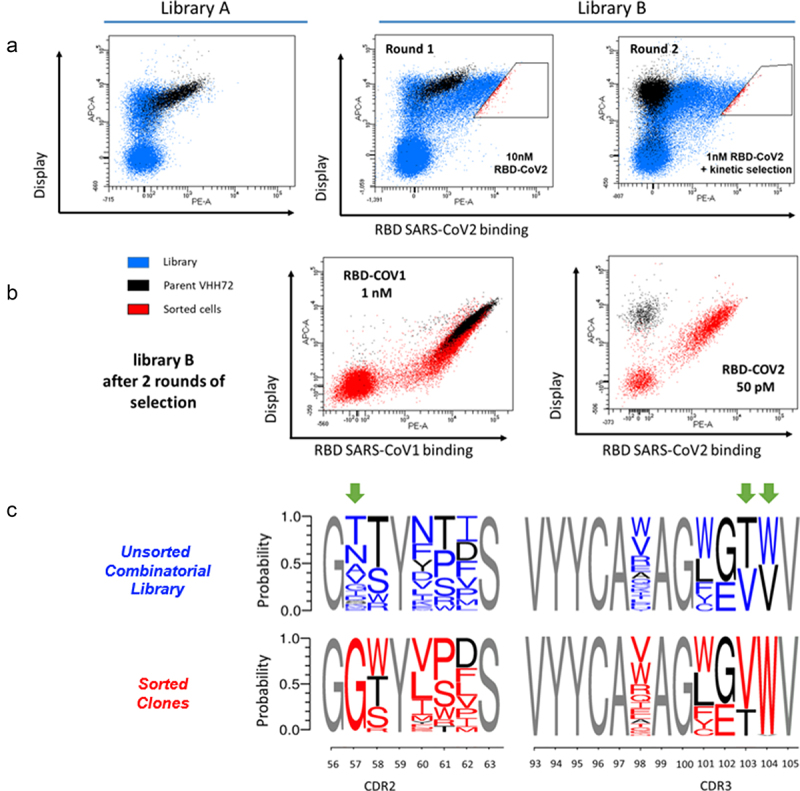
Figure 3.
Yeast Surface Display-based screening of libraries and sorting of VHH72 clones with improved binding for the RBD SARS-CoV-2 antigen. (a) Library A was incubated with 10 nM biotinylated SARS-CoV-2 RBD and analysed in FACS. Few clones with improved antigen binding were detectable. Library B was sorted twice at respective concentrations of 10 nM and 1 nM biotinylated SARS-CoV-2 RBD. In the second round, a selection based on long dissociation times (slow koff) was performed. After equilibrium with 1 nM biotinylated antigen, an excess amount of 100 nM non-biotinylated antigen was introduced to drive dissociation and limit re-association. (b) Clones selected from library B after two rounds of FACS selection. All selected clones display strong binding to both SARS-CoV-1 and SARS-CoV-2 RBD antigens. (c) Sequence logos of clones contained in library B before sorting (in red) and after two rounds of FACS selection for improved affinity (in blue).
Alt Text: Yeast Surface Display-based screening of libraries and sorting of VHH72 clones with improved binding for the RBD SARS-CoV-2 antigen (a) Cytometry dot-plot figures obtained from the analysis of yeast expressing combinatorial libraries of VHH72. (b) Clones selected from library B display strong binding to both SARS-CoV-1 and SARS-CoV-2 RBD antigens. (c) Sequence logos of clones representing the frequency of amino acid substitutions in unsorted library and in sorted library. Positions 56, 103 and 104 demonstrate a substantial shift in frequencies.