Abstract
The diffuse-type gastric cancer (DGC) is a subtype of gastric cancer (GC) associated with low HER2 positivity rate and insensitivity to chemotherapy and immune checkpoint inhibitors. Here, we identify urokinase-type plasminogen activator receptor (uPAR) as a potential therapeutic target for DGC. We have developed a novel anti-uPAR monoclonal antibody, which targets the domains II and III of uPAR and blocks the binding of urokinase-type plasminogen activator to uPAR. We show that the combination of anti-uPAR and anti–Programmed cell death protein 1 (PD-1) remarkably inhibits tumor growth and prolongs survival via multiple mechanisms, using cell line–derived xenograft and patient-derived xenograft mouse models. Furthermore, uPAR chimeric antigen receptor–expressing T cells based on the novel anti-uPAR effectively kill DGC patient–derived organoids and exhibit impressive survival benefit in the established mouse models, especially when combined with PD-1 blockade therapy. Our study provides a new possibility of DGC treatment by targeting uPAR in a unique manner.
Novel uPAR-targeted strategies that can enhance anti–PD-1 efficacy in diffuse-type gastric cancer are developed.
INTRODUCTION
Gastric cancer (GC) is the third leading cause of cancer-related deaths and ranks fifth for incidence worldwide, and in particular, the incidence rates are highest in Eastern Asia (1, 2). The Lauren classification distinguishes two major subtypes of GC, intestinal and diffuse types (3). Diffuse-type GC (DGC) accounts for approximately 30% of GC cases and is more commonly observed in younger patients (4). It is an aggressive form of GC that is composed of poorly differentiated cells. These cells lack intercellular adhesion, often exhibit scattered signet-ring cell morphology, and spread in the upper layers of the stomach wall instead of protruding into the lumen (3). Because of its rapid disease progression, delayed diagnosis, high metastatic potential, and chemoresistance, DGC is featured with poor prognosis (5). Previously, we showed that the postoperative 5-year overall survival (OS) rate of DGC was only 17.8% in a cohort study (6). In addition, despite an overall declining incidence of GC, an increasing incidence of DGC in recent years has been reported (7). Therefore, DGC still remains a major global health problem and poses a challenge for researchers and physicians.
According to the updated national comprehensive cancer network (NCCN) guidelines (8), either anti-Human epidermal growth factor receptor 2 (HER2) antibody (trastuzumab) or anti–PD-1 antibody (nivolumab) combined with chemotherapy has been recommended as the preferred regimens in first-line therapy for the treatment of HER2 overexpression–positive and HER2 overexpression–negative GC, respectively. In addition, another anti–PD-1 antibody (pembrolizumab) combined with trastuzumab and chemotherapy is also listed as an alternative recommended regimen for HER2 overexpression–positive GC. The phase 3 Trastuzumab for Gastric Cancer (ToGA) trial found that trastuzumab plus chemotherapy significantly improved OS from 11.1 to 13.8 months in HER2 overexpression–positive patients, compared with chemotherapy alone (9). Moreover, the phase 3 CheckMate 649 study involved adult patients with previously untreated, unresectable, and non–HER2-positive GC and showed that nivolumab plus chemotherapy resulted in a significant improvement in OS of patients with high expression of Programmed death ligand 1 (PD-L1), compared with chemotherapy alone (10). However, the use of anti-HER2 antibody is limited to patients with HER2 overexpression, but the HER2 positivity rate in patients with DGC is only 2 to 6% (11, 12), and the phase 3 ATTRACTION-2 study showed a decreased survival benefit in patients with DGC after anti–PD-1 monotherapy [hazard ratio (HR), 0.82; 95% confidence interval (CI), 0.57 to 1.17], compared with patients with intestinal-type GC (HR, 0.59; 95% CI, 0.41 to 0.85) (13). Therefore, it is still urgent to identify other potential targets that are specifically overexpressed in DGC for the development of alternative therapeutic strategies.
The urokinase-type plasminogen activator receptor (uPAR) is the cell surface receptor for the extracellular serine protease urokinase-type plasminogen activator (uPA, also known as urokinase). uPAR not only regulates the proteolysis of the extracellular matrix (ECM) by engaging uPA for activation of the plasminogen activation system but also initiates intracellular signalings involved in cell adhesion, migration, proliferation, and survival by interacting with other partners (e.g., integrins) (14, 15). A number of studies have found that uPAR is highly expressed in various human cancers (16) and that it has pleiotropic activities in the development of cancer (15, 17). ATN-658, a mouse monoclonal antibody (mAb) against human uPAR, was demonstrated to strongly reduce primary tumor growth or tumor metastasis in breast, ovarian, prostate, and colorectal cancers (18–23). ATN-658 does not inhibit uPA binding to uPAR; however, it can bind to the domain III (DIII) of uPAR, thereby blocking uPAR interaction with integrins that are known to play a role in cell motility and proliferation (22). A recent study showed that uPAR-targeted chimeric antigen receptor–expressing T (CAR-T) cells extended the survival of mice with lung adenocarcinoma that were treated with a senescence-inducing combination of drugs, demonstrating the therapeutic potential of uPAR CAR-T cells for senescence-associated diseases and solid tumors (24).
Here, we identify uPAR as a potential therapeutic target for DGC and develop a novel anti-uPAR mAb. It specifically targets the DII-DIII region of uPAR, blocks uPA binding to uPAR, and inhibits GC cell proliferation, migration, and adhesion in vitro. The combination therapy consisting of anti-uPAR and anti–PD-1 shows impressive antitumor effects in GC cell line–derived xenograft (CDX) and DGC patient–derived xenograft (PDX) mouse models via multiple mechanisms. Furthermore, we show that uPAR CAR-T cells effectively kill uPAR-expressing cells and DGC patient–derived organoids in vitro. Treatment with uPAR CAR-T cells along with anti–PD-1 shows remarkable antitumor efficacy, enhancing survival of the CDX and PDX mice. Our study offers promising therapeutic strategies to combat DGC.
RESULTS
uPAR is overexpressed in DGC and is a potential therapeutic target
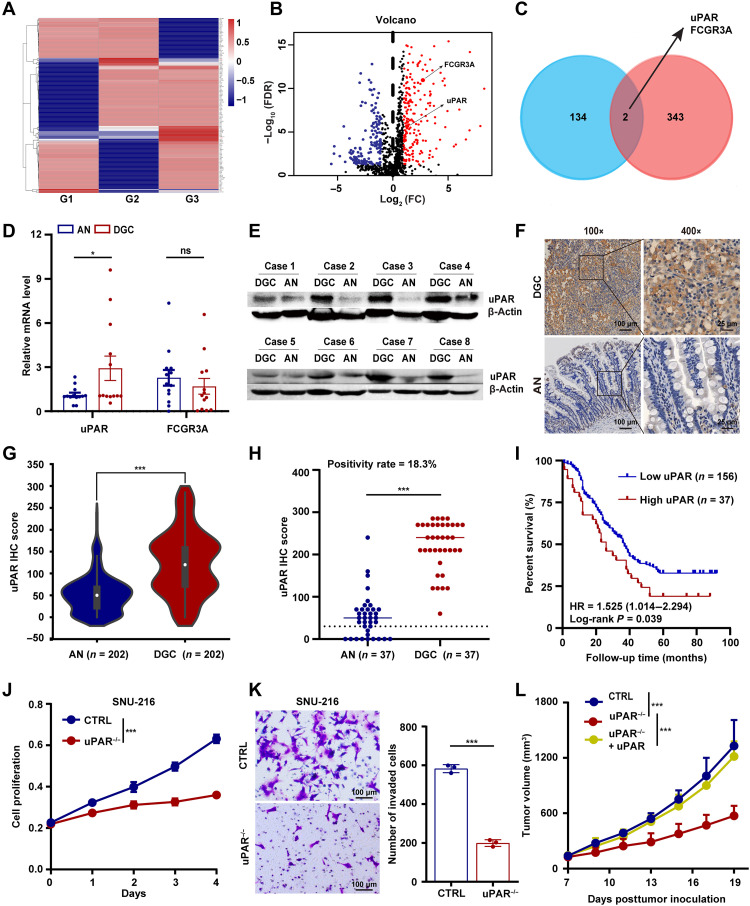
To identify cell surface antigens that are specifically overexpressed in DGC with therapeutic potential, we first performed a label-free quantitative proteomic analysis of membrane proteins differentially expressed between tumor and adjacent normal tissues from 15 patients with DGC. A total of 136 proteins were repeatedly found up-regulated in DGC tumors in at least two independent replicates (Fig. 1A and fig. S1A). Then, by analyzing the gene expression profile of GC from The Cancer Genome Atlas (TCGA) database and the data from the Immonology Database and Analysis Portal (ImmPort), we identified a total of 345 immune-related genes in GC (Fig. 1B). Comparison of the two sets of proteins in Fig. 1 (A and B) identified two common ones, uPAR and FCGR3A (Fig. 1C), indicating that they are immune-related proteins up-regulated in DGC.
Fig. 1. uPAR is overexpressed in DGC and is a potential therapeutic target.
(A) Heat map showing membrane protein expression specifically detected in the tumors but not in the adjacent normal tissues of patients with DGC (n = 5 per group, 3 groups: G1 to G3). (B) Volcano plot of immune-related genes in GC. FDR, false discovery rate; FC, fold change. (C) Venn diagram showing the overlap between (A) and (B). (D) mRNA expression of uPAR and FCGR3A in patients with DGC (n = 13) detected by qRT-PCR. AN, adjacent normal tissues. (E) Immunoblotting of uPAR in the tumor and corresponding adjacent normal tissues from patients with DGC (n = 8). (F and G) uPAR protein level detected by tissue microarrays (TMAs)–based immunohistochemistry (IHC). Representative images of uPAR staining (F) and the statistical analysis of uPAR IHC score (G) of 202 DGC specimens obtained from 623 GC cases. (H) uPAR positivity rate in patients with DGC (37 of 202) defined by IHC score of 3+ with ≥10% of the cells showing positive reactivity. (I) Kaplan-Meier plot of the DGC patient OS. (J and K) Growth curves (J) and transwell invasion assay (K) of SNU-216 control (CTRL) and uPAR−/− cells. (L) Tumor growth curves of NSG mice carrying wild-type, uPAR−/−, or uPAR−/− complemented with uPAR MKN-45 CDXs. Data are expressed as the means ± SEM (ns, nonsignificant; *P < 0.01 and ***P < 0.001).
To further validate these results, we first performed real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) to determine the transcription levels of the two candidate genes. We found that the mRNA expression of uPAR, but not FCGR3A, was significantly elevated in DGC tumors compared with adjacent normal tissues (Fig. 1D). Therefore, we chose to focus on uPAR for further analysis. Bioinformatic analysis using the tumor-immune system interactions database (TISIDB) database showed that the mRNA level of uPAR was positively correlated with that of several immunosuppressive genes, such as PD-L1, in GC (fig. S1B). This implies that uPAR may be a potential target for combination therapy with anti–PD-1/PD-L1 antibodies. Then, we performed Western blotting and found that uPAR protein levels were also up-regulated in DGC tumor compared with adjacent normal tissues (Fig. 1E). Furthermore, we analyzed uPAR expression in 202 paired DGC and adjacent normal tissues using tissue microarrays (TMAs)–based immunohistochemistry (IHC). We found that uPAR was highly expressed in DGC samples (Fig. 1, F and G). Using the criteria for defining HER2 positivity in GC (IHC score of 3+ with ≥10% of the cells showing positive reactivity) (8), we showed that the percentage of uPAR-positive DGC patients was 18.3% (Fig. 1H), while the percentage of HER2-positive DGC patients was only 5.0% (fig. S1C), which is concordant with the data from previous studies (11, 12). Of the 202 patients with DGC corresponding to Fig. 1G, 193 had available follow-up information. We found that high expression of uPAR was positively correlated with worse OS in the 193 patients, indicating that uPAR is an independent prognostic factor (Fig. 1I). Moreover, we examined uPAR expression in 18 human tissues using IHC. We detected almost no expression of uPAR in these tissues (fig. S1D), demonstrating that uPAR is not abundantly expressed in vital organs.
