Abstract
Rubia cordifolia L. is a widely used traditional medicine in the Indian sub-continent and Eastern Asia. In the present study, the aqueous leaf extract of the R. Cordifolia was used to fabricate silver nanoparticles (RC@AgNPs), following a green synthesis approach. Effect of temperature (60 °C), pH (8), as well the concentration of leaf extract (2 ml) and silver nitrate (2 mM) were optimized for the synthesis of stable RC@AgNPs. The phytofabrication of nanosilver was validated by UV–visible spectral analysis, which displayed a distinctive surface plasmon resonance peak at 432 nm. The effective functional molecules as capping and stabilizing agents, and responsible for the conversion of Ag+ to nanosilver (Ag0) were identified using the FTIR spectra. The spherical RC@AgNPs with an average size of ~ 20.98 nm, crystalline nature, and 61% elemental composition were revealed by TEM, SEM, XRD, and. EDX. Biogenic RC@AgNPs displayed a remarkable anticancer activity against B16F10 (melanoma) and A431 (carcinoma) cell lines with respective IC50 of 36.63 and 54.09 µg/mL, respectively. Besides, RC@AgNPs showed strong antifungal activity against aflatoxigenic Aspergillus flavus, DNA-binding properties, and DPPH and ABTS free radical inhibition. The presented research provides a potential therapeutic agent to be utilized in various biomedical applications.
Subject terms: Biological techniques, Biotechnology, Materials science, Nanoscience and technology
Introduction
Nanotechnology is an emerging paradigm of the twenty-first century, which deals with the manipulation of materials at the nanoscale and exploration of their versatile properties. Nanoparticles (NPs) have unique thermal, electric, chemical, magnetic, optical, and physical properties, unlike their bulk materials due to possessing quantum effects and high surface-to-volume ratio1,2. The use of NPs in biomedical science is well noticeable for several applications such as bio-imaging, cancer therapy, bio-detection of pathogens, drug and gene delivery, catalysis, biosensors, etc.3,4. Silver nanoparticles (AgNPs) are preferred over other metal-NPs in biomedical industries because of their tremendous antimicrobial applications5–7. AgNPs are used enormously in drug carriers, diagnostic technologies, coating of biomaterials and medical devices, tissue engineering and regeneration materials, challenging health-care approaches, and performance-enhanced therapeutic alternatives8. Although AgNPs have a promising future in nanoscience and biomedicine, significant efforts are needed to comprehend the intricate processes behind their biological interactions and pernicious effects9.
The common physical procedures for NPs’ synthesis, like attrition and pyrolysis, have various drawbacks, including imprecise surface formation, low fabrication rate, high manufacturing cost, and significant energy requirements. Other physicochemical methods (i.e. etching, chemical reduction, sol–gel technique, electro-explosion, laser ablation, etc.) include the use of toxic chemicals and the formation of poisonous substances10. The biological methods of NPs’ synthesis are gaining attention because of being non-hazardous, economical, eco-friendly, and devoid of the drawbacks of physicochemical processes. Phytosynthesis of NPs using leaves, stems, fruits, flowers, seeds, bark, and roots of the plants are highly cost-effective when compared to the microbial synthesis using bacteria, fungus, and algae. The tedious and expensive process of microbial culture and problems of contamination are the discouraging factors associated with microbe-mediated NP synthesis11–13. The plant-mediated synthesis of NPs is a simple, fast, reliable, economical, eco-friendly, and one-step method. The plant extracts possess various secondary metabolites viz. alkaloids, terpenoids, flavonoids, saponins, anthraquinones, tannins, etc., and are found to be responsible for the reduction of metal ion, and synthesis of NPs.
The global burden of cancer is rising with 18.1 million active cases and 9.6 million cancer deaths as reported in GLOBOCAN 201814. Certain AgNPs have been shown to possess anticancer, anti-inflammatory, anti-oxidant, and antimicrobial activities15–17.
Rubia cordifolia L., commonly known as ‘Indian madder’ or ‘Manjishta’ is a perennial climbing herb, belonging to the family: Rubiaceae18. The root of the plant is widely used in traditional medicine in India, Japan, Korea, and China for treating tuberculosis, wounds, menoxenia, and rheumatism. R. Cordifolia has some anthraquinones and anthraquinone derivatives to support various therapeutic properties, including anti-fungal, anti-oxidant, anti-inflammatory, anti-bacterial, anti-tumor, and anti-cancer activities18–21. Copious literature is available on the uses of the root, whereas, leaves are usually discarded for no use. No study has been undertaken to explore the therapeutic properties of leaves.
The study aimed to synthesize and characterize biogenic AgNPs from aqueous leaf extract of R. Cordifolia, as well as to explore their anticancer, DNA-binding, antifungal, and antioxidant properties.
Material and methods
Plant materials
R. cordifolia leaves were collected in January 2019 from the forest area of Amarkantak, Madhya Pradesh, India. The identification of R. cordifolia was certified by a subject expert, and a voucher specimen (DOB/07/RC/120/2019) was deposited in the Botany department, IGNTU, Amarkantak, MP, India. The collection of the plant material and related studies complies with relevant institutional, national, and international guidelines and legislation.
Chemicals
All the chemicals [Silver nitrate (AgNO3), CT-DNA, Tris buffer, potato dextrose broth (PD-B), potato dextrose agar (PDA), 2,2-Diphenyl-1-picrylhydrazyl (DPPH) and 2,2-Azino-bis-3-ethylbenzothiazoline-6-sulphonic acid (ABTS)] used were AR grade and purchased from Himedia (Mumbai, India). Double distilled water (DDW) was used throughout the experiments.
Preparation of leaf extract
Approximately, 10 g of leaves of R. cordifolia (Fig. 1) were weighed through the analytical balance, washed twice with DDW, and dried at room temperature. The chopped leaves were placed in a 250 mL conical flask with 100 mL of DDW, boiled for half an hour at 70 0C, and filtered twice using Whatman No. 1 filter paper. The filtrate was kept at 40C for further use.
Figure 1.

Plant of R. cordifolia L.
Phytochemical analysis
The principal phytochemicals in R. cordifolia leaf extract (RCLE) were examined following the procedure of Chandraker et al.22. Furthermore, for the detection of free and combined anthraquinones, Borntrager’s test was adopted following Ukwubile et al.23.
Optimized synthesis of AgNPs
Several factors, including temperature, pH, AgNO3 concentration, and RCLE, were optimized to synthesize saturated and stable R. cordifolia-mediated RC@AgNPs. To optimize the suitable temperature, four reaction mixtures of 1 mL of RCLE and 9 mL of AgNO3 (1.0 mM) were mixed and incubated at four different temperatures, viz., 20, 40, 60, and 80 °C for 60 min. To optimize the pH, the above experimental process was repeated at different pHs (2, 4, 6, 7, and 8). Four different volumes of RCLE (9.5, 9, 8.5, and 8 mL) were treated with corresponding volumes (0.5, 1.0, 1.5, and 2.0 mL) of 1 mM AgNO3 solution to find the optimum concentration of RCLE. Similarly, to optimize AgNO3 concentration, 0.2, 0.5, 1.0, and 2.0 mM of 1 mL AgNO3 were treated individually with 9 mL of RCLE. All the four experimental set-ups were incubated for 60 min and analyzed thereafter with UV–visible absorption spectroscopy.
Characterization of RC@AgNPs
Phytosynthesis of RC@AgNPs was confirmed by UV–visible spectroscopy (Shimadzu UV-1800). To investigate the nanosize and structure of RC@AgNPs, transmission electron microscopic (TEM-Technai G20, FEI) and X-Ray Diffraction (XRD) [Bruker-D8 at 30 kV and 20 mA current with Cu K (I = 1.54 A)] studies were performed. The topology and elemental composition of RC@AgNPs were determined by Energy Dispersive X-ray linked Scanning Electron Microscopy (SEM) (EVO 18; Carl Zeiss, Germany). The average size (size distribution), dispersity, and stability (zeta potential) of RC@AgNPs in the aqueous medium were calculated by Anton Paar (Litesizer 500). Fourier Transform Infra-Red (FTIR) spectra of RCLE and RC@AgNPs were obtained from Thermo Scientific Nicolet 5.
