Abstract
The mammalian neocortex is a highly organized six-layered structure with four major cortical neuron subtypes: corticothalamic projection neurons (CThPNs), subcerebral projection neurons (SCPNs), deep callosal projection neurons (CPNs), and superficial CPNs. Here, careful examination of multiple conditional knockout model mouse lines showed that the transcription factor FOXG1 functions as a master regulator of postmitotic cortical neuron specification and found that mice lacking functional FOXG1 exhibited projection deficits. Before embryonic day 14.5 (E14.5), FOXG1 enforces deep CPN identity in postmitotic neurons by activating Satb2 but repressing Bcl11b and Tbr1. After E14.5, FOXG1 exerts specification functions in distinct layers via differential regulation of Bcl11b and Tbr1, including specification of superficial versus deep CPNs and enforcement of CThPN identity. FOXG1 controls CThPN versus SCPN fate by fine-tuning Fezf2 levels through diverse interactions with multiple SOX family proteins. Thus, our study supports a developmental model to explain the postmitotic specification of four cortical projection neuron subtypes and sheds light on neuropathogenesis.
The Forkhead transcription factor FOXG1 spatiotemporally controls the subtype specification of cortical projection neurons.
INTRODUCTION
The mammalian cortical projection neurons are grossly classified into two main groups, the corticofugal neurons (CFuNs) and the callosal neurons (CPNs). CFuNs are further divided into layer 6 (L6) corticothalamic neurons (CThPNs) that project to the thalamus, and L5 subcerebral neurons (SCPNs) that project to the brainstem and the spinal cord (1). Clinical research has established that dysfunction of CFuNs results in perceptual-motor dysfunctions common to diverse developmental disorders (2–4). CPNs including deep and superficial CPNs function in connecting the two cerebral hemispheres and coordinate many advanced brain functions (5–7). During neurogenesis, cortical projection neurons are generated in sequential but partially overlapping waves. Deep layer neurons including CThPNs, SCPNs, and deep CPNs arise at early corticogenesis, whereas neurons positioned in more superficial layers are produced later (8). Understanding the mechanisms underlying cortical subtype specification will almost certainly help resolve the etiopathology of numerous neurological disorders.
The postmitotic acquisition and maintenance of subtype identities are coordinated by the sequential activation/repression of gene expression programs; these programs are largely mediated by stage- and subtype-specific transcription factors. Four transcription factors—TBR1 (T-box transcription factors Tbr1), FEZF2 (the zinc finger transcription factors Fezf2)/BCL11B (B-cell leukemia/lymphoma 11B), and SATB2 (chromatin-remodeling protein Satb2)—have been reported as crucial for postmitotic specification during neurogenesis; these proteins respectively determine the identities of CThPNs, SCPNs, and deep/superficial CPNs (9–11). During early corticogenesis, the respective levels of initially coexpressed regulators in newborn cortical neurons (including TBR1, FEZF2, BCL11B, and SATB2) are subsequently altered as the distinct neuron subtypes are specified (8, 12). Moreover, it has been shown that these regulators can physically interact with each other to shape the final subtype identity. Disruption of any one of these regulators leads to aberrant cortical projection neuron subtype identities (13–19). Multiple studies have demonstrated that various SOX family members are also required for specifying cortical neuron subtypes (20–22). Although there have been true breakthrough advances in our understanding in recent years, much remains unknown about the activation/repression transcriptional networks that spatiotemporally control the proper development of postmitotic cortical projection neurons.
FOXG1, a member of the Forkhead-box family of transcription factors, has been linked to a broad array of developmental processes (23–28). Clinically, patients with FOXG1-related syndrome suffer from severe mixed dyskinesia and cognitive deficits and exhibit serious corpus callosum dysplasia (29), strongly suggesting a possible role of FOXG1 in subtype specification. In the present study, we used a combination of conditional genetic disruption at postmitotic stages, cell tracing, immunostaining, in situ RNA hybridization, and chromatin immunoprecipitation (ChIP) methods, which enabled our demonstration of FOXG1 as a major spatiotemporal regulator of postmitotic projection neuron subtype specification. This protein variously controls both induction and repression programs, doing so in both developmental stage–specific and neuron subtype–specific manners.
RESULTS
Dynamic expression of transcription factors defines impactful regulatory windows during postmitotic cortical neuron specification
To investigate postmitotic specification of cortical projection neurons, we first carefully profiled the TBR1, BCL11B, and SATB2 levels during embryonic day 12.5 (E12.5) to postnatal day 0 (P0). At E12.5, TBR1 and BCL11B were highly coexpressed at the cortical plate (CP); this TBR1highBCL11Bhigh pattern suggests that these neurons have acquired CFuN characteristics after exiting the cell cycle (fig. S1A). From E14.5 onward, we observed gradual decreases for TBR1 expression in SCPNs and for BCL11B expression in CThPNs, respectively, leading to L5 BCL11BhighTBR1low SCPN and L6 BCL11BlowTBR1high CThPN patterns at P0 (fig. S1A).
Consistent with a previous report (14), SATB2 was not detectable at the E12.5 cortex (fig. S1, B and C). At E14.5, we found that SATB2 was mainly expressed in deep CPNs most present in the intermediate zone (IZ), as well as in a small proportion in the CP. These SATB2+ deep CPNs coexpressed low levels of BCL11B and TBR1, exhibiting a SATB2highTBR1lowBCL11Blow pattern (fig. S1, B and C). By E16.5, most of the deep CPNs had already migrated to the deep layer of the CP, and this migration was accompanied by a gradual down-regulation of BCL11B and TBR1. Many superficial CPNs expressing SATB2 were present in both the superficial layer and the IZ, and it was notable that BCL11B was undetectable in superficial CPNs at this stage (fig. S1, B and C).
At P0, SATB2 was highly expressed in both deep and superficial CPNs, and these cells all coexpressed a low level of TBR1 (fig. S1, B and C). BCL11B was undetectable in the majority of P0 CPNs (fig. S1, B and C). As summarized in fig. S1D, these dynamic expression patterns in each of the four cortical projection neuron subtypes suggest that acquisition of CFuN identity is already initiated at E12.5, that further specification of CFuNs toward CThPNs or SCPNs is apparently initiated around E14.5, and that distinct mechanisms function to specify the two CPN subtypes.
We also detected a possible role of FOXG1 in postmitotic subtype specification. That is, we observed strong expression of FOXG1 in CFuNs at all tested stages (i.e., matching expression patterns of BCL11B and TBR1) (fig. S2, A and B), as well as FOXG1 coexpression with SATB2 in developing deep and superficial CPNs (fig. S2C). Quantification of fluorescence intensity showed that at E14.5, FOXG1 expression level in SATB2+ deep CPNs was lower than that in BCL11B+ CFuNs in the CP (fig. S2D). As development proceeded, at E16.5, the level of FOXG1 in SATB2+ deep CPNs became higher than that in BCL11B+ SCPNs in the deep layer (fig. S2E).
FOXG1 is required for both deep and superficial postmitotic CPN identity and for progression beyond the CFuN state
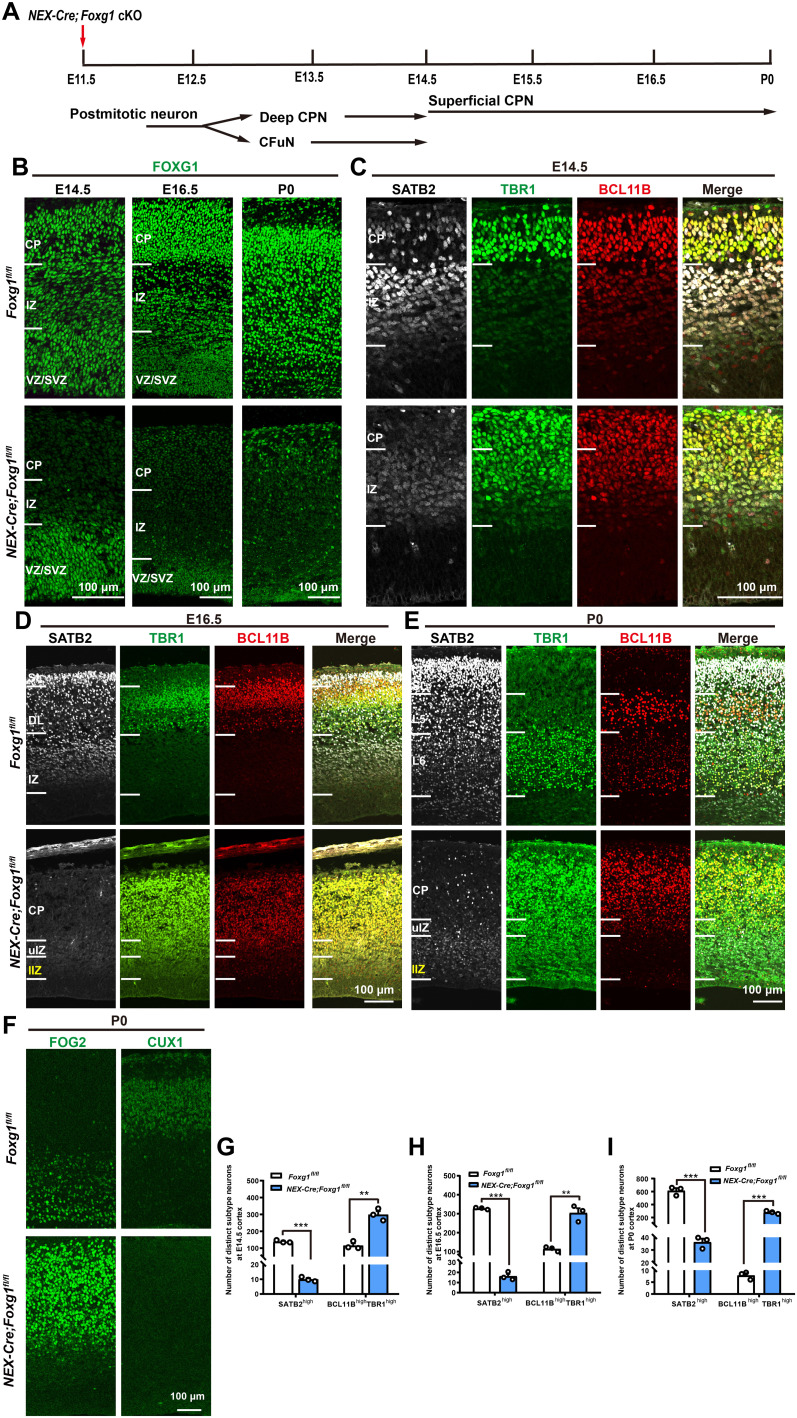
Next, Foxg1 was deleted by crossing the Foxg1fl/fl line with NEX-Cre, in which CRE (Cyclization Recombination Enzyme)–mediated recombination occurs in postmitotic cortical neurons from E11.5 onward (Fig. 1A) (30). FOXG1 was highly expressed in control but was almost undetectable in NEX-Cre;Foxg1 conditional knockout (cKO) postmitotic neurons (Fig. 1B), supporting efficient deletion. For control mice at E14.5, the CP was extensively populated with TBR1highBCL11Bhigh CFuNs as well as a small proportion of SATB2highTBR1lowBCL11Blow deep CPNs. Rich populations of deep CPNs were located in the IZ (Fig. 1C). In contrast, for the NEX-Cre;Foxg1 cKO mice, TBR1highBCL11Bhigh CFuNs populated the entire cortex including the CP and the IZ. The SATB2 levels in these mice were also notably reduced, and no SATB2highTBR1lowBCL11Blow deep CPNs were detectable (Fig. 1C). Quantitative analysis of each subtype throughout the entire cortex revealed a marked and significant decrease in the number of SATB2high CPNs in the NEX-Cre;Foxg1 cKO mice, and this was accompanied by a remarkably increased number of BCL11BhighTBR1high CFuNs (Fig. 1G), results suggesting that Foxg1 deletion after E11.5 causes extensive loss of deep CPNs.
Fig. 1. Deletion of Foxg1 from E11.5 onward leads to lost CPN identity and an arrested CFuN state.
(A) Schematic of Foxg1 disruption strategy in postmitotic neurons at E11.5. (B) Immunostaining showing that FOXG1 was efficiently disrupted. (C) Triple immunostaining against SATB2, TBR1, and BCL11B at E14.5, revealing markedly decreased SATB2 levels at the CP and IZ, and increased levels of BCL11B and TBR1 in NEX-Cre;Foxg1 cKO IZ. (D) Triple immunostaining against SATB2, TBR1, and BCL11B at E16.5, showing that the NEX-Cre;Foxg1 cKO cortex has many neurons expressing low SATB2 levels in the IZ. The whole CP and upper IZ were all occupied with TBR1highBCL11Bhigh neurons. In the lower IZ, TBR1 but not BCL11B levels were increased in NEX-Cre;Foxg1 cKO mice. (E) Triple immunostaining against SATB2, TBR1, and BCL11B at P0 showing that in the NEX-Cre;Foxg1 cKO cortex, few SATB2high CPNs were detected and that most postmitotic neurons in the CP were arrested at the TBR1highBCL11Bhigh CFuN stage. In IZ, TBR1 but not BCL11B was obviously up-regulated in SATB2low superficial CPNs. (F) Immunostaining against FOG2 and CUX1 at P0, showing that the level of FOG2 was increased while the CUX1 level was remarkably reduced in the CP of NEX-Cre;Foxg1 cKO mice. (G to I) Quantitative analysis showing a reduction in SATB2high CPNs but an increase in BCL11BhighTBR1high CFuNs at E14.5 (G), a remarkable reduction in SATB2high CPNs yet an increase in BCL11BhighTBR1high CFuNs at E16.5 (H), and reduced SATB2high CPN numbers but increased BCL11BhighTBR1high CFuN numbers at P0 (I). Data are presented as means ± SEM; n = 3, multiple Student’s t test with Bonferroni correction. **P < 0.01; ***P < 0.001. uIZ, upper intermediate zone; lIZ, lower intermediate zone; VZ, ventricular zone; SVZ, subventricular zone; DL, deep layer; SL, superficial layer.
At E16.5, SATB2high CPNs were present in both the superficial and deep layers in control mice (Fig. 1D). In NEX-Cre;Foxg1 cKO mice, few SATB2high CPNs were observed throughout the entire cortex, and only a very low level of SATB2 was detected in the IZ (Fig. 1D). Previous studies have reported that Foxg1 deletion leads to migration defects and a decrease in the number of SATB2high CPNs (24, 25); so, it is plausible that the SATB2low neurons we detected in the NEX-Cre;Foxg1 cKO IZ might represent a mixed population comprising both deep and superficial CPNs. Consistent with this idea, note that we observed increased TBR1 levels in both the upper IZ (where putative deep CPNs are positioned) and the lower IZ (superficial CPN site), whereas BCL11B accumulation was only detected in the upper IZ but not the lower IZ (Fig. 1D). Last, this examination of the NEX-Cre;Foxg1 cKO cortex revealed that the significant reduction of SATB2high CPNs was accompanied by a significant increase in BCL11BhighTBR1high CFuNs in the entire cortex (Fig. 1H). These findings collectively demonstrate that FOXG1 is required for both deep and superficial CPN identities. Moreover, it is clear that Foxg1 deletion results in the accumulation of both TBR1 and BCL11B in deep CPNs, but only results in TBR1 accumulation in superficial CPNs.
