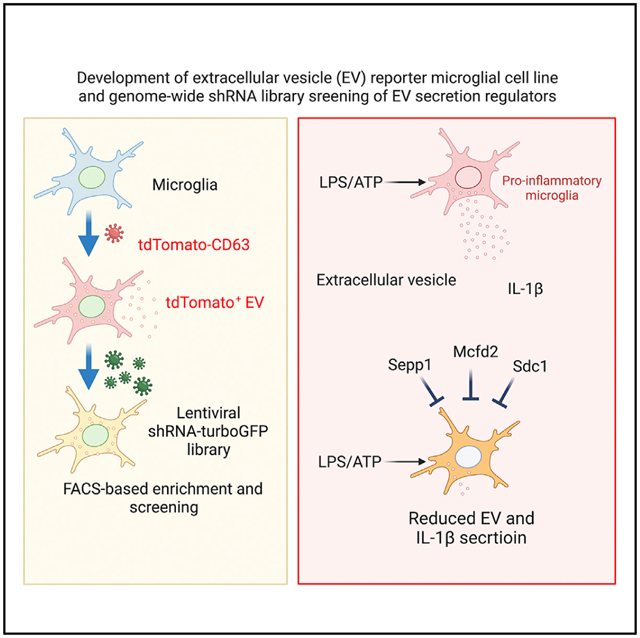
SUMMARY
Activated microglia release extracellular vesicles (EVs) as modulators of brain homeostasis and innate immunity. However, the molecules critical for regulating EV production from microglia are poorly understood. Here we establish a murine microglial cell model to monitor EV secretion by measuring the fluorescence signal of tdTomato, which is linked to tetraspanin CD63. Stimulation of tdTomato+ cells with ATP induces rapid secretion of EVs and a reduction in cellular tdTomato intensity, reflecting EV secretion. We generate a GFP+ tdTomato+ cell library expressing TurboGFP and barcoded short hairpin RNAs for genome-wide screening using next-generation sequencing. We identify Mcfd2, Sepp1, and Sdc1 as critical regulators of ATP-induced EV secretion from murine microglia. Small interfering RNA (siRNA-based) silencing of each of these genes suppresses lipopolysaccharide- and ATP-induced inflammasome activation, as determined by interleukin-1β release from primary cultured murine microglia. These molecules are critical for microglial EV secretion and are potential therapeutic targets for neuroinflammatory disorders.
Graphical Abstract
In brief
Ruan et al. report genome-wide shRNA library screening on a tdTomato-CD63+ microglia cell model to identify factors implicated in the process of extracellular vesicle (EV) release. The molecules Sepp1, Mcfd2, and Sdc1 are critical for ATP-induced secretion of EV and EV-associated interleukin-1β from murine microglia.
INTRODUCTION
Microglia, the primary innate immune cells of the central nervous system (CNS), continuously survey their microenvironment using highly dynamic processes and interact with neurons to regulate their activity (Clayton et al., 2017; Yeh and Ikezu, 2019). Intercellular communication between microglia and neurons has been reported to not only maintain brain homeostasis but also to act as a necessary pathological intermediate in the process of neurodegenerative disease (Szepesi et al., 2018). In particular, microglia function as key regulators of the neuroinflammatory response, which has been widely recognized as a common feature of neurodegenerative disorders such as Alzheimer’s disease, Parkinson’s disease, frontotemporal lobar dementia, and amyotrophic lateral sclerosis (Hickman et al., 2018; Bartels et al., 2020). Upon stimulation with environmental cues (e.g., stranger and danger signals), microglia can rapidly transition from a state of surveillance to that of activity or reactivity.
Extracellular vesicles (EVs) with a 30- to 1,000-nm diameter have been reported to be released by activated microglia. Various extracellular stimuli can activate microglia to secrete a distinct population of EVs by modulating specific signaling pathways (Turola et al., 2012). Among these, extracellular ATP, a representative damage-associated molecule, has been documented as a major stimulus for vesicle shedding in microglia through purinergic receptors, including P2RX7 (Ruan et al., 2020), which also activates inflammasomes. ATP concentration rises at sites of stress and cellular injury, leading to activation of ionotropic purinergic receptors (Burnstock, 2007). Activation of P2RX7 potently increases exosome release from microglia (Bianco et al., 2005b). ATP-induced, microglia-derived EVs are rich in interleukin-1β (IL-1β), an indicator of inflammasome activation, promoting propagation of neuroinflammatory responses in the CNS (Verderio et al., 2012; Drago et al., 2017). Stimulation with lipopolysaccharide (LPS), a component of the cell wall of Gram-negative bacteria, significantly elevates EV secretion from microglia (Kumar et al., 2017). Secreted EVs have also been reported to contain several inflammatory molecules, such as tumor necrosis factor alpha (TNF-α), IL-6, and IL-1β (Yang et al., 2018a; Kumar et al., 2017). Interestingly, emerging evidence suggests that microglia-derived EVs can also mediate the spread of pathology in multiple neurodegenerative diseases, including Alzheimer’s disease (Delpech et al., 2019; You and Ikezu, 2019; DeLeo and Ikezu, 2018). In particular, EVs carry molecules such as prions, β-amyloid (Aβ) peptides (Rajendran et al., 2006), tau protein (Ruan and Ikezu, 2019), and other misfolded proteins originally generated by neurons (Delpech et al., 2019). A more recent study demonstrated that microglia use EVs to facilitate the spreading of tau proteins via phagocytosis and exocytosis (Ruan et al., 2020, 2021). Thus, microglia-derived EVs may efficiently interact with neuronal cells to deliver pathogenic molecules and inflammatory signals during disease progression.
Despite the growing interest in the role of EVs in pathogenesis, little is known about the molecular mechanisms of EV secretion from activated microglia. The biosynthesis of EVs includes contusion of the late endosomal membrane to form intraluminal vesicles (ILVs) during multivesicular body (MVB) maturation and subsequent vesicle secretion upon their fusion to the plasma membrane (Simons and Raposo, 2009). EVs are highly enriched in tetraspanins, a family of conserved membrane proteins with four transmembrane domains. Tetraspanins are involved in the recycling routes between the plasma membrane and several cellular organelles, regulating the maturation and trafficking of their associated proteins (Andreu and Yanez-Mo, 2014). Recently, proteomic analysis of EVs released by various cell types, including microglia, revealed the presence of tetraspanins CD63, CD37, CD81, and CD9 (Andreu and Yanez-Mo, 2014; Kowal et al., 2016). Therefore, these tetraspanins have been defined as exosomal marker proteins to monitor the amount of EVs produced by cells.
In the present study, we identify key inhibitors of EV release from microglia upon ATP stimulation. By creating a vector containing a CD63-tdTomato fluorescence tag and combination with a barcoded short hairpin RNA (shRNA) lentiviral library, we identified a set of 1,353 host genes that regulate the sensitivity of small EV secretion to ATP stimulation. We validated several top-ranking hits, among which Mcfd2, Sepp1, and Sdc1 were found to play a role in the process of EV secretion from microglia. The key molecules for suppressing EV secretion may serve as potential therapeutic targets for neuroinflammatory disorders, including Alzheimer’s disease.
RESULTS
Specific co-localization of the TdTomato-CD63 reporter with late endosomes
To monitor the process of EV release from cells, we constructed a lentiviral tdTomato-CD63 reporter system in a mouse microglial BV-2 cell line (Figures 1A and 1B). After permeabilization, tdTomato-CD63+ BV-2 cells were immunofluorescence labeled with the early endosome markers Rab5/EEA1 and the late endosome marker Rab7. High-resolution confocal microscopy images were captured by a Zeiss LSM confocal microscope 880 with 63× objective lens and 2.0× digital zoom. From these images, there was robust CD63-tdTomato fluorescence signal in the perinuclear area (Figure S1A). However, EEA1 or Rab5-positive early endosomes were present in the cell periphery and the perinuclear area (Figures S1A and S1G). Approximately 25.67% and 24.40% of CD63-tdTomato also labeled the EEA1 and Rab5-positive spots, respectively (Figures S1D and S1I; Table S5). Vice versa, 68.14% of CD63-tdTomato was also labeled with Rab7-positive spots enriched in the perinuclear late endosomes (Figures S1B and S1E; Table S5). These data are also consistent with the fact that the major pool of CD63 resides in late endosomes/MVBs and lysosomes. Rab7 is associated primarily with late endosomal structures, intermediate hybrids of late endosomes-lysosomes, and the lysosome. A similar co-localization between CD63 and Rab7 has been reported recently (van der Beek et al., 2022). On the contrary, few co-localizations of early endosomes with EEA1 or Rab5 and the late endosomes with Rab7 were seen (Figures S1C, S1F, S1H, and S1J) due to efficient conversion of Rab5-to-Rab7 by the Mon1-Ccz1 complex (Poteryaev et al., 2010). This experiment confirms the co-localization of CD63-tdTomato with the late endosome marker Rab7 but much less with early endosomal markers.
Figure 1. Creation of the tdTomato-CD63+ BV-2 stable cell line and characterization of EV release after ATP stimulation.
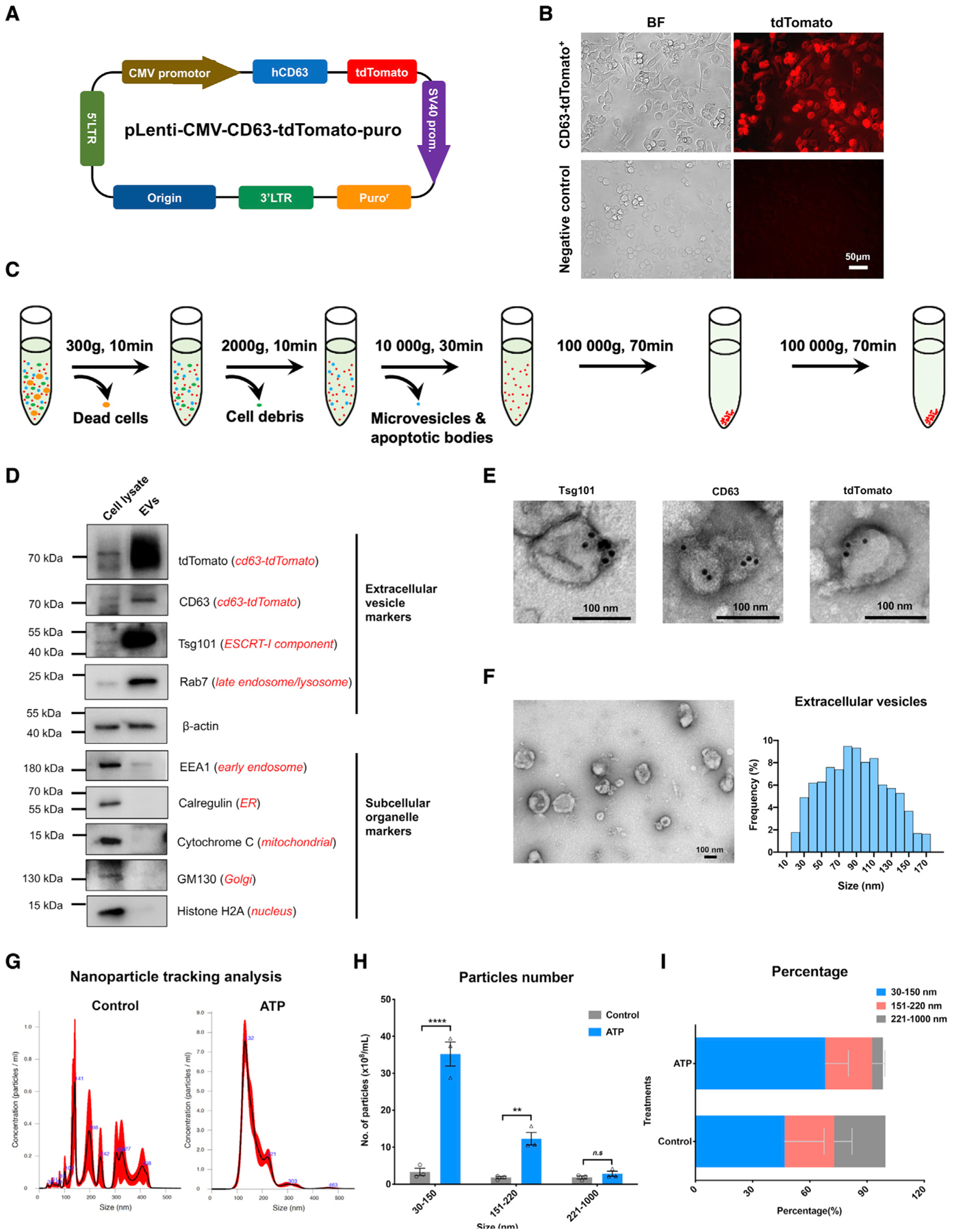
(A) Arrangement of the engineered pLenti-tdTomato-CD63 vector.
(B) Creation of the tdTomato-CD63+ reporter BV-2 stable cell line. Scale bar, 50 μm.
(C) Workflow to purify the small EV fraction from the conditioned culture medium of the tdTomato-CD63+ BV-2 stable cell line.
(D) Representative immunoblots of typical protein markers for small EVs, engineered small EV tags, and other subcellular organelles from cell lysate and purified small EV fractions of conditioned medium.
(E) Representative immunogold electron microscopy images of Tsg101, CD63, and tdTomato labeling on small EVs isolated from conditioned medium. Scale bar, 100 nm.
(F) Representative transmission electron microscopy images (left panel) and size distribution (right panel) of exosomes isolated from conditioned medium. In total, 16 images were analyzed. Scale bar, 100 nm.
(G and H) Representative size distribution of small EVs isolated from conditioned medium of tdTomato-CD63+ BV-2 cells with or without ATP stimulation by Nanosight NTA. Two-tailed Student’s t test; **p < 0.01; ****p < 0.0001; ns, no statistical significance; compared with the scramble-shRNA group; n = 3 independent experiments, each sample was recorded for 4 videos. Graphs indicate mean ± SEM.
(I) The percentage distribution of different size fractions of EVs isolated from the conditioned medium.
Monitoring EV release induced by ATP stimulation
This fluorescent EV reporter system allowed live detection of EVs secreted through the classical secretory pathway in small volumes of cell culture supernatants. BV-2 cells exhibited a reduction in cellular tdTomato-CD63 fluorescence intensity after stimulation with 5 mM ATP for 15 min (Figures S2A–S2C). This reduction was accompanied by an increase in tdTomato-CD63+ in the culture medium, as confirmed by CD63 ELISA (Figure S2D), indicating that the reduction in fluorescence intensity of tdTomato in cells served as a readout for tdTomato-CD63+ small EV secretion from BV-2 cells. To exclude the possibility that the reduction in tdTomato fluorescence intensity signal was caused by autophagy-mediated protein degradation, tdTomato-CD63+ cells were treated with the autophagy inhibitor 3-methyladenine (3-MA) before ATP stimulation, and the fluorescence signal was monitored for 1 h by the IncuCyte live-cell imaging system. The data showed that the fluorescence signal upon ATP stimulation declined independent of autophagy activation (Figures S2E and S2F). There was no obvious upregulation of microtubule-associated protein light chain 3 (LC3)-II, an autophagy activation marker, 15 min after ATP stimulation (Figure S2G). These data were consistent with a previous finding that autophagy can only be activated via multiple signaling pathways after ATP stimulation, which would require up to 2 h (Plaza-Zabala et al., 2020; Takenouchi et al., 2009). These results reflect the dynamic process of CD63+ EV release in this time window.
