Summary
NOTCH signaling is a key regulator involved in maintaining intestinal stem cell (ISC) homeostasis and for balancing differentiation. Using single-cell transcriptomics, we observed that OLFM4, a NOTCH target gene present in ISCs, is first expressed at 13 weeks post-conception in the developing human intestine and increases over time. This led us to hypothesize that the requirement for NOTCH signaling is acquired across human development. To test this, we established a series of epithelium-only organoids (enteroids) from different developmental stages and used γ-secretase inhibitors (dibenzazepine [DBZ] or DAPT) to functionally block NOTCH signaling. Using quantitative enteroid-forming assays, we observed a decrease in enteroid forming efficiency in response to γ-secretase inhibition as development progress. When DBZ was added to cultures and maintained during routine passaging, enteroids isolated from tissue before 20 weeks had higher recovery rates following single-cell serial passaging. Finally, bulk RNA sequencing (RNA-seq) analysis 1 day and 3 days after DBZ treatment showed major differences in the transcriptional changes between developing or adult enteroids. Collectively, these data suggest that ISC dependence on NOTCH signaling increases as the human intestine matures.
Keywords: intestine, enteroid, organoid, intestinal stem cell, NOTCH
Highlights
-
•
NOTCH signaling in the intestinal crypt increases as human development progresses
-
•
OLFM4 expression is a hallmark of intestinal stem cell development and maturation
-
•
Immature intestinal stem cells can be cultured in the presence of γ-secretase inhibitors
This study used single-cell transcriptomics to show that developing intestinal stem cells increase NOTCH-signaling target genes as the human intestine matures. A series of epithelium-only organoids from the developing and adult intestine were established, and functional experiments showed that immature intestinal stem cells are not perturbed by NOTCH inhibition, while more mature and adult stem cells require NOTCH for growth and stem cell maintenance.
Introduction
The single-layered epithelium lining the crypt-villus axis of the intestine undergoes continuous stem-cell renewal and differentiation. Previous work carried out in mice has shown that Notch signaling has the ability to regulate intestinal epithelial stem cell maintenance, progenitor cell proliferation, and differentiation (Carulli et al., 2015). Notch signaling plays an essential role in regulating epithelial cell fate (Fre et al., 2005; Jensen et al., 2000; Stanger et al., 2005; Pellegrinet et al., 2011; Riccio et al., 2008; van der Flier and Clevers, 2009a; VanDussen et al., 2012), with activation of Notch signaling resulting in an increased progenitor pool (Fre et al., 2005) and ablation of NOTCH signaling leading to a loss of stem cells, reduced proliferation, and an increase in secretory lineage differentiation (Pellegrinet et al., 2011; VanDussen et al., 2012; van Es et al., 2005). Much of our understanding of Notch signaling comes from studies of the adult murine intestine (Carulli et al., 2015; Tsai et al., 2014; Tian et al., 2015). In contrast, much less is known about how NOTCH functions in the human intestine or how it functions during intestinal development. Here, we interrogated NOTCH signaling in the intestine across the continuum of human development and adult human small intestine using single-cell RNA sequencing (scRNA-seq), immunofluorescence (IF), and fluorescence in situ hybridization (FISH) and isolated crypt epithelium from the human duodenum as a primary tool for functional studies.
Olfm4 expression is a robust marker for Lgr5+ crypt base columnar stem cells in mice (VanDussen et al., 2012; Chen et al., 2017). In adults, it is expressed throughout the whole small intestinal epithelium in the crypts (van der Flier and Clevers., 2009a; Schuijers et al., 2014), and Olfm4 has been validated as a direct target of Notch signaling (VanDussen et al., 2012; Tsai et al., 2014). Previous studies of the developing human intestine and pluripotent-stem-cell-derived human intestinal organoids have suggested that OLFM4 is expressed in the adult intestine but only weakly expressed in the developing intestine (Finkbeiner et al., 2015). The current study builds upon our previous observation that OLFM4 was not expressed in the crypt of the human fetal intestine at early time points but that its expression increased as developmental time progressed (Yu et al., 2021; Holloway et al., 2021). In the current study, we show that OLFM4 is absent before ∼13 weeks of fetal development (post-conception), with expression increasing as development continues. Thus, we hypothesized that the requirement for NOTCH signaling in the intestinal stem cell (ISC) compartment increases as human development progress. Using enteroid cultures and intestinal explants, we found that inhibition of γ-secretase, a key processing enzyme in NOTCH signaling, had less influence over fetal intestinal epithelial proliferation compared with adult. Changes in epithelial proliferation in enteroids established from intestinal specimens earlier than 60 days (∼8.5 weeks post-conception) are not influenced by NOTCH inhibition, whereas proliferation was affected in enteroids derived from older specimens. Taken together, this study supports a model where NOTCH signaling is uncoupled from ISC self-renewal prior to 60 days of human fetal development and where acquisition of OLFM4 expression and NOTCH signaling activity are hallmarks of ISC development and maturation.
Results
OLFM4 gene expression increases concurrently with age
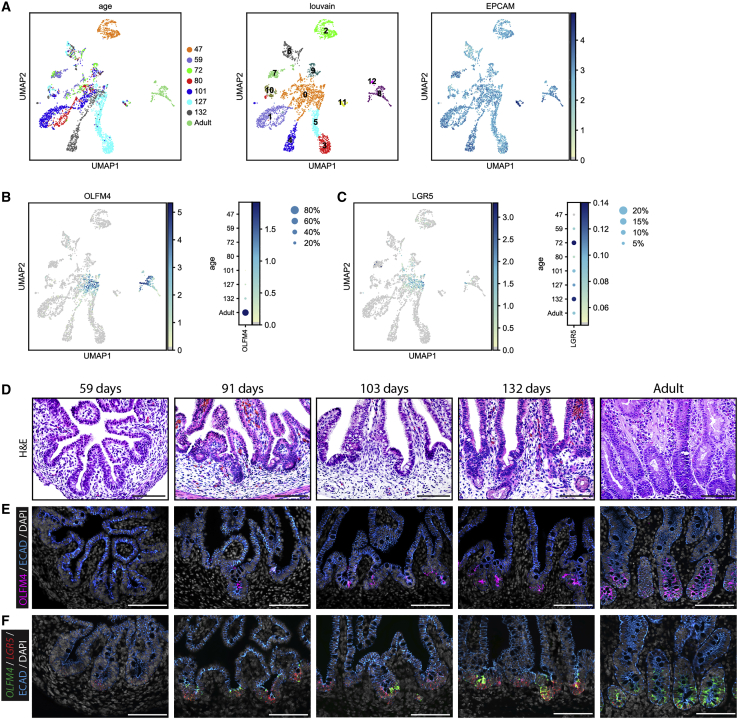
We have previously reported that acquisition of OLFM4 is a hallmark of intestinal maturation based on studies using pluripotent-stem-cell-derived human intestinal organoids grown in vitro or transplanted in vivo (Finkbeiner et al., 2015). To investigate whether this finding is recapitulated across the human lifespan, we investigated OLFM4 expression in fetal tissues from 47 to 132 days post-conception and in adult tissues using scRNA-seq of the human intestinal epithelium (Figures 1A and 1B). Whole thickness fetal duodenal samples (n = 7) and adult duodenal biopsy samples (n = 2) were dissociated and sequenced (see experimental procedures), and the intestinal epithelium was identified by EPCAM expression (Figures S1A and S1B). These EPCAM+ cell clusters were also enriched for five other specific epithelial markers and the intestinal marker CDX2 (Figures S1C and S1D). We computationally extracted and re-clustered 3,665 EPCAM+ epithelial cells (Figure 1A). By interrogating gene expression by age, we found that OLFM4 is undetected by scRNA-seq from 47 days to 72 days, and expression is detected in a very small subset of cells starting at 80 days and increases over time (Figure 1B; Table S1). In contrast, the intestinal stem and progenitor cell marker LGR5 was expressed throughout all stages examined (Figure 1C). To validate the scRNA-seq findings, we carried out histological examination (Figure 1D), OLFM4 protein staining by IF (Figure 1E), and OLFM4 and LGR5 mRNA by FISH (Figure 1F). OLFM4 expression was observed beginning at 91 days of fetal development (Figures 1E, 1F, S2A, and S2B). Consistent with scRNA-seq data, protein and mRNA expression of OLFM4 was absent at 59 days, whereas LGR5 was detected in crypt regions (Figures 1E and 1F). Weak protein and mRNA staining was detected by 91 days in the crypt regions and became more easily detectible as development progressed (Figures 1E and 1F). Protein, ISH, and FISH staining results for OLFM4 expression were confirmed in an independent time series analysis (Figure S2; see also Figure 6B).
