Abstract
Interleukin-10 (IL-10) is an immunoregulatory cytokine that possesses both anti-inflammatory and immunostimulatory properties and is frequently dysregulated in human disease. Here we used a structure-based approach to deconvolute IL-10 pleiotropy by determining the 3.5-Å cryo-EM structure of the IL-10 receptor signaling complex. The stelliform, hexameric structure shows how IL-10 and IL-10Rα form a composite surface to engage the shared signaling receptor IL-10Rβ, enabling the design of IL-10 partial agonists. IL-10 variants with a range of IL-10Rβ-binding strengths uncovered substantial differences in signaling and gene expression response thresholds across immune cell populations, providing a means of manipulating IL-10 cell-type selectivity. Some variants displayed myeloid-biased activity, suppressing monocyte and macrophage activation without stimulating inflammatory CD8+ T cell functions, thereby uncoupling the major opposing functions of IL-10. These results provide a structural and mechanistic blueprint for tuning the pleiotropic actions of IL-10.
One-sentence summary:
Cryo-EM structure of the IL-10 receptor complex enables design of myeloid-selective agonists.
Inflammation is essential for protecting organisms against infection and promoting tissue homeostasis (1). However, excessive immune cell activation can damage bystander tissues and result in organ malfunction, chronic inflammation, and autoimmune disease (2). Interleukin-10 (IL-10) is an important anti-inflammatory cytokine that plays a central role in regulating and terminating inflammatory responses (3, 4). A variety of immune cells produce IL-10 during inflammation, which in turn exerts potent anti-inflammatory effects primarily by inhibiting cytokine production and antigen presentation by activated myeloid cells (4-7). Consistent with this role, genetic loss of either IL-10 or the IL-10 receptor results in severe inflammatory bowel disease (IBD) in both mice and humans, and dysregulated IL-10 function has also been associated with cancer and chronic inflammation (4, 6-9).
Despite these important anti-inflammatory functions, IL-10 is also highly pleiotropic, eliciting diverse and seemingly opposed biological effects (10). Most notably, IL-10 potentiates inflammatory functions of activated CD8+ T-cells, enhancing production of the pro-inflammatory cytokine interferon (IFN)-γ as well as cytolytic factors such as granzyme B (11-13). Indeed, administration of exogenous IL-10 has been found to elevate serum IFN-γ levels in humans, possibly counteracting its anti-inflammatory effects (14).
IL-10 is a secreted homodimer that initiates signaling in receiver cells by engaging two copies of a heterodimeric receptor complex consisting of a high-affinity, private receptor subunit, IL-10Rα, and a low-affinity, shared receptor subunit, IL-10Rβ (4). IL-10 drives the dimerization of IL-10Rα and IL-10Rβ, resulting in the phosphorylation and activation of the transcription factor signal transducer and activator of transcription (STAT) 3 and to a lesser extent STAT1, which mediate the diverse functional effects of IL-10. Although there is significant interest in using IL-10 therapeutically, the mechanisms underlying IL-10 functional redundancy and plasticity are not fully understood. This is due, in part, to a lack of structural information for the complete IL-10 receptor signaling complex, including IL-10Rβ (15), which would enable interrogation of this axis with designed variants. The lack of structural information is principally due to the extremely low-affinity of IL-10 for IL-10Rβ, which precludes stable complex assembly in vitro (15).
Here, we engineered a stabilized IL-10 receptor signaling complex and determined its structure to a resolution of 3.5-Å by cryogenic electron microscopy (cryo-EM). The structure revealed the molecular basis for receptor engagement by IL-10, and thereby enabled the design of IL-10 variants with altered cell-type selectivity. These results uncovered a surprisingly wide range of IL-10 response thresholds across immune cell types and provide a mechanistic framework for uncoupling the anti-inflammatory and immunostimulatory actions of IL-10.
Results
Assembly and cryo-EM structure of the IL-10 signaling complex
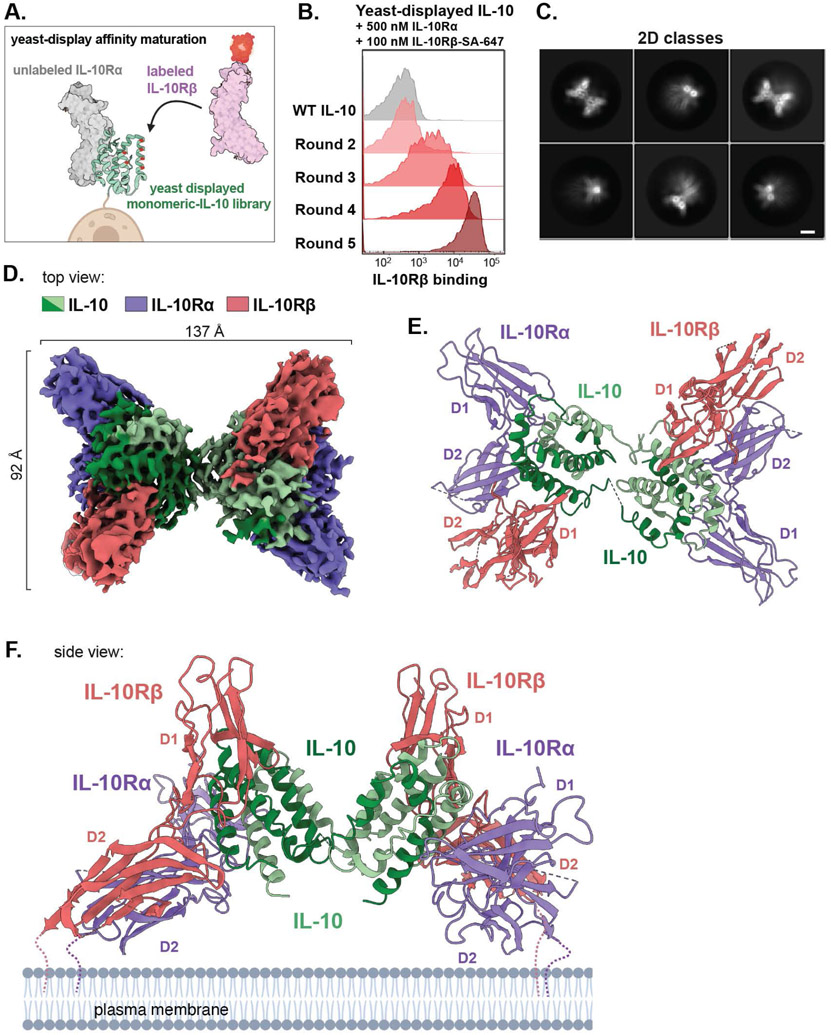
In order to stabilize assembly of the ternary IL-10–IL-10Rα-IL-10Rβ complex for structural studies, we used yeast-surface display to affinity-mature IL-10 for IL-10Rβ (Fig. 1A) (16). We first expressed a monomeric version of IL-10 (mono-IL-10) (17) on the surface of yeast and verified its ability to bind to fluorescently labelled extracellular domain (ECD) of IL-10Rα, confirming it was functionally displayed (fig. S1A). We then designed a site-directed mutagenesis library targeting two helices in IL-10 predicted to be near the IL-10Rβ-binding site (18), and carried out sequential rounds of in vitro evolution (Fig. 1B; fig. S1, B and C). After five rounds of selection, the library converged on three unique clones, each of which exhibited substantially improved binding to IL-10Rβ (fig. S1D and S2A). Clone 5.1 (“super-10”) contained four mutations relative to wild-type (WT) IL-10 and bound to IL-10Rβ in both the presence and absence of IL-10Rα, with approximate dissociation constants (KD) of 30 nM and 70 nM, respectively (fig. S2, B and C). Moreover, super-10 formed a stable 2:2:2 complex with IL-10Rα and IL-10Rβ that co-migrated over gel filtration, enabling purification of the homogenous IL-10 receptor complex (fig. S2D).