Characterization of EV secreted by cells after ATP stimulation
To characterize EV release from the tdTomato-CD63+ cell line after ATP stimulation, we used ultracentrifugation and enriched EVs from conditioned medium (Figure 1C). The engineered tdTomato-CD63 fusion protein and typical small EV markers—such as CD63 and a member of the endosomal sorting complexes required for transport (ESCRT)-I complex, Tsg101—were detected in the EV fraction (Figure 1D). MVBs and late endosomal marker Rab7 were highly enriched in the EV fraction (Figure 1D), confirming the enrichment of small EVs in the fraction. There was little contamination of cytoplasmic organelles or vesicles, as indicated by scant bands of EEA1, calregulin, cytochrome c, GM130/GOLGA2, and histone H2A in the vesicular fraction (Figure 1D). Subsequent immunogold labeling of Tsg101, CD63, and tdTomato in the EV fraction showed sizes corresponding to small EVs (Figure 1E). Using transmission electron microscopy (TEM), we found that the size of these small EVs ranged from 30–150 nm (Figure 1F). To quantitatively measure acute ATP stimulation-induced small EV release from this CD63-tdTomato+ cell line, we used the Nanosight nanoparticle tracking analysis to monitor EVs enriched from conditioned medium. It showed a single peak at 132 nm as the median size in the EV sample from the ATP-treated group but broad and lower peaks in the EV sample from the non-treated group (Figure 1G), indicating small EV release from CD63-tdTomato+ cells after ATP stimulation. The total mean number of secreted EVs after ATP treatment was 83.20 × 108 particles mL−1 compared with 11.63 × 108 particles mL−1 without stimulation (Figure 1H). Elevated EVs were mainly observed in the small EVs (size in 30–150 nm) after ATP stimulation (Figure 1H). EVs in the range of 30–150 nm accounted for 68.10% of total EVs (Figure 1I). Overall, these data show that ATP stimulation induced EV release from the CD63-tdTomato+ cell line, which could be monitored by the change in the tdTomato+ fluorescence signal.
Pooled shRNA library screening
We first optimized the promoter activity of the lentiviral shRNA library using the SMART choice promoter selection plate and then chose the vector configuration that was most effective for TurboGFP and shRNA expression in the BV-2 cell line. We examined seven promoters: human cytomegalovirus (CMV), murine CMV, human elongation factor-1α (EF1α), murine EF1α, murine phosphoglycerate kinase (PGK), human ubiquitin C (UBC), and a synthetic hybrid construct consisting of the CMV enhancer fused to the chicken β-actin (CAG) promoters. After 3 days of incubation with different titers of lentivirus, the murine EF1α promoter showed the highest expression of TurboGFP in BV-2 cells (Figure S3). We customized the Dharmacon siGENOME SMARTpool shRNA genome-wide library, comprising up to eight independent shRNA hairpins for each target gene, with the mEF1α promoter. This library contained 177,464 unique shRNAs that targeted 21,924 protein-coding genes, 1,800 non-targeting controls, and 272 common reference constructs. Each shRNA was tagged with a unique barcode, enabling subsequent identification by next-generation sequencing (NGS)-based high-throughput screening (HTS). To track lentivirus transduction and selection of shRNA clone+ cells by fluorescence-activated cell sorting (FACS), the lentiviral library expressed the TurboGFP reporter gene. After optimization and establishment of appropriate conditions, libraries were screened to identify targets for which shRNA-based knockdown of target genes retained cellular TdTomato-CD63 fluorescence intensity after ATP stimulation (Figure 2A). Briefly, stable tdTomato-CD63+ BV-2 cells were transduced with 18 lentiviral packaged shRNA pools of the shRNA genome-wide library (9,860 constructs per pool) at a low multiplicity of infection (MOI). tdTomato+ TurboGFP+ high cells were collected by FACS 4 days after infection (Figure 2B). tdTomato+ TurboGFP+ cells represented robust expression of the shRNA library and tdTomato-CD63. A total of 3.10 × 107 cells were collected from 18 sets of pooled libraries, achieving approximately 170-fold coverage of the target genes. These tdTomato+ TurboGFP+ cells were stimulated with ATP to induce EV secretion for 15 min, followed by FACS of tdTomatohigh/TurboGFP+ cells (Figures 2C and 2D), which represented enrichment of cells resistant to ATP-induced EV secretion. In total, 379,485 cells were collected from all 18 pools of the shRNA library, representing the top 1% of cells in terms of tdTomato fluorescence intensity as ATP-resistant cells. In total, 441,600 cells were collected from 18 pools of the shRNA library, representing the bottom 1% of cells in terms of tdTomato fluorescence intensity as ATP-sensitive cells. The genomic DNA was purified, and the barcoded shRNA region was amplified by two-step PCR, followed by NGS, as described in the STAR Methods.
Figure 2. Positive cells containing silencing factors acting during EV secretion.
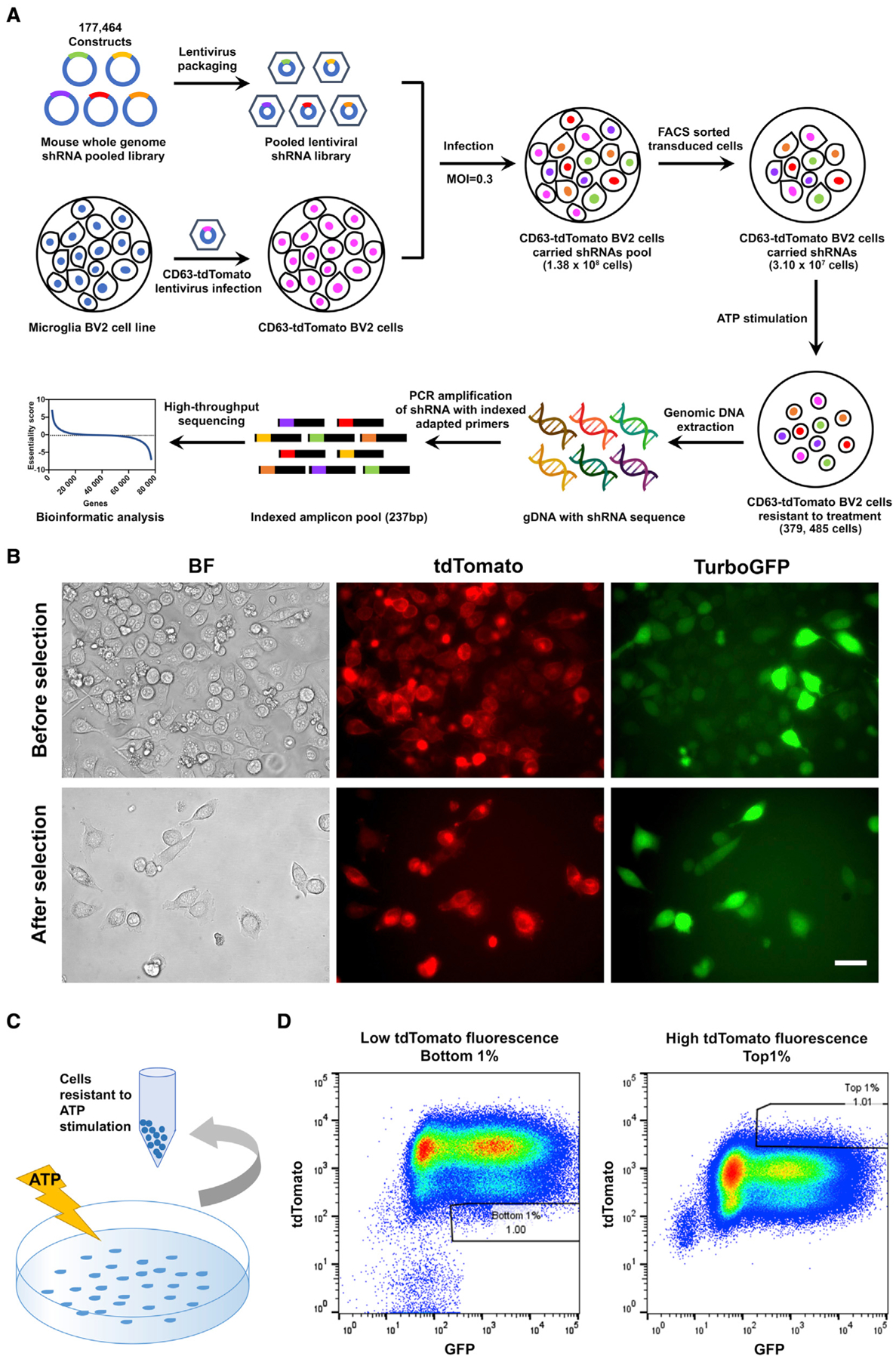
(A) Schematic of the workflow of genome-wide shRNA library screening. A genome-wide mouse shRNA library containing 177,464 constructs was packed into lentiviral particles and transduced into tdTomato-CD63 overexpressing BV-2 cells at a low multiplicity of infection (MOI). The shRNA-transduced cells were sorted by FACS to generate a mutant cell pool. Mutant cells were amplified and treated with ATP stimulation for genetic screening. Genomic DNA was extracted from the treated cells, and the shRNA fragment was amplified by PCR. The copy number of shRNAs was determined by HTS and bioinformatics analysis.
(B) Transduced tdTomato-CD63 and shRNAs lentiviral BV-2 cells sorted by FACS.
(C) The workflow of ATP stimulation and cell collection.
(D) FACS gating panels of the bottom 1% of cells representing cells sensitive to ATP stimulation for EV secretion (left) and top 1% of cells representing cells resistant to ATP stimulation for EV secretion. In total, 18 cell pools were collected.
Bioinformatic analysis and top-ranked candidates from the shRNA screen
To identify putative EV secretion silencing factors, we calculated the normalized Z score of each hit based on the differential read counts of barcoded shRNA in ATP-resistant and -sensitive cells (Figure 3A; Table S1). Bioinformatics analysis was performed to characterize the 1,353 hits identified with an enrichment Z score of >2.5. Functional annotation clustering using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics database (version 6.8) revealed a set of seven enriched functional annotation clusters (adjusted log10 p < −1.1), among which the integral components of the membrane and plasma membrane had 575 and 417 “hits,” respectively (Figure 3B; Table S2). Further analysis of this set revealed that a subset of genes (376 hits) was enriched in the olfactory pathway; however, this was beyond the scope of this study. We examined the biological relevance of the remaining subset of 616 hits with Metasacape and identified the following biological pathways: cell-to-cell adhesion via plasma membrane adhesion molecules, regulation of synapse assembly, endoplasmic reticulum-plasma membrane tethering, endomembrane system organization, and negative regulation of GTP binding protein (GTPase) activity (Figure 3C and Table S3). The results suggest that these genes were involved in cell adhesion, membrane fusion, and regulation of ATPase and GTPase cascades.
Figure 3. Identification of top-ranked candidates from the shRNA library screening.
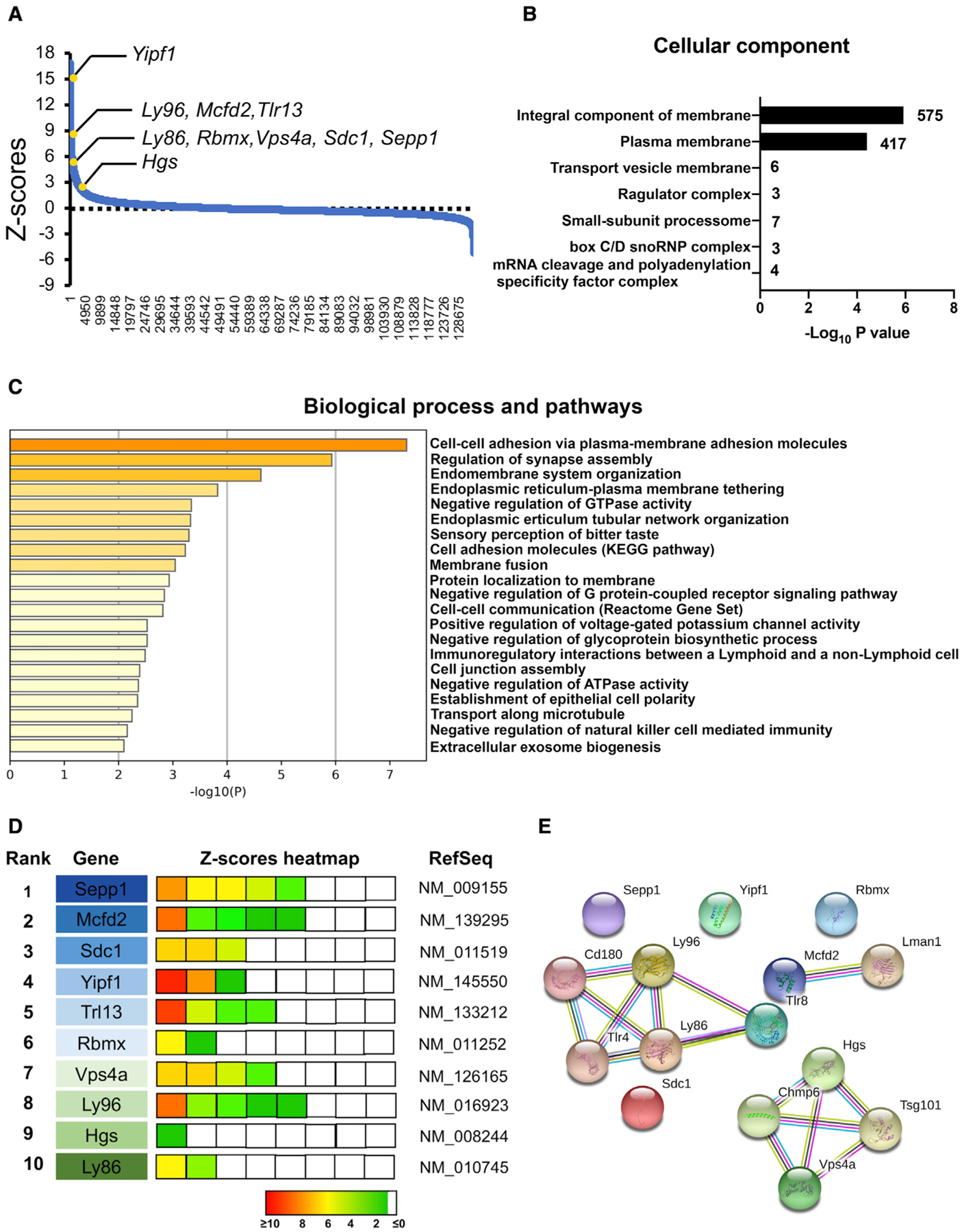
(A) Negative control normalized Z scores of all pooled RNAi screening data, ordered from most negative to positive.
(B) The seven most significantly enriched GO terms from the list of screened hits (p < 0.05) after Benjamini-Hochberg multiple-testing correction.
(C) Biological processes and pathways identified by Metascape with the candidate genes from the integral component of the membrane and plasma membrane.
(D) The 10 top-ranked candidates from the shRNA screen. The enrichment in cells sorted by FACS (Z score) of the corresponding hairpins is shown using a colored heatmap, where each square represents an independent hairpin. Crosses indicate excluded hairpins for which the read number was below a set threshold. The candidates were ranked based on the Z score for the hairpins targeting the same transcript. Factors linked to the same complexes/pathways are color coded.
(E) Candidate genes with GO terms including “secretion” and interactions by STRING association.
Validation of shRNA screening hits
The top hits were identified using multiple independent shRNA clones, eliminating the likelihood of off-target effects. Among the top-ranked hits, 10 genes (Sepp1, Mcfd2, Sdc1, Yipf1, Tlr13, Rbmx, Vps4a, Ly96, Hgs, and Ly86) were selected for further validation (Figure 3D). The top-ranked hit genes were targeted by more than three different shRNA clones and highly enriched sorted tdTomatohigh TurboGFP+ cells. Interestingly, we identified several factors associated with specific biochemically defined multi-subunit complexes and/or specific pathways (Figure 3E) by using the STRING functional protein association tool (Szklarczyk et al., 2019). Vps4a and Hgs are established molecules for ESCRT-mediated EV biogenesis (Colombo et al., 2013). Lymphocyte antigen 96 (Ly96) and Ly86 cooperate with Toll-like receptor 4 (TLR4), and Tlr13 (orthologous to human TLR8) binds and responds to the 5′-CGGAAAGACC-3′ sequence on bacterial 23S rRNA. These results suggest that TLR signaling plays a key role in EV secretion by murine microglia.