Figure 1.
OLFM4 gene expression increases concurrently with age
(A) UMAP visualizations of ages and clusters from EPCAM+ cells by scRNA-seq analysis from fetal tissue 47–132 days post-conception, corresponding to Figure S1.
(B) Feature plot and dot plot of OLFM4+ cells from extracted epithelium across time points corresponding to (A).
(C) Feature plot and dot plot of LGR5+ cells from extracted epithelium across time points corresponding to (A).
(D) H&E staining of fetal duodenum at 59, 91, 103, and 132 days post-conception and 65-year-old adult duodenum biopsy.
(E) Immunofluorescence (IF) protein staining for OLFM4 (pink) and ECAD (blue) with DAPI (gray) on fetal duodenum.
(F) Fluorescence in situ hybridization (FISH) staining for OLFM4 (green), LGR5 (red), and IF staining for ECAD (blue) with DAPI (gray) on fetal duodenum (n = 10 [n = 2 biological replicates per time point shown]).
Scale bars represent 100 μm.
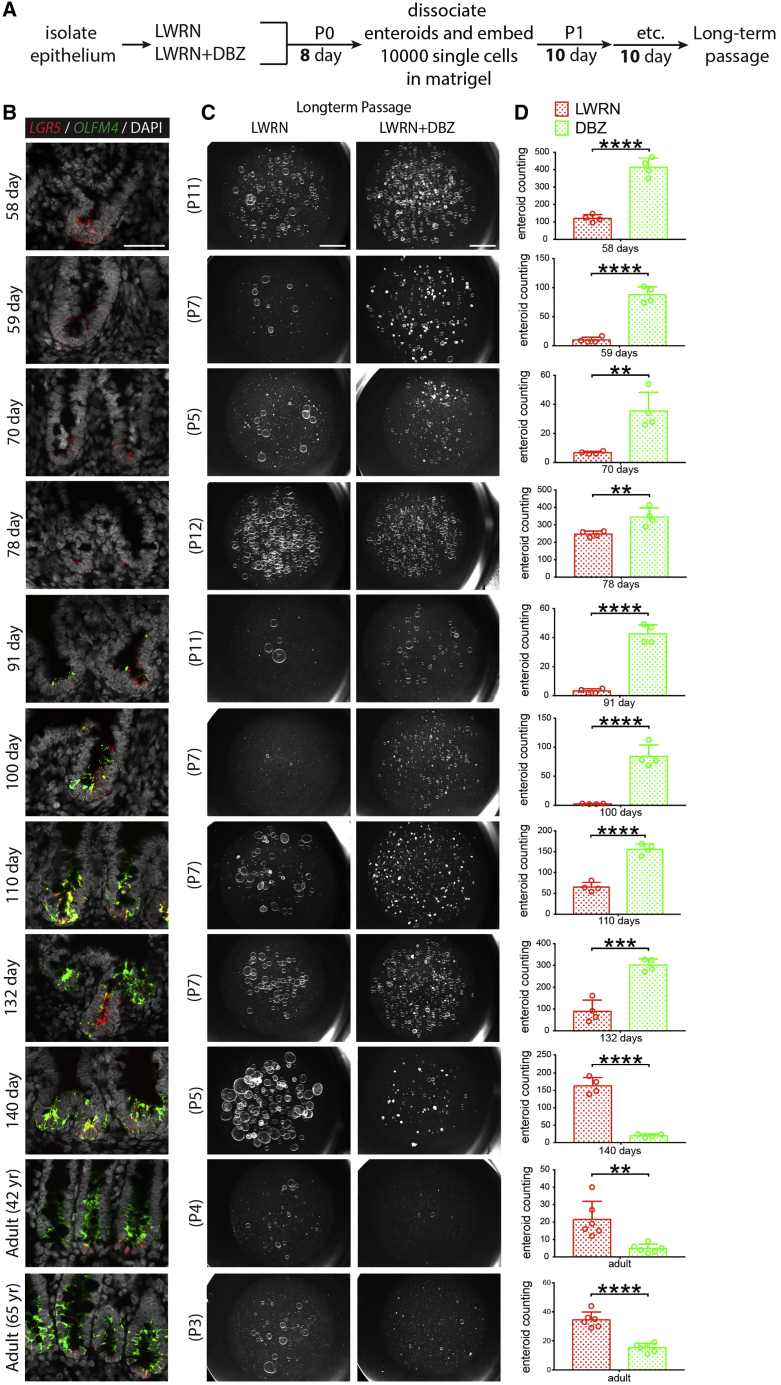
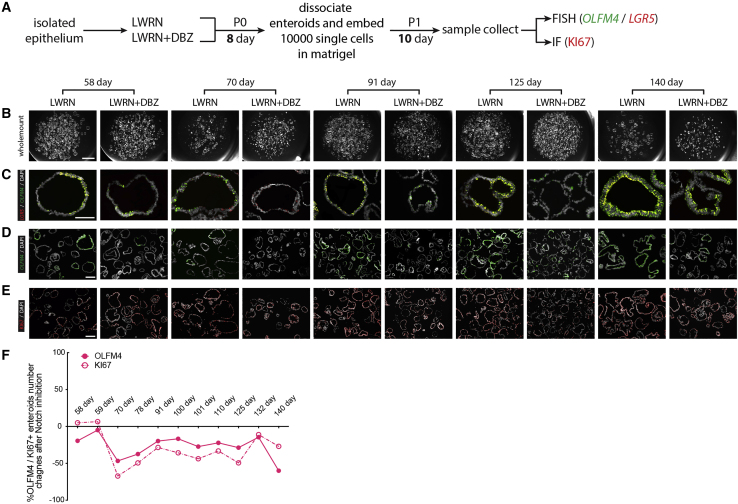
Figure 6.
Chronic γ-secretase inhibition promotes higher stem-cell survival in fetal enteroids younger than 20 weeks post-conception
(A) Experimental schematic for data presented in (B)–(D).
(B) FISH staining for OLFM4 (green) and LGR5 (red) with DAPI (gray) on fetal duodenum aged 58, 59, 70, 78, 91, 100, 110, 132, and 140 days post-conception, as well as 42- and 65-year-old adult duodenum (n = 11 [n = 1 biological replicates per time point]). Scale bar represents 50 μm.
(C) Stereomicroscope images of different passages for respective enteroids time points after LWRN and LWRN + DBZ treatments (n = 3 experimental replicates per time point from n = 1 biological replicate per time point). Scale bars represent 2 mm.