To explore the role of uPAR in GC, we first identified two GC cell lines (SNU-216 and AGS) expressing high levels of uPAR via qRT-PCR and flow cytometry (FCM). In addition, we detected a lower expression of uPAR in MKN-45, a human GC cell line commonly used to create a CDX mouse model (fig. S1, E and F). Then, we knocked out uPAR using CRISPR-Cas9 in SNU-216, AGS, and MKN-45 cells (fig. S1G). Using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and Transwell invasion assays, we showed that uPAR deficiency led to a significant reduction in cell proliferation and migration in both SNU-216 (Fig. 1, J and K) and AGS cells (fig. S1, H and I). Furthermore, we observed that uPAR knockout MKN-45 cells showed a significantly slower growth rate in the CDX mouse model compared with wild-type cells and that uPAR overexpression in the knockout cells completely rescued the defect in tumor growth (Fig. 1L and fig. S1, J and K). These results suggest that uPAR plays an oncogenic role in GC. Together, we identify uPAR as a cell surface protein that is overexpressed in DGC and is a potential target for therapeutic intervention.
Anti-uPAR antibody competes with uPA for uPAR binding and inhibits uPAR-dependent signalings in GC cells
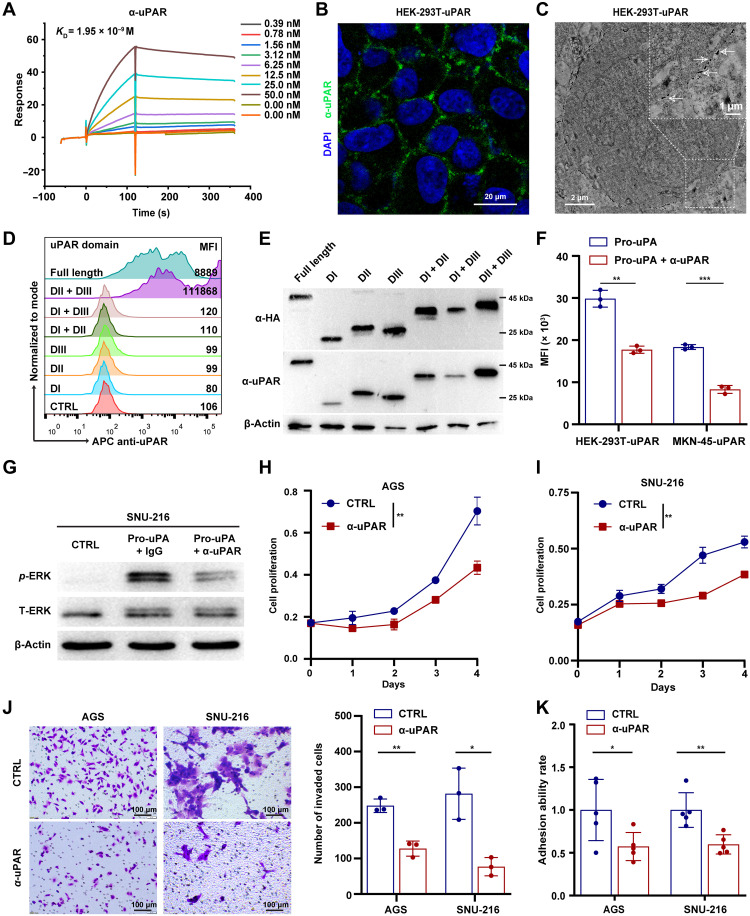
To develop antibody-based therapy for DGC, we first generated a mouse immunoglobulin G (IgG) 2a mAb with a high binding affinity for human uPAR (KD = 1.95 × 10−9 M), as shown by the surface plasmon resonance (SPR) analysis (Fig. 2A and fig. S2A). The anti-uPAR mAb was able to detect exogenous and endogenous uPAR in different cell lines, as shown by immunofluorescence (IF) and immunoelectron microscopy (Fig. 2, B and C, and fig. S2, B and C). This mAb did not cross-react with mouse uPAR (fig. S2D). In addition, we found that the mAb specifically recognized the full-length and DII-DIII region of uPAR overexpressed in human embryonic kidney (HEK) 293T cells in FCM analysis, with no detection of other truncated forms of uPAR (Fig. 2D). Consistently, the mAb seemed to have a stronger affinity in immunoblotting for the DII-DIII region of uPAR compared with other truncated forms (Fig. 2E). These results imply that the anti-uPAR mAb preferentially recognizes the DII-DIII region of uPAR in its native configuration.
Fig. 2. Anti-uPAR antibody competes with uPA for uPAR binding and inhibits uPAR-dependent signalings in GC cells.
(A) Binding affinity (KD) of the anti-uPAR mAb to human uPAR assessed by SPR. (B and C) IF (B) and immunoelectron microscopy (C) analysis of anti-uPAR binding to uPAR-expressing HEK-293T cells. DAPI, 4′,6-diamidino-2-phenylindole. (D and E) Detection of different forms of uPAR by the anti-uPAR mAb shown by FCM (D) and Western blotting (E). MFI, mean fluorescence intensity. (F) Competitive binding assay showing the ability of anti-uPAR to inhibit uPA binding to uPAR, as shown by FCM. (G) Western blotting of phosphorylated ERK (p-ERK) and total ERK (T-ERK) in SNU-216 cells pretreated with anti-uPAR or CTRL mAb after pro-uPA stimulation. (H to K) Growth curves (H and I), transwell invasion assay (J), and cell adhesion assay (K) of AGS and SNU-216 cells treated with anti-uPAR or CTRL mAb. Data are expressed as means ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001).
To investigate the influence of anti-uPAR on the uPA-uPAR system, we performed a competitive binding assay in which His-tagged pro-uPA (the zymogen form of uPA) was added to uPAR-expressing cells in the presence or absence of the anti-uPAR mAb. We found that the anti-uPAR mAb caused an approximately 50% reduction in the binding of uPA to uPAR-expressing cells (Fig. 2F). Vitronectin (VN) is an ECM protein that also interacts with uPAR and induces cell signalings (25–27). However, the anti-uPAR mAb did not affect the binding of VN to uPAR (fig. S2E). Previous studies have shown that uPA binding to uPAR induces phosphorylation and activation of extracellular signal–regulated kinase (ERK) in a few types of cancer cells (28–30), which contributes to the pleiotropic effects of uPAR on promoting cell proliferation, migration, and adhesion (14). In line with these studies, we observed an induction of ERK phosphorylation in SNU-216, AGS, and MKN-45 cells that stably express uPAR along with firefly luciferase (MKN-45-uPAR-T2A-Luc) upon uPA ligation compared to the control group, and the induction was inhibited by the anti-uPAR mAb (Fig. 2G and fig. S2, F and G). In addition, we found that treatment with the anti-uPAR mAb markedly suppressed GC cell proliferation (Fig. 2, H and I), invasion (Fig. 2J), and adhesion (Fig. 2K). Together, these results indicate that the anti-uPAR mAb competes with uPA for binding to uPAR and inhibits the protumor effects of uPAR-dependent signalings in GC cells.
Anti-uPAR alone or along with anti–PD-1 inhibits the growth of GC CDX in humanized mice
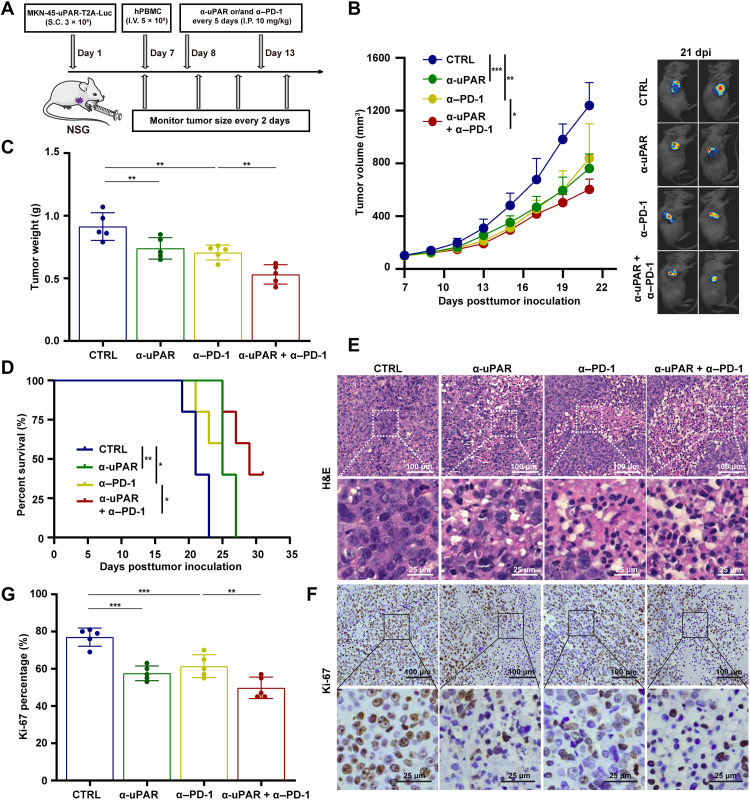
To examine the effect of anti-uPAR on GC and its potential to enhance the efficacy of anti–PD-1, we generated a CDX model using immunodeficient NOD-PrkdcscidIl2rgem1/Smoc (NSG) mice inoculated with MKN-45-uPAR-T2A-Luc cells. The expression of uPAR and PD-L1 in the stable cell line was verified by FCM (fig. S3A). When average tumor size reached 90 mm3 [approximately at day 7 posttumor inoculation (dpi)], the CDX mice were reconstituted with human peripheral blood mononuclear cells (hPBMCs). One day later, mice were treated with anti-uPAR and anti–PD-1, either alone or in combination, every 5 days. Tumor growth and body weight were monitored over the course of the experiment, starting at 7 dpi (Fig. 3A).
Fig. 3. Anti-uPAR alone or along with anti–PD-1 inhibits the growth of GC CDXs in humanized mice.
(A) Diagram of the experimental procedure. (B to D) Tumor growth curves (B), tumor weight (C) at day 21 dpi, and survival curves (D) of the MKN-45-uPAR-T2A-Luc CDX mice (n = 5 per group) injected with CTRL, anti-uPAR, anti–PD-1, or anti-uPAR plus anti–PD-1 mAbs (10 mg/kg per antibody, intraperitoneally, I.P., every 5 days starting from 8 dpi). (E to G) H&E staining (E) and Ki-67 IHC staining (F) along with the statistical analysis (G) of the antibody-treated CDX mice at 21 dpi. Data are represented as means ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001).
Human T cells (CD45+CD3+) were detectable in the periphery blood of each group at the end of the study, indicating the successful hPBMC engraftment in NSG mice (fig. S3B). We observed that tumor growth was significantly suppressed in mice treated with anti-uPAR or anti–PD-1 alone compared to the control antibody, and it was further inhibited in mice treated with the combination of two antibodies (Fig. 3, B and C). Consistently, either antibody alone or in combination significantly improved survival, with the combination therapy showing the strongest effect (Fig. 3D). In addition, we performed hematoxylin and eosin (H&E) and IHC staining of Ki-67, a cell proliferation marker (31), on the excised tumors. The results were in line with the above findings (Fig. 3, E to G). Moreover, none of the antibody treatments caused significant changes in the body weight of mice, indicating the safety and tolerability of anti-uPAR mAb alone and in combination with anti–PD-1 mAb (fig. S3C).
