Abstract
INTRODUCTION:
The ability of carbohydrate antigen 19-9 (CA19-9) to differentiate pancreatic cancer from other benign pancreatic lesions is unsatisfactory. This study explored the diagnostic value of KRAS gene mutations and plasma circulating tumor DNA (ctDNA) in patients with pancreatic cancer.
METHODS:
The prospective cohort study comprised 149 consecutive patients with solid pancreatic lesions who underwent endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA). KRAS subtype mutations were analyzed by digital droplet PCR (ddPCR) in EUS-FNA histopathology tissue samples, and blood samples were sent for plasma ctDNA analysis. The final diagnosis was based on surgical resection pathology or follow-up for at least 2 years.
RESULTS:
Adding KRAS mutation ddPCR increased the sensitivity and accuracy of EUS-FNA from 71.4% to 91.6% (P < 0.001) and 75.8% to 88.6% (P < 0.001), respectively. By comparison, the sensitivities of circulating biomarkers ctDNA and CA19-9 were only 35.2% and 71.2%. The area under the curve of the receiver operating characteristic curve (AUC) of EUS-FNA and KRAS ddPCR combination was >0.90 for distinguishing pancreatic cancer from benign lesions, whereas the AUC of EUS-FNA and CA19-9 combination was 0.83. The median survival time was significantly shorter in patients with G12D KRAS mutations than that in patients with other mutations (180 vs 240 days, P < 0.001).
DISCUSSION:
FNA tissue sample KRAS mutation analysis in tissues significantly improves the diagnostic accuracy of cyto/histopathological evaluation in EUS-FNA samples. The combination of EUS-FNA and tissue sample KRAS ddPCR provided a more accurate method for pancreatic cancer diagnosis, superior to the combination of EUS-FNA and CA19-9/ctDNA. G12D KRAS mutations in pancreatic cancer were independently associated with poor overall survival.
BACKGROUND
Pancreatic cancer is the fourth leading cause of cancer-related death globally, with more than 60,000 estimated deaths per year in China (1,2). The prognosis for patients with pancreatic cancer has hardly improved over the past 25 years, with the median survival remaining shorter than 6 months and the 5-year overall survival remaining only 9% (3).
Currently, there are no biomarkers validated for the early detection of pancreatic cancer. Carbohydrate antigen 19-9 (CA19-9) is routinely used in the pancreatic cancer screen. Still, its ability to differentiate cancerous from benign pancreatic lesions is highly limited because CA19-9 is also elevated in patients with benign pancreatic lesions (4). Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) is the current recommended first-line technique for pancreatic cancer diagnosis, staging, and sampling (5). However, EUS-FNA is an invasive procedure, and the sensitivity and accuracy of EUS-FNA can be improved further through molecular biomarker analysis of the specimen obtained (6–10). One potential biomarker is the KRAS mutation. In pancreatic cancer, the KRAS mutation is the most commonly acquired mutation with reported rates of 70–95% (11). The KRAS gene encodes the p21 RAS protein, a small guanosine triphosphatase (GTPase). This point mutation of KRAS is involved in pancreatic carcinogenesis, uncontrolled proliferation, and invasion of pancreatic cancer cells (12).
When molecular mutation detection in primary pancreatic cancer is limited by the difficulty of obtaining tumor tissues, biomarkers in circulation provide a noninvasive approach that can be more widely available (13). Recently, mutant circulating tumor DNA (ctDNA) has been explored as a biomarker. The concept underlying this approach, also called “liquid biopsies,” is that dying cancer cells release DNA into body fluids, such as plasma, so the DNA associated with cancer can be identified in those bodily fluids (14,15). ctDNA is a broadly applicable, sensitive, and specific biomarker used for various clinical and research purposes in patients with multiple different types of cancer (16). The fraction of patients with detectable plasmatic ctDNA and its concentration increase with adenocarcinoma stage (17). Therefore, detecting KRAS mutations in ctDNA can potentially be used as a diagnostic tool offering blood-based pancreatic cancer detection.
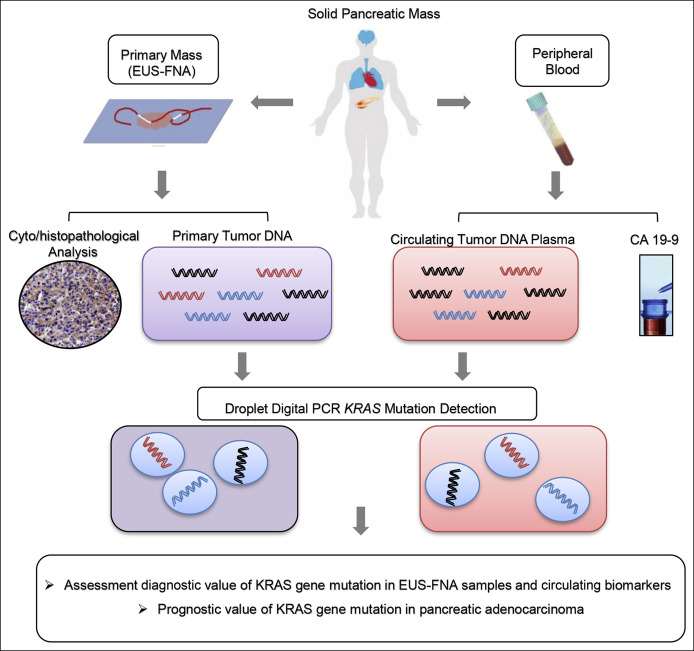
To date, it is not clear whether measurement of KRAS mutations in ctDNA or detection of KRAS mutations in EUS-FNA samples offers higher sensitivity and accuracy in the early diagnosis of pancreatic cancer. We analyzed the KRAS gene mutations in EUS-FNA samples from primary pancreatic cancer and the matched circulating biomarkers (ctDNA KRAS and CA19-9) in the same patient to address these questions. Then, we assessed the diagnostic values of KRAS gene mutations in EUS-FNA samples and plasma ctDNA and the prognostic value of KRAS gene mutations in pancreatic cancer (Figure 1). This study's primary aim was to explore the utility of detecting the KRAS gene mutation to supplement EUS-FNA evaluation of pancreatic masses. The secondary study aim was to compare the diagnostic values of KRAS mutations in EUS-FNA tissue samples with the matched circulating biomarkers (ctDNA KRAS and CA19-9) in the same patients. The tertiary study aim was to explore the predictive value of KRAS gene mutations in advanced pancreatic adenocarcinoma prognosis.
