Abstract
Background
Early prognostication of COVID-19 severity will potentially improve patient care. Biomarkers, such as TNF-related apoptosis-inducing ligand (TRAIL), interferon gamma-induced protein 10 (IP-10), and C-reactive protein (CRP), might represent possible tools for point-of-care testing and severity prediction.
Methods
In this prospective cohort study, we analyzed serum levels of TRAIL, IP-10, and CRP in patients with COVID-19, compared them with control subjects, and investigated the association with disease severity.
Results
A total of 899 measurements were performed in 132 patients (mean age 64 years, 40.2% females). Among patients with COVID-19, TRAIL levels were lower (49.5 vs 87 pg/ml, P = 0.0142), whereas IP-10 and CRP showed higher levels (667.5 vs 127 pg/ml, P <0.001; 75.3 vs 1.6 mg/l, P <0.001) than healthy controls. TRAIL yielded an inverse correlation with length of hospital and intensive care unit (ICU) stay, Simplified Acute Physiology Score II, and National Early Warning Score, and IP-10 showed a positive correlation with disease severity. Multivariable regression revealed that obesity (adjusted odds ratio [aOR] 5.434, 95% confidence interval [CI] 1.005-29.38), CRP (aOR 1.014, 95% CI 1.002-1.027), and peak IP-10 (aOR 1.001, 95% CI 1.00-1.002) were independent predictors of in-ICU mortality.
Conclusions
We demonstrated a correlation between COVID-19 severity and TRAIL, IP-10, and CRP. Multivariable regression showed a role for IP-10 in predicting unfavourable outcomes, such as in-ICU mortality.
Trial registration
Clinicaltrials.gov, NCT04655521
Keywords: COVID-19, Severity prediction, Host response, Biomarkers, TRAIL, IP-10, CRP, Point-of-care testing
Introduction
After two years into the pandemic, COVID-19 still poses an imminent global threat (Meyer et al., 2020). As of April 2022, 6.1 million people have died from or with an infection of SARS-CoV-2 (https://coronavirus.jhu.edu/, accessed April 2022) worldwide. With vaccination campaigns running slow (Papan et al., 2021), increasing numbers of vaccine breakthrough infections, and a surge of variants of concern with possible immunevasion (Juthani et al., 2021), the pandemic will most likely continue until the most dominant variants eventually become endemic (Boehm et al., 2021).
Although most infections remain asymptomatic or cause only mild symptoms, severe cases are still causing significant morbidity and mortality (Serafim et al., 2021). Such patients would benefit from early detection of risk factors for an unfavourable outcome to implement monitoring and initiate promising therapeutic approaches (Ghosn et al., 2021; REMAP-CAP Investigators et al., 2021; Rosas et al., 2021).
On the basis of current knowledge, hyperinflammation plays a critical role in disease progression and clinical deterioration (Thurner et al., 2021), suggesting that measurements of immune-based biomarkers represent promising tools for the early detection of the likelihood to deteriorate (Abers et al., 2021; Arnold et al., 2021; Vassallo et al., 2020). Because results for some of these parameters can be available within minutes through point-of-care testing, the insights provided by immune biomarkers might facilitate and accelerate diagnostic work-up (Clark et al., 2020). Ultimately, they have the potential to guide therapeutic decisions, which may lead to improved patient care and appropriate allocation of resources in return.
A novel platform (MeMed Key®) measures the circulating levels of the three host response immune proteins: TNF-related apoptosis-inducing ligand (TRAIL), interferon gamma-induced protein-10 (IP-10), and C-reactive protein (CRP). It was initially established to distinguish between a bacterial and viral etiology of infection (Oved et al., 2015; Papan et al., 2022); however, recent studies have demonstrated a wider range of application. For example, Lev et al showed that IP-10 may assist in clinical decision making in patients with COVID-19 by flagging the inflammatory response (Lev et al., 2021). This is of particular interest because TRAIL and IP-10 are virally induced markers, and interferon-mediated response is critical in severe viral infections (Papan et al., 2014), as reported also for severe courses of COVID-19 (Bastard et al., 2021, 2020; van der Wijst et al., 2021; Zhang et al., 2020).
In this study (DIRECTOR), we aimed to evaluate the different serum levels of host-immune biomarkers including TRAIL, IP-10, and CRP in patients with COVID-19 and to analyze their association with disease severity.
Methods
Study design, setting, and population
The DIRECTOR (Dynamics of the immune response to COVID-19/infection by SARS-coronavirus-2) study was a prospective, observational cohort study at the Saarland University Medical Centre in Homburg, southwest Germany. After signing written informed consent (by patients or their legal guardians), patients (age 3 months and older) who met the following inclusion criteria were enrolled: polymerase chain reaction (PCR)–confirmed infection with SARS-CoV-2 (except for controls) during the second and third pandemic wave experienced in Germany (December 2020 to July 2021). Results for patients under the age of 18 years were analyzed separately and are not reported herein. To increase the sample size, patients from the first COVID-19 wave between March and May 2020 were included retrospectively.
Control subjects were recruited mainly among the hospital staff and were divided into two groups: healthy controls (group 1) and adults with symptoms of acute respiratory tract infection (group 2), both having negative SARS-CoV-2 PCR tests.
Approval of the local ethical committee was obtained (Ärztekammer des Saarlandes, reference number 019/20), and the study was registered at clinicaltrials.gov (NCT04655521). The reporting of this study adhered to the STROBE guidelines for reporting observational cohort studies (Supplementary Table S1).
Study procedures
Nasopharyngeal swabs, blood samples, and SARS-CoV-2 RT-PCR were routinely performed on admission and regularly throughout the hospital stay to account for dynamic changes in biomarker levels over time. For performing host signature measurements, an additional serum tube was obtained.
The National Early Warning Score (NEWS) was generated for every participating patient presenting to the emergency department to objectify COVID-19 disease severity on admission. For comparability of the patient's health status in the intensive care unit (ICU), we generated the Simplified Acute Physiology Score II (SAPS II) for each in-ICU day, in which biomarker measurements were performed.
Furthermore, we conducted a follow-up telephone interview with the participants after hospital discharge. Questions included the ongoing presence of COVID-19–related symptoms and additional visits to the general practitioner due to ongoing symptoms.
Host signature measurements
Levels of TRAIL, IP-10, and CRP were measured at the study site in serum samples. After being centrifuged for 20 minutes at 4000 RPM and 20°C in a Rotixa 50 RS centrifuge, samples were measured within the next hour or intermittently frozen at -20°C. Measurements were performed on a MeMed Key® (MeMed Diagnostics, Tirat Carmel, Israel) according to the manufacturer's instructions for use. The method is based on a chemiluminescent immunoassay. The platform was placed at the central lab due to logistics but is suitable for point-of-care testing.
