Abstract
Microbial community diversity, potential microbial activity, and metal resistance were determined in three soils whose lead contents ranged from 0.00039 to 48 mmol of Pb kg of soil−1. Biomass levels were directly related to lead content. A molecular analysis of 16S rRNAs suggested that each soil contained a complex, diverse microbial community. A statistical analysis of the phospholipid fatty acids indicated that the community in the soil having the highest lead content was not related to the communities in the other soils. All of the soils contained active microbial populations that mineralized [14C]glucose. In all samples, 10 to 15% of the total culturable bacteria were Pb resistant and had MIC of Pb for growth of 100 to 150 μM.
Hazardous waste sites often contain complex mixtures of pollutants which include both organic contaminants and heavy metals (17). Microbial bioremediation of organic pollutants is a promising method of environmental cleanup. However, if the metals in soils are toxic to the microbes, removal of organic pollutants is slowed or prevented.
Many reports have shown that (i) the short-term response to toxic metals is a large reduction in microbial activity (3, 6, 15, 26, 30) and (ii) habitats that have had high levels of metal contamination for years still have microbial populations and activities that are smaller than the microbial populations and activities in uncontaminated habitats (14, 16, 18, 24). Although these observations may suggest that metal contamination of soils retards bioremediation of organic pollutants, we take a different view, that metal contamination is an extreme environment (created by humans) to which microbes can respond.
We tested this hypothesis by examining the ecology of bacteria in lead-contaminated soils (obtained from an industrial site at which lead batteries had been recycled). Our goal was to compare microbial biomass levels, metabolic potential, community diversity, and the selection pressure for Pb resistance.
Soil samples were obtained from the Avanti industrial site in Indianapolis, Ind., on 25 June 1997. Pb-contaminated soils had been excavated from this industrial site and the surrounding residential neighborhood; these soils were stored in a warehouse prior to ultimate disposal. Three samples (samples R1, R2, and R3) were obtained from the pile designated residential soils (R soils), and three samples (samples I1, I2, and I3) were obtained from the pile designated industrial soils (I soils). Agricultural soil samples (samples A1, A2, and A3) from the Purdue Agronomy Research Center (Chalmers silty clay loam, a fine silty, mixed, mesic, Typic Halaquoll) were used as the control samples (A soils). All samples were kept in plastic bags and stored at 4°C until analysis. Biological analyses were begun within 2 days of sample collection.
Pb contents were determined by inductively coupled plasma-atomic emission spectroscopy with a Perkin-Elmer model 400 ICP/AES instrument after digestion with nitric acid and hydrogen peroxide. Soil analyses were carried out by using standard techniques (22a) by A&L Great Lakes Laboratory (Ft. Wayne, Ind.).
Microbial biomass and community structure were determined by quantifying phospholipid phosphate (11) and by performing a gas chromatographic analysis of phospholipid fatty acid methyl esters (PLFAs) (28). The dry weights of soil used for extraction were approximately 4, 7, and 10 g for the A, R, and I soil samples, respectively. Identical extract volumes were analyzed by gas chromatography. The minimum detection limit for an individual fatty acid was 4 pmol g of soil−1. The PLFA data were converted to moles percentages of total fatty acids, arcsine transformed, and extracted into a correlation matrix. Principal component analysis (PCA) (Systat 8; SPSS, Chicago, Ill.) was used to discern patterns within the data (10). The PCA loading scores indicated the relative importance of individual fatty acids in the weighting along each principal component.
Nucleic acid analyses of community structure were begun by extracting DNA from 0.5 g of soil as described by Borneman et al. (5). PCR amplification and denaturing gradient gel electrophoresis (DGGE) analysis were done as described by Ovreas et al. (22). Similar amounts of DNA were loaded onto gels for the different samples.
Potential microbial activity was estimated by measuring 14CO2 evolution after the addition of 0.07 μCi of [U-14C]glucose (35 nmol; Sigma Chemical Co., St. Louis, Mo.) to 45 g of soil and incubation in Bartha flasks at a moisture content equivalent to 25% of the water-holding capacity (4).
Culturable bacteria were enumerated on PYT80 agar (10 mM MES [morpholineethanesulfonic acid] (pH 6.5), 80 mg of peptone per liter, 80 mg of yeast extract per liter, 80 mg of tryptone per liter, 15 g of washed Bacto Agar [Difco] per liter) with or without 100 μM lead nitrate. The plates were incubated at 25°C. Cells were removed from the soil by shaking 10 g of soil in 90 ml of a solution containing 4.25 g of NaCl per liter and 0.1 g of gelatin per liter for 30 min at 125 rpm.
