Abstract
Lineage analysis has been an important method for understanding the mutation patterns and the diversity of genes, such as antibodies. A mutation lineage is typically represented as a tree structure, describing the possible mutation paths. Generating lineage trees from sequence data imposes two unique challenges: (1) Types of constraints might be defined on top of sequence data and tree structures, which have to be appropriately formulated and maintained by the algorithms. (2) Enumerating all possible trees that satisfy constraints is typically computationally intractable. In this paper, we present a COnstrained Lineage Tree generation framework (COLT) that builds lineage trees from sequences, based on local and global constraints specified by domain experts and heuristics derived from the mutation processes. Our formal analysis and experimental results show that this framework can efficiently generate valid lineage trees, while strictly satisfying the constraints specified by domain experts.
Keywords: Lineage Analysis, Constrained Tree Construction, Sequence Data
I. INTRODUCTION
Lineage analysis is a widely used method for analyzing biological evolution and mutation [1]. With the development of next-gen high-throughput sequencing techniques, we can now study mutations at the molecular level via lineage structures extracted from sequences. Lineage analysis for sequence data is extremely useful for studying mutation-abundant biological mechanisms, such as the immune system. On virus infection our body can generate numerous mutated genes in a short period that direct the synthesis of various antibodies [1]. Studying this complicated mutation process can help us understand how our immune system works. Lineage analysis for sequence data has also been applied to studies on HIV and cancers. We believe it will be an essential method for many emerging applications of next-gen high-throughput sequencing techniques.
There are two major categories of algorithms for lineage tree generation, based on phylogenetic tree [2] and IgTree [3], respectively. During the early years (e.g., before 2008), phylogenetic trees have been the major tool for presenting and analyzing lineages. A phylogenetic tree generation algorithm typically employs a hierarchical clustering algorithm on the sequences to form a binary tree by progressively merging close sequence clusters. However, phylogenetic trees do not well serve the need of molecular-level studies. Specifically, its structure does not directly tell how a sequence mutates to another. It only tells the similarity between two sequences at the leaf nodes and any two groups of sequences at the internal nodes. It is also impossible to find which sequence is the origin of mutation and which sequences are the ancestors of a specific sequence.
Barak et al. [3] proposed the IgTree algorithm to address these problems. In an IgTree, each node represents a sequence. A path from the root to a leaf represents one of the possible mutation processes. The IgTree algorithm consists of two phases. With a given root, the first phase will grow the initial tree based on the minimum mutation cost assumption, where each node represents an actual sequence. The second phase will insert artificial internal nodes so that only one-base mutation happens from any parent to any of its children. The inserted artificial nodes aims to help researchers better understand the possible mutation processes.
However, the IgTree algorithm has not addressed several important features of sequence-level mutation analysis that are highly demanded by biomedical researchers. First, the IgTree algorithm assumes the root node is known (i.e., the corresponding sequence is known to be the root of mutations). This is only valid when researchers know the root sequence, for instance, by tracing back the sample that the root sequence comes from, or using the well-known germline sequence as the root. However, in many cases it is difficult to designate the root sequence of a lineage tree. In particular, mutations in the immune system can happen quickly. As a result, multiple mutated sequences can be possibly found in one sample, where the root sequence is difficult to determine. Second, when constructing a lineage tree, many constraints may have to be considered and maintained. For example, sequences from older samples cannot appear as the children of the sequences from newer samples; the sequences of isotype “IgM” in the immune system cannot be mutated from other non-“IgM” sequences. Mutations also follow a certain pattern that good mutations tend to survive and thus descendants’ mutations are unlikely to revert to ancestors’ sequences. All these unique requirements demand a new unified framework for lineage tree construction algorithm.
Scope of Research.
To address these unique challenges, we develop the COnstrained Lineage Tree generation framework for sequence data (COLT) that allows users to formulate and integrate various types of constraints, automatically determines the root sequence, and generates directed lineage trees. The developed algorithms should be efficient enough to handle a large number of sequences, and flexible enough to incorporate various types of constraints.
The basic idea is to formulate the problem as a constrained minimum spanning tree (MST) problem, where the domain-specific constraints can be mapped to sub-structures (e.g., edges and paths) in the graph. The problem is to find the MST in this directed graph that satisfies the specified constraints. The rationale using MST to model the lineage tree is that small progressive mutations are more likely to happen in nature than large dramatic ones, which can be nicely captured by MSTs.