Fig. 1. Assembly and cryo-EM structure of the IL-10 signaling complex.
(A) Schematic of yeast display-based affinity maturation of IL-10. (B) Histograms of IL-10Rβ-binding by mono-IL-10 over iterative rounds of directed evolution (SA-647, Streptavidin-Alexa Fluor 647 conjugate). (C) Representative cryo-EM 2D class averages of the stabilized IL-10–IL-10Rα–IL10Rβ complex (scale bar = 5 nm). (D) Segmented density map of the IL-10 receptor complex resolved to 3.5-Å. Map threshold used in ChimeraX set to 0.25. (E, F) Top and side views of the IL-10 receptor complex with IL-10 in green, IL-10Rα in purple, and IL-10Rβ in salmon (PDB ID: 6X93).
We then carried out single-particle cryo-EM analysis of the stabilized IL-10 receptor complex and resolved the EM density map to 3.5-Å resolution (Fig. 1, C and D; fig S3, A to F). This enabled docking and refinement of crystal structures of IL-10, IL-10Rα, and IL-10Rβ (PDB IDs: 1Y6K and 3LQM) (15, 18), yielding a molecular model of the hexameric complex (Fig. 1, E and F). Although flexibility between IL-10 subunits resulted in poorly resolved density for one ternary subcomplex, refining the map without imposing C2-symmetry yielded a high-quality map of one IL-10–IL-10Rα-IL-10Rβ trimeric subcomplex, with local resolution ranging from 3.1 Å at the ligand-receptor interface to ~5 Å at the receptor periphery (fig. S3, D and F, and fig. S4, A to C).
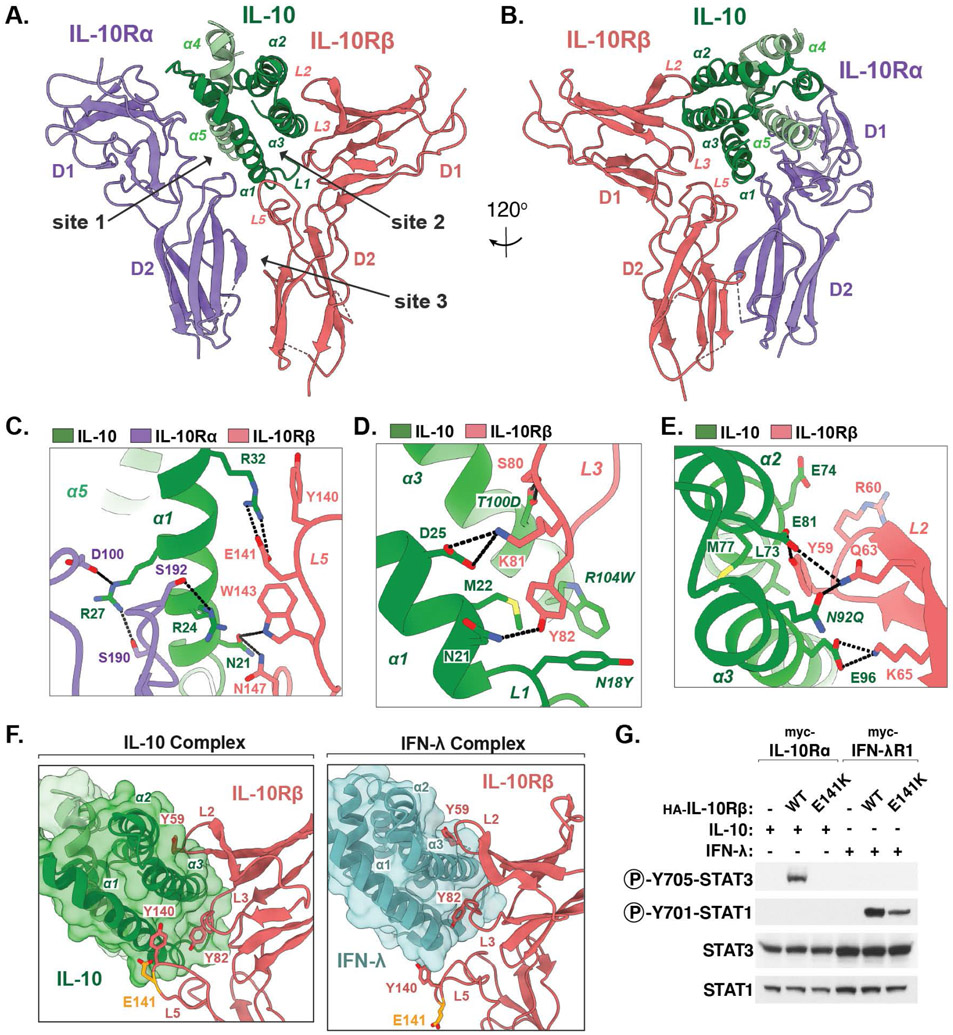
The complete IL-10 receptor complex forms an elongated star shape with two-fold symmetry centered around the domain-swapped IL-10 homodimer, which bridges two copies of the IL-10Rα–IL-10Rβ receptor unit (Fig. 1, D to F, movie S1). IL-10 and IL-10Rα form a composite interface to engage IL-10Rβ, forming three distinct contact sites (Fig. 2, A and B). At site 1, IL-10 engages IL-10Rα via an interface made up of α-helices (α)-1 and α5, each contributed by distinct IL-10 monomers, as described previously (15) (Fig. 2A). By contrast, the IL-10Rβ interface at site 2 comprises α1, α2, and α3, contributed by a single IL-10 monomer and burying 1014 Å2 of the IL-10 surface (Fig. 2, A and B). IL-10 α3 is clasped by loops (L)-2 and L3 in the D1 domain of IL-10Rβ (Fig. 2A), whereas α1 of IL-10 is positioned at the junction between IL-10Rα and IL-10Rβ, simultaneously engaging both receptor subunits (Fig. 2, A and C). Finally, site 3 comprises the receptor “stem” contacts between the D2 domains of IL-10Rα and IL-10Rβ, forming a relatively small interface burying only 259 Å2 (Fig. 2, A and B).
Fig 2: Structural basis for IL-10Rβ engagement by IL-10.
(A, B) Two views of the IL-10–IL-10Rα–IL-10Rβ ternary sub-complex, with IL-10 in green, IL-10Rα in purple, and IL-10Rβ in salmon. (C-E) Close-up views of the IL-10–IL-10Rβ binding interface. Hydrogen bonds and salt-bridges are shown as black dashed-lines. Mutated residues in affinity matured super-10 are italicized. (F) Corresponding views of the IL-10–IL-10Rβ interface and the IFN-λ–IL-10Rβ interface (PDB ID: 5T5W). The IBD-associated residue Glu141 is shown in yellow. (G) Immunoblot of lysates prepared from HEK-293T cells transiently expressing the indicated receptor constructs and stimulated with IL-10 or IFN-λ for 20 min.