Statistical analysis
To examine the levels of biomarkers including TRAIL, IP-10, and CRP in patients with COVID-19 compared with healthy controls or patients with respiratory infection but who tested negative for SARS-CoV-2, we compared each first measurement of our study population with both control groups. For assessment of the correlation between biomarker serum levels and disease severity, we evaluated the minimal and maximal (or peak) levels shown throughout the hospital stay for each patient as well. Defining parameters of disease severity were ICU admission, need for mechanical ventilation, and in-ICU mortality.
Further analyses included analyses regarding severity levels on admission and the prognosticating value of biomarkers to predict disease progression. Separate subgroup analyses on patients with early biomarker measurements (within 24 hours of hospital admission) were conducted. To address a potential bias, we compared IP-10 levels of COVID-19 patients with cancer to all other COVID-19 patients. Variables are reported as the median with interquartile range (IQR) or as the mean with standard deviation (SD). Group comparisons were performed using t-test and Mann-Whitney U-test for continuous variables, and Fisher exact test and chi-square for categorical variables. Correlation between biomarkers and continuous variables accounting for clinical severity, such as duration of hospital stay and ICU days as well as correlation with other laboratory parameters, was evaluated with Spearman rank test. Univariate and multivariable logistic regression models were developed to detect biomarkers and other variables associated with clinical severity. We performed Kaplan-Meier analyses to assess the association of biomarker levels with the outcome variables: implementation of mechanical ventilation and in-ICU death. We performed analyses using GraphPad Prism (Version 9.0.0) and SPSS (Version 28.0). The statistical significance level was set at 0.05.
Results
Patient characteristics
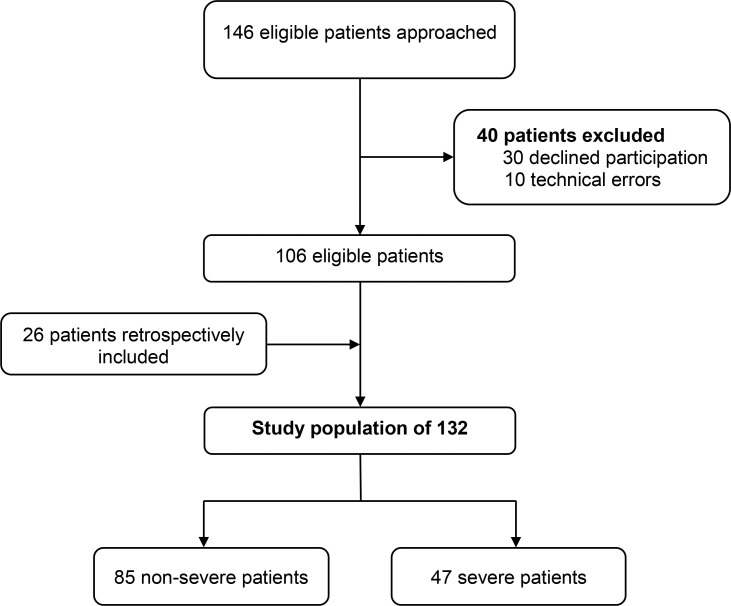
Overall, 172 eligible patients were approached to participate in the DIRECTOR study. Of these, 30 patients declined participation and in ten patients, measurements were invalid due to technical error, resulting in a total study population of 132 patients (Figure 1 ).
Figure 1.
Flow diagram of the DIRECTOR study, conducted in Saarland, Germany (2020/2021); study population comprising SARS-CoV-2-positive patients only.
The mean age was 64 years, ranging from 23-98, with 53 (40.2%) being female (Table 1 ).
Table 1.
Patient characteristics (only SARS-CoV-2-positive) for the DIRECTOR study, conducted in Saarland, Germany (2020/2021).
| Non-ICU (n=85) | ICU (n=47) | All (n=132) | P-value | |
|---|---|---|---|---|
| General | ||||
| Mean age, years (min-max) | 66 (23-98) | 64 (28-89) | 64 (23-98) | 0.519 |
| Female (n, %) | 44 (51.8) | 9 (17.0) | 53 (40.2) | <0.001 |
| Smoker (n, %) | 2 (2.4) | 0 (0.0) | 2 (1.5) | 0.493 |
| Past smoker (n, %) | 5 (5.9) | 4 (8.5) | 9 (6.8) | 0.493 |
| Hospitalization details | ||||
| Death (n, %) | 8c (9.4) | 13 (27.7) | 21 (15.9) | 0.006 |
| Duration of hospital stay, days (mean, SD) | 14.46 (11.55) | 34.19 (35.44) | 21.48 (24.83) | <0.001 |
| Ventilated (n, %) | 0 (0.0) | 29 (61.7) | 29 (22.0) | <0.001 |
| Time on ventilator, days (mean, SD) | 0.0 (0.0) | 16.13 (22.49) | 5.74 (15.42) | <0.001 |
| Time in ICU, days (mean, SD) | 0.0 (0.0) | 21.04 (34.05) | 7.49 (22.57) | <0.001 |
| Co-infection (n, %) | ||||
| Detection of bacteria | 26 (30.6) | 21 (44.7) | 47 (35.6) | 0.105 |
| Detection of fungi | 2 (2.4) | 8 (17.0) | 10 (7.6) | 0.002 |
| Other medical conditions (n, %) | ||||
| Diabetes | 22 (25.9) | 10 (21.3) | 32 (24.2) | 0.554 |
| Hypertension | 36 (42.4) | 23 (48.9) | 59 (44.7) | 0.466 |
| Cardiovascular disease | 27 (31.8) | 13 (27.7) | 40 (30.3) | 0.623 |
| Chronic lung disease | 11 (12.9) | 4 (8.5) | 15 (11.4) | 0.442 |
| Chronic renal disease | 12 (14.1) | 8 (17.0) | 20 (15.2) | 0.656 |
| Malignancy | 11 (12.9) | 2 (4.3) | 13 (9.8) | 0.109 |
| Transplant | 7 (8.2) | 3 (6.4) | 10 (7.6) | 0.700 |
| Obesity | 4 (4.7) | 20 (42.6) | 24 (18.2) | <0.001 |
| Symptoms on admission (n, %) | 69 (81.2) | 40 (85.1) | 109 (82.6) | 0.569 |
| Cougha | 29 (42.0) | 10 (25.0) | 39 (35.8) | 0.122 |
| Sore throata | 0 (0.0) | 1 (2.5) | 1 (0.9) | 0.177 |
| Dyspneaa | 44 (63.8) | 21 (52.5) | 65 (59.6) | 0.436 |
| Fevera | 39 (56.5) | 16 (40.0) | 55 (50.5) | 0.186 |
| Nauseaa | 8 (11.6) | 2 (5.0) | 10 (9.2) | 0.284 |
| Loss of smell and/or tastea | 3 (4.3) | 2 (5.0) | 5 (4.6) | 0.834 |
| Treatment (n, %) | ||||
| Remdesivir | 15 (17.6) | 1 (2.1) | 16 (12.1) | 0.009 |
| Prednisone | 8 (53.3) | 7 (14.9) | 15 (11.4) | 0.342 |
| Dexamethasone | 18 (78.3) | 5 (10.6) | 23 (17.4) | 0.126 |
| Antibiotics | 59 (59.6) | 40 (85.1) | 99 (75.0) | 0.046 |
| Renal replacement therapy | 2 (2.4) | 17 (36.2) | 19 (14.4) | <0.001 |
| Extracorporeal membrane oxygenation | 0 (0.0) | 10 (21.3) | 10 (7.6) | <0.001 |
| Follow-Up Interview (n, %) | 45 (52.9) | 11 (23.4) | 56 (42.4) | 0.001 |
| Symptomsb | 18 (40.0) | 6 (54.5) | 24 (42.9) | 0.230 |
| Repeated general practice visitsb | 20 (44.4) | 4 (36.4) | 24 (42.9) | 0.032 |
P-values are given for the comparisons between non-ICU and ICU patients.
