Abstract
Ritonavir (RTV), a pharmacoenhancer used in anti-HIV regimens, can induce liver damage. RTV is primarily metabolized by cytochrome P450 3A4 (CYP3A4) in the liver. HNF4A antisense RNA 1 (HNF4A-AS1) and HNF1A antisense RNA 1 (HNF1A-AS1) are long noncoding RNAs that regulate the expression of pregnane X receptor (PXR) and CYP3A4. This study investigated the role and underlying mechanisms of HNF4A-AS1 and HNF1A-AS1 in RTV-induced hepatotoxicity. HNF4A-AS1 and HNF1A-AS1 were knocked down by small hairpin RNAs in Huh7 and HepG2 cells. Lactate dehydrogenase and reactive oxygen species assays were performed to assess RTV-induced hepatotoxicity. Chromatin immunoprecipitation quantitative real-time polymerase chain reaction was used to detect PXR enrichment and histone modifications in the CYP3A4 promoter. HNF4A-AS1 knockdown increased PXR and CYP3A4 expression and exacerbated RTV-induced cytotoxicity, whereas HNF1A-AS1 knockdown generated the opposite phenotype. Mechanistically, enrichment of PXR and trimethylation of histone 3 lysine 4 (H3K4me3) in the CYP3A4 promoter was increased, and trimethylation of histone 3 lysine 27 (H3K27me3) was decreased after HNF4A-AS1 knockdown. However, PXR and H3K4me3 enrichment decreased after HNF1A-AS1 knockdown. Alterations in RTV-induced hepatotoxicity caused by decreasing HNF4A-AS1 or HNF1A-AS1 were reversed by knockdown or overexpression of PXR. Increased susceptibility to RTV-induced liver injury caused by the PXR activator rifampicin was attenuated by HNF4A-AS1 overexpression or HNF1A-AS1 knockdown. Taken together, these results revealed that HNF4A-AS1 and HNF1A-AS1 modulated RTV-induced hepatotoxicity by regulating CYP3A4 expression, primarily by affecting the binding of PXR and histone modification status in the CYP3A4 promoter.
SIGNIFICANCE STATEMENT
HNF4A-AS1 and HNF1A-AS1, transcribed separately from neighboring antisense genes of the human transcription factor genes HNF4A and HNF1A, were identified as long noncoding RNAs that can affect RTV-induced hepatotoxicity and susceptibility to RTV-induced hepatotoxicity caused by rifampicin exposure, mainly by affecting the expression of CY3A4 via alterations in PXR enrichment and histone modification status in the CYP3A4 promoter. This discovery provides directions for further research on the mechanisms of RTV-induced liver injury.
Introduction
Antiretroviral treatment improves the quality of life for patients living with human immunodeficiency virus (HIV)/AIDS. Nevertheless, multiple studies have shown that some antiviral drugs increase the risk of comorbidities (Mulligan, 2003; Kumar et al., 2015; Menshawy et al., 2017). Liver injury is a major cause of death among patients with HIV and is not related to AIDS (Acharya et al., 2015; Sherman et al., 2015; Maurice et al., 2017; Verna, 2017; Stanley et al., 2019). Ritonavir (RTV) is the mainstay of enhanced protease inhibitor-based regimens in antiretroviral treatment. Previous clinical studies showed that a full dose of RTV treatment may increase risk of severe hepatotoxicity (Sulkowski et al., 2000; Sulkowski, 2003).
Mounting evidence indicates the vital role of cytochrome P450 (CYP) 3A4 in the metabolism and bioactivation of RTV (Kumar et al., 1996; Denissen et al., 1997; Koudriakova et al., 1998; Gangl et al., 2002; Yao et al., 2008; Li et al., 2011). The expression of CYP3A4 can be induced or inhibited by endogenous or exogenous substances, leading to differential expression between individuals and drug interactions, thereby affecting drug effectiveness and toxicity (Hernandez et al., 2009; Tracy et al., 2016). In recent years, long noncoding RNAs (lncRNAs) have been shown to be crucial for the regulation of CYPs. Previous studies have shown that the liver enrichment transcription factor cooperatively related lncRNAs contributed to the regulation of CYP3A4. The hepatocyte nuclear factor 1α (HNF1A) antisense long noncoding RNA 1 gene (HNF1A-AS1) is located close to the HNF1A gene, and the transcribed lncRNA HNF1A-AS1 positively regulates CYP3A4 expression (Chen et al., 2018; Wang et al., 2019b). Interestingly, the lncRNA hepatocyte nuclear factor 4A antisense RNA 1 (HNF4A-AS1) negatively regulates the expression of CYP3A4 (Wang et al., 2021a). Whether HNF4A-AS1 and HNF1A-AS1 affect the hepatotoxicity of RTV by regulating CYP3A4 expression is unclear.
Histone modifications, together with DNA methylation and microRNAs, are the most extensively studied epigenetic mechanisms in mammals (Tang and Chen, 2015). In our previous studies, we found that histone modifications are involved in rifampicin (RIF) mediated induction of CYP3A4 by activating pregnane X receptor (PXR) and recruiting nuclear receptor coactivator 6 and histone acetyltransferase P300 to the CYP3A4 promoter (Yan et al., 2017). In addition, CYP3A4 is upregulated following HNF4A-AS1 knockdown due to elevated H3K4me3 enrichment in HNF4A-response elements of the CYP3A4 promoter (Wang et al., 2021a). It is unknown whether histone modification status is altered in CYP3A4-mediated RTV cytotoxicity.
Notably, clinical studies have reported hepatotoxicity in 100% of patients pretreated with RIF or efavirenz (EFV) followed by RTV-containing regimens (Nijland et al., 2008; Haas et al., 2009; Jamois et al., 2009; Schmitt et al., 2009). Both RIF and EFV are agonists of PXR, and activation of PXR upregulates the expression of CYP3A4 (Lehmann et al., 1998; Zhou, 2008; Sharma et al., 2013). Shehu et al. found that human PXR activation is essential for increasing susceptibility to RTV-induced liver injury (Shehu et al., 2019). Considering the crucial effects of HNF4A-AS1 and HNF1A-AS1 in regulating PXR expression (Chen et al., 2018; Wang et al., 2019b; Wang et al., 2021a), we hypothesized that these two lncRNAs are involved in the altered susceptibility to RTV-induced liver injury caused by RIF exposure.
In this study, we performed a series of experiments using hepatoma carcinoma cell lines to validate the role and mechanism of HNF4A-AS1 and HNF1A-AS1 in RTV-induced liver injury. HNF4A-AS1 and HNF1A-AS1 affect RTV-induced hepatotoxicity by regulating CYP3A4 expression via alteration of PXR enrichment and histone modifications status in the CYP3A4 promoter. These two lncRNAs are also involved in the altered susceptibility to RTV-induced liver injury caused by RIF exposure.
Materials and Methods
Chemicals and Reagents
Human hepatoma Huh7 and HepG2 cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences. Opti-MEM, LipofectamineTM 3,000, Pierce Lactate Dehydrogenase (LDH) Cytotoxicity Assay Kit, and TM Select Realtime PCR Master Mix (SYBR Green I) were obtained from Thermo Fisher Scientific. A reactive oxygen species (ROS) assay kit was provided by Meilun Biotechnology Co., Ltd. TRNzol reagent and PCR & DNA Cleanup Kit were provided by Tiangen Biotech Co., Ltd. RIF and RTV were purchased from Sigma-Aldrich. The antibody against CYP3A4 (ab124921) was obtained from Abcam, and that against PXR (H2019) was obtained from Santa Cruz Biotechnology. The antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (60004-1-Ig) was provided by Proteintech Group. Antibodies recognizing H3K4me3 (C42D8) and H3K27me3 (C36B11) were purchased from Cell Signaling Technology. Small hairpin RNA (shRNA) negative control (shControl) and shRNAs targeting PXR (shPXR), as well as overexpression plasmids of HNF4A-AS1 control, PXR, and pSG5, were obtained from GeneCopoeia. Oligonucleotides encoding short hairpin RNAs of HNF4A-AS1 (shHNF4A-AS1) and HNF1A-AS1 (shHNF1A-AS1) are shown in Table 1.