Structural basis for IL-10Rβ engagement by IL-10
Inspection of the IL-10–IL-10Rβ binding interface revealed that the central contact is formed by 10Rβ-Tyr82 in L3, which is inserted between IL-10 α1 and α3, facilitating Van der Waals contacts primarily with Met22 in α1 of IL-10 (Fig. 2D). This is consistent with previous mutagenesis and modeling studies suggesting that 10Rβ-Tyr82 is required for the interaction between IL-10Rβ and IL-10 (18). This key hydrophobic contact is surrounded by a network of polar and electrostatic interactions, mediated in part by IL-10 residues Asn21 and Asp25 in α1, which engage 10Rβ-Gln63 and 10Rβ-Lys81, respectively (Fig. 2D). Directly above L3, 10Rβ-Tyr59 in loop L2 similarly inserts between IL-10 α2 and α3 engaging a several residues in α2, whereas Glu96 in α3 of IL-10 engages 10Rβ-Lys65 (Fig. 2E).
The increased affinity of super-10 for IL-10Rβ appears to result primarily from the N18Y and R104W mutations, both of which are adjacent to Met22 and enhance the hydrophobicity of this central contact site accommodating 10Rβ-Tyr82 (Fig. 2D). Meanwhile, the N92Q and T100D mutations facilitate hydrogen bond contacts with 10Rβ-Gln63 and 10Rβ-Ser80, respectively, likely enhancing contacts present in the WT IL-10 complex (Fig. 2, D and E).
The hexameric IL-10 receptor complex topologically resembles the 2:2:2 IFN-γ complex (19), with the notable difference that in IL-10, the two subunits of the cytokine dimer are twisted, causing an approximately 90° rotation of the receptor pairs relative to each other (fig. S5A). This results in a significantly greater distance between receptor dimers, as well as a “flatter” profile of the overall complex relative to the IFN-γ complex (fig. S5A). Each half of the complex resembles the 1:1:1 ternary complex structure of IFN-λ bound to IFN-λR1 and IL-10Rβ, providing insights into the structural basis for receptor sharing between these two cytokines (20) (fig. S5B). In both structures, L2 and L3 of IL-10Rβ provide shape complementarity to accommodate α3 of the cytokine ligand, with aromatic residues 10Rβ-Tyr82 and 10Rβ-Tyr59 forming conserved hydrophobic anchor points surrounded by ligand specific electrostatic contacts (Fig. 2F) (20).
The IL-10 and IFN-λ complex structures differ however with respect to the conformation of L5 in IL-10Rβ, which forms a “thumb”-like protrusion that extends along the front surface of IL-10 helix α1, but forms a more minor contact with the bottom side of IFN-λ (Fig. 2, C and F). A point mutation in 10Rβ-Glu141 (E141K) within L5 was recently identified in very early onset IBD (21). Although 10Rβ-Glu141 does not contact the cytokine ligand in the context of the IFN-λ receptor, our structure showed that the negatively charged 10Rβ-Glu141 is positioned between the positively charged side chains of IL-10 residues Arg24 and Arg32 forming an apparent electrostatic contact (Fig. 2C). Consistent with this, HEK-293T cells transiently over-expressing the disease associated IL-10Rβ mutant E141K retained responsiveness to IFN-λ but not IL-10, suggesting that this mutation selectively disrupts IL-10 binding to this shared receptor subunit (Fig. 2G).
Tuning affinity for IL-10Rβ reveals differential IL-10 signaling plasticity across cell types
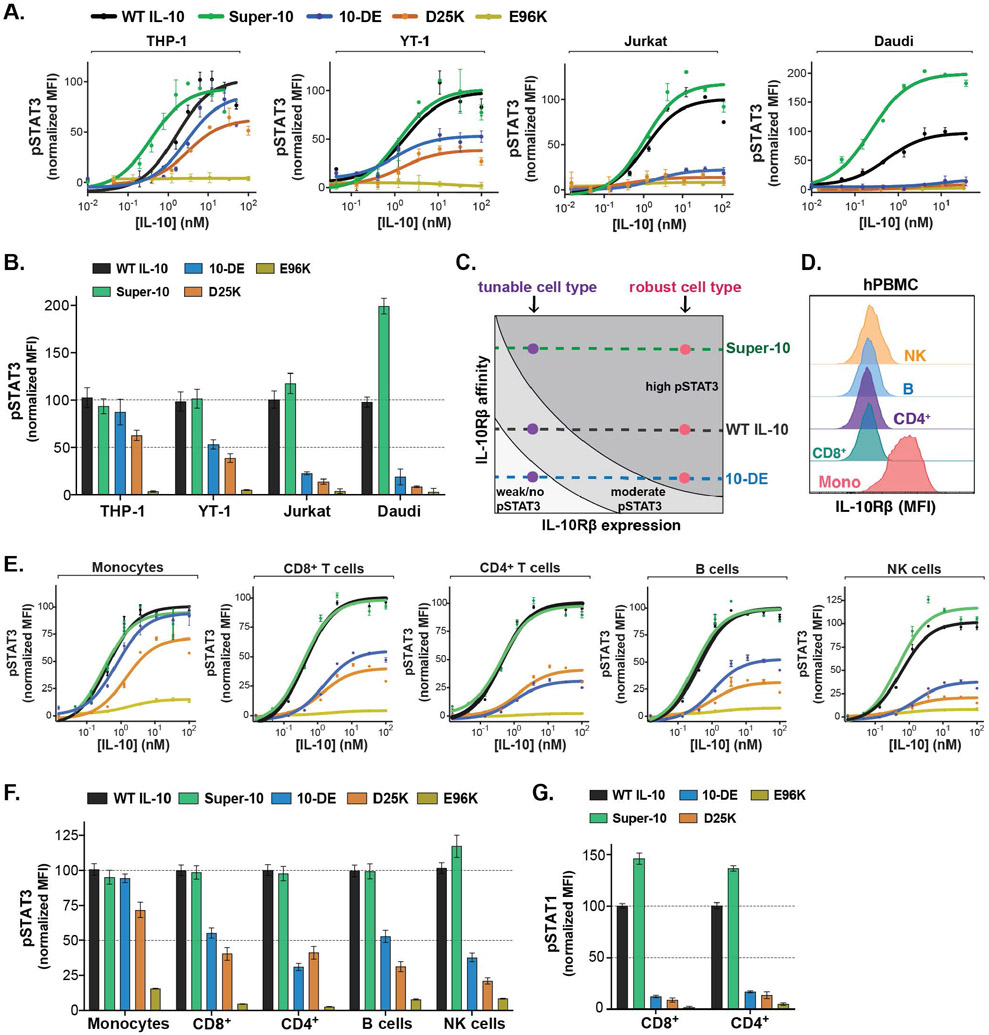
To assess the importance of the observed IL-10–IL-10Rβ contacts for IL-10 signaling, we designed a series of mutations in WT IL-10 predicted to weaken the interaction with IL-10Rβ, and then tested their ability to initiate signaling via the phosphorylation of STAT3. Mutation of Asp25 in IL-10 to alanine (D25A) or lysine (D25K) resulted in a substantial reduction in STAT3 activation in B cell-derived Daudi cells, as compared to WT IL-10 (Fig. S6A). Similarly, the IL-10 mutants E96K, D25A/E96A, N21A/R104A, and D25A/N21A/R104A all elicited negligible STAT3 phosphorylation on these cells (fig. S6A). Remarkably, however, all of these mutants with the exception of E96K elicited strong STAT3 activation in monocyte-derived THP-1 cells, with the D25A/E96A (“10-DE”) and D25K mutants showing the greatest difference in activity between the THP-1 and Daudi cell lines (fig. S6A).