Percentages only refer to patients presenting with symptoms of a respiratory tract infection on admission.
Percentages only refer to the number of patients for whom the Follow-Up Interview was completed.
Death occurred in these patients without ICU admission due to an existing patient decree.
ICU: intensive care unit.
Overall, 109 patients (82.6%) showed COVID-19–related symptoms on admission, most commonly dyspnea and fever. Only one patient remained in outpatient care after being tested positive for SARS-CoV-2. A total of 47 patients (35.6%) were admitted to the ICU, of whom 61.7% (n = 29) required mechanical ventilation and 21.3% (n = 10) were treated with extracorporeal membrane oxygenation (ECMO). In-hospital mortality was 15.9% (21 patients).
Bacteria and fungi were detected in 47 and 10 patients with COVID-19, respectively. A subset of 99 patients (75%) received antibacterial treatment, whereas glucocorticoids (prednisone or dexamethasone) were administered to 38 patients (28.8% of the study population). Remdesivir was prescribed to 16 patients (12.1%).
Telephone follow-ups were completed in 56 patients after hospital discharge. Among these patients, 24 (42.9%) were reported to still suffer from COVID-19-related symptoms (e.g., dyspnea, fatigue). Up to that time, 24 patients still required additional visits to their general practitioner.
A total of 899 measurements of host-immune biomarkers were performed in our study population, owing to an average of 6.8 measurements per patient. First measurements were conducted on average 4.5 days (SD = 6.455) after admission. The distribution of initial measurements over time is shown in Supplementary Figures S1 and S2.
The control group consisted of 27 adults (mean age 47.1 years, range 22-83; 19 [70.4%] females), composed of 19 adults in group 1 and 8 adults in group 2. The following symptoms of a respiratory tract infection were reported: sore throat n = 4, cough n = 3, fever n = 1, chest pain n = 1, and other n = 5.
Biomarker levels in patients with COVID-19 compared with controls
In the first measurements, there was a median TRAIL level of 49.5 pg/ml (IQR 68.25) in patients with COVID-19, whereas healthy adults (control group 1) attained a median level of 87 pg/ml (IQR 51). With a P-value of 0.0142, the results showed significance. Values of adults with symptoms (control group 2) ranged from 20-112 pg/ml, with a median of 73.5 pg/ml (Supplementary Figure S3).
Regarding initial levels of IP-10, patients with COVID-19 reached a median level of 667.5 pg/ml (IQR 1252.3), whereas control group 1 only reached a median level of 127 pg/ml (IQR 40) and control group 2 reached a median of 100.5 pg/ml (IQR 46.5). The P-value of <0.001 is given for the comparison between our study population and control group 1 (Supplementary Figure S3).
Initial CRP levels also showed a significant difference (P <0.001) between patients with COVID-19 (median 75.3 mg/l, IQR 117.4) and healthy controls (group 1) (median 1.6 mg/l, IQR 4.4) (Supplementary Figure S3).
Comparing controls (n = 27) and patients with COVID-19, high IP-10 (odds ratio [OR] 1.007, P = 0.001) and CRP (OR 1.144, P <0.001) serum levels were associated with a positive PCR test on SARS-CoV-2 in the univariate logistic regression model, with CRP remaining significant in the multivariable analysis (adjusted OR 1.112, P <0.001, Nagelkerke R2 0.703) (Extension of Table 2 , supplements).
Table 2.
Results of univariate and multivariable logistic regression analyses of the patient cohort recruited within the DIRECTOR study, Saarland, Germany (2020/2021). TRAIL, IP-10, and CRP indicate first measurements unless specified otherwise. Significant results are highlighted in bold.
a) All survivors of COVID-19 (n = 111) as well as in patients with COVID-19 who died in the ICU (n = 13), Admission to ICU (n = 47), Nagelkerke R2 0.404.
b) All survivors of COVID-19 (n = 111) as well as in patients with COVID-19 who died in the ICU (n = 13), Need for mechanical ventilation (n = 29), Nagelkerke R2 0.538.
c) All survivors of COVID-19 (n = 111) as well as in patients with COVID-19 who died in the ICU (n = 13), Died in the ICU (n = 13), Nagelkerke R2 0.485.
d) Subgroup of COVID-19 patients with first measurements <24 hours after admission (n = 58), Admission to ICU (n = 10), Nagelkerke R2 0.460.
