Abstract
Oligonucleotide probes, designed from genes coding for 16S rRNA, were developed to differentiate Methanosaeta concilii, Methanosarcina barkeri, and mesophilic methanogens. All M. concilii oligonucleotide probes (designated MS1, MS2, and MS5) hybridized specifically with the target DNA, but MS5 was the most specific M. concilii oligonucleotide probe. Methanosarcina barkeri oligonucleotide probes (designated MB1, MB3, and MB4) hybridized with different Methanosarcina species. The MB4 probe specifically detected Methanosarcina barkeri, and the MB3 probe detected the presence of all mesophilic Methanosarcina species. These new oligonucleotide probes facilitated the identification, localization, and quantification of the specific relative abundance of M. concilii and Methanosarcina barkeri, which play important roles in methanogenesis. The combined use of fluorescent in situ hybridization with confocal scanning laser microscopy demonstrated that anaerobic granule topography depends on granule origin and feeding. Protein-fed granules showed no layered structure with a random distribution of M. concilii. In contrast, a layered structure developed in methanol-enriched granules, where M. barkeri growth was induced in an outer layer. This outer layer was followed by a layer composed of M. concilii, with an inner core of M. concilii and other bacteria.
Anaerobic bioreactors are used to treat various organic wastes, which are ultimately converted into methane. It is generally accepted that two-thirds or more of the methane produced in an anaerobic bioreactor is derived from acetate (47). Of the many methanogenic genera, only two, Methanosaeta and Methanosarcina, are known to grow by an acetoclastic reaction, producing methane from acetate (47). Methanosaeta concilii is solely an acetoclastic bacterium and is the only mesophilic species of its genus, other species being thermophiles (43). Methanosarcina barkeri is metabolically the most versatile of all the mesophilic methanogenic bacteria isolated in pure culture, since it can form methane from H2 and CO2 (hydrogenotroph), from methanol and methylamines (methylotroph), and from acetate (acetoclast) (24, 47).
Upflow anaerobic sludge blanket (UASB) bioreactors are the most commonly used systems for wastewater anaerobic treatment. The retention of large amounts of biomass in a UASB bioreactor is due to the ability of bacterial cells to aggregate and form a granular structure which can settle and accumulate in the reactor (29). The ultrastructure of granular sludge was initially characterized by using light microscopy and scanning and transmission electron microscopy (10, 15, 23, 29, 37). Guiot and coworkers (16, 29) presented a layered conceptual model for glucose-fed granules in which Methanosaeta formed the internal core, which was surrounded by a second layer of acetogenic and hydrogenotrophic bacteria, with a peripheral layer composed predominantly of acidogenic, sulfate-reducing, and hydrogenotrophic bacteria. The layered structure of UASB granules was also observed with other substrates, such as sucrose, brewery and potato wastes, wheat- starch, and papermill wastewaters (10, 20, 37). On the other hand, no layered granular structure was observed in UASB reactors treating propionate, ethanol, glutamate, sugar refinery wastewaters, and methanol waste (7, 10). Granules at 15 and 25°C exhibited a uniform structure and were colonized predominantly by Methanosaeta-like organisms, while granules at 5°C showed a layered structure (5). These studies indicate that the bacterial composition and ultrastructure of granular sludge seem to be dependent on growth substrate and temperature (5, 40).
Oligonucleotide probes designed from 16S ribosomal DNAs (rDNAs) have been developed to identify microorganisms present in anaerobic bioreactors (15, 18, 22, 28, 42, 44, 46). Antibody probes have also been developed to characterize the topography of anaerobic granular consortia (15, 18, 22, 28, 44, 46). Recently, the fluorescent in situ hybridization (FISH) technique combined with confocal scanning laser microscopy (CSLM) has been used to analyze the spatial distribution of specifically labeled target cells present in different types of wastewater processes (2, 3, 31), but few studies have examined the relative distributions and quantifications of methanogenic populations.
The objectives of the present study were (i) to develop and validate 16S rDNA oligonucleotide probes to differentiate M. concilii and Methanosarcina barkeri at the species level and mesophilic methanogens at the group level and (ii) to use FISH coupled with CSLM to determine quantitative profiles of both bacteria in two types of anaerobic granular consortia.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. concilii GP6, Methanosarcina barkeri Fusaro, and Methanosarcina mazei were supplied by Gerish Patel (National Research Council Canada, Ottawa, Ontario, Canada). Methanospirillum hungatei and Syntrophomonas wolfei strains were supplied by Michael McInerney (University of Oklahoma, Norman). Pure bacterial strains were grown anaerobically at 35°C in serum bottles. The sources (33, 35), culture media (4, 33, 35), and trophic levels (13, 14, 41, 43) of bacteria used in this study are listed in Table 1.
TABLE 1.
