Abstract
A nucleic acid-based method for the detection of the bacterial pathogens Salmonella spp. and Listeria monocytogenes in biological waste was developed. The detection limits were less than 10 cells per ml of biological waste. The method does not include a phenol extraction step and can be easily performed in 1 to 2 days.
In many countries legislators have introduced legislation which mandates that the organic fraction of household waste, as well as other types of organic waste, be recycled. The hygienic quality of the products is of increasing importance. For environmental and health reasons, when a product is used as a fertilizer on agricultural lands or in horticulture, it has to be free of pathogenic organisms (1).
The classic detection methods for pathogenic bacteria are time- and labor-intensive. The aim of this study was to develop a method which allows rapid detection of the pathogenic bacteria Salmonella spp. (gram-negative bacteria) and Listeria monocytogenes (a gram-positive bacterium) in suspended organic waste by the PCR. Salmonella spp. (2) and L. monocytogenes (3) are both human and veterinary pathogens and are potential contaminants of organic waste. Any new detection method should require considerably less time than classic enrichment methods require, should be easily performed, and should result in high detection limits.
The problem with PCR, however, is the fact that Taq polymerase is easily inhibited by a number of substances, such as humic and fulvic acids (9), fats, proteins, and metal ions, and is also inhibited by chemicals that are required for selective enrichment of cells or DNA extraction (7). Biological waste contains humic acids, as well as other inhibitory components. Therefore, efficient extraction and purification of the DNA are crucial for successful amplification.
In this paper, we describe a method that results in rapid and sensitive detection of Salmonella spp. and L. monocytogenes in suspended organic waste and is based on single-step enrichment, DNA extraction, purification, and PCR.
Both fresh suspended organic waste and anaerobically digested suspended organic waste were used for extraction of bacterial DNA. The suspended organic waste was obtained from Kaufbeuren, Germany, and contained biological waste from households, which had been ground and suspended in water until a solids content of 10% was achieved.
Salmonella typhimurium 96 BR 385 and L. monocytogenes I HE/92/1104/666-2 (serovar 1/2a) were used as seed organisms. Each strain was grown in sterile Luria-Bertani broth at 37°C and was collected after overnight enrichment. The strains were washed once in a 0.9% NaCl solution and resuspended in this solution. Cell numbers were determined by using a Neubauer counting chamber and by plate counting. For inoculation, we used organic waste samples that did not contain Salmonella spp. or L. monocytogenes, as confirmed by classical isolation methods. One-milliliter portions of dilutions of S. typhimurium or L. monocytogenes cultures containing from 10 to 106 cells per ml were added to 9-ml portions of suspended organic waste, and the preparations were mixed by vortexing. Unseeded organic waste was used as a control.
Experiments were carried out both with freshly seeded organic waste and with samples kept for 1 week at 4°C or at room temperature in order to simulate environmental conditions.
Extraction was performed after single-step enrichment. For detection of S. typhimurium, extraction was performed after enrichment of 1 ml of suspended organic waste in 9 ml of peptone water overnight (16 h) at 37°C. For detection of L. monocytogenes, 1 ml of organic waste was enriched in 9 ml of UVM I medium (which is selective for L. monocytogenes) for 24 h at 30°C (6).
The extraction procedure was based on a protocol described previously for detection of L. monocytogenes from foods (5). After enrichment, 400 μl of the broth was mixed in a sterile 2-ml Eppendorf cap with 800 μl of lysis buffer (0.5% N-laurylsarcosine, 50 mM Tris-Cl, 25 mM EDTA; pH 8.0). The mixture was centrifuged at 20,800 × g for 5 min. The pellet was suspended in 400 μl of lysis buffer containing glycogen (0.03 μg/μl). One microliter of proteinase K (20 mg/ml) was added to the suspension, and the mixture was incubated for 1 h at 37°C. After incubation, 600 μl of an NaI solution (6 M NaI in 50 mM Tris-Cl–25 mM EDTA [pH 8.0]) and 1 ml of isopropanol were added, and the mixture was centrifuged at 20,800 × g for 5 min. The pellet was washed with 35% isopropanol, dried for a short time, and resuspended in 50 μl of sterilized water by pipetting.