Consistently, at P0, almost all of the neurons at the NEX-Cre;Foxg1 cKO CP displayed a BCL11BhighTBR1high CFuN pattern, with few SATB2high CPNs observed (Fig. 1, E and I), indicating a shift of deep CPNs toward CFuNs. Moreover, the TBR1 level was obviously increased throughout the IZ at P0, while BCL11B was only accumulated in the upper IZ, indicating that two distinct mechanisms are involved in FOXG1’s specification of deep CPNs and superficial CPNs. Moreover, TBR1highBCL11Blow CThPNs and BCL11BhighTBR1low SCPNs were not detected in the NEX-Cre;Foxg1 cKO cortex. To further characterize neuron identities, we performed immunostaining with additional subtype-specific markers at P0. We stained NEX-Cre;Foxg1 cKO brains for FOG2 (Friend of GATA-2) (reported as specifically expressed in CFuNs) (31, 32) and for the commonly used CPN marker CUX1 (cut-like homeobox 1) (33). In NEX-Cre;Foxg1 cKO brains, the level of FOG2 was significantly increased and displayed a similar abnormal accumulation pattern with TBR1 and BCL11B (Fig. 1F). Similar to SATB2, CUX1 was significantly reduced in CPNs in NEX-Cre;Foxg1 cKO mice (Fig. 1F). These data collectively support that specification of FOXG1-deficient neurons is arrested at the CFuN stage (i.e., unable to progress toward SCPN or CThPN fates).
Foxg1 deletion at E14.5 impairs the specification of cortical neurons including CThPNs, SCPNs, deep CPNs, and superficial CPNs
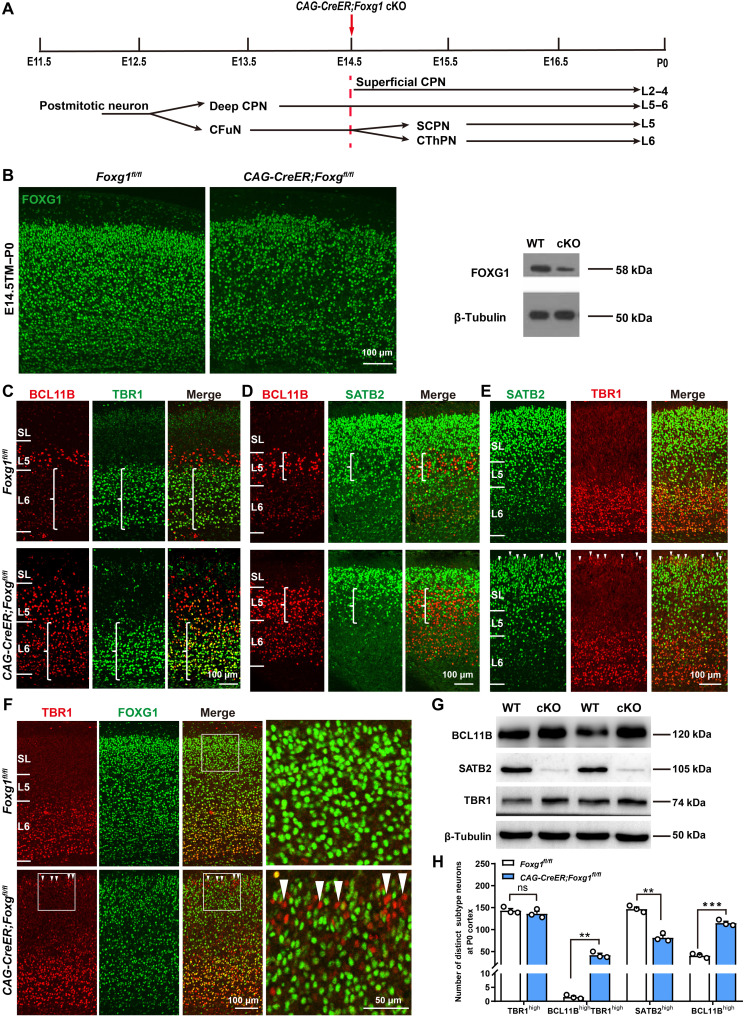
The arrested CFuN specification and lost CPNs in NEX-Cre;Foxg1 cKO mice precluded the investigation of the subsequent specification of the four neuron subtypes in these mice; we therefore used tamoxifen (TM) induction of CAG-CreER mice to disrupt Foxg1 at E14.5 (Fig. 2, A and B). At P0, compared with successful specification of L5 BCL11BhighTBR1low SCPNs and L6 TBR1highBCL11Blow CThPNs in control cortices, the TM-induced CAG-CreER;Foxg1 cKO mice did not display BCL11B down-regulation in TBR1high CThPNs, whereas TBR1 expression was normally down-regulated in Foxg1-deficient SCPNs (Fig. 2C). Consistent with these observations, quantitative analysis showed the number of TBR1high neurons in the deep layer did not differ between control and CAG-CreER;Foxg1 cKO cortices, but the number of BCL11BhighTBR1high neurons of CAG-CreER;Foxg1 cKO mice was significantly increased (Fig. 2H).
Fig. 2. Deletion of Foxg1 from E14.5 onward impairs the specification of CThPNs, deep CPNs, and superficial CPNs.
(A) Schematic of Foxg1 disruption strategy at E14.5. (B) Immunostaining and Western blot using P0 cortex, showing that Foxg1 was efficiently disrupted. (C to E) Double immunostaining at P0, showing failed BCL11B down-regulation in TBR1high CThPNs (C), decreased SATB2high deep CPNs and increased BCL11Bhigh neurons in L5 (D), and TBR1 up-regulation in SATB2low superficial CPNs in CAG-CreER;Foxg1fl/fl cKO cortices (E) (arrowheads). (F) Double immunostaining of TBR1 and FOXG1 at P0, showing up-regulation of TBR1 in FOXG1-deficient superficial CPNs (arrowheads). (G) Western blot of P0 cortex, showing increased levels of BCL11B and TBR1 and decreased levels of SATB2 in CAG-CreER;Foxg1 cKO mice. (H) Quantitative analysis showing an increase in BCL11BhighTBR1high and BCL11Bhigh neurons yet a reduction in SATB2high neurons in the deep layer at P0 in CAG-CreER;Foxg1 cKO mice. There were no obvious differences in the number of TBR1high neurons between control and CAG-CreER;Foxg1 cKO mice. Data are presented as means ± SEM; n = 3, multiple t test with Bonferroni correction. **P < 0.01; ***P < 0.001; ns, not significant. WT, wild type.
We next examined the specification of deep and superficial CPNs. Immunostaining against BCL11B was used to demarcate the superficial and deep layers. Compared with controls, the number of SATB2high CAG-CreER;Foxg1 cKO deep CPNs was significantly reduced at P0, whereas the number of BCL11Bhigh SCPNs was remarkably increased (Fig. 2, D and H). For the superficial CPNs in the cKO mice, the TBR1 level was increased (Fig. 2E). Note that staining against TBR1 and FOXG1 confirmed that these superficial CPNs were FOXG1 deficient (Fig. 2F), and Western blotting confirmed the up-regulation of BCL11B and TBR1 and down-regulation of SATB2 in P0 CAG-CreER;Foxg1 cKO cortices (Fig. 2G). Quantitative analysis of subtypes in the deep layer showed a marked decrease in the number of SATB2high CPNs that was accompanied by a nearly threefold increase in BCL11Bhigh SCPNs in the CAG-CreER;Foxg1 cKO cortex (Fig. 2H). Thus, disruption of Foxg1 at E14.5 leads to failed down-regulation of BCL11B in CThPNs, up-regulation of TBR1 in superficial CPNs, loss of deep CPNs, and an increase in the number of SCPNs.
Cell tracing reveals that Foxg1 cKO at E11.5 both forces deep CPNs into a CFuN fate and impairs superficial CPN identity
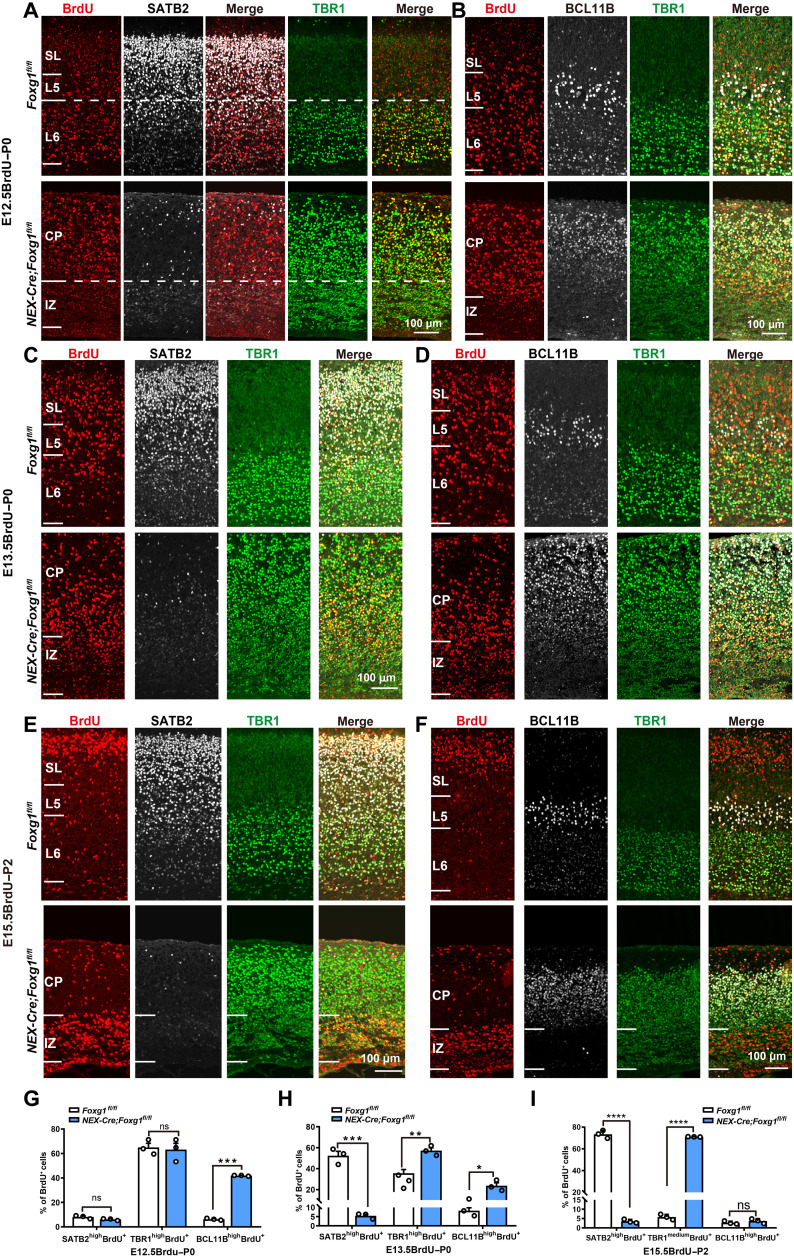
Deep CPNs emerge at the same developmental stage as CFuNs (E12.5 to E14.5) (8, 34, 35). CFuNs express BCL11B and TBR1 at high levels, while in deep CPNs, Bcl11b and Tbr1 are suppressed but Satb2 is activated. We then performed a “birth-dating” experiment in which 5-bromo-2′-deoxyuridine (BrdU) was administered to pregnant mice at E12.5 and E13.5 to label CThPNs and SCPNs/deep CPNs, respectively. The proportions of distinct subtypes specified from E12.5-born neurons were detected at P0 by quantification of TBR1highBrdU+, BCL11BhighBrdU+, and SATB2highBrdU+ neurons among total BrdU+ neurons; we detected no difference in the proportion of TBR1highBrdU+/BrdU+ neurons in the cortex of NEX-Cre;Foxg1 cKO and control mice (Fig. 3, A and G). Because only a very small number of SATB2+ CPNs are born at E12.5 (34–36), the percentages of SATB2+BrdU+ cells among total BrdU+ neurons were very low in both control and NEX-Cre;Foxg1 cKO mice (Fig. 3, A and G). Triple staining showed BCL11B was highly coexpressed in TBR1highBrdU+ neurons in NEX-Cre;Foxg1 cKO mice but not in control mice (Fig. 3B), and the proportion of BCL11BhighBrdU+/BrdU+ neurons was significantly increased, reaching a level nearly equal to that of TBR1highBrdU+/BrdU+ neurons in NEX-Cre;Foxg1 cKO mice (Fig. 3H). These findings demonstrate an arrested CFuN state upon loss of Foxg1.
Fig. 3. Cell tracing indicated deep CPNs developed into CFuNs in NEX-Cre;Foxg1 cKO mice.
(A) Immunostaining showing that most E12.5-born neurons in control mice are TBR1high CThPNs and positioned in L6; in cKO mice, TBR1high CThPNs are dispersed throughout the CP. Few SATBhighBrdU+ neurons were born at E12.5 in either control or cKO mice. (B) Failed down-regulation of BCL11B in E12.5-born TBR1highBrdU+ CThPNs in cKO CP. (C) Immunostaining tracing E13.5-born neurons, showing a significant reduction in SATB2highBrdU+ neurons and an increase in TBR1highBrdU+ neurons in cKO mice. (D) E13.5-born BrdU+ neurons have strong BCL11B expression in control L5, whereas they express high levels of both TBR1 and BCL11B in cKO mice. (E) E15.5-born neurons with strong SATB2 expression are positioned in the control superficial layer. In the cKO CP, BrdU+ neurons were restricted to the IZ and expressed some TBR1;, few of them expressed SATB2. (F) No BCL11B was detected in E15.5-born BrdU+ neurons neither in control nor in cKO mice. (G to I) Quantitative analysis showing in cKO mice an increased percentage of BCL11BhighBrdU+ neurons and unchanged percentages of SATB2high BrdU+ neurons and TBR1highBrdU+ neurons among E12.5-born neurons (G). A decreased percentage of SATB2highBrdU+ neurons and increased percentages of TBR1highBrdU+ neurons and BCL11BhighBrdU+ neurons among E13.5-born neurons (H). A sharp decrease in the percentage of SATB2high BrdU+ neurons and an increase in the percentage of TBR1mediumBrdU+ neurons among E15.5-born neurons. Few BCL11BhighBrdU+ neurons were detected in either cKO or control mice (I). Data are presented as means ± SEM; n = 3, multiple Student’s t test with Bonferroni correction. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
As for E13.5-born neurons, BCL11BhighBrdU+ and SATB2highBrdU+ neurons represented SCPNs and deep CPNs, respectively. The proportion of SATB2highBrdU+/BrdU+ neurons was sharply reduced in NEX-Cre;Foxg1 cKO mice compared with control at P0 (Fig. 3, C and H). Moreover, this reduction was accompanied by obvious increases in both the proportion of BCL11BhighBrdU+/BrdU+ neurons and the proportion of TBR1highBrdU+/BrdU+ neurons in NEX-Cre;Foxg1 cKO mice (Fig. 3, D and H). Together, these results showing that deletion of Foxg1 causes deep CPNs to develop into CFuNs provide direct evidence that Foxg1 functions to specify deep CPN identity, apparently by somehow prohibiting a CFuN developmental trajectory.