TABLE 1.
Oligonucleotides encoding shRNAs
| Oligo Set | Sequences | |
|---|---|---|
| shHNF4A-AS1 | Sense | 5′-CCGGGCACACCGGTCATATTATTGACTCGAGTCAATAATATGACCGGTGTGCTTTTTG-3′ |
| Antisense | 5′-AATTCAAAAAGCACACCGGTCATATTATTGACTCGAGTCAATAATATGACCGGTGTGC-3′ | |
| shHNF1A-AS1 | Sense | 5′-CCGGATGGAAACAGACCATCAGATTCTCGAGAATCTGATGGTCTGTTTCCATTTTTTG-3′ |
| Antisense | 5′-AATTCAAAAAATGGAAACAGACCATCAGATTCTCGAGAATCTGATGGTCTGTTTCCAT-3′ |
Cell Culture
Huh7 and HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with high sugar content under 5% CO2 at 37°C, supplemented with 10% FBS and 1% penicillin and streptomycin mixture. Cells were subcultured every 2 or 3 days and seeded into 12-, 24-, or 96-well plates until they reached 80–90% confluence before drug treatment.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted using TRNzol reagent, following the manufacturer’s instructions. A Nanodrop 2,000c spectrophotometer (Thermo Fisher Scientific) was used to analyze RNA concentrations. Total RNA was reverse transcribed using a PrimeScript RT reagent kit, and qRT-PCR was performed using the SYBR method. The primers used are listed in Table 2.
TABLE 2.
Primer sets used for qRT-PCR and ChIPqRT-PCR and ChIP
| Primer Set | Primers | Sequence | Application |
|---|---|---|---|
| GAPDH | Forward | 5′-GCACCGTCAAGGCTGAGAAC-3′ | qRT-RT-PCR |
| Reverse | 5′-TGGTGAAGACGCCAGTGGA-3′ | ||
| HNF4A-AS1 | Forward | 5′-TGGAGCTGGGATCTGACACT-3′ | qRT-RT-PCR |
| Reverse | 5′-ATGACCGGTGTGCAGTCAAG-3′ | ||
| U1 | Forward | 5′-TCCCAGGGCGAGGCTTATCCATT-3′ | qRT-RT-PCR |
| Reverse | 5′-GAACGCAGTCCCCCACTACCACAAAT-3′ | ||
| HNF4A | Forward | 5′-CGTGCTGCTCCTAGGCAA-3′ | qRT-RT-PCR |
| Reverse | 5′-GTCAAGGATGCGTATGGACAC-3′ | ||
| HNF1A-AS1 | Forward | 5′-AAAGGACCTGGGTCTGCATTTC-3′ | qRT-RT-PCR |
| Reverse | 5′-GTTGACAGGAGCAAAACTGCTAAG-3′ | ||
| HNF1A | Forward | 5′- TGGGTCCTACGTTCACCAAC-3′ | qRT-RT-PCR |
| Reverse | 5′- TCTGCACAGGTGGCATGAGC-3′ | ||
| PXR | Forward | 5′-CAACCTACATGTTCAAAGGCATC-3′ | qRT-RT-PCR |
| Reverse | 5′-ACACTCCCAGGTTCCAGTCTC-3′ | ||
| CYP3A4 | Forward | 5′-GCAGGAAAGCTCCATGCACATAG-3′ | qRT-RT-PCR |
| Reverse | 5′-GAGAAGCCAGGTTTCCATGG-3′ | ||
| Primer 1 | Forward | 5′-TGCTTTCCTCCAGCCTCT-3′ | ChIP-qRT-qPCR |
| Reverse | 5′-ATTCACCTGGGGTCAACA-3′ | ||
| Primer 2 | Forward | 5′-ATTCACCTGGGGTCAACA-3′ | ChIP-qRT-RT-PCR-qPCR |
| Reverse | 5′-AGTGAGGCTGTTGGATTGT-3′ |
Protein Sample Preparation and Western Blotting
Total proteins were obtained from Huh7 and HepG2 cells using radioimmunoprecipitation assay buffer (containing a protease inhibitor cocktail). A bicinchoninic acid method (Beyotime Institute of Biotechnology) was used to determine the protein concentrations. After separation by 10% SDS-polyacrylamide gel electrophoresis, total proteins were transferred to polyvinylidene fluoride membranes (Millipore). Membranes were blocked for 2 hours in 5% nonfat milk and then incubated with primary antibodies against CYP3A4 (Abcam), PXR (Santa), or GAPDH overnight at 4°C. The membranes were then incubated with horseradish peroxidase-labeled secondary antibodies for 2 hours. Protein bands were visualized with enhanced chemiluminescence followed by densitometry analysis using a FluorChem E system (Bio-Techne, Ltd.).
Subcellular Fractionation
Nuclear and cytoplasmic fractions of RNA were extracted from Huh7 and HepG2 cells. Cells were collected and digested with a membrane lysis buffer and NP40 (0.1%). After centrifugation, cytoplasmic (from supernatant) and nuclear (from precipitation) RNAs were extracted using TRNzol reagent. The nuclear to cytoplasmic ratios of target genes were calculated based on qRT-PCR.
shRNA Transfection
Life Technologies Lipofectamine 3000 (Thermo Fisher Scientific) was used for transfection experiments. Briefly, for gene silencing and overexpression experiments, Huh7 or HepG2 cells were seeded into 12- or 96-well plates and allowed to reach 50–70% confluence prior to transfection. Then, 1.2 or 0.1 μg of one or two plasmids overexpressing PXR, HNF4A-AS1, overexpression plasmids of HNF4A-AS1 control, and shRNAs targeting HNF4A-AS1, HNF1A-AS1, PXR, and control were mixed with Lipofectamine 3000 reagent in OPTI-MEM, and added to the culture medium with P3000. Six hours after transfection, 10% FBS was added to the culture medium. Twenty-four hours after transfection, cells were treated with different concentrations of RTV and incubated for another 24 hours before harvesting.
Lentiviral Transduction of HNF4A-AS1 and HNF1A-AS1
Lentiviral transduction was performed for the LDH experiments. The lentiviral vector pLKO.1-puro containing the gene sequences of HNF4A-AS1 and HNF1A-AS1, referred to as pLKO.1-HNF4A-AS1 and pLKO.1-HNF1A-AS1, respectively, was prepared and verified by sequencing. Lentivirus was produced according to the following steps. Briefly, the empty vector (pLKO.1), pLKO.1-HNF4A-AS1, or pLKO.1-HNF1A-AS1, together with the lentiviral packaging mix, was transfected into 293T cells using lentivirus packaging reagent. Culture media containing viral particles was harvested 48 hours after transfection and added to Huh7 or HepG2 cells. Puromycin selection (2 μg/ml) was performed following viral infection for 24 hours to select the transfected cells. The cells were then cultured in fresh medium for 48 hours.
Drug Treatments
To determine whether HNF4A-AS1 or HNF1A-AS1 influence the toxicity of RTV, Huh7 and HepG2 cells infected with pLKO.1, pLKO.1-HNF4A-AS1, or pLKO.1-HNF1A-AS1 shRNA lentivirus were seeded into 96-well plates. After 24 hours, the cells were treated with different concentrations of RTV (5, 10, 20, 40, or 80 μM) for 24 hours. To validate the effect of PXR on RTV-induced hepatotoxicity, the cells were transfected with PXR, shPXR, or their control plasmids first, and then treated with 5, 10, 20, 40, or 80 μM RTV for 24 hours. To determine the optimal pretreatment duration for RIF, HepG2 cells were seeded into 24-well plates at a density of 40,000 cells per well and treated with 10 μM RIF or 0.1% v/v DMSO for 24, 48, 72, or 96 hours before harvesting. In tests evaluating RTV-induced hepatotoxicity following RIF exposure, HepG2 cells were seeded into 96-well plates at a density of 20,000 cells per well and treated with 10 μM RIF or 0.1% v/v DMSO for 48 hours, and then treated with 5, 10, 20, 40, or 80 μM RTV and 10 μM RIF or 0.1% v/v DMSO for 24 hours, respectively. To determine the effects of HNF4A-AS1 and HNF1A-AS1 on RTV-induced hepatotoxicity induced by RIF exposure, HepG2 cells were seeded into 96-well plates and transfected with HNF4A-AS1, shHNF1A-AS1, or their control plasmids, and then treated with RIF and RTV.