(D) Manual counting of enteroid number from respective time points and passages corresponding to (C) (n = 3 experimental replicates per time point from n = 1 biological replicate per time point). All statistics were analyzed with unpaired t tests by GraphPad Prism 7.0, and data were presented as the mean ± SEM. In all figures, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
NOTCH2 and JAG1 are the major NOTCH functional components in human intestinal stem cells
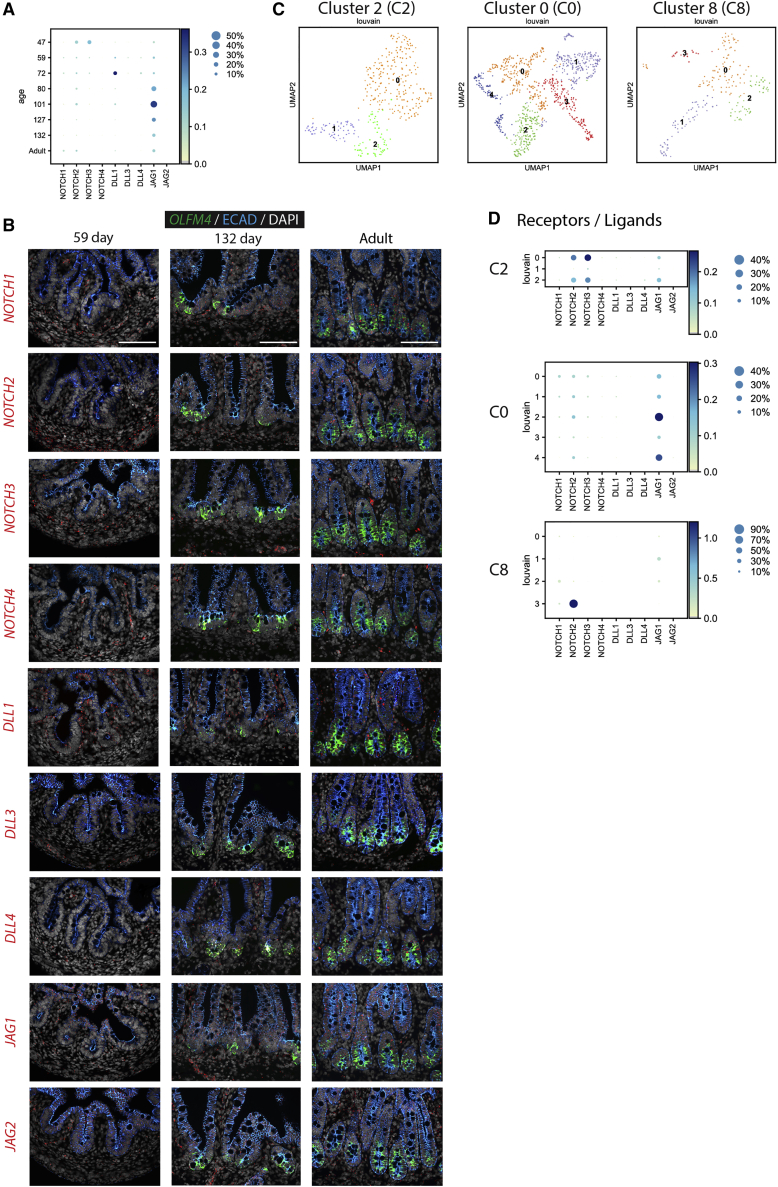
Using scRNA-seq analysis, we interrogated expression of NOTCH receptors and ligands in the primary fetal and adult duodenum (Figure 2A) and found NOTCH2 and JAG1 are expressed in all stages. NOTCH3, on the other hand, is expressed more robustly in early stages of fetal development. These results are consistent with FISH staining in fetal and adult duodenum (Figure 2B). To further investigate NOTCH components in human intestinal epithelial stem cells, cells that expressed LGR5 or OLFM4 in the epithelium were extracted (cluster 0, 2, and 8 in Figures 1A–1C). Cluster 2 (C2) corresponded to early development, possessing cells primarily from day 47 with small contribution from day 59 and day 80; cluster 0 (C0) corresponded to mid-development, possessing cells from day 59, day 72, day 80, day 101, day 127, and day 132; and cluster 8 (C8) corresponded to the adult stage, possessing only cells from the adult (Figure 2C). Again, sub-clustering revealed that NOTCH2 and JAG1 are expressed in all three groups, whereas NOTCH3 was enriched in the early-stage stem cells (C2), was weakly expressed in mid-staged stem cells, and was absent in adult stem cells (C8; Figures 2D and S3B).
Figure 2.
NOTCH2 and JAG1 are the major NOTCH functional components in human intestinal epithelial stem cells
(A) Dot plots of scRNA-seq analysis of Notch components from fetal and adult duodenum.
(B) FISH staining for OLFM4 (green); NOTCH1, NOTCH2, NOTCH3, NOTCH4, DLL1, DLL3, DLL4, JAG1, and JAG2 (red); and IF staining for ECAD (blue) with DAPI (gray) on fetal duodenum aged 59 and 132 days post-conception and adult duodenum (n = 6 [n = 2 biological replicates per time point shown]).
(C) UMAP plots of cells computationally extracted from cluster 2 (C2), cluster 0 (C0), and cluster 8 (C8) from the analysis of EPCAM+ cells in Figure 1A. All cells within each cluster were computationally extracted and included in subsequent cluster-specific analyses.
(D) Dot plots of Notch components from C2, C0, and C8.
Scale bars represent 100 μm.
Short-term γ-secretase inhibition increased stem-cell survival in fetal enteroids compared with adult enteroids
Based on previous literature showing that Olfm4 is a direct Notch signaling target in mice and our data showing increasing OLFM4 expression over developmental time in the human intestine (Figures 1 and S2), we hypothesized that OLFM4 expression may correspond to an acquisition of NOTCH signaling responsiveness in the human intestine. In order to test the requirement for NOTCH signaling function in ISCs, we isolated epithelium from fetal duodenal tissue at different stages, before and after OLFM4 expression was observed in vivo, and from adult duodenal biopsies. To block NOTCH, we added the γ-secretase inhibitor dibenzazepine (DBZ) to culture media, since γ-secretase activity is required in the final step of cleaving the NOTCH receptor following activation, allowing the NOTCH intracellular domain (NICD) to move into the nucleus, where it can exert its effects on gene transcription (Milano et al., 2004; van Es et al., 2005; Zheng et al., 2013). In order to determine optimal DBZ dose, we cultured epithelium from fetal and adult stages (142 days and 33 years) with different doses of DBZ and interrogated the NOTCH target gene HES1 by qRT-PCR (Figure S3A). After DBZ treatment for 1 day, we observed a significant decrease at the lowest dose tested (5 μM), which was used for all subsequent experiments.
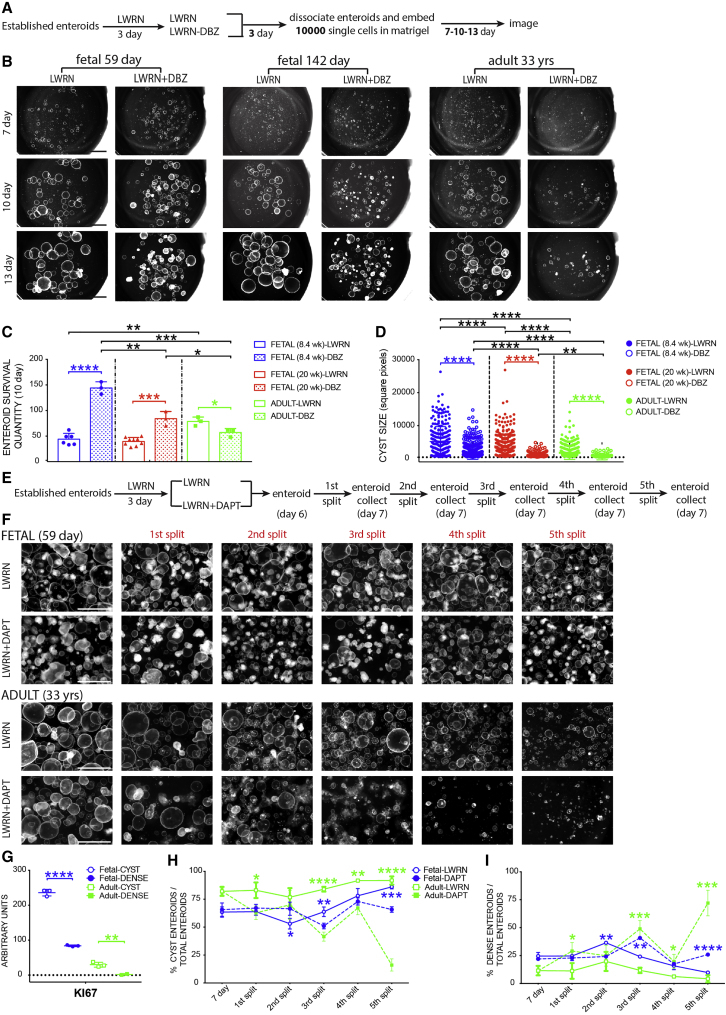
We then carried out a quantitative enteroid-forming assay on enteroids derived from three different stages (59 days, 142 days, and 33 years) to assess stem cell function following DBZ treatment (Figure 3A). Control enteroids were grown in standard “LWRN” growth media possessing WNT3A, RSPO3, NOG, and epidermal growth factor (EGF) (see experimental procedures). Three days prior to passaging, 5 μM DBZ was added to the experimental group (LWRN + DBZ). Following 3 days in control or DBZ media, enteroids were dissociated and 10,000 single cells were re-embedded in Matrigel to investigate the enteroid forming efficiency in each group (Figures 3B and 3C). Enteroids were imaged at 7, 10, and 13 days of incubation, and 10 days images were quantified for the number (Figure 3C) and size of all individual enteroids (Figure 3D) across three groups. The survival efficiency of fetal enteroids was higher in the DBZ treatment groups than in the control groups (3.3-fold higher in 59 days enteroids and 2.1-fold higher in 142 days enteroids), but the opposite was true for the adult enteroids, where enteroid forming efficiency was reduced (0.73-fold change versus control; Figure 3C). We also measured the size of cystic enteroids (omitting non-proliferative, small dense structures from the analysis; see Figure 3G) and observed a smaller size across all groups following DBZ treatment (0.39-, 0.29-, and 0.29-fold versus age-matched controls for 59- and 142-day-old fetal and adult, respectively; Figure 3D). These data suggested that short-term inhibition of NOTCH signaling has different effects on the ability of enteroids to form from single stem cells but has a similar influence on enteroid growth and size following colony formation.