Figure 1.
Schematic diagram of digital PCR analysis for KRAS mutations in EUS-FNA specimens and ctDNA samples. ddPCR analyses were performed for tumor specimens obtained from patients with pancreatic cancer using either EUS-guided FNA cytology specimens or ctDNA from blood plasma. KRAS mutations were evaluated for potential clinical utility or as prognostic indicators. ctDNA, circulating tumor DNA; ddPCR, digital droplet PCR; EUS-FNA, endoscopic ultrasound-guided fine-needle aspiration.
METHODS
Patients
The Tongji Medical College of Huazhong University of Science and Technology Review Board approved this prospective cohort study (IRB ID: TJ-C20140717) of KRAS mutation analysis in 149 consecutive patients who underwent EUS-FNA of pancreatic masses between September 2014 and May 2019 at the Endoscopy Center of Tongji Hospital. All patients provided written informed consent for procedures and tests associated with the study. The inclusion criteria were age >18 years, the presence of a solid mass lesion was confirmed by at least 1 imaging modality and was located within the pancreas, and mass size >1 cm. Exclusion criteria included (i) severe anemia (hemoglobin <60 g/L for chronic anemia and hemoglobin <70 g/L for acute anemia); (ii) pregnancy; (iii) coagulopathy (international normalized ratio >1.5), thrombocytopenia (platelet count <50,000/mm3), and acute pancreatitis within the previous 2 weeks; or (iv) intervening structures prohibiting needle access.
EUS-FNA technique
EUS-FNA was performed under deep sedation according to the principles of monitored anesthesia care. The patients were anesthetized with the intravenous administration of propofol (2 mL/kg). All patients received oxygen during the procedure, and their blood pressure and heart rate were monitored (18). All EUS procedures were performed by an experienced endosonographer (B.C.) who had completed more than 1,000 procedures. An Olympus linear echoendoscope (GF-UCT 260 or GF-UCT 240; Olympus, Tokyo, Japan) and Pentax linear echoendoscope (EG-3870UTK; Pentax, Tokyo, Japan) were used. A 22-gauge EchoTip Ultra needle (Cook Endoscopy; Cook Medical, Bloomington, IN) was advanced into lesions with real-time EUS visualization. The endosonographer maneuvered the needle back and forth 20 times within the lesion (4 passes for each patient), applying minimal negative pressure by pulling the needle stylet slowly and continuously. If no specimen was obtained, continuous suction was applied with a 5–10-mL syringe to obtain a specimen. The operator immediately evaluated the samples aspirated from all 149 patients to determine whether the specimen was sufficient according to the results of the macroscopic onsite evaluation (19). Aspirated material from each patient was separated into 2 parts: one pass for cyto/histopathological evaluation and another pass for KRAS point mutation analysis. The material for cyto/histopathological analysis was immediately fixed in 10% formalin in a standard specimen bottle, centrifuged, and then embedded in paraffin. Sections were then stained by hematoxylin and eosin and by immunohistochemical staining if necessary (Figure 2). The material for KRAS analysis was sent to laboratory for DNA extraction immediately. Total DNA was extracted from the fresh specimens using the DNeasy Blood & Tissue Kit (Qiagen), according to the manufacturer's instructions.
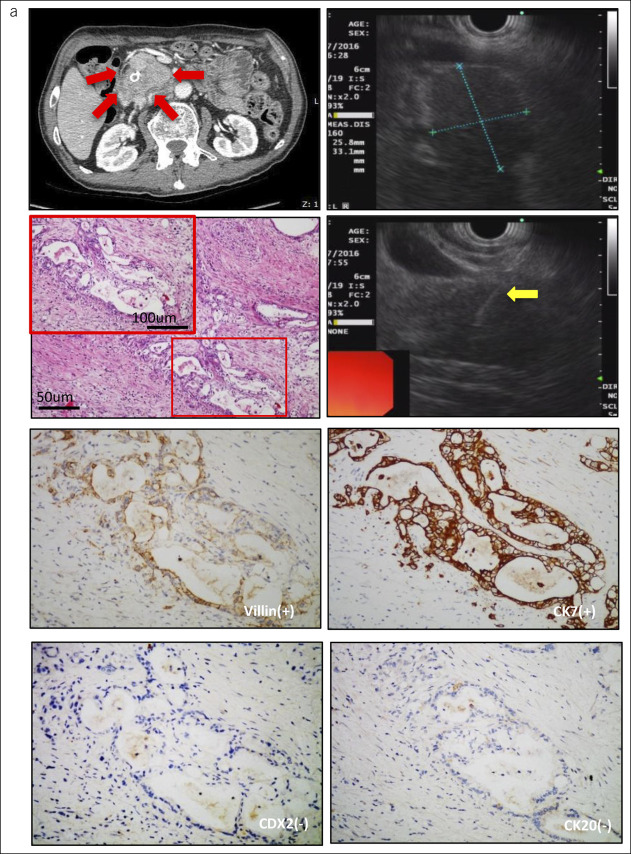
Figure 2.