Applications of RC@AgNPs
Anticancer activity
The culture and maintenance of cells were done in Dulbecco's Modified Eagle’s Medium (DMEM) with 10% Fetal Bovine Serum (FBS) and 1% antibiotic and antimitotic solution at 37 °C with 5% CO2. For the experiment, NPs were suspended in dimethylsulfoxide (DMSO) with the concentration of DMSO in the medium not exceeding 0.1% in all the treatment groups. The standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was used to assess the anticancer efficacy of RC@AgNPs against A431 (squamous cell carcinoma cell line) and B16F10 (melanoma cell line). Both the cell types were seeded at the concentration of 5000 cells/well in 96 well plates. After incubating the cells for 24 h, treatments were given to cells for 24 and 48 h with different concentrations (10, 25, 50, and 100 µg/mL) of RC@AgNPs. MTT was added to the wells, and the plate was further incubated for 4 h. The added MTT was replaced gently by 100 µl of DMSO and incubated briefly for ten minutes at 37 0C, with the absorption measured in a microplate reader at 570 nm. The percentage of cell viability was estimated using the following formula:
DNA interaction activity
Calf-thymus DNA (CT-DNA) solution was prepared with Tris buffer (pH = 7.2). Different concentrations of RC@AgNPs (0.010–0.090 nM) were treated with a concentration of CT-DNA (250 µM). The interaction between RC@AgNPs and CT-DNA has been carefully monitored in a UV–visible spectrophotometer by absorption-titration experiments.
Antifungal activity
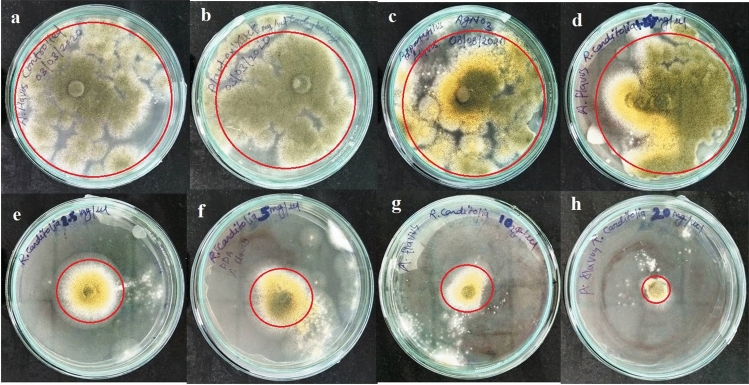
For antifungal activity, the RC@AgNPs were evaluated against a food-spoiling, saprophytic, pathogenic, aflatoxigenic, and ubiquitous fungi Aspergillus flavus. The fungal strain (No. 277) was procured from the Microbial Type Culture Collection and the Gene Bank (MTCC) of the Institute of Microbial Technology (IMTECH), Chandigarh, India. Contact assay was adopted to determine antifungal activity, following Shukla et al.24. Five different doses of RC@AgNPs (1.25, 2.5, 5.0, 10.0, and 20.0 mg) were taken in separate Petri dishes (90 mm) and 20 mL PDA was poured into each petri dish. Finally, 5 mm fungal discs taken from the A. flavus colony (One week old) were inoculated at the center of RC@AgNPs and PDA-containing plates. The inoculated Petri dishes were incubated for seven days in the dark at 27 °C to determine the inhibition of the mycelial growth of the fungi. The negative control plates without RC@AgNPs, and positive control plates with RCLE and 1 mM silver nitrate solution, separately were also incubated under the same growth conditions. The effect of RC@AgNPs, RCLE and 1 mM silver nitrate on the growth of A. flavus was tested in triplicates. After seven days, the radial colony's growth and photographic record were established.
The percentage of fungal inhibition was calculated based on the rate of radial growth inhibition, as follows:
where IRG = Inhibition of the radial growth, R1 = Control's radial growth, and R2 = Radial growth in treatments, IRG was estimated using the mean ± standard error (SE) of the triplicate data.
Antioxidant capability
The antioxidant properties of RC@AgNPs were determined using the DPPH and ABTS free radical scavenging tests described by Chandraker et al.25. Using UV–visible spectroscopy, the antioxidant potential of RC@AgNPs was tested in the presence of DPPH and ABTS free radicals, and ascorbic acid was used as a standard reference. The percentage of scavenging activity was calculated using the following formula.
Statistical analysis
OriginPro 8.5 and ImageJ software were used throughout the experiments, and all tests were performed in triplicate, with data presented in mean ± SE. Data for the MTT assays were examined using the Student's t-test, with P < 0.05 considered statistically significant.
Results and discussion
Phytochemical analysis
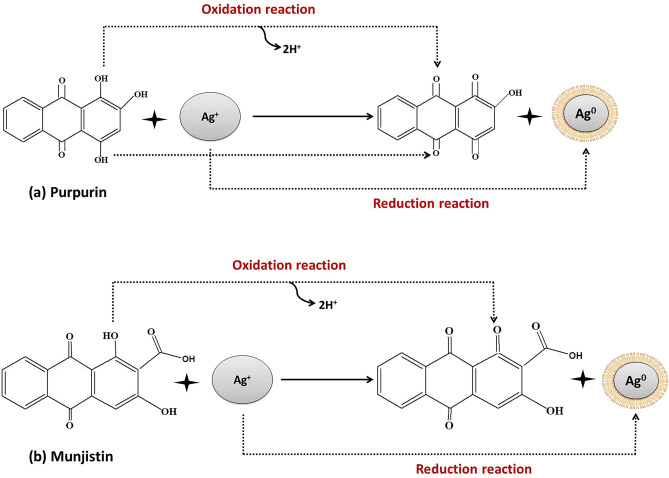
Phytochemical analysis of RCLE revealed the presence of flavonoids, tannins, phlobatannins, saponins, and anthraquinones (Table 1). Our findings are almost similar to that of previous reports where Chandrashekar et al. studied the phytoconstituents of R. cordifolia root26. These phytochemicals may act as reducing, capping, and stabilizing agents during the synthesis of AgNPs from RCLE. Phytochemicals are supposed to reduce Ag+ to Ag0 and synthesize RC@AgNPs. Some previous studies confirmed that tannins27, and saponins28 are responsible for reducing the metal ions to metallic-NPs. Anthraquinones are reported to be present in all the parts of R. cordifolia26. Purpurin (1,2,4-trihydroxyanthraquinone) and munjistin (1,3,-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid) are two predominant anthraquinones present in R. cordifolia, and supposed to be responsible for its pharmacological properties29–31.
Table 1.
Phytochemical analysis of RCLE.
| S. no | Phytochemicals | Test | Result |
|---|---|---|---|
| 1 | Alkaloids |
Mayer’s test, Wagner test Dragendroff test |
−ve −ve |
| 2 | Saponins | Foam test | + ve |
| 3 | Anthraquinones | Borntrager’s test | + ve |
| 4 | Tannins | Ferric chloride test | + ve |
| 5 | Flavanoids | Ferric chloride test and Lead acetate test | + ve |
| 6 | Phlobatannins | HCL test | + ve |
| 7 | Diterpenes | Copper acetate test | −ve |
| 8 | Steroids | Salkowski test | −ve |
| 9 | Triterpenes | Salkowski test, Liberman test | −ve |
+ Present and −Absent.
Biogenic synthesis of RC@AgNPs
Silver nanoparticles were synthesized following an eco-friendly method32. RCLE was used as a capping, stabilizing, and a reducing agent to fabricate RC@AgNPs from AgNO3. The schematic illustration of nanosilver synthesis from two major anthraquinones of RCLE is shown in Fig. 2. The change of color from light-green to reddish-brown has been considered as RC@AgNPs synthesis. Changes in the color of the reaction mixture are due to the conversion of Ag+ to Ag0, and they were easily monitored by Ultraviolet–visible spectroscopy. Figure 3 represents the UV–visible analysis of RC@AgNPs, RCLE, and silver nitrate solution. In Fig. 3 it is clearly shown that RC@AgNPs have a signature AgNPs’ absorbance peak at 432 nm, whereas, RCLE and AgNO3 do not. Another peak shown around 380 nm may originate from an interband transition33. RC@AgNPs displayed a negligible shift in λmax from 432 to 435 nm, after 5 months of synthesis, thus found to be stable.
Figure 2.
A possible mechanism showing the role of two anthraquinones in the reduction of Ag+ to nanosilver (Ag0).
Figure 3.