We model the constraints in two categories: the local ones that can be mapped to single edges and the global ones that may involve multiple edges (and nodes) such as paths. It is easy to satisfy the local constraints by removing the corresponding edges from the graph, while the global ones are difficult to maintain. We develop an undirected MST (UMST) based fast approach to generating the directed trees. Because the directed edges between a pair of vertices have the same weight in our problem, we can treat them as undirected edges first and then propagate the edge directions later. The root determining heuristic and the constraint checking and maintaining methods are applied after initial edge propagation. We show that this method is much faster than the directed MST (DMST) based approach. Our contributions can be summarized as follows.
We propose a general framework that can conveniently integrate sequence-based constraint formulation and maintenance into the lineage tree generation process.
We have developed a fast lineage tree generation algorithm that uses an iterative process to maintain the global constraints.
The algorithmic results on real datasets have been validated by domain experts. We have also conducted extensive experiments to identify the optimal setting for the framework to achieve high efficiency, scalability, and tree quality.
II. PRELIMINARIES
Sequence.
A sequence is simply a string of characters from the set {‘A’, ‘C’, ‘G’, ‘T’}, i.e., the four nucleobases, of a certain length. We use edit distance to define the similarity between sequences. For a sequence v, we use v[i] to represent the nucleobase at the position i.
Graph and Tree.
A graph G(V,E) has the vertex (or node) set V and the edge set E. Let vi (or ui) denote a vertex (i.e., a sequence) in a graph or a node in a tree, Vi a subset of vertices (nodes), ei an edge, and Ei a subset of edges. We use and to represent the size of the sets, respectively. A vertex v can also have a number of associated attributes, denoted as . A directed edge ei from the vertex ui to vi is also represented in the form , where ui is called the tail and vi the head. An edge ei is also associated with a weight denoted as wi and thus represented as a three-tuple for tail vertex u, head vertex v, and weight w. A path in the graph from u to v is represented as .
Minimum Spanning Tree (MST).
A MST algorithm is to find a tree with the minimum sum of edge costs from a weighted undirected or directed graph. The popular MST algorithms will be adopted in our framework, such as the Kruskal’s algorithm [4] for undirected graphs and the improved Edmonds’ algorithm [5] for directed graphs.
III. COLT: A FRAMEWORK FOR GENERATING CONSTRAINED LINEAGE TREES
We will first give the definition of the constraints, then present the details of the algorithm, and finally formally analyze the costs of different implementation approaches.
A. Constraints
Two types of constraints are defined in the framework.
Local Constraints.
We consider local constraints are those that can be applied to individual edges. We further classify the local constraints to the “forbidden” and “must-have” types. The forbidden ones are mapped to the edges that should be excluded from the result, while the “must-have” ones are pre-included in the MST before the MST algorithm starts.
Specifically, for an edge , a “forbidden” constraint is a boolean function that checks certain relationships between the attributes of u and v and the weight w to return true for a constraint violation. A specific example is that, in the lineage tree the sequence from a new tissue sample cannot be the parent of another sequence from an old sample. This constraint can be specified as
and if is true, the edge should be excluded. For another example, a sequence of isotype “IgM” in antibody mutation cannot be the child of another isotype, which is specified as
similarly, if f1 is true, the edge should be excluded.
Global Constraints.
Global constraints are those involving more than one edge (e.g., a path in the tree). We define such a constraint with a boolean function for a subset of vertices and edges. For example, domain experts have observed that in most cases if a mutation lineage exists, it is very unlikely that the grandchild s of u recovers from the previous mutation at the position i. This can be further extended to old ancestors of s. Formally, this constraint can be defined as
| (1) |
If is true for any v’s ancestor u, the constraint is violated.
It would be difficult to maintain such global constraints on the complete graph as we have done for local constraints. In fact, it is simply too expensive to just check constraints on the graph. For example, let’s assume the global constraints only involve two-hop paths like . Then, the number of candidates will be in the complete graph. As a result, we would like to maintain the global constraints on a generated tree instead.
Algorithm 1.