LDH Assay
An LDH release assay was used to measure the level of cell damage. After treatment with RTV, 10 μl of lysis buffer and sterile water were added to the positive control and experimental groups, respectively. Then, samples were subjected to 37°C incubation for 45 minutes. Subsequently, 50 μl of supernatant from those samples was added to 50 μl of the reaction mixture. After protecting from light for 30 minutes, the reaction was terminated with 50 μl of stop solution. LDH activity was measured using a spectrophotometer at absorbance values of 490 and 680 nm. The level of cell damage was indicated by the ratio of LDH activity between the experimental sample and the positive control.
ROS Assay
Oxidative stress in Huh7 and HepG2 cells was determined after RTV treatment using an ROS assay kit. Cells were seeded onto glass-bottom cell culture dishes (20 mm) and transfected with pLKO.1, shHNF4A-AS1, or shHNF1A-AS1. After 24 hours, the cells were treated with 20 μM RTV for 24 hours. The medium was then removed, and the cells were rinsed three times with DMEM. Diluted DCFH-DA solution (1:1,000, diluted with DMEM) was added to each dish, and cells were incubated for 30 minutes. The cells were then rinsed three times with DMEM. Images of fluorescently stained cells at 630× magnification were captured by a confocal laser scanning microscope (Leica, TCS SP8).
Chromatin Immunoprecipitation
A chromatin immunoprecipitation (ChIP) assay was performed as previously described (Wang et al., 2021b). Briefly, the cells were collected, crosslinked, and digested. A Bioruptor Pico sonication system was used to obtain sheared chromatin, which was immunoprecipitated with anti-PXR, -H3K4me3, -H3K27me3, or -IgG (as a control) antibodies overnight at 4°C. Purified DNA was analyzed by qRT-PCR to measure the levels of PXR, H3K4me3, and H3K27me3 enrichment in the CYP3A4 promoter (primer sequences shown in Table 2).
Structure Prediction
RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) was used to predict the secondary structure of HNF4A-AS1.
Data and Statistical Analysis
Data are presented as the mean ± S.D. The significance of differences between two groups was determined using a two-tailed unpaired Student’s t test. Differences among three or more groups were analyzed using one-way ANOVA. Statistical analyses were performed using SPSS (version 17.0; IBM). P < 0.05 was considered statistically significant.
Results
Human HNF4A-AS1, a Liver-enriched lncRNA with a Stable Structure, Is Mainly Located in the Nucleus
RNA features were characterized to understand the function of HNF4A-AS1. As depicted in Fig. 1A, the HNF4A-AS1 gene in the HepG2 cell line contains a specific DNase signal, suggesting that it is transcriptionally active in hepatocytes. In addition, acetylation of histone 3 lysine 27 (H3K27Ac) was enriched near to the active regulatory region of HNF4A-AS1 in various cell lines, suggesting that the transcriptional activity of HNF4A-AS1 is susceptible to environmental conditions. The sequence of HNF4A-AS1 is conserved between humans and rhesus monkeys, which is characteristic of its nonconserved lncRNA, and may represent a survival advantage acquired during environmental adaptation (Fig. 1A). RNAfold predicted that the minimum free energy (mfe) of HNF4A-AS1 was −223.01 kcal/mol, which exceeds the stability threshold for RNA structures (mfe < −80 kcal/mol) (Mohammadin et al., 2015) (Fig. 1B). In addition, the secondary structure obtained by the three algorithms (mfe, partition function, and centroid) was highly consistent (Fig. 1C), which suggested that the secondary structure of HNF4A-AS1 is stable. Moreover, the tissue distribution of HNF4A-AS1 was analyzed using RNA-sequencing data from 95 samples across 27 different human tissues in the National Center for Biotechnology Information (NCBI) database. As shown in Fig. 1D, HNF4A-AS1 was mainly concentrated in the small intestine, liver, and kidneys. This suggested that HNF4A-AS1 may be involved in regulating drug metabolism. HNF4A-AS1 is transcribed from an intergenic region on chromosome 20 and is located on the antisense strand of the HNF4A gene (Chen et al., 2020a). Nucleoplasm separation analysis of HNF4A-AS1 revealed a greater distribution of HNF4A in the nucleus compared with the cytoplasm, whereas HNF4A-AS1 was located almost exclusively in the nucleus of Huh7 and HepG2 cells (Fig. 1, E and F).
Fig. 1.
Human HNF4A-AS1, a liver-enriched lncRNA with a stable structure, is predominantly located in the nucleus. (A) Schematic representation of HNF4A-AS1 on chromosome 20 based on the University of California, Santa Cruz Genome Browser tracks showing peak signals of H3K4me1, H3K4me3, H3K27Ac, DNase, and mammalian conservation. (B) The mfe structure of HNF4A-AS1. The secondary structure of HNF4A-AS1 was predicted based on mfe (−223.01 kcal/mol). The base colors in the figure change with the entropy value. The more biased to red, the smaller the entropy of the corresponding base and the greater the probability offorming a pair. (C) A mountain plot of HNF4A-AS1. A mountain plot containing the mfe structure, partition function (pf) structure, and centroid structures. These curves are closely related, which indicates that the secondary structure of HNF4A-AS1 is stable. The height (y-axis) of the mountain plot represents the base pairs containing sequence positions in the x-axis. (D) RNA levels of HNF4A-AS1 across 27 different human tissues from NCBI database. (E and F) qRT-PCR Real-time qRT-PCR was used to reveal the enrichment levels of HNF4A-AS1 and HNF4A in the nuclei and cytoplasm of both Huh7 and HepG2 cells. U1 and GAPDH were used as nuclear and cytoplasmic markers, respectively. Data are shown as the means ± S.D. of three independent experiments.
Impact of HNF4A-AS1 and HNF1A-AS1 Knockdown on Gene Expression in Huh7 and HepG2 Cells
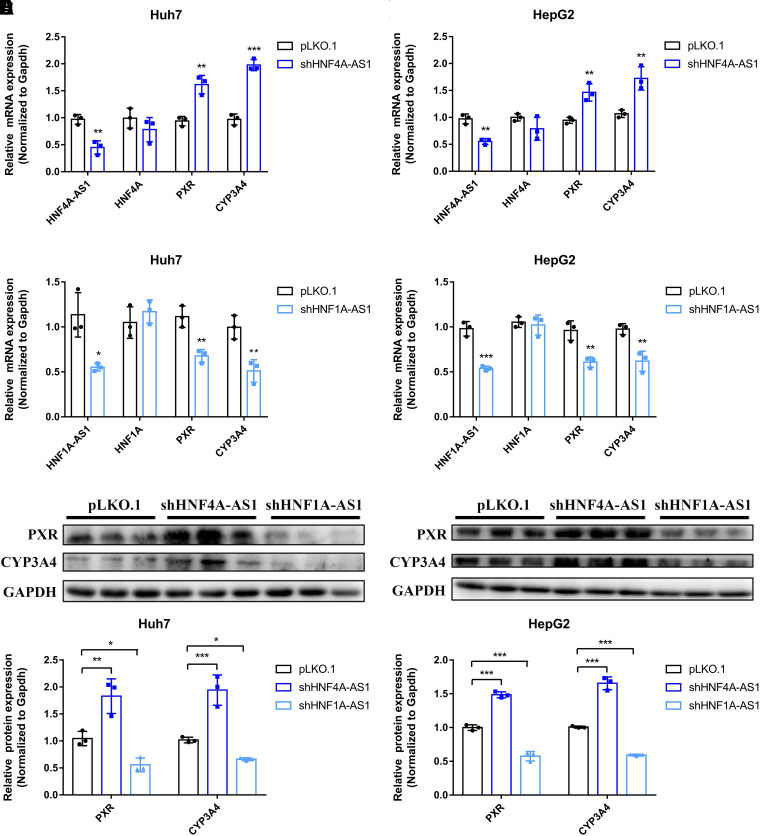
To verify the involvement of HNF4A-AS1 and HNF1A-AS1 in regulating PXR and CYP3A4 expression, shRNAs were designed to silence the expression of HNF4A-AS1 and HNF1A-AS1. The silencing efficiency of the shRNA against HNF4A-AS1 was approximately 55% and 45% in Huh7 and HepG2 cells, respectively (Fig. 2, A and B). In both Huh7 and HepG2 cells, knockdown of HNF4A-AS1 significantly increased the mRNA expression of PXR and CYP3A4 but did not affect that of HNF4A (Fig. 2, A and B). After knockdown of HNF1A-AS1, the expression of HNF1A-AS1 decreased to 55% and 54% in Huh7 and HepG2 cells, respectively (Fig. 2, C and D). As an antisense noncoding RNA of HNF1A, HNF1A-AS1 silencing did not affect the mRNA expression of HNF1A. Notably, the mRNA expression levels of PXR and CYP3A4 were markedly decreased following HNF1A-AS1 knockdown in both Huh7 and HepG2 cells (Fig. 2, C and D). PXR and CYP3A4 protein expression was detected after knockdown of HNF4A-AS1 and HNF1A-AS1. As seen in Fig. 2E, the protein expression of PXR and CYP3A4 was elevated 1.8- and 1.9-fold following HNF4A-AS1 knockdown, whereas the values were reduced to 0.53- and 0.65-fold following HNF1A-AS1 knockdown in Huh7 cells. In HepG2 cells with HNF4A-AS1 knockdown, the protein expression of PXR and CYP3A4 was 1.5- and 1.6-fold higher, respectively, compared with the control group, whereas HNF1A-AS1 knockdown led to lower expression of PXR and CYP3A4 protein (0.58- and 0.59-fold, respectively) (Fig. 2F). These results indicated that HNF4A-AS1 and HNF1A-AS1 play opposing roles in regulating the expression of PXR and CYP3A4.
