Summary
The Argonaute proteins (AGOs) are well known for their role in post-transcriptional gene silencing in the microRNA (miRNA) pathway. Here we show that in mouse embryonic stem cells, AGO1&2 serve additional functions that go beyond the miRNA pathway. Through the combined deletion of both Agos, we identified a specific set of genes that are uniquely regulated by AGOs but not by the other miRNA biogenesis factors. Deletion of Ago2&1 caused a global reduction of the repressive histone mark H3K27me3 due to downregulation at protein levels of Polycomb repressive complex 2 components. By integrating chromatin accessibility, prediction of transcription factor binding sites, and chromatin immunoprecipitation sequencing data, we identified the pluripotency factor KLF4 as a key modulator of AGO1&2-regulated genes. Our findings revealed a novel axis of gene regulation that is mediated by noncanonical functions of AGO proteins that affect chromatin states and gene expression using mechanisms outside the miRNA pathway.
Keywords: Argonautes, KLF4, CTCF, H3K27me3, PRC2, integrative analysis
Graphical abstract

Highlights
-
•
AGO1&2 regulate a specific set of genes in mESCs, independently of the miRNA pathway
-
•
PRC2 proteins are downregulated in Ago2&1_KO mESCs, leading to H3K27me3 global loss
-
•
AGO1&2 regulate gene expression through the pluripotency factor KLF4
Müller et al. report that AGO1&2 are required for the regulation of a specific set of genes in mESCs, independently of their roles in the miRNA pathway. Integration of multiple omics datasets revealed the pluripotency factor KLF4 as a key modulator of AGO1&2-regulated genes.
Introduction
The Argonaute (AGO) proteins are well known for their cytoplasmic role in the microRNA (miRNA) pathway, where they are key players involved in miRNA-mediated translational inhibition of target mRNAs (Meister, 2013; Müller et al., 2020). However, several noncanonical functions, which are not directly linked to the cytoplasmic miRNA-mediated post-transcriptional gene silencing, have been described for the AGO proteins. For instance, several studies have reported nuclear functions for the AGO proteins, such as gene silencing and activation, alternative splicing, chromatin organization, and double-strand break repair (Meister, 2013). Noticeably, most of these functions have been reported in human cancer cell lines and possible noncanonical functions in other contexts, such as mouse early development, are only just starting to be understood. In fact, one study reported nuclear localized AGO2 in mouse embryonic stem cells (mESCs) and described a role for AGO2 in post-transcriptional gene silencing within the nucleus (Sarshad et al., 2018). We also recently demonstrated that nuclear AGO1 is linked to the proper distribution of heterochromatin at pericentromeric regions in mESCs (Müller et al., 2022).
In mESCs, of the four AGO proteins (AGO1–4), only AGO1&2 are robustly expressed (Müller et al., 2020). The deletion of either one of them does not affect the viability of the cells nor their potential to differentiate into the three embryonic germ layers (Ngondo et al., 2018). Upon Ago2 depletion in mESCs, AGO1 protein levels are increased and it forms complexes with miRNAs normally loaded in AGO2, indicating a compensation of AGO2 loss by AGO1 and a redundancy of the two protein functions (Ngondo et al., 2018). However, Ago2 knockout (KO) mESCs cannot differentiate toward the extraembryonic endoderm, and this defect could not be rescued by overexpressing AGO1 (Ngondo et al., 2018). Thus, despite their overlapping functions in miRNA-mediated translational inhibition, AGO1 and AGO2 also show differences in their functional repertoire, raising the possibility for additional specialized functions in early embryonic development.
Here, by comparing the gene expression profiles of mESC lines depleted of key regulators of the miRNA pathway, we show that a larger number of genes is specifically differentially expressed (DE) in Ago2&1_KO mESCs (Agos-sDEGs), and functionally associated with the positive regulation of RNA metabolic processes. The loss of Ago2&1 in mESCs caused a global reduction of the repressive histone mark H3K27me3 due to downregulation of some Polycomb repressive complex 2 (PRC2) components at the protein level. By integrating chromatin accessibility, prediction of transcription factor (TF) binding sites, and chromatin immunoprecipitation sequencing (ChIP-seq) data, we identified the pluripotency factor KLF4 as a key modulator of Agos-sDEGs and that AGO1&2 regulate KLF4 levels. In summary, our findings revealed a novel axis of gene regulation in mESCs that is mediated by noncanonical functions of AGO1&2, and affects chromatin states and gene expression through the regulation of Polycomb components and TF expression using mechanisms outside the miRNA pathway.
