Abstract
Background:
Heart transplantation provides a significant improvement in survival and quality of life for patients with end-stage heart disease, however many recipients experience different levels of graft rejection that can be associated with significant morbidities and mortality. Current clinical standard-of-care for the evaluation of heart transplant acute rejection (AR) consists of routine endomyocardial biopsy (EMB) followed by visual assessment by histopathology for immune infiltration and cardiomyocyte damage. We assessed whether the sensitivity and/or specificity of this process could be improved upon by adding RNA sequencing (RNA-seq) of EMBs coupled with histopathological interpretation.
Methods:
Up to six standard-of-care, or for-cause EMBs, were collected from 26 heart transplant recipients from the prospective observational Clinical Trials of Transplantation (CTOT)-03 study, during the first 12-months post-transplant and subjected to RNA-seq (n=125 EMBs total). Differential expression and random-forest-based machine learning were applied to develop signatures for classification and prognostication.
Results:
Leveraging the unique longitudinal nature of this study, we show that transcriptional hallmarks for significant rejection events occur months before the actual event and are not visible using traditional histopathology. Using this information, we identified a prognostic signature for 0R/1R biopsies that with 90% accuracy can predict whether the next biopsy will be 2R/3R
Conclusions:
RNA-seq-based molecular characterization of EMBs shows significant promise for the early detection of cardiac allograft rejection.
Keywords: Transcriptomics, Cardiac Allograft, Acute Rejection
1. Introduction
Over the last three decades advances in immunosuppression therapies and patient management have yielded substantial gains in survival rates for heart transplant recipients, however, 5-year survival rates remain at only ~74%1-3. One of the major obstacles to extending short- and long-term allograft survival is a lack of robust minimally-invasive biomarkers to diagnose and prognosticate acute rejection (AR) early enough to prevent irreversible damage to the allograft1,4.
While there have been significant advances in non-invasive cell-free nucleic acid-based diagnostics including several large-scale evaluations by the CARGO and GRAfT teams5-10,the current diagnostic standard for AR following cardiac transplantation still entails histopathological evaluation of the allograft by endomyocardial biopsy (EMB) using international standards such as The International Society for Heart & Lung Transplantation (ISHLT) 2013 grading system11,12. While these highly invasive procedures have become safer and more standardized in the last few decades13, procedural risks still remain, and inter-observer variability in EMB readings greatly impact interpretation14. Furthermore, as an individual’s immune response is dynamic over time, successive biopsies are needed to capture antiallograft immunity. A fundamental limitation of for-cause biopsies, which is a biopsy performed when a patient clinically manifests symptoms of rejection, is that allograft injury and irreversible damage may already have occurred, and patients who develop acute rejection (AR) are at higher risk of developing chronic allograft vasculopathy (CAV) which can progress to a number of comorbidities including allograft loss3.
Increasingly, large-scale molecular profiling by sequencing or other methods has become an integral molecular diagnostic tool used to address clinical problems in other specialties including oncology and infectious disease15,16. In the transplant setting, these techniques also show significant promise. A previous messenger RNA (mRNA) array-based expression study characterizing antibody-mediated rejection (AbMR) versus non-AbMR heart transplant recipient EMBs demonstrated AbMR molecular pathways characterized by endothelial activation with microcirculatory inflammation from monocytes–macrophages and NK cells17. Additional recent work by Xiu et al. utilized prior expression array datasets to identify signatures associated with t-cell mediated and antibody-mediated rejection18. While these prior studies have advanced our knowledge of rejection-associated transcriptional regulation, array-based expression platforms have a large of number of limitations versus RNA-seq including smaller dynamic range and inability to detect novel transcripts and splicing isoforms19,20. In this current study, we performed the first RNA-sequencing (RNA-seq) study on 125 longitudinal EMB samples prospectively collected as part of the Clinical Trials of Transplantation (CTOT)-03 study with the aim of assessing the sensitivity and specificity of acute rejection diagnoses. The prospective nature of the CTOT-03 study design also allowed us to assess the ability of RNA-seq to prognosticate AR events.
2. Materials and Methods
Clinical Trial of Transplantation (CTOT-03) Study:
The CTOT-03 study is described in detail elsewhere (NCT:00531921) but in brief it is a prospective observational cohort study designed to test associations of proinflammatory pathways of allo-immune response and injury in thoracic (heart and lung) and abdominal (kidney and liver) allografts. The study aims include testing associations between mRNA expression and subsequent incidence of acute rejection and expression of genes involved in cell mediated immunity in recipients of kidney, liver, lung and heart transplants. For the purposes of this study, RNA-seq data were generated from 125 EMBs from 26 CTOT-03 patients recruited from the University of Pennsylvania and the University of Wisconsin, to assess effects of heart allograft gene expression and the relationship with acute rejection.