To validate the effect of selected genes on EV release from microglia, we transduced three independent shRNA clones of each of the 10 genes in the tdTomato-CD63+ BV-2 cell line and then stimulated them with ATP. The conditioned medium was collected for isolation of the exosomal fraction or measurement of CD63 by ELISA (Figure 4A). Highly efficient knockdown of targeted genes was achieved by all shRNA clones (Figures 4B–4I, mRNA graph) except for Hgs (Figure 4J) and Ly86 (Figure S4). The nanoparticle tracking analysis of the isolated exosomal fraction identified two shRNA clones (sh2 and sh3) targeting Sepp1 and showed a consistent and significant reduction in EV secretion in ATP-stimulated BV-2 cells (Figure 4B). The amount of CD63 secreted in the culture medium was also significantly reduced by the Sepp1 shRNA sh3 clone (Figure 4B). We also observed that at least one of three shRNA clones showed a significant decrease in the number of EVs, as measured by nanoparticle tracking analysis (NTA), after silencing of Mcfd2, Yipf1, Trl13, or Ly96, which were also shown by the amount of CD63+ EV secretion by ELISA (Figures 4C, 4E, 4F, and 4I). In contrast, silencing of Sdc1, Rbmx1, and Vps4a had no effect on ATP-induced EV secretion (Figures 4D, 4G, and 4H). The lack of effect on EV secretion by silencing of Sdc1 or Vps4a was unexpected because these are established molecules for EV biogenesis. It is possible that EV biogenesis is less dependent on the classic ESCRT machinery in BV-2 cells. We observed significant toxicity after silencing of Hgs in BV-2 cells, and the surviving cells showed a lack of Hgs silencing, suggesting that Hgs is essential for BV-2 survival and is not compatible with the silencing experiment. There was much less of an effect of “hit” genes on EV release under physiological conditions after gene silencing (Figure S5).
Figure 4. Modulation of EV secretion by silencing selected gene targets.
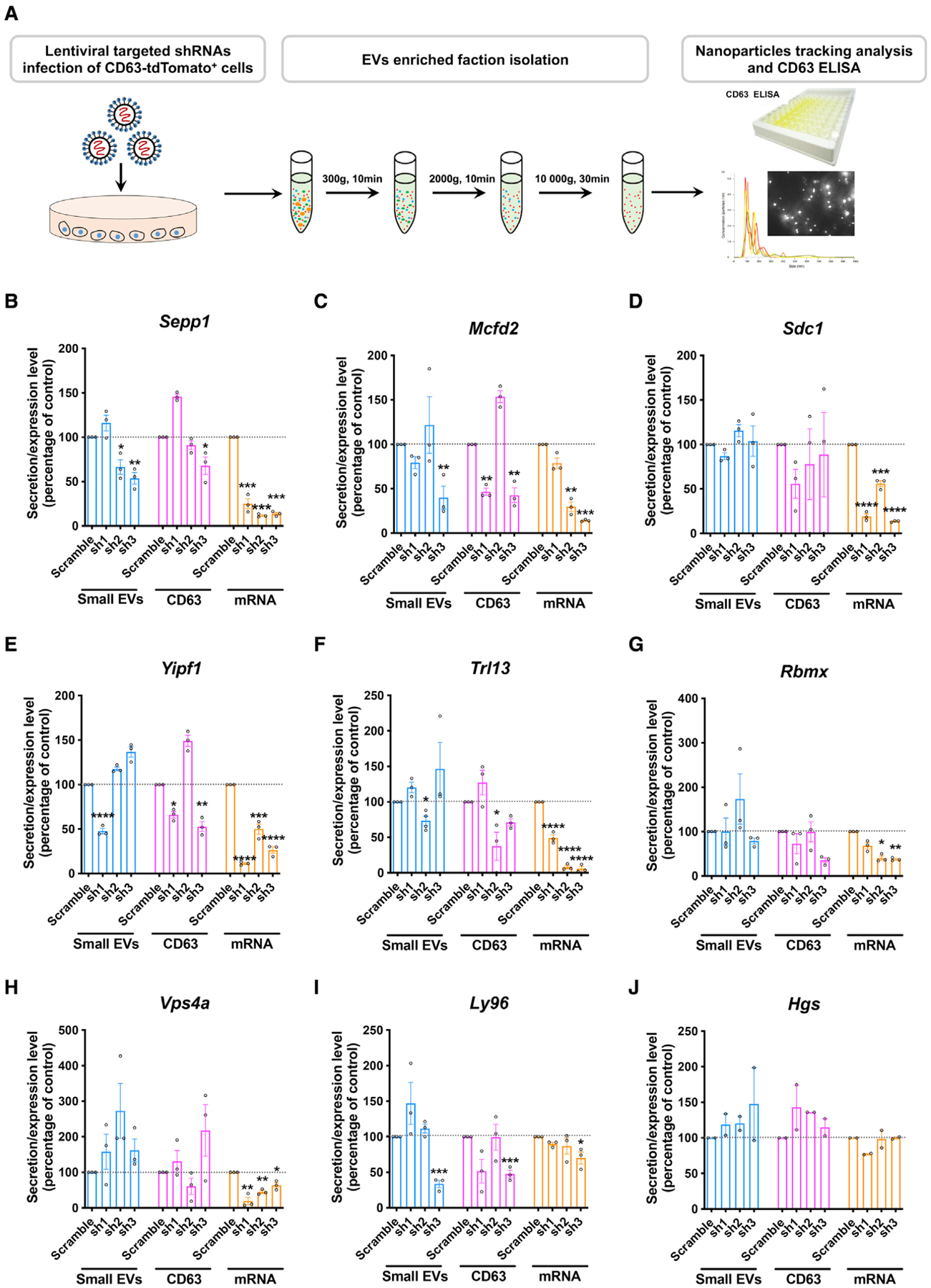
(A) Workflow to validate a potential “hit” by NTA and CD63 ELISA.
(B–J) EV and CD63 secretion and specific mRNA levels in cells transduced with lentiviral shRNA clones targeting Sepp1 (B), Mcfd2 (C), Sdc1 (D), Yipf1 (E), Tlr13 (F), Rbmx (G), Vps4a (H), Ly96 (I), and Hgs (J). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the scramble shRNA group, as determined by one-way ANOVA (alpha = 0.05) and Tukey’s post hoc test. n = 3 independent experiments, a technical duplicate analysis for each sample in CD63 ELISA and qRT-PCR. Graphs indicate mean ± SEM.
Silencing of Mcfd2, Sepp1, and Sdc1 inhibits EV secretion from primary microglia
Upon microbe exposure, microglia rapidly act as the first line of defense and trigger a pro-inflammatory response. Stimulation with LPS activates TLR4 signaling and elevates EV secretion from microglia (Yang et al., 2018b). To validate these candidate genes in murine microglia under LPS stimulation, primary cultured microglia were treated with siRNA to silence Sepp1, Mcfd2, or Sdc1. Although the target genes were efficiently silenced, there was no obvious morphology change of microglia 3 days after siRNA transduction (Figures 5A–5D). Strikingly, the number of EVs and total particles secreted by microglia were significantly reduced because of silencing of Sepp1 or Mcfd2 (Figures 5B and 5C). These findings indicate that these two genes are essential factors for EV secretion from microglia induced by LPS and ATP. On the other hand, we also observed a similar reduction effect of Sdc1 siRNA (Figure 5D), which suggested a difference in lentivirus transduction of shRNA in BV-2 cells. This inconsistent result between the two cell models may be due to Sdc1 not having been significantly downregulated by these three shRNAs packaged in the lentivirus. We successfully silenced Ly86 using siRNA in primary microglia (Figure S4D), which was not achieved in BV-2 cells. However, there was no obvious effect on EV secretion induced by ATP stimulation (Figures S4C, S4E, and S4F). We also examined whether there were any changes in EV composition after silencing of the target molecules. For that purpose, EVs were enriched by qEV-based size fractionation and examined for representative EV markers (CD63, Tsg101, and flotillin) and tdTomato. There were no obvious changes in these markers after adjusting the total protein load for each lane (Figure S6), suggesting no qualitative changes in EV composition by targeting Sepp1, Mcfd2, or Sdc1.
Figure 5. Validation of EVs secreted by Mcfd2, Sepp1, and Sdc1 knockdown primary cultured murine microglia.
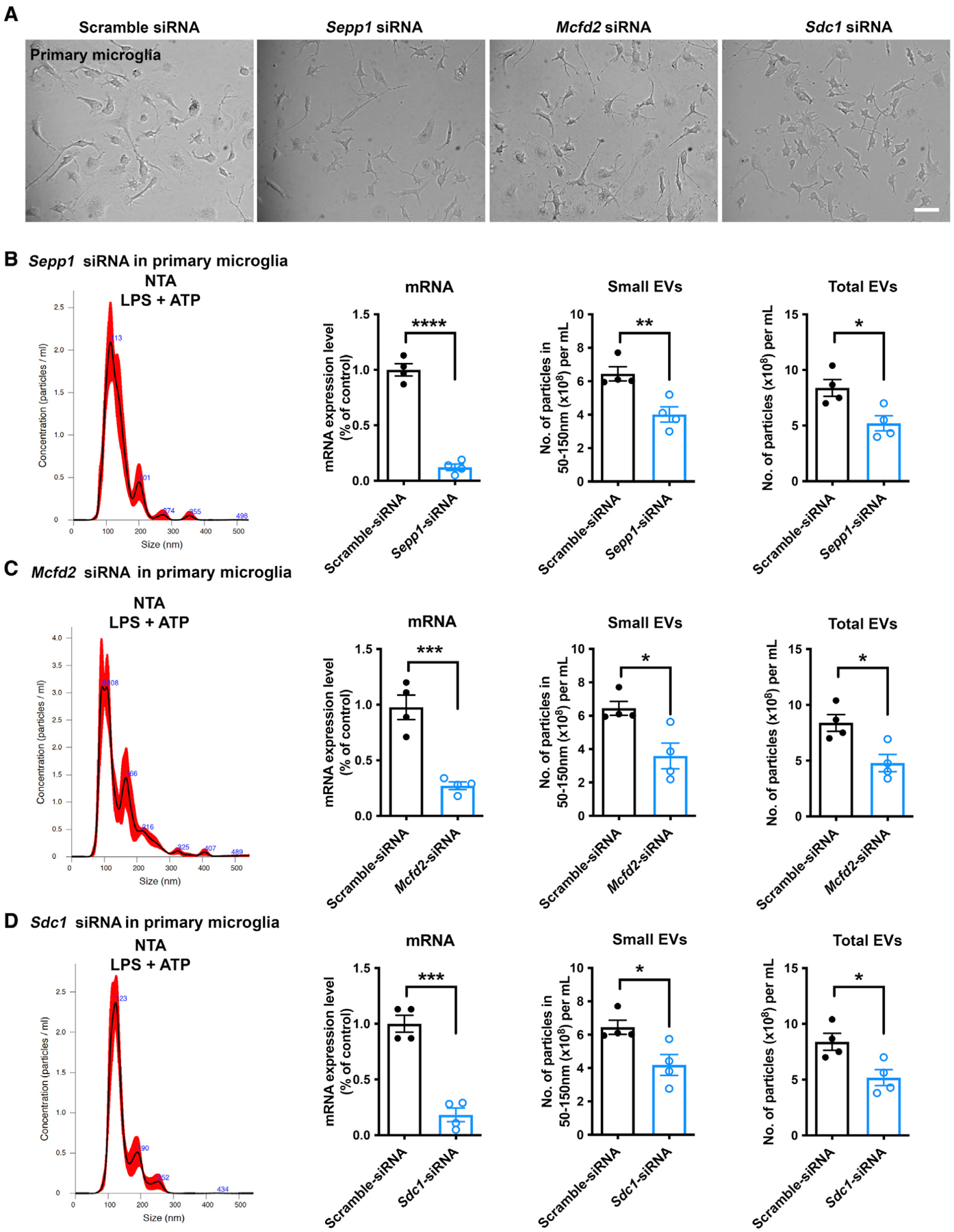
(A) Primary microglia transfected with siRNA targeting Sepp1, Mcfd2, and Sdc1.
(B–D) Particle distribution, specific mRNA levels, and EV and total particle secretion in primary microglia transduced with siRNAs targeting Sepp1 (B), Mcfd2 (C), and Sdc1 (D). Two-tailed Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the scramble shRNA group; n = 4 independent experiment, each sample recorded with 4 videos. Graphs indicate mean ± SEM.
For the other pathological conditions, we evaluated the effect on EV release under two additional different stimulation conditions: tau aggregates and polyinosinic-polycytidylic acid (poly [I:C]). The most recent study from our lab suggested that tau aggregate stimulation induces neurodegenerative microglia and significantly increases secretion of EV from microglia in APPNL-G-F/NL-G-F knockin mouse brains (Clayton et al., 2021). We also tested poly(I:C), which is known to enhance EV secretion (Srinivasan et al., 2017; Hong et al., 2021). Treatment of the target gene-silenced BV-2 reporter cell line created by shRNA lentivirus infection with tau aggregates significantly induced small and total EVs (Figures S7A and S7B). Poly(I:C) treatment, however, did not show significant enhancement of EV secretion at any fraction. Interestingly, tau aggregate-induced secretion of small and total EVs was diminished by silencing of Sepp1, Mcfd2, or Sdc1 (Figures S7A and S7B). Unexpectedly, poly(I:C)-induced total EV secretion was significantly increased under the Sepp1 silencing condition (Figure S7B), although its level was not significantly increased compared with scramble shRNA-treated control microglia. This suggest additional Sepp1-mediated regulation of EV secretion, which is different from its effect on small EVs. These findings suggest small EV-specific regulation of EV secretion by Sepp1, Mcfd2, and Scd1 in microglia.
Reduction in IL-1β release from primary microglia
IL-1β is a crucial pro-inflammatory cytokine that acts as an end product of inflammasome activation, caspase-1 activation, and cleavage of pro-IL-1β. IL-1β is located in the cytosol, although a proportion remains in vesicles and is protected from clearance (Rubartelli et al., 1990; MacKenzie et al., 2001). These IL-1β-containing vesicles have been considered MVBs or endolysosomal systems in myeloid cells (Andrei et al., 1999). It has been proposed that a fraction of cellular IL-1β is targeted to lysosomes for degradation and that this fraction can be secreted by triggering lysosome exocytosis and, thus, secretion of IL-1β. Therefore, a fraction of cellular IL-1β targeted for degradation can be secreted by EVs into the extracellular environment. EVs, including EV-associated IL-1β, can be stimulated by LPS (Qu et al., 2007; Fitzgerald et al., 2018). Thus, we tested whether microglial LPS-induced IL-1β secretion could be affected by silencing Sepp1, Mcfd2, or Sdc1. Primary cultured murine microglia were stimulated with LPS for 4 h, followed by stimulation with ATP for 15 min and collection of conditioned medium for evaluation of EV-associated IL-1β secretion (Figure 6A). Compared with the untreated control, there was a significant increase in EV-associated IL-1β levels after LPS + ATP stimulation (Figures 6B–6D). Strikingly, knockdown of Sepp1, Mcfd2, or Sdc1 significantly reduced IL-1β in microglial EVs (Figures 6B–6D). There was significant induction of Il1b and Il18 gene expression by LPS but no effect on their expression in cells after gene silencing of Sepp1, Mcfd2, or Sdc1, suggesting that the reduction in IL-1β secretion is not due to the reduction of Il1b gene expression by gene silencing (Figures 6E–6G). These data demonstrate that Sepp1, Mcfd2, and Sdc1 were involved in ATP-induced EV secretion, and that LPS + ATP induced EV-associated IL-1β secretion from microglia.
Figure 6. IL-1β secretion in EVs from primary murine microglia.