Fig. 2.
Impact of HNF4A-AS1 and HNF1A-AS1 knockdown on the expression of genes in Huh7 and HepG2 cells. (A and B) mRNA expression of HNF4A-AS1, HNF4A, PXR, and CYP3A4. (C and D) mRNA expression of HNF1A-AS1, HNF1A, PXR, and CYP3A4. (E and F) The protein expression of PXR and CYP3A4. Data are shown as the means ± S.D. of three independent experiments. Statistical analyses were performed using an unpaired Student’s t test or one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001 versus pLKO.1 group.
HNF4A-AS1 and HNF1A-AS1 Had Opposite Effects on RTV-induced Cytotoxicity in Hepatoma Cells
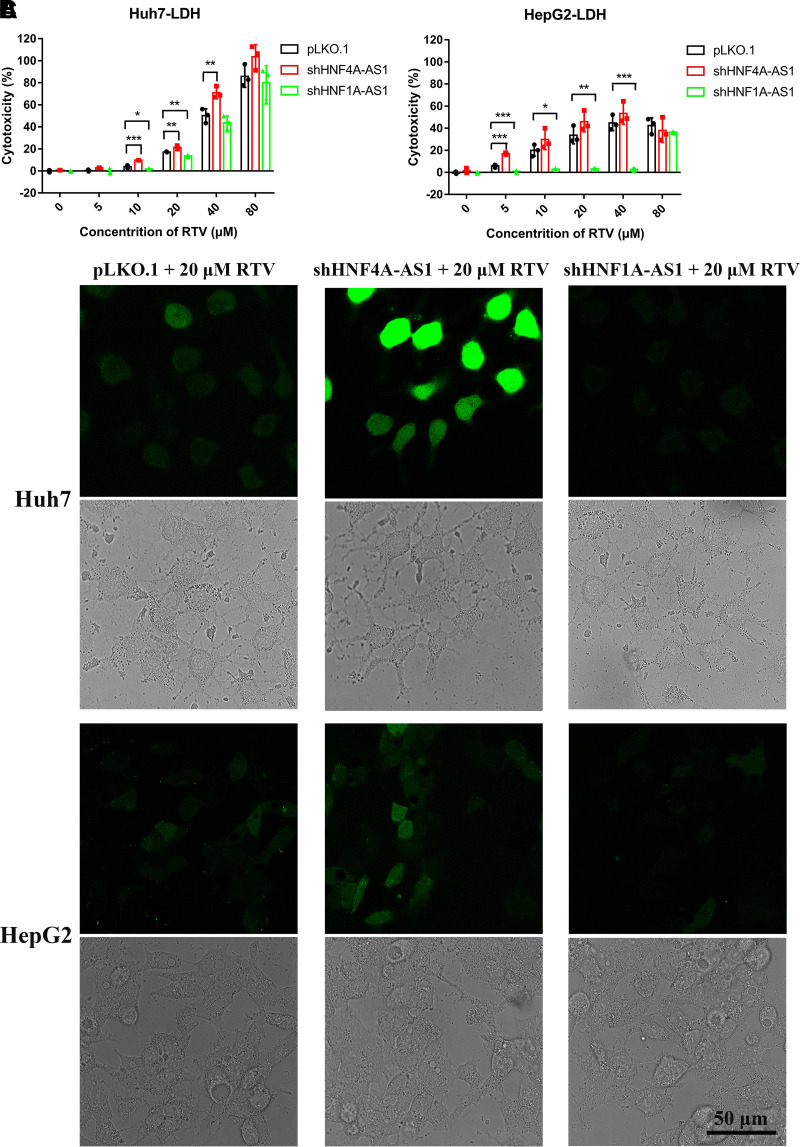
An LDH release assay was performed to determine the extent of cell damage. Huh7 and HepG2 cells were infected with viruses containing shHNF4A-AS1, shHNF1A-AS1, or pLKO.1. Cells were treated with different concentrations of RTV (0, 5, 10, 20, 40, or 80 μM) for 24 hours. As seen in Fig. 3, A and B, knockdown of HNF4A-AS1 and HNF1A-AS1 without RTV treatment did not aggravate cell damage. RTV-induced cytotoxicity was affected following the knockdown of the two lncRNAs. Specifically, knockdown of HNF4A-AS1 exacerbated RTV-induced hepatotoxicity at concentrations of 10, 20, and 40 μM, whereas knockdown of HNF1A-AS1 led to lower RTV cytotoxicity at concentrations of 10 and 20 μM in Huh7 cells (Fig. 3A). In HepG2 cells, after HNF4A-AS1 knockdown, an increase in cell damage was found due to the treatment of 5 μM RTV. However, as shown in Fig. 3B, HNF1A-AS1 knockdown significantly reduced the cytotoxicity of RTV at concentrations lower than 80 μM. Interestingly, when treated with 80 μM RTV, there were no significant differences in cytotoxicity among the three groups of Huh7 and HepG2 cells (Fig. 3, A and B).
Fig. 3.
Changes in RTV-induced hepatotoxicity following the knockdown of HNF4A-AS1 or HNF1A-AS1 in Huh7 and HepG2 cells. (A and B) Cell damage assessed by the LDH release assay after RTV treatment. (C) ROS production analyzed through fluorescence intensity in different groups of cells after treatment with 20 μM RTV. Scale bar, 50 μm. Data are shown as the means ± S.D. of three independent experiments. Statistical analyses were performed using a one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001 versus pLKO.1 group.
Excessive production of intracellular ROS is a characteristic of oxidative stress and is usually associated with drug-induced liver injury. In this study, ROS production was analyzed in Huh7 and HepG2 cells. As shown in Fig. 3C, compared with the other groups, the green fluorescence was more intense in cells transfected with shHNF4A-AS1 plasmids receiving 20 μM RTV treatment, which suggested the higher levels of ROS. Conversely, green fluorescence was weaker in cells transfected with shHNF1A-AS1 plasmids receiving 20 μM RTV treatment, indicating lower levels of ROS. These findings were consistent with the LDH test results. These results indicated that lncRNAs HNF4A-AS1 and HNF1A-AS1 have opposing effects on RTV-induced cytotoxicity. Considering that the expression of CYP3A4 mRNA was higher in HepG2 than in Huh7 cells (data not shown), the mechanism through which HNF4A-AS1 regulated CYP3A4 in subsequent experiments is mainly discussed considering Huh7 cells, whereas the mechanism related to HNF1A-AS1 is mainly discussed considering HepG2 cells.