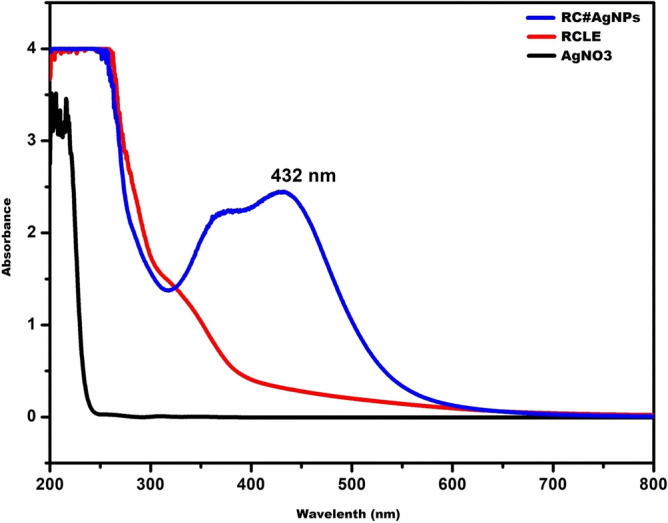
UV–visible analysis of RC@AgNPs, RCLE, and silver nitrate.
Due to surface plasmon resonance (SPR), AgNPs tend to exhibit a characteristic absorption maximum (λmax) in the 400–500 nm range34. The absorption maximum of Justicia adhatoda mediated AgNPs was 463 nm35, Sonchus arvensis mediated AgNPs was 440 nm36, Equisetum arvense mediated AgNPs was 488 nm37, and Withania coagulans mediated AgNPs were 445 nm38, SPR is a cumulative oscillation of conduction electrons triggered by incoming light at the interface between (-)ive and ( +)ive materials. The condition is regulated by the capping agents on Ag-surface. Plants have variations in their phytochemical constituents, and thus, varied functional groups act as capping, reducing, and stabilizing agents. Therefore, different plant-mediated AgNPs show variable λmax, size, and stability.
Optimization of biogenic RC@AgNPs
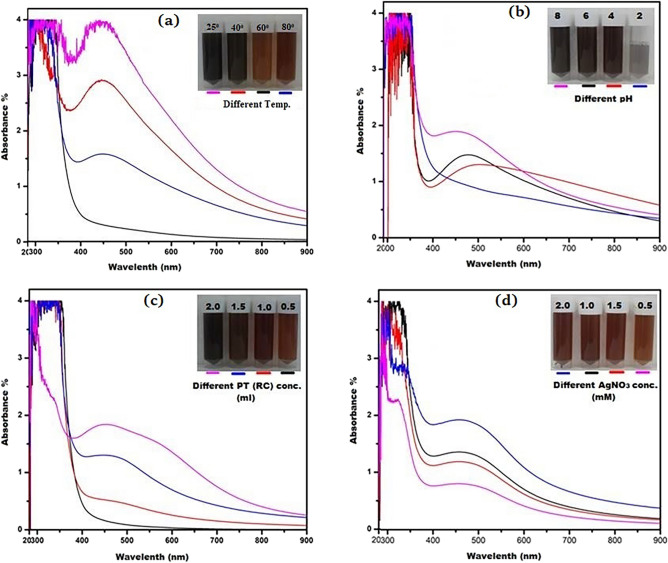
The yield of NPs is influenced by the reaction conditions of the biogenic synthesis process. Various reaction parameters explicitly alter the size distribution and reaction rate of the NPs synthesis. Figure 4 shows the alterations in the color of the reaction mixture under several experimental conditions39.
Figure 4.
(a) UV–vis spectra of RC@AgNPs: at different temperatures of 25, 40, 60, and 80 ºC; (b) at different pH of 2, 4, 6, and 8; (c) at different amount of RCLE i.e. 0.5, 1.0, 1.5 and 2 ml; (d) at different AgNO3 concentrations of 0.5, 1.0, 1.5, and 2.0 mM. (Except Fig. 4c, the remaining experiments had the same concentration of RCLE; Similarly, except Fig. 4d, the concentration of AgNO3 was the same in the rest of the experiments).
The reaction temperature was indeed a vital aspect that expressively related to the rate of biogenic synthesis of RC@AgNP. To investigate the effect of temperature on the phytosynthesis of RC@AgNPs, 1 mL of RCLE was treated with 9 mL of 2 mM AgNO3 in 4 different vials and kept at temperatures ranging from 20 °C to 80 °C. According to the findings, increasing the temperature in the reaction mixture increased the biogenic synthesis of RC@AgNPs (Fig. 4a). The considerable RC@AgNP synthesis was observed at 60 °C, even though absolute RC@AgNP synthesis was reported at 80 °C. It shows that the entire synthesis of RC@AgNPs may be achieved at temperatures of 60 °C and 80 °C, where RCLE phytochemicals conduct optimal reduction and stabilisation40. The absorbance peak at 80 °C seems distorted due to the accumulation of RC@AgNPs generated by high-temperature biogenic production.
The pH value is always crucial for any reaction. In the biogenic synthesis of AgNPs, pH is essential. The color of the reaction medium, the strength of the SPR peak, and the shape and size of the NPs were investigated to be pH-dependent. RC@AgNPs absorbance peak rises with increasing pH, and the maximum fabrication of RC@AgNPs occurred at pH 8. In our investigation, the acidic media with pH 2 and 4 had a lower absorption peak than pH 6 and 8. As a result, we concluded that an alkaline pH of 8 is ideal for the synthesis of RC@AgNPs (Fig. 4b). According to our observations, absorption increases with rising pH, indicating that an alkaline condition is preferable to an acidic one for the synthesis of NPs. During the synthesis of NPs, the change in the pH of the reaction solution affects the ability of the phytochemicals to reduce silver ions. Vanaja et al. also proposed the increase in absorption when pH rises, and it can sometimes affect the form, size, and production of NPs41. Qian et al. and Handayani et al. found similar results with Pometia pinnata (Matoa) leaf extract and Epicoccum nigrum (fungus) mediated green synthesis of AgNPs, that an alkaline medium is more pertinent than an acidic medium42,43. Sintubin et al. suggested that higher pH enhances the race between H+ and Ag+ for bond formation with phytochemicals; thus, the higher pH gave better results than the lower pH for the synthesis of AgNPs44.
The RCLE is another key component in the biosynthesis of RC@AgNPs. The absorption spectra of RC@AgNPs under visible range were established using various RCLE concentrations (0.5, 1, 1.5, and 2 mL), where the AgNO3 concentrations were held at 2 mM constant. Figure 4c indicates no absorption spectrum at 0.5 RCLE concentrations. On increasing the concentration of RCLE, absorption peaks were observed, and at 2.0 mL RCLE concentration, maximum absorbance was found. The increasing concentration of biomolecules involved in metal reduction has enhanced the synthesis of environmental-friendly NPs. Similar results were also found with the Pongamia pinnata leaf extract45 and Citrullus lanatus fruit rind extracts46.
The concentration of AgNO3 also affects the phytosynthesis of AgNPs, significantly. Multiple concentrations (0.2, 0.5, 1, and 2 mM) of AgNO3 solution were used to determine their effect on RC@AgNPs synthesis. AgNO3 concentration of 2.0 mM results in maximum RC@AgNPs synthesis, with an absorbance peak at 432 nm (Fig. 4d). Poor peaks at lower concentrations of 0.2 and 0.5, and 1.0 mM AgNO3 could be due to the deficient availability of Ag+ ions in the reaction mixture.
Characterization of RC@AgNPs
XRD analysis
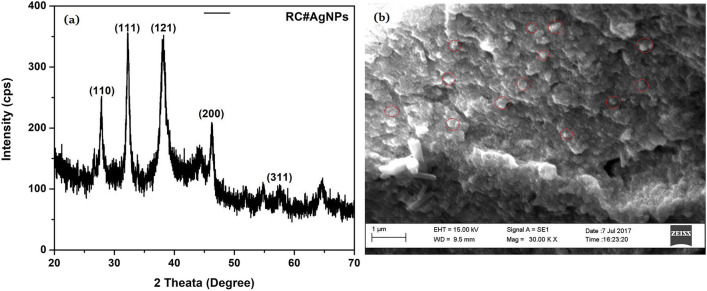
The crystalline nature of the particles was confirmed with XRD. Figure 5a shows an XRD pattern of the AgNPs synthesized by using RCLE. The XRD pattern showed many Bragg reflections based on Ag's face-centered cubic (fcc) structure. The nanosize of RC@AgNPs was confirmed after matching the obtained XRD spectrum with the standard (JCPDS file no. 84–0713). The peaks at 2 theta values of 27.85°, 32.19°, 38.19°, 46.24°, and 57.52°, demonstrate the (110), (111), (121), (200), and (311) Bragg's reflections, which may be indexed based on the fcc structure of metallic silver47. XRD results thus confirmed the crystalline nature of RC@AgNPs.