Results
AGO1&2 regulate the expression of a class of genes in mESCs that do not depend on the miRNA pathway
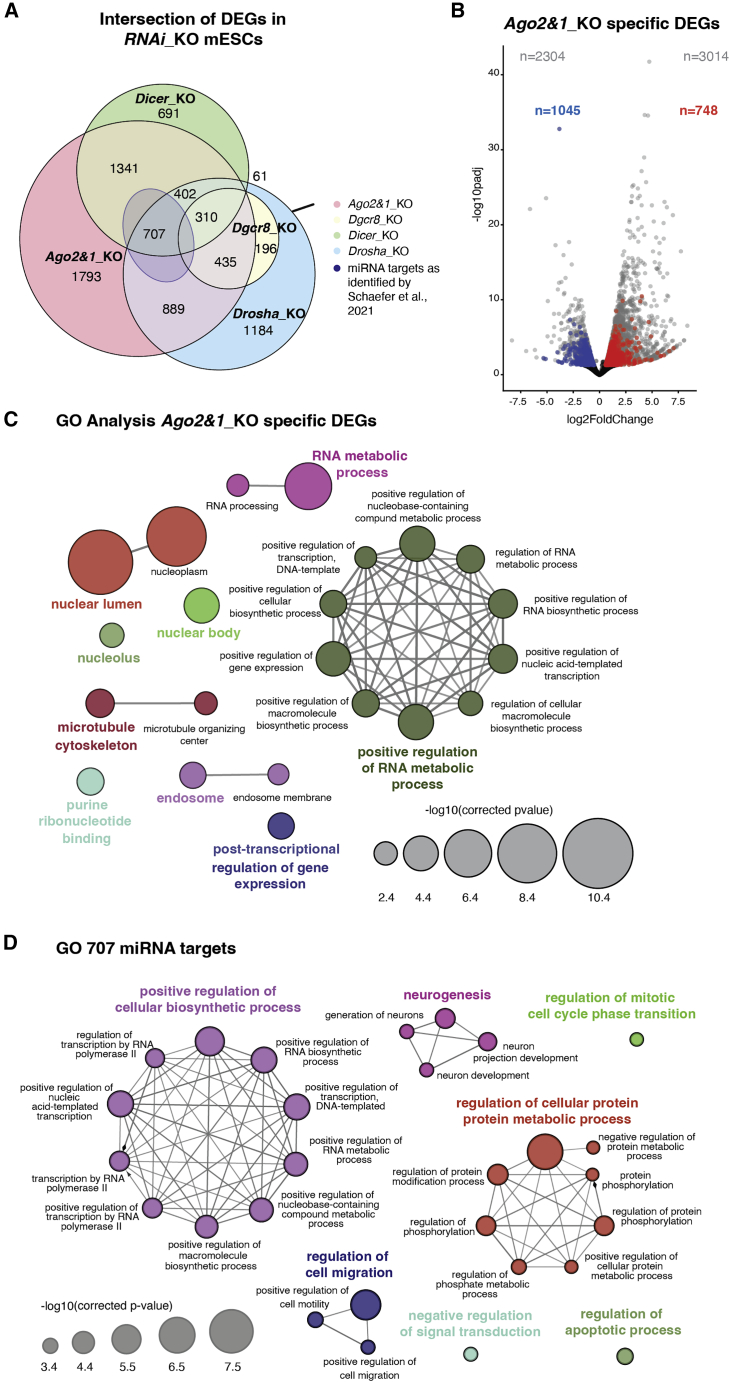
In order to assess the consequences of the loss of the AGO proteins on mESC gene expression, we integrated available RNA-sequencing (RNA-seq) data from (Schäfer et al., 2021) (Table S1). Previous transcriptomics analyses in multiple miRNA_KO and wild-type (WT) mESCs identified many differentially expressed genes (DEGs) in all these mutants (Schäfer et al., 2021). Here, we intersected the Ago2&1_KO DEGs with DEGs from other miRNA_KO mESCs. We identified 1,793 Agos-sDEGs, which are not significantly altered in any other miRNA_KO lines (Figure 1A), ruling out the possibility of direct regulation by miRNAs for these DEGs. This observation was surprising, as Ago2&1_KO mESCs grow as flat-shaped colonies (Figure S1A), have a normal expression of pluripotency factors at the RNA level (Figure S1B), and present a strong proliferation defect (Figure S1C) and an accumulation in the G1 phase of the cell cycle (Figure S1D), all phenotypes being similar to those observed in other miRNA_KO mESC lines (Bodak et al., 2017a).
Figure 1.