Histopathological features in a representative patient with pancreatic adenocarcinoma. (a) A representative patient with pancreatic cancer (2.58 × 3.31 cm) in computed tomography (red arrow) and EUS-FNA (yellow arrow), which expressed villin(+), Ki67(+), CDX2(−), and CD20(−). EUS-FNA, endoscopic ultrasound-guided fine-needle aspiration.
Extraction of cell-free ctDNA in plasma
Each collected blood sample (5 mL) was centrifuged at 3,500 rpm for 15 minutes at 4°C within 3 hours. Then, it was stored in plasma at −80°C for further use. DNA was extracted from the plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen), according to the manufacturer's instructions. The DNA quantity was assessed by using the Qubit dsDNA HS (high sensitivity) Assay Kit (Thermo Fisher), according to the manufacturer's instructions.
Digital PCR
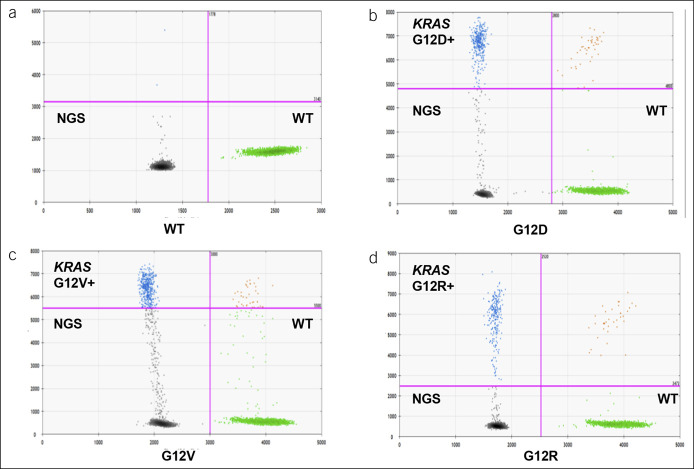
The sample was partitioned into 20,000 droplets by using droplet digital PCR (ddPCR) (QX200; Bio-Rad, Hercules, CA) (20,21). The DNA was concentrated and distributed among these droplets randomly. The authors tested for 3 types of KRAS mutations (G12V, G12R, and G12D) with PrimePCR products (Bio-Rad) for ddPCR (cat 1863115, 1863112, and 1863113) because these KRAS mutations encompass nearly 90% KRAS mutations in pancreatic cancer (12) (as shown in Figure 3a–d). Reactions were performed in 20 mL of reaction serum, which consisted of extracted DNA (5 mL), target primer mix (FAM) (1 mL), reference primer/probe mix (HEX) (1 mL), KRAS mutation droplet PCR supermix (10 mL), and distilled water (3 mL). PCR reactions were run on C1000 Touch thermal cycler incubating plates (Bio-Rad) at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds, followed by 10-minute incubation at 98°C. Negative controls without serum ctDNA showed no positive signal. All samples were analyzed in duplicate, and variations were set at <5%. The detection rate was set at >0.001%. Schematic and flowdiagram of digital PCR in EUS-guided FNA cytology and ctDNA specimen analyses is shown in Figure 1.
Figure 3.
Digital PCR KRAS mutation analysis of ctDNA in representative patients with pancreatic adenocarcinoma. (a–d) Distribution of droplets is visualized using a heat map. Droplet threshold are shown as pink lines. Signal detected in the channel 1 represents DNA positive for a KRAS 12 (G12D, G12V, and G12R) mutation. Signal detected in channel 2 represents DNA positive for KRAS WT amplification in which ctDNA mutants were nondetectable. ctDNA, circulating tumor DNA; NGS, next-generation sequencing; WT, wild type.
Final diagnosis
The final diagnosis was based on a pathological examination of a surgical resection specimen (29 patients) or clinical/imaging follow-up for at least 2 years when surgical resection was not indicated because of a benign diagnosis or malignant advanced or metastasized disease. 86.7% (91/105) patients with pancreatic cancer were unresectable (III–IV stage). If signs of malignancy were absent at the end of follow-up (disease regression or no evidence of disease progression), those patients were diagnosed as nonmalignant pancreatic masses. The case was considered malignant if clinical/imaging follow-up indicated the progression or metastasis of lesions with malignant symptoms, such as weight loss, anemia, or death. Pancreatic ductal adenocarcinoma and pancreatic acinar cell carcinoma were defined as malignant diseases. Chronic pancreatitis, autoimmune pancreatitis, and pancreatic tuberculosis were defined as nonmalignant diseases. Samples that were considered malignant or suspicious for malignancy were categorized as positive for malignancy, whereas samples that were considered benign or atypical were categorized as negative for malignancy (19,22).
Statistical analysis
Continuous variables are expressed as medians and ranges. Incidences and concordance between groups were compared by using the Fisher exact test or McNemar test where appropriate. All analyses were performed with SAS version 9.2. A P value of 0.05 was considered statistically significant.
