Abstract
GeneMatcher (genematcher.org) is a tool designed to connect individuals with an interest in the same gene. Now used around the world to create collaborations and generate the evidence needed to support novel disease gene identification, GeneMatcher is a founding member of the Matchmaker Exchange (MME; matchmakerexchange.org) and strongest possible advocate for global data sharing including those in resource‐limited environments. As of October 1, 2021, there are 12,531 submitters from 94 countries who have submitted 58,134 submissions with 13,498 unique genes in the database. Among these genes, 8970 (64%) have matched at least once and the total number of matches is 378,806, growing by about 10,000 per month. GeneMatcher submitters increase by 80–120 each month and submissions grow by >800 per month, while unique genes and gene matches continue to grow steadily at rate of about 80 per month. The number of genes without a match peaked at 4371 in February of 2019 and despite the increase in the number of new submissions, the number of unique genes without a match continues to slowly decline, currently standing at 4,016. All submissions in GeneMatcher are available for matching across the MME.
Keywords: data sharing, GeneMatcher, international, Matchmaker Exchange, rare disease
GeneMatcher is the leading resource for connecting individuals with an interest in the same gene from around the world to create collaborations and generate the evidence needed to support novel disease gene identification. GeneMatcher is a founding member of the Matchmaker Exchange (matchmakerexchange.org) and all GeneMatcher submissions are open to matching from any connected node. The thousands of unique gene matches have already generated over 500 publications.
1. OVERVIEW
GeneMatcher (Sobreira Schiettecatte, Boehm, et al., 2015) was launched in September 2013 and initially used primarily by researchers. In late 2014 a large commercial lab in the US, GeneDx, followed by others started depositing their likely pathogenic variants in genes not yet implicated in disease. This resulted in a spike in the number of genes and subsequent matches (Figure 1). After some delay, numerous publications citing GeneMatcher followed.
Figure 1.
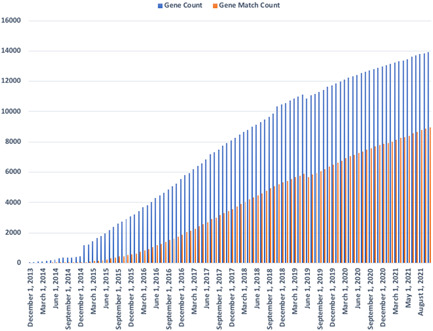
The growth of the number of unique genes and unique gene matches over time
A recent analysis (Wohler, 2021) described the number and types of genes, matches, and publications in GeneMatcher from 2013 to 2020. The same paper introduced VariantMatcher (variantmatcher.org), which is a queriable collection of rare variants and phenotypes from > 6000 individuals (affected and family members) enrolled in various projects.
GeneMatcher is a founding member of the Matchmaker Exchange (MME) (Philippakis et al., 2015), having worked with other sites to create the original API (Buske et al., 2015). Today it is part of a system built for one‐sided matching in which repositories of rare disease patient and family data can be queried for genes and variants of interest (Wohler, 2021 and Rodrigues et al., 2022).
The remarkable success of GeneMatcher can be attributed to its ease of use. A person wishing to enter a candidate gene or genes into GeneMatcher needs only to create an account, register an identifier for the case, and enter a human gene by symbol, which is then cross‐referenced to an Entrez and Ensembl Gene ID. The maximum number of allowed candidate genes per case is 10. While this is all that is required, a GeneMatcher entry can also include the genomic location of a variant and its genomic build, its zygosity, the mode of inheritance, and the presumed functional effect of the variant. In addition, the phenotypic features, based on PhenoDB (Hamosh et al., 2013), and the diagnosis, based on OMIM phenotype number (Amberger et al., 2015), can be added. If information beyond the gene symbol is entered the user can decide whether to match only on gene (which is required) or add genomic location, OMIM (omim.org) Phenotype number, or phenotypic features to the match criteria. In addition, the submitter can choose to restrict matches to researchers, healthcare providers, or patients/families. Once these selections have been made, the submitter can choose to look for matches in any of the Matchmaker exchange (MME) connected nodes, including DECIPHER (Foreman et al., 2022), PhenomeCentral (Osmond et al., 2022), seqr (Pais et al., 2022), MyGene2 (Chong et al., 2016), IRUD (Adachi et al., 2017), Patient Matcher (Rasi et al., 2022), RD‐Connect (this issue), and ModelMatcher (Harnish et al., 2022). If phenotypic features were included in a submission, they are converted to mapped HPO (hpo.jax.org) terms (Wohler et al., 2021) when sent across the MME API. A submission that matches immediately generates an email to all submitters with a match and includes the information provided with each submission. Follow‐up after matching is at the discretion of the submitters.