Anti-uPAR alone or in combination with anti–PD-1 improves survival of humanized mice bearing PDXs
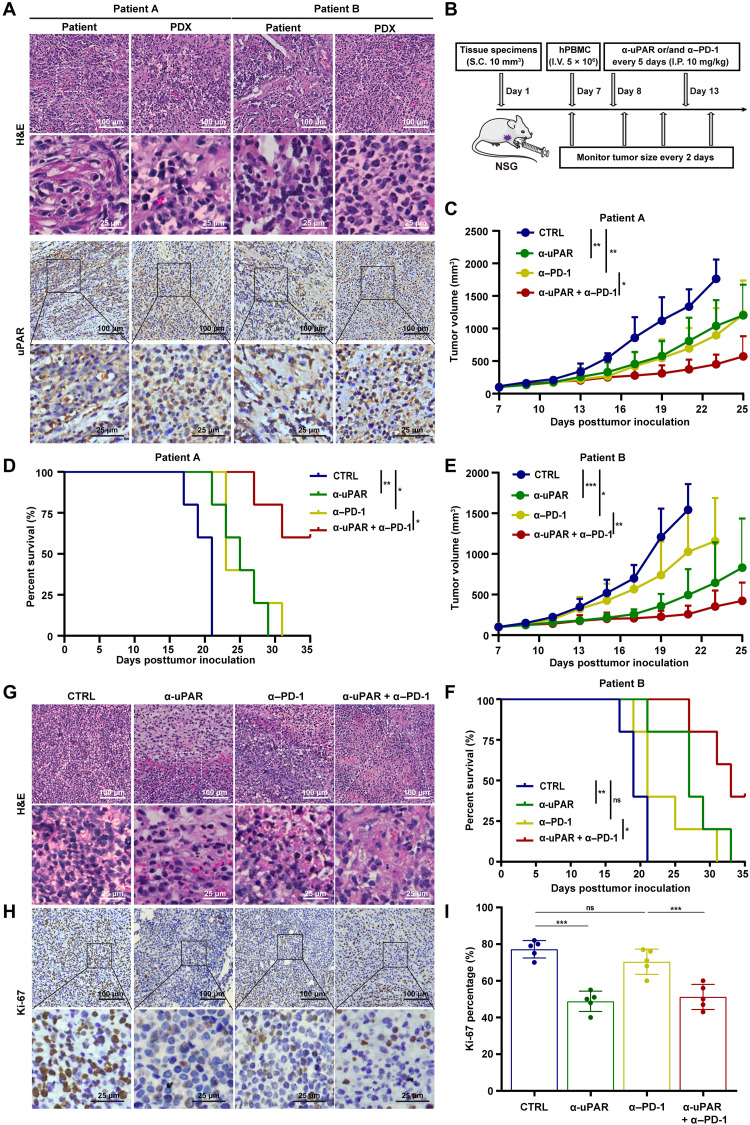
To further evaluate the efficacy of anti-uPAR alone and in combination with anti–PD-1 in GC, we established a DGC PDX model in humanized NSG mice (32). Fresh tumor specimen was obtained from a patient with DGC (patient A or B; the clinical characteristics of both patients were shown in table S1) and serially transplanted into NSG mice to produce sufficient amount of primary tumor tissues. The third generation of serially transplanted mice were used for experiments when average tumor size reached 90 mm3 (approximately at 7 dpi). First, we observed that fresh tumor specimen from the DGC patient and the derived PDX in the mice displayed similar H&E and uPAR staining patterns (Fig. 4A), implying that the PDX model retained key characteristics of the patient’s tumor. Then, hPBMC engraftment, antibody treatments, and monitoring of tumor growth and body weight were conducted as mentioned above (Fig. 4B).
Fig. 4. Anti-uPAR alone or in combination with anti–PD-1 improves survival of humanized mice bearing PDXs.
(A) H&E staining and uPAR IHC staining of fresh tumors from patients with DGC and PDX from the third generation of serially transplanted mice. (B) Diagram of the experimental procedure. (C to F) Tumor growth (C and E) and survival curves (D and F) of antibody-treated mice bearing the PDX derived from patient A or B (n = 5 per group, 10 mg/kg per antibody, I.P., every 5 days starting from 8 dpi). (G to I) H&E staining (G) and Ki-67 IHC staining (H) with data analysis (I) of the antibody-treated PDX mice (corresponding to patient A) at 21 dpi. Data are represented as means ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001).
We observed that the growth of the PDX derived from patient A was significantly hindered by anti-uPAR or anti–PD-1 alone compared to the control antibody, and it was further impeded by the combination of two antibodies (Fig. 4C). The combination therapy markedly improved survival. While all five mice in each of the other groups died within 35 dpi, three of five mice in the combination therapy group survived beyond 35 dpi (Fig. 4D). Similar results were seen in mice bearing PDX derived from patient B (Fig. 4, E and F). In line with these studies, we found that the combination therapy caused a strong inhibition of cell proliferation, as shown by H&E and IHC stainings (Fig. 4, G to I). In addition, none of the antibody treatments caused significant changes in body weight (fig. S4). Together, these results demonstrate that the combination therapy consisting of anti-uPAR and anti–PD-1 significantly inhibits tumor growth and improves survival in the PDX mouse model.
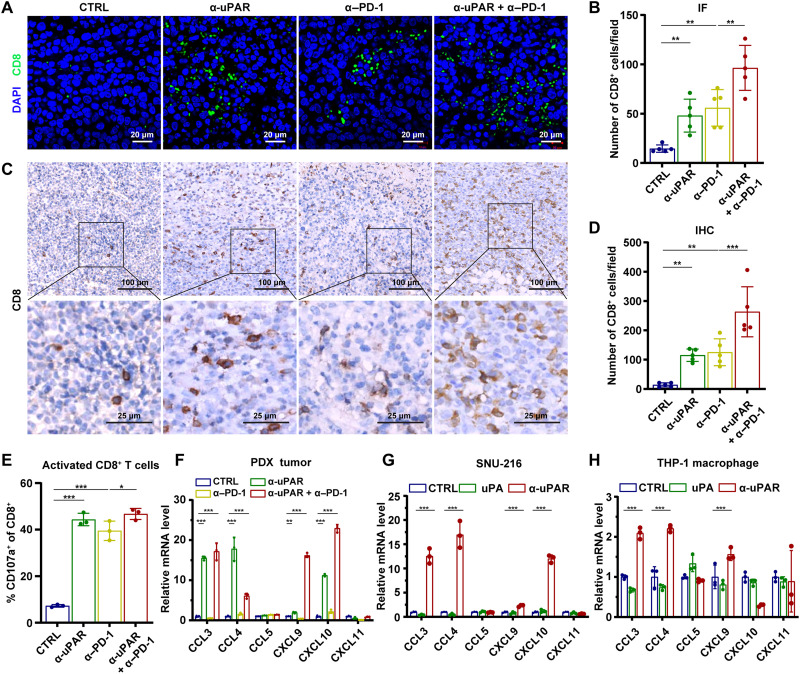
Anti-uPAR alone or together with anti–PD-1 enhances the infiltration of cytotoxic CD8+ T cells
To better understand the mechanisms underlying the antitumor effects of the anti-uPAR mAb, we first analyzed major immune cell subsets in the CDX tumors. It has been reported that anti–PD-1 antibody can increase the infiltration of CD8+ T cells into the tumor (33). Our IF and IHC assays showed that either anti-uPAR or anti–PD-1 alone caused a significant increase in the numbers of CD8+ tumor-infiltrating lymphocytes (TILs), and the combination therapy showed an additive effect (fig. S5, A to D). Furthermore, our FCM analysis found that these antibody treatments, especially the combination therapy, increased the frequency of activated cytotoxic T cells and M1 macrophages while reducing the frequency of regulatory T cells, neutrophils, and M2 macrophages in the CDX tumors (fig. S5, E to I). The ability of anti-uPAR alone or along with anti–PD-1 to modulate immune cell infiltrations could contribute to its antitumor activity.
M1 and M2 macrophages are considered antitumor and protumor, respectively (34). Neutrophils have also been shown to have cancer-promoting effects (35, 36). However, we did not observe any influence of myeloid leukocyte depletion on tumor growth in the CDX model (fig. S5, J and K). Thus, we chose to focus on the influence of anti-uPAR on T cell infiltration. Similar to our findings with the CDX tumors, IF and IHC assays using the PDX tumors (derived from the DGC patient A) also showed an increase in the tumor infiltration of CD8+ T cells following antibody treatments (Fig. 5, A to E). In line with this result, we found that expression of several chemokines (CCL3, CCL4, CXCL9, and CXCL10) that regulate T cell infiltration was significantly up-regulated in the PDX tumors treated with anti-uPAR alone or in combination with anti–PD-1 (Fig. 5F). Furthermore, we performed in vitro experiments using SNU-216 GC cells and Tohoku Hospital Pediatrics-1 (THP-1) macrophages. Treatment with anti-uPAR, but not with uPA, induced the production of CCL3, CCL4, CXCL9, and CXCL10 in these cell lines (Fig. 5, G and H). These data suggest that anti-uPAR alone or along with anti–PD-1 can induce the production of specific T cell–associated chemokines from both tumor cells and macrophages, and this process cannot be triggered by uPA.
Fig. 5. Anti-uPAR alone or together with anti–PD-1 enhances the infiltration of cytotoxic CD8+ T cells.
(A and B) IF staining (A) and quantification (B) of CD8+ T cells in the PDX mice (corresponding to patient A) at 21 dpi. (C and D) CD8 IHC staining and data analysis of the same PDX as in (A and B). (E) Frequency of activated cytotoxic T cells (CD45+CD8+CD107a+) in the patient A–derived PDX treated with different antibodies at 21 dpi, as shown by FCM. (F to H) mRNA expression levels of T cell–associated chemokines in the PDX tumor (F), SNU-216 cells (G), and THP-1 macrophages (H) after different treatments, as shown by qRT-PCR. Data are represented as means ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001).
In addition, using in vitro assays, we found that the anti-uPAR mAb strongly induced antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) (fig. S5, L and M). Together, these data suggest that anti-uPAR alone or along with anti–PD-1 can promote favorable antitumor immune responses via provoking cytotoxic T cell infiltration and inducing ADCC and CDC.