A dedicated research EMB was collected, at the same time, for up to six standard-of-care and/or for-cause timepoints in the first 12-months post-transplant, and these fresh-frozen EMBs were preserved immediately in RNAlater storage buffer (Thermo Fisher) at −80°C. A representative hematoxylin-eosin (H&E)- stained slide for each clinical biopsy was centralized in each of the two CTOT-03 sites and graded using International Society of Heart and Lung Transplantation consensus definitions. A dedicated Fresh-frozen research EMBs was collected from the donor allograft at the day of transplant (Day 0) and at one week, two weeks, one month, three months, six months and one year post-transplant. For-cause research fresh-frozen EMB timepoints were also collected. A dedicated Fresh-frozen research EMBs was collected from the donor allograft at the day of transplant (Day 0) and at one week, two weeks, one month, three months, six months and one year post-transplant. For-cause research fresh-frozen EMB timepoints were also collected. The EMBs were further independently assessed by two blinded pathologists to arrive at a consensus rejection status grade with adjudication where required. The distribution of grades across the sample cohort is shown in Table 1.
Table 1.
Demographics and Clinical Features of CTOT03 Heart Transplant Participants
| Patients without ACR | Recipients with 2R EMBs | |
|---|---|---|
| Number of patients | 19 | 7 |
| Donors | ||
| Age (Median[IQR]) | 37 (21 - 52) | 21 (16 - 32) |
| Gender (Male n, %) | 16 (84%) | 6 (85%) |
| Race | ||
| African American (n, %) | 2 (11%) | 0 (0%) |
| White (n, %) | 16(84%) | 5 (71%) |
| Unknown (n, %) | 1 (5%) | 2 (29%) |
| Recipients | ||
| Age (Median[IQR]) | 60 (53 - 61) | 56 (46 - 58) |
| Gender (Male n, %) | 16 (84%) | 5 (71%) |
| Indications for Heart Transplant | ||
| Idiopathic | 2 (11%) | 3 (43%) |
| Ischemic | 13 (68%) | 2 (29%) |
| Other | 4 (21%) | 2 (29%) |
| Ancestry | ||
| African American (n, %) | 2 (11%) | 1 (14%) |
| White (n, %) | 16(84%) | 5(71%) |
| Unknown (n, %) | 1 (5%) | 1 (14%) |
| Study site (HUP, n, %) | 12 (63%) | 3 (43%) |
| Time to 1st ACR (>=2R)(Median[IQR]) | NA | 58 [9 - 165] |
Tissue extraction:
Flash-frozen EMB tissues in RNAlater buffer were homogenized by rotor-stator homogenizer (TissueRuptor, Qiagen), and RNA was purified from the homogenized lysate using RNeasy Blood and Tissue kits (Qiagen). RNA quantity and quality were assessed on a BioAnalyzer workstation (Agilent) and by Qubit fluorometer (Thermo Fisher). Whole transcriptome libraries were prepared using TruSeq Stranded Total RNA Gold library preparation kits (Illumina) and were multiplexed for RNA sequencing on a HiSeq 2500 instrument (Illumina).
Data analysis:
For RNA-seq libraries, raw data were demultiplexed and converted to FASTQ using bcl2fastq2 (Illumina). Reads were mapped and quantified to the Ensembl hg38 human reference genome build using the Salmon 21. Salmon quants were read into R and rolled up to gene level using the tximport package. Initial quality control checks determined 9 samples failed sequencing due to low read counts and were removed from downstream analyses. Quality metrics for all samples are detailed in Table S1. The raw feature counts for the remaining 116 samples were normalized using the edgeR package in R and differentially expressed genes were calculated using the exact test function in edgeR with false-discovery rate correction (q<0.01). Pathway analysis was performed using DAVID pathway analysis tools22,23, utilizing biological pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) Immune cell deconvolution was conducted on the contrasts of interest via xCell24. Predictor models for classification of rejection and preceding rejection contrasts were constructed using the randomForest package in R. We utilized random forest, a machine learning approach for classification of covariates or biomarkers that optimizes across a large number of decision trees, and is particularly useful for classification across high-dimensional datasets such as RNA-seq. The random forest models were subjected to 10-fold cross validation in which each respective model was trained on 55% of the samples and tested for classification of rejection grade or preceding-rejection on the remaining 45% of samples which were sampled without replacement. Due to vastly more non-rejection samples in the cohort, for the random forest training and test sets the non-rejection samples were down-sampled without replacement to twice the number of rejection samples. Similarly, for the preceding-rejection classifier the preceding-non-rejection samples were down-sampled to twice the number of preceding-rejection samples. The random forest classifier models were evaluated after extracting the true positive, true negative, false positive, and false negative values for each round of cross validation. The models were assessed using metrics of accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), F1 score, false positive rate (FPR), false negative rate (FNR), false discovery rate (FDR), and Matthews correlation coefficient (MCC). MCC is a metric ranging from -1 to 1 to evaluate the quality of classification with 1 representing a perfect classification with agreement of actual and predicted values in all four categories of the confusion matrix and -1 representing a very poor classification with a high rate of disagreement between actual and predicted values that could be generated by random. Accuracy of classification was calculated on the test set by obtaining the fraction for the number of correct predictions out of the total number of samples in the test set. Accuracy of the rejection grade and preceding rejection signatures respectively were assessed through ten rounds of cross-validation with resampling of the training and testing sets each round. An average for each metric was calculated from the ten rounds of cross validation for each respective random forest model. Area under the curve (AUC) metrics and receiver-operating characteristic (ROC) curve plots were generated using the ROCR package in R.