Figure 5.
(a) XRD spectra, and (b) SEM images of biogenic synthesized RC@AgNPs.
SEM and EDX analysis
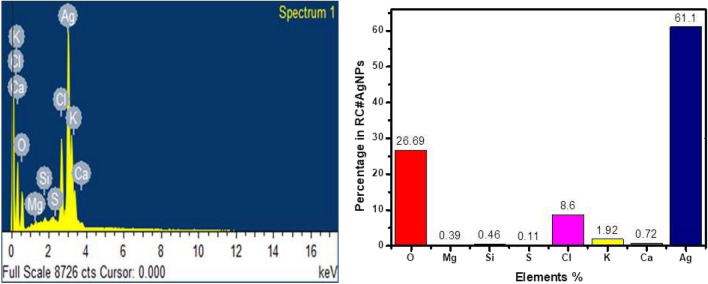
SEM image (Fig. 5b) shows the granular appearance of RC@AgNPs. Different elements' composition in RC@AgNPs is confirmed by EDX analysis at 3 keV. In Fig. 6a the EDX spectrum reveals a high elemental peak, showing metallic silver. According to the quantitative assessment, metallic silver has a more significant weight, 61.1%, while O, Mg, Si, S, Cl, K, and Ca have 26.69, 0.39, 0.46, 0.11, 8.6, 1.92, and 0.72%, respectively (Fig. 6b).
Figure 6.
(a) EDX spectrum of RC@AgNPs synthesized from RCLE, and (b) Quantitative estimation of EDX data of RC@AgNPs.
TEM analysis
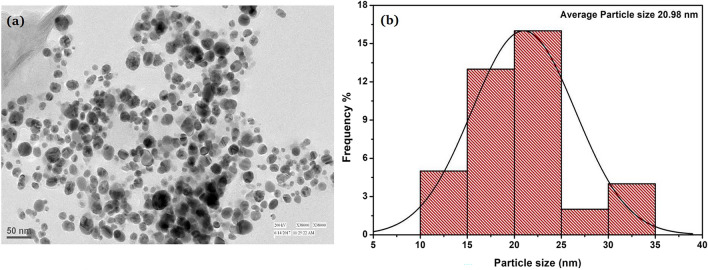
Figure 7a shows TEM images of RC@AgNPs that illustrate the formation of isotropic spherical AgNPs. The particle size distribution histogram of RC@AgNPs using TEM images revealed an average particle size of 20.98 nm (Fig. 7b). This particle size spectrum was obtained using spectroscopy-based SPR and XRD.
Figure 7.
(a) TEM images of RC@AgNPs, and (b) Particle size distribution histogram of RC@AgNPs from the TEM images.
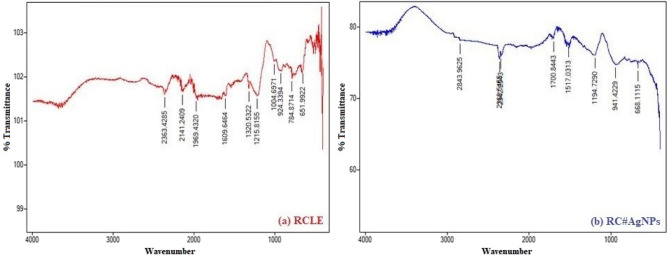
FT-IR spectral analysis
FT-IR analysis of RCLE and RC@AgNPs was done to reveal functional groups present in the extract of R. cordifolia and present on the surface of AgNPs. Table 2 shows the wavenumber and interpretation of possible functional groups of RC@AgNPs and RCLE. Figure 8a–b shows the FT-IR spectrum of RC@AgNPs and RCLE. The leaf extract contains various phytochemicals, which might be responsible for the reduction and stabilization of Ag+ to Ag0 and form RC@AgNPs. A similar pattern of the result was found with Ageratum houstonianum extracts and resulting NPs22.
Table 2.
FT-IR analysis of RCLE and RC@AgNPs; it’s probable functional groups.
| RCLE wavenumbers | Probable function groups | RC@AgNPs wavenumbers | Probable function groups |
|---|---|---|---|
| 651.9922 | Aromatic-H bending | 668.1115 | Aromatic-H bending |
| 784.8714 | N–H bend | ||
| 924.3394 | C=C–H bending | 941.4229 | C=C–H bending |
| 1004.6971 | O–C stretching | 1194.7290 | C–O stretch |
| 1215.8155 | O–C stretch | 1517.0313 | N–H bend |
| 1320.5322 | O–H bend | 1700.8443 | C=O stretching |
| 1609.6464 | N–H bend | 2342.2543 | C–H stretching |
| 1969.4320 | Polystyrene | 2843.9625 | N+–H stretch stretch |
| 2141.2409 | –N–C=O stretch |
Figure 8.
FTIR spectra of biogenic synthesized, (a) RC@AgNPs, and (b) RCLE.
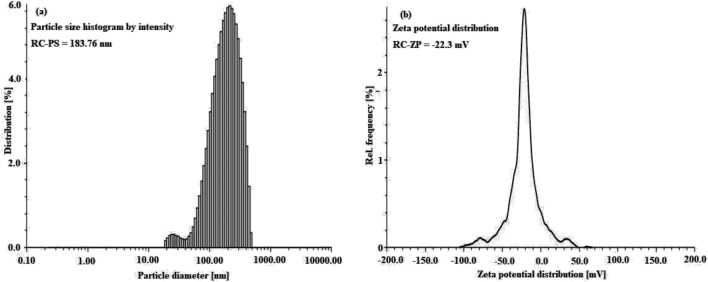
Zeta particle size and zeta potential of RC@AgNPs
Dynamic light scattering (DLS) and Laser Doppler Electrophoresis (LDE) measurements were carried out to determine the particle size distribution and zeta potential of RC@AgNPs, respectively in an aqueous solution. The resulting average zeta particle size is 183.76 nm, with a polydispersity index (PDI) of 26.2, as shown in Fig. 9a. Because DLS measurements were dependent on the hydrodynamic radius of the NPs in the aqueous environment, the grain size distribution was found to be greater than the normal NPs reported by TEM and XRD studies. The related zeta potential of RC@AgNPs was found to be −22.3 mV in Fig. 9b, indicating that RC@AgNPs are remarkably stable. Kokila et al. found similar DLS and zeta potential results with Carica papaya peel extract mediated biosynthesized AgNPs48.
Figure 9.
(a) Zeta particle size, and (b) Zeta potential of RC@AgNPs.