We also performed a birth-dating experiment at E15.5 to trace superficial CPNs in NEX-Cre;Foxg1 cKO mice at P2. In the control cortices, SATB2highBrdU+ CPNs populated the superficial layer (Fig. 3E). However, there were no SATB2highBrdU+ CPNs in NEX-Cre;Foxg1 cKO cortices, and E15.5-born BrdU+ neurons mainly accumulated in the IZ, with SATB2 staining very weak (Fig. 3E). To confirm the arrest of superficial CPNs in the IZ, we costained against SATB2 and PAX6 or TBR2 in NEX-Cre;Foxg1 cKO cortices. As expected, no colocalization for SATB2 with any of these two proteins was detected (fig. S4, A and B). Moreover, costaining showed that the majority of E15.5-born BrdU+ cells did not express PAX6 or TBR2 in the NEX-Cre;Foxg1 cKO IZ, demonstrating that these cells arrested in the IZ were not intermediate progenitors (fig. S4, C and D). Together, our results provide direct evidence that Foxg1 is required to specify superficial CPN identity. In the NEX-Cre;Foxg1 cKO IZ, the TBR1 level in BrdU+ neurons was increased to an intermediate level. Moreover, the proportion of TBR1mediumBrdU+/BrdU+ neurons in NEX-Cre;Foxg1 cKO cortices was significantly increased compared with control mice (Fig. 3, E and I). Thus, lacking Foxg1, TBR1 is up-regulated in E15.5-born superficial CPNs. Note that BCL11B was undetectable in E15.5-born BrdU+ neurons in either control or NEX-Cre;Foxg1 cKO mice (Fig. 3, F and I), suggesting that FOXG1’s regulation of superficial CPN specification may not involve BCL11B.
We next deleted Foxg1 at E12.5 using in utero electroporation to deliver pNeuroD1-Cre-GFP (37) into the Foxg1fl/fl mice. In addition to postmitotic neurons, NeuroD1 promoter can also drive gene expression in a subset of intermediate progenitors (38); thus, at E18.5, green fluorescent protein (GFP) labeled both E12.5 postmitotic deep neurons and superficial CPNs, which derived from intermediate progenitors (fig. S3, A and B). In pNeuroD1-Cre-GFP;Foxg1fl/fl cortices, one group of GFP+ neurons was detected near the deep layer in which SATB2 was undetectable, while both BCL11B and TBR1 were accumulated (fig. S3, A and B); the other group of GFP+ neurons that represents superficial CPNs with migration defects was arrested in the lower IZ. Moreover, SATB2 was undetectable in these neurons, and only TBR1 but not BCL11B was up-regulated (fig. S3, A and B). These results are in line with the observations in NEX-Cre;Foxg1 cKO mice (Fig. 1, C to E) and further suggest that Foxg1 functions to specify both deep and superficial CPN identities and controls the CFuN developmental trajectory.
Cell tracing reveals that Foxg1 cKO at E14.5 both increases BCL11B accumulation in CThPNs and forces deep CPNs into an SCPN fate
To trace the developmental trajectories of deep CPNs and CFuNs toward CThPNs or SCPNs from E14.5 onward, we performed cell tracing experiments with CAG-CreER;Foxg1 cKO mice. BrdU was administered at E12.5 or E13.5 to label CThPNs and SCPNs/deep CPNs, respectively, followed by TM induction at E14.5 to ensure that Foxg1 was postmitotically disrupted in BrdU+ neurons. Among BrdU+ neurons born at E12.5, in CAG-CreER;Foxg1 cKO mice, TBR1highBrdU+ CThPNs abnormally expressed high-level BCL11B (fig. S5A); the proportion of BCL11Bhigh BrdU+/BrdU+ neurons was significantly high in cKO compared with the control cortex, and the proportion of TBR1highBrdU+/BrdU+ neurons was comparable (fig. S5C). This result directly demonstrated that FOXG1 may repress BCL11B in CThPNs.
Among BrdU+ neurons born at E13.5, the number of BCL11BhighBrdU+ neurons was remarkably increased (fig. S5B), and this increase was accompanied by a decrease in the proportion of SATB2highBrdU+/BrdU+ neurons (fig. S5D). Thus, SCPNs increased at the expense of deep CPNs in CAG-CreER;Foxg1 cKO mice. We detected no differences in the proportion of TBR1highBrdU+/BrdU+ neurons between the two genotypes (fig. S5D). The increase in SCPNs was confirmed by staining with an antibody against the SCPN marker protein kinase C–γ (PKC-γ) (fig. S6A). Compared to control brains, the number of PKC-γ+ SCPNs in CAG-CreER;Foxg1 cKO brains was remarkably increased (fig. S6B). We next used retrograde tracing to characterize SCPNs by injecting DiI (Dioctadecyl-3,3,3’,3’-Tetramethylindocarbocyanine Perchlorate) into the medulla pyramid, where the corticospinal tracts are known to pass through (2, 39), which also successfully confirmed the increase in SCPNs in CAG-CreER;Foxg1 cKO brains (fig. S6, C and D). Together, these results demonstrate that deletion of Foxg1 from E14.5 promotes the deep CPNs born at E13.5 to develop into SCPNs.
Corpus callosum, corticothalamic tracts, and corticospinal tracts do not form in NEX-Cre;Foxg1 cKO brains lacking Foxg1 at E11.5
The observed loss of CPN identity and arrested CFuN specification resulting from Foxg1 disruption from E11.5 would be expected to manifest in projection phenotypes. We conducted an extensive series of labeling experiments, which ultimately confirmed that NEX-Cre;Foxg1 cKO brains displayed a complete loss of the corpus callosum (fig. S7A), a lack of corticothalamic axons in the dorsal thalamus (fig. S7B), and a loss of the corticospinal tracts in the pons (fig. S7C) and the medulla (fig. S7D). A previous study reported that FOXG1 can form a complex with Rp58 to regulate axon projection (25); it seems plausible that the projection deficits we observed here may result from the combined consequences of dysregulated axonal projection and neuron subtype specification.
CAG-CreER;Foxg1 cKO brains lacking Foxg1 at E14.5 have thicker corticospinal tracts and a thinner callosal callosum
Corresponding to that deep CPNs developed into SCPNs upon disruption of Foxg1 at E14.5, the CAG-CreER;Foxg1 cKO brains displayed obviously thicker corticospinal tracts (fig. S8, A and B) and an aberrantly thin corpus callosum (fig. S8C). These mice also had many fewer corticothalamic axons in the striatum and thalamus (fig. S8, D and E), further demonstrating impaired specification of CThPNs upon disruption of Foxg1 at E14.5.
Thus, our results demonstrate how Foxg1 deletion at E11.5 causes the loss of both deep and superficial CPNs that is accompanied by conversion of deep CPNs into CFuNs. Moreover, disruption of normal CFuN specification precludes further developmental progression toward SCPNs or CThPNs. Loss of Foxg1 at E14.5 caused conversion of deep CPNs to SCPNs, increased levels of TBR1 but not BCL11B in superficial CPNs, and failed BCL11B down-regulation during CThPN specification. Beyond establishing that FOXG1 activates both deep and superficial CPN identities, these results support the existence of two mechanisms through which FOXG1 regulates the specification of deep and superficial CPNs.
FOXG1 functions as an activator of Fezf2 transcription
Previous studies have shown that the precisely controlled transcription of Fezf2 is crucial to the proper specification of deep layer cortical projection neurons, and FEZF2 protein is known to function as an upstream regulator to promote Bcl11b transcription in SCPNs (13, 15, 17, 18). During CThPN specification, the levels of both FEZF2 and BCL11B must be reduced, whereas high FEZF2 and BCL11B levels are retained in SCPNs (16, 39, 40).
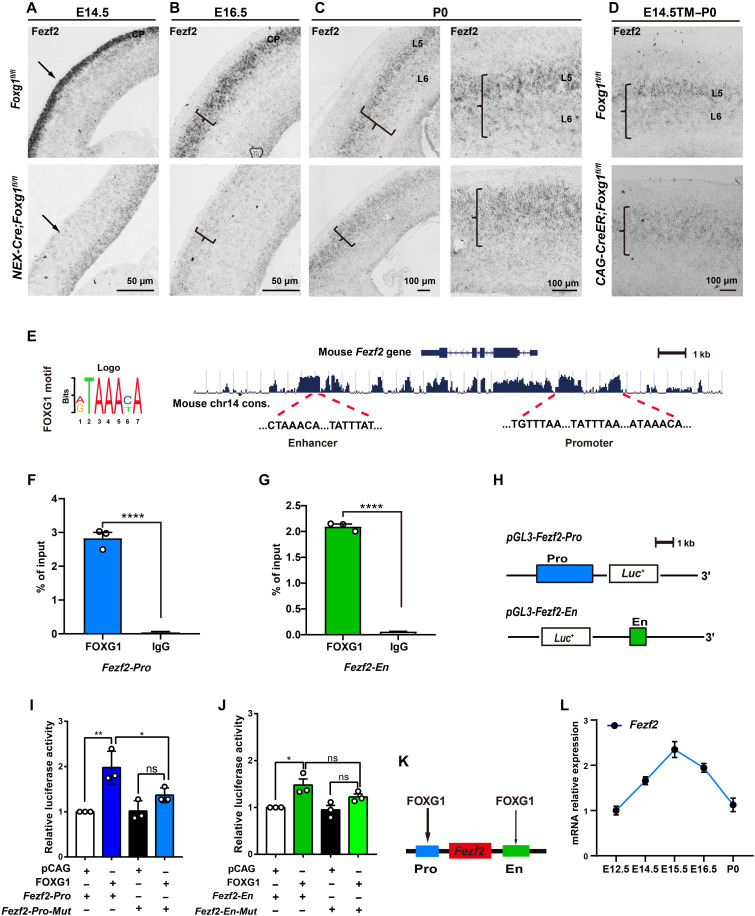
We performed in situ hybridization, which showed that Fezf2 is strongly expressed at the CP at E14.5 and E16.5 (Fig. 4, A and B). Furthermore, Fezf2 exhibited its expected SCPNhighCThPNlow expression pattern in control cortices at P0 (Fig. 4C). Unexpectedly, and apparently inconsistent with the significant increase in BCL11B that we observed in the NEX-Cre;Foxg1 cKO cortex, Fezf2 expression was almost undetectable at E14.5 (Fig. 4A). In addition, although a strong reduction in the NEX-Cre;Foxg1 cKO cortex was still evident at E16.5, the elevated Fezf2 level at this developmental stage did indicate that some slight restoration or compensatory expression was initiated by this point in the NEX-Cre;Foxg1 cKO brains (Fig. 4B). We detected a further restoration of Fezf2 expression at P0; however, rather than the SCPNhighCThPNlow pattern of control mice, the NEX-Cre;Foxg1 cKO cortex displayed a uniform Fezf2 expression pattern in the CP (Fig. 4C). Such a uniform pattern is consistent with the failed specification of CThPNs that we initially observed (Fig. 1E). Further supporting this, the same uniform Fezf2 expression pattern was also observed in the CAG-CreER;Foxg1 cKO cortex at P0 (Fig. 4D).
Fig. 4. FOXG1 directly promotes Fezf2 transcription.
(A to D) In situ RNA hybridization showing significantly decreased Fezf2 mRNA level in NEX-Cre;Foxg1 cKO CPs at E14.5 (A) and E16.5 (B). At P0, Fezf2 mRNA expression displayed a SCPNhighCThPNlow pattern in the control but a uniform expression pattern at the NEX-Cre;Foxg1 cKO CPs (C). Similarly, Fezf2 mRNA also displayed a uniform expression pattern in the CAG-CreER;Foxg1 cKO cortex at P0 (D). (E) Motif analyses and prediction of the FOXG1-binding site(s) at the Fezf2 locus. (F and G) ChIP-qPCR of cortex samples at E14.5, showing enriched FOXG1 occupancies at both the promoter and enhancer sites of the Fezf2 locus. (H) Strategy for constructing the vector for the Fezf2 promoter and enhancer for luciferase assays. (I and J) Luciferase assay showing the activation of FOXG1 at the Fezf2 promoter site (I), while showing very weak activation at the Fezf2 enhancer site (J). Deletion of the FOXG1-binding motifs from the Fezf2 promoter reduced the extent of FOXG1-mediated activation of the luciferase reporter. (K) Summary model for FOXG1’s regulation of Fezf2 expression via binding at the Fezf2 promoter and/or enhancer sites. (L) qPCR of cortex samples to monitor Fezf2 mRNA levels over the course of cortical development from E12.5 to P0. Data are presented as means ± SEM; (F and G) unpaired Student’s t test and (I and J) one-way analysis of variance (ANOVA) followed by Tukey-Kramer post hoc test. *P < 0.05, **P < 0.01; ****P < 0.0001.
Two putative FOXG1-binding sites at the Fezf2 locus have been previously reported, one within the promotor and the other within a downstream enhancer (Fig. 4E) (41). To explore the spatiotemporal regulation of Fezf2 transcription during SCPN and CThPN specification, we performed ChIP–quantitative polymerase chain reaction (qPCR) at E14.5 with an anti-FOXG1 antibody in the control cortex, seeking to measure FOXG1 enrichment at the two putative binding sites. The enrichment at both the promotor and enhancer sites was quite high (Fig. 4, F and G). Luciferase assays showed an obvious activation of FOXG1 at the Fzef2 promotor, and this activation was weakened when FOXG1-binding motifs were deleted (Fig. 4, H and I). Very weak activation was observed on the enhancer site, and point mutations in FOXG1-binding motifs at the Fezf2 enhancer did not obviously affect its activity (Fig. 4J). These results indicate that the promoter site but not the enhancer site serves as a strong direct regulatory element for FOXG1 (Fig. 4K). We next assessed Fezf2 mRNA expression from E12.5 to P0 and detected a peak at E15.5 (Fig. 4L), a transcription trend coincident with that the FEZF2 protein was highly expressed in all CFuNs but was later down-regulated during the specification of CThPNs (Fig. 4, A to C). Collectively, these results support that FOXG1 functions as an activator of Fezf2 transcription.