IRB approval:
Institutional review board (IRB) approval and informed written consent from both recipients and organ donor proxies were obtained prior to the recruitment of subjects under the CTOT-03 study (NCT:00531921).
3. Results
3.1. A gene expression signature is associated with AR in EMBs
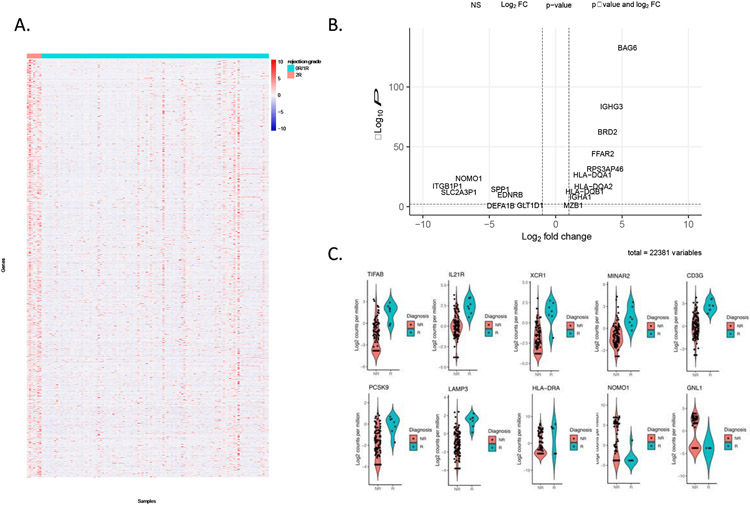
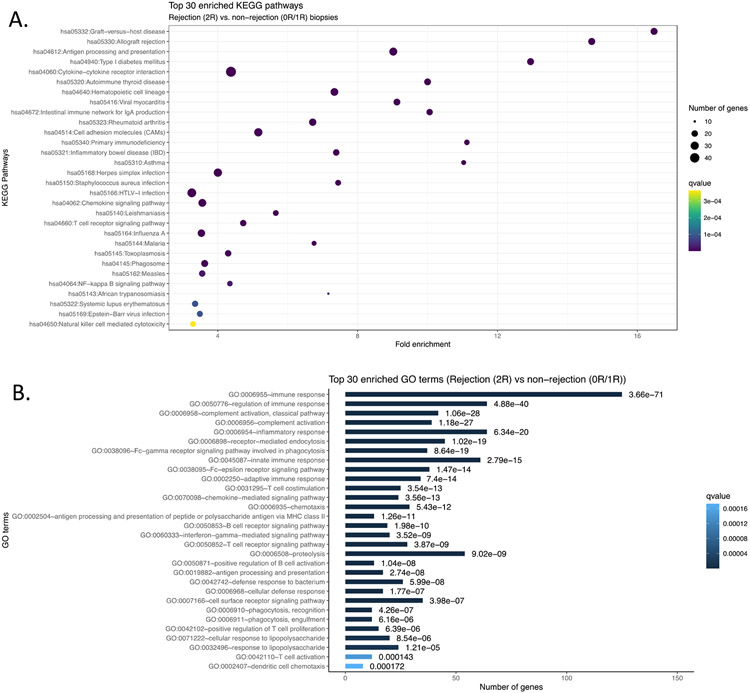
1.1 Longitudinal fresh-frozen EMBs were available from 50 heart transplant recipients who were enrolled in the CTOT-03 study (see Methods). From this cohort, we selected a subset of 26 patients that had dense sampling of EMBs over the study period for allograft profiling by RNA-seq. In total 125 tissue samples were profiled by RNA-seq. We first assessed expression differences between histopathological-determined rejection (Grade 2R or 3R) and non-rejection states (Grade 0R or 1R) as defined by ISHLT grading. Across the 26 CTOT-03 heart transplant recipients, there were 59 EMBs graded as 0R, 59 graded as 1R and seven EMBs were graded as 2R rejection episodes. We performed a transcriptome-wide Fisher’s exact test and pathway analyses to assess differences between 0R/1R and 2R samples. From these data, we observed 1079 genes significantly different between 0R/1R and 2R (FDR q<0.01, Benjamini-Hochberg correction) (Figure 1A & B and Table S2). Of note, a number of these differentially expressed genes have previously been linked to transplant-associated phenotypes, including elevated PCSK9 association with immunosuppressive therapy25 and HLA-DRA associations in peripheral blood with AR26 (Figure 1C). Genes downregulated in the AR setting included NOMO1, which is involved in inhibiting cardiomyocyte differentiation27, FOSB which is a known respond molecule to cardiac injury28, and EDNRB, a gene which is associated with cardiac stress tolerance29-31. To further elucidate functional pathways associated with AR, we performed pathway analyses using the DAVID tool, which assesses whether a differentially expressed gene list is statistically enriched for predefined sets of genes based on biological function or other properties22,23,32 (Figure 2A and B). The majority of significant pathways between 0R/1R and 2R/3R biopsies were associated with immune responses including KEGG cytokine-cytokine receptor interaction (q<1.54x10−15) and a number of pathways significantly associated with transplant rejection including the KEGG Graft-Versus-Host Disease pathway (q<6.37x10−21) and KEGG Allograft Rejection (q<1.31x10−19). For the latter pathway, significant genes comprised most facets of AR including donor and recipient antigen presentation, T helper cell mediated immune activation and both cytotoxic CD8+ T cells (CTL) and macrophage derived donor tissue destruction. As the majority of significant differences between healthy and AR EMBs were associated with immune-related pathways, we next assessed which classes of immune cells were activated in AR tissues. We performed immune cell deconvolution of the RNA-seq dataset using the xCell algorithm, which estimates the abundance of specific immune cell subtypes in a sample based on the RNA expression levels of cell-specific markers24. Consistent with pathway analysis, AR EMBs were enriched for CD8+ naive T-cells (q < 0.05).