Applications of RC@AgNPs
Anticancer activity
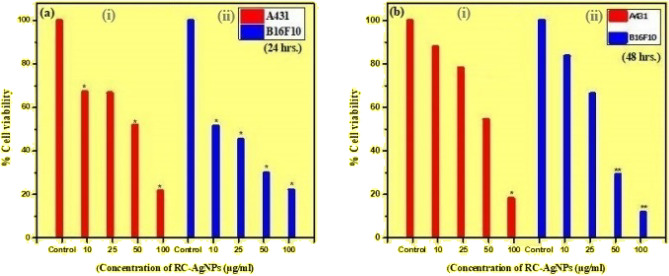
MTT assay was performed on A431 and B16F10 cells to determine the cytotoxic activity of RC@AgNPs. In this assay, yellow color dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) is reduced by the mitochondrial enzyme succinate dehydrogenase, which leads to the formation of purple-blue formazan crystals. Since it is a colorimetric measurement, the absorbance is recorded for the results. Higher the cytotoxicity of NPs lower will be the viability of cancer cells (human squamous cell carcinoma: A431 and mouse melanoma: B16F10). The results of the in vitro assays suggest the cytotoxic activity of RC@AgNPs against the A431 and B16F10 cells (Fig. 10). In the results, it was found that there was a concentration-dependent decrease in the cell viability of both A431 and B16F10 cells at both time points. In the case of A431 cells, after 24 h of treatment, there was a 10%, 20%, 50%, and 80% decrease in cell viability at 10, 20, 50, and 100 µg/mL of concentration, respectively (Fig. 10a,i). After 48 h, the decrease was around 50 and 80% at higher concentrations of 50 and 100 µg/mL (Fig. 10b,i). In B16F10 cells after 24 h of treatment the decrease in cell viability was 20%, 30%, 70% and 90% at 10, 20, 50 and 100 µg/mL of concentration respectively (Fig. 10a,ii). Treatment with RC@AgNPs for 48 h caused a 50% decrease at a lower concentration of 10 µg/mL and a higher concentration of 100 µg/mL, causing a remarkable ~ 80% decrease in the cell viability of B16F10 cells (Fig. 10b,ii). The IC50 value of RC@AgNPs against A431 and B16F10 cells were 54.09 and 36.63 µg/mL, respectively. There are some reports on the cytotoxic activity of AgNPs against different cancer cell lines. Mainly, the activity is attributed to the oxidative stress induced by AgNPs and induction of apoptosis via caspase-dependent pathway20. In a previous investigation, Bryophyllum pinnatum mediated BP-AgNPs also displayed cytotoxicity against the same cell lines49. However, after an incubation period of 24 h, the respective IC50 values of BP-AgNPs against A431 and B16F10 were 59.5 and 96.6 µg/ml, almost double the IC50 values of RC@AgNPs. Cucurbita maxima, Moringa oleifera, and Acorus calamus mediated AgNPs were also found to inhibit A431 cell lines, but their respective IC50 values (82.39, 83.57, and 78.58 μg/ml) were higher than that of RC@AgNPs50. Similarly, Indigofera hirsuta fabricated AgNPs displayed lesser cytotoxicity (IC50 value: 80.9 μg/ml) against B16F1051.
Figure 10.
(a) A431 (cell carcinoma squamous line) and (b) B16F10 (cell melanoma line) viability percentages after incubation at different times (24 and 48 h) with varying concentrations of biogenic RC@AgNPs from RCLE.
Few reports are published on the antiproliferative activity of R. cordifolia. Cytotoxic activity of ZnO and CeO2 NPs from R. cordifolia leaf extract has been reported against MG-63, a human osteosarcoma cell line52. Cytotoxic activity of methanolic plant extracts of R. cordifolia is also reported against Hela and Hep-2 cell lines, with respective IC50 values of 23.12 and 11.92 µg/Ml53. R. cordifolia has shown antiproliferative activity against a wide range of cancer cells, such as human colon carcinoma (HT-29), human breast carcinoma (MCF-7), and human liver carcinoma (HepG2) cell lines, and human colon carcinoma (HT-29) 54. The probable mechanism for the antiproliferative activity might be inhibition of the DNA synthesis. R. cordiofolia can inhibit the incorporation of [3H] thymidine and c-fos gene expression, and the c-fos gene is responsible for the proliferation and differentiation of cells55.
Further, in a study done by Adwankar and Chitnis, they isolated a pure compound RC-18 from R. cordifolia, which was found to have antitumor activity against different types of in vivo solid tumor models (B16 melanoma)53. The cyclic hexapeptides of R. cordifolia were also reported for their anticancer activity by inhibiting the process of protein synthesis. It does so by binding to the 80 s subunit of the ribosome and thus inhibiting the binding of aminoacyl-tRNA and translocation of peptidyl-tRNA. Secondary metabolites found in R. cordifolia L., such as purpurin and munjistin have antitumor activity20. They may be contributing to the anticancer activity of R. cordifolia against different types of cancer18–20.
DNA interaction capability
This is one of the most frequently used techniques to investigate the interactions between CT-DNA and NPs. This study relies on interaction to change or shift the maximum absorption of AgNPs. According to Topală et al., the hypochromic, hypsochromic (blue shift), and bathochromic (redshift) effects are caused by intercalative binding mode, whereas hyperchromism is caused by groove binding (minor and major), electrostatic interactions, and hydrogen bonding56. Chandraker et al. proposed that the stability of CT-DNA be tested at 15-min intervals before introducing NPs, and the absorption peak be studied for 1 hour25. Therefore, UV–Visible titration analyses were performed to examine the interaction between CR@AgNPs and CT-DNA.
To study the interactive effect of CR@AgNPs with CT-DNA, the varied concentration of CR@AgNPs (0.010 to 0.090 nM) was used. UV–visible spectra of CT-DNA at increasing concentrations of CR@AgNPs and constant CT-DNA 250 µM are shown in Fig. 11. The results indicate that when RC@AgNPs are introduced, the absorption at 253 nm increases without a detectable change in the value of λmax. According to Rahban et al. alteration in the secondary structure of DNA is due to hyperchromic effects57. The RC@AgNPs absorption spectra also showed a bathochromic effect from 453 to 463 nm. The hyperchromic effect in CT-DNA absorption spectra and the bathochromic effect in CR@AgNPs absorption spectra indicate a strong interaction between both. Only a few research on the interaction between NPs and DNA have been reported58,59. According to a prior study, the N7 atoms of guanine and adenine bases are likely contact sites in CT-DNA with NPs, while the N3 atoms of cytosine and thymine bases are involved in hydrogen bonding25.
Figure 11.
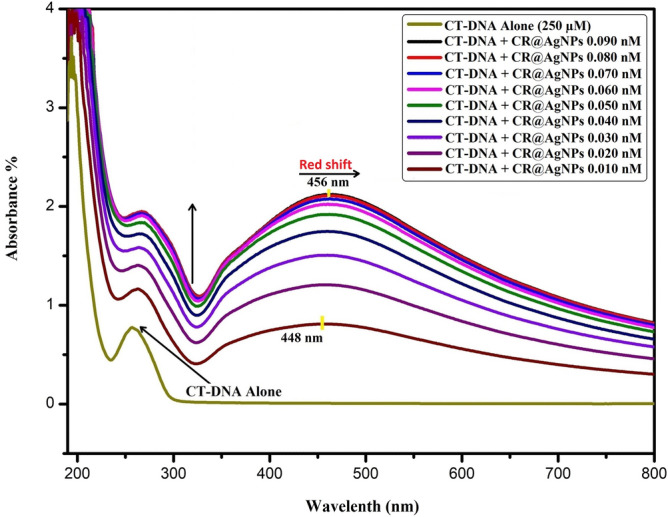
UV–Visible absorption spectra of interaction between CT-DNA (250 μM) and varying concentrations of RC@AgNPs.
Antifungal activity of CR@AgNPs
Aspergillus flavus is a serious pathogen causing preharvest and postharvest infections of nuts, cereals, and legumes. Aflatoxins, produced by certain strains of A. flavus can lead to neutropenia, immunosuppression, acute hepatitis, and hepatocellular carcinona60. The antifungal activity of the phytofabricated RC@AgNPs was assessed by observing the radial growth of the mycelial colony in all the treatments. Figure 12 depicts the effect of RC@AgNP inhibition at various doses against A. flavus over a six-day incubation period. In our research, we found that the concentration of RC@AgNPs had a direct impact on A. flavus mycelia growth. When the concentrations of RC@AgNPs increased, the inhibition of radial growth (IRG) increased as well (Fig. 13). The color, shape, texture, form, and density of the fungal colony in treated plates differed from the control group. At a maximum dose of 1 mg/mL, the IRG of RC@AgNPs against A. flavus was 84.48%, whereas, at the minimum (0.0625 mg/mL), the IRG was recorded 37.17%. The IRG was found to be 21.59, 11.26, 42.71, 48.42, and 55.39% against RCLE, AgNO3, 0.125, 0.25, and 0.5 mg/mL concentration, respectively.
Figure 12.
Antifungal activity of RC@AgNPs against: A. flavus. The rows represent (a) 0 (control), (b) RCLE, (c) AgNO3, and different concentrations of RC@AgNPs: (d) 0.0625, (e) 0.125, (f) 0.25, (g) 0.5, and (h) 1 mg/mL in PDA.
Figure 13.