Ago2&1_KO mESCs display a distinct transcriptomic profile
(A) Venn diagram representing the overlap of differentially expressed genes (DEGs) from different miRNA_KO mESCs (Dgcr8_KO, Drosha_KO, Dicer_KO, Ago2&1_KO) and the 707 miRNA targets from Schäfer et al. (2021). Numbers indicate the gene set sizes of different overlaps. Ago2&1_KO mESCs have 1,793 specific DEGs (Table S1).
(B) Volcano plot showing the Ago2&1_KO DEGs and the 1793 specific DEGs. The full set of Ago2&1_KO DEGs is shown in gray. Highlighted in red are the upregulated (748) and in blue the downregulated (1,045) Agos_sDEGs.
(C) GO analysis on the 1,793 Agos_sDEGs. The GO analysis has been performed with ClueGO (Bindea et al., 2009). The size of the circles corresponds to their p value.
(D) GO analysis on the 707 specific miRNA target genes predicted in Schäfer et al. (2021). The GO analysis has been performed with ClueGO. The size of the circles corresponds to their p value. See also Figure S1.
More than half (1045) of the Agos-sDEGs were downregulated and 748 were upregulated (Figure 1B and Table S1). Since a loss of miRNA-mediated repression leads to increased target levels, the high number of downregulated genes again argues against an miRNA-mediated regulation and implies that they might be attributed to AGO-specific functions. Importantly, the deficiency of both AGO1&2 in mESCs caused this distinct transcriptomic profile, as single Ago1_KO and Ago2_KO mESCs only had a few DEGs, as previously reported (Figures S1E, S1F, and Table S1) (Ngondo et al., 2018). Further, the overlap between the single Ago1_KO and Ago2_KO DEGs was rather small, which supports only a partial compensatory role for AGO1 and AGO2 functions (Figure S1G).
In order to understand which pathways are affected in mESCs upon combined loss of AGO1&2, we performed gene ontology (GO) analysis of Agos-sDEGs and found that they were enriched in processes linked to nuclear processes, RNA metabolism, and positive transcriptional gene regulation (Figure 1C). Interestingly, these processes differed from the same analysis performed on the previously reported 707 miRNA target genes (Schäfer et al., 2021), which were enriched for processes linked to the regulation of biosynthetic and metabolic processes as well as cell cycle regulation (Figure 1D). Finally, the enrichment of specific processes was to a great extent due to the genes downregulated in Ago2&1_KO mESCs, as shown by the GO analysis performed separately for up- and downregulated genes (Figures S1H and S1I).
Thus, the function of AGO proteins is not only related to post-transcriptional gene silencing, but also to functions that are probably independent of the miRNA pathway.
Combined loss of AGO1&2 in mESCs causes global loss of H3K27me3
To determine how the combined depletion of Ago2&1 affects gene expression in mESCs, we intersected Ago2&1_KO RNA-seq and ENCODE histone ChIP-seq from mESCs with the same genetic background (Figure 2A). We analyzed the abundance of repressive and active histone marks (H3K9me3 & H3K27me3; H3K9ac, H3K36me3, & H3K4me3) and, enhancer marks (H3K4me1 & H3K27ac) at up- and downregulated Agos-sDEGs. As control, we measured these modifications at predicted functional miRNA targets and expressed genes in mESCs. We detected only minor differences for active histone marks and an enrichment of H3K4me1, H3K27ac, and H3K27me3 at upregulated Agos-sDEGs compared with downregulated genes and expressed genes (Figure 2A). Further, H3K27me3 at Agos_sDEGs rather correlated with the loss of AGO proteins as opposed to miRNA target genes, which are rather enriched in active histone marks (Figure 2A).
Figure 2.
Integration of the Ago2&1_KO transcriptome with histone modification datasets
(A) Heatmap showing average histone modification signals at gene regions as derived from ENCODE datasets for five different gene groups; up- and downregulated Agos_sDEGs, 707 predicted functional miRNA target genes from Schäfer et al. (2021), mESC-expressed and -annotated genes. Histone marks were annotated with their previously described predominant functions (Bannister and Kouzarides, 2011): repressive (H3K27me3, H3K9me3), enhancing (H3K4me1, H3K27ac), activating (H3K36me3, H3K4me3). Columns were individually Z score normalized.
(B) Representative WBs for H3K27me3, H3K9me3, H3K4me3, H3K27ac, and H3 in WT and Ago2&1_KO mESCs out of n = 3 independent experiments. Tubulin (TUB) and Coomassie were used as a loading control. Quantification for each individual experiment is shown below the blot.