Figure 1. Gene expression differences between rejection and non-rejection specimens.
A) Heatmap of differentially-expressed genes between AR and non-rejection tissues. B) Volcano plot for the AR and NR comparison. C) Violin plots for selected differentially expressed genes between AR and NR.
Figure 2. Pathway assessment of AR versus NR genes.
A) KEGG pathway enrichment for AR versus NR genes. B) Gene Ontology enrichment for AR versus NR differentially expressed genes.
We assessed overlap of the AR signature with other previously identified signatures. This includes the antibody mediated rejection (AbMR) signature previously identified in Loupy et al. 17 as well as t-cell mediated rejection (TCMR) and AbMR signatures curated from large datasets by Xiu et al.18. For the former, we observed that ~30% of genes in the Loupy et al. signature, as well as ~20% of the genes in the TCMR signature, are present in our AR signature (Figure S1A). Furthermore, ~36% of the genes in the AbMR signature from Xiu et al. (Figure S1B) are present in our AR signature, indicating that while a subset of the characterized genes are novel, there is concordance with prior studies in this area.
3.2. A gene expression classifier for 0R/1R vs 2R acute rejection
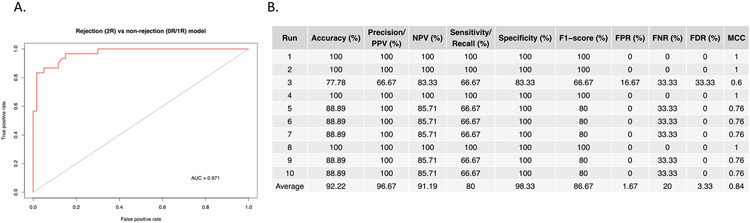
As gene expression profiling revealed robust differences between AR and non-rejection EMBs, we next asked whether the differential gene expression signature could be used to classify samples as molecular AR or non-rejection. To do this, we constructed a predictor model using the Random Forest method to classify AR grade from the differentially expressed genes between 0R/1R and 2R/3R samples. The AR predictor model was trained on 55% of the samples, sampled with replacement and tested for classification of rejection grade on the remaining 45% of samples. Overall performance assessed using the area under the curve (AUC) of 0.971 and was plotted using a receiver-operating characteristic (ROC) curve (Figure 3A&B), which illustrates the AR signature’s robust performance as a classifier. Classification accuracy averages 92.2%, with a positive predictive value (PPV) of 96.7%., a negative predictive value of 91.2% and a sensitivity, and specificity of 80% and 98.33%, respectively. While it will be important to evaluate this classifier across additional independent datasets, these results suggest that RNA-seq is a highly accurate methodology for classifying rejection and non-rejection specimens.