Silencing of HNF4A-AS1 or HNF1A-AS1 Altered the Levels of PXR Enrichment and Histone Modification Status in the CYP3A4 Promoter
ChIP-qRT-PCR experiments were conducted using Huh7 cells transfected with shHNF4A-AS1 or HepG2 cells transfected with shHNF1A-AS1. Two pairs of primers were designed to evaluate the enrichment of PXR, H3K4me3, and H3K27me3 in the PXR response element (PXRE) regions of the CYP3A4 promoter (Fig. 4A). Notably, compared with the control group, knockdown of HNF4A-AS1 resulted in a 10.8-fold enrichment of PXR (at −7881 to −7760 bp) and a 2.6- and 3.8-fold enrichment of H3K4me3 (at −7881 to −7760 bp and −263 to −108 bp, respectively) (Fig. 4, B and C). The enrichment of H3K27me3 in both regions was reduced to 15% and 31%, respectively (Fig. 4D). Conversely, in cells with HNF1A-AS1 knockdown, the enrichment of PXR was reduced to 0.06% and 0.08% in the two regions, respectively (Fig. 4E), and H3K4me3 was reduced to 36% (from −7881 to −7760 bp) (Fig. 4F). There was no significant change in H3K27me3 enrichment following HNF1A-AS1 knockdown (Fig. 4G). The data demonstrated that HNF4A-AS1 and HNF1A-AS1 could regulate CYP3A4 expression accompanied by alterations in PXR enrichment and histone modification status in the CYP3A4 promoter.
Fig. 4.
Knockdown of HNF4A-AS1 or HNF1A-AS1 altered the enrichment levels of PXR and histone modification status in the CYP3A4 promoter of Huh7 or HepG2 cells. (A) Schematic locations of the PXRE regions and ChIP-qRT-PCR primers in the CYP3A4 promoter. ChIP and qRT-PCR (normalized to input) assays were performed. (B–D) The enrichment levels of PXR, H3K4me3, and H3K27me3 in Huh7 cells transfected with shHNF4A-AS1 or pLKO.1. (E–G) The enrichment levels of PXR, H3K4me3, and H3K27me3 in HepG2 cells transfected with shHNF1A-AS1 or pLKO.1. Data are shown as the means ± S.D. of three independent experiments. Statistical analysis was performed using an unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001 versus pLKO.1 group. TSS, transcriptional start site.
PXR Is Involved in HNF4A-AS1 and HNF1A-AS1-mediated Expression of CYP3A4 and RTV-induced Hepatotoxicity
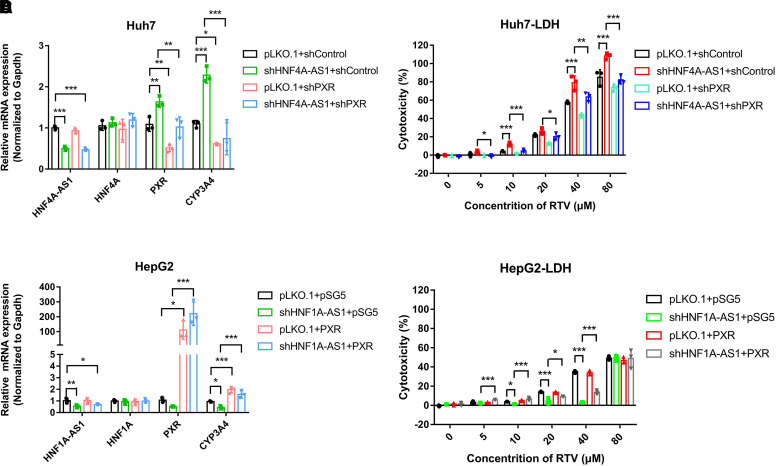
In Huh7 cells, silencing of HNF4A-AS1 upregulated the mRNA expression of PXR and CYP3A4, whereas knockdown of PXR prevented these effects (Fig. 5A). In addition, the increased cytotoxicity of different concentrations of RTV caused by HNF4A-AS1 knockdown was offset by the knockdown of PXR (Fig. 5B). In HepG2 cells, knockdown of HNF1A-AS1 attenuated the expression of PXR and CYP3A4 in the mRNA level, which was restored by overexpression of PXR (Fig. 5C). The reduced cytotoxicity of RTV resulting from knockdown of HNF1A-AS1 was reversed by overexpression of PXR at concentrations of 5 and 10 μM. When the RTV concentration was increased to 20 and 40 μM, overexpression of PXR partially reversed the protective effect of shHNF1A-AS1 (Fig. 5D). These findings indicated the involvement of PXR in the HNF4A-AS1- and HNF1A-AS1-mediated expression of CYP3A4 and RTV-induced hepatotoxicity.
Fig. 5.
PXR is involved in the HNF4A-AS1- and HNF1A-AS1-mediated expression of CYP3A4 and RTV-induced hepatotoxicity. qRT-RT-PCR and LDH assays showing the mRNA expression of HNF4A-AS1, HNF4A, PXR, and CYP3A4, and RTV-induced cell damage (A and B) in Huh7 cells transfected with shHNF4A-AS1 or pLKO.1 as well as those co-transfected with shPXR or shcontrol, (C and D) in HepG2 cells transfected with shHNF1A-AS1 or pLKO.1, and those co-transfected with PXR or pSG5. Data are shown as the means ± S.D. of three independent experiments. ANOVA was used to compare the differences among groups. *P < 0.05, **P < 0.01, ***P < 0.001.
HNF4A-AS1 and HNF1A-AS1 Are Involved in the Increased Susceptibility to RTV-induced Hepatotoxicity Caused by RIF Exposure in HepG2 Cells
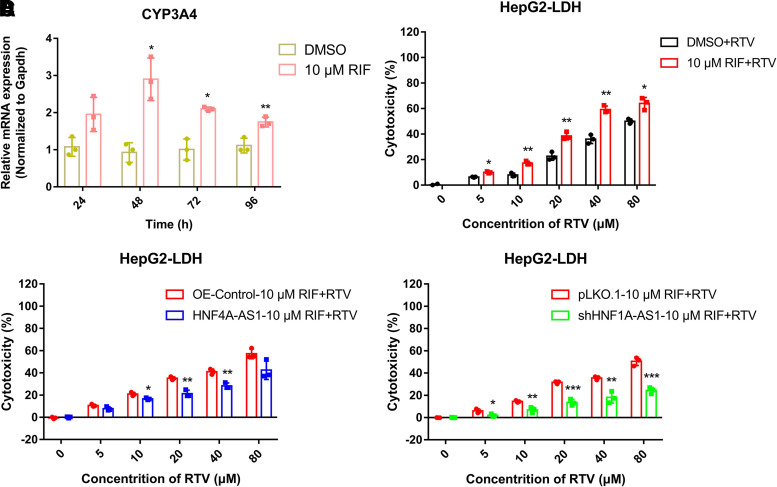
In the concentration–response experiment, 10 μM RIF significantly upregulated the expression of PXR and CYP3A4 (data not shown). The optimal induction effect of 10 μM RIF on CYP3A4 expression occurred at 48 hours (2.9-fold) in HepG2 cells (Fig. 6A). To investigate the roles of PXR and CYP3A4 in RTV-induced cytotoxicity, HepG2 cells were pretreated with 10 μM RIF for 48 hours, followed by treatment with different concentrations of RTV. The results of the LDH experiment showed that the cytotoxicity of RTV at different concentrations was significantly increased in groups pretreated with RIF (Fig. 6B). Specifically, the increased cytotoxicity of RTV caused by pretreating with RIF was significantly reduced by overexpression of HNF4A-AS1 in HepG2 cells (Fig. 6C). Similarly, knockdown of HNF1A-AS1 inhibited the increased RTV cytotoxicity caused by pretreating with RIF (Fig. 6D). These results suggested that the increased susceptibility to RTV-induced hepatotoxicity caused by RIF exposure was attenuated by HNF4A-AS1 overexpression or HNF1A-AS1 knockdown.
Fig. 6.