RESULTS
Patients' characteristics
The authors prospectively evaluated 157 patients included in this study, of which 5 patients withdrew and 3 patients were lost to follow-up. Therefore, the authors analyzed 149 patients, including 105 patients with pancreatic cancer (age 58.38 ± 11.01 years; men 69/105 [65.71%]) (14 cases of whom had TNM stage I–II cancers and 91 of whom had stage III–IV cancers) and 44 cases with nonmalignant pancreatic masses (age 52.66 ± 13.81 years; men 36/44 [81.82%]) (Table 1). These 105 pancreatic cancer cases consisted of 102 pancreatic ductal adenocarcinoma cases and 3 diagnosed acinar cell carcinoma cases (Table 1). These nonmalignant 44 cases consisted of 26 autoimmune pancreatitis, 10 chronic pancreatitis, and 3 pancreatic tuberculosis. The final diagnoses were based on evaluating surgical pathology (n = 29) and clinical courses after 2-year follow-up (n = 105). One case of chronic pancreatitis with a KRAS G12D mutation was found to be malignant during follow-up.
Table 1.
Patients' characteristics (N = 149)
| Characteristics | Pancreatic cancera (n = 105) | Nonmalignant pancreatic massb (n = 44) |
| Age, mean ± SD | 58.38 ± 11.01 | 52.66 ± 13.81 |
| Sex, n (%) | ||
| Male | 69 (65.71) | 36 (81.82) |
| Female | 36 (34.29) | 8 (18.18) |
| Location of mass, n (%) | ||
| Head/uncinate | 70 (66.67) | 38 (86.36) |
| Body/tail | 35 (33.33) | 6 (13.64) |
| CA19-9, n (%) | ||
| ≥37 U/mL | 75 (71.43) | 10 (22.73) |
| <37 U/mL | 30 (28.57) | 34 (77.27) |
| CEA, n (%) | ||
| ≥10 ng/mL | 32 (30.48) | 10 (22.73) |
| <10 ng/mL | 73 (69.52) | 34 (77.27) |
| Surgery, n (%) | ||
| Yes | 24 (22.86) | 5 (11.36) |
| No | 81 (77.14) | 39 (88.64) |
CA19-9, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen.
Pancreatic cancer including 102 ductal adenocarcinoma and 3 diagnosed acinar cell carcinoma.
Nonmalignant pancreatic mass including 26 autoimmune pancreatitis, 10 chronic pancreatitis, and 3 pancreatic tuberculosis.
Diagnostic value of KRAS gene mutations with EUS-FNA specimens in pancreatic adenocarcinoma
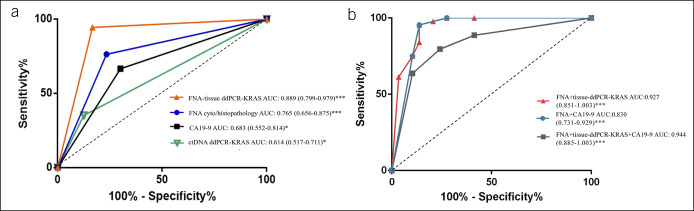
Next, we analyzed the KRAS gene mutations in codon 12 in EUS-FNA samples from primary pancreatic adenocarcinoma (n = 105, Figure 2, a representative patient) using the digital droplet polymerase chain reaction (ddPCR, Figure 3a–d). The sensitivity, specificity, PPV, NPV, accuracy, and ROC AUC of the EUS-FNA alone were 71.4%, 86.4%, 92.6%, 55.9%, 75.8%, and 0.765, respectively (Table 2 and Figure 4a), whereas these values of the EUS-FNA with ddPCR KRAS mutation analysis were 91.6%, 80.9%, 92.5%, 79.1%, 88.6%, and 0.927, respectively (Table 2 and Figure 4b). The sensitivity and accuracy of EUS-FNA diagnosis were increased from 71.4% to 91.6% (P < 0.001) and 75.8% to 88.6% (P < 0.001), respectively, when KRAS mutation ddPCR analysis was added to standard EUS-FNA assessment. Our study demonstrated that KRAS mutation analysis significantly improves the sensitivity and accuracy of EUS-FNA in pancreatic adenocarcinoma. Detecting KRAS gene mutations in FNA samples thus improved the diagnostic accuracy of pancreatic adenocarcinoma with EUS-FNA compared with currently available methods.
Table 2.
Overall accuracy of circulating biomarkers in comparison of EUS-FNA for the diagnosis of pancreatic cancer (n = 105)
| Primary tumor | Circulating biomarkers | ||||
| EUS-FNA | EUS-FNA + ddPCR (KRAS) | ctDNA (KRAS) | CA19-9 | EUS-FNA + CA19-9 | |
| Sensitivity (%) | 71.4 | 91.6 | 35.2 | 71.2 | 79.6 |
| Specificity (%) | 86.4 | 80.9 | 88.6 | 78.3 | 75.9 |
| PPV (%) | 92.6 | 92.5 | 88.1 | 91.2 | 85.4 |
| NPV (%) | 55.9 | 79.1 | 36.4 | 46.7 | 71.0 |
| Accuracy (%) | 75.8 | 88.6 | 51.0 | 73.0 | 79.5 |
| ROC AUC | 0.765 | 0.889 | 0.614 | 0.683 | 0.830 |
CA19-9, carbohydrate antigen 19-9; ctDNA, circulating tumor DNA; EUS-FNA, endoscopic ultrasound-guided fine-needle aspiration; NPV, negative predictive value; PPV, positive predictive value; ROC AUC, area under the curve of the receiver operating characteristic curve; combination: the combination of EUS-FNA and CA19-9.