Sources, cultivation conditions, and trophic levels in anaerobic mesophilic fermentors of pure bacterial strains
| Strain no. | Strain | Source (reference) | Culture medium (reference)a or strain | Trophic nature(s) and/or location |
|---|---|---|---|---|
| 1 | Methanosarcina barkeri Fusaro | G. B. Patel (35) | Medium 3 (4) | Acetoclast, hydrogenotroph, methanotroph |
| 2 | Methanosarcina acetovirans C2A | OCM 95 | Medium 3 (4) | Acetoclast, methanotroph, found in sediments |
| 3 | Methanosarcina mazei | G. B. Patel (35) | Medium 3 (4) | Acetoclast, hydrogenotroph, methanotroph |
| 4 | Methanosarcina vacuolata | ATCC 35090 | ATCC 1043 | Acetoclast, hydrogenotroph, methanotroph |
| 5 | Methanosarcina sp. strain WH1 | DSM 4659 | Medium 3 (4) | Found in anaerobic sands |
| 6 | Methanosaeta concilii GP6 | G. B. Patel (35) | Aa medium (35) | Acetoclast |
| 7 | Methanospirillum hungatei | M. J. McInerney (33) | Basal medium (33) | Hydrogenotroph |
| 8 | Methanobrevibacter arboriphilus | ATCC 33747 | ATCC 1045 | Hydrogenotroph |
| 9 | Methanogenium bourgense | ATCC 43281 | ATCC 1340 | Hydrogenotroph, formate degrader |
| 10 | Methanocorpusculum aggregans MSt | OCM 21 | MG medium from OCM | Hydrogenotroph, formate degrader |
| 11 | Methanobacterium formicicum MF | OCM 55 | MS medium from OCM | Hydrogenotroph, formate degrader |
| 12 | Methanobacterium wolfei | ATCC 43096 | ATCC 1045 | Hydrogenotroph |
| 13 | Methanobacterium espanolae | OCM 178 | MG medium from OCM | Hydrogenotroph |
| 14 | Methanobacterium bryantii M.o.H. | OCM 110 | MS medium from OCM | Hydrogenotroph |
| 15 | Syntrophomonas wolfei | M. J. McInerney (33) | Basal medium (33) | Hydrogen-producing acetogen |
| 16 | Clostridium aceticum | ATCC 35044 | ATCC 1612 | Acidogen |
| 17 | Clostridium populeti | ATCC 35295 | ATCC 1345 | Acidogen |
Aa, acetic acid; MG, Methanogenium; MS, :Methanosarcina; OCM, Oregon Collection of Methanogens, Portland, Oreg.
Anaerobic granule enrichment conditions.
The anaerobic granules used in this study were obtained from an industrial UASB bioreactor (Champlain Industries, Cornwall, Ontario, Canada) that treats food production wastewater rich in proteins and from lab-scale UASB enrichment processes (9). Three types of enriched anaerobic granular consortia were examined in the slot blot analysis: (i) Methanosaeta-enriched consortia, (ii) Methanosarcina-enriched consortia, and (iii) syntroph-enriched consortia. Details of the enrichment procedure were described previously (9).
DNA extraction from pure bacterial cultures and from anaerobic granules.
Several DNA extraction methods were examined (6, 32, 34, 36, 39). A modified version of the Johnson technique (21) resulted in the highest quantity and quality of DNA (data not shown). An overnight incubation at 37°C after addition of 10 to 30 mg of dry lysozyme (Sigma, St. Louis, Mo.) was required to initiate lysis of pure cultures. Anaerobic granular consortia were homogenized aseptically with a cell homogenizer (Kinematica PT10-35; Brinkmann Instruments, Westbury, N.Y.). Lysis of more resilient anaerobic granular consortia was initiated by freezing cells in a dry ice-ethanol bath for 10 min and thawing in a water bath at 70°C for 15 min. A five-cycle freeze-thaw procedure of anaerobic granules was performed prior to lysozyme addition. Lysis was completed as described by Johnson (21), proteins were precipitated (39), and the DNA was dissolved in RNase-containing Tris-EDTA (TE) buffer and stored at −20°C until slot blots were performed.
Design and synthesis of the oligonucleotide probes.
The nucleotide sequences of 16S rDNAs from methanogenic bacteria were retrieved from GenBank (National Center for Biotechnology Information, Bethesda, Md.). Oligonucleotide probes of 15 to 22 bases in length (Table 2) were designed with the Gene Works program (IntelliGenetics Inc., Mountain View, Calif.). All oligonucleotides were synthesized on a Biosearch 8750 DNA synthesizer (Biosystems, Cambridge, Mass.) (8). The oligonucleotides were purified with SepPak cartridges (Waters, Milford, Mass.), as described by the manufacturer.
TABLE 2.
Oligonucleotide sequences and specificities
| Target bacterium or bacteria | Oligonucleotide probe (reference) | Oligonucleotide probe sequence (5′–3′) | Length (bases) | Gene Works specificities | Specificity or specificities tested here with pure bacterial strains |
|---|---|---|---|---|---|
| Methanosaeta concilii | MS1 | CCGGATAAGTCTCTTGA | 17 | Methanothrix soehngenii, Methanosaeta concilii | Methanosaeta concilii |
| MS2 | CTGAATGAGAGCGCTTTCTTT | 21 | Methanothrix soehngenii, Methanosaeta concilii | Methanosaeta concilii | |
| MS5 | GGCCACGGTGCGACCGTTGTCG | 22 | Methanothrix soehngenii, Methanosaeta concilii, Methanothrix sp. strain CALS-1, Methanothrix sp. strain PTT, Methanosaeta thermoacetophila | Methanosaeta concilii | |
| MX825 (38) | TCGCACCGTGGCCGACACCTAGC | 23 | Methanosaeta thermoacetophila, Methanothrix soehngenii, Methanosaeta concilii | Methanosaeta concilii | |
| Methanosarcina barkeri | MB1 | TTTGGTCAGTCCTCCGG | 17 | Methanosarcina frisius, Methanosarcina sp., Methanosarcina acidovorans, Methanosarcina thermophila, Methanosarcina barkeri | Methanosarcina barkeri, Methanosarcina mazei, Methanosarcina acetivorans, Methanosarcina sp. strain WH1 |
| MB3 | CCAGACTTGGAACCG | 15 | Methanosarcina frisius, Methanosarcina acidovorans, Methanosarcina thermophila, Methanosarcina barkeri | Methanosarcina barkeri, Methanosarcina mazei, Methanosarcina acetivorans | |
| MB4 | TTTATGCGTAAAATGGATT | 19 | Methanosarcina thermophila, Methanosarcina barkeri | Methanosarcina barkeri | |
| MS821 (38) | CGCCATGCCTGACACCTAGCCAGC | 24 | Methanosarcina frisius, Methanosarcina sp., Methanosarcina acidovorans, Methanosarcina thermophila, Methanosarcina barkeri, Methanoholophilus mahii, Methanophilus sp. | Methanosarcina barkeri Methanosarcina sp. strain WH1, Methanosarcina mazei, Methanosarcina acetivorans | |
| Mesophilic methanogens | MG3 | CTCCTTGCACACACCGCCC | 19 | All methanogens + Halobacterium spp., Pyrococcus spp., Sulfolobus spp., Homo sapiens, Thermofilum spp., Pyrobaculum spp. | All mesophilic methanogens tested |
Slot blot analysis.