Figure 3.
Short-term γ-secretase inhibition increases stem cell survival in fetal enteroids compared with adult enteroids
(A) Experimental schematic for data presented in (B)–(D).
(B) Stereomicroscope images of enteroids derived from respective fetal ages and adult after LWRN and LWRN + DBZ treatments for 7, 10, and 13 days.
(C) Quantification of enteroid survival after 10 days of treatments corresponding to (B) for all three age groups (n = 3–9 experimental replicates [indicated by individual data points carried out on n = 1 biological replicate]).
(D) Measurements of surface area of individual enteroid size using ImageJ after 10 days of treatments corresponding to (B) for all three age groups.
(E) Experimental schematic for data presented in (F)–(I).
(F) Stereomicroscope images for the passages of enteroids derived from fetal tissue (59 days post-conception) and adult (33 years old) on LWRN and LWRN + DAPT treatments (n = 4 [two biological replicates per each time point]).
(G) Real-time PCR of KI67 of CYST and DENSE enteroids corresponding to (F) (n = 3 experimental replicates on n = 1 biological replicate).
(H) Quantification of the percentage of cystic enteroids to total enteroids over five passages corresponding to (F) (n = 3 experimental replicates on n = 1 biological replicate).
(I) Quantification for percentage of DENSE enteroids to total enteroids over five passages corresponding to (F) (n = 3 experimental replicates on n = 1 biological replicate).
Scale bars represent 2 mm. All statistics were analyzed with unpaired t tests by GraphPad Prism 7.0, and data were presented as the mean ± SEM. In all figures, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Fetal enteroids passage more robustly under chronic γ-secretase treatment
In order to further interrogate the different requirements for NOTCH signaling in the fetal or adult ISCs, we carried out long-term, bulk-enteroid passaging under chronic γ-secretase inhibition by adding 5 μM DAPT to the normal growth media (Figure 3E). Comparing across five passages, we found that adult enteroids were lost by the fifth and terminal passage in the DAPT treatment group, whereas cystic fetal enteroids were still growing robustly at passage 5 (Figure 3F). However, in both groups, we observed that DAPT treatment led to a mixture of cystic structures and small dense structures. To determine the difference between these structures, we manually split fetal and adult enteroids into cystic or dense structures and assayed KI67 by qRT-PCR (Figure 3G). We observed that, in both fetal and adult enteroids, cysts had significantly higher KI67 expression when compared with dense structures. Dense-structured enteroids appear to have differentiated into enterocytes in both fetal and adult groups, as they express higher levels of DPP4 and IAP compared with cystic enteroids, but there was no difference in apoptosis-regulated genes (BCL2, NLRP1, and APAF1) between cystic and dense enteroids (Figure S3C). We further quantified both phenotypes (Figures 3H and 3I) and found that, at the fifth split, γ-secretase inhibition led to a 75% reduction in cystic enteroids in the adult group compared with adult controls, while there was only a 21% reduction in the DAPT-treated fetal group compared with fetal controls (Figure 3H). Accordingly, at the fifth passage, there was an increase of 68% and 16% in dense structures in adult and fetal enteroids when compared with age-matched controls, respectively (Figure 3I). Taken together, these results suggest that enteroid formation and long-term maintenance in the adult are more sensitive to γ-secretase inhibition than are fetal enteroids.
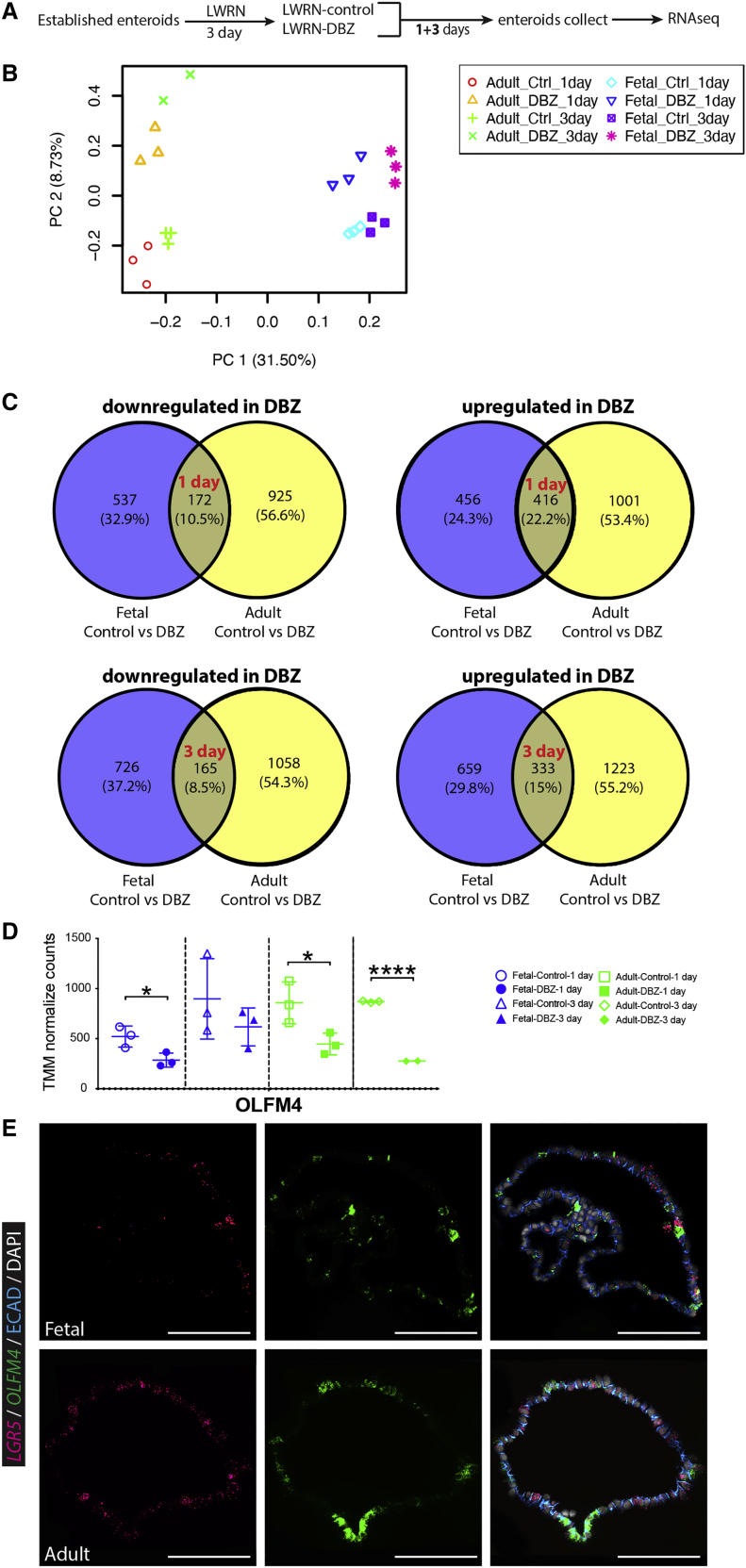
Fetal and adult human enteroids show major transcriptional differences in response to γ-secretase inhibition
Given the different functional responses to γ-secretase inhibition by fetal enteroids compared with adult enteroids, we further investigated the influence of γ-secretase inhibition on gene expression. Adult (33 years) and fetal (59 days) enteroids were maintained after bulk passaging in LWRN growth media for 3 days, at which time DBZ was added to experimental groups and RNA was collected for bulk RNA-seq after 1 and 3 days (Figure 4A). Principal-component analysis (PCA) showed that 31.5% of gene expression variability (i.e., PC1) was based on maturity of the enteroid (fetal or adult; Figure 4B). We attributed 8.73% of gene expression variability (i.e., PC2) to whether enteroids were treated with DBZ, with control and DBZ treatment groups clustering together, although we noted that control and DBZ-treated fetal enteroids showed less spread along the PC2 axis than adult enteroids. When we examined differentially expressed genes (p < 0.05) that were down- or up-regulated in fetal and adult enteroids treated with DBZ (control versus DBZ; Table S2), we found that only a small proportion of the total transcriptional changes induced in fetal or adult were shared. After 1 day of DBZ treatment, only 10.5% of downregulated genes were shared between fetal and adult enteroids compared with 89.5% of transcriptional changes that were unique to one group or the other (fetal 32.9%; adult 56.6%; Figure 4C). Similar trends were observed after 3 days of DBZ treatment and were consistent across down- and up-regulated genes (Figure 4C). In all comparisons, we observed that a larger number of genes were differentially expressed in either direction (up or down) in adult compared with fetal enteroids (Figure 4C). Collectively, these gene expression data support functional data (Figure 3), showing that there are differences in the functional and phenotypic response of adult and fetal enteroids to γ-secretase inhibition.
