaturation step at 95°C for 15 s, followed by 40 cycles of 59°C for 10 s and 72°C for 20 s. Each sample was analyzed in duplicate, and each assay contained no-template and no-primer controls. 2−ΔΔCT values were calculated by comparing the change of ZIKV signal to that of RPL32 and normalizing ratios at 2 h following infection.
ZIKV replicon.
Standard and 20°C-adapted C6/36 cells were plated at a density of 1 × 106 cells/mL in 96-well plates and were incubated at their respective temperatures overnight (2 plates of each cell type). Cells were transfected with a plasmid, a gift from Andres Merits (University of Tartu), encoding a CMV-driven ZIKV replicon in which the structural proteins were replaced by nanoluciferase (72) or a control replicon containing a GDD-AAA mutation in NS5 preventing ZIKV-induced amplification. Eight micrograms of plasmid was mixed with Lipofectamine 3000 per the manufacturer’s instructions and distributed to 14 wells on each plate (56 wells in total). The transfection was performed at the temperature in which the cells were maintained for 2 h. The medium was then replaced, and two plates were placed in the opposite-temperature incubators. At the indicated time points, duplicate wells from each plate were lysed in Nano-Glo substrate (Promega), and the luminescence was measured in a Glomax Explorer (Promega) plate reader.
Immunocytochemistry.
C6/36 cells were plated at a density of 6 × 105 cells/mL in a 24-well plate containing a glass coverslip and incubated with ZIKV (high MOI, >10) or CHIKV (MOI, 1). Plates were shifted to the 20°C incubator at the indicated time points, and cells were harvested 48 h following infection. Cells were fixed with 4% paraformaldehyde overnight at 4°C. After fixation, all staining steps occurred at 4°C. The cells were washed three times in 1× PBS, permeabilized with 0.1% Triton X-100 in 1× PBS for 5 min, and blocked with 0.1% Triton X-100 in 1× PBS–2% FBS for 15 min. Cells were incubated with mouse monoclonal anti-dsRNA antibody (rJ2; catalog no. MABE1134, EMD Millipore) diluted in a blocking solution (1:60) for 1 h, followed by three washes in a blocking solution. Alexa Fluor Plus 594-conjugated goat anti-mouse antibody (Invitrogen) was diluted in a blocking solution (1:100) and incubated for 30 min while protected from the light. The secondary antibody was removed, and 0.1% Hoechst dye in a blocking solution was incubated for 15 min in the dark. The cells were washed three times with 1× PBS, and coverslips were mounted onto slides with ProLong gold antifade reagent (Molecular Probes). Samples were visualized using a 60× oil immersion lens on a Nikon A1R confocal microscope.
Statistical analysis.
Statistical analyses were performed using Prism 9 for macOS (GraphPad Software, Inc.). All data are presented as the mean ± standard error of the mean (SEM), and the number of biological replicates for each experiment is indicated in the figure legends. When significance was calculated for viral titers, all data were first log transformed. All multiple comparisons were done using either ordinary one-way or two-way analysis of variance (ANOVA) with application of Bonferroni’s correction or Dunnett’s correction for comparison to control or mock infection. A P value of less than 0.05 was considered to be statistically significant, and statistical differences are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
ACKNOWLEDGMENTS
We thank the University of Texas Medical Branch Arbovirus Reference Collection for providing the viruses and Ted Ross for providing C6/36 cells. We thank James Barber at the CVM Cytometry Core Facility for technical support on the confocal microscope and flow cytometer. We are grateful to Christine Reitmayer, Sarah Lumley, and Luke Alphey for help with reagents. We give special thanks to Courtney Murdock for helpful discussions during the course of this work. We gratefully thank the members of the Brindley lab for thoughtful comments on the project and manuscript.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Contributor Information
Melinda A. Brindley, Email: mbrindle@uga.edu.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI. 2012. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis 6:e1760. 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 3.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 2016. WHO statement on the first meeting of the International Health Regulations (2005) (IHR 2005) Emergency Committee on Zika virus and observed increase in neurological disorders and neonatal malformations. http://www.who.int/mediacentre/news/statements/2016/1st-emergency-committee-zika/en/. Accessed July 19, 2017.