Crude DNA extracts were purified by using the Wizard PCR preps DNA purification system (Promega, Madison, Wis.) as described by the manufacturer by using 2-ml syringes to pass the extract through the minicolumn. The DNA was eluted by using 50 μl of hot (80°C) TE buffer (pH 8.0). For detection of S. typhimurium, the purified extract was used directly for PCR. For successful amplification of DNA recovered from organic waste seeded with L. monocytogenes, the extract had to be passed through a second minicolumn.
PCR assays were performed in 100-μl reaction mixtures. In the case of S. typhimurium, the primer pair consisting of LHNS-531 (5′-TACCAAAGCTAAACGCGCAGCT-3′) and RHNS-682 (5′-TGATCAGGAAATCTTCCAGTTGC-3′) was utilized for amplification of a 0.152-kbp region of the hns gene as previously described (4). The PCR was performed by using 3 mM Mg2+.
The primer pair consisting of LL5 (5′-AACCTATCCAGGTGCTC-3′) and LL6 (5′-CTGTAAGCCATTTCGTC-3′), which is specific for the hlyA gene for listeriolysin O, was used for L. monocytogenes, as previously reported (8). The Mg2+ concentration used was 1.5 mM.
For all amplification reactions, 0.2 μl of Taq DNA polymerase (Sigma) was used, and 1 μl of the purified extract was added to the assay mixtures. If a second PCR was performed in order to enhance the sensitivity, 1 μl of the amplified PCR product was used as a template in a second PCR. As a positive control, 1 μl of a pure culture of S. typhimurium or L. monocytogenes grown overnight in Luria-Bertani medium was utilized in the PCR. Sterile water was used as a negative control.
PCR products were identified by estimating sizes after agarose gel electrophoresis on a 1.3% agarose gel (100 mA, 80 V) and ethidium bromide staining.
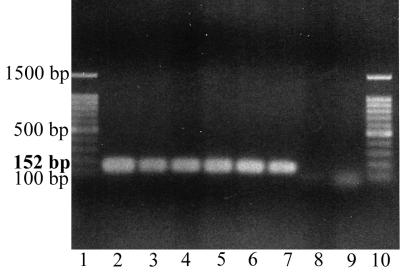
After enrichment overnight (16 h) in peptone water at 37°C, as few as 10 cells of S. typhimurium per ml of suspended organic waste could be detected easily after extraction, purification with one minicolumn, PCR, and gel electrophoresis (Fig. 1).
FIG. 1.
Agarose gel electrophoresis of PCR-amplified DNA from the hns gene of S. typhimurium following enrichment in peptone water overnight at 37°C and DNA extraction. The crude extract was purified with one minicolumn, and one PCR was performed. Lanes 1 and 10, 100-bp ladder, used as a size marker; lane 2, pure culture of S. typhimurium (positive control); lane 3, organic waste seeded with 105 cells/ml; lane 4, organic waste seeded with 104 cells/ml; lane 5, organic waste seeded with 103 cells/ml; lane 6, organic waste seeded with 102 cells/ml; lane 7, organic waste seeded with 10 cells/ml; lane 8, unseeded organic waste; lane 9, water (negative control).
For L. monocytogenes, enrichment for 24 h in UVM I broth at 30°C, DNA extraction, and purification with two minicolumns facilitated the recovery of sufficient DNA from a preparation containing 102 L. monocytogenes cells per ml of organic waste for PCR amplification and visualization on an agarose gel after ethidium bromide staining. When a second PCR was performed, as few as 10 cells could be detected (Fig. 2).
FIG. 2.