HNF4A-AS1 and HNF1A-AS1 are involved in the altered susceptibility to RTV-induced hepatotoxicity caused by RIF exposure in HepG2. (A) qRT-RT-PCR assay showing the mRNA expression of CYP3A4 in HepG2 cells after 10 μM RIF treatment of 24, 48, 72, or 96 hours. LDH assays indicating (B) RTV-induced cell damage in HepG2 cells following pretreatment with RIF for 48 hours, and those transfected with (C) HNF4A-AS1 and (D) shHNF1A-AS1. Data are shown as the means ± S.D. of three independent experiments. Statistical analyses were performed using an unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001 versus NC group.
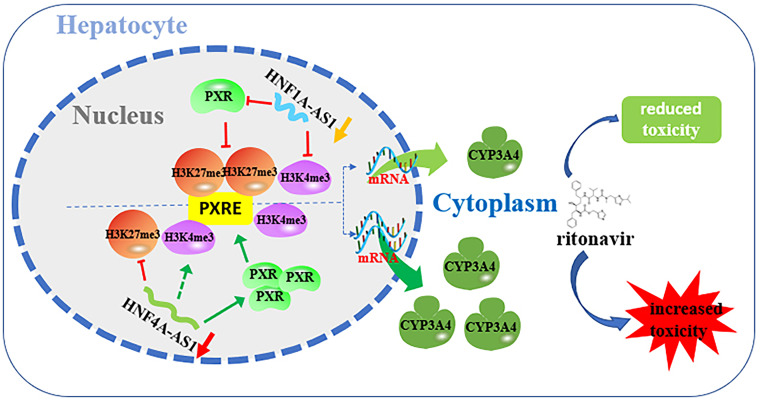
Overall, these results indicated that knockdown of HNF4A-AS1 upregulated the expression of CYP3A4 by increasing the expression of PXR, promoting the enrichment of PXR and H3K4me3, and attenuating the enrichment of H3K27me3 in the CYP3A4 promoter, ultimately leading to increased cytotoxicity of RTV. In contrast, knockdown of HNF1A-AS1 downregulated the expression of CYP3A4 by reducing the expression of PXR and attenuating the enrichment of PXR and H3K4me3 in the CYP3A4 promoter, resulting in minor cytotoxicity of RTV (Fig. 7). Moreover, HNF4A-AS1 and HNF1A-AS1 were involved in the increased susceptibility to RTV-induced hepatotoxicity caused by RIF exposure. These findings suggest the potential value of HNF4A-AS1 and HNF1A-AS1 in research on RTV-induced liver injury.
Fig. 7.
Mechanisms underlying the effects of HNF4A-AS1 and HNF1A-AS1 on CYP3A4 expression and RTV-induced hepatotoxicity. As the key cofactors in gene regulation, HNF4A-AS1 and HNF1A-AS1 affect the PXR enrichment and histone modification status in the PXRE regions of the CYP3A4 promoter, resulting in changes in CYP3A4 expression and RTV-induced hepatotoxicity.
Discussion
Ritonavir is an essential drug for antiretroviral therapy; however, it may induce hepatotoxicity through as yet unknown mechanisms. Notably, there is a high correlation between the lncRNAs and drug metabolism pathways (Chen et al., 2018; Wang et al., 2019b; Wang et al., 2021a). The present study highlights the involvement of the lncRNAs HNF4A-AS1 and HNF1A-AS1 in RTV-induced hepatotoxicity.
Human HNF4A-AS1 is transcribed from an intergenic region on chromosome 20, which contains four exons. In the current study, HNF4A-AS1 was observed to carry a specific DNase signal in the HepG2 cell line, suggesting that HNF4A-AS1 is active in the transcription of hepatocytes. It is worth noting that most DNase I hypersensitive sites (DHSs) are mapped to regulatory elements, such as promoters and enhancers. However, in the current study, the specific DHS was found in the intron region. We carefully examined the possible regulatory elements near the HNF4A-AS1 gene in the UCSC genome browser and found a strong enhancer signal coincident with the location of this DHS. Whether and how this enhancer acts as a remote regulatory element that participates in the regulation of HNF4A-AS1 gene requires further research. Additionally, histone modification H3K27Ac was enriched close to the active regulatory region of HNF4A-AS1 in various cell lines, suggesting that the transcriptional activity of HNF4A-AS1 is susceptible to environmental conditions. This is consistent with our previous results showing that HNF4A-AS1 mRNA expression is elevated approximately 2.8-fold after RIF exposure (data not shown). The high expression of HNF4A-AS1 in human small intestine, liver, and kidney suggests its potential role in drug metabolism. LncRNAs exist in both the cytoplasm and nucleus, and the mechanisms through which they regulate gene expression vary depending on their localization (Chen, 2016). The results of cytoplasmic and nuclear separation revealed that HNF4A-AS1 is predominantly localized in the nucleus of hepatoma cells, indicating its importance in the transcriptional regulation of downstream genes. As we recently reported, HNF4A-AS1 negatively regulates the expression of PXR and constitutive androstane receptor (CAR), as well as CYPs in Huh7 cells (Wang et al., 2021a). The strong correlation between HNF4A-AS1 and CYP3A4 was confirmed in the current study. In both Huh7 and HepG2 cells, the expression of PXR and CYP3A4 was increased after knockdown of HNF4A-AS1.
HNF1A-AS1 is another lncRNA that is transcribed from the neighboring antisense of the human HNF1A gene. HNF1A-AS1 (located on human chromosome 12) is 39.04 kb long and contains two exons and one intron. The detailed characterization about the secondary structure and tissue and organ distribution of HNF1A-AS1 has been reported earlier (Chen et al., 2020a). As shown here and in previous studies (Chen et al., 2018; Wang et al., 2019b), knockdown of HNF1A-AS1 markedly reduced the mRNA expression of PXR and CYP3A4, which was in contrast to the results observed following the knockdown of HNF4A-AS1.
The expression and activities of DMEs (including CYPs) are the main factors affecting drug efficacy and adverse drug reactions. Interestingly, a previous study in HepaRG cells revealed that HNF4A-AS1 and HNF1A-AS1 can regulate the expression of acetaminophen-metabolizing CYPs, thus affecting susceptibility to acetaminophen-induced liver injury (Chen et al., 2020b). Notably, the bioactivation of RTV is mainly catalyzed by CYP3A4 (Kumar et al., 1996; Denissen et al., 1997; Koudriakova et al., 1998; Gangl et al., 2002; Yao et al., 2008; Li et al., 2011). Shehu et al. revealed that isopropylthiazole ring-open metabolites of RTV were significantly increased in PXR- and CYP3A4-humanized mouse models pretreated with RIF (Shehu et al., 2019), whereas the ring-open metabolites of thiazole derivatives can result in liver injury through further oxidizing (Mizutani and Suzuki, 1996; Ji et al., 2007), which showed that human PXR and CYP3A4 play a crucial role in RTV-induced hepatotoxicity. HNF4A-AS1 and HNF1A-AS1 expression was significantly correlated with PXR and CYP3A4 expression in hepatoma cells, further supporting the potential roles of these two lncRNAs in RTV-induced hepatotoxicity. In the present study, our data suggested that knockdown of HNF4A-AS1 significantly increased the cytotoxicity of different concentrations of RTV; however, knockdown of HNF1A-AS1 reduced the cytotoxicity of RTV in both Huh7 and HepG2 cells. These results elucidated an important feature of the two lncRNAs in the regulation of CYP3A4 expression and RTV-induced hepatotoxicity.
Researchers have verified the emerging roles of histone modifications in the regulation of CYPs (Yan et al., 2017; Wang et al., 2019a; Pande et al., 2020). In the present study, knockdown of HNF4A-AS1 increased H3K4me3 and reduced H3K27me3, whereas knockdown of HNF1A-AS1 resulted in reduced H3K4me3 enrichment in the PXRE regions of the CYP3A4 promoter. Both H3K4me3 and H3K27me3 are important epigenetic markers of gene transcription, indicating gene activation and silencing, respectively. Therefore, alterations in histone modification status caused by knockdown of HNF4A-AS1 and HNF1A-AS1 were considered to be factors affecting the expression of CYP3A4. LncRNAs exert regulatory functions by interacting with other molecules via four mechanisms: decoy, signal, guide, and scaffold (Wang and Chang, 2011). For example, long intergenic noncoding RNA (lincRNA) HOTAIR acts as a scaffold to specify the histone modification patterns on target genes by providing binding surfaces for histone modification enzymes (Tsai et al., 2010). Hence, we deduce that HNF4A-AS1 and HNF1A-AS1 may serve as scaffolds for the binding of diverse histone modification enzymes to the CYP3A4 promoter, thus regulating CYP3A4 expression and RTV-induced hepatotoxicity. However, further study is needed to clarify the accurate mechanism of action.