- 5.Mordecai EA, Caldwell JM, Grossman MK, Lippi CA, Johnson LR, Neira M, Rohr JR, Ryan SJ, Savage V, Shocket MS, Sippy R, Stewart Ibarra AM, Thomas MB, Villena O. 2019. Thermal biology of mosquito-borne disease. Ecol Lett 22:1690–1708. 10.1111/ele.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winokur OC, Main BJ, Nicholson J, Barker CM. 2020. Impact of temperature on the extrinsic incubation period of Zika virus in Aedes aegypti. PLoS Negl Trop Dis 14:e0008047. 10.1371/journal.pntd.0008047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambrechts L, Paaijmans KP, Fansiri T, Carrington LB, Kramer LD, Thomas MB, Scott TW. 2011. Impact of daily temperature fluctuations on dengue virus transmission by Aedes aegypti. Proc Natl Acad Sci USA 108:7460–7465. 10.1073/pnas.1101377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao FZ, Zhang Y, Deng YQ, He S, Xie HG, Zhou XN, Yan YS. 2014. The effect of temperature on the extrinsic incubation period and infection rate of dengue virus serotype 2 infection in Aedes albopictus. Arch Virol 159:3053–3057. 10.1007/s00705-014-2051-1. [DOI] [PubMed] [Google Scholar]
- 9.Mbaika S, Lutomiah J, Chepkorir E, Mulwa F, Khayeka-Wandabwa C, Tigoi C, Oyoo-Okoth E, Mutisya J, Ng'ang'a Z, Sang R. 2016. Vector competence of Aedes aegypti in transmitting Chikungunya virus: effects and implications of extrinsic incubation temperature on dissemination and infection rates. Virol J 13:114. 10.1186/s12985-016-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mordecai EA, Cohen JM, Evans MV, Gudapati P, Johnson LR, Lippi CA, Miazgowicz K, Murdock CC, Rohr JR, Ryan SJ, Savage V, Shocket MS, Stewart Ibarra A, Thomas MB, Weikel DP. 2017. Detecting the impact of temperature on transmission of Zika, dengue, and chikungunya using mechanistic models. PLoS Negl Trop Dis 11:e0005568. 10.1371/journal.pntd.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellone R, Failloux AB. 2020. The role of temperature in shaping mosquito-borne viruses transmission. Front Microbiol 11:584846. 10.3389/fmicb.2020.584846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adelman ZN, Anderson MA, Wiley MR, Murreddu MG, Samuel GH, Morazzani EM, Myles KM. 2013. Cooler temperatures destabilize RNA interference and increase susceptibility of disease vector mosquitoes to viral infection. PLoS Negl Trop Dis 7:e2239. 10.1371/journal.pntd.0002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murdock CC, Paaijmans KP, Bell AS, King JG, Hillyer JF, Read AF, Thomas MB. 2012. Complex effects of temperature on mosquito immune function. Proc Biol Sci 279:3357–3366. 10.1098/rspb.2012.0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepock JR. 2005. How do cells respond to their thermal environment? Int J Hyperthermia 21:681–687. 10.1080/02656730500307298. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci USA 110:6795–6799. 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. 2013. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J Virol 87:7585–7592. 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Sun L, Rossmann MG. 2015. Temperature dependent conformational change of dengue virus. Curr Opin Virol 12:109–112. 10.1016/j.coviro.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudlacek ST, Premkumar L, Metz SW, Tripathy A, Bobkov AA, Payne AM, Graham S, Brackbill JA, Miley MJ, de Silva AM, Kuhlman B. 2018. Physiological temperatures reduce dimerization of dengue and Zika virus recombinant envelope proteins. J Biol Chem 293:8922–8933. 10.1074/jbc.RA118.002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackermann M, Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem 276:39926–39937. 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 20.Meyer A, Freier M, Schmidt T, Rostowski K, Zwoch J, Lilie H, Behrens SE, Friedrich S. 2020. An RNA thermometer activity of the West Nile virus genomic 3'-terminal stem-loop element modulates viral replication efficiency during host switching. Viruses 12:104. 10.3390/v12010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostyuchenko VA, Lim EX, Zhang S, Fibriansah G, Ng TS, Ooi JS, Shi J, Lok SM. 2016. Structure of the thermally stable Zika virus. Nature 533:425–428. 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- 22.Xie X, Yang Y, Muruato AE, Zou J, Shan C, Nunes BT, Medeiros DB, Vasconcelos PF, Weaver SC, Rossi SL, Shi PY. 2017. Understanding Zika virus stability and developing a chimeric vaccine through functional analysis. mBio 8:02134-16. 