Figure 3. Clinical utility of acute rejection genes.
A). ROC curve for the AR signature as a classifier between rejection and non-rejection biopsies with calculated AUC. B) Performance statistics for Random Forest classification using the AR signature. Abbreviated metrics correspond to the following: positive predictive value (PPV), negative predictive value (NPV), false positive rate (FPR), false negative rate (FNR), false discovery rate (FDR) and Matthews correlation coefficient (MCC).
3.3. A prognostic signature for acute rejection
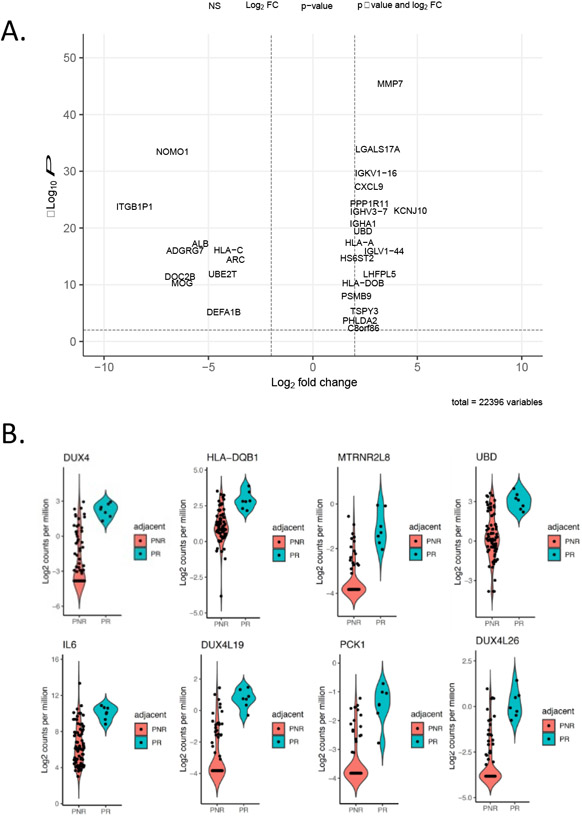
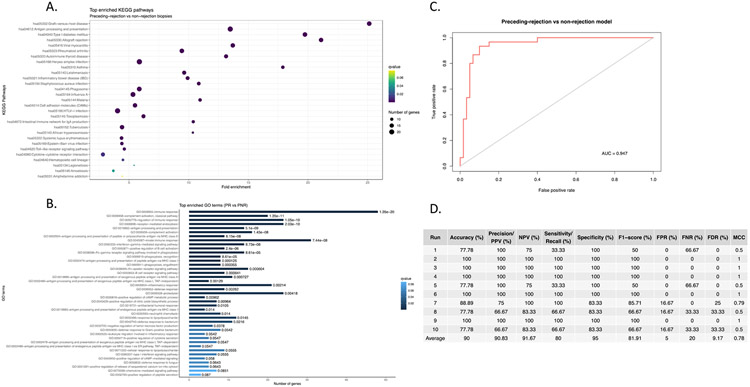
A subset of rejection-associated gene expression exhibited relatively stable high expression in patients that would later experience a rejection event, especially in non-rejection timepoints that immediately preceded a 2R rejection event (Figure 1A). As such we hypothesized that biopsies adjudicated as non-rejection by traditional histopathological grading exhibit molecular features of early acute rejection as determined by RNA-seq and thus could be prognostic for later acute rejection. In order to test this hypothesis, we divided the set of 0R/1R EMBs into two groups: those immediately preceding a 2R/3R rejection EMB (labeled “preceding-rejection”, PR) and those 0R/1R EMBs not preceding a 2R/3R EMB (labeled “preceding-non-rejection”, PNR). We also expanded the analysis to include the entire transcriptome to avoid excluding any genes that may be specifically associated with the PR group. From this, we observed 528 transcripts that were significantly differentially expressed between the PR and the PNR samples (q<0.01) (Figure 4 and Table S3). Intriguingly, top genes upregulated preceding rejection included the double homeobox transactivator DUX4 as well as two similar pseudogenes DUX4L19 and DUX4L26 (Figure 4B). Pathways associated with PR included immune/inflammatory pathways such as cytokine receptor interactions (KEGG q<0.01) as well as genes involved in the inflammatory response (GO, q<0.0022) (Figure 5A and B). Of note, these signals were detected across a variety of time intervals between PR and 2R biopsies, with an average time of 88 days, a minimum of nine days and a maximum of 168 days (Table S4).