FOXG1 promotes transcription of multiple Sox family members
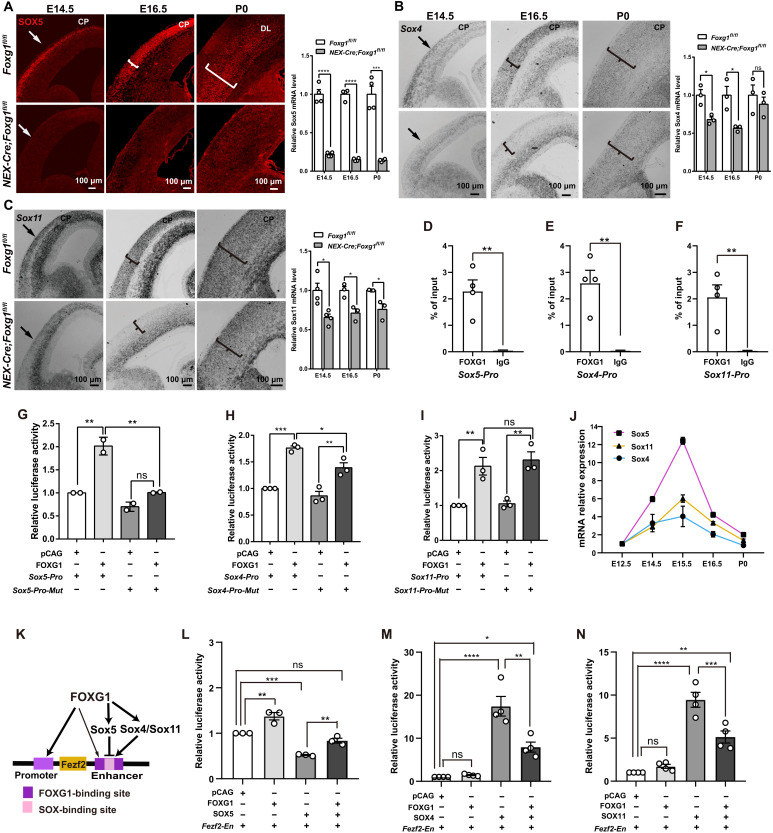
It has been reported that SOX5, a member of the SOXD (the N-terminal Sry-related HMG box subfamily D) subfamily, controls CThPN specification by down-regulating Fezf2, while SOX4 and SOX11 [members of the SOXC (the N-terminal Sry-related HMG box subfamily C) subfamily] are required to retain a high FEZF2 level in SCPNs (20–22). Thus, the arrested development of CFuNs that we observed in Foxg1 cKO mice suggests the possible involvement of SOX members. We conducted immunostaining, which showed that SOX5 was strongly expressed at the CP at E14.5 in control mice, after which its expression was confined to SCPNs and CThPNs (E16.5 to P0) (Fig. 5A), consistent with the reported role of this SOXD subfamily member in SCPN and CThPN specification, little if any SOX5 was detected in the NEX-Cre;Foxg1 cKO cortex (Fig. 5A).
Fig. 5. FOXG1 promotes the transcription of Sox members and coordinates SOXs to precisely control the level of FEZF2.
(A) Immunostaining and qPCR showing significantly decreased SOX5 in NEX-Cre;Foxg1 cKO compared with the control mice during E14.5 to P0. (B and C) In situ hybridization and qPCR showing that Sox4 (B) and Sox11 (C) were markedly decreased at E14.5 and E16.5 but were elevated at P0 in the NEX-Cre;Foxg1 cKO mice. (D to F) ChIP-qPCR of cortex samples from E14.5, showing strong enrichment of FOXG1 at promoter sites of Sox5, Sox4, and Sox11 loci. (G to I) Luciferase assay showing the activation of FOXG1 on Sox5 (G), Sox4 (H), and Sox11 (I). Point mutations in FOXG1-binding motifs in the Sox5 promoter and deletion of the FOXG1-binding motifs from the Sox4 promoter caused reduced reporter activation (G and H); no reduction was detected upon its deletion from the Sox11 promoter (I). (J) qPCR analysis showing mRNA levels of Sox members. (K) Exploratory model about FOXG1’s regulation of Fezf2 transcription mediated via binding competition with SOX proteins. The thin arrow indicates the weak activation of FOXG1 at the Fezf2 enhancer site. (L to N) Luciferase assay showing the competition between FOXG1 and SOX members. Data were presented as means ± SEM; (D to F) unpaired Student’s t test and (G to I and L to N) one-way ANOVA followed by Tukey-Kramer post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Furthermore, in situ hybridization experiments showed that the SOXC subfamily members Sox4 and Sox11 were strongly expressed in control postmitotic neurons at the CP during E14.5 to P0 (Fig. 5, B and C). In contrast, the NEX-Cre;Foxg1 cKO mice displayed remarkably decreased Sox4 and Sox11 expression levels, specifically in postmitotic neurons from which Foxg1 was deleted (Fig. 5, B and C). It bears emphasis that both SOX4 and SOX11 levels were somewhat restored at the CP at P0 compared with the E14.5 to E16.5 levels (Fig. 5, B and C). Thus, the expression trends we observed for SOXC subfamily members are temporally consistent with the reported function of these proteins in maintaining FEZF2 levels in SCPN and CThPN specification and with our observations of FEZF2 levels in the NEX-Cre;Foxg1 cKO cortex (Fig. 4, A to C).
We next examined whether FOXG1 directly transcriptionally activates Sox family genes. A bioinformatics analysis using the UCSC Genome Browser indicated apparent conservation of putative FOXG1-binding sequences (42) in the promoter regions of Sox family members. We conducted ChIP-qPCR and found that FOXG1 was highly enriched at the promoter regions of Sox5, Sox4, and Sox11 in the E14.5 control cortex (Fig. 5, D to F). Luciferase assays showed obvious activations of FOXG1 at the Sox4, Sox5, and Sox11 promotors (Fig. 5, G to I). Deletion of FOXG1-binding motifs from the Sox4 promotor and point mutations at the Sox5 promotor weakened the activations of FOXG1 (Fig. 5, G and H). However, the luciferase activity was not obviously changed when FOXG1-binding motifs were deleted from the Sox11 promotor (Fig. 5I), suggesting that FOXG1 regulates Sox11 transcription through coordinating with some as-yet-unknown partner. qPCR analysis of whole control cortex samples showed that mRNA expression levels for these three Sox members were gradually increased from E12.5 onward, reaching a peak around E15.5 (Fig. 5J). These results establish that FOXG1 directly binds to the promoters of Sox5 and Sox4 and positively regulates their transcription, while activation of Sox11 apparently involves other regulatory mechanisms.
The proper Fezf2 dosage required to specify SCPN versus CThPN fate is precisely controlled through FOXG1 and SOX competition
Previous studies have demonstrated that SOX5 and SOX4/SOX11 competitively bind to an enhancer at the Fezf2 locus: This leads not only to FEZF2 down-regulation in CThPNs but also to maintenance of high FEZF2 levels in SCPNs during specification (20). It was therefore highly conspicuous when we found that the SOX-binding motif at the Fezf2 locus was embedded within multiple FOXG1-binding motifs present in the same enhancer domain (Fig. 5K). This finding strongly suggested the possibility that FOXG1 may compete with SOX members to control the precise expression of FEZF2. Pursuing this hypothesis, we generated overexpression vectors for Sox5, Sox4, and Sox11. Subsequent luciferase reporter assays with these Sox vectors and the Fezf2-En vector revealed that transfection with Sox5 led to a 47.9 ± 1.2% decrease in reporter activity, consistent with SOX5 as a repressor of Fezf2 transcription. Simultaneous transfection with the reporter vector alongside both the pCAG-Foxg1 and pCAG-Sox5 vectors significantly increased reporter activity compared with the Sox5 vector alone (0.84 ± 0.03 versus 0.52 ± 0.01), indicating that competitive binding of FOXG1 at the enhancer element of the Fezf2 locus can alleviate SOX5-mediated repression of Fezf2 transcription (Fig. 5L).
Conversely, transfection with the Sox4 or Sox11 vectors resulted in 17- and 9-fold increases in reporter activity (compared with cells harboring the reporter vector alone), findings demonstrating the strong Fezf2 transcriptional activation function of these SOXC subfamily proteins (Fig. 5, M and N). We found that simultaneous transfection of the Foxg1 vector with the Sox4 or Sox11 vectors reduced reporter activity significantly (only eight- and fourfold over the reporter vector alone cells) (Fig. 5, M and N). Thus, these results are, on the one hand, consistent with the idea of competitive FOXG1 versus (general) SOX protein binding at the Fezf2 enhancer. On the other hand, these results also indicate that the strength of FOXG1’s transcriptional activation impacts—specifically at the enhancer region of Fezf2—is substantially weaker than the induction effects of Sox4 and Sox11. These results, when viewed alongside our earlier findings about the much stronger activation from the promoter-localized FOXG1-binding site relative to the enhancer-localized FOXG1-binding site, unveil a complex regulatory network for controlling Fezf2 expression to specify CThPN and SCPN fate.
FOXG1 directly activates Satb2 and represses both Tbr1 and Bcl11b
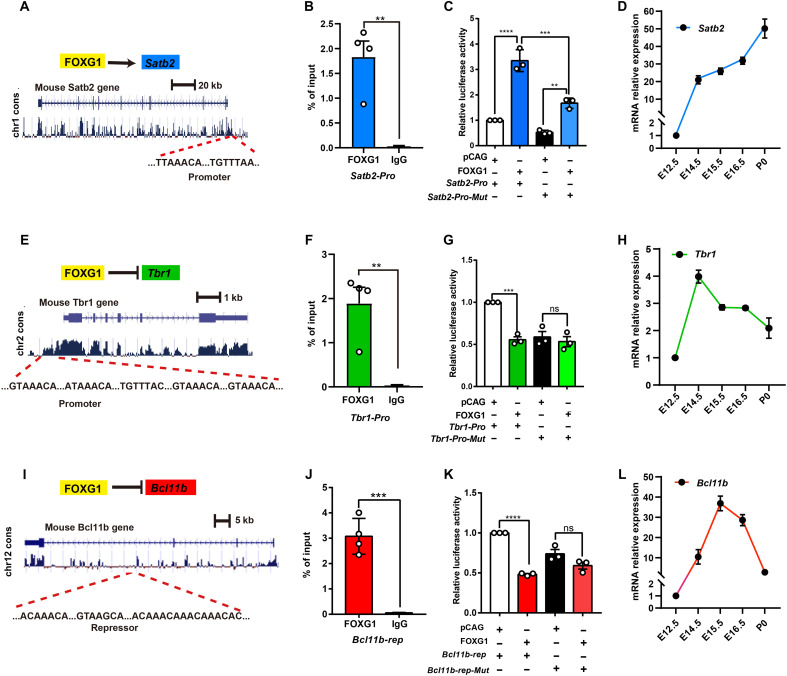
Considering the significant reduction of SATB2 we observed in both deep and superficial CPNs in both NEX-Cre;Foxg1 cKO and CAG-CreER;Foxg1 cKO mice (Figs. 1, C to E, and 2, D and E), we analyze the sequence of the Satb2 locus and identified a putative consensus FOXG1-binding site within the promoter (Fig. 6A). We next performed ChIP-qPCR and detected a strong FOXG1 enrichment in the E14.5 control cortex (Fig. 6B). Luciferase assays demonstrated that FOXG1 can directly activate Satb2, and deletion of binding motifs weakened the activation (Fig. 6C). qPCR analysis revealed a continuous increase in Satb2 mRNA expression during E12.5 to P0 (Fig. 6D), the developmental window when a majority of CPNs are produced and specified (5). Thus, FOXG1 acts as an activator of Satb2.
Fig. 6. FOXG1 directly promotes Satb2 but suppresses Tbr1 and Bcl11b.
(A) Motif analyses and prediction of FOXG1-binding site at Satb2 locus. (B) ChIP-qPCR with cortex samples from E14.5, showing high FOXG1 occupancy at the promotor site of the Satb2. (C) Luciferase assay showing that FOXG1 directly activates Satb2 by binding at the promoter site, and deletion of FOXG1-binding motifs from Satb2 promoter weakened its activation. (D) qPCR with cortex sample–assessed Satb2 mRNA from E12.5 to P0. (E) Motif analyses and prediction of FOXG1-binding site at Tbr1 locus. (F) ChIP-qPCR with cortex samples from E14.5, showing high FOXG1 occupancy at the promotor site of the Tbr1. (G) Luciferase assay showing that FOXG1 directly represses Tbr1 transcription by binding at the promoter site; deletion of FOXG1-binding motifs from the Tbr1 promoter weakened the extent of FOXG1’s repression of transcription. (H) qPCR with cortex sample–assessed Tbr1 mRNA from E12.5 to P0. (I) Motif analyses and prediction of FOXG1-binding site at Bcl11b locus. (J) ChIP-qPCR with cortex samples from E14.5, showing high FOXG1 occupancy at the repressor site of Bcl11b. (K) Luciferase assay showing that FOXG1 directly represses Bcl11b transcription by binding at the repressor site; deletion of FOXG1-binding motifs from Bcl11b repressor element weakened its repression. (L) qPCR with cortex sample–assessed Bcl11b mRNA from E12.5 to P0. Data were presented as means ± SEM; (B, F, and J) unpaired Student’s t test and (C, G, and K) one-way ANOVA followed by Tukey-Kramer post hoc test. **P < 0.01; ***P< 0.001; ****P < 0.0001.
It is reported that during the acquisition of superficial competence, FOXG1 may derepress Fezf2 through repressing Tbr1 (43). In the present study, Tbr1 was up-regulated in both deep and superficial CPNs in NEX-Cre;Foxg1 cKO and CAG-CreER;Foxg1 cKO mice. ChIP-qPCR revealed strong FOXG1 occupancy at the Tbr1 promoter at E14.5 (Fig. 6, E and F). Luciferase assays revealed that FOXG1 functions as a direct repressor of Tbr1 transcription, and deletion of FOXG1-binding motifs weakened the repression on the Tbr1 promotor (Fig. 6G). qPCR analysis showed that Tbr1 mRNA level was high during E12.5 to E14.5 but decreased afterward (Fig. 6H). These results support that FOXG1 may directly inhibit Tbr1 transcription in both deep and superficial CPNs.
Although BCL11B has been reported as a downstream target of FEZF2 signaling (13), disruption of Fezf2 resulted in loss of BCL11B. However, ectopic expression of Fezf2 by in utero electroporation was unable to induce BCL11B expression (13), suggesting that Bcl11b is also controlled via an as-yet-unknown FEZF2-independent pathway. To date, there is no evidence confirming that FEZF2 can directly influence BCL11B accumulation. We also found that BCL11B is massively up-regulated in CThPNs and deep CPNs at P0 in CAG-CreER;Foxg1 cKO mice; recall that these mice have severely decreased FEZF2 levels, again suggesting that Bcl11b may be controlled via an unknown FEZF2-independent pathway.
Pursuing this FEZF2-independent speculation, we identified multiple putative FOXG1-binding motifs within a distal 50-kb conserved region of the Bcl11b locus (Fig. 6I). ChIP-qPCR analysis revealed a high enrichment in the binding site (Fig. 6J). Luciferase assays showed that FOXG1 negatively regulates Bcl11b transcription by binding to the distal repressor site (Fig. 6K). qPCR showed that the Bcl11b mRNA level reached a peak at E15.5 and then rapidly declined (Fig. 6L). These results demonstrate that FOXG1 strongly represses Bcl11b transcription.