GeneMatcher also allows submission of candidate genes being investigated in model organisms (which are entered through the human orthologue, but species is designated). These submissions have led to successful matches and publications as the linking of human cases with an animal model with a similar phenotype generates functional data that strengthens the evidence for the new gene‐phenotype association. The vast majority of animal models submitted are in the mouse (Mus musculus – 210 or 73.9% of submissions with an organism other than human); however, 11 other organisms are represented including zebrafish (D. rerio – 27 or 9.5%), fruit fly (D. melanogaster – 24 or 8.5%), nematodes (C. elegans – 8 or 2.8%) and others (1–3 or 0.4%–1.1%) (genematcher.org/statistics). These 284 nonhuman submissions comprise 346 genes.
2. INTERNATIONAL PARTICIPATION
While individuals from 94 countries have created an account in GeneMatcher (Figure 2), submissions originate from only 80 countries (Figure 3). Perhaps account creators from the other 14 countries thought that GeneMatcher was searchable and did not have a case to enter to match or chose not to. Among the 80 countries with submissions, 45 are a high income (HI), 20 are upper middle income (UMI), 10 are lower middle income (LMI), and three are low income (LI) countries as defined by the World Bank. Figure 3 shows submissions by country. Remarkably, five of the largest 20 countries by population: Nigeria (LMI), Bangladesh (LMI), Ethiopia (LI), Philippines (LMI), and DR Congo (LI)) do not have a single submission in GeneMatcher. Figure 4 shows submissions by country versus population and organized by World Bank economic status. The 15 countries with the largest number of submissions, which each number > 600, include USA, France, Germany, Netherlands, Italy, United Kingdom, Canada, China, Israel, Switzerland, Spain, Australia, India, Belgium, and Denmark in order from largest to smallest. The three most populous countries in the world: China, India, and the US rank 5, 16, and 1, respectively in a number of submitters, and 9, 14, and 1, respectively in a number of submissions. Iran is the outlier lower‐middle‐income country with more than 147 submissions and Iceland and Estonia have the highest number of submissions per capita. Despite the socioeconomic challenges, there are submissions from Burkina Faso, South Sudan, and Yemen, all low‐income countries.
Figure 2.
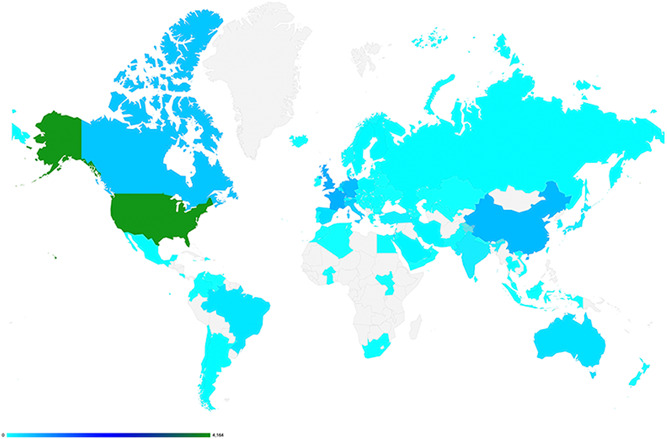
A heat map of the submitters by country as of October 1, 2021. Individuals from 94 countries have created accounts in GeneMatcher
Figure 3.