Sketch of COLT Framework
| 1: W ← compute the distance matrix for each pair of sequences, where wij is the weight of the edge eij |
| 2: ← the “forbidden” constraints; |
| 3: ← the “must-have” constraints; |
| 4: ← global constraints; |
| 5: E0 ← all edges of the complete graph; |
| 6: ; // valid edges |
| 7: ; // initial set of edges for the MST |
| 8: if E1 contains cycles then |
| 9: report error and return; |
| 10: end if |
| 11: repeat |
| 12: E2 ← generate_UMST (E0, E1) |
| 13: T ← generate_directed_tree (E2); |
| 14: E3 ← check_global_constraints(T, ); // return a set of edges to be removed |
| 15: |
| 16: until // all constraints are satisfied |
| 17: return T |
B. Framework Design
With the definitions of constraints, we are ready to discuss our framework in more details. Assume there are N sequences. Let represent the sequences, and eij represent the edge . The algorithm first removes the “forbidden” edges and initializes the MST algorithm with the “must-have” edges. Then, it enters the iterations, each of which will generates the MST, derives the directed lineage tree, and checks the global constraints, until all global constraints are met. Algorithm 1 gives the sketch of the framework.
C. Tree Generation With UMST
Since the directed edges between a pair of vertices have the same weight in our problem, we can apply undirected MST algorithm first, and assign the edge directions later. In the following, we describe the method of assigning edge directions and determining the root.
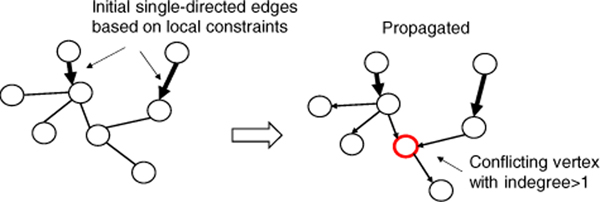
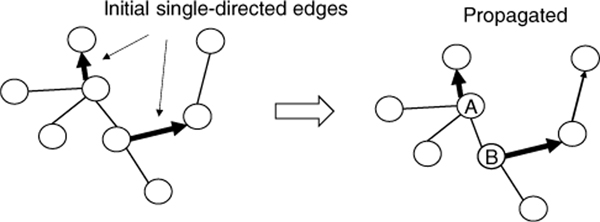
To grow a directed tree from a UMST, we start propagating directions from the existing single-directed edges in the UMST. Edge propagation is an iterative process. With a direction-determined edge , for any neighbor of v, say w, we can decide the direction . It continues by checking the newly reached nodes and so on, until no more nodes can be expanded. The result of propagation has to be validated so that no vertex is the head of more than one edge. Figure 1 and 2 show an invalid result and a valid one, respectively.
Figure 1.
An invalid result after direction propagation
Figure 2.
A valid edge propagation result. Node A is chosen as the root.
As Figure 2 shows, after the initial propagation the directions of some bidirectional edges are still undetermined, which will be finally determined after the root is selected. We use the following heuristics to rank the candidate nodes and determine the root node.
For a single-directed edge , v cannot be the root;
The remaining valid candidate nodes are ranked by their outdegrees in the tree; the node of the highest outdegree becomes the root; for tied top candidates, randomly select one as the root.
In Figure 2, the nodes A-E are the candidates of root. Since the node A’s outdegree 4 is the highest among all candidates’, A is chosen as the root. As the outdegree is mapped to the diversity of mutations started from this node, this algorithm prefers the root as the center of most diversified mutations. Once the root is selected, the same propagation process is applied starting from the root. The following proposition says that this algorithm will not leave any edge direction undetermined.
Proposition 1:
With a valid initially propagated tree, once the root is selected and direction propagation is applied, every edge’s direction will be uniquely determined.
The proof is straightforward and thus we skip the details due to the space limitation.
D. Maintaining Global Constraints
The algorithm checks the global constraints after the directed tree is formed. It first traverses the tree to setup the root-to-node mutation profile for each node. Then, another traversal is conducted to check any node v with its parent node’s and grandparent node’s mutation profile to see whether the constraint is violated. A node’s mutation profile M is an array of the sequence length, where each element M[i] records the bases having shown in the path from the root to the node in the position i. If a violation is detected, the algorithm will try to break the path using a certain policy. We will test two policies in experiments. (1) Choose the end edge to break. The intuition is to maintain the stability of the generated tree so that the iterations can converge quickly. (2) Choose the largest-weight edge in the path to break. The intuition is to maintain the overall cost of the tree as small as possible. We will use experiments to evaluate the validity of these methods. The primary goal is to obtain minimum-cost and valid lineage trees that do not break one lineage to multiple1.