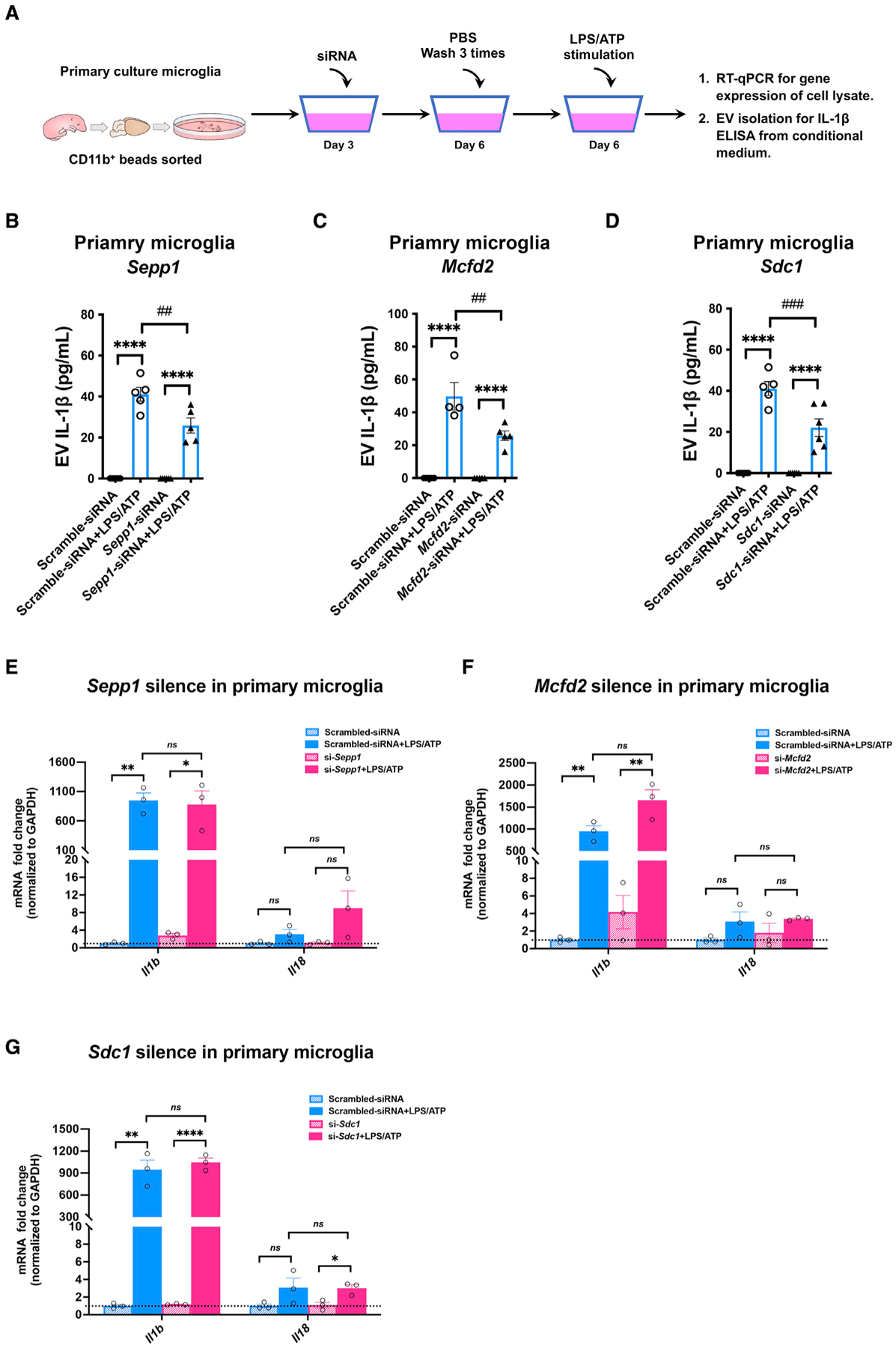
(A) The workflow for measuring pro-inflammatory cytokine IL-1β secretion from microglia after siRNA silencing.
(B–D) EV-associated IL-1β from primary microglia transduced with siRNAs targeting Sepp1 (B), Mcfd2 (C), and Sdc1 (D). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control group; ##p < 0.01, ###p < 0.001 compared with the scramble group; determined by two-way ANOVA (alpha = 0.05) and Tukey’s post hoc test; n = 4–5 independent experiments. Graphs indicate mean ± SEM.
(E–G) mRNA expression of Il1b and Il18 in primary cultured murine microglia after siRNA-based silencing of Sepp1 (E), Sdc1 (F), and Mcfd2 (G); treated with vehicle or LPS/ATP; and tested for mRNA expression of Il1b and Il18. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control group; determined by two-way ANOVA (alpha = 0.05) and Tukey’s post hoc test; n = 3 independent experiments. Graphs indicate mean ± SEM.
DISCUSSION
The present genetic screening was designed to identify regulators of ATP-induced EV secretion from microglia. Specifically, the tdTomato-CD63 reporter system expressed in the BV-2 microglial cell line was successfully developed and utilized within the critical time window for cell sorting after ATP stimulation. CD63 is known to be localized to ILVs in MVBs and is enriched on the outer leaflet of EVs (Hurwitz et al., 2018). The fact that we identified multiple genes of different functional families linked to specific pathways indicates that our shRNA-based gene screening was successful. Our screening procedure was developed after extensive optimization with several promoters driving gene expression. It was important to ensure that the number of cells transduced with a given shRNA was sufficient to reliably detect over-representation in the selected populations, particularly given that lentiviral transduction of microglia is relatively inefficient.
There is a precedent of guide RNA (gRNA)-based CRISPR-Cas9 genome editing cell library screening using bEXOmiRs as reporters of EV release (Lu et al., 2018). This is based on K562, a human myelogenous leukemia cell line that is not compatible with microglia-specific cell library screening. The advantage of gRNA-based editing is the potential for complete deletion of the target if it is a homozygotic disruption. The disadvantage of the platform is that it is not compatible with essential genes for cell survival or proliferation. The advantage of shRNA-based screening is that it does not disrupt the genomic sequence of the target genes and can target essential genes for survival or proliferation; a disadvantage is incomplete silencing of the target genes. Lu et al. (2018) focused on identification of genes involved in EV secretion without stimulation, whereas our study is more focused on ATP-induced EV secretion. This may also result in identification of different genes in our screening.
Sepp1, Mcfd2, and Sdc1 as significant regulators of EV secretion from microglia
The validation of top-ranked hits identified three factors, Sepp1, Mcfd2, and Sdc1, as playing a key role in EV secretion from LPS + ATP-stimulated microglia. Sepp1 encodes selenoprotein P, a secreted heparin-binding glycoprotein, most notably present in the brain and testes (Olson et al., 2007). As the most abundant selenoprotein in cerebrospinal fluid, Sepp1 plays a central role as a brain selenium supplier (Kuhbacher et al., 2009). Sepp1 binds to apolipoprotein E receptor 2, a member of the lipoprotein receptor family, using the clathrin-dependent endocytosis pathway and mediates delivery of selenium from hepatocytes to the brain (Kurokawa et al., 2012). Sepp1 knockout mice showed decreased brain and neurological dysfunction, recapitulating the phenotype of mice raised on a diet without selenium (Hill et al., 2004). Interestingly, Sepp1 is secreted from hepatocytes as an exosomal component and is protected from plasma kallikrein-mediated degradation (Saito et al., 2004). The Sepp1 gene is one of the IL-10-inducible genes and is associated with alternative activation of myeloid cells (Bosschaerts et al., 2008). Thus, Sepp1 may be involved in microglial activation for phagocytosis, EV synthesis, and secretion in a direct or indirect manner. In an in vitro blood-brain barrier reconstitution model consisting of mouse brain microvascular endothelial cells and neuroblastoma cells, exosomal Sepp1-supplied selenium was utilized to synthesize other selenoproteins in endothelial and neuronal cells (Jin et al., 2020). The major cholesterol carrier apolipoprotein E potentially plays a role in exosomal Sepp1 secretion and transport in hepatocytes (Jin et al., 2020). Another 11-kDa selenoprotein, selenoprotein K, is involved in Ca2+ flux in LPS-activated macrophages, leading to enhanced migration and phagocytosis of microglia. (Huang et al., 2011; Meng et al., 2019) Thus, Sepp1 might have a similar effect on microglia, and silencing of Sepp1 may lead to microglia resistance to the ATP-induced Ca2+ influx for exocytosis of EVs.
Mcfd2 is a helix-loop-helix structural domain protein that forms a calcium-dependent heteromeric complex with lectin mannose binding 1 (Lman1), which is essential for cargo formation in transport of the coagulation factors FV and FVIII (Nyfeler et al., 2006). The Mcfd2-Lman1 complex and cargo proteins are transported from the endoplasmic reticulum-Golgi intermediate compartment to the cis-Golgi. This is a well-studied transport pathway of glycosylated FV and FVIII (Guy et al., 2008; Zhang et al., 2005), specifically regarding sites of vesicle budding within the endoplasmic reticulum lumen. Interestingly, EVs secreted from hepatocellular carcinomas contain Mcfd2, as determined by mass spectroscopy (He et al., 2015). Our data suggest that Mcfd2 also participates in EV secretion from microglia. In the CNS, the functional role of Mcfd2 is poorly understood, although it may function as a potential autocrine and paracrine mechanism for neural stem cell maintenance (Liu et al., 2013). Because it is predominantly expressed in mouse microglia and endothelial cells (Zhang et al., 2014), further investigation is needed to understand the exact mechanisms by which Mcfd2 mediates EV secretion in these cell types.
Sdc1 is the most widely studied member of four structurally related cell surface heparan sulfate proteoglycans that serve as co-receptors for several ligand molecules to regulate their signaling and distribution. Sdc1 mediates the biogenesis of EVs in tumor cells. (Choi et al., 2007; Welton et al., 2010) Formation of EVs is driven by the interaction of the Sdc1 cytoplasmic domain with syntenin and programmed cell death 6-interacting protein to form a complex that facilitates budding of ILVs within endosomal membranes (Baietti et al., 2012). This mechanism depends on Src-mediated Sdc1 endocytosis and requires phospholipase D2 and ADP-ribosylation factor 6 GTPase activity (Durcin et al., 2017). Increasing the levels of heparinase by upregulation or exogenous application in tumor cells results in increased secretion of EVs that are retained in their cargo, Sdc1, and haparanase (Thompson et al., 2013). A more recent study has demonstrated that EV production is significantly reduced upon knockdown of Sdc1 and Sdc4 in MCF-7 cells (Roucourt et al., 2015), a commonly used breast cancer cell line. Although we could not suppress EV secretion from BV-2 cells by shRNA lentiviral infection, we did observe a reduction in EV secretion from primary murine microglia after siRNA-based silencing of Sdc1, indicating its role in EV secretion.
IL-1β secretion upon LPS/ATP stimulation
EVs have been suggested to regulate inflammatory signals and modulate intercellular communication (Yoon et al., 2014; Delpech et al., 2019). Studies have shown that microglia release EVs containing IL-1β upon exposure to ATP derived from astrocytes (Bianco et al., 2005a). The present study demonstrated that silencing of the genes Sepp1, Mcfd2, and Sdc1 can dampen EV-associated IL-1β secretion without affecting the expression of Il1b, although each molecule appears to act on different exocytotic or vesicular transport machinery in the microglia.
Limitation of the study
The study has some limitations. First, it is undetermined whether this reflects down-regulation of IL-1β in EVs or associated with EVs. Our attempt to differentiate them by size exclusion chromatography-based EV isolation was unsuccessful because of insufficient sensitivity of the commercially available IL-1β ELISA kit. Further study will be necessary to distinguish the location of EV-associated IL-1β. IL-18 is another cytokine known to be generated by cleavage of pro-IL-18 by caspase-1 and secreted in a similar pathway as IL-1β. We were unable to detect EV-associated IL-18 using the commercially available ELISA kit; this will need to be examined using a more sensitive assay platform. Investigation of EV secretion from microglia involved in the inflammatory process may broaden our understanding of the roles of microglia in neurodegenerative diseases as well as their potential for therapeutic manipulation.
We cannot exclude the possibility that other potential genes were not identified because of inefficient silencing efficiency of the target genes, lack of expression of the genes of interest, toxicity because of gene silencing, or incomplete coverage of the shRNA libraries. Our screening system also has a potential pitfall because the threshold for collecting tdTomatohigh TurboGFP+ cells was arbitrary, and other potential candidate genes may have been missed. Possible sources of false positive screening results include silencing of genes that repress the murine EF1α promoter for tdTomato-CD63 expression, although those genes were not selected in our top hits. Our validation assays segregated these genes from the identified genes, but it is possible that our screening did not identify all potential candidates.
Conclusions
This study reports the successful development of a tdTomato-CD63+TurboGFP+ BV-2 cell library of barcoded shRNA clones for genome-wide screening. This was used to discover important mechanisms underlying EV secretion after ATP− or ATP+ LPS-induced microglial activation. We also identified important host targets for regulating EV and IL-1β release from microglia. We found that Sepp1, Mcfd2, and Sdc1 are required for EV secretion from primary cultured microglia and their involvement in IL-1β secretion in EVs upon LPS/ATP stimulation. These molecules are potential therapeutic targets for ameliorating EV- and IL-1β-mediated disease progression in neuroinflammatory disorders.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Any additional information and requests for reagents should be directed to and will be fulfilled by the lead contact, Tsuneya Ikezu (ikezu.tsuneya@mayo.edu).
Materials availability
Reagents used in this study are publicly available or available from the lead contact upon request.