To generate a more complete picture of how altering the stability of the IL-10–IL-10Rβ interaction influences IL-10 signaling across cell types, we compared the activity of WT IL-10, super-10, and the partial agonists D25A/E96A (10-DE) and D25K across a panel of four cell lines representing different classes of immune cells. Whereas 10-DE and D25K elicited no activity in either Daudi or T cell-derived Jurkat cells, they elicited ~50% activity in natural killer (NK) cell-derived YT-1 cells and nearly full STAT3 activation in myeloid THP-1 cells (Fig. 3, A-C; table S2). By contrast, super-10 showed substantially higher Emax relative to WT IL-10 in both Daudi and Jurkat cells, but similar signaling as WT IL10 in YT-1 and THP-1 cells (Fig. 3, A-C; table S2). Thus, the ratio of super-10 to 10-DE STAT3 Emax varied considerably, from ~1:1 in THP-1 cells to over 20:1 in Daudi cells, representing substantial differences in signaling plasticity between these cell lines.
Fig. 3. Tuning affinity for IL-10Rβ reveals differential IL-10 signaling plasticity across cell types.
(A) Dose-response curves for phospho-STAT3 in cell lines stimulated with WT or mutant IL-10, analyzed by flow cytometry. Data are shown as a percent of maximal WT IL-10 mean fluorescent intensity (MFI) (n≥2, N≥3). (B) Normalized Emax values for phospho-STAT3 calculated from sigmoidal dose-response curves (mean ± SEM). (C) Diagram illustrating how IL-10Rβ expression and ligand affinity influence maximal STAT3 activation. (D) Representative histograms showing IL-10Rβ surface expression in human PBMCs analyzed by flow cytometry. (E) Dose-response curves for phospho-STAT3 in human PBMCs stimulated with WT or mutant IL-10, analyzed by flow cytometry. Data are shown as a percent of maximal WT IL-10 mean fluorescent intensity (MFI) (n≥2, N=3). (F) Normalized Emax values for phospho-STAT3 calculated from sigmoidal dose-response curves (mean ± SEM). (G) Phospho-STAT1 activation in primary immune cells treated with 10 nM of WT or mutant IL-10, analyzed by flow cytometry. (mean ± SEM, n=3, N=2).
Comparison of IL-10Rα and IL-10Rβ surface expression across these diverse cell lines revealed that the level of IL-10Rα and IL-10Rβ correlated well with the relative strength of STAT3 signaling induced by the engineered IL-10 variants (fig. S6, D and E). Thus, the extent of STAT3 activation at saturating IL-10 concentration can be modulated by a combination of receptor expression and ligand affinity (Fig. 3C). The finding that super-10 can increase the signaling Emax relative to WT IL-10 on certain cell lines suggests that assembly of the IL-10 signaling complex on these cells is limited by low IL-10R expression, which can be compensated for by enhancing the affinity of the IL-10–IL-10Rβ interaction. Conversely, other cell types can maintain robust STAT3 responses even to the low-affinity IL-10 partial agonists such as 10-DE and D25K, due to their elevated IL-10R expression (Fig. 3C). This observation suggests the presence of a functional window that can be exploited with engineered agonists to achieve narrowed IL-10 cell-type specificity.
Analysis of primary human peripheral blood mononuclear cells (PBMCs) revealed high IL-10Rα and IL-10Rβ expression on primary monocytes, and low but present expression on NK, B, and T cells, suggesting that monocytes may exhibit more robust IL-10 signaling responses (Fig. 3D). Indeed, despite vast differences in affinity for IL-10Rβ, 10-DE, D25K, and super-10 all induced similar levels of STAT3 activation as WT IL-10 in primary monocytes (Fig. 3, E and F; table S3). However, 10-DE and D25K exhibited substantially reduced STAT3 activation in T cells, B cells, and NK cells relative to WT IL-10 (Fig. 3, E and F; table S3). By contrast, super-10 elicited stronger STAT3 activation than WT IL-10 in NK cells, and substantially stronger STAT1 activation in primary CD4+ and CD8+ T cells, whereas 10-DE and D25K induced negligible STAT1 phosphorylation in these cells (Fig. 3, E to G, table S3). Thus, compared to WT IL-10, the partial agonists 10-DE and D25K exhibit pronounced monocyte-biased signaling activity in primary human PBMCs, whereas the high-affinity super-10 variant displayed enhanced activity only on T cells and NK cells.
Engineered IL-10 variants retain anti-inflammatory functions of IL-10
IL-10 elicits both anti-inflammatory and immunostimulatory effects in vivo due to its ability to initiate signaling in multiple immune cell types (4). The anti-inflammatory effects of IL-10 are primarily due to suppression of monocyte and macrophage activity, whereas IL-10 promotes pro-inflammatory IFN-γ production via stimulation of CD8+ T-cells (10). We therefore asked whether natural differences in IL-10 signaling plasticity could be exploited to tune the functional pleiotropy of IL-10 in primary immune cell populations.
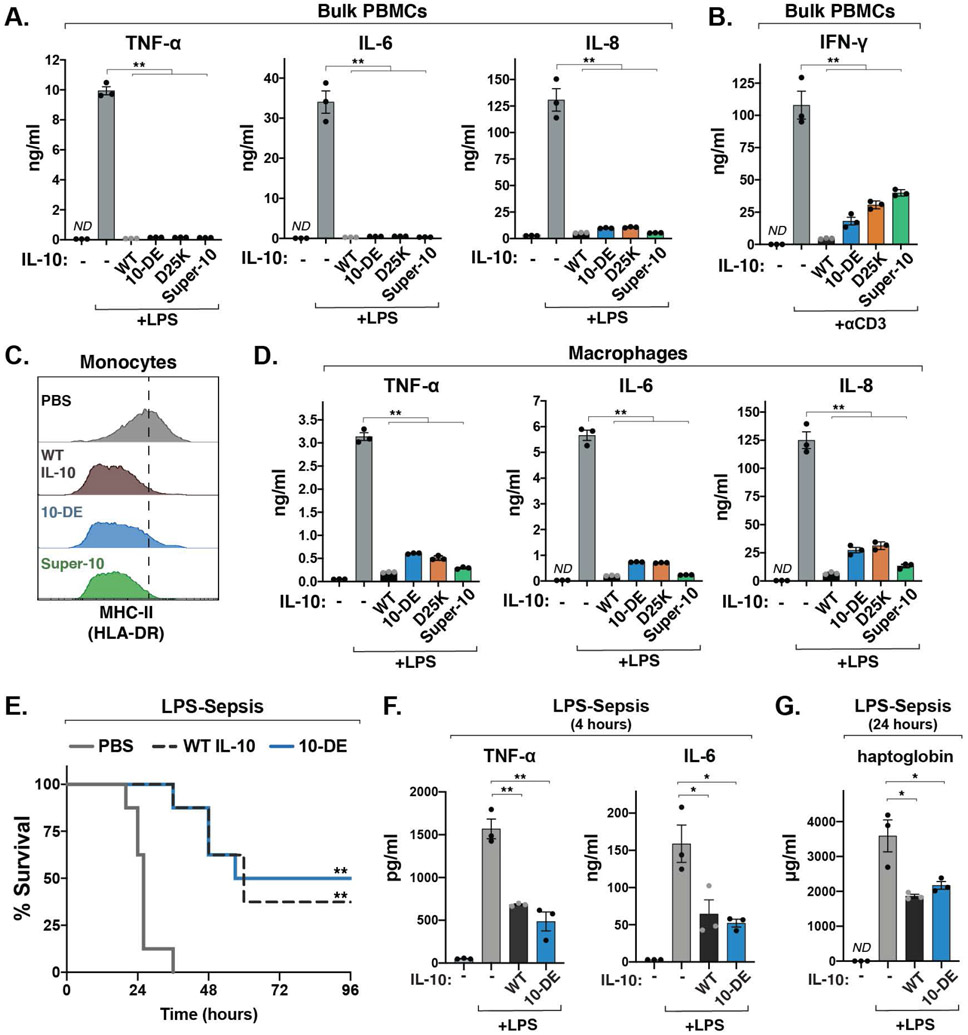
To test whether the level of STAT3 activation induced by 10-DE and D25K in monocytes was sufficient to drive anti-inflammatory functions of IL-10, we stimulated bulk human PBMCs with bacterial lipopolysaccharide (LPS) in the presence or absence of these variants. Both 10-DE and D25K inhibited production of the pro-inflammatory cytokines IL-1β, IL-6, and TNF-α, as well as the chemokine IL-8, to a similar extent as WT IL-10 and super-10 (Fig. 4A; fig. S8A). In addition, 10-DE and D25K retained the ability to suppress IFN-γ production in bulk PBMCs activated with anti-CD3 antibodies (Fig. 4B), and also downregulated surface expression of MHC class II and the co-stimulatory ligand CD86 on peripheral monocytes (Fig. 4C; fig. S8, B and C). Importantly, 10-DE and D25K also strongly abrogated LPS-induced IL-6, IL-8, and TNF-α production in isolated monocyte-derived macrophages (Fig. 4D), the key target for IL-10 in autoimmune and autoinflammatory diseases (6, 10).