It is easy to verify that if a common UMST algorithm (e.g., the Kruskal’s algorithm) with complexity is used, the overall complexity of the COLT algorithm is , where d is the length of the sequence and t is the number of iterations.
IV. EXPERIMENTS
The experiments’ goal is twofold: (1) test the algorithms on the real datasets and validate the results, (2) evaluate different framework settings to find the best one and understand the actual cost distribution with simulated datasets.
Implementation.
We implemented the algorithms with C++. The local constraints include the time ordering and the Ig* dependency. The global constraint is the path-based mutation rule. These constraints have been discussed in Section III-A. The sequences in the real datasets are obtained from an open-source sequence processing pipeline (SeqPrep:github.com/jstjohn/SeqPrep) and some in-house processing tools. Edit distance is used for computing pairwise sequence distances. We also implemented a version of DMST-based algorithm for comparison, using the optimized implementation of Edmonds’ algorithm (edmonds-alg.sourceforge.net). For each node, the DMST-based algorithm generates the directed tree using that node as the root and then checks the global constraints. The final tree is selected among the valid ones using the optimal root selection heuristic.
Datasets.
Two real datasets were from our recent study on human antibody repertoire for the malaria disease [6]. These datasets are used to validate the generated lineage trees. We will compare the trees manually constructed by domain experts and those generated by our algorithm.
We also generate a bunch of simulated sequence datasets for performance and tree-quality evaluation. The process of generating sequences mimics the mutation process with a preset depth of mutation tree d (i.e., the longest path from the root to the leaves) and the upper bound of mutations m (i.e., the number of mutations is randomly chosen in the range [1,m]). The algorithm will start with a seed sequence from a real dataset to form the sequence pool. In each iteration, a sequence is randomly drawn from the pool and then randomly mutated, satisfying the restriction of mutation depth and upper bound, and also the local and global constraints defined in Section III-A. The new sequence is then added to the pool for future iterations. In our experiments we use d = 5 and m = 10 to generate all the simulated datasets.
Validation with Real Datasets.
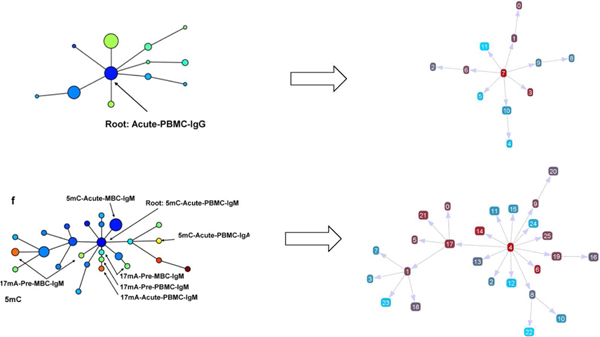
These two real datasets are quite small, having 12 and 26 sequences, respectively. Thus, they are appropriate for domain experts to manually analyze the mutations and construct the lineage trees. Figure 3 shows the comparison between the manually constructed trees (left) and the automatically generated trees (right). For the first dataset the manually constructed tree is identical to the automatically generated one. For the second dataset, the three internal nodes on the backbone (i.e., the major mutation path) match exactly, while only a couple of leaf nodes are placed to different branches. The generated tree also leads to the same analytical result for the domain problem. Thus, the domain experts consider both are valid lineages.
Figure 3.
A comparison between manually constructed trees (left) and automatically generated ones (right). The automatically generated trees are visualized with an in-house lineage tree visualization tool.
Results on Simulated Datasets.
The second set of experiments focuses on the time costs of algorithms and the quality of generated trees. Batches of simulated datasets are used for evaluation. For each number of sequences, we randomly generate multiple sets of sequences and obtain the average values and standard deviations. We will first look at the effect of the edge breaking methods for maintaining the global constraints, and then compare the UMST-based method with the more expensive DMST-based method. Finally, we will investigate the cost distribution over the major components in the framework.
Since our goal is to achieve minimum cost lineage trees that satisfy all the specified constraints, we use the sum of edge weights as the quality of the tree and aim to find valid trees with smaller sums as possible. According to our method of generating the simulated datasets, each dataset exists one valid single lineage tree that contains all the sequences. Trees that do not contain all the sequences are considered invalid results.