Data and code availability
Next generation sequencing data have been deposited at the Sequence Read Archive / BioProject and are publicly available upon publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to re-analyze data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-early endosome antigen A11 (EEA1) | Santa Cruz Biotechnology | sc-137130: RRID:AB_2246349 |
| Mouse monoclonal anti-Rab5 | BD Biosciences | 610725: RRID:AB_398048 |
| Rabbit Monoclonal anti-Rab7 | Cell Signaling Technology | 9367: RRID:AB_1904103 |
| Rabbit polyclonal anti-CD63 | Biorbyt | orb229829 |
| Rabbit anti-mouse bridging antibody | Abcam | ab6709; RRID:AB_956006 |
| Mouse monoclonal anti-CD63 | BioLegend | 143901; RRID:AB_11203908 |
| Rabbit Polyclonal anti-tdTomato | Thermofisher scientific | TA150128 |
| Mouse monoclonal anti-Tsg101 (Y16J) | Santa Cruz Biotechnology | sc-101254: RRID:AB_1130856 |
| Mouse monoclonal anti-calregulin | Santa Cruz Biotechnology | sc-373863; RRID:AB_10915425 |
| Mouse monoclonal anti-cytochrome C | Santa Cruz Biotechnology | sc-13560; RRID:AB_627383 |
| Mouse monoclonal anti-GM130 | Santa Cruz Biotechnology | sc-53296; RRID:AB_629538 |
| Mouse monoclonal anti-Flotillin | Santa Cruz Biotechnology | sc-74566; RRID:AB_2106563 |
| Rabbit Polyclonal anti-Histone H2A.Z | Cell Signaling Technology | 2718; RRID:AB_659840 |
| Rabbit Polyclonal anti-LC3B antibody | Cell Signaling Technology | 4108S; RRID:AB_2137703 |
| Mouse monoclonal anti-β-actin | Sigma-Aldrich | A1978; RRID:AB_476692 |
| AlexaFluor-647 Donkey anti-rabbit | Thermofisher scientific | A-31573; RRID:AB_2536183 |
| AlexaFluor-488 Donkey anti-mouse | Thermofisher scientific | R37114; RRID:AB_2556542 |
| Goat anti-rabbit IgG HRP-linked antibody | Cell Signaling Technology | 7074; RRID:AB_2099233 |
| Horse anti-mouse IgG, HRP-linked antibody | Cell Signaling Technology | 7076; RRID:AB_330924 |
| Bacterial and virus strains | ||
| Agilent SURE 2 Supercompetent Cells | Agilent technologies | 200152 |
| pLenti-tdTomato-CD63 | This paper | N/A |
| pLenti-puro vector | Addgene | Plasmid #39481; RRID:Addgene_39481 |
| SMARTvector Mouse Whole Genome Lentiviral shRNA Pooled Library mEF1a-GFP | Horizon Discovery Ltd. | V3SM8631 |
| SMARTchoice Promoter Selection Plate | Horizon Discovery Ltd. | SP-001000-01 |
| pLKO.1-puro-CMV-tGFP- Ly86-Clo.1 | Sigma-Aldrich | TRCN0000101268 |
| pLKO.1-puro-CMV-tGFP- Ly86-Clo.2 | Sigma-Aldrich | TRCN0000101269 |
| pLKO.1-puro-CMV-tGFP- Ly86-Clo.3 | Sigma-Aldrich | TRCN0000101266 |
| pLKO.1-puro-CMV-tGFP-Ly96-Clo.1 | Sigma-Aldrich | TRCN0000066224 |
| pLKO.1-puro-CMV-tGFP-Ly96-Clo.2 | Sigma-Aldrich | TRCN0000326822 |
| pLKO.1-puro-CMV-tGFP-Ly96-Clo.3 | Sigma-Aldrich | TRCN0000326824 |
| pLKO.1-puro-CMV-tGFP-Vps4A-Clo.1 | Sigma-Aldrich | TRCN0000287397 |
| pLKO.1-puro-CMV-tGFP-Vps4A-Clo.2 | Sigma-Aldrich | TRCN0000101820 |
| pLKO.1-puro-CMV-tGFP-Vps4A-Clo.3 | Sigma-Aldrich | TRCN0000101822 |
| pLKO.1-puro-CMV-tGFP-MCFD-Clo.1 | Sigma-Aldrich | TRCN0000196018 |
| pLKO.1-puro-CMV-tGFP-MCFD-Clo.2 | Sigma-Aldrich | TRCN0000183944 |
| pLKO.1-puro-CMV-tGFP-MCFD-Clo.3 | Sigma-Aldrich | TRCN0000183004 |
| pLKO.1-puro-CMV-tGFP-Trl13-Clo.1 | Sigma-Aldrich | TRCN0000066034 |
| pLKO.1-puro-CMV-tGFP-Trl13-Clo.2 | Sigma-Aldrich | TRCN0000368665 |
| pLKO.1-puro-CMV-tGFP-Trl13-Clo.3 | Sigma-Aldrich | TRCN0000360480 |
| pLKO.1-puro-CMV-tGFP-Sepp1-Clo.1 | Sigma-Aldrich | TRCN0000120685 |
| pLKO.1-puro-CMV-tGFP-Sepp1-Clo.2 | Sigma-Aldrich | TRCN0000352521 |
| pLKO.1-puro-CMV-tGFP-Sepp1-Clo.3 | Sigma-Aldrich | TRCN0000340550 |
| pLKO.1-puro-CMV-tGFP- Hgs-Clo.1 | Sigma-Aldrich | TRCN0000088689 |
| pLKO.1-puro-CMV-tGFP- Hgs-Clo.2 | Sigma-Aldrich | TRCN0000088691 |
| pLKO.1-puro-CMV-tGFP- Hgs-Clo.3 | Sigma-Aldrich | TRCN0000088692 |
| pLKO.1-puro-CMV-tGFP- Yipf1-Clo.1 | Sigma-Aldrich | TRCN0000126345 |
| pLKO.1-puro-CMV-tGFP- Yipf1-Clo.2 | Sigma-Aldrich | TRCN0000126346 |
| pLKO.1-puro-CMV-tGFP- Yipf1-Clo.3 | Sigma-Aldrich | TRCN0000126348 |
| pLKO.1-puro-CMV-tGFP- Rbmx-Clo.1 | Sigma-Aldrich | TRCN0000104042 |
| pLKO.1-puro-CMV-tGFP- Rbmx-Clo.2 | Sigma-Aldrich | TRCN0000438339 |
| pLKO.1-puro-CMV-tGFP- Rbmx-Clo.3 | Sigma-Aldrich | TRCN0000434600 |
| pLKO.1-puro-CMV-tGFP- SDC-Clo.1 | Sigma-Aldrich | TRCN0000302195 |
| pLKO.1-puro-CMV-tGFP- SDC-Clo.2 | Sigma-Aldrich | TRCN0000302270 |
| pLKO.1-puro-CMV-tGFP- SDC-Clo.3 | Sigma-Aldrich | TRCN0000311084 |
| pLKO.1-puro-CMV-tGFP-scramble | Sigma-Aldrich | Non-Target shRNA Control |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Thermofisher scientific | D1306; RRID:AB_2629482 |
| Adenosine 5 -triphosphate | Sigma-Aldrich | A6419-5G |
| Tris-HCl | Sigma-Aldrich | 10812846001 |
| EDTA | Sigma-Aldrich | E9884-100G |
| NaCl | Sigma-Aldrich | S9888-500G |
| 3-Methyladenine | Sigma-Aldrich | M9281 |
| Chloroquine | Sigma-Aldrich | C6628 |
| Tau-410, (2N3R) | rPeptide | T-1002-2 |
| Tau-441, (2N4R) | rPeptide | T-1001-2 |
| Critical commercial assays | ||
| CD63 ELISA | LifeSpan Biosciences | LS-F7104 |
| IL-1β ELISA | Abcam | ab197742 |
| Experimental models: Cell lines | ||
| BV2 cell line | CABRI | ICLC ATL03001; RRID:CVCL_0182 |
| HEK293FT cell line | Invitrogen | R70007 |
| Experimental models: Organisms/strains | ||
| Mouse: CD-1® IGS Mouse | Charles River Laboratories | strain code 022 |
| Oligonucleotides | ||
| Accell Non-targeting siRNA | Horizon Discovery Ltd. | D-001910-01-50 |
| Accell Mouse Mcfd2 (193813) siRNA - SMARTpool | Horizon Discovery Ltd. | E-052885-00-0020 |
| Accell Mouse Selenop (20363) siRNA - SMARTpool | Horizon Discovery Ltd. | E-041618-00-0020 |
| Accell Mouse Sdc1 (20969) siRNA -SMARTpool | Horizon Discovery Ltd. | E-044225-00-0020 |
| Accell Mouse ly86 (17084) siRNA -SMARTpool | Horizon Discovery Ltd. | E-044084-00-0005 |
| Ly86 TaqMan probes | Thermofisher scientific | Mm00440240_m1 |
| Ly96 TaqMan probes | Thermofisher scientific | Mm01227593_m1 |
| Vps4a TaqMan probes | Thermofisher scientific | Mm00459928_m1 |
| Mcfd2 TaqMan probes | Thermofisher scientific | Mm00454818_m1 |
| Hgs TaqMan probes | Thermofisher scientific | Mm00468635_m1 |
| Sdc1 TaqMan probes | Thermofisher scientific | Mm00448918_m1 |
| Sepp1 TaqMan probes | Thermofisher scientific | Mm00486048_m1 |
| Rbmx TaqMan probes | Thermofisher scientific | Mm00726735_s1 |
| Tlr8 TaqMan probes | Thermofisher scientific | Mm04209873_m1 |
| Yipf1 TaqMan probes | Thermofisher scientific | Mm00523828_m1 |
| Gapdh TaqMan probes | Thermofisher scientific | Mm99999915_g1 |
| IL-1β Forward: GAAATGCCACCTTTTGACAGTG | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| IL-1β Reverse: RTGGATGCTCTCATCAGGACAG | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| IL18 Forward: GTGAACCCCAGACCAGACTG | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| IL18 Reverse: CCTGGAACACGTTTCTGAAAGA | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| GAPDH Forward: TGGCCTTCCGTGTTCCTAC | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| GAPDH Reverse: GAGTTGCTGTTGAAGTCGCA | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| Illumina-adapted SMARTvector Indexing PCR primer Kit A | Horizon Discovery Ltd. | PRM7668 |
| Illumina-adapted SMARTvector Indexing PCR primer Kit B | Horizon Discovery Ltd. | PRM10186 |
| Recombinant DNA | ||
| tdTomato-CD63 | This paper | Synthesized by GenScript Biotech Inc |
| Software and algorithms | ||
| (Fiji is just) ImageJ 2.0.0 | National Institutes of Health | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8 | GraphPad Software Inc. | |
| DAVID Bioinformatic Resources 6.8 | National Institute of Health | https://david.ncifcrf.gov |
| STRING functional protein association tool | Szklarczyk et al. (2019) | http://string-db.org/ |
| Metascape | Zhou et al., 2019 | http://metascape.org/ |
| BioRender | BioRender | https://www.BioRender.com |
| Other | ||
| DMEM Medium | ThermoFisher scientific | 11965118 |
| Fetal Bovine Serum | ThermoFisher scientific | 10082147 |
| GlutaMAX™ Supplement | ThermoFisher scientific | 35050061 |
| Phosphate buffered saline (PBS) | ThermoFisher scientific | 10-010-031 |
| HBSS, calcium, magnesium, no phenol red | ThermoFisher scientific | 4025134 |
| HBSS, no calcium, no magnesium, no phenol red | ThermoFisher scientific | 14175095 |
| Lipopolysaccharide | Sigma-Aldrich | L3024 |
| 0.45 μm Millex-HV filter | Millipore Sigma | SLHV033RS |
| Medical Millex-GP Syringe Filter Unit, 0.22 μm | Millipore Sigma | SLGPM33RS |
| Puromycin | ThermoFisher scientific | A1113803 |
| DNeasy Blood & Tissue Kit | Qiagen | 69504 |
| Phusion Hot Start II DNA Polymerase | Thermo Fisher Scientific | F549L |
| Ultrapure UltraPure Agarose | Thermo Fisher Scientific | 16500500 |
| Gel Extraction Kit | Denville | CM-510-250 |
| Trizol | Thermo Fisher Scientific | 15596026 |
| SuperScript™ IV VILO™ Master Mix kit | Thermo Fisher Scientific | 11766050 |
| 2X TaqMan Gene Expression Master Mix | Life Technologies | 4369016 |
| Applied Biosystems™ SYBR™ Green PCR Master Mix | Applied Biosystems | 4309155 |
| CD11b bead | Miltenyi Biotec | 130-049-601 |
| Neural Tissue Dissociation Kit | Miltenyi Biotec | 130-093-231 |
| MACS column | Miltenyi Biotec | 30-042-201 |
| Accell siRNA delivery media | Horizon Discovery Ltd. | B-005000-100 |
| Murine monocyte colony-stimulating factor | BioVision | 4238 |
| Poly(I:C) | InvivoGen | tlrl-pic |
| Heparin | Stemcell Technologies | 07980 |
| RIPA buffer | Thermo Fisher Scientific | 89900 |
| Protease / phosphatase inhibitor cocktail | Thermo Fisher Scientific | PI78443 |
| 4–20% SDS–PAGE precast gels | Bio-Rad | 4561093 |
| 4x Laemmli Sample Buffer | Bio-Rad | 1610747 |
| Nitrocellulose membranes | Bio-Rad | 162-0090 |
| Normal Donkey serum | Sigma-Aldrich | 9663-10ML |
| Triton-X 100 | Sigma-Aldrich | T9284-100ML |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 |
| Next Generation Sequencing Dataset | SRA / BioProject | PRJNA826851 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethical approval and consent to participate
All animal studies and procedures were performed in accordance with the Boston University and Mayo Clinic institutional animal care and use committee (IACUC) approved protocols. There is no human subjects involved in this study.
Animals
E18 pregnant female mice (CD-1) were directly purchased from Charles River Laboratories (strain code 022) for delivering pups (P0 to P3) for the primary microglial culture.
Cell lines
Murine microglia BV2 cell lines (cat. no. ICLC ATL03001, CABRI) were grown at 37°C in Dulbecco’s minimal essential medium (DMEM) (ThermoFisher scientific, #11965118) supplemented with 10% Fetal Bovine Serum (FBS, Invitrogen, #10082147), 2 mM GlutaMAX™ Supplement (ThermoFisher scientific, #35050061). HEK293FT cells (Invitrogen, #R70007) were grown at 37°C in DMEM medium supplemented with 10% FBS. Cell lines were authenticated by short tandem repeat profiling and tested periodically for the presence of mycoplasma.
METHOD DETAILS
pLenti-tdTomato-CD63 vector construction and lentivirus production
The tdTomato DNA sequence, fused to the human CD63 gene (Entrez gene ID 967), was synthesized by GenScript Biotech Inc., and cloned into the pLenti-puro vector (Addgene, #39481) to create the pLenti-tdTomato-CD63 vector. Lentivirus particles of pLenti-tdTomato-CD63 were generated by transfection of HEK293FT cells with a transducing vector and the packaging vectors pLP1, pLP2, and pLP/VSV-G as previously described (Holehonnur et al., 2015). The supernatant containing virus particles was collected 48 and 72 h after transfection, filtered through a 0.45 μm Millex-HV filter (Millipore Sigma, SLHV033RS), concentrated by ultracentrifugation at 100,000 × g for 2 h, and stored at −80°C until use.
Establishment of tdTomato-CD63 expressing BV-2 cell line for shRNA screening
BV-2 cells were infected with pLenti-tdTomato-CD63 lentiviral particles and incubated at 37°C overnight; medium was replaced following incubation. The medium was changed daily, and at 48 h post-infection, puromycin (Invitrogen, 2 μg/mL) was added to the medium. After 2–3 d, puromycin-selected cells were plated onto a 96-well plate at limiting cell dilutions and incubated with higher concentrations of puromycin (2–5 μg/mL). Single cell with tdTomato fluorescence that grew into individual colonies were transferred to a new 24-well plate. The stable tdTomato-CD63+ BV-2 cell line was then expanded and maintained with puromycin.
Lentivirus transduction with the pooled shRNA library
The pooled shRNA library was constructed with the Horizon Discovery Ltd. SMART vector lentiviral shRNA constructs expressing shRNAs targeting a genome-wide library. The library encompassed 18 pools (9,860 constructs per pool) targeting 21,924 genes (8 shRNA designs per gene). These included 34 protein-coding genes, such as Actb, Gapdh, Lamb1, and Ppib, and 100 non-targeting shRNA controls in each pool. All constructs in the library were in the constitutive promoter of mEF1α with the fluorescent reporter TurboGFP, which has been validated as the best promoter in the BV-2 line (Figure S3). For the pooled library shRNA lentivirus particles, tdTomato-CD63+ BV-2 cells were transduced at a low MOI (0.3 TU/cell) to produce a population of cells with one integration event per cell. Following lentiviral transduction, cells successfully carrying the fluorescent TurboGFP were purified using fluorescence activated cell sorting (FACS) Beckman Coulter MoFlo Astrios (around 21.09% of positive cells of all 18 pools). Most cells that remained after selection carried a single copy of the inducible shRNA. Another population with a low fluorescent signal in tdTomatolow (TurboGFPlow) was collected as an uninfected negative control for future comparison. TurboGFPhigh cells were then expanded in culture for additional 2–3 d to obtain at least the number of cells that corresponds to the desired number of lentiviral integrants. Cells were then used to perform the screening as described below.
Lentiviral shRNA-transduced cell library screening
The pooled shRNA lentiviral library was screened in tdTomato-CD63+ BV-2 cells, as summarized in Figure 1. After viral integration, cells were stimulated with 5 mM ATP for 15 min and then harvested for FACS. To maximize the potential for phenotypic effects in the final analysis, we collected an average representation of the top 1% of tdTomatohigh (TurboGFPhigh) cells per pool, representing the cells resistant to ATP stimulation. In addition, we ensured that the cells were always in log growth during the screening (<70% confluence) to minimize changes in shRNA representation caused by localized restriction of cell growth due to over-confluence. After performing FACS, cells were collected by centrifugation at 5000g for 5 min, and the cell pellet was snap-frozen in a dry-ice/ethanol bath and stored at −80°C until use.