The specificities of the oligonucleotide probes were examined by slot blot analysis by a modified method of Raskin et al. (38). Extracted DNA was denatured by boiling it in TE buffer (pH 8.0) for 8 min, instead of by glutaraldehyde denaturation as previously suggested (38). Boiling DNA in TE buffer gave the best 32P-signals and was the most effective DNA denaturation method, compared to incubation in 2% glutaraldehyde at room temperature for 10 minutes (38) and to boiling in a 1.6 M NaOH–40 mM EDTA solution for 10 min (data not shown). Extracted DNA was filtered on Zeta Probe nylon membranes (Bio-Rad Laboratories, Hercules, Calif.) with a Minifold II Slot Blot system (Schleicher & Schuell, Keene, N.H.). Slot blots were cross-linked twice with a UV Stratalinker 1800 (Stratagene, La Jolla, Calif.). Membranes were air dried and stored at −20°C. Purified oligonucleotide probes (8 pmol) were 5′ end labeled with a solution containing 5 μl of [γ-32P]ATP (4,500 Ci/mmol; NEN Research Products, Dupont Inc., Markham, Ontario, Canada), 6.67 U of T4 DNA kinase (New England Biolabs, Beverly, Mass.), and 2 μl of 10× kinase buffer (New England Biolabs) and brought to a volume of 20 μl with sterile deionized H2O. The labeling reaction was performed at 37°C for 2 h. Slot blots were prehybridized at 40°C for 2 h in 40 ml of Zeta Probe hybridization solution to which 5% dextran sulfate had been added and hybridized overnight at 40°C with the 32P-labeled oligonucleotide. Hybridized membranes were rinsed twice in Zeta Probe wash 1 at 40°C for 60 min, and final wash conditions in Zeta Probe wash 2 were optimized for each oligonucleotide (Table 3). Probed membranes were exposed to X-ray films (Kodak X-Omat AR and BioMax MR; Eastman Kodak Co., Rochester, N.Y.) at −80°C for 7 to 14 days.
TABLE 3.
Hybridization final-wash conditions
| Oligonucleotide probe | Final-wash temp (°C) | No. of final washes (30 min each) |
|---|---|---|
| MX825 | 59 | 4 |
| MS1 | 40 | 3 |
| MS2 | 42 | 4 |
| MS5 | 55 | 3 |
| MS821 | 60 | 2 |
| MB1 | 45 | 3 |
| MB3 | 45 | 2 |
| MB4 | 42 | 3 |
| MG3 | 40 | 3 |
In situ hybridization of anaerobic granular consortia with fluorescence-labeled probes.
Fresh and frozen granules (2 ml) from the inoculum and from Methanosarcina-enriched consortia were gently washed twice with 5 ml of phosphate-buffered saline (PBS). Granules were fixed overnight at 4°C in fresh 4% paraformaldehyde–PBS (5 ml) and washed twice with PBS (17). Fixed granules were dehydrated at room temperature in increasing concentrations of ethanol (50, 70, 95, and 100%), in pure xylenes, and then in an equal amount of xylenes in Paraplast wax overnight (19). Granules were embedded in Paraplast wax at 60°C for 3 h (19). Paraplast blocks were cut at room temperature with a Histostat rotary microtome (model 820; Reichert/Jung, Buffalo, N.Y.) into 8-μm-thick sections. The microtome sections were transferred onto poly-l-lysine-coated microscope slides (Polysciences, Warrington, Pa.), subbed with 1% gelatin, dried overnight at 42°C, deparaffinated, and air dried (19).
In situ hybridization was performed with (i) the complementary sequences of MB4 that had been 5′ end labeled with FAM–N-hydroxysuccinimide (NHS; a succinimidyl ester of carboxyfluorescein) or with Cy5 (indodicarbocyanine), synthesized by Medicorps (Montreal, Quebec, Canada), and with (ii) MS5 that had been 5′ end labeled with TAMRA NHS (tetramethyl rhodamine carboxylic acid), synthesized by University Core DNA Services (Calgary, Alberta, Canada). Purification of the probes was performed by high-performance liquid chromatography. A volume of 10 μl of hybridization solution (35% deionized formamide, 0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate, 25 ng of each fluorescence-labeled probe) was added to each granule section, and the mixture was incubated at 46°C for 2 h (30). Sections were rinsed with washing buffer (20 mM Tris, 0.01% sodium dodecyl sulfate, 40 mM NaCl, 5 mM EDTA) and incubated in washing buffer at 48°C for 20 min. Sections were carefully rinsed with ultrapure H2O, air dried, and covered with fade retardant in PBS (Molecular Probes Inc., Eugene, Oreg.) prior to application of a glass coverslip.
Confocal laser microscopy and staining.