Figure 4.
Diagnostic performance of EUS-FNA, CA19-9, ctDNA, and FNA specimen KRAS mutation detection either alone or in combination for distinguishing patients with pancreatic cancer (n = 105) from control patients (n = 44). KRAS mutation was determined using ddPCR. ROC curves: X-axis, 1-specificity; Y-axis, sensitivity. CA19-9, carbohydrate antigen 19-9; ctDNA, circulating tumor DNA; ddPCR, digital droplet PCR; EUS-FNA, endoscopic ultrasound-guided fine-needle aspiration; AUC, area under the curve.
Diagnostic value of KRAS gene mutations in ctDNA and serum CA19-9
Aiming for a noninvasive method for detecting KRAS mutations, the authors also conducted a KRAS mutation analysis of ctDNA in all matched plasma samples. The concordance of results obtained from EUS-FNA samples and plasma samples was evaluated. As shown in Table 2 and Figure 4, the sensitivity, accuracy, and ROC AUC of KRAS mutations in EUS-FNA samples were 91.6%, 88.6%, and 0.889, whereas the respective values of KRAS mutation in ctDNA were 35.2%, 51.0%, and 0.683. In the pancreatic adenocarcinoma group, KRAS gene mutations were found in 88 (83.8%) of 105 cases in primary cancer, whereas only 37 cases (35.2%) of KRAS mutation were found in ctDNA (P < 0.001, χ2 test, Table 2). The accuracy of noninvasive ctDNA KRAS in detecting pancreatic adenocarcinoma was thus not as high as EUS-FNS KRAS mutation analysis (P < 0.0001, Table 2). Although ctDNA KRAS mutation detection showed a tendency of higher positivity in patients with Ⅲ/Ⅳ pancreatic cancer than in patients withⅠ/Ⅱ pancreatic cancer, there was no significant difference in sensitivity between different stages (28.6% vs 42.0%, P = 0.506) (see Supplementary Figure S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A794). The sensitivity, accuracy, and ROC AUC of CA19-9 alone were 71.2%, 73.0%, and 0.683. In addition, the ROC AUC of EUS-FNA and CA19-9 combination was 0.830, which was still inferior to the combination of EUS-FNA and tissue sample ddPCR (Figure 4).
Prognostic value of KRAS gene mutations in pancreatic adenocarcinoma
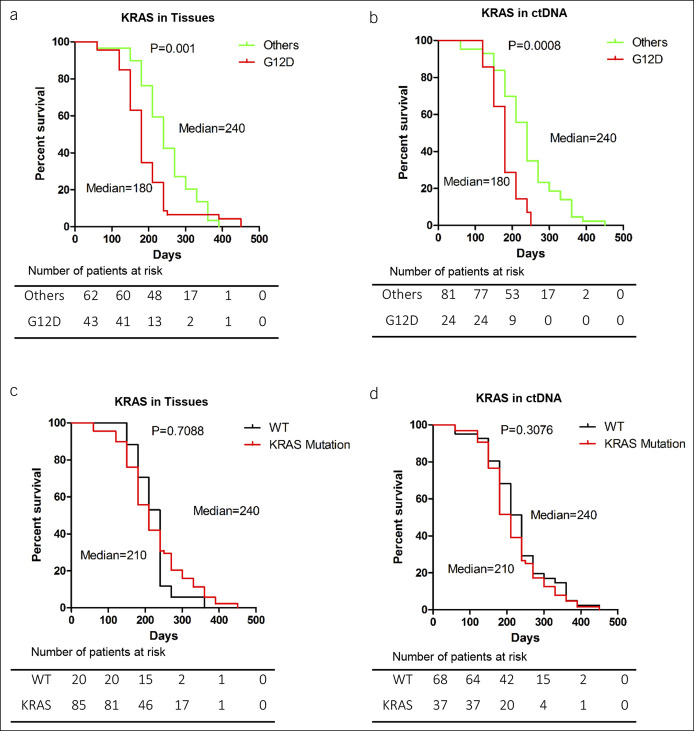
The authors analyzed the effect of G12D, G12V, and G12R mutations in the following Kaplan–Meier study. The median survival time (MST) was significantly shorter in patients with G12D mutations (180 days) compared with patients with other mutations (240 days) in their EUS-FNA tissue samples and ctDNA samples (log-rank test, P = 0.001 and P = 0.0008, respectively) (Figure 5a,b). By contrast, the MST was not found to be significantly different between the patients with wild-type KRAS (240 days) and those with KRAS mutations (210 days) in their EUS-FNA tissue samples and ctDNA sample (log-rank test, P = 0.7088 and P = 0.3076, respectively) (Figure 5c,d).
Figure 5.
Overall survival of patients. (a and b) Survival rates of patients with KRAS G12D mutations (red line) and other mutations or wild-type (green line). The mutations were detected with either EUS-FNA or ctDNA samples (P = 0.001 and P = 0.0008, respectively). (c and d) Survival rates of patients with wild-type KRAS (black line) and in those with KRAS mutations (red line). There was no significant difference of the MSTs between EUS-FNA and plasma ctDNA groups (P = 0.7088 and P = 0.3076, respectively). *P < 0.05, ***P < 0.001. ctDNA, circulating tumor DNA; EUS-FNA, endoscopic ultrasound-guided fine-needle aspiration; MST, median survival time; WT, wild type.