| Variable | OR (95% CI) | P-value | aOR (95% CI) | P-value |
|---|---|---|---|---|
| a) Outcome: ICU admission | ||||
| Male Sex | 4.114 (1.75-9.654) | 0.001 | 3.529 (1.32-9.434) | 0.012 |
| Obesity | 18.272 (5.024-66.45) | <0.001 | 12.883 (3.22-51.59) | <0.001 |
| TRAIL | 0.989 (0.98-0.998) | 0.012 | 1.001 (0.991-1.01) | 0.883 |
| IP-10 | 1.00 (1.00-1.00) | 0.722 | 1.00 (0.999-1.00) | 0.633 |
| CRP | 1.01 (1.05-1.015) | <0.001 | 1.009 (1.001-1.016) | 0.018 |
| b) Outcome: Mechanical ventilation | ||||
| Male Sex | 3.804 (1.338-10.82) | 0.012 | 3.007 (0.819-11.03) | 0.097 |
| Obesity | 21.014 (6.933-63.69) | <0.001 | 14.311 (3.904-52.47) | <0.001 |
| TRAIL | 0.976 (0.962-0.991) | 0.001 | 0.999 (0.984-1.014) | 0.863 |
| IP-10 | 1.00 (1.00-1.001) | 0.156 | 1.00 (1.00-1.001) | 0.660 |
| CRP | 1.016 (1.009-1.022) | <0.001 | 1.014 (1.005-1.024) | 0.002 |
| c) Outcome: Death in ICU | ||||
| Male Sex | 2.189 (0.57-8.403) | 0.254 | 2.996 (0.361-24.88) | 0.310 |
| Obesity | 6.927 (2.061-23.28) | 0.002 | 4.582 (0.814-25.80) | 0.084 |
| TRAIL | 0.989 (0.97-1.004) | 0.989 | 1.014 (0.993-1.035) | 0.188 |
| IP-10 | 1.001 (1.00-1.001) | <0.001 | 1.00 (0.999-1.001) | 0.871 |
| CRP | 1.013 (1.005-1.021) | <0.001 | 1.015 (1.002-1.027) | 0.018 |
| Minimal TRAIL | 0.958 (0.922-0.997) | 0.033 | 0.979 (0.929-1.032) | 0.429 |
| Maximal IP-10 | 1.001 (1.00-1.001) | <0.001 | 1.001 (1.00-1.001) | 0.020 |
| d) Subgroup, Outcome: ICU admission | ||||
| Male Sex | 3.267 (0.752-14.19) | 0.114 | 3.159 (0.48-20.93) | 0.233 |
| Obesity | 22.5 (4.02-125.95) | <0.001 | 23.192 (3.19-168.36) | 0.002 |
| TRAIL | 0.989 (0.97-1.004) | 0.157 | 1.002 (0.984-1.02) | 0.848 |
| IP-10 | 1.00 (1.00-1.001) | 0.507 | 1.00 (0.999-1.001) | 0.609 |
| CRP | 1.01 (1.00-1.02) | 0.045 | 1.011 (0.995-1.026) | 0.180 |
aOR: adjusted odds ratio (multivariable); CI: confidence interval CRP: C-reactive protein; ICU: intensive care unit; IP-10: interferon gamma-induced protein 10; OR: odds ratio (univariate); TRAIL: TNF-related apoptosis-inducing ligand.
Biomarkers correlate with disease severity
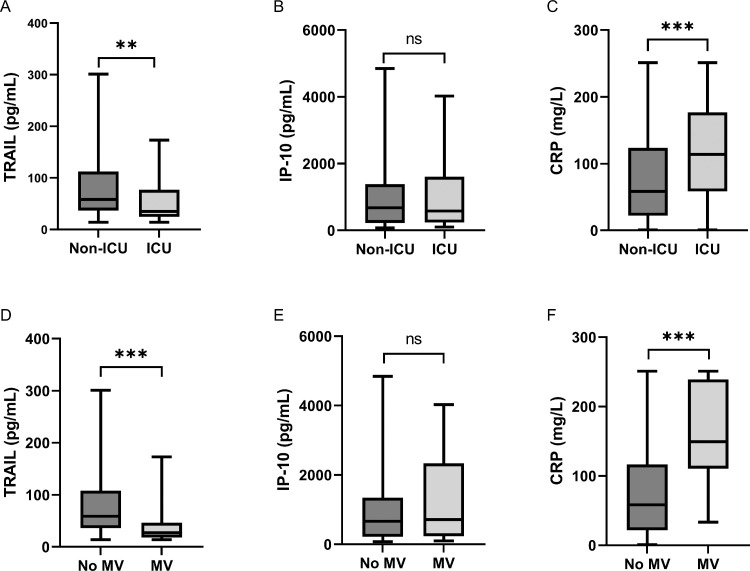
Serum levels of TRAIL showed an inverse correlation and of CRP a positive correlation with different parameters of disease severity. Regarding the first measurement, patients admitted to the ICU showed significantly lower TRAIL levels (P = 0.0032). The median level was 35 pg/ml (IQR 52.7), whereas the median TRAIL level of patients remaining in the normal care unit (NCU) was 58 pg/ml (IQR 76) (Figure 2 A). Furthermore, CRP levels were significantly higher in patients developing a severe course of the disease (P = 0.0003 for ICU admission, P <0.001 for mechanical ventilation), comparing a median of 58.3 mg/l (IQR 101.8) and 58.3 mg/l (IQR 94.7) for non-severe COVID-19 with a median of 113.8 mg/l (IQR 118.3) and 149.2 mg/l (IQR 128.6) for severe COVID-19, respectively (Figure 2C and F). There was no significant difference in these groups regarding initial IP-10 measurements (Figure 2B and E).
Figure 2.
Differential expression of biomarkers between non-severe and severe COVID-19.
Comparisons of (A) first TRAIL measurements, (B) first IP-10 measurements, and (C) first CRP measurements between patients who were admitted to the ICU (severe COVID-19, n = 47) and patients who remained in the NCU (Non-ICU = non-severe COVID-19, n = 85). Further comparisons (D-F) regarding patients requiring mechanical ventilation (MV, n = 29) and patients who did not (no MV, n = 103). The black line denotes the median of each group. The box identifies values between 25 and 75 percentiles, whereas the whiskers indicate minimal and maximal serum levels. P-values are defined as follows: ns: nonsignificant, *: P ≤0.05, **: P ≤0.01, ***: P ≤0.001. CRP: C reactive protein; ICU: intensive care unit; IP-10: interferon gamma-induced protein 10; TRAIL: TNF-related apoptosis-inducing ligand.