A Bio-Rad MRC 1000 CSLM with a krypton-argon laser system mounted on a Nikon Microphot-SA microscope was used to image thin sections of the anaerobic inoculum (for treating protein-rich wastewater) and Methanosarcina-enriched granular consortia. The microscope was equipped with a variety of objective lenses. The CSLM was operated as described previously (25, 27) to collect single and serial thin sections in the xy plane.
Sections were imaged at two scales of observation: (i) at low resolution with a 10× 0.18-numerical-aperture lens objective in combination with a laser intensity of 100%, pinhole setting of 7.0, and gain of 1,500, with sections being collected through Kalman filtration, and (ii) at higher resolution with a 60× 1.4-numerical-aperture oil immersion lens objective (Nikon Corporation, Chiyoda-ku, Tokyo, Japan), laser intensity of 10 to 30%, pinhole of 1.2, and gain of 1,100, with Kalman filtration. The 10× lens provided a full scan of the entire hybridized granule section, whereas the 60× lens allowed detailed examination of granules along transects across the sectioned granule material. Images were collected by two- and three-channel imaging procedures, where carboxyfluorescein (FAM NHS [green])-labeled probes were visualized by excitation at 488 nm and emission at 522/32, tetramethyl rhodamine (TAMRA NHS [red])-labeled probes were visualized by excitation at 568 nm and emission at 605/32, and Cy5 (far red)-labeled probes were visualized by excitation at 647 nm and emission at 680/32 (26). Sections were also scanned for autofluorescence signals at the same wavelengths. After completion of observations, hybridized sections were perfused with the general nucleic acid stain Syto 9 (Molecular Probes) and the same microscope locations were reexamined at 10× and 60× lens objectives to obtain information on the total numbers of bacterial cells.
Image analysis and processing.
Digital image analysis of the CSLM thin sections in each of the channels was used to determine the bacterial cell area (biomass) binding each of the rRNA probes and Syto 9. The software package NIH Image, version 1.61, was used for all analyses. CSLM images were thresholded to form a binary (black-and-white) image, with the object, i.e., the bacterial cell, being defined prior to measurement of the object area. Details of discrimination, object recognition, and measurement may be found in the work of Lawrence et al. (25, 27). Results of object area analyses for an image series were then plotted versus granule location, and ratios of cell area bound by each probe were calculated to assess treatment effects. Digital images obtained by CSLM were also used to construct two- and three-color digital images of the stained granule sections with the software NIH Image and Adobe Photoshop. The colors green, red, and blue were assigned to the green-, red-, and far-red-wavelength images, respectively.
RESULTS AND DISCUSSION
Optimization of slot blotting conditions.
The slot blotting conditions published by Raskin et al. (38) were modified by using Zeta Probe hybridization solutions and by optimizing the final wash conditions. Efficiencies of the hybridization solutions with and without poly(A) from the Raskin et al. protocol (38) were compared with those of the Zeta Probe hybridization solutions. The Zeta Probe solutions gave stronger hybridization signals (results not shown) and were therefore used for all subsequent hybridizations. Final wash conditions such as temperature, duration, and frequency of washes were optimized for each oligonucleotide probe (Table 3). Additional washes, compared to the number recommended by Raskin et al. (38), were required for MX825 and MS821 to increase specificity, probably because different hybridization conditions were used.
Specificities of the designed oligonucleotide probes.
The designed oligonucleotide probes were aligned with DNA sequences available in GenBank with the BLAST program (1) to assess their theoretical specificities and to compare their specificities to those of the MX825 and MS821 probes designed by Raskin et al. (38). All Methanosaeta-specific oligonucleotide probes (MS1, MS2, MS5, and MX825) had 100% homology to M. concilii and Methanothrix soehngenii 16S ribosomal gene sequences, which indicates that these probes may show equivalent specificities (Table 2). The Methanosarcina-specific oligonucleotide probe MB4 was 100% homologous to Methanosarcina barkeri and Methanosarcina thermophila only, which indicates that this probe may be specific to Methanosarcina barkeri found in mesophilic granules. The other Methanosarcina-specific oligonucleotide probes (MB1, MB3, and MS821) had 100% homology to other Methanosarcina spp., and MS821 had 100% homology to two non-Methanosarcina species. The mesophilic methanogen oligonucleotide probe (MG3) had 100% homology to all methanogen 16S rDNA sequences but was also homologous to other species that are not normally found in anaerobic digesters (Table 2).