Fig. 4. Engineered IL-10 variants retain anti-inflammatory functions of IL-10.
(A) Levels of IL-6, IL-8, and TNF-α from bulk human PBMCs treated with LPS and the indicated IL-10 variant, measured by ELISA. (mean ± SEM, n=3, N=3, **P<0.01, two-sided Student’s t test; ND, not detectable). (B) Levels of IFN-γ from bulk PBMCs stimulated with anti-CD3 antibody for 72 hours, alone or in combination with the indicated IL-10 variants, measured by ELISA. (mean ± SEM, n=3, N=3, **P<0.01, two-sided Student’s t test). (C) Representative histograms of MHC class II surface expression on human peripheral monocytes activated with LPS alone or in combination with the indicated IL-10 variant, analyzed by flow cytometry (n=5, N=2). (D) Levels of IL-6, IL-8, and TNF-α produced by primary human monocyte-derived macrophages stimulated with LPS, measured by ELISA. (mean ± SEM, n=3, N=3, **P<0.01, two-sided Student’s t test). (E) Survival after intraperitoneal injection of LPS (15 mg/kg) in combination with PBS, WT IL-10 or 10-DE (n=8 mice, N=2, **P <0.01, log-rank Mantel-Cox test). (H, I) Levels of TNF-α, IL-6, and haptoglobin in mouse serum following injection of LPS (4 mg/kg) and the indicated IL-10 variant, measured by ELISA (mean ± SEM, n=3, N=2, *P<0.05. **P<0.01, two-sided Student’s t test).
Finally, we assessed whether the myeloid-biased activity of 10-DE was sufficient to suppress systemic inflammation in a mouse model of LPS-induced sepsis. Importantly, a single injection of 10-DE significantly promoted survival in this model, to the same extent as WT IL-10 (Fig. 4E). Moreover, 10-DE blunted the elevation of TNF-α, IL-6, and haptoglobin, key markers of systemic inflammation (Fig. 4, F and G).
Myeloid-biased IL-10 variants have reduced capacity to promote inflammatory T cell functions
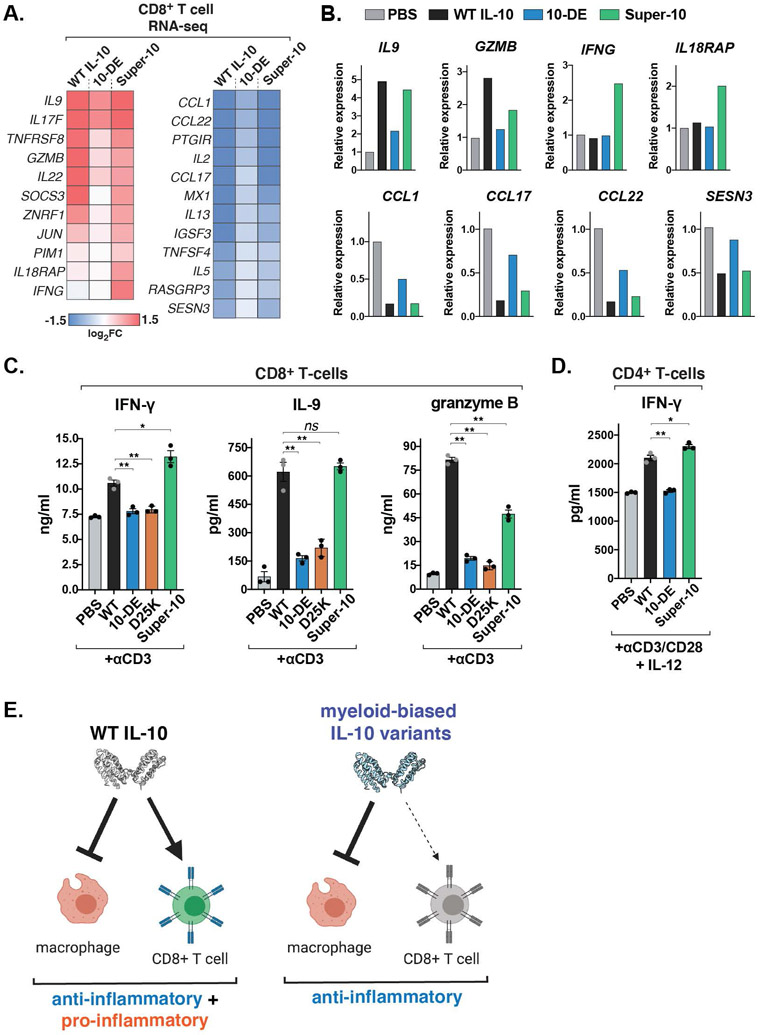
IL-10 is also known to induce pro-inflammatory IFN-γ production in some contexts, and recent studies have suggested that this occurs primarily through the potentiation of CD8+ T-cell activity (11-14). In order to analyze the effects of our engineered IL-10 variants on CD8+ T-cells, we first performed RNA sequencing (RNA-seq) analysis of TCR-stimulated CD8+ T-cells cultured with WT IL-10, super-10, or 10-DE for twenty-four hours (fig. S9, A and B). In this acute treatment setting, IL-10-induced genes were enriched for pro-inflammatory factors, such as IL9, IL17F, and GZMB, whereas genes downregulated by IL-10 were enriched for chemokines including CCL1, CCL17, and CCL22 (Fig. 5, A and B).
Fig. 5. Myeloid-biased IL-10 variants have reduced capacity to promote inflammatory T cell functions.
(A) Heatmaps of differentially expressed genes in activated CD8+ T cells treated with the indicated IL-10 variants for 24 hours, analyzed by RNA-seq (n=2 biological replicates). (B) Fold change of select IL-10-regulated genes in activated CD8+ T cells treated with the indicated IL-10 variants for 24 hours, analyzed by RNA-seq (n=2 biological replicates). (C) Levels of IFN-γ, IL-9, and granzyme B from activated human CD8+ T cells cultured with the indicated IL-10 variants, measured by ELISA. (mean ± SEM, n=3, N=3, ns P>0.05, *P<0.05, **P<0.01, two-sided Student’s t test). (D) Levels of IFN-γ from isolated CD4+ T cells polarized to Th1 cells in the presence or absence of the indicated IL-10 variant for 5 days, measured by ELISA (mean ± SEM, n=3, N=2 *P<0.05. **P<0.01, two-sided Student’s t test). (E) Schematic depicting myeloid-biased IL-10 agonists decoupling the pro- and anti-inflammatory functions of IL-10.