Figure 4. Characterization of non-rejection biopsies: preceding rejection versus preceding non-rejection.
A) Volcano plot of gene expression differences for PR vs PNR biopsies. B) Volcano plots of top differentially expressed genes between PR and PNR.
Figure 5. Functional characterization of biopsies preceding rejection.
A) KEGG pathway enrichment for PR versus PNR gene expression. B) GO pathway enrichment for PR versus PNR biopsies. C) ROC curve for the PR classifier. D) Performance statistics for the PR classifier.
We also examined whether these signals persisted after treatment. Non-rejection biopsies immediately following a treated 2R rejection event were distinctly different from other non-rejection biopsies, and by differential gene and pathway expression exhibited higher expression for many of the same genes and pathways associated with Grade 2R events (Figure S2). Specifically, pathways associated with antigen presentation and processing as well as graft versus host disease remained the top overexpressed pathways in these non-rejection biopsies. Thus, while histopathological grading does not indicate AR in these biopsies, gene expression profiles indicate residual AR after treatment.
To evaluate whether gene expression profiling of PR samples could act as an early prognostic marker of acute rejection, we generated a classification model of PR versus non-rejection biopsies using a Random Forest approach. Briefly, the set of 0R/1R PR and PNR biopsies were randomly sampled without replacement into 55% training and 45% test sets. A predictor model was built using the 528 differentially expressed genes between the PR and PNR EMB samples. The model was validated through ten rounds of cross validation with resampling of the training and test sets each round, and classification accuracy is shown via ROC plot in Figure 5C with an AUC of 0.947. Overall statistics show an average classification accuracy of 90% with 90.8% precision (Figure 5D). The NPV was 91.7%, sensitivity was 80% and specificity was 95%. In conclusion, the classifier exhibits strong performance in assessing whether the biopsy is preceding a rejection or non-rejection timepoint, a capability that may be of significant utility in a clinical setting.
4. Discussion
One and five-year survival rates for heart transplant recipients have remained static over the last decade in part due to acute allograft rejection. Although there are differences in reporting accuracies acute rejection is thought to occurs in approximately 30% of heart allograft recipients in the first year post-transplant alone33-35. A key focus in improving mortality rates is more rapid and accurate AR diagnosis and intervention, however this can be challenging given the invasive nature of EMB as well as the sensitivity and specificity of histopathological evaluation. Other studies have characterized array-based gene expression changes, in a cross-sectional manner, during acute allograft rejection. In this study we have performed the first large-scale longitudinal characterization which shows that a subset of these transcriptional differences are stable and precede the actual histopathological rejection event in some cases several months before the acute rejection EMB. As such these represent novel putative biomarkers for AR, with higher sensitivity than conventional histopathology. The current gold-standard histological assessment of H&E stains of EMBs is imperfect due to various factors including intra- and inter-pathologist variability in histological classification, or where histological rejection is evident in one EMB for a given recipient but not evident in additional independent EMB from the same timepoint. These limitations impact the sensitivity and specificity and thus the PPV and NPVs of minimally invasive assays such as donor-derived, cell-free (dd-cf)-DNA profiling. Rigorous agnostic assessment and validation of molecular rejection signatures through RNA profiling will thus create a better histological gold standard upon which dd-cf-DNA profiling can be compared against. In a clinical setting, such approaches may ultimately allow for early detection of AR in heart transplant recipients prior to diagnosis by conventional EMB histopathology, thus allowing for more rapid intervention, limiting irreversible graft injury and improving overall outcomes. While evaluation across larger patient cohorts is needed, we hypothesize that integrating gene expression analysis as a standard step in the histopathological evaluation of EMBs will ultimately reduce the number of biopsies needed (i.e. if the prognostic signature is negative, the care team may be able to delay a subsequent biopsy).