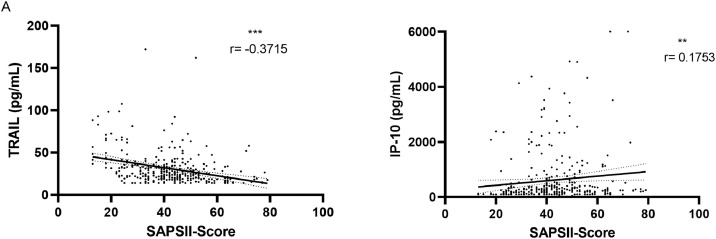
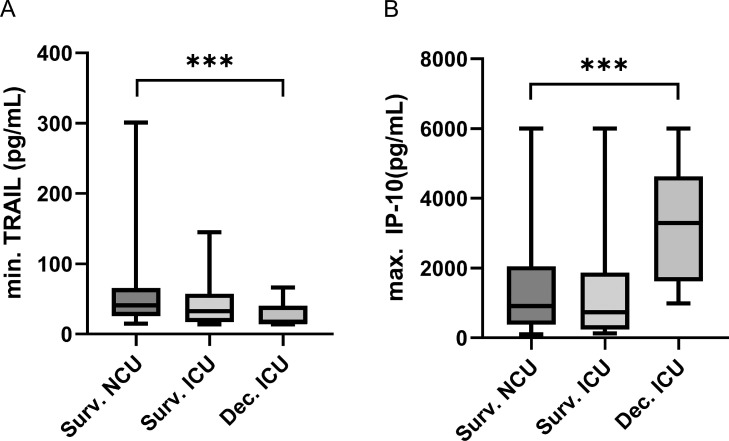
Notably, low TRAIL levels on patients’ first measurements also correlated with a longer ICU stay (r = -0.6264, P <0.001) (Supplementary Figure S4) and showed weak correlations with duration of hospital stay (r = -0.2532, P = 0.0034) (Supplementary Figure S4) and SAPS II scores for patients in the ICU (r = -0.3715, P <0.001) (Figure 3 A). In addition, we found a significant difference in minimal TRAIL levels (P <0.001) between patients who died in the ICU (n = 13, median 17 pg/ml, IQR 26) compared with survivors of COVID-19 in the NCU (n = 77, median 41 pg/ml, IQR 40) (Figure 4 A). Minimal TRAIL levels were found, on average, 8.08 days (SD = 8.171) after hospital admission and, on average, 19 days (SD = 22) before death for patients who died in the ICU.
Figure 3.
Correlation of biomarkers with SAPS II score.
Correlation of all (A) TRAIL and (B) IP-10 measurements conducted in the ICU (n = 340) with the Simplified Acute Physiology II score. Significance levels, Spearman correlation coefficients (r), and simple linear regression lines are shown. P-values are defined as follows: ns: nonsignificant, *: P ≤0.05, **: P ≤0.01, ***: P ≤0.001. ICU: intensive care unit; IP-10: interferon gamma-induced protein 10; SAPS II: Simplified Acute Physiology Score II; TRAIL: TNF-related apoptosis-inducing ligand.
Figure 4.
Differential expression of biomarkers between survivors of COVID-19 and in the ICU deceased patients with COVID-19.
Comparisons of (A) minimal TRAIL levels and (B) maximal IP-10 levels between survivors of COVID-19 (NCU n = 77, ICU n = 34) and later in the ICU deceased patients with COVID-19 (n = 13). The black line denotes the median of each group. The box identifies values between 25 and 75 percentiles, whereas the whiskers indicate minimal and maximal serum levels. P-values are defined as follows: ns: nonsignificant, *: P ≤0.05, **: P ≤0.01, ***: P ≤0.001. ICU: intensive care unit; IP-10: interferon gamma-induced protein 10; NCU: normal care unit; TRAIL: TNF-related apoptosis-inducing ligand.
Examining maximal IP-10 levels, these were significantly higher in patients who later died in the ICU compared with survivors in the NCU (p< 0.001). Patients who later died showed a median maximal level of 3295 pg/ml (IQR 3010), whereas survivors only had a median level of 909 pg/ml (IQR 1679) (Figure 4B). Maximal IP-10 levels were present, on average, 6.91 days (s = 8.787) after admission and for patients who died in the ICU, 21 days (s = 24) before death. Levels of all IP-10 measurements in the ICU (n = 340) also showed a weak correlation with the SAPS II score (r = 0.1753, P = 0.0012) (Figure 3B). Of note, CRP levels, but not IP-10 levels, showed a significant correlation with length of hospital or ICU stay (Supplementary Figures S5 and S6). The dynamic changes of biomarker levels per patient are shown in the Supplemental Material (Supplementary Figures 6-13). Of note, detailed analyses of intraindividual biomarker analyses (Supplementary Figures S13 and S14) showed that all deceased patients had very low TRAIL levels one day before death.
Biomarkers in prediction of disease severity
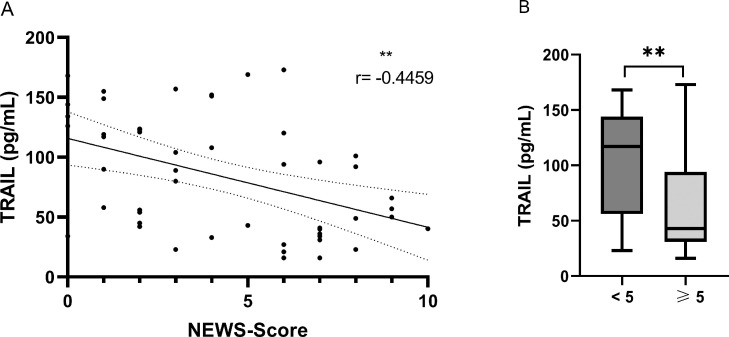
In 58 patients who presented at the emergency department, the first biomarker measurements were available within 24 hours of admission. A correlation of low levels of TRAIL with high NEWS results was shown for this subgroup (r = -0.4459, P = 0.0012) (Figure 5 ). We found no significant correlation between IP-10 and the NEWS (Supplementary Figure S15).
Figure 5.
Correlation of TRAIL with NEWS-Score on admission.
(A) Correlation of the New Early Warning Score of patients with COVID-19 presenting to the hospital's emergency department with initial TRAIL measurements conducted less than 24 hours after admission (n = 50). Significance level, Spearman correlation coefficient (r), and simple linear regression line are shown.
(B) Comparison of initial TRAIL levels (measurements conducted less than 24 hours after admission) between patients presenting to the emergency department with a New Early Warning Score lower than 5 (n = 27) and patients with a New Early Warning Score of five or higher (n = 23). The black line denotes the median of each group. The box identifies values between 25 and 75 percentiles, whereas the whiskers indicate minimal and maximal serum levels.