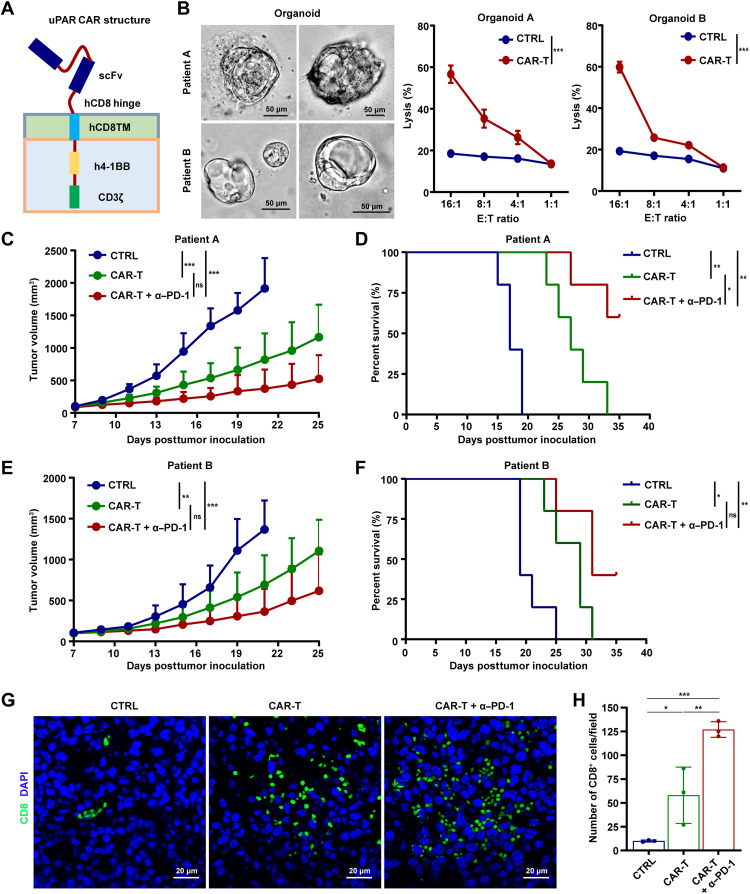
uPAR CAR-T cells alone or in combination with PD-1 blockade suppress GC growth and prolong survival
Next, we sought to develop CAR-T cells based on the novel anti-uPAR mAb and assess the anticancer efficacy of uPAR CAR-T cells, alone and in combination with anti–PD-1 mAb, against GC. We constructed a second-generation CAR consisting of a single-chain variable fragment (scFv) derived from the anti-uPAR mAb, human CD8–derived hinge and transmembrane domains, and human 4-1BB costimulatory and CD3ζ signaling domains (Fig. 6A). Our IF assay showed that the T cells transduced with uPAR CAR lentivirus interacted with uPAR-expressing HEK-293T cells (fig. S6A), indicating the successful expression of scFv in uPAR CAR-T cells and their ability to recognize target cells. In addition, we found that uPAR CAR-T cells effectively lysed wild-type AGS cells while having no effect on uPAR−/− AGS cells (fig. S6B). High killing activity of uPAR CAR-T cells was also observed with MKN-45-uPAR-T2A-Luc cells (fig. S6C). These results show uPAR-dependent cytotoxicity of the CAR-T cells in vitro. Furthermore, the antitumor efficacy of uPAR CAR-T cells in vivo was determined using the established CDX mouse model. We found that uPAR CAR-T cells potently curbed tumor growth and enhanced survival, compared with untransduced T cells. This antitumor effect of uPAR CAR-T cells was further significantly boosted by cotreatment with anti–PD-1 (fig. S6, D to F). As expected, higher concentrations of interferon-γ (IFN-γ) and granzyme B were detected in the serum of uPAR CAR-T cell–treated mice than the control mice, indicative of an activated immune response of the infused CAR-T cells (fig. S6G).
Fig. 6. uPAR CAR-T cells alone or in combination with PD-1 blockade suppress GC growth and prolong survival.
(A) Schematic diagram of uPAR CAR. (B) Representative images of DGC patient–derived organoids and killing efficiency of organoids by uPAR CAR-T cells determined by a lactate dehydrogenase (LDH)–based cytotoxicity assay, using untransduced T cells as a negative control. E:T, effector cells:target cells. (C to F) Tumor growth (C and E) and survival curves (D and F) of mice bearing the PDX derived from patient A or B (n = 5 per group; uPAR CAR-T or CTRL cells 2 × 106, I.V., only once at 7 dpi; anti–PD-1 mAb 10 mg/kg, I.P., every 5 days starting from 8 dpi). (G and H) IF staining (G) and quantification (H) of cytotoxic CD8+ T cells in the PDX derived from patient A at 21 dpi. Data are represented as means ± SEM (*P < 0.05, **P < 0.01, and ***P < 0.001).
By using DGC patient–derived organoids and the established PDX mouse model, we further examined the antitumor activities of uPAR CAR-T cells. We found that uPAR CAR-T cells effectively lysed the organoids derived from DGC patients A and B (Fig. 6B). Treatment with uPAR CAR-T cells alone remarkably suppressed tumor growth and promoted survival in mice bearing the PDX derived from the DGC patient (A or B), and the antitumor efficacy was even stronger when combining uPAR CAR-T cells with the anti–PD-1 mAb (Fig. 6, C to F). Consistent with these findings, our IF assay revealed a significant increase in the number of CD8+ TILs in the uPAR CAR-T cell–treated mice compared with the control mice, and cotreatment with anti–PD-1 further enhanced the infiltration of these cytotoxic T cells into the tumor (Fig. 6, G and H). Expression of molecules associated with T cell exhaustion (37), such as PD-1, lymphocyte activation gene 3 (LAG-3), and T cell immunoglobulin and mucin domain–containing 3 (TIM-3), was generally decreased in the tumor-infiltrating CAR-T cells compared with untransduced T cells, especially in the presence of the anti–PD-1 mAb (fig. S7A). Induction of IFN-γ and granzyme B was detected in the serum of mice treated with uPAR CAR-T cells alone or in combination with anti–PD-1 (fig. S7B). Besides, no significant difference in body weight was observed among all the groups tested (fig. S7C). These data demonstrate that uPAR CAR-T cells alone strongly inhibit GC growth and prolong survival, and the antitumor effects are further augmented when combined with PD-1 blockade.
DISCUSSION
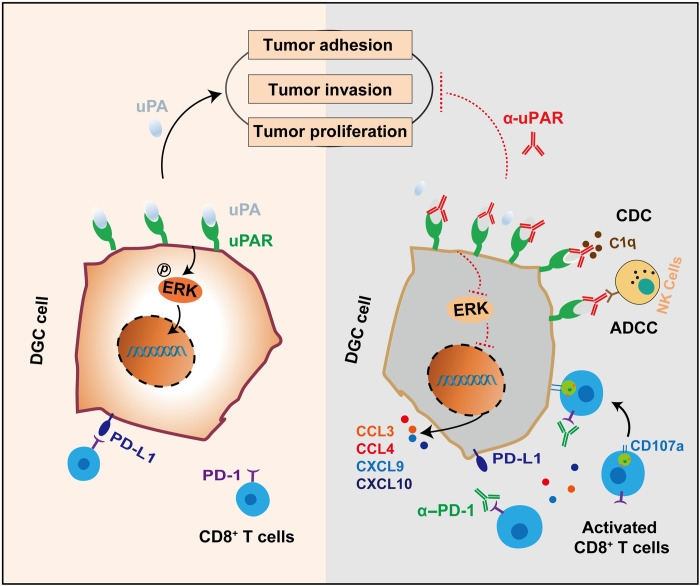
DGC is a subtype of GC with poor prognosis and few targeted treatment options. Here, we identified uPAR as a potential therapeutic target for DGC and developed therapeutic strategies based on a novel anti-uPAR mAb. We demonstrated the capability of therapeutic strategies targeting uPAR alone, and particularly in combination with PD-1 blockade, to inhibit tumor growth and promote survival, using GC cell lines and organoids, as well as humanized CDX and PDX mouse models. In addition, we showed that the anti-uPAR mAb exerts antitumor effects via multiple mechanisms (Fig. 7). Our findings offer promising therapeutic strategies for the treatment of DGC.
Fig. 7. Therapeutic strategies targeting uPAR potentiate anti–PD-1 efficacy in DGC.
We identify uPAR as an immune-related cell surface protein that is specifically overexpressed in DGC with a relatively high positivity rate and a promising therapeutic potential. Our anti-uPAR mAb targets the DII-DIII region of uPAR and blocks uPA binding to its receptor, thereby inhibiting uPAR-dependent ERK activation and downstream signalings involved in cell proliferation, migration, and adhesion. Moreover, the anti-uPAR mAb stimulates infiltration of cytotoxic CD8+ T cells, induces robust ADCC and CDC, and augments the antitumor efficacy of PD-1 blockade therapy.
DGC is highly aggressive and resistant to chemotherapy. Immunotherapies targeting HER2 and PD-1 have been recommended by NCCN guidelines for the use in unresectable locally advanced, recurrent, or metastatic GC (8). However, the use of anti-HER2 therapy is limited to patients exhibiting overexpression of HER2 (IHC score of 3+ with ≥10% of cells showing positive reactivity), but the positivity rate of HER2 is only 2 to 6% in DGC, and the objective response rate with anti–PD-1 monotherapy in GC is only 11.2% (ATTRACTION-2), which is even lower in DGC (13). Therefore, identification of novel targets for DGC treatment is of significant importance. Here, we screened out uPAR as an immune-related cell surface protein that is overexpressed in the tumors, but not in the matching adjacent normal tissues, of 15 patients with DGC using membrane proteomics combined with gene expression analyses. Our screening result is consistent with a recent study, which performed a comprehensive proteomic analysis of DGC, and uPAR/PLAUR was among their list of proteins up-regulated in DGC tumors (38). Earlier studies have documented that uPAR is expressed in many types of human cancers, including breast cancer (39), non–small cell lung cancer (40), ovarian cancer (21), and GC (41). High expression of uPAR was particularly found in poorly differentiated GC (42). In our study, we further validated uPAR overexpression in DGC using Western blotting, qRT-PCR, and IHC staining. In line with previous studies, our IHC analysis showed that the positivity rate of HER2 in patients with DGC was 5.0%, and under the same scoring criteria, the positivity rate of uPAR was 18.3%. In addition, we found that high uPAR expression was as an independent prognostic factor for poor OS in DGC. The high positivity rate and prognostic value of uPAR indicate that it could be a promising alternative target for DGC treatment.
uPAR is the cell surface receptor for uPA, an extracellular serine protease. uPAR recruits uPA to the cell surface where activation of the proteolytic activity of uPA is accelerated, which, in turn, stimulates the activity of the plasminogen activation system, thereby regulating ECM proteolysis. Apart from this, uPAR can initiate various intracellular signalings, independently of uPA proteolytic activity, to regulate cell adhesion, migration, invasion, proliferation, and survival. The pleiotropic functions of uPAR play a critical role in the development of types of cancers (43). In line with these studies, our findings showed that uPAR knockout significantly suppressed proliferation and migration of GC cells in vitro and in vivo. Moreover, we developed a novel anti-uPAR mAb with strong affinity for human uPAR, which also suppressed proliferation and migration, as well as adhesion, of GC cells in vitro. These data further support the oncogenic role of uPAR and suggest that the anti-uPAR mAb acts as an antagonist of uPAR-dependent cell signalings. Consistently, we showed that the anti-uPAR mAb strongly blocked uPA binding to uPAR and inhibited the downstream activation of ERK. Previously, activation of ERK by uPAR has been shown to promote cell proliferation (29) and motility (28), which seems to be dependent on uPA binding to its receptor and does not require uPA proteolytic activity. The binding sites of uPA are known to be located in all three domains of uPAR (15). Here, we observed that the anti-uPAR mAb preferentially targeted the DII-DIII region of uPAR. This could partly explain its blocking function and also indicates its difference from the previously reported ATN-658 mAb directing to the DIII of uPAR.
The overexpression of uPAR in tumors and its pleiotropic activities in cancer progression make it an attractive therapeutic target (44). A number of studies have generated uPAR-targeted antibodies (18–23, 45), CAR-T cells (24, 46), peptides (47), and small molecules (48) for cancer diagnosis and treatment. Here, we assessed the efficacy of anti-uPAR alone and along with anti–PD-1 against GC in vivo using immune-humanized mice bearing CDX or PDX. By monitoring the volume, weight, and pathology of tumors, we showed that anti-uPAR alone already caused a significant suppression of growth of CDX or PDX in the mice, while the combination with anti-uPAR and anti–PD-1 further enhanced the antitumor efficacy. Currently, PDX models have been considered as the most clinically relevant in vivo cancer models as they preserve the key characteristics of the patient’s tumor, and drug response of PDX has a high degree of translatability to the patient (32, 49, 50). In our study, we further showed that anti-uPAR monotherapy and particularly the combination therapy with anti-uPAR and anti–PD-1 markedly prolonged survival of the PDX mice, underscoring the potential of the anti-uPAR mAb for DGC treatment.