Figure 4.
Fetal and adult human enteroids show major transcriptional differences in response to γ-secretase inhibition
(A) Experimental schematic for data presented in (B)–(D).
(B) Principal-component analysis (PCA) with PC1 showing maturity of the enteroids and PC2 showing treatments of the enteroids.
(C) Gene expression differentiation was analyzed by Venny 2.1.0 between control and DBZ treatments.
(D) TMM normalized counts of OLFM4 on LWRN-control and LWRN-DBZ for fetal (59 days post-conception) and adult (33 years old) enteroids (n = 3 experimental replicates on n = 1 biological replicate per time point).
(E) FISH staining for OLFM4 (green), LGR5 (pink), and IF staining for ECAD (blue) with DAPI (gray) on fetal (59 days post-conception) and adult (33 years old) enteroids cultured in LWRN (n = 4 [n = 2 biological replicates per time point]). Scale bars represent 100 μm.
All statistics were analyzed with unpaired t tests by GraphPad Prism 7.0, and data are presented as the mean ± SEM. In all figures, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
When interrogating RNA-seq trimmed mean of the M-values (TMM) normalized count data, we unexpectedly observed that OLFM4 was detected in 59-day fetal intestinal enteroids (Figure 4D). This is in contrast to what was observed in in vivo fetal intestinal scRNA-seq data (Figure 1). In addition, OLFM4 was reduced after DBZ treatment in fetal enteroids (1 day of treatment) and adult enteroids (1 day and 3 days of treatment). These data suggested that the in vitro culture environment may lead to precocious OLFM4 expression. To further interrogate the finding that OLFM4 is expressed in fetal enteroids in vitro, we carried out FISH in adult and 59 days fetal enteroids for OLFM4 and LGR5 and co-stained for ECAD (IF; Figure 4E). Consistent with bulk RNA-seq data, we observed OLFM4 expression in 59 days and adult enteroids in vitro. These results suggested that the artificial in vitro niche does not fully mimic the in vivo environment and leads to early OLFM4 expression.
Reduced self-renewal of fetal epithelium cultured with γ-secretase inhibitors coincides with the onset of OLFM4 expression
Because we observed ectopic OLFM4 expression in established fetal enteroids, we carried out a series of inhibition experiments on freshly isolated epithelium from multiple fetal time points prior to the onset of in vivo OLFM4 expression and after in vivo OLFM4 expression (Figure 5). For these experiments, the intestinal epithelium was isolated, placed in Matrigel, and immediately cultured in LWRN (control) or LWRN + DBZ (Figure 5A). Passage 0 (P0) enteroids were established for 8 days, at which time they were dissociated into single cells and passaged (10,000 cells/well). Passaged P1 enteroids were allowed to re-establish for 10 days prior to histological assessment for OLFM4 and LGR5 mRNA expression and KI67 protein expression (representative stages shown in Figure 5B). Enteroids from all stages were established at P0 and successfully single cell passaged to P1 in control and DBZ growth conditions (Figures 5B and S4B). Consistent with previous observations (Figure 4E), we found that OLFM4 was expressed at P1 in all cases (Figures 5C and S4C); however, DBZ culture conditions had reduced OLFM4 expression compared with controls (Figures 5C, 5D, S4C, and S4D). Interestingly, while DBZ treatment reduced OLFM4 expression at all stages, proliferation as assessed by KI67 immunofluorescence was not influenced in early cultures (58 and 59 days), relative to culture starting at 70 days and onward (Figures 5E, 5F, and S4E). These data show that ectopic OLFM4 expression is induced in the in vitro culture environment but that DBZ-induced changes in proliferation are not observed unless in vitro grown enteroids are obtained from intestine that is greater than 70 days post-conception, further suggesting that the early intestinal epithelium is not as sensitive to NOTCH inhibition as later time points, coinciding with OLFM4 expression in vivo.
Figure 5.
Reduced self-renewal of epithelium cultured under γ-secretase inhibition coincides with the onset of OLFM4 expression
(A) Experimental schematic for data presented in (B)–(F).
(B) Stereomicroscope images of P1 enteroids derived from fresh epithelium of fetal tissues aged 58, 70, 91, 125, and 140 days post-conception after LWRN and LWRN + DBZ treatments for 10 days. Scale bar represents 2 mm.
(C) FISH staining for OLFM4 (green) and LGR5 (red) with DAPI (gray) on respective time points corresponding to (B). Scale bar represents 100 μm.
(D) FISH staining for OLFM4 (green) with DAPI (gray) on respective time points corresponding to (B) and (C). Scale bar represents 200 μm.
(E) IF staining for KI67 (red) with DAPI (gray) on respective time points corresponding to (B)–(D) (n = 5 [n = 1 biological replicate per time point]). Scale bar represents 200 μm.
(F) Manual quantification of the percent changes of OLFM4HIGH and KI67HIGH enteroids compared with total enteroids with respective time points corresponding to (D) and (E) (n = 5 [n = 1 biological replicate per time point]; the other time points [n = 6] are present in Figure S4).
Chronic γ-secretase inhibition promotes higher stem-cell survival in fetal enteroids younger than 20 weeks post-conception
We extended experiments where enteroids were established in LWRN or LWRN + DBZ (Figure 5) to include long-term passaging under chronic γ-secretase inhibition. Enteroids were established as described (Figure 5A) from 58, 59, 70, 78, 91, 100, 110, 132, and 140 days specimens as well as from adult specimens (Figure 6). Enteroids were disassociated into single cells, and 10,000 single cells were re-embedded in Matrigel and cultured for 10 days before the next split; this single-cell passaging was carried out until the terminal passage, a time when either LWRN- or DBZ-treated cultures failed to expand further, or at a time where there appeared to be no difference in control versus DBZ treatment following chronic treatment (Figure 6C). Matched primary tissue was also fixed for interrogation via FISH, and OLFM4 and LGR5 expression in primary tissue was interrogated for each enteroid line that was established (Figure 6B). We observed that 58–132 days enteroids cultured in DBZ could be passaged longer (range P5–P12) than enteroids older than 140 days (range P3–P5; Figure 6C), and the number of established enteroids at the final passage for the DBZ groups was significantly higher than those of the control groups for fetal enteroids younger than 140 days (Figure 6D). In contrast, the number of surviving enteroids for 140 days and adult enteroids was lower in the DBZ treatment groups. We did not find a linear relationship between passage number and fetal age. However, we did find that the younger fetal enteroids were able to attain a higher terminal passage than those of both the older fetal and adult enteroids (Figures S4F–S4G'). These data suggest that NOTCH and γ-secretase are dispensable in early stages, but as stem cells age, this dependency increases, concomitant with the onset of OLFM4 expression. It is possible that younger fetal stem cells possess mechanisms to preserve long-term survival and proliferation under prolonged γ-secretase inhibition that older fetal and adult stem cells lack.