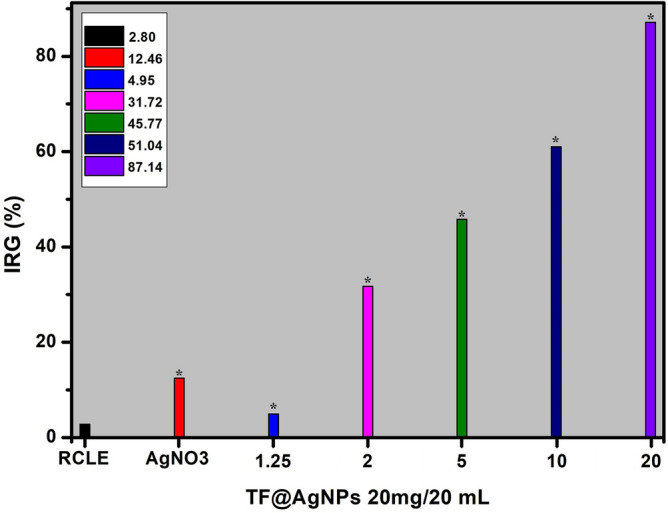
IRG (%) of RC@AgNPs against A. flavus, (*) indicates significant differences (P < 0.05) in comparison to their control.
Very few reports are available on the fungitoxicity of AgNPs against aflatoxigenic A. flavus. Kigellia pinnata bark extract mediated AgNPs were used for the antifungal activity following the well-diffusion method61. Corallocarpus epigaeus mediated AgNPs showed only 46% inhibition of A. flavus at the same concentration we used62. Antifungal activities of Ougeinia oojeinensis and Juniperus procera mediated AgNPs were evaluated against other Aspergillus spp. (A. niger and A. fumigatus), respectively63,64. Jaffri and Ahmad followed a disc-diffusion assay to determine the fungitoxicity of prunosynthetic AgNPs against four Aspergillus spp40. Bocate et al. similarly studied the impact of AgNPs on various Aspergillus spp., but they used fungi-mediated AgNPs deploying Fusarium oxysporum and an antilipemic drug, simvastatin60.
Antioxidant activity
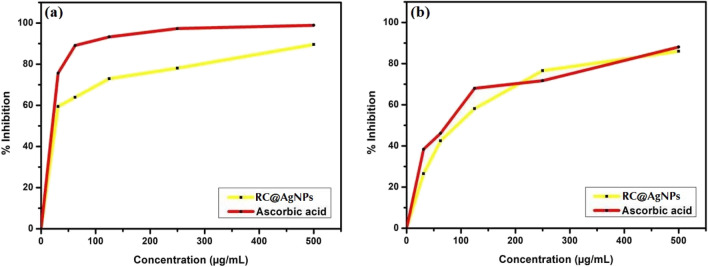
in-vitro antioxidant activity of RC@AgNPs was assessed following DPPH and ABTS assays. Both the assays are comparatively quick and sensitive for analyzing the antioxidant activity of a particular substance compared to others25. The scavenging impact of RC@AgNPs on DPPH (Fig. 14a) and ABTS (Fig. 14b) free radicals was noticed in a dose-dependent manner. DPPH is a strong oxidant with an absorption wavelength range of 515–520 nm. between 515–520 nm. DPPH has a purple-blue color that changes into a bright yellow or colorless when it reacts with a substance having strong; stable free radical scavenging property65. The intensity of color transition is dependent on the total dose and nature of the sample. The DPPH radical inhibition of RC@AgNPs was 59.43 ± 0.296 to 89.58 ± 0.221% in the concentration range of 31.25 to 500 µg/mL. The same concentration of the standard reference (ascorbic acid) showed 75.64 ± 0.08 to 98.94 ± 0.03% inhibition (Fig. 14a). Similarly, at concentrations of 31.25 to 500 mg/mL of RC@AgNPs, ABTS free radical inhibition ranged from 26.54 ± 0.05 to 85.96 ± 0.10%. Ascorbic acid, on the other hand, was inhibited from 38.39 ± 0.05 to 88.09 ± 0.1% (Fig. 14b). AgNPs mediated by Prosopis farcta fruit extract, Cucumis prophetarum leaf extract, and blackcurrant pomace extract have similar DPPH free radical scavenging capabilities66–68. The capacity of the NPs to quench neutral and cationic radicals was examined, indicating that the RC@AgNPs may generate stable neutral radicals by DPPH and free cation radicals from ABTS. The antioxidant mechanism was distinct in both of the assays used. The DPPH test demonstrates AgNPs' ability to transfer electrons and neutralize reactive DPPH free radicals in the reaction medium69. The ABTS assay identifies cationic-free radical scavenging activities utilizing both el- and H+ transport mechanisms70.
Figure 14.
Antioxidant activity of RC@AgNPs, (a) DPPH assay, (b) ABTS assay.
Conclusions
R. cordifolia-mediated AgNPs were synthesized following a green synthesis and eco-friendly approach. The biogenic AgNPs are formed during the reduction of Ag+ ions by RCLE. The phytosynthesized RC@AgNPs were spherical with an average size of ~ 20.98 nm. Different biomedical activities (DNA-binding, antifungal, and antioxidant) of RC@AgNPs were explored, and it was found to have significant cytotoxic activity against skin cancer cell lines, A431 and B16F10. This is the first report on R. Cordifolia leaf-mediated biocompatible and bio-fabricated AgNPs. Based on the remarkable biological activities, RC@AgNPs is hereby recommended for its uses in biomedical applications with elaborated research. The study motivates further therapeutic research in the fields of cancer and antifungal agents.
Acknowledgements
The authors gratefully acknowledge the necessary instrumental (SEM, XRD) facilities of UGC-DAE Consortium for Scientific Research, Indore; and Sophisticated Analytical Instrumentation Facility, AIIMS, New Delhi for TEM. The authors are thankful to the Department of pharmacy, IGNTU for providing Zetasizer facility, and Central Instrumentation Facility, IGNTU, Amarkantak for FTIR.
Author contributions
S.K.C. and F.K. synthesized the RC@AgNPs. The applications of the AgNPs were investigated by S.K.C., F.K., and M.L. The anticancer experiment was performed and reviewed by P.D., S.K.C., and R.P.S. The manuscript was written and reviewed by S.C. and R.S. All the experiments were supervised under R.S. All authors contributed to the submitted version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chandraker SK, Ghosh MK, Lal M, Shukla R. A review on plant-mediated synthesis of silver nanoparticles, their characterization and applications. Nano Express. 2021;2(2):022008. doi: 10.1088/2632-959X/ac0355. [DOI] [Google Scholar]
- 2.Khan I, Saeed K, Khan I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019;12(7):908–931. doi: 10.1016/j.arabjc.2017.05.011. [DOI] [Google Scholar]
- 3.Patra JK, et al. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 2018;16:71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasrollahzadeh M, et al. Valorisation of fruits, their juices and residues into valuable (nano) materials for applications in chemical catalysis and environment. Chem. Rec. 2020;20:1338–1393. doi: 10.1002/tcr.202000078. [DOI] [PubMed] [Google Scholar]
- 5.McQuillan JS, Groenaga Infante H, Stokes E, Shaw AM. Silver nanoparticle enhanced silver ion stress response in Escherichia coli K12. Nanotoxicology. 2012;6(8):857–866. doi: 10.3109/17435390.2011.626532. [DOI] [PubMed] [Google Scholar]
- 6.Mijnendonckx K, et al. Antimicrobial silver: Uses, toxicity and potential for resistance. Biometals. 2013;26(4):609–621. doi: 10.1007/s10534-013-9645-z. [DOI] [PubMed] [Google Scholar]
- 7.Pareek V, Gupta R, Panwar J. Do physico-chemical properties of silver nanoparticles decide their interaction with biological media and bactericidal action? A review. Mater. Sci. Eng. C. 2018;90:739–749. doi: 10.1016/j.msec.2018.04.093. [DOI] [PubMed] [Google Scholar]
- 8.Wong KK, Liu X. Silver nanoparticles-the real “silver bullet” in clinical medicine? Med. Chem. Commun. 2010;1:125–131. doi: 10.1039/C0MD00069H. [DOI] [Google Scholar]
- 9.Burdușel AC, et al. Biomedical applications of silver nanoparticles: An up-to-date overview. Nanomaterials. 2018;8(9):1. doi: 10.3390/nano8090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thakkar KN, Mhatre SS, Parikh RY. Biological synthesis of metallic nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2010;6:257–262. doi: 10.1016/j.nano.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Kaur P. Biosynthesis of nanoparticles using eco-friendly factories and their role in plant pathogenicity: A review. Biotechnol. Res. Innov. 2018;2:63–73. doi: 10.1016/j.biori.2018.09.003. [DOI] [Google Scholar]