10.1128/mBio.02134-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slon Campos JL, Marchese S, Rana J, Mossenta M, Poggianella M, Bestagno M, Burrone OR. 2017. Temperature-dependent folding allows stable dimerization of secretory and virus-associated E proteins of Dengue and Zika viruses in mammalian cells. Sci Rep 7:966. 10.1038/s41598-017-01097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tesla B, Demakovsky LR, Mordecai EA, Ryan SJ, Bonds MH, Ngonghala CN, Brindley MA, Murdock CC. 2018. Temperature drives Zika virus transmission: evidence from empirical and mathematical models. Proc Biol Sci 285:20180795. 10.1098/rspb.2018.0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persaud M, Martinez-Lopez A, Buffone C, Porcelli SA, Diaz-Griffero F. 2018. Infection by Zika viruses requires the transmembrane protein AXL, endocytosis and low pH. Virology 518:301–312. 10.1016/j.virol.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Zhang D, Li C, Zheng Z, Fu M, Ni F, Liu Y, Du T, Wang H, Griffin GE, Zhang M, Hu Q. 2020. Characterization of Zika virus endocytic pathways in human glioblastoma cells. Front Microbiol 11:242. 10.3389/fmicb.2020.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohkuma S, Poole B. 1978. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA 75:3327–3331. 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiasny K, Fritz R, Pangerl K, Heinz FX. 2011. Molecular mechanisms of flavivirus membrane fusion. Amino Acids 41:1159–1163. 10.1007/s00726-009-0370-4. [DOI] [PubMed] [Google Scholar]
- 29.Rinkenberger N, Schoggins JW. 2019. Comparative analysis of viral entry for Asian and African lineages of Zika virus. Virology 533:59–67. 10.1016/j.virol.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watts DM, Burke DS, Harrison BA, Whitmire RE, Nisalak A. 1987. Effect of temperature on the vector efficiency of Aedes aegypti for dengue 2 virus. Am J Trop Med Hyg 36:143–152. 10.4269/ajtmh.1987.36.143. [DOI] [PubMed] [Google Scholar]
- 31.Turell MJ. 1993. Effect of environmental temperature on the vector competence of Aedes taeniorhynchus for Rift Valley fever and Venezuelan equine encephalitis viruses. Am J Trop Med Hyg 49:672–676. 10.4269/ajtmh.1993.49.672. [DOI] [PubMed] [Google Scholar]
- 32.Rohani A, Wong YC, Zamre I, Lee HL, Zurainee MN. 2009. The effect of extrinsic incubation temperature on development of dengue serotype 2 and 4 viruses in Aedes aegypti (L.). Southeast Asian J Trop Med Public Health 40:942–950. [PubMed] [Google Scholar]
- 33.Zouache K, Fontaine A, Vega-Rua A, Mousson L, Thiberge JM, Lourenco-De-Oliveira R, Caro V, Lambrechts L, Failloux AB. 2014. Three-way interactions between mosquito population, viral strain and temperature underlying chikungunya virus transmission potential. Proc Biol Sci 281:20141078. 10.1098/rspb.2014.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reisen WK, Fang Y, Martinez VM. 2006. Effects of temperature on the transmission of West Nile virus by Culex tarsalis (Diptera: Culicidae). J Med Entomol 43:309–317.2.0.CO;2]. 10.1093/jmedent/43.2.309. [DOI] [PubMed] [Google Scholar]
- 35.Samuel GH, Adelman ZN, Myles KM. 2016. Temperature-dependent effects on the replication and transmission of arthropod-borne viruses in their insect hosts. Curr Opin Insect Sci 16:108–113. 10.1016/j.cois.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mojica KD, Brussaard CP. 2014. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol Ecol 89:495–515. 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 37.Pujhari S, Brustolin M, Macias VM, Nissly RH, Nomura M, Kuchipudi SV, Rasgon JL. 2019. Heat shock protein 70 (Hsp70) mediates Zika virus entry, replication, and egress from host cells. Emerg Microbes Infect 8:8–16. 10.1080/22221751.2018.1557988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chavez-Salinas S, Ceballos-Olvera I, Reyes-Del Valle J, Medina F, Del Angel RM. 2008. Heat shock effect upon dengue virus replication into U937 cells. Virus Res 138:111–118. 10.1016/j.virusres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Chu JJ, Leong PW, Ng ML. 2005. Characterization of plasma membrane-associated proteins from Aedes albopictus mosquito (C6/36) cells that mediate West Nile virus binding and infection. Virology 339:249–260. 10.1016/j.virol.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 40.Li XD, Deng CL, Yuan ZM, Ye HQ, Zhang B. 2020. Different degrees of 5'-to-3' DAR interactions modulate Zika virus genome cyclization and host-specific replication. J Virol 94:e01602-19. 10.1128/JVI.01602-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zachariassen KE. 1991. Hypothermia and cellular physiology. Arctic Med Res 50(Suppl 6):13–17. [PubMed] [Google Scholar]