The expression changes induced by super-10 largely matched the pattern of observed with WT IL-10, albeit with less upregulation of some IL-10 target genes, such as GZMB, IL22, and SOCS3, and enhanced induction of others such as IFNG and IL18RAP (Fig. 5, A and B), potentially reflecting the increased STAT1 activation elicited by super-10 compared to WT IL-10 in these cells (Fig. 3G). By contrast, the partial agonist 10-DE elicited a pattern of substantially weaker transcriptional changes across both upregulated and downregulated IL-10 target genes, including reduced expression of pro-inflammatory genes such as GZMB, IL9, IL17F, and IL22 (Fig. 5, A and B). Consistent with this, both 10-DE and D25K failed to potentiate granzyme B, IFN-γ, and IL-9 production by activated CD8+ T cells, in contrast to WT IL-10 and super-10 (Fig. 5C). Similarly, WT IL-10 and super-10 also enhanced the production of IFN-γ by Th1-polarized CD4+ T cells, whereas 10-DE had no effect (Fig. 5D). Thus, although the myeloid-biased IL-10 variants retained the ability to suppress monocyte and macrophage activation, they showed substantially reduced capacity to potentiate inflammatory T cell activity, thereby uncoupling the major anti-inflammatory and pro-inflammatory functions of IL-10 (Fig. 5E).
Discussion
Together, these data reveal the structural basis for receptor engagement by IL-10, providing important insights into the mechanisms underlying IL-10 signaling plasticity and functional pleiotropy. Most notably, the structure shows in molecular detail how IL-10 interacts with its shared, low-affinity receptor subunit IL-10Rβ, enabling the structure-guided design of mutations that perturb this interface. Analysis of these engineered IL-10 variants revealed that the plasticity of IL-10 signaling varied significantly across cell types, correlating with the level of IL-10Rβ expression. Moreover, these differences could be exploited with engineered ligands by modulating the affinity of IL-10 for IL-10Rβ, resulting in altered cell type specificity.
These results suggest a model in which a limiting factor for IL-10 signaling is formation of the ternary IL-10–IL10Rα–IL-10Rβ complex, which can be influenced both by the affinity of the IL-10–IL-10Rβ interaction, as well as the abundance of IL-10Rβ on the cell surface (Fig. 3C). As a result, high expression of IL-10Rβ can compensate for low ligand affinity, resulting in robust STAT3 activation across a range of ligand affinities, as observed in myeloid cells (Fig. 3, A-F). Conversely, cells with low IL-10Rβ expression are far more sensitive to reductions in the affinity of IL-10 for IL-lORb, but they also show enhanced signaling beyond that induced by WT IL-10 in response to increased IL-10Rβ affinity, as in the case of super-10. These features were observed in multiple lymphocyte populations, in terms of phosphorylation of both STAT3 and, to an even greater extent, STAT1 (Fig. 3, A-G).
Our results also showed that the myeloid-biased signaling induced by engineered IL-10 partial agonists can uncouple important aspects of IL-10 function. The differential IL-10 receptor expression between myeloid and lymphocyte populations may therefore represent a natural mechanism to provide functional specificity to otherwise pleiotropic IL-10 responses, a feature that can be exploited using engineered agonists as shown here. The partial agonists 10-DE and D25K retained the anti-inflammatory functions of IL-10, consistent with their capacity to activate STAT3 in these cells to a similar extent as WT IL-10 (Fig. 3, E and F; Fig. 4, A-G). By contrast, these variants had significantly diminished capacity to potentiate inflammatory IFN-γ production by both CD4+ and CD8+ T cells, consistent with their reduced ability to activate both STAT3 and STAT1 in these cells (Fig. 3, F and G; Fig. 5A-D). Meanwhile, super-10 elicited stronger induction of IFN-γ by both CD4+ and CD8+ T cells relative to WT IL-10, despite eliciting a similar STAT3 response (Fig. 3F; Fig. 5, A-D). This finding is likely explained by the observation that super-10 elicited substantially stronger STAT1 activation in these cells (Fig. 3G), consistent with STAT1 having been suggested to mediate the IFN-γ-inducing effects of IL-10 (12).
The natural ability of IL-10 to potently inhibit the production of pro-inflammatory cytokines like IL-6 and TNF-α in vivo makes it a promising therapeutic candidate for the treatment of several autoimmune diseases, including Rheumatoid Arthritis, IBD, and Type 1 diabetes (10). However, the observation that IL-10 administration elevates IFN-γ levels in humans has presented a barrier to the use of IL-10 in these contexts (14). The ability of engineered IL-10 partial agonists to suppress inflammatory monocyte and macrophage activation without stimulating IFN-γ production by CD4+ and CD8+ T cells therefore has significant implications for the clinical use of IL-10 in these settings, providing a blueprint for unlocking the full therapeutic potential of IL-10.
Materials and Methods:
Protein production and purification
For yeast-binding studies, affinity maturation, and SPR, the ECDs of human IL-10Rα (residues 20-235) and IL-10Rβ (residues 20-220) were cloned into the pAcGP67a baculoviral vector with an N-terminal gp67 signal peptide, C-terminal 3C cleavage site followed by a biotin-acceptor peptide tag (BAP tag, GLNDIFEAQKIEW) and 6xHis tag. The proteins were purified from the supernatant of baculovirus infected Hi5 cells 72 hours after infection and purified with Ni-NTA resin (Qiagen) followed by size-exclusion chromatography (SEC) on a Superdex 200 column (GE). IL-10Rβ ECD was site-specifically biotinylated at the C-terminal BAP tag using BirA ligase and re-purified by SEC.
For structural studies, a glyco-mutant version the IL-10Rβ ECD (N49Q, N68Q, N102Q, N161Q) (18) was cloned into the pAcGP67a baculoviral vector with an N-terminal gp67 signal peptide and C-terminal 6xHis tag, expressed and purified as described above. Affinity matured IL-10 (super-10, clone 5.1) and the IL-10Rα ECD were cloned into the pD649 mammalian expression vector. The proteins were purified from the supernatant of Expi-293 cells (Thermo) with Ni-NTA resin (Qiagen) followed by SEC on a Superdex 200 column (GE) in HEPES buffered saline (HBS). Following SEC, the individual proteins were incubated overnight at a 1:1:1.2 molar ratio of IL-10:IL-10Rα:IL-10Rβ and re-purified by SEC on a Superdex 200 column (GE). For signaling and functional experiments, IL-10 variants were cloned into the pD649 mammalian expression vector and purified from Expi-293 cells as described above.
Yeast display, library assembly, and affinity maturation
A monomeric IL-10 variant (17) containing a C-terminal Myc-tag was displayed on the surface of Saccharomyces cerevisiae strain EBY100 as a C-terminal fusion to Aga2 using the pCT302 vector. A mutant mono-IL-10 library containing seven randomized residues at the predicted IL-10Rβ contact site was generated by primer assembly PCR using degenerate codons (fig. S1D). Electroporation, rescue and expansion of the yeast library were performed as described previously (16).
Library selection was conducted as described previously with small modifications (16). Briefly, the initial selections (rounds 1-4) were conducted using magnetic-activated cell sorting (MACS). In round 5, library selection was performed using two-color fluorescence-activated cell sorting (FACS) to normalize apparent affinity to protein expression on the cell surface. Alexa Fluor 647+ and Alexa Fluor 488+ yeast were purified using a SH800S Cell Sorter (Sony Biotechnology). ~4×106 cells from the post-round 5 library were used to extract library DNA using the Zymoprep Yeast Plasmid Miniprep II Kit (Zymo Research), according to the manufacturer’s instructions. The extracted DNA was transformed into DH5α Escherichia coli and plated to sequence the individual clones. To measure relative binding affinities, individual clones were displayed on the yeast surface and incubated with 500 nM IL-10Rα and increasing concentrations of biotinylated IL-10Rβ. The cells were then stained with streptavidin-Alexa Fluor 647 for 15 min. and analyzed on a CytoFLEX Flow Cytometer (Beckman Coulter).