Cell type–specific disruptions/overexpression establish that FOXG1 controls cortical neuron subtype–specific gene expression programs
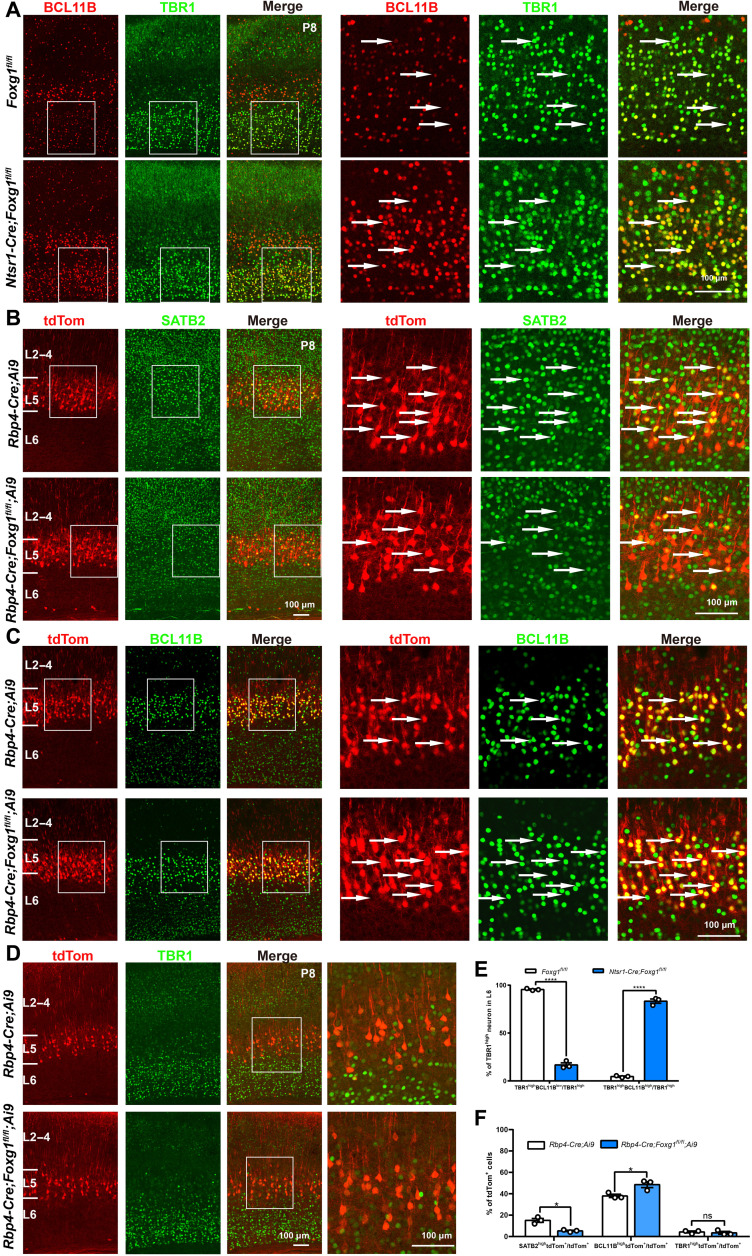
To experimentally confirm FOXG1’s repression of Bcl11b in CThPNs, we used Ntsr1-Cre mice (44)—which enable specific cKO of Foxg1 in CThPNs after E14.5—to detect the effects of Foxg1 deletion on CThPN specification. At P8 when the specification of cortical projection neuron subtypes is finished, we detected failed down-regulation of BCL11B in Ntsr1-Cre;Foxg1 cKO CThPNs (Fig. 7, A and E).
Fig. 7. Cell type–specific disruption further demonstrated that FOXG1 controls subtype-specific gene expression programs.
(A) Double immunostaining against TBR1 and BCL11B at P8. Control mice have low BCL11B levels in CThPNs. Ntsr1-Cre;Foxg1 cKO mice feature CThPN-specific disruption of Foxg1, and these cells had high BCL11B levels. (B) Double immunostaining against tdTom and SATB2 at P8. Rbp4-Cre;Ai9;Foxg1 cKO mice feature disruption of Foxg1 from both deep CPNs and SCPNs, which are labeled by tdTom. Compared with control mice, SATB2hightdTom+ deep CPNs remarkably reduced in Rbp4-Cre;Ai9;Foxg1 cKO mice. (C) Double immunostaining against tdTom and BCL11B at P8, showing that BCL11BhightdTom+ SCPNs increased in Rbp4-Cre;Ai9;Foxg1 cKO mice. (D) Double immunostaining against tdTom and TBR1 at P8. There was no difference in TBR1 levels in tdTom+ neurons in Rbp4-Cre;Ai9 or Rbp4-Cre;Ai9;Foxg1 cKO animals, a finding indicating that FOXG1 does not affect TBR1 expression in deep CPNs or SCPNs. (E) Quantitative analysis of neurons in L6, showing the decreased percentages of TBR1highBCL11Blow neurons in TBR1high neurons and the increased percentage of TBR1highBCL11Bhigh neurons in Ntsr1-Cre;Foxg1 cKO mice at P8. (F). Quantitative analysis of neurons in L5, showing the decreased percentages of SATB2hightdTom+ neurons in tdTom+ neurons and the increased percentage of BCL11BhightdTom+ neurons in Rbp4-Cre;Foxg1 cKO mice at P8. Few TBR1hightdTom+ neurons were observed in both control and Rbp4-Cre;Foxg1 cKO mice, and no obvious changes were detected in Rbp4-Cre;Foxg1 cKO mice. Data are presented as means ± SEM; n = 3, multiple Student’s t test with Bonferroni correction. *P < 0.05; ****P < 0.0001.
We next examined Rbp4-Cre;Ai9;Foxg1 cKO mice—in which Foxg1 is specifically knocked out after E14.5 in deep CPNs and SCPNs, which are both labeled by a tdTomato reporter—seeking to experimentally confirm two FOXG1 functions after E14.5: (i) that FOXG1 activates Satb2 but represses Bcl11b in deep CPNs and (ii) that FOXG1 has no impact on the Tbr1 in deep CPNs and SCPNs. At P8, many tdTom+ neurons in the control mice were SATB2+, while the number of SATB2+tdTom+ was significantly decreased in Rbp4-Cre;Ai9;Foxg1 cKO mice (Fig. 7B). Meanwhile, a large number of the tdTom+ neurons in the Rbp4-Cre;Ai9;Foxg1 cKO mice were BCL11Bhigh (Fig. 7C). Quantification analysis showed that the percentage of SATB2+tdTom+ deep CPNs among tdTom+ neurons was obviously decreased in Rbp4-Cre;Ai9;Foxg1 cKO mice compared with that of control mice, while the percentage of BCL11B+tdTom+ SCPNs among tdTom+ neurons was increased (Fig. 7F). We did not detect obvious changes in TBR1 expression in tdTom+ neurons between the control and Rbp4-Cre;Ai9;Foxg1 cKO mice (Fig. 7, D and F). Together, these results confirm that lacking Foxg1 causes deep CPNs to develop into SCPNs and show that FOXG1 activates Satb2 but represses Bcl11b in deep CPNs. Note that no aberrant accumulation of TBR1 was observed in tdTom+ neurons of Rbp4-Cre;Ai9;Foxg1 cKO mice (Fig. 7, D and F), further supporting our conclusion that FOXG1 does not repress Tbr1 in deep CPNs or SCPNs after E14.5.
To further examine the role of FOXG1 in controlling the specification of CFuNs versus deep CPNs, we overexpressed Foxg1 by crossing NEX-Cre with a CAG-loxp-stop-loxp-Foxg1-IRES-EGFP mouse line (fig. S9, A and E) (45): We found an obvious increase in the SATB2 level in both BCL11B+ SCPNs and TBR1+ CThPNs, in which Foxg1 was overexpressed when measured on the basis of fluorescence intensity (fig. S9, B, E, and F). We found that the BCL11B level was slightly decreased in Foxg1-overexpressing SCPNs; no obvious changes were detected in the TBR1 level in Foxg1-overexpressing CThPNs (fig. S9, C to F). Because we did not detect obvious accumulation of BCL11B in Foxg1-deficient SCPNs and considering that SATB2 is demonstrated to directly repress Bcl11b (14, 36), here the decreased level of BCL11B might be caused by the increased SATB2 level in Foxg1-overexpressing SCPNs. Together, our results show that FOXG1 is capable for the induction of Satb2 in both CThPNs and SCPNs but is insufficient for the repression of (i) Tbr1 in CThPNs and (ii) Bcl11b in SCPNs.
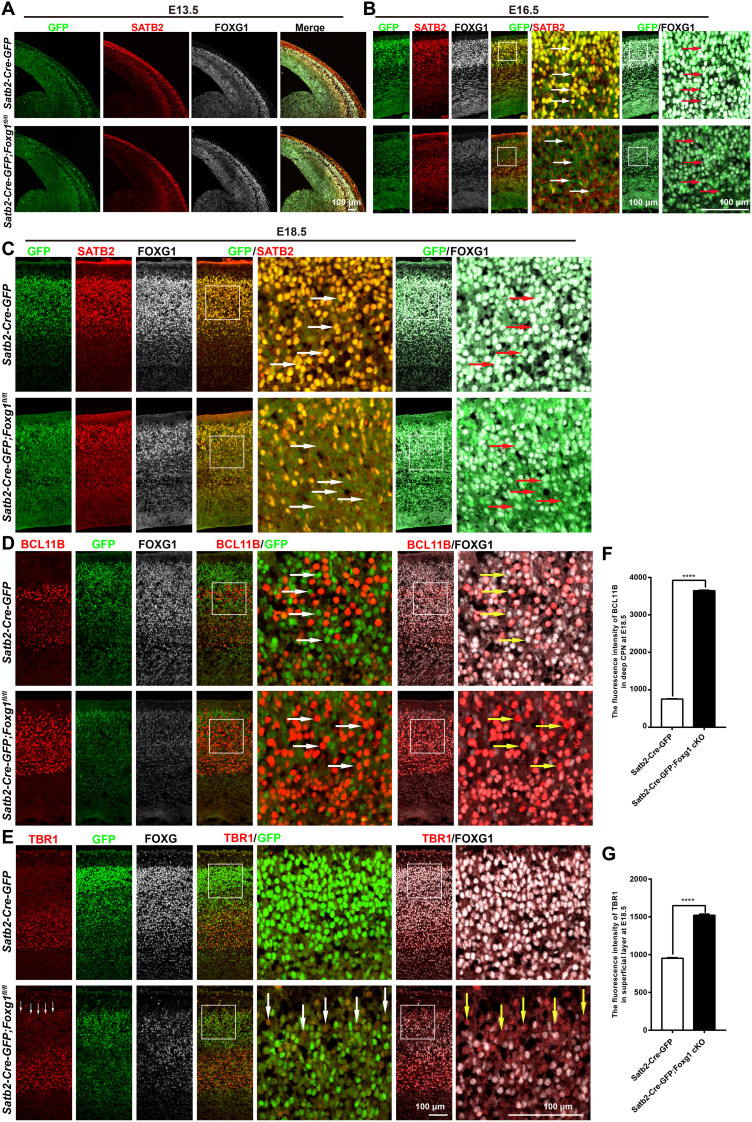
To experimentally further confirm that (i) FOXG1 activates Satb2 in both developing deep and superficial CPNs, (ii) FOXG1 represses Tbr1 in superficial CPNs, and (iii) FOXG1 represses Bcl11b in deep CPNs, we used a Satb2-Cre-IRSE-GFP line in which GFP cDNA was introduced into Satb2 locus (46) to delete Foxg1 in CPNs. We first examined the expression pattern of GFP during the time window of E13.5 to E18.5 and found that GFP exhibited the same expression pattern as SATB2 in Satb2-Cre-IRSE-GFP mice, thereby confirming that GFP+ neurons represent both SATB2+ deep CPNs and superficial CPNs (Fig. 8, A to C).
Fig. 8. Disruption of Foxg1 in CPNs.
(A) Triple immunostaining for GFP, SATB2, and FOXG1 at the E13.5 cortex, showing that very few GFP+SATB2+ CPNs existed in the CP in both control and Satb2-Cre-GFP;Foxg1 cKO mice. The level of FOXG1 was comparable between control and Satb2-Cre-GFP;Foxg1 cKO mice. SATB2 level was also comparable. (B and C) Triple immunostaining for GFP, SATB2, and FOXG1 at the E16.5 and E18.5 cortices, showing that GFP was strongly coexpressed in both of SATB2+ deep CPNs and superficial CPNs (arrows) in control mice. In Satb2-Cre-GFP;Foxg1 cKO mice, the levels of both FOXG1 and GFP were significantly decreased, and SATB2 was also severely reduced in GFPweakFOXG1weak neurons (arrows). (D) Triple immunostaining for GFP, BCL11B, and FOXG1 at the E18.5 cortex. BCL11B was not expressed in many GFPstrongFOXG1strong deep CPNs in the control mice (arrows). In the Satb2-Cre-GFP;Foxg1 cKO mice, BCl11B significantly accumulated in GFPweakFOXG1weak neurons in the deep layer (arrows). (E) Triple immunostaining for GFP, TBR1, and FOXG1 at the E18.5 cortex, showing a slight increase in TBR1 in GFPweakFOXG1weak neurons in the superficial layer in the Satb2-Cre-GFP;Foxg1 cKO mice (arrows). (F) Quantification of fluorescence intensity, showing that BCl11B was significantly increased in GFPweakFOXG1weak neurons of the deep layer in the Satb2-Cre-GFP;Foxg1 cKO mice. (G) Quantification of fluorescence intensity, showing that TBR1 level was increased in GFPweakFOXG1weak neurons, which populated in the superficial layer. Data were presented as means ± SEM; unpaired Student’s t test. ****P < 0.0001.
We then explored the efficiency of Foxg1 deletion and the identities of Foxg1-deficient neurons in Satb2-Cre-GFP;Foxg1 cKO brains. At E13.5, a time point when very few of deep CPNs appeared in the CP, we found the FOXG1 level was not obviously changed in GFP+ neurons in the CP in Satb2-Cre-GFP;Foxg1 cKO mice, suggesting that Foxg1 was not obviously disrupted at E13.5 in the developing cortex. No obvious change in the SATB2 level was detected at this time point either (Fig. 8A).
At E16.5, we found the expression level of FOXG1 was significantly decreased in the majority of GFP+ neurons in Satb2-Cre-GFP;Foxg1 cKO cortices (Fig. 8B). Note that expression of GFP was also severely decreased. Because GFP was introduced into the Satb2 locus in the Satb2-Cre-GFP line, GFP expression is likely to be regulated by the activation of FOXG1 on the Satb2 promotor as well, which could result in this observed decrease in GFP. It seemed that Foxg1 was not disrupted in a small number of CPNs in which high levels of FOXG1, GFP, and SATB2 remained (Fig. 8B). Similar results were obtained at E18.5 in Satb2-Cre-GFP;Foxg1 cKO mice (Fig. 8C). We then examined the identities of GFPweakFOXG1weak neurons at E18.5. The expression level of SATB2 was significantly decreased throughout Satb2-Cre-GFP;Foxg1 cKO cortices (Fig. 8C), and this was accompanied by an obvious accumulation of BCl11B in the deep layer and a slight increase in TBR1 in the superficial layer (Fig. 8, D and E). Quantification of fluorescence intensity showed that TBR1 level was increased in GFPweakFOXG1weak neurons, which populated in the superficial layer (Fig. 8G), similar to the observations in CAG-Cre;Foxg1 cKO mice. In GFPweakFOXG1weak neurons in the deep layer, BCl11B was significantly increased (Fig. 8F). Together, these results reveal that FOXG1 activates Satb2 in both deep and superficial CPNs, represses Tbr1 in superficial CPNs, and represses Bcl11b in deep CPNs. Collectively, these various cell type–specific disruption and overexpression experiments establish that FOXG1 controls postmitotic projection neuron subtype specification in both developmental stage–specific and neuron subtype–specific manners, doing so by regulating both induction and repression programs (Fig. 9).