The strong concordance observed between histopathological rejection grade and gene expression profiles may have significant clinical implications as a method for molecular characterization of EMBs to improve the diagnosis of acute rejection. While the current study represents the largest collection of EMB RNA-seq profiles ever assembled, the study is still limited by modest sample size. Additionally, while the EMBs were adjudicated by a centralized pathology protocol, differences in standard of care patient treatment protocols and patient characteristics between the CTOT-03 study sites may introduce subtle biases. We are currently expanding the size and diversity of our cohort by including additional EMB patient datasets across multiple heart transplant study sites within the International Genetics & Translational Research in Transplantation Network36. The larger cohort size will increase the number of clinically significant samples available to generate larger training and testing datasets. Additionally, while our approach has shown utility on frozen EMBs, most diagnostic pathology is currently practiced on formalin-fixed paraffin-embedded (FFPE) specimens. As such ongoing work is focused on translating these results to FFPE. Along with clinical translation, a key focus will be on mechanistic study of genes and molecular pathways identified here in order to gain a deeper understanding of the biological underpinnings of AR, including the coordinated timing of early and late rejection targets. Follow-up studies planned by our teams include utilizing a combination of multi-omic, single cell and spatial methods to identify specific cell-types, cell-cell interactions and pathway regulation that comprise AR1,37. Advances in machine learning (ML) of TCMR grading of EMBs has also advanced rapidly38, and such agnostic approaches can also be combined with molecular and conventional histology to improve accuracy. The next iterations of these ML algorithms include AbMR assessment. As these studies have yet to be attempted at a large-scale in the heart transplant setting, we expect that this will provide a wealth of data that may inform the development of novel diagnostics and/or treatments for improving post-transplant survival rates.
In conclusion, we show that RNA-seq reveals a wealth of information regarding the molecular effects of AR in EMBs, most specifically the activation of a multi-faceted immune response involving a wide variety of immune pathways. These signals can be used to accurately classify rejection and non-rejection EMBs and may ultimately be utilized to inform AR diagnosis. Of note, lower levels of these signals already exist in non-rejection biopsies that precede AR diagnosis and can be developed into a highly accurate prognostic classifier that could improve early detection of AR. While this work represents comprehensive transcriptome profiling of 125 prospectively collected biopsies from longitudinal time courses, we expect these efforts will require additional validation and refinement to facilitate clinical deployment. As already evident in oncology and other clinical settings, we expect that next-generation sequencing will soon become an integral tool in transplant biology diagnostic workflows.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIAID (R01AI144522) and CTOT-03 (U01AI063589). BDP was supported by fellowships from the NHLBI (1T32HL098049) and NIDDK (1F32DK100072). Sequencing support was provided by the Stanford Center for Genomics and Personalized Medicine (Grant 1S10OD020141-01).
Glossary
- AbMR
antibody-mediated rejection
- AR
acute rejection
- AUC
area under the curve
- CAV
chronic allograft vasculopathy
- CTOT
Clinical Trials of Transplantation
- dd-cf-DNA
donor-derived cell free DNA
- EMB
endomyocardial biopsy
- FDR
false discovery rate
- FNR
false negative rate
- FPR
false positive rate
- GO
Gene Ontology
- H&E
hematoxylin and eosin
- IRB
institutional review board
- ISHLT
International Society for Heart and Lung Transplantation
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MCC
Matthews correlation coefficient
- ML
machine learning
- mRNA
messenger RNA
- NPV
negative predictive value
- PNR
preceding non-rejection
- PPV
positive predictive value
- PR
preceding rejection
- RNA-seq
RNA sequencing
- ROC
receiver-operating characteristic
- TCMR
t-cell mediated rejection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keating BJ, Pereira AC, Snyder M, Piening BD. Applying genomics in heart transplantation. Transpl Int. 2018;31(3):278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathier MA, McNamara DM. Management of the Patient After Heart Transplant. Curr Treat Options Cardiovasc Med. 2004;6(6):459–469. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm MJ. Long-term outcome following heart transplantation: current perspective. J Thorac Dis. 2015;7(3):549–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labarrere CA, Jaeger BR. Biomarkers of heart transplant rejection: the good, the bad, and the ugly! Translational research : the journal of laboratory and clinical medicine. 2012;159(4):238–251. [DOI] [PubMed] [Google Scholar]