The immune-humanized mouse model (immunodeficient mice engrafted with hPBMCs) used in our study is a fast and simple method for mouse humanization and has been widely used in a variety of cancer research (51, 52). Meanwhile, it has a limitation due to its susceptibility to xenogeneic graft–versus–host disease (GvHD), which leads to loss of body weight and early death (53). Although clinical signs of severe GvHD were not observed in any of the mice in our study, this disease limits the experimental window and prevents the study on long-time efficacy of anti-uPAR. In addition, xenogenic tumor models (including CDX and PDX models) could have compromised interactions among immune cells and between the host and the tumor due to the species barrier. To rule out the influence of GvHD and overcome these limitations, we will develop antibodies targeting murine uPAR and syngeneic immunocompetent mouse models of GC, including an autochthonous GC model (54), in our future study. We believe that these tools will be very useful for us to further evaluate the long-time efficacy, potential toxicity, and functional mechanisms of anti-uPAR antibodies.
Furthermore, we showed that the anti-uPAR mAb exerts its antitumor function via multiple mechanisms. (i) As mentioned above, the anti-uPAR mAb alone inhibited GC cell proliferation, migration, and adhesion through inhibition of uPA-uPAR–mediated cell signalings. (ii) The anti-uPAR mAb induced robust ADCC and CDC in our in vitro assays. Considering the high abundance of uPAR in DGC, it is very likely that the anti-uPAR mAb could promote the killing of tumor cells by inducing ADCC and CDC in vivo. In future studies, we will develop humanized mAb based on the current mouse monoclonal anti-uPAR, which will have lower immunogenicity, longer serum half-life, and better human effector functions (55). (iii) The anti-uPAR mAb caused a significant increase in the tumor infiltration of cytotoxic CD8+ T cells. First, this could be partly explained by the production of specific T cell–associated chemokines (CCL3, CCL4, CXCL9, and CXCL10) in the tumor following anti-uPAR treatment. GC cells and macrophages treated with anti-uPAR are likely the cellular source of the four specific chemokines as shown by our in vitro experiments, although we did not rule out other possible cell types. uPA-dependent activities appear not to be required for this process, because cells treated with uPA did not produce any T cell–associated chemokines. The detailed mechanism underlying the chemokine production still needs to be further explored. Second, the ability of anti-uPAR to reduce tumor cell adhesion could also help immune cells to gain access to the tumor microenvironment. (iv) The combination therapy consisting of anti-uPAR and anti–PD-1 had additive effects on immune activation and tumor growth inhibition.
Myeloid leukocytes play important and complex roles in cancer development (34, 36). A recent study showed that heteromerization of uPA and plasminogen activator inhibitor-1 (PAI-1), two key components of the uPA-uPAR system, promotes breast cancer progression by attracting tumorigenic neutrophils (35). In addition, M1 and M2 macrophages are considered to have antitumor and protumor function, respectively (34). In our study, treatment with anti-uPAR altered the tumor infiltration of neutrophils and macrophages in the CDX model. Yet, the exact role of these myeloid leukocytes in the pathogenesis of DGC and their contribution to the antitumor efficacy of anti-uPAR remain to be further elucidated, for instance, by using anti-mouse uPAR antibodies along with syngeneic immunocompetent mouse models of GC.
Two previous studies have constructed uPAR CAR-T cells and investigated their therapeutic potential for the treatment of ovarian cancer and senescence-associated diseases (24, 46). In our study, the second-generation uPAR CAR-T cells that we generated on the basis of the novel anti-uPAR mAb effectively killed uPAR-expressing cell lines and DGC patient–derived organoids. uPAR CAR-T cells alone and in combination with PD-1 blockade significantly inhibited GC growth and enhanced survival in both the CDX and PDX models. In addition, we found that uPAR CAR-T cells were more resistant to T cell exhaustion when compared with untransduced T cells, especially in the presence of the anti–PD-1 antibody. Therefore, combination therapy consisting of uPAR CAR-T cells and anti–PD-1 could be another promising strategy for DGC treatment.
In conclusion, we demonstrate that targeting uPAR along with PD-1 can significantly inhibit tumor growth and improve survival in DGC via multiple mechanisms. Our findings provide promising strategies that could complement the current use of HER2- and PD-1–targeted therapies for the treatment of DGC.
MATERIALS AND METHODS
Cells and culture conditions
Human GC cell lines—NCI-N87, HGC-27, MKN-45, and AGS—were obtained from the Chinese Academy of Medical Sciences (China). The human GC cell line SNU-216 was obtained from the Korean Cell Line Bank. HEK-293T cells were obtained from the American Type Culture Collection. THP-1 was a gift from Z. Jiang (Peking University). AGS, HGC-27, and HEK-293T were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 mg/ml). NCI-N87, MKN-45, SNU-216, and THP-1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 mg/ml). All cell lines were validated by short tandem repeat DNA fingerprinting and were tested for mycoplasma contamination. hPBMCs of healthy donors were collected at Lanzhou University Second Hospital under the protocols approved by the Human Research Ethics Committee (2020A-052). CD3+ T cells were enriched via negative selection using the EasySep Human T Cell Isolation Kit (STEMCELL Technologies, 17951) and then cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 mg/ml), and recombinant human interleukin-2 (IL-2; 10 ng/ml; Peprotech, 200-02-100). Cells were maintained in a humidified incubator at 37°C with 5% CO2.
Animals
M-NSG (NOD-PrkdcscidIl2rgem1/Smoc) mice lacking mature T, B, and natural killer cells were purchased from the Shanghai Model Organisms Center Inc. (NM-NSG-001). All mice were maintained under specific pathogen–free conditions. Animal care and experiments were carried out under the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. This study has been approved by the Laboratory Animal Ethics Committee of Lanzhou University Second Hospital (D2020-36).
Human DGC specimens
All specimens were acquired from patients under the protocols approved by the Human Research Ethics Committee of Lanzhou University Second Hospital (2020A-052). Informed consent was obtained from all patients. A total of 623 pairs of GC and adjacent normal tissues were constructed into TMAs as formalin-fixed and paraffin-embedded blocks and then analyzed by IHC analysis. A total of 18 normal organ tissues were used for IHC staining of uPAR. A total of 15 and 13 pairs of DGC and adjacent normal tissues were used for membrane proteomics analysis and gene expression analysis by qRT-PCR and Western blot, respectively.
Membrane proteomics
Tumor and adjacent normal tissues obtained from patients with DGC (n = 15) were randomly divided into six groups, with each group containing either tumor or adjacent normal tissues pooled from five patients. Membrane proteins were extracted and purified from the six groups of tissues and then analyzed via mass spectrometry–based label-free quantitative proteomics. A total of 32,187 peptides and 4293 proteins were identified. Among these proteins, a total of 136 proteins were repeatedly found to be expressed in DGC tumors, but not in adjacent normal tissues, in at least two independent biological replicates.
Bioinformatics analysis of gene expression profiles in DGC
Genes up-regulated in GC were retrieved from TCGA database (https://cancergenome.nih.gov), which includes 375 GC tumor samples and 32 normal samples. Immune-related genes in human cancer were retrieved from the ImmPort (http://immport.org). Immune-related genes up-regulated in GC were obtained from the overlap between the above two datasets. Correlation between uPAR and immunosuppressive genes was analyzed using the TISIDB database (http://cis.hku.hk/TISIDB).
Generation of anti-uPAR mAb
To generate mAbs against human uPAR, 6-week-old C57BL/6 mice were immunized thrice with 1 × 107 HEK-293T cells stably expressing uPAR (HEK-293T-uPAR) via subcutaneous multipoint injection. Serum samples were collected by retroorbital bleeding for monitoring of antibody titers. Five days before splenectomy for generation of monoclonal antibodies, mice were reboosted by subcutaneous multipoint injection of 2 × 106 HEK-293T-uPAR cells. mAbs were then prepared by Microfluid Based Antibody Tech (Huazhitiancheng Inc.). The anti-uPAR mAb used in this study was selected on the basis of its high affinity for binding to human uPAR.
Surface plasmon resonance
The binding affinity of the anti-uPAR mAb to uPAR was analyzed by SPR using a Biacore T200 system (GE Healthcare). The anti-uPAR mAb was diluted in phosphate-buffered saline (PBS) into different concentrations, ranging from 0.39 to 50 nM, and was allowed to pass over the immobilized human uPAR at the flow rate of 30 μl/min. The association and dissociation time were 120 and 240 s, respectively. The kinetics of antibody-antigen interactions were measured on the basis of the steady-state affinity fit model according to the manual of the Biacore T200 evaluation software.
Immunoelectron microscopy
HEK-293T-uPAR-T2A-Luc or SNU-216 cells (1 × 107) were prepared and incubated with the colloidal gold–conjugated anti-uPAR mAb (10 μg/ml) in PBS at 37°C for 30 min. Cells were pelleted by centrifugation at 250g for 15 min at 4°C. The supernatant was removed, and 1 ml of ice-cold 2% glutaraldehyde was added. Following fixation for 1 hour on ice, cells were processed for ultramicrotomy and viewed under a transmission electron microscope (Hitachi 7800, Japan).
Generation of uPAR CAR
The plasmid was constructed by stepwise Gibson assembly (New England Biolabs, E2611L) using the backbone as previously described (56). The amino acid sequence of the scFv specific for human uPAR was obtained from the heavy- and light-chain variable regions of the anti-uPAR mAb that we generated. A second-generation CAR, consisting of the anti-uPAR scFv, human CD8–derived hinge and transmembrane domains, and human 4-1BB costimulatory and CD3ζ signaling domains, was constructed.
Lentiviral vector production
HEK-293T cells were used for lentiviral vector production. To generate HEK-293T-uPAR-T2A-Luc, HEK-293T-uPAR-T2A-mCherry, and MKN-45-uPAR-T2A-Luc stable cell lines and uPAR−/− MKN-45 cells rescued with uPAR overexpression, pseudotyped lentiviruses were produced by cotransfecting 1 μg of pLenti-CMV-uPAR-T2A-Luc, 1 μg of pDD, and 0.5 μg of plasmid encoding the glycoprotein of the vesicular stomatitis virus (pVSV-G) into HEK-293T cells in a 3.5-cm dish. Cell culture supernatants containing the lentiviruses were collected 48 hours after transfection, filtered through a 0.45-μm nonpyrogenic filter (JET BlOFIL, FPV403150), and immediately used to transduce target cells in the presence of polybrene (10 mg/ml; Solarbio, H8761). Transduced cells were selected with puromycin (2 μg/ml; Thermo Fisher Scientific, A1113803) for 5 to 7 days to generate stable cell lines. To generate uPAR knockout SNU-216, AGS, and MKN-45 cell lines, pLenti-CRISPR-V2, pSPAX2, and pMD2G were used, and the transduction and selection of knockout cell lines were performed in a similar way as mentioned above. The sequences of guide RNAs used for CRISPR-Cas9–mediated uPAR knockout were listed in table S2.
To produce lentiviruses for CAR-T cell transduction, five dishes of HEK-293T cells (6 × 106 cells per 10-cm dish) were seeded. After 18 hours, 7 μg of Lenti-CAR plasmid and the lentiviral packing plasmids psPAX2 (5 μg) and pMD2.G (3.5 μg) were transfected into each dish using 500 μl of opti-MEM medium (Gibco, 31985062) containing 30 μl of Lipofectamine 2000 (Thermo Fisher Scientific, 11668019). After 6 hours, fresh DMEM medium containing 10% FBS was replaced into each dish. Sixty hours after transfection, cell culture media were harvested from all dishes and centrifuged at 1000g, 4°C for 10 min to pellet cell debris. The supernatant was filtered through a 0.45-μm low protein-binding membrane, and then the lentiviruses were concentrated into DMEM containing 10% FBS plus 1% bovine serum albumin (Solarbio, H1130).