Cryo-EM specimen preparation and data collection
The hexameric IL-10–IL-10Rα–IL-10Rβ complex was concentrated to 12 mg/ml and mixed with 0.01% w/v fluorinated octyl maltoside (fOM, Anatrace, O310F) immediately prior to specimen preparation. 2.5 microliters (μl) of the sample was applied to glow-discharged 300 mesh gold grids (Quantifoil R1.2/1.3). Grids were glow-discharged using an easiGlow (PELCO) at 15 mA and 0.39 mBar for 40 s. Cryo-EM data were collected at the CryoEM facility at the HHMI Janelia Research Campus. Images were collected on a Titan Krios operated at 300 kV equipped with a Gatan imaging filter and K3 camera in correlated double sampling super-resolution mode at a nominal magnification of 81,000X, corresponding to a physical pixel size of 1.078 A. The dataset was collected using SerialEM (22) with defocus range set between −0.8 and −2.0 μm and beam-image shift to collect nine movies from nine holes per stage shift and focusing.
Cryo-EM Data processing and 3D reconstruction
A total of 9413 raw image stacks were subjected to motion correction with binning (2 x) to a pixel size of 1.078 Å and dose weighting with default parameters using MotionCor2 (23). Dose-weighted micrographs were imported into the cryoSPARC software package (24) for all subsequent processing steps. Reference-free particle picking on a small subset of data was used to generate 2D classes for reference-based picking. A total of 6,701,298 “particles” were picked initially, on which extensive 2D classification was performed. After ab initio reconstruction and heterogeneous refinement for 3D classification, the final reconstruction contained 86,725 particles. We then used non-uniform refinement in combination with local refinement with a “fulcrum” (i.e. a pivot point between two flexibly connected sub-volumes, used by CryoSPARC to re-calculate the local angular search grid for improved accuracy) on the center of the complex, which yielded a 3.49-Å resolution reconstruction determined by gold-standard Fourier shell correlation using the 0.143 criterion.
Model building and refinement
Models derived from crystal structures of IL-10 (PDB ID: 1Y6K), IL-10Rα (PDB ID: 1Y6K), and IL-10Rβ (PDB ID: 3LQM) were docked into the EM map using UCSF Chimera X (26). Iterative rounds of manual rebuilding were carried out in COOT (27) for one ternary subcomplex, followed by iterative rounds of real space refinement using PHENIX with secondary structure restraints and Ramachandran restraints (28, 29) first for the trimeric subcomplex complex alone, followed by refinement of the full hexameric complex with the addition of NCS restraints, with a resolution of 3.9 Å used in refinement and resolution factor of 0.25. Due to the poorly resolved density for much of the second trimeric complex, parts of chains E, F, and D were truncated to poly-Ala. Model statistics for both the trimeric and hexameric models were generated using the comprehensive validation (cryo-EM) module in PHENIX (28, 29) and are listed in Table S1. All structure figures were made using ChimeraX (26).
Signaling assays in human cell lines
For phospho-flow cytometry experiments, THP-1, YT-1, Jurkat, and Daudi cells were plated in 96-well plates and stimulated with WT or mutant IL-10 for 20 min. at 37°C, followed by fixation with paraformaldehyde (Electron Microscopy Sciences) for 10 minutes at room temperature. The cells were permeabilized for intracellular staining by treatment with ice-cold methanol (Fisher) for 30 min. at −20°C. The cells were then incubated with Alexa Fluor 647 conjugated Anti-Stat3 (pY705) antibody (1:50, BD, clone 4/P-STAT3) for 1 hour at room temperature in autoMACS buffer (Miltenyi).
Phospho-flow signaling assays in human PBMC
Healthy donor derived human Peripheral blood mononuclear cells (PBMC) were isolated from the LRS chambers and stored as frozen aliquots. Cells were stimulated with varying concentrations of WT or mutant IL-10 and incubated at 37°C for 20 min. The PBMCs were then fixed with paraformaldehyde (Electron Microscopy Sciences) for 10 min. at room temperature, and permeabilized for intracellular staining by treatment with ice cold methanol (Fisher) for at least 30 min. at −20°C. The cells were washed with autoMACS buffer (Miltenyi), incubated with human TruStain FcX (Biolegend), and then stained with the following antibodies: CD3 Pacific Blue (1:50, BD, clone UCHT1), CD4 PerCP-Cy5.5 (1:20, BD, clone SK3), CD20 PerCp-Cy5.5 (1:20, BD, clone H1), CD33 PE-Cy7 (1:50, BD, clone P67.6), and pSTAT-3 AlexaFluor 488 (1:50, BD, clone 4/P-STAT3) or pSTAT-1 AlexaFluor 488 (1:50, BD, clone 4a). Data was acquired using CytoFlex flow cytometer instrument (Beckman Coulter) and analyzed using FlowJo software (BD).
Stimulation of human PBMCs, monocytes, and macrophages
PBMCs were thawed in warm media, washed twice and resuspended in complete RPMI at 1×107 cells/ml. For stimulation with lipopolysaccharides (LPS), cells were plated at 500 μl/well in a 24-well culture dish and rested for 2 hours at 37°C. Cells were then incubated with 1 ng/ml of LPS (Sigma) alone or with 10 nM IL-10 (WT or mutant) for 24 hours. For T cell activation in the context of bulk PBMCs, cells were plated at 500 μl/well in a 24-well tissue culture plate pre-coated with anti-CD3 antibody (clone OKT1) for 72 hours. For cytokine measurements, cell supernatants were isolated and levels of TNF-α (Biolegend), IL-6 (R&D), IL-8 (R&D), IL-1β (Biolegend) and IFN-γ (Biolegend) were measured by ELISA as per the manufacturer’s instructions.
For monocyte HLA-DR and CD86 surface expression analysis, PBMCs were washed and resuspended in autoMACS buffer, incubated with human TruStain FcX (Biolegend) and stained with CD 14 PE-Cy7 (1:50, BD, clone M5E2), HLA-DRBV605 (1:50, Biolegend, clone L243), and CD86 APC (1:50, clone GL-1) antibodies for 30 minutes at 4°C. Fluorescent intensities were measured using CytoFlex flow cytometer instrument (Beckman Coulter) and analyzed using FlowJo software (BD). Cells were gated on live cells based on forward versus side scatter profiles, followed by monocyte specific gating (FSChiCD14+).
For analysis of primary human macrophages, Monocytes were isolated from human PBMCs using MACS LS columns (Miltenyi) in combination with human monocyte (CD14+) isolation kit (Miltenyi). Isolated monocytes were plated in a 12-well tissue culture dish and incubated in human M-CSF at 20 ng/ml. Cells were incubated for 24 hours at 37°C to allow monocytes to adhere to the plate. Media was aspirated to remove non-adherent cells, and cells were incubated in fresh media containing 20 ng/ml M-CSF at 37°C for an additional 5 days. Cells were then stimulated with 1 ng/ml of LPS alone or with 10 nM IL-10 (WT or mutant) for 24 hours. Cell supernatants were isolated and levels of TNF-α (Biolegend), IL-6 (R&D), and IL-8 (R&D) were measured by ELISA as per the manufacturer’s instructions.