P-values are defined as follows: ns: nonsignificant, *: P ≤0.05, **: P ≤0.01, ***: P ≤0.001. ICU: intensive care unit; NEWS: New Early Warning Score; TRAIL: TNF-related apoptosis-inducing ligand.
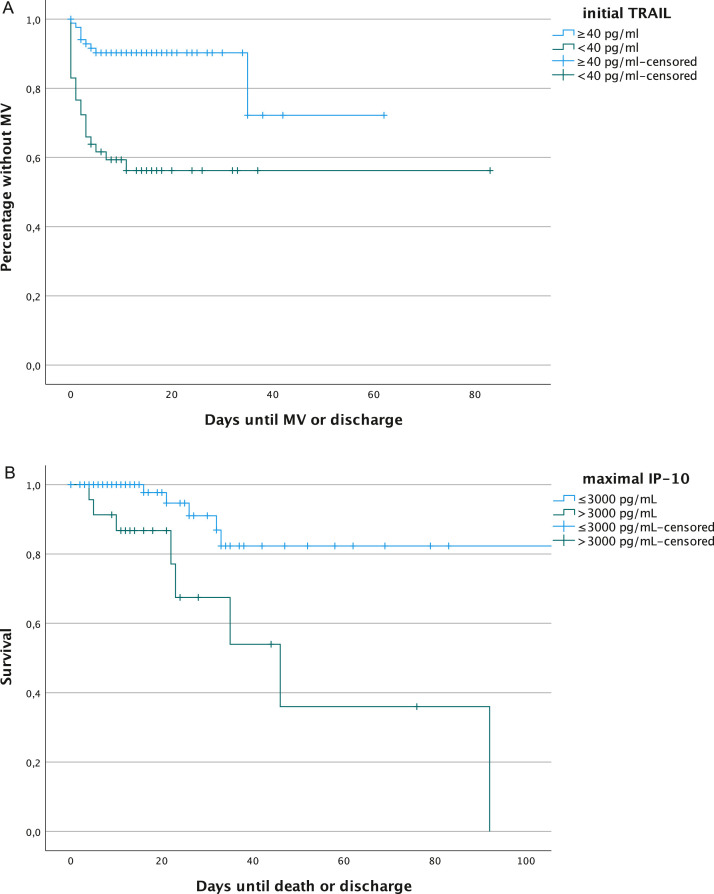
Regarding the univariate logistic regression model, low initial TRAIL levels and male sex were associated with increased likelihood of required mechanical ventilation, with an OR of 0.976 (P = 0.001) and 3.804 (P = 0.012), respectively. This association was also found in Kaplan-Meier analysis, using a cut-off for initial TRAIL of 40 pg/ml, which yielded a significantly higher probability of mechanical ventilation in patients with initial TRAIL levels of <40 pg/ml (logrank test x2 18.8, P <0.001) (Figure 6 ). In the multivariable logistic regression model, obesity (OR 14.311, 95% CI 3.904-52.47) and CRP (OR 1.014, 95% CI 1.005-1.024) were significantly related to the clinical outcome mechanical ventilation (Table 2b). For the outcome death in ICU, CRP and peak IP-10 were significant in the multivariable regression model (Table 2c). IP-10 also yielded a significant difference in the Kaplan-Meier survival analysis for patients with peak IP-10 levels of >3000 pg/ml (logrank rest x2 10.5, P = 0.001) (Figure 6).
Figure 6.
Kaplain-Meier analyses of (A) the relation between initial TRAIL with cut-off 40 pg/ml for the outcome variable mechanical ventilation and (B) the relation between maximal IP-10 cut-off 3000 pg/ml and death in the ICU (maximum set at 100 days). ICU: intensive care unit; IP-10: interferon gamma-induced protein 10; MV: mechanical ventilation; TRAIL: TNF-related apoptosis-inducing ligand.
Similarly, for the subgroup with early biomarker measurement (within first 24 hours), only obesity was associated with ICU admission (Table 2d).
Additional results
We found weak correlations of TRAIL and IP-10 levels on patients’ first measurements with other laboratory parameters that were measured on the same day (Supplementary Table S2).
There was no statistically significant difference in initial IP-10 levels in patients with COVID-19 with cancer (n = 13, median 872 pg/ml, IQR 2299) and all other patients with COVID-19 (n = 119, median 631 pg/ml, IQR 1126, P = 0.173).
Discussion
In this study, we assessed the levels of TRAIL, IP-10, and CRP in patients with COVID-19, as well as their association with disease severity. We showed that IP-10 may be a valuable biomarker in detecting patients with a higher mortality risk, alongside obesity and CRP. Low TRAIL levels related to high NEWS on admission, which considers different vital parameters (respiratory rate, oxygen saturation, temperature, systolic blood pressure, heart rate) as well as level of consciousness and need for supplemental oxygen (Tirkkonen et al., 2019). In addition, patients with an initial TRAIL level <40 pg/ml had a significant association with mechanical ventilation in the Kaplan-Meier analysis, and similarly low TRAIL levels were observed in patients who died. However, TRAIL failed to show a significant association with any of the adverse outcomes in the multivariable regression model. The discrepancies in the aforementioned analyses can be explained by the important role of the time factor in Kaplan-Meier analyses. The correlation to a high NEWS and the results shown in the discussed Kaplan-Meier analysis highlight TRAIL's potential use for risk stratification in the emergency department. As levels were extremely low in all patients shortly before death, further exploration of TRAIL's capability for monitoring purposes seems reasonable as well. Even among survivors of COVID-19, the levels decreased as hospital stay increased. Accordingly, increasing IP-10 levels would be expected, but this could not be shown in our study.
For measurements in the ICU, a correlation of TRAIL and IP-10 with the SAPS II score was found. SAPS II scores convert to the probability of hospital mortality (Le Gall et al., 1993). These results confirm the findings of Chen and colleagues and Yang et al regarding IP-10’s association with disease severity (Chen et al., 2020; Yang et al., 2020).
Our revelations support the suitability of the analyzed immune-based biomarkers TRAIL, IP-10, and CRP to be included in risk assessment models. Early detection of risk factors as well as of clinical deterioration may help in clinical decision making. Identifying patients at risk could reduce the administration of expensive monoclonal antibodies or antivirals in unnecessary cases as well as protect non-vulnerable patients from hospitalization and invasive measures, both contributing to better patient outcome and better use of scarce resources.
Especially because a shift in the coming patient population is to be expected owing to vaccination of individuals who are at high risk, patient characteristics (e.g., comorbidities, age, sex) should not be used as the sole tool for risk stratification even though male sex and obesity were independently associated with poor outcome in this and other studies (Palaiodimos et al., 2020; Simonnet et al., 2020). Therefore, prediction models that combine several variables and features seem reasonable.