Furthermore, univariate analysis demonstrated that the G12D KRAS mutation in both EUS-FNA tissue samples (hazard ratio [HR], 1.94; 95% confidence interval [CI], 1.12–3.36, P < 0.0001) and ctDNA (HR, 1.579; 95% CI, 1.383–3.520, P < 0.0005) was a significant factor for poor survival. Multivariate analysis demonstrated that the G12D mutation in both EUS-FNA tissue samples (HR, 1.495, 95% CI, 1.325–1.753, P = 0.0010) and ctDNA (HR, 1.417, 95% CI, 1.199–2.870, P = 0.0199) was independently associated with poor overall survival (Table 3). The following factors were analyzed as possible risk factors for survival: age, sex, TNM stage, location of masses, tumor size, carcinoembryonic antigen, CA19-9, and KRAS mutations (G12D, G12V, and G12R). In univariate analysis, both baseline CA19-9 and ctDNA KRAS were associated with overall survival, which was expected given the known positive correlation between the 2 variables.
Table 3.
Prognostic factors for overall survival by univariate and multivariate analyses in pancreatic cancer (n = 105)
| Univariate analysis | Multivariate analysis | |
| HR (95% CI); P | HR (95% CI); P | |
| Age (>65 yr) | 1.00 (0.62–1.63); 0.97 | NS |
| Sex (male) | 0.84 (0.56–1.27); 0.41 | NS |
| TNM stage (III–IV/I–II) | 1.35 (0.74–2.48); 0.33 | NS |
| Location of mass (head, uncinate/body, and tail) | 1.28 (0.85–1.93); 0.23 | NS |
| Tumor size (>20 mm) | 1.26 (0.69–2.31); 0.45 | NS |
| CEA (>10 ng/mL) | 0.64 (0.42–0.99); 0.51 | NS |
| CA19-9 (>37 U/mL) | 1.75 (1.15-2.76); 0.04a | NS |
| EUF-FNA KRAS | 0.85 (0.52–1.39); 0.52 | NS |
| G12D KRAS | 1.94 (1.12-3.36); <0.0001b | 1.495 (1.325-1.753); 0.0010b |
| G12V KRAS | 1.23 (0.80–1.90); 0.35 | NS |
| G12R KRAS | 0.43 (0.29–0.66); 0.02a | NS |
| ctDNA KRAS | 0.540 (0.379–1.417); 0.3559 | NS |
| G12D KRAS | 1.579 (1.383–3.520); 0.0005b | 1.417 (1.199–2.870); 0.0199a |
| G12V KRAS | 0.993 (0.393–2.508); 0.9881 | NS |
| G12R KRAS | 0.296 (0.149–0.589); 0.5276 | NS |
CA19-9, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen; CI, confidence interval; HR, hazard ratio; NS, no significance.
aP<0.05; bP<0.01.
DISCUSSION
Our study explored 3 crucial concerns on the diagnosis of pancreatic cancer. First, we evaluated whether the use of KRAS mutation analysis is able to improve the efficacy of pancreatic cancer diagnosis in EUS-FNA samples. Although intraepithelial neoplasia-originated cancer may take up to 10 years to develop, it only takes approximately 1 year for the pancreatic adenocarcinoma to progress from stage I to stage IV (23). Currently, cyto/histopathological evaluation of EUS-FNA samples is one of the most useful methods for the early diagnosis of pancreatic masses. However, previous studies have shown that the sensitivity, specificity, and accuracy of EUS-FNA in the diagnosis of pancreatic cancer range from 64 to 94%, 71% to 100%, and 78% to 95%, respectively (24–27), indicative of low stability. Our findings demonstrated that ddPCR analysis of KRAS mutations significantly improved the cyto/histopathological sensitivity and accuracy in EUS-FNA samples over currently available methods.
Second, we compared noninvasive ctDNA and EUS-FNA in detecting pancreatic cancer and found that the accuracy of KRAS mutation analysis through noninvasive ctDNA KRAS is inferior to that of KRAS mutation analysis in EUS-FNA samples. These findings indicated that KRAS mutations in plasma ctDNA, also termed as “liquid biopsy,” might not be sufficient to complement the use of EUS-FNA in the diagnosis of pancreatic cancer. In addition, KRAS mutation ddPCR analysis in tissues significantly improves cyto/histopathological sensitivity and accuracy in EUS-FNA samples. In conclusion, combining KRAS mutation ddPCR and cyto/histopathological analysis in EUS-FNA samples might be more clinically valuable than blood tests for diagnosing pancreatic cancer.
To date, cyto/histopathological evaluation of EUS-FNA samples is one of the most useful methods for the early diagnosis of pancreatic masses. Conversely, ctDNA is derived from apoptosis and necrosis of tumor cells, characteristic of advanced-stage disease (28). Very few studies have reported the concordance of results between tumor and plasma samples for pancreatic cancer. In this sense, it is challenging to assess the actual diagnostic sensitivity and specificity of previous analyses. In this study, we demonstrated that the sensitivity, specificity, and accuracy of KRAS mutations in EUS-FNA samples were 71.4%, 86.4%, and 75.8%. Because these values of the KRAS mutation in ctDNA were 35.2%, 88.6%, and 51.0%, the accuracy of KRAS mutation analysis in EUS-FNA samples was higher than that in a noninvasive blood-based ctDNA KRAS mutation in detecting pancreatic cancer. Identification of early driver mutations in blood samples with “liquid biopsy” (16) does not alter cyto/histopathological evaluation of EUS-FNA samples. However, detection of circulating biomarkers combination (ctDNA and CA19-9) is also promising, which may complement other diagnostic techniques to diagnose pancreatic cancer.