Agarose gel electrophoresis of PCR-amplified DNA from the hlyA gene of L. monocytogenes following enrichment in UVM I broth for 24 h at 30°C and DNA extraction. Crude DNA extract was purified with two minicolumns. Lanes 1 and 12, 100-bp ladder, used as a size marker; lane 2, pure culture of L. monocytogenes (positive control); lane 3, organic waste seeded with 105 cells/ml; lane 4, organic waste seeded with 104 cells/ml; lane 5, organic waste seeded with 103 cells/ml; lane 6, organic waste seeded with 102 cells/ml; lane 7, organic waste seeded with 10 cells/ml; lane 8, organic waste seeded with 10 cells/ml, second PCR; lane 9, unseeded organic waste; lane 10, unseeded organic waste, second PCR; lane 11, water (negative control).
For both species, the detection limits were the same for freshly seeded samples and for samples incubated after inoculation for 1 week at 4°C or at room temperature.
Enrichment of L. monocytogenes cultures for 24 h in nonselective peptone water resulted in a detection level of 106 cells per ml of organic waste (data not shown). The higher detection level for L. monocytogenes despite a longer period of enrichment can be explained by the longer generation time of this organism and indicates that selective enrichment is necessary for detection of low numbers of cells of this species. Moreover, since L. monocytogenes is a gram-positive bacterium, L. monocytogenes cells are more difficult to extract, which makes DNA recovery less efficient.
The fact that the DNA extract had to be purified with a second minicolumn in order to detect L. monocytogenes can be explained by the presence of inhibitory substances in UVM I medium and by the less efficient recovery of DNA, which makes the system more sensitive.
The time required for extraction of bacterial DNA, purification, PCR, and gel electrophoresis is about 1 working day. Therefore, this detection method, which includes a single enrichment step, requires considerably less time than the classic procedures used for Salmonella spp. or L. monocytogenes require, which usually take at least 4 days. Our method does not involve phenol extraction, which makes it convenient and easy to perform, and it can be performed in a laboratory with standard molecular biology equipment.
In the future, the method will be tested for its applicability to other relevant pathogenic bacteria.
Acknowledgments
This work was supported in part by BayFORREST grant F145. C.B. gratefully acknowledges an award from the Bavarian Industrial Graduate Sponsorship and Technology Transfer (BIT) Program.
We thank J.-M. Collard of the Institute of Hygiene and Epidemiology, Brussels, Belgium, for his generous gift of S. typhimurium 96 BR 385 and L. monocytogenes I HE/92/1104/666-2 (serovar 1/2a).
REFERENCES
- 1.Anonymous. LAGA-Merkblatt M 10. Qualitätskriterien und Anwendungsempfehlungen für Kompost. In: Hösel G, Bilitewski B, Schenkel W, Schnurer H, editors. Müll-Handbuch, vol. 5, No. 6856. Berlin, Germany: Erich Schmidt Verlag; 1995. pp. 39–59. [Google Scholar]
- 2.D’Aoust J. Salmonella. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 327–445. [Google Scholar]
- 3.Farber J M, Peterkin P I. Listeria monocytogenes, a foodborne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones D D, Law R, Bej A K. Detection of Salmonella spp. in oysters using polymerase reaction (PCR) and gene probes. J Food Sci. 1993;58:1191–1197. [Google Scholar]
- 5.Makino S-I, Okada Y, Maruyama T. A new method for direct detection of Listeria monocytogenes from foods by PCR. Appl Environ Microbiol. 1995;61:3745–3747. doi: 10.1128/aem.61.10.3745-3747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ralovich B. Data on enrichment and selective cultivation of listeriae. Int J Food Microbiol. 1989;8:205–217. doi: 10.1016/0168-1605(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 7.Rossen L, Norskov P, Holmstrom K, Rasmussen O F. Inhibition of PCR by components of food samples, microbiological diagnosic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 8.Thomas E J G, King R K, Burchak J, Gannon V P J. Sensitive and specific detection of Listeria monocytogenes in milk and ground beef with the polymerase chain reaction. Appl Environ Microbiol. 1991;57:2576–2580. doi: 10.1128/aem.57.9.2576-2580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai Y-L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]