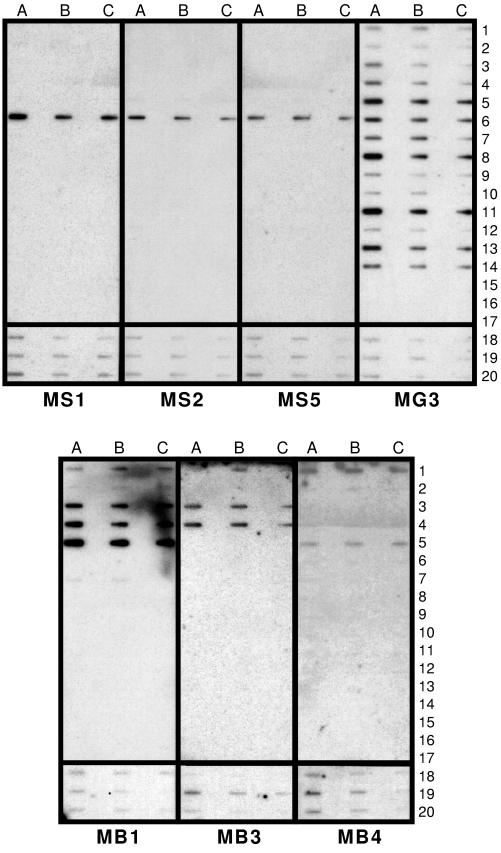
The specificities of the oligonucleotide probes were assessed by slot blot analysis, with pure bacterial strains being used as positive controls (Table 2). The chosen bacteria are normally found in anaerobic mesophilic fermentors that treat industrial and municipal sludge (Table 1). The three M. concilii-specific oligonucleotide probes (MS1, MS2, and MS5), designed in this study, hybridized only with their corresponding positive control (Fig. 1). MS5 was chosen as the most specific M. concilii oligonucleotide probe because it had the most stringent final wash temperature. The MX825 probe hybridized with Methanobacterium formicicum, a non-Methanosaeta species (data not shown). The Methanosarcina barkeri-specific probe MB4 hybridized only with M. barkeri and Methanosarcina sp. strain WH1. The MB1 probe hybridized with Methanosarcina barkeri, Methanosarcina mazei, Methanosarcina vacuolata, Methanosarcina sp. strain WH1, Methanosarcina acetovirans, and Methanospirillum hungatei. The MB3 probe hybridized with Methanosarcina barkeri, Methanosarcina mazei, and Methanosarcina vacuolata. The MS821 probe hybridized with Methanosarcina mazei, Methanosarcina vacuolata, and Methanosarcina sp. and poorly with the target species, Methanosarcina barkeri (data not shown). Based on these results, the MB4 oligonucleotide probe can be used to specifically identify Methanosarcina barkeri, since Methanosarcina sp. strain WH1 is unlikely to be found in anaerobic digesters that treat industrial and municipal sludge (43). The MB3 probe can be used to detect the presence of all Methanosarcina species in anaerobic mesophilic granular consortia. The MG3 mesophilic methanogen-specific oligonucleotide probe hybridized with all positive controls but not with the negative controls Syntrophomonas wolfei, Clostridium aceticum, and Clostridium populeti (Fig. 1). Therefore, MG3 can be used to detect mesophilic methanogens normally found in anaerobic digesters that treat industrial and municipal sludge.
FIG. 1.
Hybridization of 32P-labeled Methanosaeta spp. oligonucleotide probes (MS1, MS2, and MS5), Methanosarcina spp. oligonucleotide probes (MB1, MB3, and MB4), and a mesophilic-methanogen probe (MG3) with slot blots with 17 pure bacterial strains and three anaerobically enriched consortia. Rows 1 to 17, bacterial strains 1 to 17 presented in Table 1; row 18, Methanosaeta-enriched consortium; row 19, Methanosarcina-enriched consortium; row 20, syntrophic-methanation consortium. For pure bacterial cultures, the DNA concentrations were 1,000 ng in column A, 250 ng in column B, and 50 ng in column C. For anaerobic enriched consortia hybridized with Methanosaeta oligonucleotide probes, the DNA concentrations were 2,500 ng in column A, 500 ng in column B, and 100 ng in column C. For anaerobic enriched consortia hybridized with Methanosarcina oligonucleotide probes, the DNA concentrations were 25,000 ng in column A, 5,000 ng in column B, and 1,000 ng in column C.
Slot blotting of DNAs extracted from anaerobic granules.
Slot blots with the oligonucleotide probes were performed with total-community DNAs extracted from three types of anaerobic granular consortia: (i) Methanosaeta-enriched consortia, (ii) Methanosarcina-enriched consortia, and (iii) syntroph-enriched consortia (Fig. 1). M. concilii and Methanosarcina barkeri were present in all three types of granules, as detected by hybridization with all the designed oligonucleotide probes. The presence of bacteria of Methanosaeta spp. was expected in the Methanosaeta- and syntroph-enriched granules but unanticipated in the Methanosarcina-enriched granules. This result may suggest that although enrichment of Methanosarcina took place in the Methanosarcina-enriched granules, M. concilii continued to be present in the consortium. Similarly, the detection of Methanosarcina barkeri in Methanosaeta-enriched granules was not expected, although Methanosarcina barkeri was relatively less abundant in these granules.
FISH and CSLM.
In order to avoid autofluorescence from Methanosarcina barkeri in the green wavelength (17, 42), the MB4 probe was labeled with the Cy5 (far red) fluorochrome. The MS5 probe, specific to M. concilii, was labeled with the TAMRA NHS (red) fluorochrome. Syto 9 (green) staining was used to obtain information on the total number of bacterial cells. The colors of the digital images obtained by CSLM were blue for MB4, red for MS5, and green for Syto 9, colors assigned to differentiate between Methanosarcina barkeri, M. concilii, and the rest of the bacterial population, respectively.
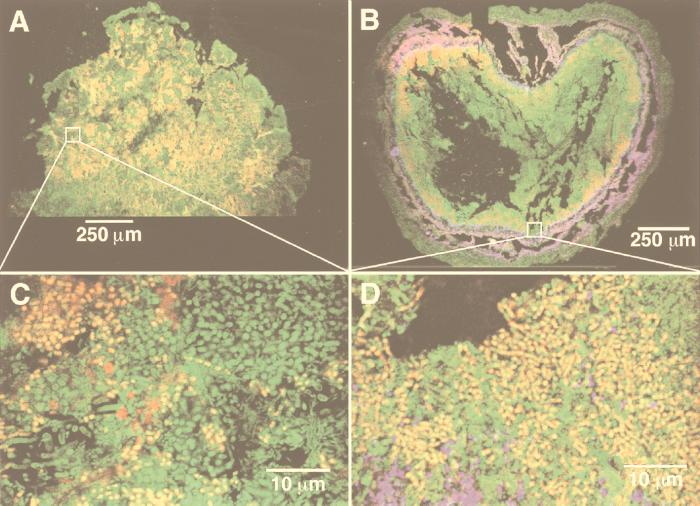
The MB4 and MS5 fluorescent probes were evaluated with two types of granules: one obtained from a protein-rich wastewater treatment plant and the other obtained from a methanol-enriched consortium. CSLM observations demonstrated how the granule structures may differ, depending on origin and feeding. In the protein-fed granule, no layered structure was observed, the granules were composed mainly of M. concilii distributed randomly throughout the granule (Fig. 2A), and Methanosarcina barkeri was essentially absent (Fig. 2C). The methanol enrichment induced the growth of Methanosarcina barkeri (Fig. 2B), and a layered structure developed during the enrichment process, where Methanosarcina barkeri was present in a second layer, just below the granule surface. The second layer was followed by an inner layer of M. concilii closely associated with Methanosarcina barkeri (Fig. 2B and D), and the centers of the enriched granules were sparsely colonized by the rest of the bacterial population, as demonstrated by Syto 9 staining.