Genomic DNA extraction, barcode amplification, and next generation sequencing
DNA was isolated from the collected cells using a DNeasy Blood & Tissue Kit (Qiagen, #69504) according to the manufacturer’s instructions. Pooled barcodes were amplified from 825 ng of genomic DNA by 50 μL reactions of PCR using Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific, #F549L) and the Illumina-adapted SMARTvector Indexing PCR primer Kit A and B (Horizon Discovery Ltd., PRM7668, PRM10186) according to the manufacturer’s instructions. PCR was performed at 98°C for 3 min followed by 35 cycles of 98°C for 10 s, 60°C for 15 s, 72°C for 15 s, and 72°C for 5 min on a T100™ Thermal Cycler (Bio-Rad, #1861096). PCR products were analyzed by electrophoresis on a 2% agarose gel, and DNA was purified using the Gel Extraction Kit (Denville, CM-510-250) following the manufacturer’s protocol. DNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific), and concentrations were adjusted to 15 ng/μL. All DNA samples showed a single peak at 237 bp, as analyzed by a 2100 Bioanalyzer Instrument together with the 2100 Expert Software and Bioanalyzer assays at the Boston University Microarray and Sequencing Resource Core. HTS of amplified barcodes was performed at the Boston University Microarray Core on the Illumina NextSeq 500 Next Generation Sequencing using the SMARTvector Read 1 Sequencing Primer, following the manufacturer’s protocol. Briefly, barcoded DNA libraries were pooled in equimolar concentrations and templates were prepared and enriched for sequencing. The library was then spiked with 40% PhiX Control to serve as an internal control and loaded onto the flow cell.
Sequencing data analysis and “hit” selection
The conversion of raw NGS data into summary files that included annotation for each identified gene and sequence frequency was performed using bioinformatics analysis following the manufacturer’s protocol (Horizon Discovery Ltd., cat#V3SM8631). Next, data were processed, edited, normalized, and transformed using Microsoft Excel and GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA). To assess the frequency distribution, read counts per barcode were log-transformed. After normalization, fold enrichment for each sequence in the tdTomato sample was calculated over the uninfected control. “Hits” were defined as shRNA sequences whose frequencies in the experimental sample were increased at least 2-fold over the maximum sequence frequency detected in the uninfected negative control. Then, the frequencies of the selected sequences were compared between the positive and negative samples. The standard scores (Z scores) were calculated as follows: z = (x − μ)/σ.
Gene process enrichment and interaction analysis
After all candidate selection lists (hits) had been assembled, genes in the respective lists were clustered in functional categories and signaling pathways using DAVID Bioinformatic Resources 6.8 (NIH) (Huang et al., 2009). Clusters of enriched genes were queried for interaction/association analysis using the STRING functional protein association tool (version 9.0; http://string-db.org/) with all available prediction methods (Szklarczyk et al., 2019). Functional and KEGG pathway enrichment analyses of selected genes were performed using online tools in Metascape (http://metascape.org/).
Lentivirus transduction and cell infection
The top 10 potential genes from the list of hits were selected to be validated by lentivirus transduction. All lentivirus particles (106 TU/mL) constructed into the pLenti-puro-CMV-TurboGFP vector were purchased from Sigma-Aldrich (Table S4) with three clones per gene. The tdTomato-CD63+ BV-2 cells were transduced with lentivirus particles. Following lentiviral transduction, cells that were transduced successfully carrying the fluorescent TurboGFP were purified using FACS. Cells were then expanded in complete culture medium to validate their effects on the secretion of EVs.
qRT-PCR analysis of mRNA knockdown of target genes
Total RNA was isolated with Trizol (Thermo Fisher Scientific, Cat #15596026) and was reverse transcribed using SuperScript™ IV VILO™ Master Mix kit (Thermo Fisher Scientific, #11766050), and the resulting cDNA was diluted before use. For qPCR, 5 μL/well of reaction system containing 2.5 μL/well of 2X TaqMan Gene Expression Master Mix (Life Technologies, #4369016) or Applied Biosystems SYBR Green PCR Master Mix (Applied Biosystem, #4309155), 0.25 μL/well of the appropriate TaqMan primer-probe set or synthesized primer oligonucleotides, and 0.25 μL/well of nuclease free water was added to the 2 μL cDNA sample in each well. After sealing the plates, mixing the wells, and centrifuging at 1,000 × g for 4 min, 40 cycles of amplification were performed according to the manufacturer’s specifications in an Applied Biosystems 7900HT quantitative reverse transcription (qRT)-PCR machine. Relative amounts of mRNA between non-target shRNA control (NTC) and query shRNA samples were calculated using the comparative Ct method (Schmittgen and Livak, 2008) with normalization to the Gapdh control Ct value.
Primary culture of murine microglia
Primary cultures of mouse microglia were prepared from CD-1 mouse P0 to P3 pups by using MACS Technology (Miltenyi Biotec) according to the man-ufacturer’s instructions. Briefly, brain cortices were dissected, freed from meninges, and dissociated by enzymatic digestion using a Neural Tissue Dissociation Kit (Miltenyi Biotec, #130-093-231). CD11b positive microglia were isolated using a magnetic CD11b bead (Miltenyi Biotec, #130-049-601) separation technique, loaded onto a MACS column (Miltenyi Biotec, #30-042-201), and subjected to magnetic separation. Isolated microglia were plated into a 48-well plate at a density of 15 × 104 cells/well. Microglia were maintained in DMEM medium (Invitrogen, 11965118) with 10% FBS (Invitrogen, 10082147) in a 5% CO2 humidified incubator. Murine monocyte colony-stimulating factor (10 ng/mL, BioVision, #4238, Milpitas, CA, USA) was added to the microglial culture after plating. After 24 h, cultured medium was replaced with fresh medium for 2 days until used for siRNA transfection. Horizon SMARTpool siRNA (premixture of 4 siRNA clones targeting target multiple regions within the transcript, 1 μM siRNA/well) for Mcdf2 (Horizon Discovery Ltd., #E-052885-00-0020), Sepp1 (Horizon Discovery Ltd., #E-041618-00-0020), Sdc1 (Horizon Discovery Ltd., #E-044225-00-0020), or the scramble siRNA control (Horizon Discovery Ltd., # D-001910-01-50) was transduced to microglia using the Accell siRNA delivery media (Horizon Discovery Ltd., #B-005000-100) for 72 h according to the manufacturer’s protocol.
Stimulation of EV release from microglia
Primary-cultured murine microglia were washed twice with DMEM to remove serum factors in the complete DMEM and stimulated with or without lipopolysaccharide (from E. coli 0111:B4, 1 μg/mL, cat. no. L2654, Sigma-Aldrich, St. Louis, MO) for 4 h. After the LPS stimulation, the cells were treated with or without 5 mM ATP (#A6419-5G, Sigma-Aldrich) for 15 min. The media were collected as conditioned media (CM) and the cells were washed twice with ice-cold PBS and stored at −80°C. Collected CM were centrifuged, filtered through 0.22-μm pore-size membrane (EMD Millipore) and performed the isolation of the EV-enriched fraction as discussed following. For the stimulation of microglia with poly(I:C), the gene targeted cell lines (tdTomato-CD63+ BV-2 cell line with Sepp1-shRNA-sh2, Mcfd2-shRNA-sh3, Sdc1-shRNA-sh3, and Scramble-shRNA, respectively) were stimulated with 20 μg/mL high-molecular-weight Poly(I:C) (InvivoGen, # tlrl-pic) for 16 h or were left untreated (Hong et al., 2021). The EV-enriched fraction was isolated from the conditioned media. For the stimulation of microglia with tau aggregates, recombinant 2N3R human tau-410 human tau (500 μg/mL, # T-1002-2, rPeptide, Bogart, GA) and 2N4R human tau-441 (500 μg/mL, # T-1001-2, rPeptide) were mixed and preincubated with 30 μM heparin (# 07980, Stemcell Technologies, Vancouver, Canada) to be aggregated for 5d at 37°C as previously described with minor modifications (Asai, 2015). The gene targeted tdTomato-CD63+ BV-2 cell lines were treated with the 10 mg/mL tau aggregates for 16 h or were left untreated. The EV-enriched fraction was isolated from the conditioned media.
Isolation of the EV-enriched fraction
The cells were washed twice with double-filtered PBS (MF-Millipore® Membrane Filter, 0.22 μm) to remove serum factors, and the medium was replaced with fresh conditioned medium containing double-filtered DMEM (MF-Millipore® Membrane Filter, 0.22 μm) without FBS. Cells were exposed to 1 μg/mL LPS (Millipore-Sigma, L2654-1MG) for 3 h followed by 5 mM ATP stimulation for 15 min. Conditional medium was collected and enriched for EVs by sequential high-speed centrifugation, as previously described (Asai, 2015). Briefly, the CM was centrifuged at 300 × g for 10 min, and the supernatant was centrifuged at 2,000 × g for 10 min at 4°C to remove cell debris. This supernatant was centrifuged at 10,000 × g for 30 min at 4°C to remove microvesicles, and the resulting supernatant was kept as an EV-enriched fraction. Using our purification method, enriched EVs under ATP stimulation were not positive for apoptotic markers (Verderio et al., 2012) or contaminated by intracellular organelles derived from damaged cells (Gabrielli, 2015). The EV-enriched fraction was collected and stored at −80°C until further use.
Nanoparticle track analysis (NTA)
NTA was performed using a NanoSight NS300 (Malvern Panalytical) equipped with a sample chamber and complementary metal–oxide–semiconductor (sCMOS) camera. EV samples were diluted in double-filtered PBS to adjust the particle numbers to the optimal range (10–100 particles/frame). The particle live-imaging settings were set according to the manufacturer’s software manual (NanoSight, NS300 User Manual, MAN0541-01-EN-00, 2017).
Live cell monitoring and analysis by IncuCyte
Well plates were imaged hourly or every two min in the IncuCyte S3 Zoom HD/2CLR time-lapse microscopy system (Sartorius) (Kowal et al., 2016) equipped with an IncuCyte Zoom 10× Plan Fluor objective (Sartorius). Imaging was performed for 1 h at 37°C. Phase images were acquired for each experiment. For fluorescence imaging, the acquisition time was 400 ms for the orange channel. Analysis parameters were set up from Basic Analyzer mode (BA; endpoint, confluence [%]). The optimized processing definitions were subsequently used for real-time image analysis.
TEM and immunoelectron microscope analyses
TEM of EVs purified from the conditioned medium was conducted as previously described (Ruan et al., 2021). Briefly, 5 μL of the EV sample was adsorbed for 1 min on a carbon-coated grid (# CF400-CU, Electron Microscopy Sciences) made hydrophilic by a 20 s exposure to a glow discharge (25 mA). Excess liquid was removed with filter paper (#1 Whatman). The grid was then floated briefly on a drop of water (to wash away phosphate or salt), blotted on filter paper, and then stained with 0.75% uranyl formate (#22451 EMS) for 15 s. After removing the excess uranyl formate with a filter paper, the grids were examined using a JEOL 1200EX TEM, and images were recorded with an AMT 2k CCD camera. For immunogold labeling on EV samples, samples were adsorbed to the grid for 5 min, blocked on 1% bovine serum albumin (BSA) for 10 min, added to 5 μL drops of primary antibody (Tsg101, Santa Cruz Biotechnology, Y16J, mouse mAb, 1:5; CD63, BioLegend, #143901, mouse mAb, 1:10; tdTomato, Thermo Fisher Scientific, TA150128, mouse mAb, 1:10) and incubated for 30 min. Samples were then washed in four drops of PBS (total 10 min) before incubation with Rabbit anti-mouse bridging antibody (1:50, Abcam ab6709) for 30 min, followed by addition of 5 nm protein A–gold (University Medical Center, Utrecht, the Netherlands) for 20 min. Grids were washed in two drops of PBS followed by four drops of water (totally a 15 min period), then stained in 0.75% uranyl formate and examined at 80 kV with a JEOL 1200EX TEM. Images were recorded with an AMT 2k CCD camera.
Western blotting
For EV samples, after lysis with TENT buffer (50mM Tris-HCl, 2mM EDTA, 150mM NaCl, 1% Triton-X 100, pH7.5), an equivalent amount of protein was loaded on 4–20% sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS–PAGE) gels (#4561093, Bio-Rad) and electro-transferred to 0.45 μm nitrocellulose membranes (#162-0090, Bio-Rad). The membranes were blocked in freshly prepared 5% skim milk diluted in TBS before being immunoblotted with specific primary antibodies (anti-rabbit CD63, Biorbyt, # orb229829), anti-Tsg101 (1:100, clone Y16J, Santa Cruz Biotechnology), tdTomato, Thermo Fisher, TA150128, mouse mAb, anti-early endosome antigen A1 (EEA1, 1:100, clone G-4, Santa Cruz Biotechnology), anti-Rab7 (1:1,000, clone D95F2, Cell Signaling Technology), anti-calregulin (1:200, clone F-4, Santa Cruz Biotechnology), anti-cytochrome C antibody (1:100, clone 7H8, Santa Cruz Biotechnology), anti-GM130 (1:100, clone NN2C10/1, Santa Cruz Biotechnology), anti-Histone H2A.Z (1:400, 2718, Cell Signaling Technology), anti-LC3 a/b antibody (1:1,000, 4108S, Cell Signaling Technology), anti-β-actin (1:2,000, A1978, Sigma-Aldrich). The membrane was further incubated with HRP-labeled secondary antibodies (Cell Signaling Technology) and scanned using a C300 digital chemiluminescent imager (Azure Biosystems). The band densities were digitally measured using ImageJ software (NIH).
Immunofluorescence
TdTomato-CD63+ BV-2 cells were seeded on the coverslips in 24-well plate one day prior to be washed with PBS three times, followed by the fixation with ice-cold 4% paraformaldehyde for 10 min. The sections were permeabilized in 0.5% Triton-X 100(#T9284-100ML, Sigma-Aldrich) /PBS, and blocked in 10% normal Donkey serum (#D9663-10ML, Sigma-Aldrich) for 30 min. Cell samples were incubated with primary antibodies against RAB7 (#D95F2, rabbit IgG, Cell Signaling Technology, 1: 50), Rab5 (#610725, mouse IgG, BD Biosciences, 1: 50), EEA1 (#sc-137130, mouse IgG, Santa cruz, 1: 50), diluted with 1% BSA, 0.025% tween-20 in PBS at 4°C for overnight. The second day, cell samples were washed and incubated in secondary antibodies (AlexaFluor-647 Donkey anti-rabbit; 1:1000, AlexaFluor-488 Donkey anti-mouse; 1:1000, for 1 h at room temperature, followed by DAPI staining (#D1306, Thermo Fisher Scientific) for 5 min. All images were captured on confocal microscopic imaging as described below.
Image capture
For the live cell images, Nikon deconvolution wide-field epifluorescence system (Nikon Instruments) and ImageJ software (version 1.43; NIH, Bethesda, MD, USA) were used to capture and crop images and merge channels into composite RGB images. In all cases, identical adjustments were applied across all the images used in the experiment for each channel. For fixed cell samples, images were captured on a Zeiss LSM confocal microscope 880 with 63x objective lens and 2.0x digital zoom using the Airy Scan super-resolution module. Images were analyzed in Imaris software based on the object based colocalization mode. The ‘Shortest distance to Spots’ was utilized to define Colocalized spots based on their distance to the other Spots object. We will name one class as ‘Coloc’, another one ‘Not coloc’, In this study, the distance within 1 μm or less was defined as colocalized. For averages and standard deviations of those colocalization quantification between each channel were attached as Table S5.