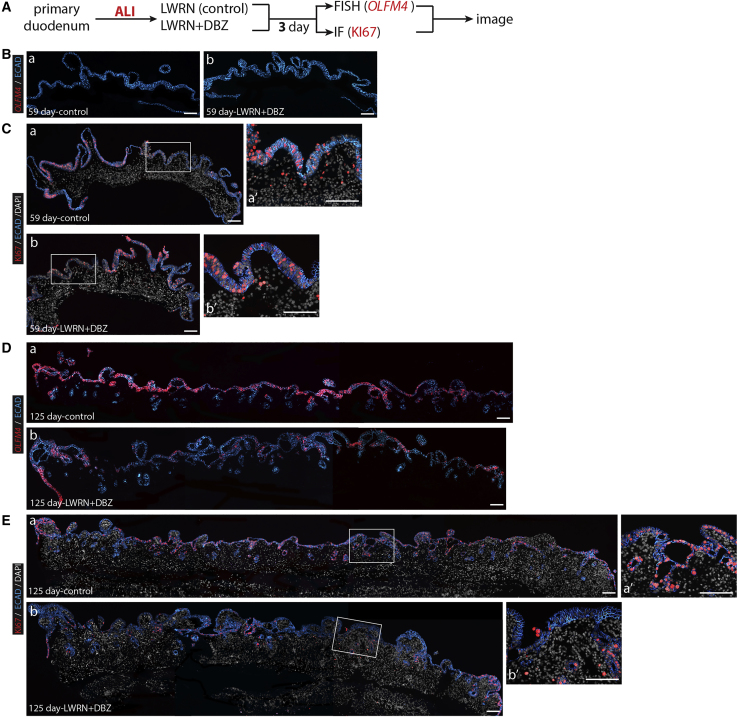
Primary fetal explant cultures respond to γ-secretase inhibition differently across time
To support findings from enteroid experiments, we carried out explant cultures where specimens were cultured in either control (LWRN) media or LWRN + DBZ (Figure 7A). In 59-day primary tissue explants, OLFM4 was not detected by FISH (Figure 7B). We observed that epithelial proliferation, visualized by KI67 immunofluorescence, was not affected by DBZ in 59-day primary fetal tissue following 3 days of explant culture on transwell membranes (air-liquid interface [ALI]; Figure 7C). In contrast, OLFM4 expression was robustly detected in 125 days control conditions and was decreased when cultured in the presence of DBZ (Figure 7D). Similarly, epithelial KI67 protein staining was decreased with γ-secretase inhibition (Figure 7E). These results were consistent with those found in enteroid experiments conducted with independent samples from the same age range.
Figure 7.
Primary fetal explant cultures respond to γ-secretase inhibition differently across fetal development
(A) Experimental schematic for data presented in (B)–(E).
(B) FISH staining for OLFM4 (red) and IF staining for ECAD (blue) with DAPI (gray) on fetal duodenum 59 days post-conception after LWRN (a) and LWRN + DBZ (b) treatments. Scale bars represent 200 μm.
(C) IF staining for KI67 (red) and ECAD (blue) with DAPI (gray) on fetal duodenum at 59 days post-conception after LWRN (a) and LWRN + DBZ (b) treatments. Scale bars represent 200 μm. (aʹ) represents high magnification of (a), and (bʹ) represents high magnification of (b) (n = 2 biological replicates). Scale bars represent 100 μm.
(D) FISH staining for OLFM4 (red) and IF staining for ECAD (blue) with DAPI (gray) on fetal duodenum at 125 days post-conception after LWRN (a) and LWRN + DBZ (b) treatments. Scale bars represent 200 μm.
(E) IF staining for KI67 (red) and ECAD (blue) with DAPI (gray) on fetal duodenum at 125 days post-conception after LWRN (a) and LWRN + DBZ (b) treatments. Scale bars represent 200 μm. (aʹ) represents high magnification of (a), and (bʹ) represents high magnification of (b) (n = 1 biological replicate). Scale bars represent 100 μm.
Discussion
The expression of OLFM4 has been found in both mouse small intestine (Dalerba et al., 2007) and human intestine, including the colon (van der Flier et al., 2009c). In this study, we found that OLFM4 expression emerges in 90-day fetal tissue with a decreasing gradient from the proximal to distal axis that also increases across developmental time (Figures 1 and S2). Previous studies have established both LGR5 (Barker and Clevers, 2007a, 2007b) and OLFM4 as ISC markers (van der Flier et al., 2009b). LGR5+ cells can be characterized as longer lived cycling stem cells, whereas OLFM4 expression is more robustly distributed in crypt base columnar (CBC) stem cells and in the transit amplifying (TA) zone (Figure 1). In addition, OLFM4 has been found to be the target gene of Notch, WNT, and the nuclear factor κB (NF-κB) pathway (Liu and Rodgers, 2016). The decreased survival of 91-day fetal enteroids as a consequence of NOTCH inhibition at P0 reveals that OLFM4 might serve as a marker to signify the beginning of NOTCH dependency on the ISC compartment.
The role of Notch signaling in ISC homeostasis has been studied extensively. In this study, we used human primary tissues and purified epithelium-only enteroids to show the distribution of Notch receptors and ligands in the intestinal epithelium and to investigate the influence of Notch signaling on survival and maintenance of ISCs across fetal development and into adulthood. Studies in mice reveal that all Notch ligands are restricted to the mesenchyme at embryonic day 13.5 (E13.5), but by E18.5, DLL1 expression is found predominantly in the proliferative intervillus zone (IVZ), and by postnatal day 25 (P25), DLL1, DLL4, and JAG1 can be detected in the crypts (Schröder and Gossler, 2002). With the exception of NOTCH4, which remains restricted to the mesenchyme, all Notch receptors can be found in the epithelium at E13.5. By E18.5, NOTCH1 and 2 are detected in the IVZ, with NOTCH1 becoming primarily restricted to the crypts at P25. NOTCH2 expression, however, remains very weak in the crypts, and both NOTCH3 and NOTCH4 are absent in the murine epithelium. Here, we identified in this study that NOTCH1, NOTCH2, and NOTCH3 are expressed in the developing human epithelium, and later during development, there is increased expression of NOTCH1 and NOTCH2 in the crypts. However, in contrast to expression patterns in mice where NOTCH1 is the major receptor for NOTCH signaling (Carulli et al., 2015), we found that NOTCH2 and NOTCH3 are most abundant in epithelial stem cells during fetal development, while lower levels of NOTCH1 and high NOTCH2 are expressed in the adult. Mechanistically, NOTCH signaling components are expressed in the early fetal ISCs despite the fact these cells are not as sensitive to DBZ treatment and despite the fact that OLFM4 is not expressed at this time, suggesting that it is not purely ligand-receptor expression that regulates signaling. Unfortunately, attempts to validate this with NICD staining were unsuccessful due to the fact that the antibodies are not very robust. In this light, it is interesting to note that early fetal epithelium expresses the highest levels of NOTCH3, which is gradually reduced over time, and that NOTCH3 has been described as an inhibitor of NOTCH1 signaling in other contexts (Beatus et al., 1999). The absence of OLFM4 expression in the early-stage samples may imply that a stromal cell or extracellular matrix component may be responsible for repressing OLFM4 in the fetal intestines.