FIG. 2.
Simultaneous in situ hybridization of UASB granules with the MS5 M. concilii-specific probe (labeled with TAMRA NHS [red]) and the MB4 Methanosarcina barkeri-specific probe (labeled with Cy5 [blue]). Low-resolution (10× lens objective) CSLM optically thin sections (xy plane) show the random distribution of M. concilii within the rest of the bacterial population (green) in the inoculum granules (A) and the layered structure of the Methanosarcina-enriched granules (B), in which Methanosarcina barkeri and M. concilii are present within the layers. Higher-resolution (60× lens objective) CSLM sections show the absence of Methanosarcina barkeri from the inoculum granules (C) and the colocalization of Methanosarcina barkeri and M. concilii within the Methanosarcina-enriched granules (D).
Relative quantification and localization of the two targeted bacteria.
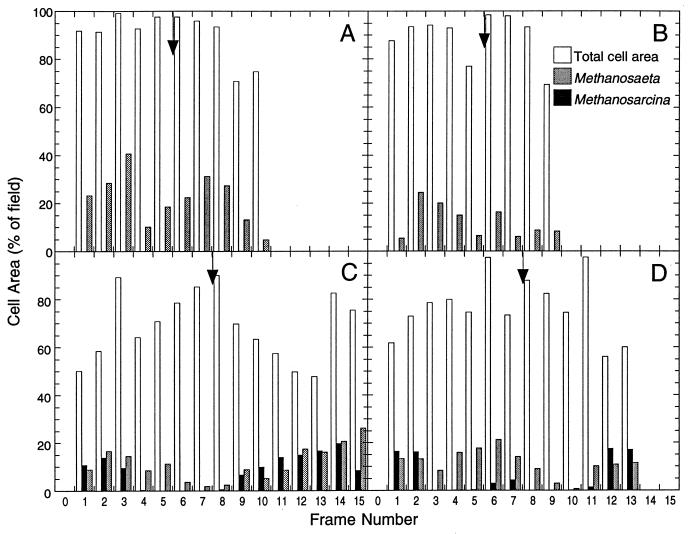
Transects across the granule centers showed that Methanosarcina barkeri was not detected in the protein-rich wastewater-fed granules and that M. concilii was randomly distributed throughout the granules (Fig. 3A and B). In the methanol-enriched granules, both Methanosarcina barkeri and M. concilii were detected in the outer layers (Fig. 3C and D). Moreover, the surface area occupied by both of these targeted bacteria was shown to have increased, in comparison to the total cell area, a result of the enrichment of the granular sludge with methanol. By combining transects across the granule centers with the cell area graphs, it was possible to observe and quantify the relative levels of abundance of both targeted bacteria.
FIG. 3.
Relative quantifications of Methanosarcina barkeri and M. concilii cell areas by CSLM digital image analysis. Simultaneous in situ hybridization was performed on 8-μm-thick sections of UASB protein-rich wastewater-fed granules (A and B) and of methanol-enriched granules (C and D) with the TAMRA (red)-labeled MS5 M. concilii-specific probe and the Cy5 (blue)-labeled MB4 Methanosarcina barkeri-specific probe. Transects across the centers of the granules were analyzed, and the cell area containing both M. concilii and Methanosarcina barkeri and the total cell area (detected with Syto 9) were determined. Data are expressed as relative cell area (percentages of the field) versus location within the granule, and the arrows indicate the center of each thin section. Frame numbers represent transects of these thin sections through the center of the granule.
Comprehensive comparison of the observed granules topography.
Results of the present study show that the bacterial organization of UASB granular consortia was dependent on the growth substrate. Anaerobic degradation is a multistep process involving hydrolysis, acidogenic fermentation, acetogenesis, and methanogenesis. Carbohydrates, for which the initial step of degradation is notably faster than subsequent steps, produce granules with a layered structure, as has been observed in UASB granules fed with a variety of carbohydrate wastes (10, 20, 29, 37). In protein-rich effluents, proteolysis and acetogenesis from amino acids are the limiting steps in the overall degradation process. When acid formation is the rate-controlling step, low and homogeneous concentrations of acetate throughout the granule are the result. Slow-growing Methanosaeta usually has a competitive advantage over Methanosarcina at low concentrations of acetate (45, 47). These conditions explain why, for granules grown on protein-rich wastewater, no layered structure was demonstrated and M. concilii was distributed randomly throughout the granule while Methanosarcina populations were practically absent. This explanation is in agreement with previous scanning electron microscopic observations where a uniform microstructure developed in glutamate-degrading granular consortia (10), in protein- and peptone-degrading granules (11), and in propionate-degrading granules (11, 15).