During the past months, several studies evaluated different laboratory parameters for such purposes. D-dimers, ferritin, procalcitonin, lactate dehydrogenase (LDH), and serum amyloid A (SAA) are some examples of markers associated with poor prognosis of COVID-19, but their accuracy remains to be validated (Huang et al., 2020; Li et al., 2020; Poggiali et al., 2020; Wang et al., 2021). Because disease progression is most likely caused by hyperinflammation, application of cytokine measurements seems reasonable for prognosticating and monitoring patients with COVID-19 (Juthani et al., 2021; Lev et al., 2021). Del Valle et al demonstrated that serum interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) outperformed other markers (Del Valle et al., 2020). However, their specificity is uncertain, and cytokine quantification as well as clinically helpful interpretation remains challenging. In addition, correlation to the primarily analyzed biomarkers was weak in this study and measurement of multiple laboratory parameters comes across as impractical (Liu et al., 2021). TRAIL, IP-10, and CRP can be measured on a single immunoassay platform, generating results within 15 minutes, overcoming these limitations. Especially their computational integration into a single numeric score is promising, with a novel clinical severity score currently being developed (Mastboim et al., 2021). Previously, Tang et al showed that plasma concentrations of IP-10 and MIG were associated with poor clinical outcome already in SARS-CoV-1 in 2005. IP-10 even acted as an independent prognostic factor for adverse outcome (Tang et al., 2005). COVID-19 appeared to be the main reason for the elevated IP-10 levels in our patient population because comorbidities such as cancer showed no significant differences in group comparisons.
Either way, a critical examination and validation of a prediction model is paramount in avoiding doing more harm than good. Additionally, defining a baseline for these models is challenging because patients can present to the emergency department at different stages of the disease.
A limitation of our study is the low number of mild, outpatient COVID-19 cases, including those who were asymptomatic, who usually do not present to the hospital. The overall cohort size may have impacted the relatively small effect sizes and the weak correlations reported herein. Furthermore, it is important to note that this study was conducted at a University hospital, thus creating a selection bias. Some patients were transferred here due to clinical deterioration, making it impossible to capture biomarkers and clinical data at an early stage of the disease. Thus, generalizability of the reported severity prediction needs further and external validation. Comparability between patients and controls is additionally hampered by the small sample size and the fact that controls were younger and more often female. Another limitation is that patients with a potentially bacterial co-infection were not separately assessed because no adjudication process was included in the study to approximate disease etiology as previously reported (van Houten et al., 2019; Papan et al., 2022). Further studies are warranted to investigate generalizability to other viral diseases, such as influenza. Besides, information about biomarker performance in patients who are vaccinated is lacking. Our choice in not giving a single definition for severe COVID-19, but rather demonstrating association of biomarkers to different parameters accounting for disease severity can be seen as a strength and as a weakness because definitions may shift in the future. In either case, further studies should externally validate the potential of TRAIL, IP-10, and CRP for predicting disease severity, with definitions fully aligned to the World Health Organization COVID-19 guidelines to allow categorization into more severity groups. Evaluation of the biomarkers’ capability for monitoring disease progression, co-infections, response to treatment, and selection of therapy could also bring an extra benefit. To verify the applicability of TRAIL, IP-10, and CRP for predicting COVID-19, further comparisons of serum levels to different infectious and non-infectious diseases would be helpful.
To conclude, TRAIL and IP-10 showed significant correlation with COVID-19 severity, and CRP and IP-10 levels were associated with adverse COVID-19 outcomes. This suggests that the inclusion of these markers into multivariable risk assessment models could be a promising tool in the management of patients with COVID-19.
Declaration of Competing Interest
The authors have no competing interests to declare.
Acknowledgments
Acknowledgments
We wish to express our gratitude to all patients and their families who participated in this study. We sincerely thank the following members of the staff for their time and dedication: Claudia Teunis, Ronald Schnell, Tim Schmidt, Jaqueline Margardt, Meytan Akman, Nina Bühler, Jochen Geitlinger, Nuran Darancik, Alexander Halfmann, and the technicians of the Centre for Infectious Diseases.
Author contributions
CP, GD, SLB, and PML conceived the study and its design, had full access to the data, and take responsibility for the integrity of the data and accuracy of the analysis. Funding acquisition was made by CP. ST, GD, DK, and CK organized and entered data. CP, ST, GD, and JG performed data analyses. All authors contributed to data interpretation. ST, GD, and CP wrote the main draft of the manuscript. All authors contributed to the final drafting of the manuscript.
Funding
This study has been funded by a 2021 CAREer Grant from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) to CP. Additionally, it was supported by a grant awarded to MeMed from the European Commission, Executive Agency for Small and Medium-sized Enterprises H2020-EIC-SMEInst-2018-2020-2 [grant number 88124]. The funding sources played no role in study design, data collection, data analysis, data interpretation, writing of the manuscript, or in the decision to submit the manuscript for publication.
Ethical approval statement
This study was approved by the ethical committee of the Ärztekammer des Saarlandes (reference number 019/20).
Data sharing
All relevant data that support the findings of the study are available within the article. Complementary data is available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.05.051.