Stimulation of human CD4+ and CD8+ T cells
CD8+ T cells were isolated from human PBMCs using MACS LS columns (Miltenyi) in combination with human CD8+ isolation kit (Miltenyi) per the manufacturer’s instructions. The potentiation of IFN-γ and granzyme B production by IL-10 was performed as described previously (13). Briefly, isolated CD8+ T cells were seeded at 2×106 cells/ml in a 12-well tissue culture dish pre-coated with anti-CD3 antibody (clone OKT1) and incubated with soluble anti-CD28 antibody (5 μg/ml, Biolegend, clone CD28.2) for 3 days. Cells were then collected and re-seeded in fresh media at 1×106 cells/ml in a 24 well tissue culture dish with or without 10 nM IL-10 (WT or mutant) and incubated for an additional 3 days. Cells were then restimulated with soluble anti-CD3 (clone OKT1, 2 μg/ml) for 4 hours. Supernatant was then isolated and levels of IFN-γ (Biolegend), IL-9 (Biolegend) and Granzyme B (R&D) were measured by ELISA as per the manufacturer’s instructions.
For transcriptomic analysis of CD8+ T cells, RNA was then isolated using the RNAeasy mini kit (Qiagen). cDNA libraries were loaded onto an Illumina NovaSeq 6000 sequencer, PE150 platform. RNA samples from two biological replicates were used for sequencing. Indexes of the reference genome was built using STAR and paired-end clean reads were aligned to the reference genome using STAR (v2.5) (31). The FPKM of each gene was calculated based on the length of the gene and reads count mapped to the gene and used to quantify relative gene expression levels (32).
For CD4+ Th1 differentiation assays, naïve CD4+ T cells were isolated using MACS LS columns (Miltenyi) in combination with human naïve CD4+ T cell isolation kit (Stem Cell Technologies). Isolated naïve CD4+ T cells were seeded at 1×106 cells/ml in a 24-well tissue culture dish pre-coated with anti-CD3 antibody (clone OKT1) and incubated with Th1 differentiation media containing soluble anti-CD28 antibody (2 μg/ml, Biolegend, clone CD28.2), anti-IL-4 antibody (1 μg/ml, Biolegend, clone MP4-25D2), IL-2 (5 ng/ml), and IL-12 (10 ng/ml) in RPMI1640 supplemented with 10% v/v FBS, NEAA, antibiotics, and 55 μM β-mercaptoethanol), with or without IL-10 (10 nM, WT or mutant). After 5 days, cell supernatant was harvested and levels of IFN-γ (Biolegend) were measured by ELISA per the manufacturer’s instructions.
LPS Sepsis
Endotoxemia was induced in 4-6 week old female C57BL/6 mice by intraperitoneal injection of 200 μl of LPS derived from E. coli O111:B4 diluted in PBS at either a lethal (15 mg/kg) or sub-lethal (4 mg/kg) dose, in combination with a single injection of 200 μl of PBS or IL-10 (WT or 10-DE, 10 μg/mouse). Survival was monitored for 120 hours. Kaplan–Meier survival curves were compared using the log-rank Mantel–Cox test. For serum cytokine measurements, blood was drawn by submandibular puncture from mice treated with a sub-lethal LPS dose at 4 hours and 24 hours post LPS injection. Serum was isolated by centrifugation at 2,000 g for 10 min. at 4°C and TNF-α (Biolegend), IL-6 (Biolegend), and haptoglobin (R&D) were measured by ELISA per the manufacturer’s instructions.
Supplementary Material
Acknowledgments:
The authors thank C. Glassman and all members of the Garcia Lab for helpful discussions. Cryo-EM data were collected at the Cryo-EM facility at the HHMI Janelia Research Campus; we thank Drs. Z. Yu, D. Mathies, and R. Yan for their support in microscope operation and data collection. Preliminary cryo-EM screening was performed at the Cryo-EM Facility at the Department of Structural Biology, Stanford University and the Stanford-SLAC Cryo-EM facility; We thank Drs. D.-H. Chen and E. Montabana for their supports in operating the facilities and user training. Most of data were processed on the SLAC cluster; We thank Dr. Yee-Ting Li for support with computing resources. Figures 1A and 5E were made using BioRender.
Funding:
K.C.G. is an investigator of the Howard Hughes Medical Institute (HHMI). R.A.S. is an HHMI Fellow of the Helen Hay Whitney Foundation. This work was supported by NIH grants 5R01CA177684, NIH R37-AI51321 and U54CA244711 to K.C.G., and by the Department of Energy, Laboratory Directed Research and Development program at SLAC National Accelerator Laboratory, under contract DE-AC02-76SF00515, to C.G. G.C.A is supported by the Stanford Medical Scientist Training Program (T32GM007365).
Footnotes
Competing interests: R.A.S. and K.C.G. are inventors on a patent application covering therapeutic IL-10 variants described here.
Data and materials availability: The cryo-EM map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-22098, and the model coordinates have been deposited in the Protein Data Bank (PDB) under accession code 6X93. Raw cryo-EM movies have been deposited to the Electron Microscopy Public Image Archive (EMPIAR) with an accession code of EMPIAR-10557. Next-generation sequencing data files were deposited to the NCBI Gene Expression Omnibus (GEO) data repository with accession number GSE160332.
References and Notes:
- 1.Medzhitov R, Origin and physiological roles of inflammation. Nature 454, 428–435 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Kotas ME, Medzhitov R, Homeostasis, inflammation, and disease susceptibility. Cell 160, 816–827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A, Interleukin-10 and the interleukin-10 receptor. Annu. Rev. of Immunol 19, 683–765 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG, Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. of Immunol 29, 71–109 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'Garra A, IL-10 inhibits cytokine production by activated macrophages. J. Immunol 147, 3815–3822 (1991). [PubMed] [Google Scholar]
- 6.Zigmond E et al. , Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Shouval DS et al. , Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W, Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt KR, Grimbacher B, IL-10 in humans: lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr. Top. Microbiol. and Immunol 380, 1–18 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Saraiva M, Vieira P, O'Garra A, Biology and therapeutic potential of interleukin-10. The J. Exp. Med 217, e20190418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii S, Shimizu K, Shimizu T, Lotze MT, Interleukin-10 promotes the maintenance of antitumor CD8(+) T-cell effector function in situ. Blood 98, 2143–2151 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Mumm JB et al. , IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer cell 20, 781–796 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Chan IH et al. , The Potentiation of IFN-gamma and Induction of Cytotoxic Proteins by Pegylated IL-10 in Human CD8 T Cells. J. Interferon Cytokine Res 35, 948–955 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Tilg H et al. , Treatment of Crohn's disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut 50, 191–195 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josephson K, Logsdon NJ, Walter MR, Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 15, 35–46 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Chao G et al. , Isolating and engineering human antibodies using yeast surface display. Nat. Protoc 1, 755–768 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Josephson K et al. , Design and analysis of an engineered human interleukin-10 monomer. J. Biol 275, 13552–13557 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Yoon SI et al. , Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure 18, 638–648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendoza JL et al. , Structure of the IFNgamma receptor complex guides design of biased agonists. Nature 567, 56–60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendoza JL et al. , The IFN-lambda-IFN-lambdaR1-IL-10Rbeta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity. Immunity 46, 379–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Y et al. , Comprehensive mutation screening for 10 genes in Chinese patients suffering very early onset inflammatory bowel disease. World J. Gastroenterol 22, 5578–5588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B and Lyumkis D, 2017. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature methods, 14(8), p.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goddard TD et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Cryst. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodin P et al. , Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1): 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortazavi A, Williams B, McCue K et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.