Abstract
Yeast strains disrupted for ATH1, which encodes vacuolar acid trehalase, have been reported to grow to higher cell densities than reference strains. We showed that the increase in cell density is due to the URA3 gene introduced as a part of the disruption and concluded that the misinterpretation is a result of not using a control strain with matching auxotrophic markers.
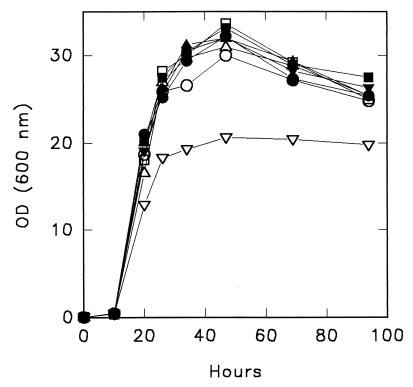
The ATH1 gene of Saccharomyces cerevisiae codes for vacuolar acid trehalase (1). Kim et al. (2) reported that disruption of this gene resulted in an almost twofold increase in the cell density attained in a yeast extract- and peptone-based medium with a high glucose concentration. These authors used URA3 as a marker gene for disruption of ATH1 and compared the ura3 ATH1 parent strain (uracil auxotroph but wild type for ATH1) with the ath1::URA3 disruptant (uracil prototroph but disrupted for ATH1). To separate the effects of uracil prototrophy and ATH1 disruption, we deleted ATH1 in a prototrophic strain (FY4) isogenic to S288C (9) by using a kanMX module as the selectable marker (8). Deletion of ATH1 was confirmed by PCR and by the inability of the mutant (DA1) to utilize trehalose as a sole carbon source (3). Growth of the strains was compared in YP medium (1% Bacto Yeast Extract [Difco], 2% Bacto Peptone [Difco]) containing 25% glucose. The cell density attained by DA1 was comparable to the cell density of FY4, which indicated that the Δath1 mutation did not affect growth under these conditions, and was greater than the cell density of FY3, an isogenic ura3-52 mutant of FY4 (9). Growth of RC47, a Ura+ transformant of FY3, was comparable to growth of FY4, confirming that the poor growth of FY3 was due to uracil auxotrophy. When the medium was supplemented with 40 mg of uracil per liter, the strain FY3 cell density was the same as the cell density of the Ura+ strains (Fig. 1). Kim et al. (2) compared the growth of the ATH1 parent strain (SEY6210), a uracil auxotroph, with the growth of the Δath1::URA3 mutant (MDY3), a uracil prototroph. Our results suggest that the increased growth which Kim et al. attributed to the Δath1 mutation is due to the uracil prototrophy of the Δath1 mutant and that differences in growth should disappear when the uracil limitation is relieved.
FIG. 1.
Growth of S. cerevisiae strains in YP medium containing 25% glucose without (open symbols) or with (solid symbols) additional uracil (40 mg/liter) at 30°C. The following strains were tested: FY4 (○) and RC47 (□), which are prototrophic for uracil and wild type for ATH1 (URA3 ATH1); DA1 (▵), which is prototrophic for uracil and deleted in ATH1 (URA3 Δath1::kanMX); and FY3 (▿), which is auxotrophic for uracil and wild type for ATH1 (ura3-52 ATH1). OD (600 nm), optical density at 600 nm.
To study the effect of disrupting a gene, ideally the disruptant should differ from the control strain only with respect to the disrupted gene. However, in many gene disruption studies the disruptant and control strains differ in two respects, namely, in the disrupted gene and in the auxotrophic marker used for the disruption. Many researchers assume that the auxotrophic marker is phenotypically neutral, which may often be true. However, if the auxotrophic supplement is limiting, the absence of control strains with matching auxotrophy can result in significant errors. The fact that uracil is present in a limiting amount in YP medium is not well appreciated; the reason for this may be the fact that in standard YPD medium containing 2% glucose the cell densities of the Ura+ and Ura− strains may not be strikingly different. For example, the final cell density of FY3 in YPD medium was about 80% of the final cell density of FY4. When the medium was supplemented with additional uracil, FY3 grew to the same cell density as FY4, implying that in standard YPD medium cells may reach the stationary phase due to starvation for uracil or glucose, depending on the auxotrophy of the strain.
Even if uracil is present in the medium, under some conditions the cells are not necessarily able to take it up (6). Starvation for a nitrogen, phosphate, or carbon source can trigger rapid degradation of uracil permease and a loss of uracil uptake (7). Thus, the physiological results may be different for uracil auxotrophs and prototrophs, even if the medium is supplemented with sufficient uracil. Similarly, auxotrophic markers involved in amino acid biosynthesis are considered ill-suited for disrupting genes in intermediary carbon metabolism, as amino acid biosynthesis is an integral part of this metabolism (5). We suggest that markers such as the kanMX module conferring G418 resistance (8) should be used as alternatives to auxotrophic markers in gene disruption studies (4).
Acknowledgments
We thank Anand K. Bachhawat for helpful discussions, Fred Winston for providing the FY series of yeast strains, and Achim Wach for providing the pFA6-kanMX4 plasmid.
Footnotes
Communication no. 007/98 of the Institute of Microbial Technology, Chandigarh, India.
REFERENCES
- 1.Destruelle M, Holzer H, Klionsky D J. Isolation and characterization of a novel yeast gene, ATH1, that is required for vacuolar acid trehalase activity. Yeast. 1995;11:1015–1025. doi: 10.1002/yea.320111103. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Alizadeh P, Harding T, Hefner-Gravink A, Klionsky D J. Disruption of the yeast ATH1 gene confers better survival after dehydration, freezing, and ethanol shock: potential commercial applications. Appl Environ Microbiol. 1996;62:1563–1569. doi: 10.1128/aem.62.5.1563-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nwaka S, Mechler B, Holzer H. Deletion of the ATH1 gene in Saccharomyces cerevisiae prevents growth on trehalose. FEBS Lett. 1996;386:235–238. doi: 10.1016/0014-5793(96)00450-4. [DOI] [PubMed] [Google Scholar]
- 4.Oliver S. A network approach to the systematic analysis of yeast gene function. Trends Genet. 1996;12:241–242. doi: 10.1016/0168-9525(96)30053-x. [DOI] [PubMed] [Google Scholar]
- 5.Pronk J T, Steensma H Y, van Dijken J P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Varela J C S, van Beekvelt C, Planta R J, Mager W H. Osmostress-induced changes in gene expression. Mol Microbiol. 1992;6:2183–2190. doi: 10.1111/j.1365-2958.1992.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 7.Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- 8.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 9.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]