We identified KRAS mutations in 35.2% of ctDNA in plasma samples from patients with pancreatic cancer, which is consistent with the data of KRAS-positive ctDNA in previous studies, specifically 39% (29), 35.3% (30), 32% (25), and 26% (31) reported previously. In our study, 37 cases (35.2%) of KRAS mutations were found in blood tests (22.9% G12D mutation, 8.6% G12V mutation, and 3.8% G12R mutation). Although the positivity rate for KRAS mutation detection in patients with advanced pancreatic cancer was even higher, no differences in sensitivity of ctDNA detection for the classification of different stage pancreatic cancer were found, possibly due to the small sample size of I/II stage patients recruited.
Finally, the efficacy of KRAS gene mutations on patients' prognosis was investigated. Previous studies indicated that ctDNA was produced by tumor cells prone to metastasis and apoptosis; thus, tumor-originated ctDNA might not be satisfying enough for early diagnosis. However, it is still considered an ideal biomarker for predicting prognosis and relapse (32). KRAS gene mutations at codon 12, including G12D (47%), G12V (37%), and G12R (11%), are known as the most common KRAS mutations in circulating DNA of patients with pancreatic cancer. However, the results are controversial regarding the types of KRAS mutation and survival. It was also demonstrated that patients with the G12V mutation had significantly better overall survival compared with patients with the G12D and G12R mutations (9,33). By contrast, other studies reported that patients with the G12V mutation possess poorer prognosis than patients with the G12D mutation (34,35). In our study, patients with KRAS G12D mutations in pancreatic adenocarcinoma had worse overall survival than those with other mutations. The heterogeneity of tumor characteristics may cause the disparity in these results (36). Multivariate analysis suggested that G12D mutations in both EUS-FNA tissue samples and ctDNA were independent risk factors of poor survival. These results may indicate that ctDNA is produced by the tumor cells prone to metastasis and apoptosis. In addition, the detection of ctDNA may make it possible to detect tumors with heterogeneity. In multivariate analysis, ctDNA KRAS mutations were significantly associated with poor overall survival, indicating that these biomarkers capture unique but complementary prognostic information.
Our study found that adding an analysis of KRAS mutations improves the efficacy of EUS-FNA as a tool to diagnose pancreatic cancer over current methods. It also found that KRAS mutations are more effectively analyzed in EUS-FNA samples than “liquid biopsy” ctDNA samples and that the presence of these mutations worsens the patients' prognosis.
However, there are several limitations of the study that should be acknowledged. The results we obtained may underestimate the survival benefits of early detection. Most of the patients recruited in this study were with III–IV stage pancreatic cancer. Future studies including a larger number of patients with I–IV stage pancreatic cancer in matched plasma and EUS-FNA samples with a larger quantity of alleles are needed to develop ctDNA analysis an early diagnostic marker in routine clinical application.
CONFLICTS OF INTEREST
Guarantor of the article: Bin Cheng, MD and Jinlin Wang, MD.
Specific author contributions: R.W. and Y.Z.: designed the study and performed data analysis. Y.Z., J.W., and Y.W.: performed acquisition of data. Y.Z.: drafted the manuscript. Q.C.: provided critical revision of the manuscript for important intellectual content. Y.Z., S.X., and Z.Z.: performed technical support. Y.D.: provided pathological verification. B.C. and L.Z.: performed study supervision. All authors have read and approved the manuscript.
Financial support: This work was supported by the National Natural Science Foundation of China Grant No. 81802427(to Z.L.), No. 81802418 (to R.W.), No. 81372352, and No. 81172063 (to B.C.).
Potential competing interests: None to report.
Availability of data and material: The data sets used and/or analyzed during the current study available from the corresponding author on reasonable request. The EUS-FNA data of patients with pancreatic cancer was upload as Table 1.
Study Highlights.
WHAT IS KNOWN
✓ Endoscopic ultrasound-guided fine-needle aspiration is the first-line technique for pancreatic cancer diagnosis, staging, and sampling.
✓ The ability of carbohydrate antigen 19-9 to differentiate pancreatic cancer from other benign pancreatic lesions is highly limited.
WHAT IS NEW HERE
✓ Digital droplet PCR KRAS analysis of fine-needle aspiration samples significantly improved the sensitivity and accuracy of pancreatic cancer diagnosis, superior to the efficacy of carbohydrate antigen 19-9 and noninvasive blood-based circulating tumor DNA.
✓ G12D KRAS mutations in pancreatic cancer were independently associated with poor overall survival.
Supplementary Material
ACKNOWLEDGMENTS
The authors sincerely thank all staff from the Tongji Hospital, Tongji Medical College, HUST, for their effort in project execution. The authors also thank the statistical staffs (Ping Yin and Chang Shu) at the Department of Epidemiology and Biostatistics, School of Public Health, Tongji Medical College, HUST, and Hepatic Surgery Centre, Tongji Hospital, Tongji Medical College, HUST, and support from Shou-jiang Tang, University of Mississippi Medical Center.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A794
Ronghua Wang and Yuchong Zhao contributed equally to this article.
Contributor Information
Ronghua Wang, Email: ronnawang@foxmail.com.
Yuchong Zhao, Email: zhaoyuchongtj@163.com.
Yun Wang, Email: 820308045@qq.com.
Zhenxiong Zhao, Email: 1813264120@qq.com.
Qian Chen, Email: chenqian@yahoo.com.