In general, the binding of transcription factors to their target genes results in the upregulation of gene expression. For example, some CYPs are induced by activation of PXR or CAR (Burk et al., 2004). This may also explain the alterations in CYP3A4 expression and RTV-induced hepatotoxicity after knockdown of HNF4A-AS1 or HNF1A-AS1 that the diverse enrichment of PXR in PXRE regions of the CYP3A4 promoter. Knockdown of HNF4A-AS1 and HNF1A-AS1 altered the expression of PXR (although the specific mechanism remains to be explored), which may indirectly affect the expression of CYP3A4. Importantly, further experiments have confirmed that silencing of HNF4A-AS1 or HNF1A-AS1 increased or decreased CYP3A4 expression and RTV-induced hepatotoxicity, whereas knockdown or overexpression of PXR prevented these effects, respectively (Fig. 5). These findings indicated that PXR plays a vital role in the regulation of CYP3A4 expression and RTV-induced hepatotoxicity via HNF4A-AS1 and HNF1A-AS1. It was noticed that overexpression of PXR did not completely reverse the protective effect of shHNF1A-AS1 in HepG2 cells treated with 20 or 40 μM RTV. This may be because, after knockdown of HNF1A-AS1, the increased cytotoxicity of RTV following transfection with PXR plasmids might not be enough to offset the effects of increased RTV concentration.
Clinical studies have reported hepatotoxicity in 100% of patients treated with RTV-containing regimens following pretreatment with RIF or EFV (Shehu et al., 2019). Our findings verified that the hepatotoxicity of RTV at different concentrations increased significantly when HepG2 cells were pretreated with RIF. Notably, the increase in RTV-induced hepatotoxicity was attenuated by HNF4A-AS1 overexpression or HNF1A-AS1 knockdown, indicating the potential roles for HNF4A-AS1 and HNF1A-AS1 in RTV-induced hepatotoxicity and RTV susceptibility induced by RIF exposure. These findings provide direction for future research on the roles and mechanisms of HNF4A-AS1 and HNF1A-AS1 under different physiologic conditions. During the time course of liver repair and regeneration after liver injury, the expression and activities of CYPs, as well as drug effectiveness and adverse drug reactions, are altered (Bao et al., 2021). Therefore, early prediction of hepatotoxicity is important for the rational use of drugs. lncRNAs in plasma and serum are stable, which causes a potential value for relevant clinical diagnosis, like plasma H19 for gastric cancer (Zhou et al., 2015) and serum LINC00161 for hepatocellular carcinoma (Sun et al., 2018). HNF4A-AS1 and HNF1A-AS1, as lncRNAs that have opposite impacts on CYP3A4 expression, may also be used to predict the effects and toxicity of drugs metabolized by CYP3A4 in the future.
The expression of DMEs is not constitutive. There may exist a regulatory loop containing HNF4A-AS1 and HNF1A-AS1 that regulates the expression of CYP3A4 to maintain homeostasis in hepatic metabolism. However, the specific mechanism needs further research. In the current study, Huh7 and HepG2 cells were used to investigate the roles of lncRNAs in RTV-induced hepatotoxicity. However, there are some limitations to hepatoma cell models. Huh7 is a highly deteriorated liver tumor cell line, whereas the HepG2 cell line is derived from a heterozygous patient carrying a CYP3A4*1G mutation, and the expression levels of nuclear receptors and DMEs are low in both cell lines. Human induced pluripotent stem cell-derived hepatocytes or primary human hepatocytes should be used to validate these findings in the future.
In summary, HNF4A-AS1 and HNF1A-AS1 were identified as lncRNAs with the ability to alter RTV-induced hepatotoxicity with contrasting effects, mainly by regulating CYP3A4 expression via alteration of PXR enrichment and histone modification status in the PXRE regions of the CYP3A4 promoter. HNF4A-AS1 and HNF1A-AS1 are also involved in the altered susceptibility to RTV-induced hepatotoxicity caused by RIF exposure.
Abbreviations
- ChIP
chromatin immunoprecipitation
- CYP
cytochrome P450
- DMEs
drug metabolizing enzymes
- EFV
efavirenz
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- H3K4me3
trimethylation of histone 3 lysine 4
- H3K27me3
trimethylation of histone 3 lysine 27
- HIV
human immunodeficiency virus
- HNF1A
hepatocyte nuclear factor 1α
- HNF1A-AS1
HNF1A antisense RNA 1
- HNF4A
hepatocyte nuclear factor 4α
- HNF4A-AS1
HNF4A antisense RNA 1
- LDH
lactate dehydrogenase
- lincRNA
long intergenic noncoding RNA
- lncRNA
long noncoding RNA
- mfe
minimum free energy
- NCBI
national center for biotechnology information
- PXR
pregnane X receptor
- PXRE
PXR response element
- qRT-RT-PCR
quantitative real-time real-time polymerase chain reaction
- RIF
rifampicin
- ROS
reactive oxygen species
- RTV
ritonavir
- shHNF1A-AS1
shRNA targeting HNF1A-AS1
- shHNF4A-AS1
shRNA targeting HNF4A-AS1
- shRNA
small hairpin RNA
Authorship Contributions
Participated in research design: X. Wang, P. Wang, Yan, Zhong, Zhang.
Conducted experiments: X. Wang, Yu, Yang, Y. Wang, Yan.
Performed data analysis: X. Wang, Yu, Yang, Zhang.
Wrote or contributed to the writing of the manuscript: X. Wang, Yu, P. Wang, Zhong, Zhang.
Footnotes
This work was supported by the National Natural Science Foundation of China [Grants 82073931 and 81773815] (to L.Z.). Xiao-bo Zhong is supported by the National Institutes of Health National Institute of General Medical Sciences [Grant R35GM140862].