Table 1 shows some of the chemical characteristics of the soils. The microbial biomass (estimated by determining phospholipid P contents) decreased as the Pb content increased. The uncontaminated A soils contained 39.2 ± 12.9 nmol of P g (dry weight) of soil−1, the R soils contained 5.4 ± 2.1 nmol of P g−1, and the I soils contained 1.2 ± 0.6 nmol of P g−1.
TABLE 1.
Chemical characteristics of soil samples
| Soil source | Cation exchange capacity (meq/100 g) | Organic matter content (%) | pH | Total Pb concn (mmol · kg of soil−1) | pPb2+a |
|---|---|---|---|---|---|
| Industrial (I soils) | 25.1 ± 0 | 2.7 ± 0.5 | 7.8 ± 0.2 | 4.8 | 8.2 |
| Residential (R soils) | 17.6 ± 1.8 | 3.5 ± 1.0 | 7.4 ± 0.1 | 3.9 | 8.9 |
| Agricultural (A soils) | 21.5 ± 0.1 | 5.0 ± 1.5 | 7.0 ± 0.1 | 0.0039 | 9.5 |
pPb2+ (the negative log of free Pb2+ activity) was calculated from the pH and the total Pb content (27).
The size of the population of the culturable organisms was only twofold lower in the Pb-contaminated soils than in the A soils (Table 2), whereas the estimated phospholipid biomasses were 8- to 30-fold lower in the Pb-contaminated soils than in the A soils. Pb-resistant bacteria were isolated from all of the soil samples. The proportion of Pb-resistant bacteria was low (<15%) and relatively independent of the level of Pb contamination in the soils (Table 2).
TABLE 2.
Viable counts of heterotrophic bacteria on PYT media with and without 100 μM Pb(NO3)2
| Incubation time (days) | A soils
|
R soils
|
I soils
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Viable counts (10−6 CFU g of soil−1)
|
% Pbr | Viable counts (10−6 CFU g of soil−1)
|
% Pbr | Viable counts (10−6 CFU g of soil−1)
|
% Pbr | ||||
| Without Pb | With Pb | Without Pb | With Pb | Without Pb | With Pb | ||||
| 2 | 23 ± 2.6 | 14 ± 2.5 | 7.0 ± 1.0 | 3.4 ± 0.4 | 48 | ||||
| 5 | 80 ± 19 | 3.7 ± 0.2 | 4.6 | 27 ± 6 | 47 ± 17 | 4.1 ± 0.1 | 8.7 | ||
| 12 | 105 ± 45 | 6 ± 2.4 | 5.7 | 37 ± 8.2 | 3.2 ± 0.6 | 8.7 | 66 ± 28 | 4.6 ± 0.1 | 7.0 |
| 19 | 119 ± 36 | 7.2 ± 2.9 | 6.0 | 43 ± 5.5 | 3.5 ± 0.5 | 8.1 | 70 ± 28 | 5.2 ± 0.2 | 7.4 |
| 26 | 127 ± 43 | 8.5 ± 3.3 | 6.7 | 46 ± 5.6 | 3.6 ± 0.4 | 7.8 | 71 ± 29 | 5.4 ± 0.3 | 7.6 |
| 34 | 127 ± 43 | 8.5 ± 3.3 | 6.7 | 47 ± 5.6 | 3.7 ± 0.5 | 7.8 | 71 ± 29 | 5.5 ± 0.05 | 7.7 |
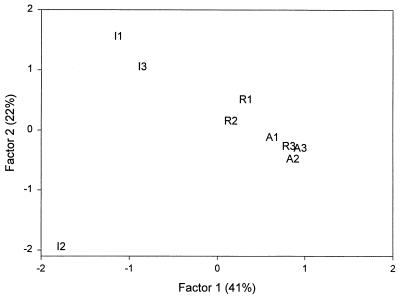
Multivariate statistical analysis (Fig. 1) of the PLFA patterns indicated that samples of the microbial community in the R soils (3.9 mmol of Pb kg−1) were more similar to samples of the microbial community in the uncontaminated A soils than to samples of the microbial community in the highly Pb-contaminated I soils. Whereas the replicate samples obtained from the A and R soils grouped fairly closely together, the replicate I soil samples were more heterogeneous. The first two principal components explained 41 and 22% of the total variance, respectively.
FIG. 1.
PCA of the PLFA profiles obtained for discrete soil samples taken from an industrial site (samples I1, I2, and I3), a residential site (samples R1, R2, and R3), and an agricultural field (samples A1, A2, and A3).