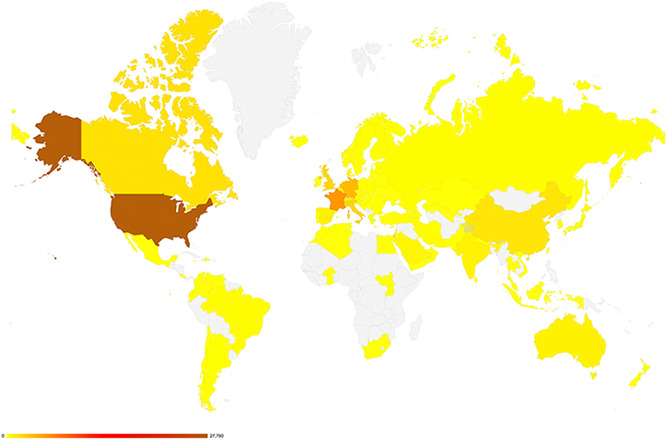
A heat map of submissions by country as of October 1, 2021. Submissions to GeneMatcher come from individuals in 80 countries
Figure 4.
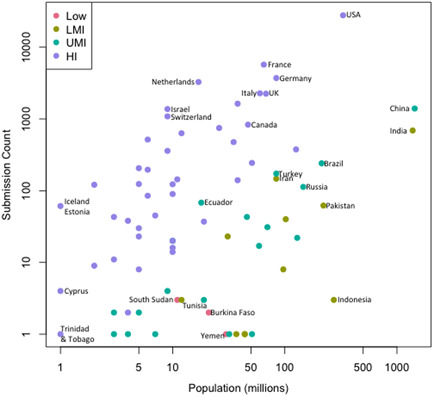
A plot of country by population size and submission number coded by economic status as defined by the World Bank (https://datatopics.worldbank.org/world-development-indicators/the-world-by-income-and-region.html), accessed on October 9, 2021. Population data are through the end of 2020 and the source is (https://www.worldometers.info/world-population/population-by-country/) accessed on October 9, 2021. Submission count is from GeneMatcher from October 1, 2021. Plots were generated using RStudio. Data were log‐transformed before analysis to normalize the data
3. PUBLICATIONS
Over 500 papers cite one of the two primary GeneMatcher publications (Sobreira, Schiettecatte, Boehm, et al., 2015; Sobreira, Schiettecatte, Valle, et al., 2015) (genematcher.org/publications). Figure 5 shows the growth per year of publications citing GeneMatcher and the proportion that describes novel disease genes that grew out of matches made in the tool. Wohler et al. (2021) performed an analysis of the proportion of genes matching and the time lag to publication of genes submitted to GeneMatcher using data through June of 2020. They reported 302 papers that described new disease genes that cited GeneMatcher, while in OMIM more than 445 new disease genes included a GeneMatcher connection (Wohler 2021). Due to the time necessary to collate clinical information on patients with matching genes and phenotypes and the increased stringency of journals now requiring functional evidence to prove causality in novel disease gene discoveries, we anticipate that many more papers will be published in the next few years based on matches already made.
Figure 5.
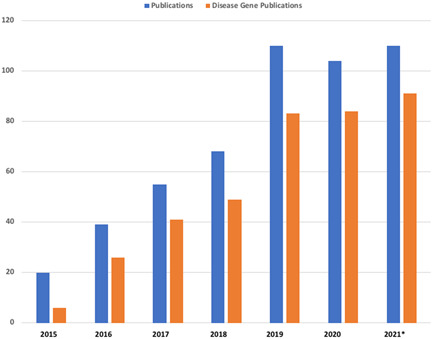
Growth in the number of publications (blue) and proportion of those reporting novel disease genes (orange) that cite GeneMatcher (genematcher.org/publications, *accessed October 9, 2021, so data for 2021 are incomplete)
4. CONCLUSIONS
GeneMatcher has proved to be an efficient and effective tool to bring together researchers, clinicians, and patients with an interest in the same gene. These matches have led to many novel discoveries and new collaborations around the world. GeneMatcher will continue to connect to other nodes as they are added to the MME. Furthermore, we encourage the submission of phenotypic information as well as variant information to expedite understanding of the validity of a gene match as a phenotype and zygosity match. While we are immensely pleased with the international participation in GeneMatcher and its carry‐over effects into the MME, we recognize that much work needs to be done and resources applied to clinically characterize and sequence individuals and their families with rare diseases around the world, so that the benefit of the advances in genomic medicine available to those in high‐income nations can accrue to all. Furthermore, the young age and large populations of lower middle income and lowincome countries suggest that there are many more novel disease genes to be identified in those populations. The biological insights and potential for therapies that follow each new disease gene discovery mandate that no gene and no patient be left behind.