Isolation, expansion, and transduction of human T cells
All blood samples were handled following the required ethical and safety procedures. hPBMCs were obtained from anonymous healthy donors and isolated by density gradient centrifugation. T cells were purified using the EasySep Human T Cell Isolation Kit and then stimulated with CD3/CD28 T cell activator Dynabeads (Gibco, 11131D) at a bead:cell ratio of 1:2 and IL-2 (10 ng/ml). After 24 hours, T cells were transduced with lentiviruses by centrifugation at 1000g, 32°C for 90 min in the presence of polybrene (4.4 μg/ml). The viral supernatant was replaced 10 hours later with fresh culture medium containing IL-2 (10 ng/ml). T cells were further expanded by replacing half of the culture medium with fresh medium every 2 days. Six days later, Dynabeads were removed, and T cells were allowed to expand for another 3 days. Transduction efficiencies were determined by FCM, and CAR-T cells were adoptively transferred into mice or used for in vitro experiments.
Cytotoxicity assays
The cytotoxicity of uPAR CAR-T cells was determined by standard lactate dehydrogenase (LDH) or firefly luciferase–based assays. For LDH assays, target tumor cells (2 × 103) were cocultured with CAR-T cells at various effector-to-target (E:T) ratios. After 8 hours, 50 μl of the supernatant from each well was transferred into a fresh 96-well flat clear bottom plate, and 50 μl of the CytoTox 96 Reagent (Promega, G1780) was added. The plate was incubated at room temperature for 30 min, 50 μl of stop solution was added into each well, and the absorbance signal at 490 nm was measured in a plate reader (Varioskan Flash, Thermo Fisher Scientific). Maximum LDH release was determined by addition of 0.2% Triton X-100. Cytotoxicity (%) was determined as [(experimental − effector spontaneous − target spontaneous)/(target maximum − target spontaneous)] × 100. For organoid cytotoxicity assays, DGC organoids were seeded on a Matrigel layer and incubated with uPAR-CAR T cells as follows. The 96-well standard culture plates were first moistened using culture medium. Subsequently, each well was evenly covered with 15 μl of undiluted Matrigel, which was allowed to solidify at room temperature for 15 min. Confluent organoids were collected, mechanically sheared, pelleted, and seeded into the precoated 96-well plate (approximately 2.5 × 103 cells per well). uPAR CAR-T cells were added at various E:T ratios and cocultured with organoids for 8 hours. Cytotoxicity was determined by the LDH assay as mentioned.
For firefly luciferase assays, target cells (2 × 103) expressing firefly luciferase were cocultured with CAR-T cells in a black-walled 96-well plate (100 μl per well) for 8 hours at various E:T ratios. Target cells alone were plated at the same cell density to determine the maximum luciferase expression, and maximum release was determined by addition of 0.2% Triton X-100. After 8 hours, 100 μl of luciferase substrate (Promega) was directly added to each well, and firefly luciferase activities were measured with a Varioskan Flash (Thermo Fisher Scientific). Cytotoxicty (%) was calculated as: [1 − (experimental relative light units)/(maximum relative light units)] × 100.
Humanized mouse tumor models
The PDX model in hPBMC-humanized NSG mice was generated as previously described (32). Briefly, fresh tumor tissues obtained from a patient with DGC (A or B; the clinical characteristics of both patients were shown in table S1) were divided into small pieces (about 10 mm3) and subcutaneously (S.C.) injected into 6-week-old NSG mice for serial transplantation. hPBMCs (5 × 106 per mouse) were intravenously (I.V.) injected into the third generation of serially transplanted NSG mice when the average tumor size reached 90 mm3 (approximately at 7 dpi). In antibody treatment studies, mice were intraperitoneally (I.P.) injected with the anti-uPAR mAb either alone or in combination with the anti–PD-1 mAb (Sintilimab, 10 mg/kg) every 5 days, starting from day 8 dpi. An isotype-matched irrelevant mAb (Proteintech, 10284-1-AP) was used as a negative control. In CAR-T cell therapy studies, mice received adoptive transfer of transduced uPAR CAR-T cells (2 × 106 per mouse, I.V. injection) only once at 7 dpi, and the combination therapy group received anti–PD-1 (10 mg/kg, I.P. injection) every 5 days, starting from 8 dpi. Untransduced human T cells were used as a negative control. Tumor volume was measured using a digital caliper, and body weight was monitored every other day, starting from 7 dpi. Tumor volume was calculated using the following formula: tumor volume = (longer diameter) × (shorter diameter)2/2. The experimental end point was defined as either death or tumor size reaching 1500 mm3. Tumor tissues were collected at the experimental end point and prepared as formalin-fixed, paraffin-embedded samples for H&E, IHC, and IF staining.
The CDX model in hPBMC-humanized NSG mice was established and used in a similar manner to that of the PDX model except for the following procedures. MKN-45 cells that stably express uPAR along with firefly luciferase (MKN-45-uPAR-T2A-Luc) were generated, suspended in 100 μl of PBS, mixed with 100 μl of Matrigel (BD Biosciences, 354230), and then injected into 6-week-old NSG mice (3 × 106 per mouse, S.C. injection). Bioluminescent imaging of the CDX mice was performed at the indicated time points using VISQUE Invivo Smart-LF (Vieworks, Korea), and tumors were harvested at the experimental end point and weighted.
Organoid culture
DGC organoids were generated as previously described (57). Briefly, tumor tissues were collected from the PDX mice, washed in human washing medium (2 mM glutamine and 10% FBS with DMEM/F-12), minced with scissors, and enzymatically digested using collagenase IV (0.1 mg/ml; Roche, 11088858001) at 37°C for 30 min. Digestion was stopped by adding human washing medium, and the digested tumor tissues were filtered through a 100-μm cell strainer (JET BlOFIL, CSS013100). Cell clusters were spun down at 250g for 5 min, resuspended in 50% Matrigel/organoid culture medium, and plated in a 50-μl drop in the middle of one well of a six-well plate (JET BlOFIL, TCP011006) precoated with 60% Matrigel. The drop was solidified by a 30-min incubation at 37°C and 5% CO2. After solid drops formed, 0.5 ml of the organoid culture medium was added to the well, and the medium was changed every 3 to 4 days.
FCM analysis
To analyze cells from the PDX model, tumors separated from mice were minced with scissors and digested with collagenase IV to generate single-cell suspensions. The obtained tumor slurry was transferred to a 15-ml centrifuge tube by passing through a 300-mesh stainless steel filter and centrifuged at 250g for 5 min. The supernatant was discarded, and cells were resuspended in 2 ml of 45% percoll solution (freshly prepared) and then centrifuged at 350g for 7 min. Red blood cells were lysed using ammonium chloride solution (STEMCELL Technologies). Cells were filtered through a 70-μm cell strainer, resuspended in PBS containing 2% FBS, stained with antibodies for 30 min in the dark, and then detected by FCM. The following antibodies were used: fluorescein isothiocyanate anti-human CD3 (BioLegend, 300452), phycoerythrin (PE) anti-human CD4 (BioLegend, 357404), allophycocyanin (APC) anti-human CD8 (BioLegend, 344722), APC-Cy7 anti-human CD45 (BD Biosciences, 557833; BioLegend, 304014), PE anti-mouse/human CD11b (BioLegend, 101208), BV421 anti-human CD66b (BioLegend, 392916), APC anti-human CD68 (BioLegend, 333810), BV421 anti-human CD86 (BioLegend, 371242), PE-Cy7 anti-human CD206 (BioLegend, 321124), BV421 anti-human FoxP3 (BioLegend, 320124), anti–PD-L1 (Abcam, ab210931), PE anti-human CD107a (BioLegend, 328608), APC anti–PD-1 (BioLegend, 367406), PE-Cy7 anti-TIM-3 (BioLegend, 345014), and PE anti-LAG-3 (BioLegend, 369306). To analyze the expression of uPAR and PD-L1 in GC cell lines, MKN-45 and MKN-45-uPAR-T2A-Luc cells were trypsinized, resuspended in PBS containing 2% FBS, and stained with the homemade anti-human uPAR mAb or anti-human PD-L1 (Abcam, ab210931) for 30 min on ice. PE goat anti-mouse IgG (BioLegend, 405307) was used as the secondary antibody. To identify the binding region of the anti-uPAR mAb in uPAR, pRK5–hemagglutinin (HA) plasmids expressing full-length or truncated forms of human uPAR were transiently transfected into different groups of HEK-293T cells. The anti-uPAR mAb was labeled with APC (Lanzbiotech, Hzh030487), and the ability of the anti-uPAR mAb to recognize each group of cells was analyzed by FCM (FACSCanto, BD Biosciences).
Competitive binding assay
HEK-293T-uPAR-T2A-Luc or MKN-45-uPAR-T2A-Luc cells were seeded into a 10-cm dish. One day later, cells were trypsinized, washed twice with staining buffer (PBS plus 2% FBS), and split into three groups. One group (1 × 106 cells in 100 μl of staining buffer) was treated with 1 μg of His-tagged pro-uPA (Sino Biological, 10815-H08H) plus 1 μg of the anti-uPAR mAb in the dark at 4°C for 30 min. The other two groups, the untreated group and the group treated with only 1 μg of His-tagged pro-uPA, were used as controls. After treatment, cells were washed thrice with the staining buffer and stained with APC-conjugated anti-His antibody (BioLegend, 362605). Pro–uPA-bound cells were detected using FCM (FACSCanto, BD Biosciences).
Immunofluorescence
Cell lines (HEK-293T-uPAR-T2A-Luc and SNU-216) cultured on a 35-mm glass-bottom microwell dish (MatTek Corporation) were fixed with 4% paraformaldehyde for 10 min. Both the cell lines and antigen-recovered paraffin section of xenografts derived from the DGC patient A were subjected to permeabilization with 0.1% Triton X-100, followed by incubation with primary antibodies, the anti-uPAR mAb, and anti-human CD8a (1:200; Sigma-Aldrich, ZRB1010), overnight at 4°C. Then, the samples were washed thrice with PBS and incubated with fluorescent-labeled secondary antibody (Abcam, ab150113). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole before mounting. Confocal fluorescence images were captured using a Zeiss LSM880 laser microscope (Plan-Apochromat 63× /1.4 oil objective).
To determine the ability of uPAR CAR-T cells to bind to HEK-293T-uPAR-T2A-mCherry cells, they were cocultured on a 35-mm glass-bottom microwell dish for 4 hours. Confocal fluorescent images were captured using a Zeiss LSM880 laser microscope.
Histological analysis
Tissue specimens obtained from patients or mice were fixed in 10% formalin, embedded in paraffin, and cut into 5-μm sections. Then, the tissues were deparaffinized and rehydrated, followed by H&E or IHC staining. The following primary antibodies were used: anti-human uPAR (1:200; Abcam, ab218106), anti-human HER2 (1:200; Abcam, ab214275), and anti-human CD8 (1:200; MXB biotechnologles, RMA-0514). The images were acquired with K-ViEWER (KFBIO). Staining intensity (0, 1, 2, and 3) and the percentage of positive cells (0 to 100%) were evaluated by a pathologist at Lanzhou University Second Hospital. The histoscore was calculated by multiplying the staining intensity and the percentage of positive cells. uPAR or HER2 overexpression positivity was defined as IHC score of 3+ with ≥10% of cells showing positive reactivity (8).
RNA extraction and real-time quantitative PCR
SNU-216 cells and THP-1 cells treated with phorbol ester (100 ng/μl) were resuspended in culture medium containing the anti-uPAR, uPA (Sino Biological, 10815-H08H-A), or control mAb (10 μg/ml, 3 × 104 cells) and cultured for 3 days. Total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen) and reversely transcribed into cDNA using a reverse transcription kit (RR037A, Takara Bio, Japan). The relative expression levels of mRNAs were determined by qRT-PCR with a LightCycler instrument (Roche) using SYBR Green dye (Takara), and the results were analyzed by the ΔΔCT method. The sequences of forward and reverse primers (10 μM) for human uPAR, CCL3, CCL4, CCL5, CXCL9, CXCL10, CXCL11, and GAPDH were listed in table S3.