References
- Acharya C, Dharel N, Sterling RK (2015) Chronic liver disease in the human immunodeficiency virus patient. Clin Liver Dis 19:1–22. [DOI] [PubMed] [Google Scholar]
- Bao Y, Phan M, Zhu J, Ma X, Manautou JE, Zhong XB (2021) Alterations of cytochrome P450-mediated drug metabolism during liver repair and regeneration after acetaminophen-induced liver injury in mice. Drug Metab Dispos 50: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk O, Koch I, Raucy J, Hustert E, Eichelbaum M, Brockmöller J, Zanger UM, Wojnowski L (2004) The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR). J Biol Chem 279:38379–38385. [DOI] [PubMed] [Google Scholar]
- Chen L, Bao Y, Jiang S, Zhong XB (2020a) The roles of long noncoding RNAs HNF1α-AS1 and HNF4α-AS1 in drug metabolism and human diseases. Noncoding RNA 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bao Y, Piekos SC, Zhu K, Zhang L, Zhong XB (2018) A transcriptional regulatory network containing nuclear receptors and long noncoding RNAs controls basal and drug-induced expression of cytochrome P450s in HepaRG cells. Mol Pharmacol 94:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang P, Manautou JE, Zhong XB (2020b) Knockdown of long noncoding RNAs hepatocyte nuclear factor 1α antisense RNA 1 and hepatocyte nuclear factor 4α Aantisense RNA 1 alters susceptibility of acetaminophen-induced cytotoxicity in HepaRG cells. Mol Pharmacol 97:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL (2016) Linking long noncoding RNA localization and function. Trends Biochem Sci 41:761–772. [DOI] [PubMed] [Google Scholar]
- Denissen JF, Grabowski BA, Johnson MK, Buko AM, Kempf DJ, Thomas SB, Surber BW (1997) Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug Metab Dispos 25:489–501. [PubMed] [Google Scholar]
- Gangl E, Utkin I, Gerber N, Vouros P (2002) Structural elucidation of metabolites of ritonavir and indinavir by liquid chromatography-mass spectrometry. J Chromatogr A 974:91–101. [DOI] [PubMed] [Google Scholar]
- Haas DWKoletar SLLaughlin LKendall MASuckow CGerber JGZolopa ARBertz RChild MJHosey L, et al. ; A5213 StudyTeam (2009) Hepatotoxicity and gastrointestinal intolerance when healthy volunteers taking rifampin add twice-daily atazanavir and ritonavir. J Acquir Immune Defic Syndr 50:290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Baldwin WS (2009) Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr Pharmacogenomics Person Med 7:81–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamois C, Riek M, Schmitt C (2009) Potential hepatotoxicity of efavirenz and saquinavir/ritonavir coadministration in healthy volunteers. Arch Drug Inf 2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T, Ikehata K, Koen YM, Esch SW, Williams TD, Hanzlik RP (2007) Covalent modification of microsomal lipids by thiobenzamide metabolites in vivo. Chem Res Toxicol 20:701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, Orza D, Marinina J, Gerber N (1998) Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab Dispos 26:552–561. [PubMed] [Google Scholar]
- Kumar GN, Rodrigues AD, Buko AM, Denissen JF (1996) Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J Pharmacol Exp Ther 277:423–431. [PubMed] [Google Scholar]
- Kumar S, Rao PS, Earla R, Kumar A (2015) Drug-drug interactions between anti-retroviral therapies and drugs of abuse in HIV systems. Expert Opin Drug Metab Toxicol 11:343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Lu J, Ma X (2011) Metabolomic screening and identification of the bioactivation pathways of ritonavir. Chem Res Toxicol 24:2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice JB, Patel A, Scott AJ, Patel K, Thursz M, Lemoine M (2017) Prevalence and risk factors of nonalcoholic fatty liver disease in HIV-monoinfection. AIDS 31:1621–1632. [DOI] [PubMed] [Google Scholar]
- Menshawy A, Ismail A, Abushouk AI, Ahmed H, Menshawy E, Elmaraezy A, Gadelkarim M, Abdel-Maboud M, Attia A, Negida A (2017) Efficacy and safety of atazanavir/ritonavir-based antiretroviral therapy for HIV-1 infected subjects: a systematic review and meta-analysis. Arch Virol 162:2181–2190. [DOI] [PubMed] [Google Scholar]
- Mizutani T, Suzuki K (1996) Relative hepatotoxicity of 2-(substituted phenyl)thiazoles and substituted thiobenzamides in mice: evidence for the involvement of thiobenzamides as ring cleavage metabolites in the hepatotoxicity of 2-phenylthiazoles. Toxicol Lett 85:101–105. [DOI] [PubMed] [Google Scholar]
- Mohammadin S, Edger PP, Pires JC, Schranz ME (2015) Positionally-conserved but sequence-diverged: identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol 15:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan K (2003) Metabolic abnormalities in patients with HIV infection. J Int Assoc Physicians AIDS Care (Chic) 2:66–74. [DOI] [PubMed] [Google Scholar]
- Nijland HM, L’homme RF, Rongen GA, van Uden P, van Crevel R, Boeree MJ, Aarnoutse RE, Koopmans PP, Burger DM (2008) High incidence of adverse events in healthy volunteers receiving rifampicin and adjusted doses of lopinavir/ritonavir tablets. AIDS 22:931–935. [DOI] [PubMed] [Google Scholar]
- Pande P, Zhong XB, Ku WW (2020) Histone methyltransferase G9a regulates expression of nuclear receptors and cytochrome P450 enzymes in HepaRG Cells at basal level and in fatty acid induced steatosis. Drug Metab Dispos 48:1321–1329. [DOI] [PubMed] [Google Scholar]
- Schmitt C, Riek M, Winters K, Schutz M, Grange S (2009) Unexpected hepatotoxicity of rifampin and saquinavir/ritonavir in healthy male volunteers. Arch Drug Inf 2:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Lau AJ, Sherman MA, Chang TK (2013) Agonism of human pregnane X receptor by rilpivirine and etravirine: comparison with first generation non-nucleoside reverse transcriptase inhibitors. Biochem Pharmacol 85:1700–1711. [DOI] [PubMed] [Google Scholar]
- Shehu AI, Lu J, Wang P, Zhu J, Wang Y, Yang D, McMahon D, Xie W, Gonzalez FJ, Ma X (2019) Pregnane X receptor activation potentiates ritonavir hepatotoxicity. J Clin Invest 129:2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman KE, Rockstroh J, Thomas D (2015) Human immunodeficiency virus and liver disease: an update. Hepatology 62:1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley TLFourman LTFeldpausch MNPurdy JZheng IPan CSAepfelbacher JBuckless CTsao AKellogg A, et al. (2019) Effects of tesamorelin on non-alcoholic fatty liver disease in HIV: a randomised, double-blind, multicentre trial. Lancet HIV 6:e821–e830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski MS (2003) Hepatotoxicity associated with antiretroviral therapy containing HIV-1 protease inhibitors. Semin Liver Dis 23:183–194. [DOI] [PubMed] [Google Scholar]
- Sulkowski MS, Thomas DL, Chaisson RE, Moore RD (2000) Hepatotoxicity associated with antiretroviral therapy in adults infected with human immunodeficiency virus and the role of hepatitis C or B virus infection. JAMA 283:74–80. [DOI] [PubMed] [Google Scholar]
- Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, Guo Z, Bai T, Dong L, Wei C, et al. (2018) Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer 9:2631–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Chen S (2015) Epigenetic regulation of cytochrome P450 enzymes and clinical implication. Curr Drug Metab 16:86–96. [DOI] [PubMed] [Google Scholar]
- Tracy TSChaudhry ASPrasad BThummel KESchuetz EGZhong XBTien YCJeong HPan XShireman LM, et al. (2016) Interindividual variability in cytochrome P450-mediated drug metabolism. Drug Metab Dispos 44:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verna EC (2017) Non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in patients with HIV. Lancet Gastroenterol Hepatol 2:211–223. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Chen S, Wang Y, Wang X, Yan L, Yang K, Zhong XB, Han S, Zhang L (2021a) The long noncoding RNA hepatocyte nuclear factor 4α antisense RNA 1 negatively regulates cytochrome P450 enzymes in Huh7 cells via histone modifications. Drug Metab Dispos 49:361–368. [DOI] [PubMed] [Google Scholar]
- Wang P, Liu G, Nie Y, Han S, Li J, Zhong XB, Zhang L (2019a) Epigenetic memory is involved in the persistent alterations of drug-processing genes in adult mice due to PCN-activated PXR during early life. Toxicol Sci DOI: 10.1093/toxsci/kfz177 [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Wang X, Wei L, Yang J, Wang Y, Chen S, Yang K, Meng X, Zhang L (2021b) DNA methylation determines the regulation of pregnane X receptor on CYP3A4 expression. Clin Exp Pharmacol Physiol 48:250–259. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yan L, Liu J, Chen S, Liu G, Nie Y, Wang P, Yang W, Chen L, Zhong X, et al. (2019b) The HNF1α-regulated lncRNA HNF1α-AS1 is involved in the regulation of cytochrome P450 expression in human liver tissues and Huh7 cells. J Pharmacol Exp Ther 368:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Wang Y, Liu J, Nie Y, Zhong XB, Kan Q, Zhang L (2017) Alterations of histone modifications contribute to pregnane X receptor-mediated induction of CYP3A4 by rifampicin. Mol Pharmacol 92:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Ma L, Humphreys WG, Zhu M (2008) Rapid screening and characterization of drug metabolites using a multiple ion monitoring-dependent MS/MS acquisition method on a hybrid triple quadrupole-linear ion trap mass spectrometer. J Mass Spectrom 43:1364–1375. [DOI] [PubMed] [Google Scholar]
- Zhou SF (2008) Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab 9:310–322. [DOI] [PubMed] [Google Scholar]
- Zhou X, Yin C, Dang Y, Ye F, Zhang G (2015) Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep 5:11516. [DOI] [PMC free article] [PubMed] [Google Scholar]