Bacterial fatty acids were the predominant fatty acids in all of the samples (16:0 in the I and R soils and 18:1ω9t in the A soils), but key differences between soils were noted. The positive loading along PCA factor 1 was driven by differences in 15:0, a15:0, 18:1ω11c, 16:1ω9t, and (to a lesser degree) Cy17:0. The negative loading on PCA factor 1 was driven primarily by levels of 18:0, 16:0, and 12:0. In general, these data suggested that the A and R soils harbored a bacterial population (i15:0, a15:0, 18:1, 16:1) that was dominated by gram-positive bacteria (i15:0, a15:0) but also contained a gram-negative bacterial component (Cy17:0). All of the soils had a weak contribution from 18:2, which suggested that there was a low-level fungal contribution to the total biomass. The I soils had a reduced contribution from actinomycetes (10Me 18:0) compared to either the R soils or the A soils. Along PCA factor 2, differences in 14:0, 15:0, 17:0, 20:0, and 20:1ω11c separated the replicate samples from the I soils. These data suggested that long-term exposure to high Pb concentrations or the subsequent effects of this exposure (for example, lower plant growth) changed the population structure.
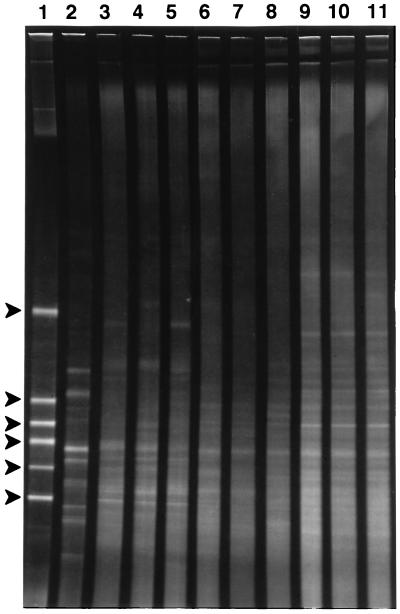
Microbial community diversity was also evaluated by DGGE analysis. Soil microbial communities are typically very diverse; therefore, DGGE produces a complex smear of amplification products (20). This result was obtained with the uncontaminated A soil samples (Fig. 2, lanes 9 through 11). However, the diversity in soils contaminated with polychlorinated biphenyl was severely reduced (Fig. 2, lane 2) (20). Both the R soils and the I soils exhibited much higher diversity than the polychlorinated biphenyl-contaminated soils. The patterns obtained for the three soil types were distinguishable; however, the patterns for replicates from the A and R soils were grossly similar, but the patterns for replicates from the I soil samples were not similar. The complexity of the PCR products precluded any quantitative analysis of the extent of the differences.
FIG. 2.
DGGE gel of 16S ribosomal DNA PCR products (bases 338 to 518 [Escherichia coli rRNA sequence numbering]) from lead-contaminated and uncontaminated soil DNA extracts. The denaturant gradient in the gel ranged from 30 to 55%. Lane 1 contained markers; the PCR products were products of (from top to bottom) Pseudomonas putida, Acinetobacter sp. strain ADP1, Comamonas acidovorans ATCC 15668, E. coli DH5a, Alcaligenes sp. strain BR40, and Comamonas testosteroni). Lane 2, soil contaminated with polycyclic aromatic hydrocarbons (700 ppm); lanes 3 to 5, three I soil samples (48 mmol of Pb kg−1); lanes 6 to 8, three R soil samples (3.9 mmol of Pb kg−1); lanes 9 to 11, three A soil samples (3.9 μmol of Pb kg−1).
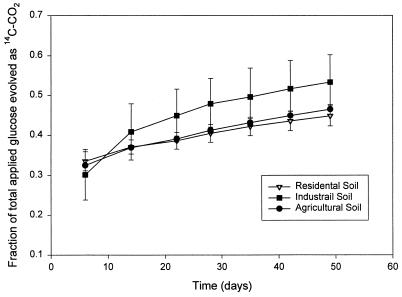
Microbial heterotrophic potential was readily detected in each sample (Fig. 3). No significant differences (P < 0.05) were noted among the three sites. This implied that the metabolic potential of a readily degradable substrate was not inhibited by lead. The potential explanations for this result include (i) the possibility that the concentration of bioavailable Pb was below toxic levels despite the high total Pb concentration and (ii) the possibility that there had been selection for Pb resistance in the microbial population.
FIG. 3.
Fraction of [14C]glucose that was recovered as 14CO2 for three soils containing different levels of lead. The bars indicate the standard errors (n = 3).