WEB RESOURCES
http://omim.org http://hpo.jax.org http://hpo.jax.org http://Genematcher.org
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
We thank Shira Ziegler, MD, PhD for her help with programming in R to generate Figure 4. Funding for GeneMatcher comes from the Sutland‐Pakula Family and from NHGRI through the Baylor‐Hopkins Center for Mendelian Genomics, 1U54HG006542.
Hamosh, A. , Wohler, E. , Martin, R. , Griffith, S. , Rodrigues, da S. E. , Antonescu, C. , Doheny, K. F. , Valle, D. , & Sobreira, N. (2022). The impact of GeneMatcher on international data sharing and collaboration. Human Mutation, 43, 668–673. 10.1002/humu.24350
DATA AVAILABILITY STATEMENT
The data that support the figures of this paper are available from the corresponding author upon request. The data contained within GeneMatcher are closed except to submitters, matching submitters, and the site administrators.
REFERENCES
- Adachi, T. , Kawamura, K. , Furusawa, Y. , Nishizaki, Y. , Imanishi, N. , Umehara, S. , Izumi, K. , & Suematsu, M. (2017). Japan's initiative on rare and undiagnosed diseases (IRUD): Towards an end to the diagnostic odyssey. European Journal of Human Genetics, 25(9), 1025–1028. 10.1038/ejhg.2017.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger, J. S. , Bocchini, C. A. , Schiettecatte, F. , Scott, A. F. , & Hamosh, A. (2015). OMIM.org: Online Mendelian Inheritance in Man (OMIM), an online catalog of human genes and genetic disorders. Nucleic Acids Research, 43(Database issue), D789–D798. 10.1093/nar/gku1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske, O. J. , Schiettecatte, F. , Hutton, B. , Dumitriu, S. , Misyura, A. , Huang, L. , Hartley, T. , Girdea, M. , Sobreira, N. , Mungall, C. , & Brudno, M. (2015). The Matchmaker Exchange API: Automating patient matching through the exchange of structured phenotypic and genotypic profiles. Human Mutation, 36(10), 922–927. 10.1002/humu.22850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J. X. , Yu, J. H. , Lorentzen, P. , Park, K. M. , Jamal, S. M. , Tabor, H. K. , Rauch, A. , Saenz, M. S. , Boltshauser, E. , Patterson, K. E. , Nickerson, D. A. , & Bamshad, M. J. (2016). Gene discovery for Mendelian conditions via social networking: De novo variants in KDM1A cause developmental delay and distinctive facial features. Genetics in Medicine, 18(8), 788–795. 10.1038/gim.2015.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J. , Brent, S. , Perrett, D. , Bevan, A. P. , Hunt, S. E. , Cunningham, F. , Hurles, M. E. , & Firth, H. V. (2022). DECIPHER: Supporting the interpretation and sharing of rare disease phenotype‐linked variant data to advance diagnosis and research. Human Mutation. 10.1002/humu.24340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh, A. , Sobreira, N. , Hoover‐Fong, J. , Sutton, V. R. , Boehm, C. , Schiettecatte, F. , & Valle, D. (2013). PhenoDB: A new web‐based tool for the collection, storage, and analysis of phenotypic features. Human Mutation, 34(4), 566–571. 10.1002/humu.22283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnish, J. M. , Li, L. , Rogic, S. , Poirier‐ Morency, Kim, G. , Undiagnosed Diseases Network, S. Y. , Boycott, K. M. , Wangler, M. F. , Bellen, H. J. , Hieter, P. , Pavlidis, P. , Liu, Z. , & Yamamoto, S. (2022). Model matcher: A scientist‐centric online platform to facilitate collaborations between stakeholders of rare and undiagnosed disease research. Human Mutation, 1–17. 10.1002/humu.24364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond, M. , Hartley, T. , Johnstone, B. , Andjic, S. , Girdea, M. , Gillespie, M. , Buske, O. , Dumitriu, S. , Koltunova, V. , Ramani, A. , Boycott, K. M. , & Brudno, M. (2022). PhenomeCentral: 7 years of rare disease matchmaking. Human Mutation. 10.1002/humu.24348 [DOI] [PubMed] [Google Scholar]