- 12.Ahmad T, et al. M. Biosynthesis, structural characterization and antimicrobial activity of gold and silver nanoparticles. Colloids Surfaces B: Biointerfaces. 2013;107:227–234. doi: 10.1016/j.colsurfb.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Wani IA, et al. Structural characterization and antimicrobial properties of silver nanoparticles prepared by inverse microemulsion method. Colloids Surf. B. 2013;101:243–250. doi: 10.1016/j.colsurfb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama K, Welchons DR. The conjugation of amyloid beta protein on the gold colloidal nanoparticles’ surfaces. Nanotechnology. 2007;18:105101. doi: 10.1088/0957-4484/18/10/105101. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro DR, et al. Silver nanoparticles: influence of stabilizing agent and diameter on antifungal activity against Candida albicans and Candida glabrata biofilms. Lett. Appl. Microbiol. 2012;54:383–391. doi: 10.1111/j.1472-765X.2012.03219.x. [DOI] [PubMed] [Google Scholar]
- 16.Bendale Y, Bendale V, Paul S. Evaluation of cytotoxic activity of platinum nanoparticles against normal and cancer cells and its anticancer potential through induction of apoptosis. Integr. Med. Res. 2017;6:141–148. doi: 10.1016/j.imr.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sisubalan N, et al. ROS-mediated cytotoxic activity of ZnO and CeO 2 nanoparticles synthesized using the R. cordifolia L. leaf extract on MG-63 human osteosarcoma cell lines. Environ. Sci. Pollut. Res. 2018;25:10482–10492. doi: 10.1007/s11356-017-0003-5. [DOI] [PubMed] [Google Scholar]
- 18.Natarajan S, et al. ISSR characterization and quantification of purpurin and Alizarin in R. cordifolia L. populations from India. Biochem. Genet. 2019;57:56–72. doi: 10.1007/s10528-018-9875-4. [DOI] [PubMed] [Google Scholar]
- 19.Mishchenko NP, et al. Chemical composition and pharmacological activity of anthraquinones from Rubia cordifolia cell culture. Pharm. Chem. J. 2007;41:605–609. doi: 10.1007/s11094-008-0021-1. [DOI] [Google Scholar]
- 20.Bhatt P, Kushwah AS. Rubia cordifolia overview: A new approach to treat cardiac disorders. Int. J. Drug Dev. & Res. 2013;5:47–54. [Google Scholar]
- 21.Zheng Z, et al. UPLC-QTOF-MS identification of the chemical constituents in rat plasma and urine after oral administration of R. cordifolia L. extract. Molecules. 2017;22:1327. doi: 10.3390/molecules22081327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandraker SK, et al. Green synthesis of copper nanoparticles using leaf extract of Ageratum houstonianum Mill. and study of their photocatalytic and antibacterial activities. Nano Express. 2020 doi: 10.1088/2632-959X/ab8e99. [DOI] [Google Scholar]
- 23.Ukwubile CA, Oise IE, Nyiayem JT. Preliminary phytochemical screening and antibacterial activity of Thaumatococcus daniellii (Benn) Benth (Marantaceae) leaf extract. J. Bacteriol. Mycol. 2017;4(2):53–57. [Google Scholar]
- 24.Shukla R, Singh P, Prakash B, Dubey NK. Antifungal, aflatoxin inhibition and antioxidant activity of Callistemon lanceolatus (Sm) Sweet essential oil and its major component 1, 8-cineole against fungal isolates from chickpea seeds. Food Control. 2012;25(1):27–33. doi: 10.1016/j.foodcont.2011.10.010. [DOI] [Google Scholar]
- 25.Chandraker SK, Lal M, Shukla R. DNA-binding, antioxidant, H2O2 sensing and photocatalytic properties of biogenic silver nanoparticles using Ageratum conyzoides L. leaf extract. RSC Adv. 2019;9:23408–23417. doi: 10.1039/C9RA03590G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar BS, et al. Characterization of Rubia cordifolia L root extract and its evaluation of cardioprotective effect in Wistar rat model. Indian J. Pharmacol. 2018;50(12):1. doi: 10.4103/ijp.IJP_418_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad T. Reviewing the tannic acid mediated synthesis of metal nanoparticles. J. Nanotechnol. 2014 doi: 10.1155/2014/954206. [DOI] [Google Scholar]
- 28.Choi Y, et al. Platycodon saponins from Platycodi radix (Platycodongrandiflorum) for the green synthesis of gold and silver nanoparticles. Nanoscale Res. Lett. 2018;13:1–10. doi: 10.1186/s11671-018-2436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishchenko NP, et al. Chemical composition and pharmacological activity of anthraquinones from Rubia cordifolia cell culture. Pharm. Chem. J. 2007;41(11):605–609. doi: 10.1007/s11094-008-0021-1. [DOI] [Google Scholar]
- 30.Gao M, et al. Simultaneous determination of purpurin, munjistin and mollugin in rat plasma by ultra high performance liquid chromatography-tandem mass spectrometry: Application to a pharmacokinetic study after oral administration of Rubia cordifolia L extract. Molecules. 2016;21(6):717. doi: 10.3390/molecules21060717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali A, Aslam M, Chaudhary SS. A review on pharmacognostic and therapeutic uses of Rubia cordifolia. J. Drug Deliv. Ther. 2020;10(6):195–202. doi: 10.22270/jddt.v10i6.4514. [DOI] [Google Scholar]
- 32.Alomar TS, et al. An eco-friendly plant-mediated synthesis of silver nanoparticles: Characterization, pharmaceutical and biomedical applications. Mater. Chem. Phys. 2020;249:123007. doi: 10.1016/j.matchemphys.2020.123007. [DOI] [Google Scholar]
- 33.Zamiri R, et al. Preparation of silver nanoparticles in virgin coconut oil using laser ablation. Int. J. Nanomedicine. 2011;6(71):71–75. doi: 10.2147/IJN.S14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashraf JM, et al. Green synthesis of silver nanoparticles and characterization of their inhibitory effects on AGEs formation using biophysical techniques. Sci. Rep. 2016;6:20414. doi: 10.1038/srep20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandraker SK, Lal M, Kumar A, Shukla R. Justicia adhatoda L. mediated green synthesis of silver nanoparticles and assessment of their antioxidant, hydrogen peroxide sensing and optical properties. Mater. Technol. 2021;1:1–11. doi: 10.1080/10667857.2021.1949525. [DOI] [Google Scholar]
- 36.Chandraker SK, et al. Colorimetric sensing of Fe3+ and Hg2+ and photocatalytic activity of green synthesized silver nanoparticles from the leaf extract of Sonchus arvensis L. New J. Chem. 2019;43:18175–18183. doi: 10.1039/C9NJ01338E. [DOI] [Google Scholar]
- 37.Das G, Patra JK, Shin HS. Biosynthesis, and potential effect of fern mediated biocompatible silver nanoparticles by cytotoxicity, antidiabetic, antioxidant and antibacterial, studies. Mater. Sci. Eng. C. 2020;114:111011. doi: 10.1016/j.msec.2020.111011. [DOI] [PubMed] [Google Scholar]
- 38.Tripathi D, Modi A, Narayan G, Rai SP. Green and cost effective synthesis of silver nanoparticles from endangered medicinal plant Withania coagulans and their potential biomedical properties. Mater. Sci. Eng. C. 2019;100:152–164. doi: 10.1016/j.msec.2019.02.113. [DOI] [PubMed] [Google Scholar]
- 39.Singh D, et al. Optimization and characterization of silver nanoparticle by endophytic fungi Penicillium sp. isolated from Curcuma longa (turmeric) and application studies against MDR E. coli and S. aureus. Bioinorg. Chem. Appl. 2014;1:1. doi: 10.1155/2014/408021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaffri SB, Ahmad KS. Augmented photocatalytic, antibacterial and antifungal activity of prunosynthetic silver nanoparticles. Artif. Cells Nanomed. Biotechnol. 2018;46:127–137. doi: 10.1080/21691401.2017.1414826. [DOI] [PubMed] [Google Scholar]
- 41.Vanaja M, et al. Phytosynthesis of silver nanoparticles by Cissus quadrangularis: influence of physicochemical factors. J. Nanostructure Chem. 2013;3(17):1. doi: 10.1186/2193-8865-3-17. [DOI] [Google Scholar]