Fig. 9. Summary model of the multiple regulatory functions of FOXG1 in spatiotemporally controlling postmitotic specification of cortical projection neurons.
“┫” indicates the dominant repression of SOX5 on Fezf2. The thick arrow indicates the dominant activation of SOX4/SOX11 on Fezf2. The thin arrow indicates the weak activation of FOXG1 at the Fezf2 enhancer site.
DISCUSSION
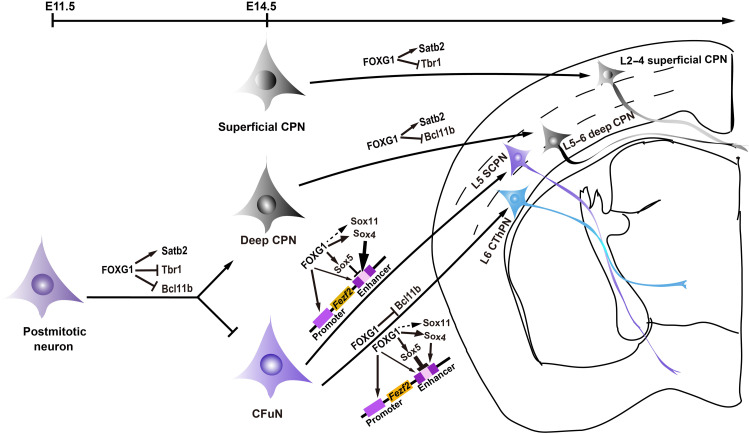
Much remains unknown about the sequential postmitotic specification of neuron subtypes during cortical development. By combining cKO mice and finely time-resolved sampling, we here show that the transcription factor FOXG1 exerts at least four major regulatory impacts during cortical neuron development. First, in postmitotic neurons before E14.5, FOXG1 directly activates Satb2 and represses both Bcl11b and Tbr1 to enforce deep CPN identity. Second, for specification of superficial CPNs and deep CPNs (after E14.5), FOXG1 still activates Satb2 and represses Tbr1 but no longer represses Bcl11b in superficial CPNs. Conversely, FOXG1 no longer represses Tbr1 in deep CPNs. Third, we found a sophisticated regulatory axis wherein FOXG1 specifies CThPN versus SCPN fate after E14.5 by fine-tuning Fezf2 levels: FOXG1 directly activates transcription of both Fezf2 and multiple Sox family genes (Sox5 and Sox4/Sox11); FOXG1 then competes with SOX proteins for binding at an enhancer site in the regulatory region of the Fezf2 locus. Fourth, we finally show that FOXG1 also directly represses Bcl11b transcription in CThPNs from E14.5 onward (Fig. 9).
Among the forkhead transcription factor family, FOXG1 represents a single subclass and consists of a proline-rich transcriptional activation domain (N-terminal) (47–49), a forkhead DNA binding domain, a Groucho-binding domain to recruit Groucho/TLE (Transducin-like Enhancer of Split-1) forming a transcriptional corepressor complex, and a JARID1B (the H3K4me2/3 histone demethylase Jarid1b)–binding domain (through which FOXG1 epigenetically represses its garget genes) (50–52). In recent years, numerous studies have demonstrated pleiotropic roles of FOXG1, ranging from cell proliferation and migration to cortical circuit specialization during telencephalon development (25, 53–55). FOXG1 exerts multiple functions, including both repression and activation roles, based on forming a complex with specific proteins crucial for gene expression or by directly binding to cis-regulatory elements of its target genes. For instance, it has been reported that FOXG1 (i) represses Coup-TFI by binding at its repressor (56), (ii) represses Wnt8b by binding to its promotor (57), and (iii) represses Robo1, Slit3, and Reelin by forming a FOXG1-RP58 complex that directly binds at Foxg1-Rp58–binding sites (25). It has also been demonstrated that FOXG1 activates Kcnh3 by binding at its enhancer (58). FOXG1 may directly activate Fgf8 (59) and Sox9 (60). Thus, FOXG1 variously controls both induction and repression programs, doing so in both developmental stage–specific and neuron subtype–specific manners.
Fate choice of deep CPNs versus CFuNs before E14.5
Deep CPNs share a common birthdate with CFuNs during early corticogenesis. These newborn neurons proceed along two distinct trajectories: toward TBR1highBCL11BhighSATB2− CFuNs or toward TBR1lowBCL11BlowSATB2high deep CPNs (8). It is known that the Satb2 transcription must be postmitotically activated and that Bcl11b and Tbr1 have to be repressed to enforce deep CPN identity (14, 36, 61). We are unaware of any postmitotic studies that have specifically investigated how deep CPN versus CFuN identity is specified before E14.5. We have data starting from E11.5, and we show that Foxg1 disruption in postmitotic neurons causes deep CPNs to develop into CFuNs. Specifically, these results support that FOXG1 specifies deep CPN identity by directly activating Satb2 and simultaneously repressing both Bcl11b and Tbr1 in these cells.
There is an apparent discrepancy between our detection of elevated BCL11B and a report of decreased BCL11B in the CP in NEX-Cre;Foxg1 cKO mice (25). Some methodological considerations bear mention. Our findings about elevated BCL11B were based on quantification of the number of BCL11B+ neurons, as well as on cell tracing using BrdU. The conclusion from Cargnin et al. (25) was based on an immunostaining image. Moreover, their observation that BCL11B+ deep layer neurons were broadly dispersed throughout the cortex is entirely in line with our findings.
It is known that CFuN specification requires postmitotic activation of Bcl11b and Tbr1 (8, 12). Moreover, Fezf2 is strongly coexpressed in TBR1highBCL11Bhigh CFuNs, and a Fezf2-Bcl11b pathway has been reported to regulate the fate choice of SCPNs versus CPNs (13, 15, 17). Several observations from our present study are quite informative when considered in light of this background knowledge. For example, our data suggest that FEZF2 may not be required for the postmitotic maintenance of TBR1highBCL11Bhigh CFuN identity, at least before E14.5: When Foxg1 was deleted in postmitotic neurons at E11.5 in the brains of our NEX-Cre;Foxg1 cKO mice, FEZF2 was undetectable in TBR1highBCL11Bhigh CFuNs before E14.5, although cells staining for this TBR1highBCL11Bhigh CFuN identity were present throughout the cortex (Fig. 4A).
FOXG1 participates in fine-scale regulation of Fezf2 transcription in CThPNs and SCPNs via both transcriptional and protein-level engagement with SOX members
Previous work has established that SCPNs and CThPNs share a common TBR1highFEZF2highBCL11Bhigh molecular profile during early corticogenesis; these cells later diverge and develop into TBR1highFEZF2lowBCL11Blow CThPNs or TBR1lowFEZF2highBCL11Bhigh SCPNs. We know that a high FEZF2 level directs CFuNs toward a SCPN fate, whereas a low level of this protein permits a CThPN fate (15–18, 39, 62). It was conspicuous we did not observe the expected FEZF2high SCPN/FEZF2low CThPN expression pattern in our cKO cortices; rather, we found that FEZF2 was expressed uniformly in both neuron subtypes, which consequently were unable to reduce the BCL11B level in CThPNs.
Here, we characterized three regulatory layers through which FOXG1 precisely regulates the extent of FEZF2 transcription. FOXG1 directly activates Fezf2 transcription. It also activates Sox5, Sox4, and Sox11, three genes known to function as upstream regulators of FEZF2 transcription. Last, we show that FOXG1 can compete with SOX proteins at the Fezf2 enhancer site. This apparently three-layer regulation of FOXG1 for Fezf2 underscores that developing neurons are apparently highly sensitive to the FEZF2 level, which must be very precisely regulated. Previous studies have demonstrated that SOX5 exerts a dominant repression influence on Fezf2 transcription in CThPNs, whereas SOX4/SOX11 drives overwhelming Fezf2 activation in SCPNs (20–22). Note that FEZF2 expression was severely reduced in NEX-Cre;Foxg1 cKO cortices before E14.5 but gradually elevated afterward. Despite this expression rebound, note that at P0, FEZF2 showed a uniform expression pattern in SCPNs and CThPNs in NEX-Cre;Foxg1 cKO cortices, rather than a SCPNhighCThPNlow pattern in the P0. Note that from E14.5 onward, both Sox4 and Sox11, which have been reported to activate Fzef2 transcription, were rebound in our cKO mice. These results suggest that (i) other regulatory mechanisms beyond Fezf2 transcription are involved from E14.5 onward, and (ii) in the process of CFuNs diverging and developing into CThPNs or SCPNs, FOXG1 is required for the down-regulation of FEZF2 in the developing CThPNs and the accumulation of FEZF2 in the developing SCPNs. It seems likely that careful experimental characterization of FOXG1 and SOX protein binding competition at the Fezf2 enhancer motif, especially around E14.5 to E16.5, as well as involvement of other mechanisms, could yield insights about the mechanism underlying SCPN versus CThPN fate.
FOXG1/BCL11B in CThPN and FOXG1/TBR1 in SCPN specification after E14.5
To achieve a TBR1highBCL11Blow CThPN profile after E14.5, BCL11B must become reduced in CThPNs. The ectopic high expression of FEZF2/BCL11B in CThPNs can reverse their progression, ultimately directing them toward a SCPN fate (16, 32, 63). Our study found that FOXG1 is a negative regulator that directly represses BCL11B in CThPNs after E14.5.
To achieve a TBR1lowBCL11Bhigh SCPN pattern after E14.5, the TBR1 level needs to be reduced in developing SCPNs (15). Previous work showed an increase in TBR1+ CThPNs in Ctip2−/− mice, suggesting that BCL11B may repress TBR1 in SCPNs (62). Although we here show that FOXG1 is a repressor of Tbr1 transcription in superficial CPNs, we found that FOXG1 does not repress Tbr1 in SCPNs. Thus, our results are congruent with the previous proposal that BCL11B may function as a transcriptional repressor of Tbr1 in SCPNs.
Specification of deep and superficial CPNs after E14.5
SATB2 is postmitotically highly expressed in both deep and superficial CPNs and contributes to specification of their identities (14, 36). In Fezf2−/− mice, a marked accumulation of SATB2 is observed and SCPNs converged to CPNs, but there is no experimental evidence that FEZF2 directly regulates Satb2 (13). The zinc finger and broad complex, tramtrack, bric-a-brac (BTB) domain–containing protein 20 (ZBTB20) was previously reported to repress CPN fate by repressing Satb2 in the isocortex (64). Here, we have identified FOXG1 as an activator of Satb2 and show that FOXG1 enforces the identities of both deep and superficial CPNs (Fig. 9). It will be interesting to explore whether FOXG1 antagonizes ZBTB20 to promote CPN identity.
Deep CPNs exhibit a SATB2highBCL11BlowTBR1low profile, whereas superficial CPNs display a SATB2highBCL11B−TBR1low profile (36, 40). In the present study, we found that after E14.5, FOXG1 directly represses Bcl11b in deep CPNs and that FOXG1 represses Tbr1 in superficial CPNs. Our study therefore reveals mechanisms through which FOXG1 instructs the specification of two neuron subtypes that are quite morphologically and functionally distinct.
Impaired cortical projection neuron subtype specification and FOXG1 syndrome
FOXG1 syndrome is a severe developmental encephalopathy characterized by autistic features including intellectual disability, limited language ability, and prominent movement disorders (65). It is also highly relevant that corpus callosum agenesis has been reported in FOXG1 syndrome (66); corpus callosum agenesis is closely related to cognitive syndromes with high-level associative dysfunction (6). We know that dysfunction of CThPNs and SCPNs results in perceptual-motor dysfunction as well as attention deficit and hyperactivity (2, 3). In the present study, extending beyond the molecular and neuronal developmental phenotypes toward direct clinical relevance, we detected multiple brain anatomy phenotypes in mice lacking Foxg1 that mirror the human disorder FOXG1 syndrome. Specifically, these mice have a severely disrupted corpus callosum (exhibiting an agenesis phenotype specifically affected by deep and superficial CPNs), as well as impaired corticothalamic tracts (related to CThPNs) and corticospinal tracts (related to SCPNs). Our study thus deepens understanding of the timing and molecular nature of the activation/repression transcription networks that control subtype specification for cortical projection neurons and sheds light on the pathogenesis FOXG1-associated dyskinesia and cognitive deficits and related neurodevelopmental disorders.
MATERIALS AND METHODS
Mice
Foxg1fl/fl mice and CAG-loxp-stop-loxp-Foxg1-IRES-EGFP mice were generated as previously described (26, 45). ROSA-Ai9-tdTomato (stock no. 007905), CAG-CreER (stock no. 004453), and Satb2-Cre-GFP mice (stock no. 030546) were purchased from the Jackson Laboratory. Ntsr1-Cre mice (MMRRC:030648-UC) and Rbp4-Cre mice (MMRRC:037128-UCD) were purchased from the Mutant Mouse Regional Resource Centers (MMRRC) (44). NEX-Cre mice were previously described (30). CAG-CreER;Foxg1fl/fl, NEX-Cre;Foxg1fl/fl, Ntsr1-Cre;Foxg1fl/fl, Rbp4-Cre;Foxg1fl/fl, and Satb2-Cre-GFP;Foxg1fl/fl mice were referred to as CAG-CreER;Foxg1 cKO, NEX-Cre;Foxg1 cKO, Ntsr1-Cre;Foxg1 cKO, Rbp4-Cre;Foxg1 cKO, and Satb2-Cre;Foxg1 cKO mice, respectively. Foxg1fl/fl mice were referred to as control mice. The day of the vaginal plug detection was assigned as E0.5. The day of birth was assigned as P0. Unless noted otherwise, all experiments were performed using mice maintained on a CD1 background. No significant differences based on sex were observed, and data were pooled between sexes. All mouse studies were approved by the Southeast University Institutional Animal Care and Use Committee and were performed in accordance with institutional and national guidelines.
TM induction
TM (Sigma-Aldrich, catalog no. T5648) was dissolved in corn oil at the concentration of 20 mg/ml. Pregnant mice were dosed with TM (2 mg per 40 g of body weight) once at E14.5.
Cell tracing and immunofluorescence
For BrdU (Sigma-Aldrich, catalog no. B5002) labeling, timed pregnant females were intraperitoneally administered with 50 mg/kg body weight at E12.5, E13.5, and E15.5. Brains of paired littermates were examined at P0 (E12.5BrdU and E13.5BrdU) or P2 (E15.5BrdU). Briefly, mice were transcardially perfused with 4% paraformaldehyde (PFA), cryoprotected in 30% sucrose, and sectioned on a cryostat (Leica, CM 3050S). Immunofluorescence staining was performed as previously described (67): Briefly, sections were permeabilized in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 for 0.5 hours, blocked with 10% calf serum for 2 hours, and incubated in primary antibody diluted in blocking solution overnight at 4°C. Sections were then incubated in secondary antibody for 2 hours at room temperature (RT). As for BrdU immunofluorescence, sections were fixed in 4% PFA for 20 min at RT after the aforementioned incubation and were treated with 2 N HCl at 37°C for 0.5 hours, rinsed three times in PBS, and then incubated with anti-BrdU antibody overnight at 4°C followed by secondary antibody incubation for 2 hours at RT. Images were captured with a confocal microscope (FluoView FV1000, Olympus).