- 42.Quinn PJ. 1988. Effects of temperature on cell membranes. Symp Soc Exp Biol 42:237–258. [PubMed] [Google Scholar]
- 43.Rossignol ED, Peters KN, Connor JH, Bullitt E. 2017. Zika virus induced cellular remodelling. Cell Microbiol 19:10.1111/cmi.12740. 10.1111/cmi.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R. 2017. Ultrastructural characterization of Zika virus replication factories. Cell Rep 18:2113–2123. 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Armas-Rillo L, Valera MS, Marrero-Hernandez S, Valenzuela-Fernandez A. 2016. Membrane dynamics associated with viral infection. Rev Med Virol 26:146–160. 10.1002/rmv.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann HH, Schneider WM, Rozen-Gagnon K, Miles LA, Schuster F, Razooky B, Jacobson E, Wu X, Yi S, Rudin CM, MacDonald MR, McMullan LK, Poirier JT, Rice CM. 2021. TMEM41B is a pan-flavivirus host factor. Cell 184:133–148.e20. 10.1016/j.cell.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog 8:e1002584. 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid DW, Campos RK, Child JR, Zheng T, Chan KWK, Bradrick SS, Vasudevan SG, Garcia-Blanco MA, Nicchitta CV. 2018. Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for co-opting host cell protein synthesis. J Virol 92:e01766-17. 10.1128/JVI.01766-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci USA 107:17345–17350. 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willard KA, Demakovsky L, Tesla B, Goodfellow FT, Stice SL, Murdock CC, Brindley MA. 2017. Zika virus exhibits lineage-specific phenotypes in cell culture, in Aedes aegypti mosquitoes, and in an embryo model. Viruses 9:383. 10.3390/v9120383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodfellow FT, Willard KA, Wu X, Scoville S, Stice SL, Brindley MA. 2018. Strain-dependent consequences of Zika virus infection and differential impact on neural development. Viruses 10:550. 10.3390/v10100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anfasa F, Siegers JY, van der Kroeg M, Mumtaz N, Stalin Raj V, de Vrij FMS, Widagdo W, Gabriel G, Salinas S, Simonin Y, Reusken C, Kushner SA, Koopmans MPG, Haagmans B, Martina BEE, van Riel D. 2017. Phenotypic differences between Asian and African lineage Zika viruses in human neural progenitor cells. mSphere 2:e00292-17. 10.1128/mSphere.00292-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheridan MA, Balaraman V, Schust DJ, Ezashi T, Roberts RM, Franz AWE. 2018. African and Asian strains of Zika virus differ in their ability to infect and lyse primitive human placental trophoblast. PLoS One 13:e0200086. 10.1371/journal.pone.0200086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shao Q, Herrlinger S, Zhu YN, Yang M, Goodfellow F, Stice SL, Qi XP, Brindley MA, Chen JF. 2017. The African Zika virus MR-766 is more virulent and causes more severe brain damage than current Asian lineage and dengue virus. Development 144:4114–4124. 10.1242/dev.156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowall SD, Graham VA, Rayner E, Hunter L, Atkinson B, Pearson G, Dennis M, Hewson R. 2017. Lineage-dependent differences in the disease progression of Zika virus infection in type-I interferon receptor knockout (A129) mice. PLoS Negl Trop Dis 11:e0005704. 10.1371/journal.pntd.0005704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merwaiss F, Filomatori CV, Susuki Y, Bardossy ES, Alvarez DE, Saleh MC. 2021. Chikungunya virus replication rate determines the capacity of crossing tissue barriers in mosquitoes. J Virol 95:e01956-20. 10.1128/JVI.01956-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira PG, Tesla B, Horácio ECA, Nahum LA, Brindley MA, de Oliveira Mendes TA, Murdock CC. 2020. Temperature dramatically shapes mosquito gene expression with consequences for mosquito-Zika virus interactions. Front Microbiol 11:901. 10.3389/fmicb.2020.