Appendix. Supplementary materials
References
- Abers MS, Delmonte OM, Ricotta EE, Fintzi J, Fink DL, de Jesus AAA, et al. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6 doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold DT, Attwood M, Barratt S, Morley A, Elvers KT, McKernon J, et al. Predicting outcomes of COVID-19 from admission biomarkers: a prospective UK cohort study. Emerg Med J. 2021;38:543–548. doi: 10.1136/emermed-2020-210380. [DOI] [PubMed] [Google Scholar]
- Bastard P, Orlova E, Sozaeva L, Lévy R, James A, Schmitt MM, et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med. 2021;218 doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm E, Kronig I, Neher RA, Eckerle I, Vetter P, Kaiser L. Geneva Centre for Emerging Viral Diseases. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021;27:1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang J, Liu C, Su L, Zhang D, Fan J, et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol Med. 2020;26:97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TW, Brendish NJ, Poole S, Naidu VV, Mansbridge C, Norton N, et al. Diagnostic accuracy of the FebriDx host response point-of-care test in patients hospitalised with suspected COVID-19. J Infect. 2020;81:607–613. doi: 10.1016/j.jinf.2020.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn L, Chaimani A, Evrenoglou T, Davidson M, Graña C, Schmucker C, et al. Interleukin-6 blocking agents for treating COVID-19: a living systematic review. Cochrane Database Syst Rev. 2021;3 doi: 10.1002/14651858.CD013881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I, Pranata R, Lim MA, Oehadian A, Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther Adv Respir Dis. 2020;14 doi: 10.1177/1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juthani PV, Gupta A, Borges KA, Price CC, Lee AI, Won CH, et al. Hospitalisation among vaccine breakthrough COVID-19 infections. Lancet Infect Dis. 2021;21:1485–1486. doi: 10.1016/S1473-3099(21)00558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall JR, Lemeshow S, Saulnier F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270:2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- Lev S, Gottesman T, Sahaf Levin G, Lederfein D, Berkov E, Diker D, et al. Observational cohort study of IP-10’s potential as a biomarker to aid in inflammation regulation within a clinical decision support protocol for patients with severe COVID-19. PLOS ONE. 2021;16 doi: 10.1371/journal.pone.0245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xiang X, Ren H, Xu L, Zhao L, Chen X, et al. Serum amyloid A is a biomarker of severe coronavirus Disease and poor prognosis. J Infect. 2020;80:646–655. doi: 10.1016/j.jinf.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BM, Martins TB, Peterson LK, Hill HR. Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: a review. Cytokine. 2021;142 doi: 10.1016/j.cyto.2021.155478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastboim NS, Angel A, Shaham O, Ber TI, Navon R, Simon E, et al. An immune-protein signature combining TRAIL, IP-10 and CRP for accurate prediction of severe COVID-19 outcome. Br Med J. 2021 doi: 10.1101/2021.06.27.21259196. [DOI] [Google Scholar]
- Meyer S, Papan C, Last K. A global health perspective on SARS-CoV-2—hazards, disaster and hope. Wien Med Wochenschr. 2020;170:357–358. doi: 10.1007/s10354-020-00769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved K, Cohen A, Boico O, Navon R, Friedman T, Etshtein L, et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLOS ONE. 2015;10 doi: 10.1371/journal.pone.0120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaiodimos L, Kokkinidis DG, Li W, Karamanis D, Ognibene J, Arora S, et al. Severe obesity, increasing age and male sex are independently associated with worse in-hospital outcomes, and higher in-hospital mortality, in a cohort of patients with COVID-19 in the Bronx, New York. Metabolism. 2020;108 doi: 10.1016/j.metabol.2020.154262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papan C, Argentiero A, Porwoll M, Hakim U, Farinelli E, Testa I, et al. A host signature based on TRAIL, IP-10, and CRP for reducing antibiotic overuse in children by differentiating bacterial from viral infections: a prospective, multicentre cohort study. Clin Microbiol Infect. 2022;28:723–730. doi: 10.1016/j.cmi.2021.10.019. [DOI] [PubMed] [Google Scholar]
- Papan C, Hagl B, Heinz V, Albert MH, Ehrt O, Sawalle-Belohradsky J, et al. Beneficial IFN-α treatment of tumorous herpes simplex blepharoconjunctivitis in dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol. 2014;133:1456–1458. doi: 10.1016/j.jaci.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Papan C, Last K, Meyer S. COVID-19: fighting the foe with Virchow. Infection. 2021;49:1069–1070. doi: 10.1007/s15010-021-01628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggiali E, Zaino D, Immovilli P, Rovero L, Losi G, Dacrema A, et al. Lactate dehydrogenase and C-reactive protein as predictors of respiratory failure in CoVID-19 patients. Clin Chim Acta. 2020;509:135–138. doi: 10.1016/j.cca.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REMAP-CAP Investigators. Gordon AC, Mouncey PR, Al-Beidh F, Rowan KM, Nichol AD, et al. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N Engl J Med. 2021;384:1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas IO, Bräu N, Waters M, Go RC, Hunter BD, Bhagani S, et al. Tocilizumab in hospitalized patients with severe Covid-19 pneumonia. N Engl J Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafim RB, Póvoa P, Souza-Dantas V, Kalil AC, Salluh JIF. Clinical course and outcomes of critically ill patients with COVID-19 infection: a systematic review. Clin Microbiol Infect. 2021;27:47–54. doi: 10.1016/j.cmi.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonnet A, Chetboun M, Poissy J, Raverdy V, Noulette J, Duhamel A, et al. High prevalence of obesity in severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity (Silver Spring) 2020;28:1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NL-S, Chan PK-S, Wong CK, To KF, Wu AK-L, Sung YM, et al. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin Chem. 2005;51:2333–2340. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurner L, Fadle N, Bewarder M, Kos I, Regitz E, Thurner B, et al. Autoantibodies against Progranulin and IL-1 receptor antagonist due to immunogenic posttranslational isoforms contribute to hyperinflammation in critically ill COVID-19. 2021. doi: 10.1101/2021.04.23.441188. [DOI]
- Tirkkonen J, Karlsson S, Skrifvars MB. National early warning score (NEWS) and the new alternative SpO2 scale during rapid response team reviews: a prospective observational study. Scand J Trauma Resusc Emerg Med. 2019;27:111. doi: 10.1186/s13049-019-0691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst MGP, Vazquez SE, Hartoularos GC, Bastard P, Grant T, Bueno R, et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021;13:eabh2624. doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houten CB, Naaktgeboren CA, Ashkenazi-Hoffnung L, Ashkenazi S, Avis W, Chistyakov I, et al. Expert panel diagnosis demonstrated high reproducibility as reference standard in infectious diseases. J Clin Epidemiol. 2019;112:20–27. doi: 10.1016/j.jclinepi.2019.03.010. [DOI] [PubMed] [Google Scholar]
- Vassallo M, Manni S, Pini P, Blanchouin E, Ticchioni M, Seitz-Polski B, et al. Patients with Covid-19 exhibit different immunological profiles according to their clinical presentation. Int J Infect Dis. 2020;101:174–179. doi: 10.1016/j.ijid.2020.09.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang LM, Pei SF, Chong YZ, Guo Y, Gao XL, et al. CRP, SAA, LDH, and DD predict poor prognosis of coronavirus disease (COVID-19): a meta-analysis from 7739 patients. Scand J Clin Lab Invest. 2021;81:679–686. doi: 10.1080/00365513.2021.2000635. [DOI] [PubMed] [Google Scholar]
- Yang Y, Shen C, Li J, Yuan J, Wei J, Huang F, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol. 2020;146 doi: 10.1016/j.jaci.2020.04.027. 119–127.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.