- 42.Qian Y, et al. Biosynthesis of silver nanoparticles by the endophytic fungus Epicoccum nigrum and their activity against pathogenic fungi. Bioprocess. Biosyst. Eng. 2013;36(11):613–1619. doi: 10.1007/s00449-013-0937-z. [DOI] [PubMed] [Google Scholar]
- 43.Handayani W, Ningrum AS, Imawan C. The role of pH in synthesis silver nanoparticles using pometia pinnata (matoa) leaves extract as bioreductor. J. Phys. Conf. Ser. 2020 doi: 10.1088/1742-6596/1428/1/012021. [DOI] [Google Scholar]
- 44.Sintubin L, et al. Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl. Microbiol. Biotechnol. 2009;84(4):741–749. doi: 10.1007/s00253-009-2032-6. [DOI] [PubMed] [Google Scholar]
- 45.Priya RS, Geetha D, Ramesh PS. Antioxidant activity of chemically synthesized AgNPs and biosynthesized Pongamia pinnata leaf extract mediated AgNPs–A comparative study. Ecotoxicol. Environ. Saf. 2016;134:308–318. doi: 10.1016/j.ecoenv.2015.07.037. [DOI] [PubMed] [Google Scholar]
- 46.Ndikau M, Noah NM, Andala DM, Masika E. Green synthesis and characterization of silver nanoparticles using Citrullus lanatus fruit rind extract. Int. J. Environ. Anal. Chem. 2017 doi: 10.1155/2017/8108504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elumalai D, Hemavathi M, Deepaa CV, Kaleena PK. Evaluation of phytosynthesised silver nanoparticles from leaf extracts of Leucas aspera and Hyptis suaveolens and their larvicidal activity against malaria, dengue and filariasis vectors. Parasite Epidemiol. Control. 2017;2:15–26. doi: 10.1016/j.parepi.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kokila T, Ramesh PS, Geetha D. Biosynthesis of AgNPs using Carica Papaya peel extract and evaluation of its antioxidant and antimicrobial activities. Ecotoxicol. Environ. Saf. 2016;134:467–473. doi: 10.1016/j.ecoenv.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 49.Chandraker SK, et al. Cytotoxic, antimitotic, DNA binding, photocatalytic, H2O2 sensing, and antioxidant properties of biofabricated silver nanoparticles using leaf extract of Bryophyllum pinnatum (Lam.) Oken. Front. Mol. Biosci. 2021;465:1. doi: 10.3389/fmolb.2020.593040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nayak D, et al. Biologically synthesised silver nanoparticles from three diverse family of plant extracts and their anticancer activity against epidermoid A431 carcinoma. J. Colloid Interface Sci. 2015;457:329–338. doi: 10.1016/j.jcis.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 51.Netala VR, et al. Biogenesis of silver nanoparticles using leaf extract of Indigofera hirsuta L. and their potential biomedical applications (3-in-1 system) Artif Cells Nanomed Biotechnol. 2018;46(1):1138–1148. doi: 10.1080/21691401.2018.1446967. [DOI] [PubMed] [Google Scholar]
- 52.Patel PR, et al. In-vitro anticancer activity of Rubia cordifolia against Hela and Hep-2 cell lines. Phytomedicine. 2010;2:44–46. [Google Scholar]
- 53.Adwankar MK, Chitnis MP. In vivo anti-Cancer activity of RC-18. Chemotherapy. 1982;28:291–293. doi: 10.1159/000238092. [DOI] [PubMed] [Google Scholar]
- 54.Tripathi YB, Shukla SD. Rubia cordifolia extract inhibits cell proliferation in A-431 cells. Phytotherapy Research: Phytother. Res. 1998;12(6):454–456. doi: 10.1002/(SICI)1099-1573(199809)12:6<454::AID-PTR331>3.0.CO;2-N. [DOI] [Google Scholar]
- 55.Adwankar MK, Chitnis MP, Khandalekar DD, Bhadsavale CG. Anti-cancer activity of the extracts of Rubia cordifolia Linn. (NSC b668893) Indian J. Exp. Biol. 1980;18(102):1. [PubMed] [Google Scholar]
- 56.Topală T, Bodoki A, Oprean L, Oprean R. Experimental techniques employed in the study of metal complexes-DNA–interactions. Exp. Tech. 2014;62(6):1. [Google Scholar]
- 57.Rahban M, Divsalar A, Saboury AA, Golestani A. Nanotoxicity and spectroscopy studies of silver nanoparticle: calf thymus DNA and K562 as targets. J. Phys. Chem. C. 2010;114:5798–5803. doi: 10.1021/jp910656g. [DOI] [Google Scholar]
- 58.Pramanik S, et al. Unraveling the interaction of silver nanoparticles with mammalian and bacterial DNA. J. Phys. Chem. 2016;120(24):5313–5324. doi: 10.1021/acs.jpcb.6b01586. [DOI] [PubMed] [Google Scholar]
- 59.Ribeiro APC, et al. Evaluation of cell toxicity and DNA and protein binding of green synthesized silver nanoparticles. Biomed. Pharmacother. 2018;101:137–144. doi: 10.1016/j.biopha.2018.02.069. [DOI] [PubMed] [Google Scholar]
- 60.Bocate KP, et al. Antifungal activity of silver nanoparticles and simvastatin against toxigenic species of Aspergillus. Int. J. Food Microbiol. 2019;291:79–86. doi: 10.1016/j.ijfoodmicro.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 61.Ravi L, Kannabiran K. Antifungal potential of green synthesized silver nanoparticles (AgNPS) from the stem bark extract of Kigelia pinnata. Res. J. Pharm. Technol. 2021;14(4):1842–1846. doi: 10.52711/0974-360X.2021.00326. [DOI] [Google Scholar]
- 62.Sowmiya K, et al. Green synthesis of silver nanoparticles using aqueous rhizome extract of Corallocarpus Epigaeus for biomedical applications. Appl. Sci. Converg. Technol. 2021;30(2):54–61. doi: 10.5757/ASCT.2021.30.2.54. [DOI] [Google Scholar]
- 63.Kumar D, Arora S, Danish M. Plant based synthesis of silver nanoparticles from Ougeinia oojeinensis leaves extract and their membrane stabilizing, antioxidant and antimicrobial activities. Mater. Today: Proc. 2019;17:313–320. doi: 10.1016/j.matpr.2019.06.435. [DOI] [Google Scholar]
- 64.Bakri MM, El-Naggar MA, Helmy EA, Ashoor MS, Ghany A. Efficacy of Juniperus procera constituents with silver nanoparticles against Aspergillus fumigatus and Fusarium chlamydosporum. BioNanoScience. 2020;10(1):62–72. doi: 10.1007/s12668-019-00716-x. [DOI] [Google Scholar]
- 65.Khorrami S, Zarepour A, Zarrabi A. Green synthesis of silver nanoparticles at low temperature in a fast pace with unique DPPH radical scavenging and selective cytotoxicity against MCF-7 and BT-20 tumor cell lines. Biotechnol. Rep. 2019;24:e00393. doi: 10.1016/j.btre.2019.e00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salari S, Bahabadi SE, Samzadeh-Kermani A, Yosefzaei F. In-vitro evaluation of antioxidant and antibacterial potential of green synthesized silver nanoparticles using Prosopis farcta fruit extract. Iran. J. Pharm. Res. 2019;18(430):1. [PMC free article] [PubMed] [Google Scholar]
- 67.Hemlata Meena PR, Singh AP, Tejavath KK. Biosynthesis of silver nanoparticles using Cucumis prophetarum aqueous leaf extract and their antibacterial and antiproliferative activity against cancer cell lines. ACS Omega. 2020;5:5520–5528. doi: 10.1021/acsomega.0c00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vorobyova V, Vasyliev G, Skiba M. Eco-friendly, “green” synthesis of silver nanoparticles with the black currant pomace extract and its antibacterial, electrochemical, and antioxidant activity. Appl. Nanosci. 2020;10:4523–4534. doi: 10.1007/s13204-020-01369-z. [DOI] [Google Scholar]
- 69.Rasheed T, Bilal M, Iqbal HM, Li C. Green biosynthesis of silver nanoparticles using leaves extract of Artemisia vulgaris and their potential biomedical applications. Colloids Surf. B. 2017;158:408–415. doi: 10.1016/j.colsurfb.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 70.Sowinska M, et al. Molecular Antioxidant Properties and In Vitro Cell Toxicity of the p-Aminobenzoic Acid (PABA) Functionalized Peptide Dendrimers. Biomolecules. 2019;9(89):1. doi: 10.3390/biom9030089. [DOI] [PMC free article] [PubMed] [Google Scholar]