- Pais, L. S. , Snow, H. , Weisburd, B. , Zhang, S. , Baxter, S. M. , DiTroia, S. , O'Heir, E. , England, E. , Chao, K. R. , Lemire, G. , Osei‐Owusu, I. , VanNoy, G. E. , Wilson, M. , Nguyen, K. , Arachchi, H. , Phu, W. , Solomonson, M. , Mano, S. , O'Leary, M. , & O'Donnell‐Luria, A. (2022). seqr: A webbased analysis and collaboration tool for rare disease genomics. Human Mutation, 1–11. 10.1002/humu.24366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis, A. A. , Azzariti, D. R. , Beltran, S. , Brookes, A. J. , Brownstein, C. A. , Brudno, M. , Brunner, H. G. , Buske, O. J. , Carey, K. , Doll, C. , Dumitriu, S. , Dyke, S. O. , den Dunnen, J. T. , Firth, H. V. , Gibbs, R. A. , Girdea, M. , Gonzalez, M. , Haendel, M. A. , Hamosh, A. , … Rehm, H. L. (2015). The Matchmaker Exchange: A platform for rare disease gene discovery. Human Mutation, 36(10), 915–921. 10.1002/humu.22858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasi, C. , Nilsson, D. , Magnusson, M. , Lesko, N. , Lagerstedt‐Robinson, K. , Wedell, A. , Lindstrand, A. , Wirta, V. , & Stranneheim, H. (2022). Patient matcher: A customizable python‐based open‐source tool for matching undiagnosed rare disease patients via the Matchmaker Exchange network. Human Mutation. 10.1002/humu.24358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, E. D. S. , Griffith, S. , Martin, R. , Antonescu, C. , Posey, J. E. , Coban‐Akdemir, Z. , Jhangiani, S. N. , Doheny, K. F. , Lupski, J. R. , Valle, D. , Bamshad, M. J. , Hamosh, A. , Sheffer, A. , Chong, J. X. , Einhorn, Y. , Cupak, M. , … Sobreira, N. (2022). Variant‐level matching for diagnosis and discovery: Challenges and opportunities. Human Mutation, 1–9. 10.1002/humu.24359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira, N. , Schiettecatte, F. , Boehm, C. , Valle, D. , & Hamosh, A. (2015). New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, A web‐based tool for linking investigators with an interest in the same gene. Human Mutation, 36(4), 425–431. 10.1002/humu.22769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira, N. , Schiettecatte, F. , Valle, D. , & Hamosh, A. (2015). GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Human Mutation, 36(10), 928–930. 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohler, E. , Martin, R. , Griffith, S. , Rodrigues, E. D. S. , Antonescu, C. , Posey, J. E. , Coban‐Akdemir, Z. , Jhangiani, S. N. , Doheny, K. F. , Lupski, J. R. , Valle, D. , Hamosh, A. , & Sobreira, N. (2021). PhenoDB, GeneMatcher and VariantMatcher, tools for analysis and sharing of sequence data. Orphanet Journal of Rare Diseases, 16(1), 365. 10.1186/s13023-021-01916-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the figures of this paper are available from the corresponding author upon request. The data contained within GeneMatcher are closed except to submitters, matching submitters, and the site administrators.