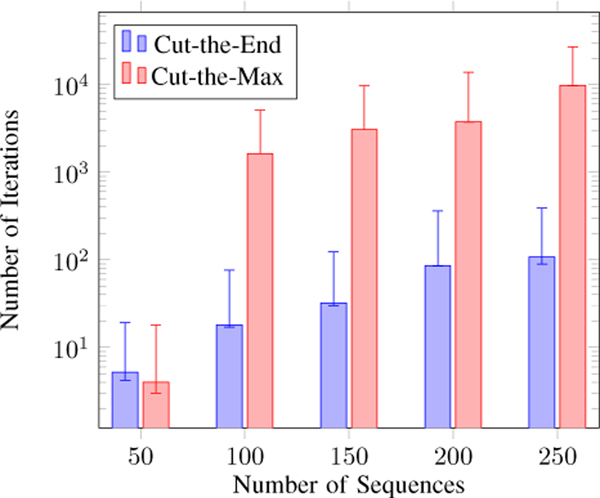
The first experiment is focused on two edge breaking methods: Cut-the-End and Cut-the-Max for maintaining the constraints. Specifically, we use the UMST method to generate trees and compare the number of iterations for each method that is proportional to the cost of the tree generation process. Figure 4 shows that Cut-the-Max has overwhelmingly larger costs than the other. Furthermore, we find that the Cut-the-Max very frequently leads to invalid trees if the dataset size is larger than 50. Among the sizes of 50, 100, 150, 200, 250 sequences, we find that it only constantly generates valid trees for datasets of size 50. Overall, it can find valid results for only 11 of 25 datasets, while the Cut-the-End can find valid results for 24 of 25. Therefore, we consider the Cut-the-Max method is not an effective method for our framework.
Figure 4.
Comparing the costs (i.e., the number of iterations) needed for the two edge-breaking methods. Y-axis is log-scale.
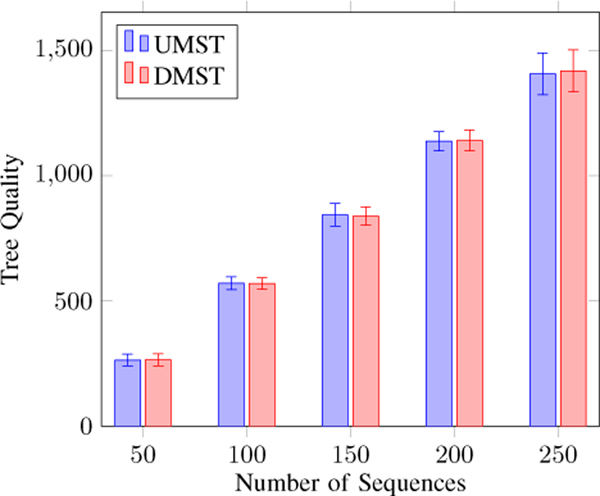
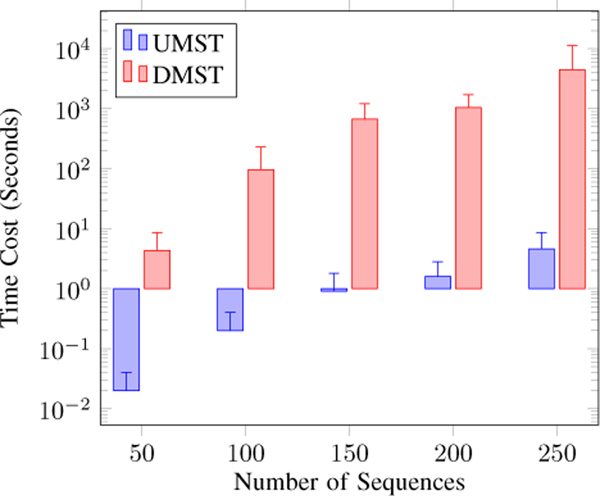
Next, we study the use of UMST or DMST as the tree generation algorithm. We use the Cut-the-End method to maintain the global constraints. Figure 5 shows that both methods can find trees of about the same quality. However, the DMST-based method is much more expensive than the UMST-based method, as shown in Figure 6. We conclude that the UMST-based method should be the method of choice for the framework.
Figure 5.
Comparing the quality of trees generated by UMST and DMST. Both generate similar quality trees.
Figure 6.
The cost comparison on the UMST and DMST methods.