CD63 and IL-1β ELISA
Cells were washed with ice-cold PBS, flash frozen in liquid nitrogen, and stored at −80°C. After lysis with RIPA buffer () containing 1X protease / phosphatase inhibitor cocktail (Thermo Fisher Scientific, PI78443), the supernatant was collected for protein concentration measurement using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, 23225) and human CD63 ELISA (LifeSpan Biosciences, LS-F7104). The exosomal fraction was isolated from the culture medium by sequential centrifugation (300 × g for 10 min, 2,000 × g for 10 min, and 10,000 × g for 30 min at 4°C), and then measured for CD63 using the same CD63 ELISA kit, or for IL-1β using the IL-1β ELISA kit (Abcam, ab197742). All ELISA applications followed the manufacturer’s protocol.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as the mean ± standard error of the mean (SEM). Two-tailed paired or unpaired Student’s t-tests were used for comparisons between the two groups. Multiple comparisons were performed using either one- or two-way analysis of variance (ANOVA), followed by Tukey’s or Dunnett’s post hoc tests. Statistical analyses were performed using Prism 8.0 (GraphPad Software). Statistical significance was set at p < 0.05.
Supplementary Material
Highlights.
Development of tdTomato-CD63 cell model for monitoring extracellular vesicle release
An shRNA screen identifies factors implicated in extracellular vesicle release
Sepp1, Mcfd2, and Sdc1 are required for microglial extracellular vesicle release
Sepp1, Mcfd2, and Sdc1 suppress EV-associated Il-1β release from microglia
ACKNOWLEDGMENTS
We thank Michelle Li, Hana Do, Shuiqiao Hu, and other members of the Laboratory of Molecular NeuroTherapeutics for technical assistance. We also thank Brian Tilton at the Flow Cytometry Core Facility at Boston University School of Medicine, Dr. Yuriy Alekseyev and Ashley LeClerc at the Microarray & Sequencing Resource Core Facility at Boston University School of Medicine, and Dr. Laura J. Lewis-Tuffin at the Cytometry and Imaging Laboratory at the Mayo Clinic Florida. This work was funded in part by Ono Pharmaceutical (to T.I.), NIH R01 AG066429 (to T.I.), RF1 AG054199 (to T.I.), R01 AG054672 (to T.I.), R01 AG067763-01A1 (to T.I.), R01 AG072719 (to T.I.), the Cure Alzheimer’s Fund (to T.I. and S.I.), and BU ADC NIH P30 AG013846 (to S.I. and Z.R.).
DECLARATIONS OF INTERESTS
T.I. had a sponsored research agreement with Ono Pharmaceutical and consults with Takeda.
ABBREVIATIONS
- Aβ
amyloid-β peptide
- ANOVA
analysis of variances
- CD11b
cluster of differentiation molecule 11b
- CD37
cluster of differentiation molecule 37
- CD63
cluster of differentiation molecule 63
- CD81
cluster of differentiation molecule 81
- CD9
cluster of differentiation molecule 9
- CMV
cytomegalovirus
- CNS
central nervous system
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DMEM
Dulbecco’s minimal essential medium
- EEA1
early endosome antigen A1
- EF1α
elongation factor-1α
- ESCRT
endosomal sorting complexes required for transport
- EV
extracellular vesicle
- FACS
fluorescence activated cell sorting
- FBS
fetal bovine serum
- GM130/GOLGA2
Golgin subfamily A member 2
- H2A
histone H2A
- Hgs
hepatocyte growth factor-regulated tyrosine kinase substrate
- HTS
high-throughput screening
- IL-1β
interleukin-1β
- IL-6
interleukin-6
- ILV
intraluminal vesicles
- LC3
microtubule-associated protein light chain 3
- Lman1
lectin mannose binding 1
- LPS
lipopolysaccharide
- Ly86
Lymphocyte Antigen 86
- Ly96
Lymphocyte Antigen 96
- MOI
multiplicity of infection
- MVB
multivesicular body
- NGS
next generation sequencing
- Mcfd2
multiple coagulation factor deficiency 2, ER Cargo Receptor Complex Subunit
- NIH
National Institute of Health
- P2RX7
purinergic receptor P2X 7
- PCR
polymerase chain reaction
- PGK
phosphoglycerate kinase
- qRT-PCR
quantitative reverse transcription PCR
- Rab7
Ras-related protein Rab7
- Rbmx
RNA Binding Motif Protein X-Linked
- tdTomato
tandem dimer tomato
- sCMOS
scientific Complementary metal–oxide–semiconductor
- Sdc1
syndecan 1
- Sepp1
selenoprotein P
- SDS–PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- shRNA
short hairpin RNA
- siRNA
silencing RNA
- TLR4
Toll-like receptor 4
- Tlr13
Toll-like receptor 13
- TNF-α
tumor necrosis factor-α
- Tsg101
tumor susceptibility gene 101
- TurboGFP
turbo green fluorescent protein
- UBC
ubiquitin C
- Vps4a
vacuolar protein sorting 4 homolog A
- Yipf1
Yip1 Domain Family Member 1
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110791.
REFERENCES
- Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, and Rubartelli A (1999). The secretory route of the leaderless protein interleukin 1beta involves exocytosis of endolysosome-related vesicles. Mol. Biol. Cell 10, 1463–1475. 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu Z, and Yanez-Mo M (2014). Tetraspanins in extracellular vesicle formation and function. Front Immunol. 5, 442. 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai H, et al. (2015). Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci 18, 1584–1593. 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol 14, 677–685. 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Bartels T, De Schepper S, and Hong S (2020). Microglia modulate neurodegeneration in Alzheimer’s and Parkinson’s diseases. Science 370, 66–69. 10.1126/science.abb8587. [DOI] [PubMed] [Google Scholar]
- van der Beek J, de Heus C, Liv N, and Klumperman J (2022). Quantitative correlative microscopy reveals the ultrastructural distribution of endogenous endosomal proteins. J. Cell Biol 221, e202106044. 10.1083/jcb.202106044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco F, Pravettoni E, Colombo A, Schenk U, Moller T, Matteoli M, and Verderio C (2005a). Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J. Immunol 174, 7268–7277. 10.4049/jimmunol.174.11.7268. [DOI] [PubMed] [Google Scholar]
- Bianco F, Pravettoni E, Colombo A, Schenk U, Moller T, Matteoli M, and Verderio C (2005b). Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J. Immunol 174, 7268–7277. 10.4049/jimmunol.174.11.7268. [DOI] [PubMed] [Google Scholar]
- Bosschaerts T, Guilliams M, Noel W, Herin M, Burk RF, Hill KE, Brys L, Raes G, Ghassabeh GH, De Baetselier P, et al. (2008). Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J. Immunol 180, 6168–6175. 10.4049/jimmunol.180.9.6168. [DOI] [PubMed] [Google Scholar]
- Burnstock G (2007). Physiology and pathophysiology of purinergic neurotransmission. Physiol. Rev 87, 659–797. 10.1152/physrev.00043.2006. [DOI] [PubMed] [Google Scholar]
- Clayton KA, Van Enoo AA, and Ikezu T (2017). Alzheimer’s disease: the role of microglia in brain homeostasis and proteopathy. Front. Neurosci 11, 680. 10.3389/fnins.2017.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, and Gho YS (2007. Dec). Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res 6, 4646–55. 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- Clayton K, Delpech JC, Herron S, Iwahara N, Ericsson M, Saito T, Saido TC, Ikezu S, and Ikezu T (2021). Plaque associated microglia hyper-secrete extracellular vesicles and accelerate tau propagation in a humanized APP mouse model. Mol. Neurodegener 16, 18. 10.1186/s13024-021-00440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, and Raposo G (2013). Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci 126, 5553–5565. 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- DeLeo AM, and Ikezu T (2018). Extracellular vesicle biology in Alzheimer’s disease and related tauopathy. J. Neuroimmune Pharmacol 13, 292–308. 10.1007/s11481-017-9768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpech JC, Herron S, Botros MB, and Ikezu T (2019). Neuroimmune crosstalk through extracellular vesicles in health and disease. Trends Neurosci. 42, 361–372. 10.1016/j.tins.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago F, Lombardi M, Prada I, Gabrielli M, Joshi P, Cojoc D, Franck J, Fournier I, Vizioli J, and Verderio C (2017). ATP modifies the proteome of extracellular vesicles released by microglia and influences their action on astrocytes. Front. Pharmacol 8, 910. 10.3389/fphar.2017.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcin M, Fleury A, Taillebois E, Hilairet G, Krupova Z, Henry C, Truchet S, Trotzmuller M, Kofeler H, Mabilleau G, et al. (2017). Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracellular Vesicles 6, 1305677. 10.1080/20013078.2017.1305677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald W, Freeman ML, Lederman MM, Vasilieva E, Romero R, and Margolis L (2018). A system of cytokines encapsulated in ExtraCellular vesicles. Sci. Rep 8, 8973. 10.1038/s41598-018-27190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli M, et al. (2015). Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep 16, 213–220. 10.15252/embr.201439668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy JE, Wigren E, Svard M, Hard T, and Lindqvist Y (2008). New insights into multiple coagulation factor deficiency from the solution structure of human MCFD2. J. Mol. Biol 381, 941–955. 10.1016/j.jmb.2008.06.042. [DOI] [PubMed] [Google Scholar]
- He M, Qin H, Poon TCW, Sze SC, Ding XF, Co NN, Ngai SM, Chan TF, and Wong N (2015). Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 36, 1008–1018. 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, and El Khoury J (2018). Microglia in neurodegeneration. Nat. Neurosci 21, 1359–1369. 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KE, Zhou J, McMahan WJ, Motley AK, and Burk RF (2004). Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J. Nutr 134, 157–161. 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- Holehonnur R, Lella SK, Ho A, Luong JA, and Ploski JE (2015). The production of viral vectors designed to express large and difficult to express transgenes within neurons. Mol Brain 8, 12. 10.1186/s13041-015-0100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Lee J, Vu TH, Lee S, Lillehoj HS, and Hong YH (2021). Immunomodulatory effects of poly(I:C)-stimulated exosomes derived from chicken macrophages. Poult. Sci 100, 101247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Z, Hoffmann FW, Norton RL, Hashimoto AC, and Hoffmann PR (2011). Selenoprotein K is a novel target of m-calpain, and cleavage is regulated by Toll-like receptor-induced calpastatin in macrophages. J Biol Chem. 286, 34830–34838. 10.1074/jbc.M111.265520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz SN, Cheerathodi MR, Nkosi D, York SB, and Meckes DG Jr. (2018). Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of epstein-barr virus LMP1. J. Virol 92, e01969–17. 10.1128/JVI.01969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Chung YW, Jung MK, Lee JH, Ko KY, Jang JK, Ham M, Kang H, Pack CG, Mihara H, et al. (2020). Apolipoprotein E-mediated regulation of selenoprotein P transportation via exosomes. Cell Mol. Life Sci 77, 2367–2386. 10.1007/s00018-019-03287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, and Thery C (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U S A 113, E968–E977. 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhbacher M, Bartel J, Hoppe B, Alber D, Bukalis G, Brauer AU, Behne D, and Kyriakopoulos A (2009). The brain selenoproteome: priorities in the hierarchy and different levels of selenium homeostasis in the brain of selenium-deficient rats. J. Neurochem 110, 133–142. 10.1111/j.1471-4159.2009.06109.x. [DOI] [PubMed] [Google Scholar]
- Kumar A, Stoica BA, Loane DJ, Yang M, Abulwerdi G, Khan N, Kumar A, Thom SR, and Faden AI (2017). Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J. Neuroinflammation 14, 47. 10.1186/s12974-017-0819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa S, Hill KE, McDonald WH, and Burk RF (2012). Long isoform mouse selenoprotein P (Sepp1) supplies rat myoblast L8 cells with selenium via endocytosis mediated by heparin binding properties and apolipoprotein E receptor-2 (ApoER2). J. Biol. Chem 287, 28717–28726. 10.1074/jbc.M112.383521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhao B, Chen YC, You DW, Liu R, Rong MQ, Ji WZ, Zheng P, and Lai R (2013). Multiple coagulation factor deficiency protein 2 contains the ability to support stem cell self-renewal. Faseb J. 27, 3298–3305. 10.1096/fj.13-228825. [DOI] [PubMed] [Google Scholar]
- Lu A, Wawro P, Morgens DW, Portela F, Bassik MC, and Pfeffer SR (2018). Genome-wide interrogation of extracellular vesicle biology using barcoded miRNAs. Elife 7, e41460. 10.7554/eLife.41460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, and Surprenant A (2001). Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity 15, 825–835. 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- Meng XL, Chen CL, Liu YY, Su SJ, Gou JM, Huan FN, Wang D, Liu HS, Ben SB, and Lu J (2019). Selenoprotein SELENOK Enhances the Migration and Phagocytosis of Microglial Cells by Increasing the Cytosolic Free Ca2+ Level Resulted from the Up-Regulation of IP3R. Neuroscience. 406, 38–49. 10.1016/j.neuroscience.2019.02.029. [DOI] [PubMed] [Google Scholar]
- Nyfeler B, Zhang B, Ginsburg D, Kaufman RJ, and Hauri HP (2006). Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic 7, 1473–1481. 10.1111/j.1600-0854.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- Olson GE, Winfrey VP, NagDas SK, Hill KE, and Burk RF (2007). Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J. Biol. Chem 282, 12290–12297. 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- Plaza-Zabala A, Sierra-Torre V, and Sierra A (2020). Assessing autophagy in microglia: a two-step model to determine Autophagosome formation, degradation, and net turnover. Front Immunol. 11, 620602. 10.3389/fimmu.2020.620602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, and Spang A (2010). Identification of the switch in early-to-late endosome transition. Cell 141, 497–508. 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Qu Y, Franchi L, Nunez G, and Dubyak GR (2007). Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol 179, 1913–1925. 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, and Simons K (2006). Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. U S A 103, 11172–11177. 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucourt B, Meeussen S, Bao J, Zimmermann P, and David G (2015). Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 25, 412–428. 10.1038/cr.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z, and Ikezu T (2019). Tau secretion. Adv. Exp. Med. Biol 1184, 123–134. 10.1007/978-981-32-9358-8_11. [DOI] [PubMed] [Google Scholar]