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chouin-Carneiro T, David MR, de Bruycker Nogueira F, Dos Santos FB, Lourenço-de-Oliveira R. 2020. Zika virus transmission by Brazilian Aedes aegypti and Aedes albopictus is virus dose and temperature-dependent. PLoS Negl Trop Dis 14:e0008527. 10.1371/journal.pntd.0008527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onyango MG, Bialosuknia SM, Payne AF, Mathias N, Kuo L, Vigneron A, DeGennaro M, Ciota AT, Kramer LD. 2020. Increased temperatures reduce the vectorial capacity of Aedes mosquitoes for Zika virus. Emerg Microbes Infect 9:67–77. 10.1080/22221751.2019.1707125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blagrove MSC, Caminade C, Diggle PJ, Patterson EI, Sherlock K, Chapman GE, Hesson J, Metelmann S, McCall PJ, Lycett G, Medlock J, Hughes GL, Della Torre A, Baylis M. 2020. Potential for Zika virus transmission by mosquitoes in temperate climates. Proc Biol Sci 287:20200119. 10.1098/rspb.2020.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murrieta RA, Garcia-Luna SM, Murrieta DJ, Halladay G, Young MC, Fauver JR, Gendernalik A, Weger-Lucarelli J, Rückert C, Ebel GD. 2021. Impact of extrinsic incubation temperature on natural selection during Zika virus infection of Aedes aegypti and Aedes albopictus. PLoS Pathog 17:e1009433. 10.1371/journal.ppat.1009433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerbois M, Fernandez-Salas I, Azar SR, Danis-Lozano R, Alpuche-Aranda CM, Leal G, Garcia-Malo IR, Diaz-Gonzalez EE, Casas-Martinez M, Rossi SL, Del Río-Galván SL, Sanchez-Casas RM, Roundy CM, Wood TG, Widen SG, Vasilakis N, Weaver SC. 2016. Outbreak of Zika virus infection, Chiapas State, Mexico, 2015, and first confirmed transmission by Aedes aegypti mosquitoes in the Americas. J Infect Dis 214:1349–1356. 10.1093/infdis/jiw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faria NR, Azevedo R, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami F, Macedo FLL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, Medeiros DBA, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, Simith DB, Messina JP, Abade L, Lourenço J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, Lemos PDS, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Júnior J, Mir D, Bello G, Delatorre E, Khan K, et al. 2016. Zika virus in the Americas: early epidemiological and genetic findings. Science 352:345–349. 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6:e1477. 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lay Mendoza MF, Acciani MD, Levit CN, Santa Maria C, Brindley MA. 2020. Monitoring viral entry in real-time using a luciferase recombinant vesicular stomatitis virus producing SARS-CoV-2, EBOV, LASV, CHIKV, and VSV glycoproteins. Viruses 12:1457. 10.3390/v12121457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mainou BA, Zamora PF, Ashbrook AW, Dorset DC, Kim KS, Dermody TS. 2013. Reovirus cell entry requires functional microtubules. mBio 4:e00405-13. 10.1128/mBio.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramakrishnan MA. 2016. Determination of 50% endpoint titer using a simple formula. World J Virol 5:85–86. 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.González M, Martín-Ruíz I, Jiménez S, Pirone L, Barrio R, Sutherland JD. 2011. Generation of stable Drosophila cell lines using multicistronic vectors. Sci Rep 1:75. 10.1038/srep00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schultz MJ, Tan AL, Gray CN, Isern S, Michael SF, Frydman HM, Connor JH. 2018. Wolbachia wStri blocks Zika virus growth at two independent stages of viral replication. mBio 9:e00738-18. 10.1128/mBio.00738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Araujo RV, Feitosa-Suntheimer F, Gold AS, Londono-Renteria B, Colpitts TM. 2020. One-step RT-qPCR assay for ZIKV RNA detection in Aedes aegypti samples: a protocol to study infection and gene expression during ZIKV infection. Parasit Vectors 13:128. 10.1186/s13071-020-4002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dzaki N, Azzam G. 2018. Assessment of Aedes albopictus reference genes for quantitative PCR at different stages of development. PLoS One 13:e0194664. 10.1371/journal.pone.0194664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mutso M, Saul S, Rausalu K, Susova O, Žusinaite E, Mahalingam S, Merits A. 2017. Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J Gen Virol 98:2712–2724. 10.1099/jgv.0.000938. [DOI] [PubMed] [Google Scholar]