Previous studies have implied that Notch signaling is essential for maintaining ISC homeostasis alongside Wnt signaling (Nakamura et al., 2007); however, NOTCH’s influence on human fetal and adult ISCs in the context of WNT has not been addressed. Here, our studies suggest that NOTCH signaling is not essential for stem cell renewal during early fetal development. Consistent with the different requirement developmental dependencies in NOTCH, our data show that blocking NOTCH activity influences WNT activity differently in adult and fetal enteroids. In adults, the WNT target gene AXIN2 (Jho et al., 2002; Lustig et al., 2001) is reduced, while expression of DKK1, a negative regulator of WNT signaling (Niida et al., 2004), is increased based on RNA-seq data (Figure S4H). In contrast, AXIN2 expression was not influenced by NOTCH inhibition in 58-day enteroids (Figure S4I). Recent studies in the mouse have established precedent for context-dependent plasticity in the stem cell niche, leading to altered stem-cell regulation. For example, injury of the adult stem cell domain through infection or inflammatory damage can lead to loss of canonical stem cell signature genes, including Lgr5 and Olfm4. Following loss of these stem cell genes, a “fetal reversion” of the gene signature occurs, where adult cells acquire a gene signature that is similar to that of the fetal gut (Nusse et al., 2018; Yui et al., 2018). In these cases, loss of Wnt target genes (i.e., Lgr5) and Notch target genes (i.e., Olfm4) suggests that context-specific situations exist (i.e., development or disease), where the requirements for Notch or Wnt are different from the homeostatic state. This also further highlights the extreme plasticity of the ISC niche that has been demonstrated in injury contexts, such as when ISCs are ablated. Thus, the work conducted here may yield important insights from development that can be used to understand disease and regeneration in the adult.
Overall, we show in this study that the Notch pathway regulates the human ISC compartment differently across early and late stages of fetal development as well as in the adult.
Experimental procedures
Human tissue
Normal, de-identified human fetal intestinal tissue was obtained from the University of Washington Laboratory of Developmental Biology. Human adult biopsies were collected via endoscopy from the duodenum under protocols HUM00041845 and HUM00039121. All human tissue used in this work was de-identified and conducted in accordance with the University of Michigan Institutional Review Board regulations.
Isolating, establishing, and maintaining human fetal enteroids
Fresh human fetal epithelium was isolated and maintained as previously described (Tsai et al., 2018, Supplemental Protocol 1). Once enteroids were established, healthy cystic enteroids were manually selected under a stereoscope, bulk passaged through a 30G needle, and embedded in Matrigel (Corning; 354234). For single-cell passaging, healthy cystic enteroids were manually selected under a stereoscope and dissociated with TrypLE Express (Gibco; 12605-010) at 37°C before filtering through 40-μm cell strainers. Cells were then counted using a hemocytometer (Thermo Fisher Scientific) and embedded in Matrigel.
Enteroid media composition
Control culture media consisted of 50% LWRN conditioned media (Miyoshi and Stappenbeck, 2013) and 50% human 2× basal media (Advanced DMEM/F12 [Gibco; 12634-028]; Glutamax 4 mM [Gibco; 35050-061]; HEPES 20 mM [Gibco; 15630-080]; N2 Supplement [2×; Gibco; 17502-048], B27 Supplement [2×; Gibco; 17504-044]; penicillin-streptomycin [2×; Gibco; 15140-122]; N-acetylcysteine [2 mM; Sigma; A9165-25G]; nicotinamide [20 mM; Sigma; N0636-06Z1]). NOTCH inhibition media was control media with the addition of 5 μM DBZ (Apexbio Technology; YO-01027).
Single-cell dissociation
To dissociate human fetal tissue to single cells, fetal duodenum was first dissected using forceps and a scalpel in a Petri dish filled with cold 1× Hank’s balanced salt solution (HBSS) (with Mg2+ and Ca2+). Whole-thickness intestine was cut into small pieces and transferred to a 15-mL conical tube with 1% BSA in HBSS. Dissociation enzymes and reagents were used from the Neural Tissue Dissociation Kit (Miltenyi Biotec; 130-092-628), and all incubation steps were carried out in a refrigerated centrifuge pre-chilled to 10°C unless otherwise stated. All tubes and pipette tips used to handle cell suspensions were pre-washed with 1% BSA in 1× HBSS to prevent adhesion of cells to the plastic. Tissue was treated for 15 min at 10°C with mix one and then incubated for 10-min increments at 10°C with mix two interrupted by agitation by pipetting with a P1000 pipette until fully dissociated. Cells were filtered through a 70-μm filter coated with 1% BSA in 1× HBSS, spun down at 500 × g for 5 min at 10°C and resuspended in 500 μL 1× HBSS (with Mg2+ and Ca2+) with 1 mL Red Blood Cell Lysis buffer (Roche; C755C08). The cell mixture was placed on a rocker for 15 min in the cold room (4°C) before being spun down (500 × g for 5 min at 10°C) and washed twice by suspension in 2 mL of HBSS + 1% BSA, followed by centrifugation. Cells were counted using a hemocytometer and then spun down and resuspended to reach a concentration of 1,000 cells/μL and kept on ice. Single-cell libraries were immediately prepared on the 10× Chromium at the University of Michigan Sequencing Core facility with a target of 5,000 cells. The same protocol was used for single-cell dissociation of healthy cystic enteroids manually collected under a stereoscope. A full, detailed protocol of tissue dissociation for scRNA-seq can be found at www.jasonspencelab.com/protocols.
Primary tissue collection, fixation, and paraffin processing
Human fetal intestine tissue samples were collected as 1-cm fragments for each section of duodenum, jejunum, and ileum and fixed for 24 h at room temperature in 10% neutral buffered formalin (NBF). This was followed by three washes in Ultra-Pure Distilled Water (Invitrogen; 10977-015) for a total of 3 h. Tissue was dehydrated by an alcohol series diluted in Ultra-Pure Distilled Water (Invitrogen; 10977-015). Tissue was incubated for 1 h each solution: 25% methanol, 50% methanol, 75% methanol, 100% methanol followed by equilibration in 100% ethanol for 1 h, and then 70% ethanol prior to tissue processing. Tissue was processed into paraffin blocks in an automated tissue processor (Leica ASP300) with 1-h changes overnight.
Histology, immunofluorescence, in situ hybridization, and multiplex FISH
Immunofluorescence was carried out as previously described (Dye et al., 2015; Rockich et al., 2013) using antibodies outlined in Table S3. Paraffin blocks were sectioned to generate 5-μm-thick sections within 1 week prior to performing ISH. Slides were baked for 1 h in a 60°C dry oven the night before. ISH was performed using the RNAscope 2.0 HD detection kit and with commercially available mRNA probes outlined in Table S3, according to the standard protocol provided. All incubations were performed at 40°C in a HybEZ hybridization system oven (Advanced Cell Diagnostics; 310010). FISH protocol was performed according to the manufacturer’s instructions (ACD; RNAscope multiplex fluorescent manual protocol; 323100-USM) and with commercially available mRNA probes outlined in Table S3. Immediately following the horseradish peroxidase (HRP) blocking for the C2 channel of the FISH, slides were washed three times for 5 min in PBS, followed by standard IF protocol.
Quantification of human enteroids
Both cyst number and cyst size were quantified using images of cultures taken under a dissection scope. Cyst number was determined by manually counting and marking the structures present in each well. Cyst size was established by using the two-dimensional surface area of each cyst as an indicator of overall size. Cysts were assigned a numerical identifier before being analyzed with ImageJ (Schneider et al., 2012). Using either the “oval selections” or “freehand selections” tool, the circumference of every cyst was traced in numerical order and the “measure” function was used to quantify the area of each selection in square pixels.
Bulk RNA sequencing
RNA was isolated from enteroids using the MagMAX-96 Total RNA Isolation Kit according to the manufacturer’s protocol. RNA concentration and purity were assessed using the Nanodrop spectrophotometer and bioanalyzed using Agilent RNA 6000 Nano Kit (260/280 > 1.9 and 260/230 > 1.8, RNA integrity number [RIN] > 8). Samples were stored at −80°C. All reads were aligned to an index of transcripts from human genes within the Ensembl GRCh38 and quantified using Kallisto (Bray et al., 2016). Gene-level data generated from Kallisto were used for TMM normalization in edgeR to create normalized data matrix of pseudocounts (Robinson and Oshlack, 2010). PCA and sample clustering were done in R using the “cluster” and Bioconductor “qvalue” packages (Storey and Helmy, 2019).
Air-liquid interface (ALI) tissue culture
Fresh human fetal duodenum was dissected into 0.5-cm segments to be cultured on a transwell membrane in a 24-well plate. We added 600 μL LWRN medium with or without DBZ to the bottom of the transwell and cultured for 3 days with medium changes every day. Tissues were collected for IF and FISH staining using the same procedures described above.