- 5.Crespo-Leiro MG, Stypmann J, Schulz U, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. European heart journal. 2016;37(33):2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crespo-Leiro MG, Stypmann J, Schulz U, et al. Performance of gene-expression profiling test score variability to predict future clinical events in heart transplant recipients. BMC cardiovascular disorders. 2015;15:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6(1):150–160. [DOI] [PubMed] [Google Scholar]
- 8.Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362(20):1890–1900. [DOI] [PubMed] [Google Scholar]
- 9.De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Science translational medicine. 2014;6(241):241ra277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agbor-Enoh S, Shah P, Tunc I, et al. Cell-Free DNA to Detect Heart Allograft Acute Rejection. Circulation. 2021;143(12):1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry GJ, Angelini A, Burke MM, et al. The ISHLT working formulation for pathologic diagnosis of antibody-mediated rejection in heart transplantation: evolution and current status (2005-2011). The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2011;30(6):601–611. [DOI] [PubMed] [Google Scholar]
- 12.Berry GJ, Burke MM, Andersen C, et al. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2013;32(12):1147–1162. [DOI] [PubMed] [Google Scholar]
- 13.Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2005;24(11):1710–1720. [DOI] [PubMed] [Google Scholar]
- 14.Crespo-Leiro MG, Zuckermann A, Bara C, et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplantation. 2012;94(11):1172–1177. [DOI] [PubMed] [Google Scholar]
- 15.Yohe S, Thyagarajan B. Review of Clinical Next-Generation Sequencing. Arch Pathol Lab Med. 2017;141(11):1544–1557. [DOI] [PubMed] [Google Scholar]
- 16.Ilyas M Next-Generation Sequencing in Diagnostic Pathology. Pathobiology. 2017;84(6):292–305. [DOI] [PubMed] [Google Scholar]
- 17.Loupy A, Duong Van Huyen JP, Hidalgo L, et al. Gene Expression Profiling for the Identification and Classification of Antibody-Mediated Heart Rejection. Circulation. 2017;135(10):917–935. [DOI] [PubMed] [Google Scholar]
- 18.Xiu MX, Liu YM, Wang WJ. Investigation of hub genes and immune status in heart transplant rejection using endomyocardial biopsies. Journal of cellular and molecular medicine. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nature methods. 2017;14(4):417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simha V, Qin S, Shah P, et al. Sirolimus Therapy Is Associated with Elevation in Circulating PCSK9 Levels in Cardiac Transplant Patients. J Cardiovasc Transl Res. 2017;10(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation. 2008;86(2):192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Xu C, Yang R, et al. Silencing of nodal modulator 1 inhibits the differentiation of P19 cells into cardiomyocytes. Experimental cell research. 2015;331(2):369–376. [DOI] [PubMed] [Google Scholar]
- 28.Alfonso-Jaume MA, Bergman MR, Mahimkar R, et al. Cardiac ischemia-reperfusion injury induces matrix metalloproteinase-2 expression through the AP-1 components FosB and JunB. American journal of physiology Heart and circulatory physiology. 2006;291(4):H1838–1846. [DOI] [PubMed] [Google Scholar]
- 29.Stobdan T, Zhou D, Ao-Ieong E, et al. Endothelin receptor B, a candidate gene from human studies at high altitude, improves cardiac tolerance to hypoxia in genetically engineered heterozygote mice. Proc Natl Acad Sci U S A. 2015;112(33):10425–10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paradis A, Xiao D, Zhou J, Zhang L. Endothelin-1 promotes cardiomyocyte terminal differentiation in the developing heart via heightened DNA methylation. Int J Med Sci. 2014;11(4):373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubin LJ. Endothelin-1 and the pulmonary vascular response to altitude: a new therapeutic target? Circulation. 2006;114(13):1350–1351. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Zheng X, et al. Extracting biological meaning from large gene lists with DAVID. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al]. 2009;Chapter 13:Unit 13 11. [DOI] [PubMed] [Google Scholar]
- 33.Eisen HJ, Tuzcu EM, Dorent R, et al. Everolimus for the prevention of allograft rejection and vasculopathy in cardiac-transplant recipients. N Engl J Med. 2003;349(9):847–858. [DOI] [PubMed] [Google Scholar]
- 34.Kobashigawa JA, Miller LW, Russell SD, et al. Tacrolimus with mycophenolate mofetil (MMF) or sirolimus vs. cyclosporine with MMF in cardiac transplant patients: 1-year report. Am J Transplant. 2006;6(6):1377–1386. [DOI] [PubMed] [Google Scholar]
- 35.Patel JK, Kobashigawa JA. Should we be doing routine biopsy after heart transplantation in a new era of anti-rejection? Curr Opin Cardiol. 2006;21(2):127–131. [DOI] [PubMed] [Google Scholar]
- 36.International G, Translational Research in Transplantation N. Design and Implementation of the International Genetics and Translational Research in Transplantation Network. Transplantation. 2015;99(11):2401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khachatoorian Y, Khachadourian V, Chang E, et al. Noninvasive biomarkers for prediction and diagnosis of heart transplantation rejection. Transplant Rev (Orlando). 2021;35(1):100590. [DOI] [PubMed] [Google Scholar]
- 38.Peyster EG, Arabyarmohammadi S, Janowczyk A, et al. An automated computational image analysis pipeline for histological grading of cardiac allograft rejection. European heart journal. 2021;42(24):2356–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.