Granules that convert single substrates, such as formate and acetate, directly into methane appear to produce uniform microstructures, with one methanogen species predominating (11). However, in this study, a layered structure was formed in the anaerobic granules enriched with methanol. Bacteria other than Methanosarcina and Methanosaeta occupied the outermost layer, Methanosarcina barkeri was present in the underlying layer, followed by a layer of M. concilii and an inner core composed of M. concilii and other bacteria. Since methanol is both a methanogenic and an acetogenic substrate, bacteria found in the outermost layer were probably methanol consumers other than Methanosarcina, such as homoacetogens. Homoacetogens are known to out-compete methanogens when methanol and cobalt concentrations are relatively high (over 2 g/liter and 0.05 mg/liter, respectively) and inorganic carbon is readily available (9, 12), conditions that were used in this study to produce the methanol-enriched granules. Since methanol is usually degraded rapidly, a decreasing methanol concentration gradient is likely to occur due to diffusional limitations. This methanol gradient probably created a niche more favorable to Methanosarcina than to the homoacetogens in the underlying layer. Finally, the acetate produced from the conversion of methanol by the homoacetogens diffused along a decreasing gradient, explaining why the obligate acetoclastic M. concilii was also present in significant numbers on the inside of the Methanosarcina barkeri layer and randomly distributed in the core of the granule.
In conclusion, 16S rDNA oligonucleotide probes were developed to differentiate M. concilii, Methanosarcina barkeri, and mesophilic methanogens present in anaerobic mesophilic fermentors. The specificities of the MS5 and MB4 oligonucleotide probes, targeting the 16S rRNA sequences of M. concilii and Methanosarcina barkeri, respectively, were demonstrated by slot blot analysis with DNAs extracted from pure bacterial cultures and from anaerobic granular consortia. The MB3 Methanosarcina-specific oligonucleotide probe detected all tested mesophilic Methanosarcina species. The MG3 oligonucleotide probe detected all tested mesophilic methanogens normally found in anaerobic digesters. FISH with the species-specific probes MS5 and MB4 combined with CSLM was used to identify and quantify the specific relative abundance of M. concilii and Methanosarcina barkeri in two different types of granules. The present study is the first attempt to simultaneously visualize the spatial distribution and quantify the relative abundance of these two important methanogenic bacteria, at the species level, in anaerobic granules. This novel approach may be an extremely versatile method to monitor population dynamics in UASB processes.
ACKNOWLEDGMENTS
We acknowledge David Juck, Michelle Manuel, Tara Norcott, Alberto Mazza, Lyle Whyte, Gabrielle Préfontaine, Daniel Dignard, Josée Ash, and Joanne Magoon from the Biotechnology Research Institute. We thank George D. W. Swerhone from the National Water Research Institute for his confocal scanning laser microscopy technical expertise.
Footnotes
Publication 41841 of the National Research Council Canada.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R, Lemmer H, Wagner M. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol Ecol. 1998;25:205–215. [Google Scholar]
- 3.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K-H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch W E, Fox G E, Magrum C J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banik G C, Ellis T G, Dague R R. Structure and methanogenic activity of granules from an ASBR treating dilute wastewater at low temperatures. Water Sci Technol. 1997;36:149–156. [Google Scholar]
- 6.Beji A, Izard D, Gavini F, Leclerc H, Leseine-Delstanche M, Krembel J. A rapid chemical procedure for isolation and purification of chromosomal DNA from Gram-negative bacilli. Anal Biochem. 1987;162:18–23. doi: 10.1016/0003-2697(87)90005-4. [DOI] [PubMed] [Google Scholar]
- 7.Bhatti Z I, Furukawa K, Fujita M. Treatment performance and microbial structure of a granular consortium handling methanolic waste. J Ferment Bioeng. 1993;76:218–223. [Google Scholar]
- 8.Chatellier J, Mazza A, Brousseau R, Vernet T. Codon-based combinatorial alanine scanning site-directed mutagenesis: design, implementation, and polymerase chain reaction screening. Anal Biochem. 1995;229:282–290. doi: 10.1006/abio.1995.1414. [DOI] [PubMed] [Google Scholar]
- 9.El-Mamouni R, Guiot S R, Leduc R, Costerton J W. Characterization of different microbial nuclei as potential precursors of anaerobic granulation. J Biotechnol. 1995;39:239–249. [Google Scholar]
- 10.Fang H H P, Chui H K, Li Y Y. Microbial structure and activity of UASB granules treating different wastewaters. Water Sci Technol. 1994;30:87–96. [Google Scholar]
- 11.Fang H H P, Chui H K, Li Y Y. Effect of degradation kinetics on the microstructure of anaerobic biogranules. Water Sci Technol. 1995;32:165–172. [Google Scholar]
- 12.Florencio L, Field J A, Lettinga G. Importance of cobalt for individual trophic groups in an anaerobic methanol-degrading consortium. Appl Environ Microbiol. 1994;60:227–234. doi: 10.1128/aem.60.1.227-234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia J L. Taxonomy and ecology of methanogens. FEMS Microbiol Rev. 1990;87:297–308. [Google Scholar]
- 14.Gottschalk G, Braun M. Revival of the name Clostridium aceticum. Int J Syst Bacteriol. 1981;31:476. [Google Scholar]
- 15.Grotenhuis J T C, Smit M, Plugge C M, Xu Y, van Lammeren A A M, Stams A J M, Zehnder A J B. Bacteriological composition and structure of granular sludge adapted to different substrates. Appl Environ Microbiol. 1991;57:1942–1949. doi: 10.1128/aem.57.7.1942-1949.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guiot S R, Pauss A, Costerton J W. A structured model of the anaerobic granule consortium. Water Sci Technol. 1992;25:1–10. [Google Scholar]