Data code and availability
Sequencing data generated and used by this study are deposited at EMBL-EBI ArrayExpress. Datasets for human fetal intestine are available at ArrayExpress: E-MTAB-9489. Datasets for bulk RNA sequencing (fetal and adult control, DBZ treatment) are available at ArrayExpress: E-MTAB-11619. Code used to process raw data can be found at https://github.com/jason-spence-lab/Tsai_Wu_2020
Author contributions
Project conceptualization, Y.-H.T., A.W., and J.R.S.; experimental design, Y.-H.T., A.W., and J.R.S.; experiments and data collection, Y.-H.T., A.W., M.C., E.M.H., and S.H.; essential material reagents, I.G. and P.D.R.H.; data analysis and interpretation, Y.-H.T., A.W., J.H.W., M.C., E.M.H., S.H., and J.R.S.; writing manuscript, Y.-H.T., A.W., and J.R.S.; editing manuscript, all authors.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We thank Judy Opp and the University of Michigan Advanced Genomics Core for their expertise operating the 10× Chromium single-cell capture platform and sequencing expertise. We would also like to thank the University of Washington Laboratory of Developmental Biology staff. This work was supported by the Intestinal Stem Cell Consortium (U01DK103141 to J.R.S.), a collaborative research project funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institute of Allergy and Infectious Diseases (NIAID). This work was also supported by the NIAID Novel Alternative Model Systems for Enteric Diseases (NAMSED) consortium (U19AI116482 to J.R.S.). This project has been made possible in part by grant number CZF2019-002440 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, and by the University of Michigan Center for Gastrointestinal Research (UMCGR) (NIDDK 5P30DK034933). The University of Washington Laboratory of Developmental Biology was supported by NIH award number 5R24HD000836 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). M.C. was supported by the Training Program in Organogenesis (NIH-NICHD T32 HD007505). E.M.H. was supported by the Training in Basic and Translational Digestive Sciences Training Grant (NIH-NIDDK 5T32DK094775), the Cellular Biotechnology Training Program Training Grant (NIH-NIGMS 2T32GM008353), and the Ruth L. Kirschstein Predoctoral Individual National Research Service Award (NIH-NHLBI F31HL146162).
Published: April 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.03.007.
Supporting citations
The following references appear in the Supplemental information: Blondel et al., 2008; McInnes et al., 2018; Wolf et al., 2018.
Supplemental information
References
- Barker N., Clevers H. Tracking down the stem cells of the intestine: strategies to identify adult stem cells. Gastroenterology. 2007;133:1755–1760. doi: 10.1053/j.gastro.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Beatus P., Lundkvist J., Öberg C., Lendahl U. The Notch 3 intracellular domain represses Notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. doi: 10.1242/dev.126.17.3925. [DOI] [PubMed] [Google Scholar]
- Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 2008;2008:1–12. [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev. Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Srinivasan T., Tung K., Belmonte J.M., Wang L., Murthy P.K.L., Choi J., Rakhilin N., King S., Varanko A.K., et al. A Notch positive feedback in the intestinal stem cell niche is essential for stem cell self-renewal. Mol. Syst. Biol. 2017;13:1–16. doi: 10.15252/msb.20167324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Dylla S.J., Park I.-K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:179–184. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.-H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4:1–25. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Hill D.R., Altheim C.H., Dedhia P.H., Taylor M.J., Tsai Y.-H., Chin A.M., Mahe M.M., Watson C.L., Freeman J.J., et al. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Reports. 2015;4:1140–1155. doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Holloway E.M., Czerwinski M., Tsai Y.H., Wu J.H., Wu A., Childs C.J., Walton K.D., Sweet C.W., Yu Q., Ian Glass I., et al. Mapping development of the human intestinal niche at single-cell resolution. Cell Stem Cell. 2021;28:568–580.e4. doi: 10.1016/j.stem.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J., Pedersen E.E., Galante P., Hald J., Scott Heller R., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jho E., Zhang T., Domon C., Joo C.-K., Freund J.-N., Costantini F. Wnt/β-Catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Rodgers G.P. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev. 2016;35:201–212. doi: 10.1007/s10555-016-9624-2. [DOI] [PubMed] [Google Scholar]
- Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Rarsten U., van de Wetering M., Clevers H., Schlag P.M., Birchmeier W., et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell Biol. 2001;22:1182–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 2018;3:861. [Google Scholar]
- Milano J., McKay J., Dagenais C., Foster-Brown L., Pognan F., Gadient R., Jacobs R.T., Zacco A., Greenberg B., Ciaccio P.J. Modulation of Notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 2004;82:341–358. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013;12:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tsuchiya K., Watanabe M. Crosstalk between Wnt and Notch signaling in intestinal epithelial cell fate decision. J. Gastroenterol. 2007;42:705–710. doi: 10.1007/s00535-007-2087-z. [DOI] [PubMed] [Google Scholar]
- Niida A., Hiroko T., Kasai M., Furukawa Y., Nakamura Y., Suzuki Y., Sugano S., Akiyama T. DKK1, a negative regulator of Wnt signaling, is a target of the β-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- Nusse Y.M., Savage A.K., Marangoni P., Rosendahl-Huber A.K.M., Landman T.A., de Sauvage F.J., Locksley R.M., Klein O.D. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 2018;559:109–113. doi: 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrinet L., Rodilla V., Liu Z., Chen S., Koch U., Espinosa L., Kaestner K.H., Kopan R., Lewis J., Radtke F. Dll1- and Dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240.e7. doi: 10.1053/j.gastro.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio O., van Gijn M.E., Bezdek A.C., Pellegrinet L., van Es J.H., Zimber-Strobl U., Strobl L.J., Honjo T., Clevers H., Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;25:1–9. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich B.E., Hrycaj S.M., Shih H.P., Nagy M.S., Ferguson M.A.H., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. U S A. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder N., Gossler A. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr. Patterns. 2002;2:247–250. doi: 10.1016/s1567-133x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- Schuijers J., van der Flier L.G., van Es J., Clevers H. Robust cre-mediated recombination in small intestinal stem cells utilizing the Olfm4 locus. Stem Cell Reports. 2014;3:234–241. doi: 10.1016/j.stemcr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger B.Z., Datar R., Charles Murtaugh L., Melton D.A. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. U S A. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey E.E., Helmy A.S. Optimized preprocessing and machine learning for quantitative Raman spectroscopy in biology. J. Raman Spectrosc. 2019;50:958–968. [Google Scholar]
- Tian H., Biehs B., Chiu C., Siebel C.W., Wu Y., Costa M., de Sauvage F.J., Klein O.D. Opposing activities of notch and wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-H., Czerwinski M., Wu A., Dame M.K., Attili D., Hill E., Colacino J.A., Nowacki L.M., Shroyer N.F., Higgins P.D.R., et al. A method for cryogenic preservation of human biopsy specimens and subsequent organoid culture. Cell Mol. Gastroenterol. Hepatol. 2018;6:218–222. doi: 10.1016/j.jcmgh.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-H., Vandussen K.L., Sawey E.T., Wade A.W., Kasper C., Rakshit S., Bhatt R.G., Stoeck A., Maillard I., Crawford H.C., et al. ADAM10 regulates notch function in intestinal stem cells of mice. Gastroenterology. 2014;147:822–834.e13. doi: 10.1053/j.gastro.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., Haegebarth A., Stange D.E., van de Wetering M., Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I., et al. Transcription factor Achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud Å., et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:1–5. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Kilik U., Holloway E.M., Tsai Y.H., Hamel C., Wu A., Wu J.H., Czerwinski M., Childs C.J., He Z., et al. Charting human development using a multi-endodermal organ atlas and organoid models. Cell. 2021;184:3281–3298.e22. doi: 10.1016/j.cell.2021.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Azzolin L., Maimets M., Pedersen M.T., Fordham R.P., Hansen S.L., Larsen H.L., Guiu J., Alves M.R.P., Rundsten C.F., et al. YAP/TAZ-Dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell. 2018;22:35–49. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.H., Li F.D., Tian C., Ren H.L., Du J., Li H.H. Notch γ-secretase inhibitor dibenzazepine attenuates angiotensin II-induced abdominal aortic aneurysm in ApoE knockout mice by multiple mechanisms. PLoS One. 2013;8:1–14. doi: 10.1371/journal.pone.0083310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.