Yaqi Duan, Email: duanyaqi@tjh.tjmu.edu.cn.
Si Xiong, Email: xiongsi@tjh.tjmu.edu.cn.
Zhou Luan, Email: luanzhoutj@163.com.
Jinlin Wang, Email: 417661238@qq.com.
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;70(4):313. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4.Pereira SP, Oldfield L, Ney A, et al. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol 2020;5(7):698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Xie K, Wolff R, et al. Pancreatic cancer. Lancet 2004;363(9414):1049–57. [DOI] [PubMed] [Google Scholar]
- 6.Trisolini E, Armellini E, Paganotti A, et al. KRAS mutation testing on all non-malignant diagnosis of pancreatic endoscopic ultrasound-guided fine-needle aspiration biopsies improves diagnostic accuracy. Pathology 2017;49(4):379–86. [DOI] [PubMed] [Google Scholar]
- 7.Bournet B, Souque A, Senesse P, et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with KRAS mutation assay to distinguish pancreatic cancer from pseudotumoral chronic pancreatitis. Endoscopy 2009;41(6):552–7. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Gao J, Ren Y, et al. Detection of KRAS gene mutations in endoscopic ultrasound-guided fine-needle aspiration biopsy for improving pancreatic cancer diagnosis. Am J Gastroenterol 2011;106(12):2104–11. [DOI] [PubMed] [Google Scholar]
- 9.Ogura T, Yamao K, Sawaki A, et al. Clinical impact of K-ras mutation analysis in EUS-guided FNA specimens from pancreatic masses. Gastrointest Endosc 2012;75(4):769–74. [DOI] [PubMed] [Google Scholar]
- 10.Karstensen JG, Vilmann P. Endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic lesions: Striving for perfection. Endoscopy 2018;50(5):466–8. [DOI] [PubMed] [Google Scholar]
- 11.Bournet B, Buscail C, Muscari F, et al. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur J Cancer 2016;54:75–83. [DOI] [PubMed] [Google Scholar]
- 12.Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017;546(7659):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen JD, Javed AA, Thoburn C, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci U S A 2017;114(38):10202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haber DA, Velculescu VE. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4(6):650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368(13):1199–209. [DOI] [PubMed] [Google Scholar]
- 16.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6(224):224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietrasz D, Pecuchet N, Garlan F, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res 2017;23(1):116–23. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Chen Q, Wu X, et al. Role of endoscopic ultrasound-guided fine-needle aspiration in evaluating mediastinal and intra-abdominal lymphadenopathies of unknown origin. Oncol Lett 2018;15(5):6991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Wu X, Yin P, et al. Comparing endoscopic ultrasound (EUS)-guided fine needle aspiration (FNA) versus fine needle biopsy (FNB) in the diagnosis of solid lesions: Study protocol for a randomized controlled trial. Trials 2016;17:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brychta N, Krahn T, von Ahsen O. Detection of KRAS mutations in circulating tumor DNA by digital PCR in early stages of pancreatic cancer. Clin Chem 2016;62(11):1482–91. [DOI] [PubMed] [Google Scholar]
- 21.Gevensleben H, Garcia-Murillas I, Graeser MK, et al. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin Cancer Res 2013;19(12):3276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Chen Q, Wang J, et al. Comparison of modified wet suction technique and dry suction technique in endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) for solid lesions: Study protocol for a randomized controlled trial. Trials 2018;19(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinugasa H, Nouso K, Miyahara K, et al. Detection of K‐ras gene mutation by liquid biopsy in patients with pancreatic cancer. Gastroenterology 2015;148(4):2271–80. [DOI] [PubMed] [Google Scholar]
- 24.Cheng B, Zhang Y, Chen Q, et al. Analysis of fine-needle biopsy vs fine-needle aspiration in diagnosis of pancreatic and abdominal masses: A prospective, multicenter, randomized controlled trial. Clin Gastroenterol Hepatol 2018;16(8):1314–21. [DOI] [PubMed] [Google Scholar]
- 25.Uemura T, Hibi K, Kaneko T, et al. Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J Gastroenterol 2004;39(1):56–60. [DOI] [PubMed] [Google Scholar]
- 26.Mukai S, Itoi T, Ashida R, et al. Multicenter, prospective, crossover trial comparing the door-knocking method with the conventional method for EUS-FNA of solid pancreatic masses (with videos). Gastrointest Endosc 2016;83(6):1210–7. [DOI] [PubMed] [Google Scholar]
- 27.Berzosa M, Villa N, El-Serag HB, et al. Comparison of endoscopic ultrasound guided 22-gauge core needle with standard 25-gauge fine-needle aspiration for diagnosing solid pancreatic lesions. Endoscopic Ultrasound 2015;4(1):28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dabritz J, Preston R, Hanfler J, et al. K-ras mutations in the plasma correspond to computed tomographic findings in patients with pancreatic cancer. Pancreas 2012;41(2):323–5. [DOI] [PubMed] [Google Scholar]
- 30.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28(4):741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earl J, Garcia-Nieto S, Martinez-Avila JC, et al. Circulating tumor cells (CTC) and KRAS mutant circulating free DNA (cfDNA) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015;15:797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16(7):409–24. [DOI] [PubMed] [Google Scholar]
- 33.Bournet B, Muscari F, Buscail C, et al. KRAS G12D mutation subtype is A prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol 2016;7:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015;121(13):2271–80. [DOI] [PubMed] [Google Scholar]
- 35.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br J Cancer 2001;85(5):692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013;501(7467):328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.