The level of Pb resistance in cultures isolated from each of the soil samples was determined by measuring the MIC for growth. Colonies were picked at random from the agar plates that lacked Pb. In each sample, there were Pb-sensitive isolates which were inhibited by 12.5 μM lead nitrate (the lowest concentration tested). There were several A soil strains which had an MIC of 100 μM. Six of the I soil strains had an MIC of <12.5 μM, two had an MIC of 100 μM, and two had an MIC of 150 μM. Thus, the levels of lead resistance were similar (MIC, 100 to 150 μM) for all samples. The Pb-resistant isolates were all gram-positive bacteria that were members of the genus Bacillus, coryneforms, or actinomycetes.
The lead-contaminated soils contained lower levels of microbial biomass than the uncontaminated soils. However, this was not necessarily a direct effect of lead toxicity because the soils did not have plant communities and, therefore, the lack of organic matter input could have reduced the microbial populations. Kuperman and Carreiro (18) reported that soils contaminated with heavy metals had significantly less plant biomass, and this affected the microbial population levels in the soils. Metabolically active bacteria were present in the contaminated soils. Similar results obtained by using other activity measures have been reported previously (2, 7, 9). Other workers (14) reported that they observed reduced (but detectable) microbial activity in metal-contaminated soils.
Of significant interest is the fact that the microbial community at the highly contaminated site was different than the microbial communities in the other soils. This effect has been reported previously for soils experimentally contaminated with metals (12, 13) and for soils exposed for more than 20 years (23).
The presence of Pb-resistant microbes in the soils was also assessed by isolating cultures; however, only a small fraction of bacteria can be cultured (8), so our results may not be representative of the entire microbial community. It was surprising to find that the proportion of the culturable community that was Pb resistant was not much higher in the Pb-contaminated soils than in the A soils. Other studies which have focused on the culturable fraction of the microbial community have indicated that from 10 to 100% of the bacteria in habitats contaminated for extended periods of time are metal resistant (19, 25).
The Pb resistance levels were similar (100 to 150 μM) for isolates from all soils. The total Pb levels were 100- to 1,000-fold higher in the contaminated soils than in the uncontaminated soils. However, much of the Pb may have been unavailable due to reactions with minerals (29), soil organic matter (21), or inorganic anions. The predicted activity of free Pb2+ ions in a soil solution (27), based on pH and total Pb content, indicated that even in the I soils, free Pb2+ ions were present at submicromolar concentrations (Table 1).
Analyses of both lipids and nucleic acids were used to assess community diversity. These approaches were complementary. PLFA analysis yielded a numerical estimate of similarity. This estimate could be improved by broader sampling of Pb-contaminated sites. However, it is difficult to conduct a more detailed analysis of the microbial community on the basis of PLFA contents beyond insights gained from a few biomarker fatty acids. Complex microbial communities found in soil produce complex patterns on DGGE gels; thus, it is difficult to obtain quantitative estimates of similarity in these systems. However, nucleic acid analysis can be used to perform more detailed phylogenetic analyses (1) in future studies.
Acknowledgments
We thank Tom Sweeney and Paul Steadman for providing access to the soil samples and Judy Lindell for technical assistance.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoyama M, Nagumo T. Effects of heavy metal accumulation in apple orchard soils on microbial biomass and microbial activities. Soil Sci Plant Nutr. 1997;43:601–612. [Google Scholar]
- 3.Barnhart C L, Vestal R. Effect of environmental toxicant on metabolic activity of natural microbial communities. Appl Environ Microbiol. 1983;46:970–977. doi: 10.1128/aem.46.5.970-977.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartha R, Pramer D. Features of a flask and method for measuring the persistence and biological effects of pesticides in soil. Soil Sci. 1965;100:68–70. [Google Scholar]
- 5.Borneman J, Skroch P W, O’Sullivan K M, Plus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capone D G, Reese D, Kiene R P. Effects of metals on methanogenesis, sulfate reduction, carbon dioxide evolution, and microbial biomass in anoxic salt marsh sediments. Appl Environ Microbiol. 1983;45:1586–1591. doi: 10.1128/aem.45.5.1586-1591.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dìaz-Raviña M, Bååth E. Thymidine and leucine incorporation into bacteria from soils experimentally contaminated with heavy metals. Appl Soil Ecol. 1996;3:225–234. [Google Scholar]