Immunoblotting
Cells (2 × 106) or tissues (20 mg) were washed twice with cold PBS, and whole cell lysates were extracted using 100 μl of lysis buffer (PBS, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) containing phosphatase inhibitor cocktail (Sigma-Aldrich, P0114-10KU). After lysis, cell debris were removed by centrifugation (3000g, 10 min). For detection of ERK phosphorylation, serum-starved SNU-216, AGS, or MKN-45-uPAR-T2A-Luc cells were washed with 50 mM glycine-HCl and 100 mM NaCl (pH 3.0) for 3 min to remove surface-bound endogenous uPA and neutralized with 0.5 M Hepes and 0.1 M NaCl (pH 7.5) for 10 min on ice. Cells were pretreated with anti-uPAR (10 μg/ml) or control human IgG for 1 hour at 37°C. Pro-uPA (10 nM) was added and incubated with cells at 37°C for 5 min to initiate ERK activation. After incubation, cells were lysed and blotted for phospho-ERK (Cell Signaling Technology, 4370T) and total ERK (Cell Signaling Technology, 4695T). To identify the binding region of the anti-uPAR mAb in uPAR, pRK5-HA plasmids expressing full-length or truncated forms of human uPAR were transiently transfected into different groups of HEK-293T cells. After 36 hours, cell lysates were run on SDS–polyacrylamide gel electrophoresis gels and immunoblotted with anti-HA antibody (Lanzbiotech, HZTA0151), the anti-uPAR mAb, or anti–β-actin antibody (Proteintech, 66009-1). Horseradish peroxidase–conjugated goat anti-mouse IgG antibody (Proteintech, SA00001-1) was used as the secondary antibody.
Enzyme-linked immunosorbent assays
Proteins levels of human IFN-γ and granzyme B in the serum of CDX or PDX mice were measured using human enzyme-linked immunosorbent assay kits (Thermo Fisher Scientific, KAC1231 and BMS2027) according to the manufacturer’s instructions. The isotype of the anti-uPAR mAb was determined using mouse mAb isotyping reagents (Sigma-Aldrich, ISO2).
Cell proliferation assay
Wild-type and uPAR−/− human GC cells (SNU-216 and AGS) were plated into a 96-well plate (2 × 103 per well) and cultured for 4 days. For assessment of antibody function, the anti-uPAR mAb or control mAb (final concentration, 10 μg/ml) was added into wild-type cells, which were then cultured for 4 days. Cell viability at the indicated time points was measured using the CellTiter 96 AQueous One Solution Cell Proliferation Assay [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) Promega], according to the manufacturer’s instructions. Absorbance at 490 nm was read using a plate reader (Multiskan FC, Thermo Fisher Scientific).
Transwell invasion assay
Transwell invasion assay was performed as previously described (58). Briefly, GC cells (2 × 105) were carefully seeded into the upper chamber of a transwell plate (Corning 3422) precoated with Matrigel (BD Biosciences, 354248). Serum-free medium was added into the upper chamber, while medium containing 10% FBS was added into the lower chamber. After 48 hours of incubation, the cells that did not invade through the Matrigel were removed by wiping gently with a cotton swab. Invaded cells were fixed with 4% paraformaldehyde, stained with crystal violet (Solarbio, C8470), and imaged using a microscope (Olympus IX53). Data were processed by ImageJ software (v1.49).
Cell adhesion assay
GC cells were trypsinized, washed twice with cold PBS, resuspended in culture medium containing the anti-uPAR or control mAb (10 μg/ml, 3 × 104 cells), and incubated at room temperature for 30 min. Then, the cells of each group were seeded into a 96-well plate (5 × 103 cells per well) in triplicate and incubated at 37°C with 5% CO2 for 6 hours. After washing once with PBS, cells were fixed with 4% paraformaldehyde and counted.
ADCC assay
To assess ADCC, an FcγRIIIa reporter assay was performed and optimized. Jurkat FcγRIIIa (158V) effector cells were developed as previously described (59). Target (SNU-216, 1.5 × 104 per well) and effector cells (1.5 × 105 per well) were cocultured in a 96-well plate. The anti-uPAR mAb was serially diluted with dilution medium (RPMI 1640 with 1% low IgG FBS) and added into different wells. Anti-HER2 (trastuzumab, Roche) and isotype-matched mAbs were used as positive and negative controls, respectively. Cultures were incubated at 37°C with 5% CO2 for 6 hours. Bright-Glo Luciferase Assay System (Promega) reagent was added, and relative luciferase units were measured using a plate reader (SpectraMax). The abilities of antibodies to induce ADCC were quantified as half maximal effective concentration (EC50) values (micrograms per milliliter).
CDC assay
The CDC (60) of the anti-uPAR mAb was measured as follows. SNU-216 cells were trypsinized, washed with PBS, resuspended in Opti-MEM, and filtered through a 70-μm cell strainer to remove aggregated cells. Cells were then counted and seeded into a clear U-bottom, 96-well tissue culture-treated plate (5 × 104 cells per well). Complement source (human AB serum, diluted 1:4, 50 μl; Sigma-Aldrich, H4522) and the anti-uPAR mAb (final concentration, 10 μg/ml) were added into each well and incubated with cells for 3 hours at 37°C. Anti-HER2 and isotype-matched mAbs were used as positive and negative controls, respectively. After incubation, 5 μl of propidium iodide (BioLegend, 421301) was added into each well, and the percentage of dead cells was analyzed by FCM.
Statistical analysis
SPASS 24.0 (IBM, Armonk, NY, USA) and GraphPad Prism 8.0 (San Diego, CA, USA) were used for statistical analyses and graphing. Unpaired two-tailed t test or paired t test was used for comparison of two groups. One-way analysis of variance followed by a Tamhane’s T2 or least significant difference post hoc test was used for comparing multiple groups. For the survival studies, the Kaplan-Meier analysis and log-rank (Mantel-Cox) test were used. Data were shown as means ± SEM. Statistical significance was denoted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
We thank all members in the Cuiying Biomedical Research Center, Department of General Surgery, and the Second Clinical Medical College of Lanzhou University Second Hospital. We thank H. Sun (Lanzhou University Second Hospital), J. Guan (Lanzhou University Second Hospital), H. Yang (Lanzhou University Second Hospital), and K. Li (Lanzhou Yahua Biotech Co. Ltd.) for critical discussions on the manuscript. We thank S. Zheng (the National Institute of Biological Sciences, Beijing, China) for technical support.
Funding: This work was supported by National Natural Sciences Foundation of China (32170729), Fundamental Research Funds of the Central Universities (lzujbky-2019-cd06), major project granted from Gansu Provincial Science and Technology Department (21ZD4FA015), and Cuiying Scientific and Technological Innovation Program of the Lanzhou University Second Hospital (CY2017-ZD03).
Author contributions: Z.J., Q.W., and J.Y. conceived the study. X.J. and W.C. performed bioinformatics analysis. H.Z., H.Y., B.L., Z. Yu, and W.C. performed human GC clinical sample collection and clinical data acquisition. Q.W., L.Q., Y.W., and T.Z. generated anti-uPAR mAb and uPAR CAR-T cells. X.J., W.C., and L.W. performed the organoids. L.Q., L.W., J.Z., Z.Y., W.C., Y.W., and F.W. designed and performed functional experiment. L.Q., L.W., H.Z., J.Z., Z.Y., W.C., and F.W. designed and performed mouse experiments. L.W., J.Q., Z.Y., and X.G. performed the IHC and H&E staining. L.Q., L.W., J.Z., F.W., and W.S. performed the IF, immunoelectron microscopy, and immunoblotting. J.Z., L.W., and W.C. performed the FCM and RT-PCR. J.Y., L.Q., and Z.J. contributed to the writing and editing of the manuscript.
Competing interests: Z.J., Q.W., L.Q., H.S., L.W., Y.W., J.Z., Z.Y., F.W., and W.C. are inventors on four patent applications related to this work filed by Lanzhou University Second Hospital and Lanzhou Huazhitiancheng Biotechnologies (nos. 202210129119.3, 202210129110.2, PCT/CN2022/076086, and PCT/CN2022/076087, filed 11 February 2022). The authors declare they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
REFERENCES AND NOTES
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A., Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Lauren P., The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma, An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 64, 31–49 (1965). [DOI] [PubMed] [Google Scholar]
- 4.Lordick F., Janjigian Y. Y., Clinical impact of tumour biology in the management of gastroesophageal cancer. Nat. Rev. Clin. Oncol. 13, 348–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J. H., Chang K. K., Yoon C., Tang L. H., Strong V. E., Yoon S. S., Lauren histologic type is the most important factor associated with pattern of recurrence following resection of gastric adenocarcinoma. Ann. Surg. 267, 105–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J., Li L., Zhang G., Zhou H., Yu Z., Jiao Z., Clinical study on surgical method and prognosis in diffuse-type advanced gastric cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban 41, 151–157 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Henson D. E., Dittus C., Younes M., Nguyen H., Albores-Saavedra J., Differential trends in the intestinal and diffuse types of gastric carcinoma in the United States, 1973-2000: Increase in the signet ring cell type. Arch. Pathol. Lab. Med. 128, 765–770 (2004). [DOI] [PubMed] [Google Scholar]
- 8.NCCN Guidelines Version 4.2021 Gastric Cancer. http://nccn.org. (2021).