In situ hybridization analyses
Digoxigenin (DIG) UTP (uridine 5′-triphosphate)–labeled riboprobes were used. The primers for probes are seen in table S1. Probes were obtained by PCR amplification. In situ hybridization was performed as previously described (45). Simply, E14.5 brains were dissected out and immersed in 4% diethyl pyrocarbonate (DEPC)–PFA overnight; E16.5 and P0 brains were transcardially perfused with 4% DEPC-PFA and fixed in 4% DEPC-PFA overnight. Brains were then cryoprotected in 30% sucrose/DEPC-PBS at 4°C. Brain sections were hybridized with digoxigenin-labeled antisense RNA probes at 65°C overnight, incubated with anti–digoxigenin–alkaline phosphatase antibody for 2 hours at RT, and then subjected to color development.
Western blotting analyses
Western blotting was performed according to a standard protocol (68). Cortex extracts at P0 were separated on 8% SDS–polyacrylamide gel electrophoresis gel and transferred to nitrocellulose membranes. The membranes were blocked for 2 hours in 10% nonfat dry milk containing 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.1% Tween 20 and incubated with primary antibodies overnight at 4°C followed by secondary horseradish peroxidase–linked anti-rabbit immunoglobulin G (IgG) incubation for 2 hours at RT. Proteins were detected using chemiluminescence SuperSignal West Dura kit (Thermo Fisher Scientific).
Retrograde tracing
For in vivo retrograde tracing, DiI (Invitrogen, catalog no. 1818465) was injected into the pyramid region of the medulla at P2 live mice under ice anesthesia (39). Brains were collected at P20.
RNA isolation, reverse transcription, and qPCR analysis
RNA isolation, reverse transcription, and qPCR were performed as previously described (69). Dorsal cortices were dissected from control mice over the developmental stages of E12.5 to P0. Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN, 74104) and was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, RR047A). The qPCRs were performed using the SYBR Green fluorescent master mix (Roche, catalog no. 04707516001) on the Step One-Plus Real-Time PCR System (Applied Biosystems). Relative mRNA expression levels were normalized with the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers for qPCR were designed with Integrated DNA Technologies (IDT; https://sg.idtdna.com) and are listed in table S1. At least three brains from three different litters were used, and samples were run in three duplicates of each sample. All of values were presented as means ± SEM.
Identification of FOXG1-binding sites
The binding sites of FOXG1 at Fezf2 promotor and enhancer, and Tbr1 promotor have been previously reported by Eckler et al. (41) and by Hanashima and co-workers (43). For picking up binding sites at transcription factors of Sox5, Sox4, Sox11, Satb2, and Bcl11b, we first scored a 10-kb upstream region of each transcription start site to predict its promoter domain using BDGP (Berkeley Drosophila Genome Project) (www.fruitfly.org/seq_tools/promoter.html) and Promoter 2.0 (https://services.healthtech.dtu.dk/service.php?Promoter-2.0). Next, we evaluated the conservation of the consensus binding sites of RYAAAYA of FOXG1 at each putative promotor locus across species at the UCSC database (http://genome.ucsc.edu/) as previously reported (25, 43, 70). Third, we scored the binding motifs in each putative promoter by JASPAR (https://jaspar.genereg.net/); high-score binding motifs were selected and were further compared with the ChIP-seq data reported by Cargnin et al. (25). Last, promotor domains with high-score binding motifs that were validated by the FOXG1-bound ChIP-seq peak data were selected for further ChIP-qPCR and luciferase assays. As for Bcl11b, we identified a repressor site containing FOXG1-binding motifs using the VISTER Enhancer Browser (http://enhancer.lbl.gov/), which we compared with the ChIP-seq data reported by Cargnin et al. (25).
ChIP-qPCR analyses
ChIP was performed as described previously using an EZ-Magna ChIP A/G kit (Millipore, catalog no. 17-10086) (71). Briefly, dorsal cortices from E14.5 were mechanically homogenized, and samples were cross-linked with 1% formaldehyde for 15 min and quenched with 125 mM glycine. The chromatin was sheared into 200 to 1000 base pairs for immunoprecipitation using 3 μg of rabbit antibodies against FOXG1, and an equal amount of rabbit IgG was applied as negative control. One percent unimmunoprecipitated input DNA served as internal control. After cross-linking was reversed, immunoprecipitated DNA and input DNA were purified for qPCR analyses. The primers for qPCR were designed with IDT and are listed in table S1. All experiments were run in three duplicates and repeated at least three independent times. ChIP-qPCR data were normalized to the amount of chromatin input. The percentage of input was calculated as follows: % of input = 1% × 2−ΔCt (C[T] IP sample − C[T] input sample). C[T] = CT = average threshold cycle of PCR. The values were presented as means ± SEM.
Luciferase assay
The luciferase assay was performed as previous described (22, 71). Overexpression vectors of pCAG-Foxg1 (mouse; NM_008241), pCAG-Sox4 (mouse; NM_009238), pCAG-Sox5 (mouse; AB006330), and pCAG-Sox11 (mouse; NM_009234) were customized from Synbio Technologies (www.synbio-tech.com; Beijing, China). Reporter vectors of pGL3-Fezf2-Pro (chr14:13177700-13181294), pGL3-Fezf2-En (chr14:13170122-13170950), pGL3-Sox4-Pro (crh13:29045352-29047551), pGL3-Sox5-Pro (crh6:144157589-144160688), pGL3-Sox11-Pro (crh12:28027000-2809583), pGL3-Satb2-Pro (chr1:57026666-57030006), and pGL3-Bcl11b-rep (chr12: 109189361-109192204) were customized from GENEWIZ (www.Genewiz.com; Nanjing, China). pGL3-Tbr1-Pro (chr2: 61638039-61642764) was homemade, in which the promoter fragment of Tbr1 amplified by PCR was subcloned into the pGL3 basic vector between 5′ Nhe I and 3′ Hind III. CE Design V1.04 was used for primer design (Vazyme, Nanjing, China). Homologous recombination was performed with the ClonExpress II One Step Cloning Kit (Vazyme, catalog no. C112-01). The reporters in which the FOXG1-binding motifs were deleted/point mutated were generated using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme, catalog no. C214-01). Deletion/mutation information is presented in table S3. All of the constructs were verified by sequencing.
Neuro2a cells [RRID (Research Resource Identifiers): CVCL0470; The Cell Bank of the Chinese Academy of Science, Shanghai, China] were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Thermo Fisher Scientific, catalog no. 11320082) supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, catalog no. 10099141C), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Millipore, catalog no. 516104-M). A total of 1 × 10−5 cells per cell were seeded for transfection. The transfection complex was prepared including the pCAG vector (control) or overexpression vectors (250 ng per cell), control or mutation reporter vector (200 ng per cell), pRL-SV40 plasmid containing Renilla luciferase gene (25 ng per cell), and Lipofectamine 2000 (2.5 μl per cell) (Thermo Fisher Scientific, catalog no. 11668-019). For normalization of luciferase activity, the pRL-SV40 vector encoding Renilla luciferase was used as an internal control reporter and was used in combination with any experimental reporter vector to cotransfect mammalian cells. The cells were harvested 24 hours after transfection. Firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, E1910). The ratio of Firefly luciferase readouts and Renilla luciferase readout was calculated to represent the activity of the reporter vector. Three replicate wells were made for each transfection. Each experiment was repeated at least three times, and all results are shown as means ± SEM.
In utero electroporation
For in utero electroporation, timed-pregnant Foxg1fl/fl females (E12.5) were deeply anesthetized, and embryos were surgically manipulated as described previously (72). Plasmids encoding GFP only (pNeuroD1-IRES-GFP) or Cre-GFP (pNeuroD1-Cre-HA- IRES-GFP) (2 μg/μl) (37) were injected directly into the lateral ventricles and electroporated with paddle electrodes across the cerebrum. Electroporation was performed using Tweezertrodes (diameter of 5 mm; BTX) with five pulses (30 V for E12.5 embryos) for 50-ms duration and 950-ms intervals using a square-wave pulse generator (ECM830, BTX). The uterus was then returned to the abdominal cavity, and the inner skin and outer skin were sutured. Surgically manipulated pregnant mice were then put on an electric heating plate (50°C) until they awoke. Progenies were euthanized at E18.5 for analysis. Three pairs of control and Foxg1 knockdown brains were analyzed.
Quantification of histological analyses
The confocal images were acquired by FluoView FV1000 confocal microscopy (Olympus) with 20× objective lens. A minimum of three successive coronal sections crossing the somatosensory cortex were selected. Quantitative analyses of neurons were consistently performed in the same area in the somatosensory cortex by direct comparisons of brain sections. At least three brains of each genotype from three different litters were used. The experiments of cell counting were all cross-quantified blindly (i.e., the investigator was unaware of the genotypes). All results are shown as means ± SEM, except indicated otherwise. GraphPad Prism 7 software was used for statistical analyses, and multiple t test with Bonferroni correction and unpaired Student’s t test were used for two-group comparisons. The following convention was used: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
In Fig. 1 (F to H), to measure SATB2high and BCL11BhighTBR1high cells in the control and NEX-Cre;Foxg1 cKO cortices, a square box spanning the marginal zone to the white matter of the somatosensory cortex was overlaid, and all SATB2high and BCL11BhighTBR1high neurons within the box were quantified. In detail, cells in a box of 180 μm by 360 μm at E14.5, 180 μm by 420 μm at E16.5, and 300 μm by 500 μm at P0 were quantified with Image-Pro Plus software (Media Cybernetics).
In Fig. 2H, to measure TBR1high, BCL11BhighTBR1high, SATBhigh, and BCL11Bhigh neurons at P0 in control and CAG-CreER;Foxg1 cKO, a box of 175 μm by 500 μm in the somatosensory cortex was overlaid. The upper boundary of BCL11B+ neurons was used to demarcate the superficial and deep layers, and neurons in the deep layer within the box were counted.
In Fig. 3 (G to I) and fig. S5 (C and D), cell tracing was performed to detect subtype specification. Only the first-generation wave of BrdU+ neurons with strong and homogeneous nuclear labeling was counted. BrdU+, SATBhighBrdU+, BCL11BhighBrdU+, and TBR1highBrdU+ neurons in the entire cortex were counted in a box of 175 μm by 500 μm at P0 (for E12.5BrdU and E13.5BrdU labeling) and a box of 175 μm by 600 μm at P2 (for E15.5BrdU labeling). The proportions of SATBhigh BrdU+, BCL11Bhigh BrdU+, and TBR1high BrdU+ neurons among total BrdU+ neurons were calculated.
In Fig. 7E, to measure TBR1highBCL11Blow and TBR1highBCL11Bhigh neurons in control and Ntsr1-Cre;Foxg1 cKO cortices, a box of 300 μm by 800 μm in the somatosensory cortex was overlaid. All neurons in L6 in this box were quantified. In Fig. 7F, to measure tdTom+, SATB2+tdTom+, BCL11B+tdTom+, and TBR1+tdTom+ neurons in control and Rbp4-Cre;Foxg1 cKO cortices, a box of 300 μm by 800 μm in the somatosensory cortex was overlaid. All tdTom+, SATB2+tdTom+, BCL11B+tdTom+, and TBR1+tdTom+ neurons in this box were quantified, and the ratios of SATB2+tdTom+, BCL11B+tdTom+, and TBR1+tdTom+ neurons in tdTom+ neurons were calculated.
In fig. S3C, all GFP+, SATB2+GFP+, BCL11B+GFP+, and TBR1+GFP+ neurons in the cortices of three pairs of control and Foxg1 knockdown brains were measured, and the ratios of SATB2+GFP+, BCL11B+GFP+, and TBR1+GFP+ neurons in GFP+ neurons were calculated. In fig. S6, to measure PKC-γ+ neurons in control and CAG-CreER;Foxg1 cKO brains at P8, a box of 300 μm by 800 μm was overlaid, and all labeled neurons within the box were quantified.
Quantification of fluorescence intensity
Immunostaining was performed as described above. Sections were imaged with the same confocal acquisition parameters at the same level for each of the individual protein assessed with immunostaining. The mean fluorescence intensities were analyzed using the FV10-ASW4.2 software, with the same settings. All results are shown as means ± SEM. GraphPad Prism 7 software was used for statistical analyses, and unpaired Student’s t test was used for two-group comparisons. The following convention was used: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
In fig. S2, the mean fluorescence intensities of FOXG1 in deep SATB2high CPNs and BCL11Bhigh CFuNs in CP were measured. At least 200 neurons at E14.5 and E16.5 were measured. Two control brains were analyzed at each time point.
In fig. S9, the mean fluorescence intensities of FOXG1, SATB2, BCL11B, or TBR1 in SCPNs and CThPNs were measured. Two pairs of E18.5 brains from control versus NEX-Cre;CAG-Foxg1 TG mice were analyzed. At least 200 neurons for each population were analyzed.
In Fig. 8, the mean fluorescence intensities of BCL11B in deep CPNs and TBR1 in superficial CPNs were measured. Two pairs of E18.5 brains from control versus Satb2-Cre;Foxg1 cKO mice were used, and at least 450 neurons were analyzed for each population.
Statistical analysis for ChIP-qPCR and luciferase assay
The data of ChIP-qPCR were analyzed with unpaired Student’s t test for two-group comparisons [Figs. 4 (F and G), 5 (D to F), and 6 (B, F, and J)]. The data of the luciferase assay were analyzed with one-way analysis of variance (ANOVA) followed by Tukey-Kramer post hoc test for multiple-group cotransfection tests [Figs. 4 (I and J), 5 (D to I and L to N), and 6 (C, G, and K)].
Supplementary Material
Acknowledgments
We thank M. Fang, C. Lin, and J. Li at Southeast University, as well as S. J. Pleasure at UCSF for insightful comments and suggestions. We thank D. Zhang at Peking University for gift of pNeuroD1 plasmid. We thank Y. Wei and L. Liu for assistance with laboratory and animal care, and other members of the laboratory for discussions.
Funding: The National Natural Science Foundation of China grant 31930045, major projects of the Ministry of Science and Technology of China 2021ZD0202304, and National Natural Science Foundation of China grant 81870899.