- Ruan Z, Delpech JC, Venkatesan Kalavai S, Van Enoo AA, Hu J, Ikezu S, and Ikezu T (2020). P2RX7 inhibitor suppresses exosome secretion and disease phenotype in P301S tau transgenic mice. Mol. Neurodegener 15, 47. 10.1186/s13024-020-00396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z, Pathak D, Venkatesan Kalavai S, Yoshii-Kitahara A, Muraoka S, Bhatt N, Takamatsu-Yukawa K, Hu J, Wang Y, Hersh S, et al. (2021). Alzheimer’s disease brain-derived extracellular vesicles spread tau pathology in interneurons. Brain 144, 288–309. 10.1093/brain/awaa376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A, Cozzolino F, Talio M, and Sitia R (1990). A novel secretory pathway for interleukin-1-beta, a protein lacking a signal sequence. Embo J. 9, 1503–1510. 10.1002/j.1460-2075.1990.tb08268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Sato N, Hirashima M, Takebe G, Nagasawa S, and Takahashi K (2004). Domain structure of bi-functional selenoprotein P. Biochem. J 381, 841–846. 10.1042/BJ20040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Simons M, and Raposo G (2009). Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol 21, 575–581. 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Su M, Ravishankar S, Moore J, Head P, Dixon JB, and Vannberg F (2017). TLR-exosomes exhibit distinct kinetics and effector function. Sci. Rep 7, 41623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szepesi Z, Manouchehrian O, Bachiller S, and Deierborg T (2018). Bidirectional microglia-neuron communication in health and disease. Front Cell Neurosci. 12, 323. 10.3389/fncel.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi T, Nakai M, Iwamaru Y, Sugama S, Tsukimoto M, Fujita M, Wei J, Sekigawa A, Sato M, Kojima S, et al. (2009). The activation of P2X7 receptor impairs lysosomal functions and stimulates the release of autophagolysosomes in microglial cells. J. Immunol 182, 2051–2062. 10.4049/jimmunol.0802577. [DOI] [PubMed] [Google Scholar]
- Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, and Sanderson RD (2013). Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem 288, 10093–10099. 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turola E, Furlan R, Bianco F, Matteoli M, and Verderio C (2012). Microglial microvesicle secretion and intercellular signaling. Front. Physiol 3, 149. 10.3389/fphys.2012.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderio C, Muzio L, Turola E, Bergami A, Novellino L, Ruffini F, Riganti L, Corradini I, Francolini M, Garzetti L, et al. (2012). Myeloid microvesicles are a marker and therapeutic target for neuroinflammation. Ann. Neurol 72, 610–624. 10.1002/ana.23627. [DOI] [PubMed] [Google Scholar]
- Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, and Clayton A (2010. Jun). Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics 9, 1324–38. 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Boza-Serrano A, Dunning CJR, Clausen BH, Lambertsen KL, and Deierborg T (2018a). Inflammation leads to distinct populations of extracellular vesicles from microglia. J. Neuroinflammation 15, 168. 10.1186/s12974-018-1204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Boza-Serrano A, Dunning CJR, Clausen BH, Lambertsen KL, and Deierborg T (2018b). Inflammation leads to distinct populations of extracellular vesicles from microglia. J. Neuroinflammation 15, 168. 10.1186/s12974-018-1204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H, and Ikezu T (2019). Transcriptional and epigenetic regulation of microglia in health and disease. Trends Mol. Med 25, 96–111. 10.1016/j.molmed.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YJ, Kim OY, and Gho YS (2014). Extracellular vesicles as emerging intercellular communicasomes. Bmb Rep. 47, 531–539. 10.5483/BMBRep.2014.47.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, and Ikezu T (2019). Emerging roles of extracellular vesicles in neurodegenerative disorders. Neurobiol. Dis 130, 104512. 10.1016/j.nbd.2019.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kaufman RJ, and Ginsburg D (2005). LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. J. Biol. Chem 280, 25881–25886. 10.1074/jbc.M502160200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen KN, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of Glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. 10.1523/Jneurosci.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next generation sequencing data have been deposited at the Sequence Read Archive / BioProject and are publicly available upon publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to re-analyze data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-early endosome antigen A11 (EEA1) | Santa Cruz Biotechnology | sc-137130: RRID:AB_2246349 |
| Mouse monoclonal anti-Rab5 | BD Biosciences | 610725: RRID:AB_398048 |
| Rabbit Monoclonal anti-Rab7 | Cell Signaling Technology | 9367: RRID:AB_1904103 |
| Rabbit polyclonal anti-CD63 | Biorbyt | orb229829 |
| Rabbit anti-mouse bridging antibody | Abcam | ab6709; RRID:AB_956006 |
| Mouse monoclonal anti-CD63 | BioLegend | 143901; RRID:AB_11203908 |
| Rabbit Polyclonal anti-tdTomato | Thermofisher scientific | TA150128 |
| Mouse monoclonal anti-Tsg101 (Y16J) | Santa Cruz Biotechnology | sc-101254: RRID:AB_1130856 |
| Mouse monoclonal anti-calregulin | Santa Cruz Biotechnology | sc-373863; RRID:AB_10915425 |
| Mouse monoclonal anti-cytochrome C | Santa Cruz Biotechnology | sc-13560; RRID:AB_627383 |
| Mouse monoclonal anti-GM130 | Santa Cruz Biotechnology | sc-53296; RRID:AB_629538 |
| Mouse monoclonal anti-Flotillin | Santa Cruz Biotechnology | sc-74566; RRID:AB_2106563 |
| Rabbit Polyclonal anti-Histone H2A.Z | Cell Signaling Technology | 2718; RRID:AB_659840 |
| Rabbit Polyclonal anti-LC3B antibody | Cell Signaling Technology | 4108S; RRID:AB_2137703 |
| Mouse monoclonal anti-β-actin | Sigma-Aldrich | A1978; RRID:AB_476692 |
| AlexaFluor-647 Donkey anti-rabbit | Thermofisher scientific | A-31573; RRID:AB_2536183 |
| AlexaFluor-488 Donkey anti-mouse | Thermofisher scientific | R37114; RRID:AB_2556542 |
| Goat anti-rabbit IgG HRP-linked antibody | Cell Signaling Technology | 7074; RRID:AB_2099233 |
| Horse anti-mouse IgG, HRP-linked antibody | Cell Signaling Technology | 7076; RRID:AB_330924 |
| Bacterial and virus strains | ||
| Agilent SURE 2 Supercompetent Cells | Agilent technologies | 200152 |
| pLenti-tdTomato-CD63 | This paper | N/A |
| pLenti-puro vector | Addgene | Plasmid #39481; RRID:Addgene_39481 |
| SMARTvector Mouse Whole Genome Lentiviral shRNA Pooled Library mEF1a-GFP | Horizon Discovery Ltd. | V3SM8631 |
| SMARTchoice Promoter Selection Plate | Horizon Discovery Ltd. | SP-001000-01 |
| pLKO.1-puro-CMV-tGFP- Ly86-Clo.1 | Sigma-Aldrich | TRCN0000101268 |
| pLKO.1-puro-CMV-tGFP- Ly86-Clo.2 | Sigma-Aldrich | TRCN0000101269 |
| pLKO.1-puro-CMV-tGFP- Ly86-Clo.3 | Sigma-Aldrich | TRCN0000101266 |
| pLKO.1-puro-CMV-tGFP-Ly96-Clo.1 | Sigma-Aldrich | TRCN0000066224 |
| pLKO.1-puro-CMV-tGFP-Ly96-Clo.2 | Sigma-Aldrich | TRCN0000326822 |
| pLKO.1-puro-CMV-tGFP-Ly96-Clo.3 | Sigma-Aldrich | TRCN0000326824 |
| pLKO.1-puro-CMV-tGFP-Vps4A-Clo.1 | Sigma-Aldrich | TRCN0000287397 |
| pLKO.1-puro-CMV-tGFP-Vps4A-Clo.2 | Sigma-Aldrich | TRCN0000101820 |
| pLKO.1-puro-CMV-tGFP-Vps4A-Clo.3 | Sigma-Aldrich | TRCN0000101822 |
| pLKO.1-puro-CMV-tGFP-MCFD-Clo.1 | Sigma-Aldrich | TRCN0000196018 |
| pLKO.1-puro-CMV-tGFP-MCFD-Clo.2 | Sigma-Aldrich | TRCN0000183944 |
| pLKO.1-puro-CMV-tGFP-MCFD-Clo.3 | Sigma-Aldrich | TRCN0000183004 |
| pLKO.1-puro-CMV-tGFP-Trl13-Clo.1 | Sigma-Aldrich | TRCN0000066034 |
| pLKO.1-puro-CMV-tGFP-Trl13-Clo.2 | Sigma-Aldrich | TRCN0000368665 |
| pLKO.1-puro-CMV-tGFP-Trl13-Clo.3 | Sigma-Aldrich | TRCN0000360480 |
| pLKO.1-puro-CMV-tGFP-Sepp1-Clo.1 | Sigma-Aldrich | TRCN0000120685 |
| pLKO.1-puro-CMV-tGFP-Sepp1-Clo.2 | Sigma-Aldrich | TRCN0000352521 |
| pLKO.1-puro-CMV-tGFP-Sepp1-Clo.3 | Sigma-Aldrich | TRCN0000340550 |
| pLKO.1-puro-CMV-tGFP- Hgs-Clo.1 | Sigma-Aldrich | TRCN0000088689 |
| pLKO.1-puro-CMV-tGFP- Hgs-Clo.2 | Sigma-Aldrich | TRCN0000088691 |
| pLKO.1-puro-CMV-tGFP- Hgs-Clo.3 | Sigma-Aldrich | TRCN0000088692 |
| pLKO.1-puro-CMV-tGFP- Yipf1-Clo.1 | Sigma-Aldrich | TRCN0000126345 |
| pLKO.1-puro-CMV-tGFP- Yipf1-Clo.2 | Sigma-Aldrich | TRCN0000126346 |
| pLKO.1-puro-CMV-tGFP- Yipf1-Clo.3 | Sigma-Aldrich | TRCN0000126348 |
| pLKO.1-puro-CMV-tGFP- Rbmx-Clo.1 | Sigma-Aldrich | TRCN0000104042 |
| pLKO.1-puro-CMV-tGFP- Rbmx-Clo.2 | Sigma-Aldrich | TRCN0000438339 |
| pLKO.1-puro-CMV-tGFP- Rbmx-Clo.3 | Sigma-Aldrich | TRCN0000434600 |
| pLKO.1-puro-CMV-tGFP- SDC-Clo.1 | Sigma-Aldrich | TRCN0000302195 |
| pLKO.1-puro-CMV-tGFP- SDC-Clo.2 | Sigma-Aldrich | TRCN0000302270 |
| pLKO.1-puro-CMV-tGFP- SDC-Clo.3 | Sigma-Aldrich | TRCN0000311084 |
| pLKO.1-puro-CMV-tGFP-scramble | Sigma-Aldrich | Non-Target shRNA Control |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Thermofisher scientific | D1306; RRID:AB_2629482 |
| Adenosine 5 -triphosphate | Sigma-Aldrich | A6419-5G |
| Tris-HCl | Sigma-Aldrich | 10812846001 |
| EDTA | Sigma-Aldrich | E9884-100G |
| NaCl | Sigma-Aldrich | S9888-500G |
| 3-Methyladenine | Sigma-Aldrich | M9281 |
| Chloroquine | Sigma-Aldrich | C6628 |
| Tau-410, (2N3R) | rPeptide | T-1002-2 |
| Tau-441, (2N4R) | rPeptide | T-1001-2 |
| Critical commercial assays | ||
| CD63 ELISA | LifeSpan Biosciences | LS-F7104 |
| IL-1β ELISA | Abcam | ab197742 |
| Experimental models: Cell lines | ||
| BV2 cell line | CABRI | ICLC ATL03001; RRID:CVCL_0182 |
| HEK293FT cell line | Invitrogen | R70007 |
| Experimental models: Organisms/strains | ||
| Mouse: CD-1® IGS Mouse | Charles River Laboratories | strain code 022 |
| Oligonucleotides | ||
| Accell Non-targeting siRNA | Horizon Discovery Ltd. | D-001910-01-50 |
| Accell Mouse Mcfd2 (193813) siRNA - SMARTpool | Horizon Discovery Ltd. | E-052885-00-0020 |
| Accell Mouse Selenop (20363) siRNA - SMARTpool | Horizon Discovery Ltd. | E-041618-00-0020 |
| Accell Mouse Sdc1 (20969) siRNA -SMARTpool | Horizon Discovery Ltd. | E-044225-00-0020 |
| Accell Mouse ly86 (17084) siRNA -SMARTpool | Horizon Discovery Ltd. | E-044084-00-0005 |
| Ly86 TaqMan probes | Thermofisher scientific | Mm00440240_m1 |
| Ly96 TaqMan probes | Thermofisher scientific | Mm01227593_m1 |
| Vps4a TaqMan probes | Thermofisher scientific | Mm00459928_m1 |
| Mcfd2 TaqMan probes | Thermofisher scientific | Mm00454818_m1 |
| Hgs TaqMan probes | Thermofisher scientific | Mm00468635_m1 |
| Sdc1 TaqMan probes | Thermofisher scientific | Mm00448918_m1 |
| Sepp1 TaqMan probes | Thermofisher scientific | Mm00486048_m1 |
| Rbmx TaqMan probes | Thermofisher scientific | Mm00726735_s1 |
| Tlr8 TaqMan probes | Thermofisher scientific | Mm04209873_m1 |
| Yipf1 TaqMan probes | Thermofisher scientific | Mm00523828_m1 |
| Gapdh TaqMan probes | Thermofisher scientific | Mm99999915_g1 |
| IL-1β Forward: GAAATGCCACCTTTTGACAGTG | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| IL-1β Reverse: RTGGATGCTCTCATCAGGACAG | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| IL18 Forward: GTGAACCCCAGACCAGACTG | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| IL18 Reverse: CCTGGAACACGTTTCTGAAAGA | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| GAPDH Forward: TGGCCTTCCGTGTTCCTAC | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| GAPDH Reverse: GAGTTGCTGTTGAAGTCGCA | PrimerBank | https://pga.mgh.harvard.edu/primerbank/ |
| Illumina-adapted SMARTvector Indexing PCR primer Kit A | Horizon Discovery Ltd. | PRM7668 |
| Illumina-adapted SMARTvector Indexing PCR primer Kit B | Horizon Discovery Ltd. | PRM10186 |
| Recombinant DNA | ||
| tdTomato-CD63 | This paper | Synthesized by GenScript Biotech Inc |
| Software and algorithms | ||
| (Fiji is just) ImageJ 2.0.0 | National Institutes of Health | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8 | GraphPad Software Inc. | |
| DAVID Bioinformatic Resources 6.8 | National Institute of Health | https://david.ncifcrf.gov |
| STRING functional protein association tool | Szklarczyk et al. (2019) | http://string-db.org/ |
| Metascape | Zhou et al., 2019 | http://metascape.org/ |
| BioRender | BioRender | https://www.BioRender.com |
| Other | ||
| DMEM Medium | ThermoFisher scientific | 11965118 |
| Fetal Bovine Serum | ThermoFisher scientific | 10082147 |
| GlutaMAX™ Supplement | ThermoFisher scientific | 35050061 |
| Phosphate buffered saline (PBS) | ThermoFisher scientific | 10-010-031 |
| HBSS, calcium, magnesium, no phenol red | ThermoFisher scientific | 4025134 |
| HBSS, no calcium, no magnesium, no phenol red | ThermoFisher scientific | 14175095 |
| Lipopolysaccharide | Sigma-Aldrich | L3024 |
| 0.45 μm Millex-HV filter | Millipore Sigma | SLHV033RS |
| Medical Millex-GP Syringe Filter Unit, 0.22 μm | Millipore Sigma | SLGPM33RS |
| Puromycin | ThermoFisher scientific | A1113803 |
| DNeasy Blood & Tissue Kit | Qiagen | 69504 |
| Phusion Hot Start II DNA Polymerase | Thermo Fisher Scientific | F549L |
| Ultrapure UltraPure Agarose | Thermo Fisher Scientific | 16500500 |
| Gel Extraction Kit | Denville | CM-510-250 |
| Trizol | Thermo Fisher Scientific | 15596026 |
| SuperScript™ IV VILO™ Master Mix kit | Thermo Fisher Scientific | 11766050 |
| 2X TaqMan Gene Expression Master Mix | Life Technologies | 4369016 |
| Applied Biosystems™ SYBR™ Green PCR Master Mix | Applied Biosystems | 4309155 |
| CD11b bead | Miltenyi Biotec | 130-049-601 |
| Neural Tissue Dissociation Kit | Miltenyi Biotec | 130-093-231 |
| MACS column | Miltenyi Biotec | 30-042-201 |
| Accell siRNA delivery media | Horizon Discovery Ltd. | B-005000-100 |
| Murine monocyte colony-stimulating factor | BioVision | 4238 |
| Poly(I:C) | InvivoGen | tlrl-pic |
| Heparin | Stemcell Technologies | 07980 |
| RIPA buffer | Thermo Fisher Scientific | 89900 |
| Protease / phosphatase inhibitor cocktail | Thermo Fisher Scientific | PI78443 |
| 4–20% SDS–PAGE precast gels | Bio-Rad | 4561093 |
| 4x Laemmli Sample Buffer | Bio-Rad | 1610747 |
| Nitrocellulose membranes | Bio-Rad | 162-0090 |
| Normal Donkey serum | Sigma-Aldrich | 9663-10ML |
| Triton-X 100 | Sigma-Aldrich | T9284-100ML |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 |
| Next Generation Sequencing Dataset | SRA / BioProject | PRJNA826851 |