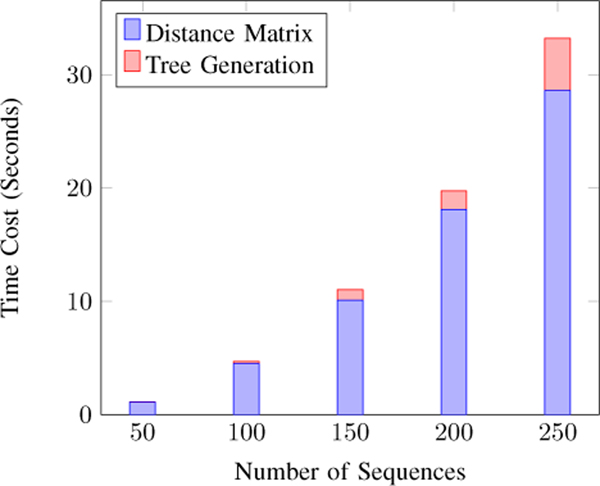
Finally, we consider the cost distribution of the two major components in the framework: distance matrix computation and tree generation. We use the UMST-based method to generate the trees and the Cut-the-End method to maintain the global constraints. Figure 7 shows that with the efficient UMST-based method the cost of computing distance matrix will become the bottleneck in analyzing a practical number of sequences.
Figure 7.
Overall cost distribution. Computing distance matrix is still dominating.
V. CONCLUSION
Automatically generating lineage trees is appealing to many sequence-based biomedical studies. However, it presents several unique challenges, especially for generating trees under constraints. In this paper, we establish the constraint-based lineage tree generation framework COLT that can adapt to different types of constraints (e.g., local or global) and generate rooted and directed lineage trees. The algorithm uses the heuristic that evolutions in nature are more likely driven by a series of small mutations rather than large dramatic ones, which is modeled with the minimum spanning tree (MST) of the complete sequence-sequence mutation graph. We develop an iterative UMST-based algorithm to maintain the global constraints. Experimental results show that the proposed approach is efficient and can generate high-quality lineage trees.
Acknowledgment.
This study is partially supported by NIH grant R00AG040149, Cancer prevention and Research Institute of Texas grant R1120, Welch Foundation grant F1785, Damon Runyon-Rachleff Innovation Award DRR-32-15, and NSF grant 1245847.
Footnotes
Note that we assume the set of sequence under study forms one lineage. For the cases where multiple lineage may exist, the best way is to apply clustering first and then apply our framework on each cluster of sequences.
Contributor Information
Keke Chen, Data Intensive Analysis and Computing Lab Department of Computer Science and Engineering Wright State University, Dayton, Ohio, USA.
Venkata Sai Abhishek Gogu, Data Intensive Analysis and Computing Lab Department of Computer Science and Engineering Wright State University, Dayton, Ohio, USA.
Di Wu, Systems Immunology Lab Department of Biomedical Engineering University of Texas at Austin, Austin, Texas, USA.
Jiang Ning, Systems Immunology Lab Department of Biomedical Engineering University of Texas at Austin, Austin, Texas, USA.
REFERENCES
- [1].Jiang N et al. , “Lineage structure of the human antibody repertoire in response to influenza vaccination,” Science Translational Medicine, vol. 5, no. 171, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fitch WM and Margoliash E, “Construction of phylogenetic trees,” Science, vol. 155, pp. 279–284, 1967. [DOI] [PubMed] [Google Scholar]
- [3].Barak M, Zuckerman NS, Edelman H, Unger R, and Mehr R, “Igtree: Creating immunoglobulin variable region gene lineage trees,” Journal of Immunological Methods, vol. 338, no. 1–2, p. 6774, 2008. [DOI] [PubMed] [Google Scholar]
- [4].Cormen TH, Stein C, Rivest RL, and Leiserson CE, Introduction to Algorithms, 2nd ed. McGraw-Hill Higher Education, 2001. [Google Scholar]
- [5].Gabow HN, Galil Z, Spencer T, and Tarjan RE, “Efficient algorithms for finding minimum spanning trees in undirected and directed graphs,” Combinatorica, vol. 6, no. 2, pp. 109–122, 1986. [Google Scholar]
- [6].Wu D, Qu M, He C, Wendell B, Hunicke-Smith S, Ren P, and Jiang N, “Accurate and quantitative immune repertoire sequencing using unique molecular identifiers,” Manuscript under review, 2016. [Google Scholar]