- 9.Bang Y. J., Van Cutsem E., Feyereislova A., Chung H. C., Shen L., Sawaki A., Lordick F., Ohtsu A., Omuro Y., Satoh T., Aprile G., Kulikov E., Hill J., Lehle M., Ruschoff J., Kang Y. K.; G. A. T. I. To , Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Janjigian Y. Y., Shitara K., Moehler M., Garrido M., Salman P., Shen L., Wyrwicz L., Yamaguchi K., Skoczylas T., Bragagnoli A. C., First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 398, 27–40 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner M., Hollmen M., Junttila T. T., Kapanen A. I., Tommola S., Soini Y., Helin H., Salo J., Joensuu H., Sihvo E., Elenius K., Isola J., Amplification of HER-2 in gastric carcinoma: Association with Topoisomerase IIalpha gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann. Oncol. 16, 273–278 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Van Cutsem E., Bang Y. J., Feng-Yi F., Xu J. M., Lee K. W., Jiao S. C., Chong J. L., Lopez-Sanchez R. I., Price T., Gladkov O., Stoss O., Hill J., Ng V., Lehle M., Thomas M., Kiermaier A., Ruschoff J., HER2 screening data from ToGA: Targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 18, 476–484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang Y. K., Boku N., Satoh T., Ryu M. H., Chao Y., Kato K., Chung H. C., Chen J. S., Muro K., Kang W. K., Yeh K. H., Yoshikawa T., Oh S. C., Bai L. Y., Tamura T., Lee K. W., Hamamoto Y., Kim J. G., Chin K., Oh D. Y., Minashi K., Cho J. Y., Tsuda M., Chen L. T., Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 2461–2471 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Smith H. W., Marshall C. J., Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 11, 23–36 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Li Santi A., Napolitano F., Montuori N., Ragno P., The urokinase receptor: A multifunctional receptor in cancer cell biology. Therapeutic Implications. Int. J. Mol. Sci. 22, 4111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamoto J., Mizokami Y., Takahashi K., Matsuoka T., Matsuzaki Y., The effects of cyclooxygenase2-prostaglandinE2 pathway on Helicobacter pylori-induced urokinase-type plasminogen activator system in the gastric cancer cells. Helicobacter 13, 174–182 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Semina E. V., Rubina K. A., Shmakova A. A., Rysenkova K. D., Klimovich P. S., Aleksanrushkina N. A., Sysoeva V. Y., Karagyaur M. N., Tkachuk V. A., Downregulation of uPAR promotes urokinase translocation into the nucleus and epithelial to mesenchymal transition in neuroblastoma. J. Cell. Physiol. 235, 6268–6286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer T. W., Liu W., Fan F., Camp E. R., Yang A., Somcio R. J., Bucana C. D., Callahan J., Parry G. C., Evans D. B., Boyd D. D., Mazar A. P., Ellis L. M., Targeting of urokinase plasminogen activator receptor in human pancreatic carcinoma cells inhibits c-Met- and insulin-like growth factor-I receptor-mediated migration and invasion and orthotopic tumor growth in mice. Cancer Res. 65, 7775–7781 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Van Buren G. II, Gray M. J., Dallas N. A., Xia L., Lim S. J., Fan F., Mazar A. P., Ellis L. M., Targeting the urokinase plasminogen activator receptor with a monoclonal antibody impairs the growth of human colorectal cancer in the liver. Cancer 115, 3360–3368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabbani S. A., Ateeq B., Arakelian A., Valentino M. L., Shaw D. E., Dauffenbach L. M., Kerfoot C. A., Mazar A. P., An anti-urokinase plasminogen activator receptor antibody (ATN-658) blocks prostate cancer invasion, migration, growth, and experimental skeletal metastasis in vitro and in vivo. Neoplasia 12, 778–788 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny H. A., Leonhardt P., Ladanyi A., Yamada S. D., Montag A., Im H. K., Jagadeeswaran S., Shaw D. E., Mazar A. P., Lengyel E., Targeting the urokinase plasminogen activator receptor inhibits ovarian cancer metastasis. Clin. Cancer Res. 17, 459–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X., Cai Y., Wei Y., Donate F., Juarez J., Parry G., Chen L., Meehan E. J., Ahn R. W., Ugolkov A., Identification of a new epitope in uPAR as a target for the cancer therapeutic monoclonal antibody ATN-658, a structural homolog of the uPAR binding integrin CD11b (αM). PLOS ONE 9, e85349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahmood N., Arakelian A., Khan H. A., Tanvir I., Mazar A. P., Rabbani S. A., uPAR antibody (huATN-658) and Zometa reduce breast cancer growth and skeletal lesions. Bone Res. 8, 18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amor C., Feucht J., Leibold J., Ho Y. J., Zhu C., Alonso-Curbelo D., Mansilla-Soto J., Boyer J. A., Li X., Giavridis T., Kulick A., Houlihan S., Peerschke E., Friedman S. L., Ponomarev V., Piersigilli A., Sadelain M., Lowe S. W., Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y., Waltz D. A., Rao N., Drummond R. J., Rosenberg S., Chapman H. A., Identification of the urokinase receptor as an adhesion receptor for vitronectin. J. Biol. Chem. 269, 32380–32388 (1994). [PubMed] [Google Scholar]
- 26.Kjoller L., Hall A., Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J. Cell Biol. 152, 1145–1158 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madsen C. D., Ferraris G. M., Andolfo A., Cunningham O., Sidenius N., uPAR-induced cell adhesion and migration: Vitronectin provides the key. J. Cell Biol. 177, 927–939 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen D. H., Catling A. D., Webb D. J., Sankovic M., Walker L. A., Somlyo A. V., Weber M. J., Gonias S. L., Myosin light chain kinase functions downstream of Ras/ERK to promote migration of urokinase-type plasminogen activator-stimulated cells in an integrin-selective manner. J. Cell Biol. 146, 149–164 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D., Ghiso J. A. A., Estrada Y., Ossowski L., EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell 1, 445–457 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Tang C.-H., Hill M. L., Brumwell A. N., Chapman H. A., Wei Y., Signaling through urokinase and urokinase receptor in lung cancer cells requires interactions with β1 integrins. J. Cell Sci. 121, 3747–3756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menon S. S., Guruvayoorappan C., Sakthivel K. M., Rasmi R. R., Ki-67 protein as a tumour proliferation marker. Clin. Chim. Acta 491, 39–45 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo M., Amant F., Biankin A. V., Budinska E., Byrne A. T., Caldas C., Clarke R. B., de Jong S., Jonkers J., Maelandsmo G. M., Roman-Roman S., Seoane J., Trusolino L., Villanueva A., Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 4, 998–1013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tumeh P. C., Harview C. L., Yearley J. H., Shintaku I. P., Taylor E. J., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., West A. N., Carmona M., Kivork C., Seja E., Cherry G., Gutierrez A. J., Grogan T. R., Mateus C., Tomasic G., Glaspy J. A., Emerson R. O., Robins H., Pierce R. H., Elashoff D. A., Robert C., Ribas A., PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galdiero M. R., Garlanda C., Jaillon S., Marone G., Mantovani A., Tumor associated macrophages and neutrophils in tumor progression. J. Cell. Physiol. 228, 1404–1412 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Uhl B., Mittmann L. A., Dominik J., Hennel R., Smiljanov B., Haring F., Schaubächer J. B., Braun C., Padovan L., Pick R., Canis M., Schulz C., Mack M., Gutjahr E., Sinn P., Heil J., Steiger K., Kanse S. M., Weichert W., Sperandio M., Lauber K., Krombach F., Reichel C. A., uPA-PAI-1 heteromerization promotes breast cancer progression by attracting tumorigenic neutrophils. EMBO Mol. Med. 13, e13110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong S., Dong L., Cheng L., Neutrophils in cancer carcinogenesis and metastasis. J. Hematol. Oncol. 14, 173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si J., Shi X., Sun S., Zou B., Li Y., An D., Lin X., Gao Y., Long F., Pang B., Hematopoietic progenitor kinase1 (HPK1) mediates T cell dysfunction and is a druggable target for T cell-based immunotherapies. Cancer Cell 38, 551–566.e11 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Ge S., Xia X., Ding C., Zhen B., Zhou Q., Feng J., Yuan J., Chen R., Li Y., Ge Z., Ji J., Zhang L., Wang J., Li Z., Lai Y., Hu Y., Li Y., Li Y., Gao J., Chen L., Xu J., Zhang C., Jung S. Y., Choi J. M., Jain A., Liu M., Song L., Liu W., Guo G., Gong T., Huang Y., Qiu Y., Huang W., Shi T., Zhu W., Wang Y., He F., Shen L., Qin J., A proteomic landscape of diffuse-type gastric cancer. Nat. Commun. 9, 1012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeBeau A. M., Duriseti S., Murphy S. T., Pepin F., Hann B., Gray J. W., VanBrocklin H. F., Craik C. S., Targeting uPAR with antagonistic recombinant human antibodies in aggressive breast cancer. Cancer Res. 73, 2070–2081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duriseti S., Goetz D. H., Hostetter D. R., LeBeau A. M., Wei Y., Craik C. S., Antagonistic anti-urokinase plasminogen activator receptor (uPAR) antibodies significantly inhibit uPAR-mediated cellular signaling and migration. J. Biol. Chem. 285, 26878–26888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heiss M. M., Allgayer H., Gruetzner K. U., Funke I., Babic R., Jauch K. W., Schildberg F. W., Individual development and uPA-receptor expression of disseminated tumour cells in bone marrow: A reference to early systemic disease in solid cancer. Nat. Med. 1, 1035–1039 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Plebani M., Herszènyi L., Carraro P., De Paoli M., Roveroni G., Cardin R., Tulassay Z., Naccarato R., Farinati F., Urokinase-type plasminogen activator receptor in gastric cancer: Tissue expression and prognostic role. Clin. Exp. Metastasis 15, 418–426 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Mahmood N., Mihalcioiu C., Rabbani S. A., Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): Diagnostic, prognostic, and therapeutic applications. Front. Oncol. 8, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montuori N., Pesapane A., Rossi F. W., Giudice V., De Paulis A., Selleri C., Ragno P., Urokinase type plasminogen activator receptor (uPAR) as a new therapeutic target in cancer. Transl. Med. UniSa 15, 15–21 (2016). [PMC free article] [PubMed] [Google Scholar]
- 45.Rabbani S. A., Gladu J., Urokinase receptor antibody can reduce tumor volume and detect the presence of occult tumor metastases in vivo. Cancer Res. 62, 2390–2397 (2002). [PubMed] [Google Scholar]
- 46.Wang L., Yang R., Zhao L., Zhang X., Xu T., Cui M., Basing on uPAR-binding fragment to design chimeric antigen receptors triggers antitumor efficacy against uPAR expressing ovarian cancer cells. Biomed. Pharmacother. 117, 109173 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Carriero M. V., Bifulco K., Minopoli M., Lista L., Maglio O., Mele L., Di Carluccio G., De Rosa M., Pavone V., UPARANT: A urokinase receptor-derived peptide inhibitor of VEGF-driven angiogenesis with enhanced stability and in vitro and in vivo potency. Mol. Cancer Ther. 13, 1092–1104 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Mani T., Wang F., Knabe W. E., Sinn A. L., Khanna M., Jo I., Sandusky G. E., Sledge G. W. Jr., Jones D. R., Khanna R., Pollok K. E., Meroueh S. O., Small-molecule inhibition of the uPAR.uPA interaction: Synthesis, biochemical, cellular, in vivo pharmacokinetics and efficacy studies in breast cancer metastasis. Bioorg. Med. Chem. 21, 2145–2155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao H., Korn J. M., Ferretti S., Monahan J. E., Wang Y., Singh M., Zhang C., Schnell C., Yang G., Zhang Y., High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Pompili L., Porru M., Caruso C., Biroccio A., Leonetti C., Patient-derived xenografts: A relevant preclinical model for drug development. J. Exp. Clin. Cancer Res. 35, 189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Q., Ghafoor A., Mian I., Rathkey D., Thomas A., Alewine C., Sengupta M., Ahlman M. A., Zhang J., Morrow B., Steinberg S. M., Pastan I., Hassan R., Enhanced efficacy of mesothelin-targeted immunotoxin LMB-100 and anti-PD-1 antibody in patients with mesothelioma and mouse tumor models. Sci. Transl. Med. 12, eaaz7252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang A., Ren Z., Tseng K. F., Liu X., Li H., Lu C., Cai Y., Minna J. D., Fu Y. X., Dual targeting of CTLA-4 and CD47 on Treg cells promotes immunity against solid tumors. Sci. Transl. Med. 13, eabg8693 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holguin L., Echavarria L., Burnett J. C., Novel humanized peripheral blood mononuclear cell mouse model with delayed onset of graft-versus-host disease for preclinical HIV research. J. Virol. 96, e0139421 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim W., Chu T. H., Nienhuser H., Jiang Z., Del Portillo A., Remotti H. E., White R. A., Hayakawa Y., Tomita H., Fox J. G., Drake C. G., Wang T. C., PD-1 signaling promotes tumor-infiltrating myeloid-derived suppressor cells and gastric tumorigenesis in mice. Gastroenterology 160, 781–796 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson A. L., Dhimolea E., Reichert J. M., Development trends for human monoclonal antibody therapeutics. Nat. Rev. Drug Discov. 9, 767–774 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., Smith H. O., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Lee S. H., Hu W., Matulay J. T., Silva M. V., Owczarek T. B., Kim K., Chua C. W., Barlow L. J., Kandoth C., Williams A. B., Bergren S. K., Pietzak E. J., Anderson C. B., Benson M. C., Coleman J. A., Taylor B. S., Abate-Shen C., McKiernan J. M., Al-Ahmadie H., Solit D. B., Shen M. M., Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marshall J., Transwell(®) invasion assays. Methods Mol. Biol. 769, 97–110 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Parekh B. S., Berger E., Sibley S., Cahya S., Xiao L., LaCerte M. A., Vaillancourt P., Wooden S., Gately D., Development and validation of an antibody-dependent cell-mediated cytotoxicity-reporter gene assay. MAbs 4, 310–318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duensing T. D., Watson S. R., Complement-dependent cytotoxicity assay. Cold Spring Harb. Protoc. 2, prot093799 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3