- 8.Faegri A, Torsvik V, Goksoyr J. Bacterial and fungal activities in soil: separation of bacteria and fungi by a rapid fractionated centrifugation technique. Soil Biol Biochem. 1977;9:105–112. [Google Scholar]
- 9.Filcheva E, Cheshire M V, Campbell C D, McPhail D B. Effect of heavy metal contamination on the rate of decomposition of sewage sludge and microbial activity. Appl Geochem. 1996;11:331–333. [Google Scholar]
- 10.Findlay R H. The use of phospholipid fatty acids to determine microbial community structure. In: Akkermans A D L, Van Elsas J D, De Bruijn F, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1–17. [Google Scholar]
- 11.Findlay R H, King G M, Watling L. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl Environ Microbiol. 1989;55:2888–2893. doi: 10.1128/aem.55.11.2888-2893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frostegård Å, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frostegård Å, Tunlid A, Bååth E. Changes in microbial community structure during long-term incubation in two soils experimentally contaminated with metals. Soil Biol Biochem. 1996;28:55–63. [Google Scholar]
- 14.Hutchinson T C, Symington M S. Persistence of metal stress in a forested ecosystem near Sudbury, 66 years after closure of the O’Donnell roast bed. J Geochem Explor. 1997;58:323–330. [Google Scholar]
- 15.Jonas R B, Gilmour C G, Stoner D L, Weir M M, Tuttle J H. Comparison of methods to measure acute metal and organometal toxicity to natural aquatic microbial communities. Appl Environ Microbiol. 1984;47:1005–1011. doi: 10.1128/aem.47.5.1005-1011.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandeler E, Kampichler C, Horak O. Influence of heavy metals on the functional diversity of soil microbial communities. Biol Fertil Soils. 1996;23:299–306. [Google Scholar]
- 17.Kovalick W. Strategies 2000. Proceedings of the 4th World Congress of Chemical Engineering. Frankfurt, Germany: Dechema; 1992. Perspectives on risks of soil pollution and experience with innovative remediation technologies; pp. 281–295. [Google Scholar]
- 18.Kuperman R G, Carreiro M M. Soil heavy metal concentrations, microbial biomass and enzyme activities in a contaminated grassland ecosystem. Soil Biol Biochem. 1997;29:179–190. [Google Scholar]
- 19.Margesin R, Schinner F. Bacterial heavy metal-tolerance—extreme resistance to nickel in Arthrobacter spp. strains. J Basic Microbiol. 1996;36:269–282. [Google Scholar]
- 20.Nakatsu, C. H., V. Torsvik, and L. Ovreas. Unpublished data.
- 21.Nriagu J O. The biogeochemistry of lead in the environment. Part B. Biological effects. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1978. [Google Scholar]
- 22.Ovreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Page A L, Miller R H, Keeney D R. Methods of soil analysis, 2nd ed., pt. 1. Physical and mineralogical methods. Madison, Wis: Soil Science Society of America; 1982. [Google Scholar]
- 23.Pennanen T, Frostegård Å A, Fritze H, Bååth E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl Environ Microbiol. 1996;62:420–428. doi: 10.1128/aem.62.2.420-428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roane T M, Kellogg S T. Characterization of bacterial communities in heavy metal contaminated soils. Can J Microbiol. 1996;42:593–603. doi: 10.1139/m96-080. [DOI] [PubMed] [Google Scholar]
- 25.Sabry S A, Ghozlan H A, Abou-Zeid D-M. Metal tolerance and antibiotic resistance patterns of a bacterial population isolated from sea water. J Appl Microbiol. 1997;82:245–252. doi: 10.1111/j.1365-2672.1997.tb02858.x. [DOI] [PubMed] [Google Scholar]
- 26.Said W A, Lewis D L. Quantitative assessment of the effects of metals on microbial degradation of organic chemicals. Appl Environ Microbiol. 1991;57:1498–1503. doi: 10.1128/aem.57.5.1498-1503.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauve S, McBride M B, Hendershot W H. Speciation of lead in contaminated soils. Environ Pollut. 1997;98:149–155. [Google Scholar]
- 28.Tunlid A, White D. Soil biochemistry. Vol. 7. New York, N.Y: Dekker; 1992. Biochemical analysis of biomass, community structure, nutritional status, and metabolic activities of microbial communities in soil; pp. 229–262. [Google Scholar]
- 29.Undabeytia T, Morillo E, Maqueda C. Adsorption of Cd and Zn on montmorillonite in the presence of a cationic pesticide. Clay Miner. 1996;31:485–490. [Google Scholar]
- 30.Vives-Rego J, Vaque D, Martinez J. Effect of heavy metals and surfactants on glucose metabolism, thymidine incorporation and exoproteolytic activity in sea water. Water Res. 1986;20:1411–1415. [Google Scholar]