- 17.Harmsen H J M, Kengen H M P, Akkermans A D L, Stams A J M, de Vos W M. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl Environ Microbiol. 1996;62:1656–1663. doi: 10.1128/aem.62.5.1656-1663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howgrave-Graham A R, Macario A J L, Wallis F M. Quantitative analysis and mapping of micro-organisms in anaerobic digester granules using a combination of transmission electron microscopy with immunotechnology. J Appl Microbiol. 1997;83:587–595. [Google Scholar]
- 19.Janssen K. In situ hybridization and immunohistochemistry. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1996. pp. 14.0.1–14.2.8. [Google Scholar]
- 20.Jianrong Z, Jicui H, Xiasheng G. The bacterial numeration and an observation of a new syntrophic association for granular sludge. Water Sci Technol. 1997;36:133–140. [Google Scholar]
- 21.Johnson J L. Isolation and purification of nucleic acids. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. West Sussex, England: John Wiley & Sons; 1991. pp. 1–20. [Google Scholar]
- 22.Koornneef E, Macario A J L, Grotenhuis J T C, de Macario E C. Methanogens revealed immunologically in granules from five upflow anaerobic sludge blanket (UASB) bioreactors grown on different substrates. FEMS Microbiol Ecol. 1990;73:225–230. [Google Scholar]
- 23.Kosaric N, Blaszczyk R, Orphan L, Valladares J. The characteristics of granules from upflow anaerobic sludge blanket reactors. Water Res. 1990;24:1473–1477. [Google Scholar]
- 24.Koster I W. Microbial, chemical, and technological aspects of the anaerobic degradation of organic pollutants. In: Wise D L, editor. CRC biotreatment systems. Boca Raton, Fla: CRC Press; 1988. pp. 285–316. [Google Scholar]
- 25.Lawrence J R, Korber D R, Wolfaardt G M, Caldwell D E. Analytical imaging and microscopy techniques. In: Hurst C J, Knudsen G R, McInerney M, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C: ASM Press; 1996. pp. 29–51. [Google Scholar]
- 26.Lawrence J R, Neu T R, Swerhone G D W. Application of multiple parameter imaging for the quantification of algal, bacterial and exopolymer components of microbial biofilms. J Microbiol Methods. 1998;32:253–261. [Google Scholar]
- 27.Lawrence J R, Wolfaardt G M, Neu T R. The study of biofilms using confocal laser scanning microscopy. In: Wilkinson M H F, Schut F, editors. Digital image analysis of microbes: imaging, morphometry, fluorometry and motility techniques and applications. Sussex, United Kingdom: John Wiley and Sons Ltd.; 1997. pp. 431–465. [Google Scholar]
- 28.Macario A J L, Visser F A, van Lier J B, de Macario E C. Topography of methanogenic subpopulations in a microbial consortium adapting to thermophilic conditions. J Gen Microbiol. 1991;137:2179–2189. [Google Scholar]
- 29.MacLeod F A, Guiot S R, Costerton J W. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl Environ Microbiol. 1990;56:1598–1607. doi: 10.1128/aem.56.6.1598-1607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 31.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 32.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 33.McInerney M J, Bryant M P, Pfennig N. Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens. Arch Microbiol. 1979;122:129–135. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- 34.Meakin S A, Nash J H E, Murray W D, Kennedy K F J, Sprott G D. A generally applicable technique for the extraction of restrictable DNA from methanogenic bacteria. J Microbiol Methods. 1991;14:119–126. [Google Scholar]
- 35.Patel G B. Characterization and nutritional properties of Methanothrix concilii sp. nov., a mesophilic acetoclastic methanogen. Can J Microbiol. 1984;30:1383–1396. [Google Scholar]
- 36.Pitchner D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 37.Quarmby J, Forster C F. A comparative study of the internal architecture of anaerobic granular sludges. J Chem Technol Biotechnol. 1995;63:60–68. [Google Scholar]
- 38.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schmidt J E, Ahring B K. Granulation in thermophilic upflow anaerobic sludge blanket (UASB) reactors. Antonie Leeuwenhoek. 1995;68:339–344. doi: 10.1007/BF00874144. [DOI] [PubMed] [Google Scholar]
- 41.Sleat R, Mah R A. Clostridium populeti sp. nov., a cellulolytic species from a woody-biomass digester. Int J Syst Bacteriol. 1985;35:160–163. [Google Scholar]
- 42.Sorensen A H, Torsvik V L, Torsvik T, Poulsen L K, Ahring B K. Whole-cell hybridization of Methanosarcina cells with two new oligonucleotide probes. Appl Environ Microbiol. 1997;63:3043–3050. doi: 10.1128/aem.63.8.3043-3050.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staley J T, Bryant M P, Pfennig N, Holt J G. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams & Wilkins; 1989. [Google Scholar]
- 44.Visser F A, van Lier J B, Macario A J L, de Macario E C. Diversity and population dynamics of methanogenic bacteria in granular consortium. Appl Environ Microbiol. 1991;57:1728–1734. doi: 10.1128/aem.57.6.1728-1734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zehnder A J, Ingvorsen K, Marti T. Proceedings of the 2nd International Symposium on Anaerobic Digestion. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1982. Microbiology of methane bacteria; pp. 45–68. [Google Scholar]
- 46.Zellner G, Macario A J L, de Macario E C. A study of three anaerobic methanogenic bioreactors reveals that syntrophs are diverse and different from reference organisms. FEMS Microbiol Ecol. 1997;22:295–301. [Google Scholar]
- 47.Zinder S H. Physiological ecology of methanogens. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry and genetics. New York, N.Y: Chapman & Hall; 1993. pp. 128–206. [Google Scholar]