Author contributions: Conceptualization: C.Z. and J.L. Methodology: C.Z. and J.L. Investigation: J.L., M.Y., M.S., B.L., K.Z., C.S., R.B., B.Y., R.B., B.Z., Z.Z., W.F., K.W., M.Z., and J.H. Visualization: J.L. Supervision: C.Z. Writing—original draft: C.Z. and J.L. Writing—review and editing: C.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Correction (12 July 2022): The authors inadvertently provided the incorrect labels in their Fig. 3H, Fig. 5A-C, and Fig. 7E-F. The figures have been replaced in the PDF and HTML (full text).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S3
REFERENCES AND NOTES
- 1.Lodato S., Shetty A. S., Arlotta P., Cerebral cortex assembly: Generating and reprogramming projection neuron diversity. Trends Neurosci. 38, 117–125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welniarz Q., Dusart I., Roze E., The corticospinal tract: Evolution, development, and human disorders. Dev. Neurobiol. 77, 810–829 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Yuste R., Hawrylycz M., Aalling N., Aguilar-Valles A., Arendt D., Armañanzas R., Ascoli G. A., Bielza C., Bokharaie V., Bergmann T. B., Bystron I., Capogna M., Chang Y. J., Clemens A., de Kock C. P. J., DeFelipe J., Dos Santos S. E., Dunville K., Feldmeyer D., Fiáth R., Fishell G. J., Foggetti A., Gao X., Ghaderi P., Goriounova N. A., Güntürkün O., Hagihara K., Hall V. J., Helmstaedter M., Herculano-Houzel S., Hilscher M. M., Hirase H., Hjerling-Leffler J., Hodge R., Huang J., Huda R., Khodosevich K., Kiehn O., Koch H., Kuebler E. S., Kühnemund M., Larrañaga P., Lelieveldt B., Louth E. L., Lui J. H., Mansvelder H. D., Marin O., Martinez-Trujillo J., Chameh H. M., Mohapatra A. N., Munguba H., Nedergaard M., Němec P., Ofer N., Pfisterer U. G., Pontes S., Redmond W., Rossier J., Sanes J. R., Scheuermann R. H., Serrano-Saiz E., Staiger J. F., Somogyi P., Tamás G., Tolias A. S., Tosches M. A., García M. T., Wozny C., Wuttke T. V., Liu Y., Yuan J., Zeng H., Lein E., A community-based transcriptomics classification and nomenclature of neocortical cell types. Nat. Neurosci. 23, 1456–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avram M., Brandl F., Bauml J., Sorg C., Cortico-thalamic hypo- and hyperconnectivity extend consistently to basal ganglia in schizophrenia. Neuropsychopharmacology 43, 2239–2248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fame R. M., MacDonald J. L., Macklis J. D., Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 34, 41–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown W. S., Paul L. K., The neuropsychological syndrome of agenesis of the corpus callosum. J. Int. Neuropsychol. Soc. 25, 324–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Leon Reyes N. S., Bragg-Gonzalo L., Nieto M., Development and plasticity of the corpus callosum. Development 147, (2020). [DOI] [PubMed] [Google Scholar]
- 8.Greig L. C., Woodworth M. B., Galazo M. J., Padmanabhan H., Macklis J. D., Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata M., Gulden F. O., Sestan N., From trans to cis: Transcriptional regulatory networks in neocortical development. Trends Genet. 31, 77–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paolino A., Fenlon L. R., Suarez R., Richards L. J., Transcriptional control of long-range cortical projections. Curr. Opin. Neurobiol. 53, 57–65 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Rouaux C., Bhai S., Arlotta P., Programming and reprogramming neuronal subtypes in the central nervous system. Dev. Neurobiol. 72, 1085–1098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leone D. P., Srinivasan K., Chen B., Alcamo E., McConnell S. K., The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 18, 28–35 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B., Wang S. S., Hattox A. M., Rayburn H., Nelson S. B., McConnell S. K., The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 105, 11382–11387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcamo E. A., Chirivella L., Dautzenberg M., Dobreva G., Fariñas I., Grosschedl R., McConnell S. K., Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Molyneaux B. J., Arlotta P., Hirata T., Hibi M., Macklis J. D., Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47, 817–831 (2005). [DOI] [PubMed] [Google Scholar]
- 16.McKenna W. L., Betancourt J., Larkin K. A., Abrams B., Guo C., Rubenstein J. L. R., Chen B., Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J. Neurosci. 31, 549–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J. G., Rasin M. R., Kwan K. Y., Sestan N., Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 102, 17792–17797 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B., Schaevitz L. R., McConnell S. K., Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 102, 17184–17189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arlotta P., Molyneaux B. J., Chen J., Inoue J., Kominami R., Macklis J. D., Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Shim S., Kwan K. Y., Li M., Lefebvre V., Sestan N., Cis-regulatory control of corticospinal system development and evolution. Nature 486, 74–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai T., Jabaudon D., Molyneaux B. J., Azim E., Arlotta P., Menezes J. R. L., Macklis J. D., SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron 57, 232–247 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Kwan K. Y., Lam M. M. S., Krsnik Ž., Kawasawa Y. I., Lefebvre V., Šestan N., SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. U.S.A. 105, 16021–16026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xuan S., Baptista C. A., Balas G., Tao W., Soares V. C., Lai E., Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14, 1141–1152 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi G., Fishell G., Dynamic FoxG1 expression coordinates the integration of multipolar pyramidal neuron precursors into the cortical plate. Neuron 74, 1045–1058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cargnin F., Kwon J.-S., Katzman S., Chen B., Lee J. W., Lee S.-K., FOXG1 orchestrates neocortical organization and cortico-cortical connections. Neuron 100, 1083–1096.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian C., Gong Y., Yang Y., Shen W., Wang K., Liu J., Xu B., Zhao J., Zhao C., Foxg1 has an essential role in postnatal development of the dentate gyrus. J. Neurosci. 32, 2931–2949 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanashima C., Li S. C., Shen L., Lai E., Fishell G., Foxg1 suppresses early cortical cell fate. Science 303, 56–59 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Seoane J., Le H. V., Shen L., Anderson S. A., Massague J., Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Vegas N., Cavallin M., Maillard C., Boddaert N., Toulouse J., Schaefer E., Lerman-Sagie T., Lev D., Magalie B., Moutton S., Haan E., Isidor B., Heron D., Milh M., Rondeau S., Michot C., Valence S., Wagner S., Hully M., Mignot C., Masurel A., Datta A., Odent S., Nizon M., Lazaro L., Vincent M., Cogné B., Guerrot A. M., Arpin S., Pedespan J. M., Caubel I., Pontier B., Troude B., Rivier F., Philippe C., Bienvenu T., Spitz M. A., Bery A., Bahi-Buisson N., Delineating FOXG1 syndrome: From congenital microcephaly to hyperkinetic encephalopathy. Neurol Genet 4, e281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goebbels S., Bormuth I., Bode U., Hermanson O., Schwab M. H., Nave K. A., Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44, 611–621 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Lu J. R., McKinsey T. A., Xu H., Wang D. Z., Richardson J. A., Olson E. N., FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 19, 4495–4502 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galazo M. J., Emsley J. G., Macklis J. D., Corticothalamic projection neuron development beyond subtype specification: Fog2 and intersectional controls regulate intraclass neuronal diversity. Neuron 91, 90–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cubelos B., Sebastián-Serrano A., Beccari L., Calcagnotto M. E., Cisneros E., Kim S., Dopazo A., Alvarez-Dolado M., Redondo J. M., Bovolenta P., Walsh C. A., Nieto M., Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron 66, 523–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Britanova O., Akopov S., Lukyanov S., Gruss P., Tarabykin V., Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 21, 658–668 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Dennis D. J., Wilkinson G., Li S., Dixit R., Adnani L., Balakrishnan A., Han S., Kovach C., Gruenig N., Kurrasch D. M., Dyck R. H., Schuurmans C., Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc. Natl. Acad. Sci. U.S.A. 114, E4934–E4943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Britanova O., de Juan Romero C., Cheung A., Kwan K. Y., Schwark M., Gyorgy A., Vogel T., Akopov S., Mitkovski M., Agoston D., Šestan N., Molnár Z., Tarabykin V., Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Wei C., Sun M., Sun X., Meng H., Li Q., Gao K., Yue W., Wang L., Zhang D., Li J., RhoGEF trio regulates radial migration of projection neurons via its distinct domains. Neurosci. Bull. 38, 249–262 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W. L., Frost A., Polleux F., The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han W., Kwan K. Y., Shim S., Lam M. M. S., Shin Y., Xu X., Zhu Y., Li M., Šestan N., TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc. Natl. Acad. Sci. U.S.A. 108, 3041–3046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedogni F., Hodge R. D., Elsen G. E., Nelson B. R., Daza R. A. M., Beyer R. P., Bammler T. K., Rubenstein J. L. R., Hevner R. F., Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc. Natl. Acad. Sci. U.S.A. 107, 13129–13134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckler M. J., Larkin K. A., McKenna W. L., Katzman S., Guo C., Roque R., Visel A., Rubenstein J. L. L., Chen B., Multiple conserved regulatory domains promote Fezf2 expression in the developing cerebral cortex. Neural Dev. 9, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao W., Lai E., Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron 8, 957–966 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Toma K., Kumamoto T., Hanashima C., The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J. Neurosci. 34, 13259–13276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong S., Doughty M., Harbaugh C. R., Cummins A., Hatten M. E., Heintz N., Gerfen C. R., Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci. 27, 9817–9823 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B., Zhou K., Wu X., Zhao C., Foxg1 deletion impairs the development of the epithalamus. Mol. Brain 11, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarvie B. C., Chen J. Y., King H. O., Palmiter R. D., Satb2 neurons in the parabrachial nucleus mediate taste perception. Nat. Commun. 12, 224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassel-Duby R., Hernandez M. D., Yang Q., Rochelle J. M., Seldin M. F., Williams R. S., Myocyte nuclear factor, a novel winged-helix transcription factor under both developmental and neural regulation in striated myocytes. Mol. Cell. Biol. 14, 4596–4605 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bredenkamp N., Seoighe C., Illing N., Comparative evolutionary analysis of the FoxG1 transcription factor from diverse vertebrates identifies conserved recognition sites for microRNA regulation. Dev. Genes Evol. 217, 227–233 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Xie X., Stubbington M. J. T., Nissen J. K., Andersen K. G., Hebenstreit D., Teichmann S. A., Betz A. G., The regulatory T cell lineage factor Foxp3 regulates gene expression through several distinct mechanisms mostly independent of direct DNA binding. PLOS Genet. 11, e1005251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao J., Lai E., Stifani S., The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol. Cell. Biol. 21, 1962–1972 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcal N., Patel H., Dong Z., Belanger-Jasmin S., Hoffman B., Helgason C. D., Dang J., Stifani S., Antagonistic effects of Grg6 and Groucho/TLE on the transcription repression activity of brain factor 1/FoxG1 and cortical neuron differentiation. Mol. Cell. Biol. 25, 10916–10929 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan K., Shaw A. L., Madsen B., Jensen K., Taylor-Papadimitriou J., Freemont P. S., Human PLU-1 has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 278, 20507–20513 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Hanashima C., Shen L., Li S. C., Lai E., Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J. Neurosci. 22, 6526–6536 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumamoto T., Toma K. I., Gunadi, McKenna W. L., Kasukawa T., Katzman S., Chen B., Hanashima C., Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression. Cell Rep. 3, 931–945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyoshi G., Ueta Y., Natsubori A., Hiraga K., Osaki H., Yagasaki Y., Kishi Y., Yanagawa Y., Fishell G., Machold R. P., Miyata M., FoxG1 regulates the formation of cortical GABAergic circuit during an early postnatal critical period resulting in autism spectrum disorder-like phenotypes. Nat. Commun. 12, 3773 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou P. S., Miyoshi G., Hanashima C., Sensory cortex wiring requires preselection of short- and long-range projection neurons through an Egr-Foxg1-COUP-TFI network. Nat. Commun. 10, 3581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith R., Huang Y. T., Tian T., Vojtasova D., Mesalles-Naranjo O., Pollard S. M., Pratt T., Price D. J., Fotaki V., The transcription factor Foxg1 promotes optic fissure closure in the mouse by suppressing Wnt8b in the Nasal Optic Stalk. J. Neurosci. 37, 7975–7993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vezzali R., Weise S. C., Hellbach N., Machado V., Heidrich S., Vogel T., The FOXG1/FOXO/SMAD network balances proliferation and differentiation of cortical progenitors and activates Kcnh3 expression in mature neurons. Oncotarget 7, 37436–37455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martynoga B., Morrison H., Price D. J., Mason J. O., Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Liu F., Hon G. C., Villa G. R., Turner K. M., Ikegami S., Yang H., Ye Z., Li B., Kuan S., Lee A. Y., Zanca C., Wei B., Lucey G., Jenkins D., Zhang W., Barr C. L., Furnari F. B., Cloughesy T. F., Yong W. H., Gahman T. C., Shiau A. K., Cavenee W. K., Ren B., Mischel P. S., EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol. Cell 60, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buttner N., Johnsen S. A., Kugler S., Vogel T., Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 7042–7047 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srinivasan K., Leone D. P., Bateson R. K., Dobreva G., Kohwi Y., Kohwi-Shigematsu T., Grosschedl R., McConnell S. K., A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. U.S.A. 109, 19071–19078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomassy G. S., de Leonibus E., Jabaudon D., Lodato S., Alfano C., Mele A., Macklis J. D., Studer M., Area-specific temporal control of corticospinal motor neuron differentiation by COUP-TFI. Proc. Natl. Acad. Sci. U.S.A. 107, 3576–3581 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen J. V., Thomassen M., Mollgard K., Noraberg J., Jensen N. A., Zbtb20 defines a hippocampal neuronal identity through direct repression of genes that control projection neuron development in the isocortex. Cereb. Cortex 24, 1216–1229 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Hou P. S., hAilin D. O., Vogel T., Hanashima C., Transcription and beyond: Delineating FOXG1 function in cortical development and disorders. Front Cell Neurosci 14, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kortum F., Das S., Flindt M., Morris-Rosendahl D. J., Stefanova I., Goldstein A., Horn D., Klopocki E., Kluger G., Martin P., Rauch A., Roumer A., Saitta S., Walsh L. E., Wieczorek D., Uyanik G., Kutsche K., Dobyns W. B., The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J. Med. Genet. 48, 396–406 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan Y., Jiang Z., Sun D., Li Z., Pu Y., Wang D., Huang A., He C., Cao L., Cyclin-dependent kinase 18 promotes oligodendrocyte precursor cell differentiation through activating the extracellular signal-regulated kinase signaling pathway. Neurosci. Bull. 35, 802–814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J., Zhang X., Zhou Y., Wu B., Tan Z. Y., Up-regulation of P2X7 receptors contributes to spinal microglial activation and the development of pain induced by BmK-I. Neurosci. Bull. 35, 624–636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen W., Ba R., Su Y., Ni Y., Chen D., Xie W., Pleasure S. J., Zhao C., Foxg1 regulates the postnatal development of cortical interneurons. Cereb. Cortex 29, 1547–1560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai S., Li J., Zhang H., Chen X., Guo M., Chen Z., Chen Y., Structural basis for DNA recognition by FOXG1 and the characterization of disease-causing FOXG1 mutations. J. Mol. Biol. 432, 6146–6156 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Du A., Wu X., Chen H., Bai Q.-R., Han X., Liu B., Zhang X., Ding Z., Shen Q., Zhao C., Foxg1 directly represses Dbx1 to confine the POA and subsequently regulate ventral telencephalic patterning. Cereb. Cortex 29, 4968–4981 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Yang F., Hu A., Guo Y., Wang J., Li D., Wang X., Jin S., Yuan B., Cai S., Zhou Y., Li Q., Chen G., Gao H., Zheng L., Tong Q., p113 isoform encoded by CUX1 circular RNA drives tumor progression via facilitating ZRF1/BRD4 transactivation. Mol. Cancer 20, 123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S3