(C) H3K27me3 ChIP-seq heatmaps of the TSS for all annotated mouse transcripts. Shown is one replicate per condition/experiment for ENCODE WT (Davis et al., 2018; The ENCODE Project Consortium et al., 2013), WT and Ago2&1_KO samples. Shown transcript regions are divided using k-means clustering (k = 4, Table S2). Shown are ±5 kb from the TSS.
(D) Genome browser view of a region derived from transcripts from cluster_1. ChIP-seq coverage signals are shown for the three samples from (C) along with annotated genes in that region (bottom).
(E) Cumulative Distribution Function (CDF) plot showing the differential expression in Ago2&1_KO versus WT of genes associated with the different ChIP peak clusters identified in the ENCODE and WT datasets. The x axis represents the log2FoldChange of the Ago2&1_KO versus WT RNA-seq and the y axis the cumulative proportion over the full set of log2FoldChanges for each cluster.
(F) Genome browser view of two example genes from cluster_1 (Cldn6 and Lefty2) showing both ChIP-seq and RNA-seq coverages for WT and Ago2&1_KO (one sample per experiment and condition). The y axis represents the deposition of H3K27me3 (for the ChIP-seq in green) or the expression levels of the RNA-seq in gray. See also Figure S2.
To determine whether the combined loss of AGO1&2 affects H3K27me3 levels and gene expression, we first measured and compared the global levels of several histone marks in Ago2&1_KO and WT mESCs by Western blotting (WB) (Figure 2B). We observed a drastic reduction of H3K27me3 signal in Ago2&1_KO compared with WT mESCs, whereas the levels of other modified histones, such as H3K9me3, H3K27ac, and H3K4me3, were not particularly affected. These results indicated that the combined loss of AGO1&2 globally decreased H3K27me3 levels. PRC2 is the complex involved in the deposition of H3K27me3 mark (Margueron and Reinberg, 2011). It was recently reported in cancer cells that miRNAs reinforce the repression of PRC2 transcriptional targets through independent and feedforward regulatory networks (Shivram et al., 2019). In addition, several members of the PRC2 complex have been shown to be directly regulated by miRNAs in Drosophila (Kennerdell et al., 2018), but not in mESCs deleted for Dicer, excluding a direct regulation by miRNAs (Graham et al., 2016). In order to assess the integrity of the PRC2 complex in Ago2&1_KO mESCs and the potential contribution of miRNA regulation, we measured the expression of three PRC2-components, SUZ12, EZH2, and JARID2, in miRNA_KO mESC lines. SUZ12 and JARID2, and to a lesser extent also EZH2, were specifically downregulated at the protein level (Figure S2A), but not at the RNA level (Figure S2B), in Ago2&1_KO compared with WT mESCs. The analysis of RNA levels of other PRC2 members showed no significant changes (Figure S2B). In contrast, and consistent with previous results (Graham et al., 2016), we did not observe significant changes in PRC2 members at protein and mRNA levels in the other miRNA_KO mESC lines. These results indicated that AGO1&2 globally regulate H3K27me3 levels by affecting the protein levels of key components of the PRC2 complex using mechanisms that are independent of the canonical miRNA pathway.
We next analyzed whether the observed differential H3K27me3 levels might affect gene expression. We performed H3K27me3 ChIP-seq in WT and Ago2&1_KO mESCs (Figure 2C) and compared it with available H3K27me3 ChIP-seq data from ENCODE (Davis et al., 2018; The ENCODE Project Consortium et al., 2013). We found a clear overlap between the two WT ChIP-seq data (Figures 2C and 2D). Consistent with WB analysis, ChIP-seq data confirmed a genome-wide loss of H3K27me3 in Ago2&1_KO mESCs. In order to determine whether H3K27me3 loss correlates with changes in the expression of the Ago2&1_KO DEGs, we clustered ChIP-seq levels at transcript regions using k-means (k = 4) clustering (Table S2) (Ramírez et al., 2016). The first cluster (cluster_1) represents the genes most strongly enriched in H3K27me3 in WT and highly overlaps with known bivalent genes in mESCs (Asenjo et al., 2020) (Figures 2C and S2C, and Table S2), while the second, third, and fourth clusters represent transcripts with minor H3K27me3 levels.
We expected the loss of the repressive histone mark H3K27me3 to lead to an observable upregulation of associated genes. Indeed, genes from clusters with strong loss in H3K27me3 levels (clusters 1 and 2) showed a tendency to be upregulated (Figures 2E and 2F). In contrast, genes, not marked with H3K27me3 (clusters 3 and 4), showed no enrichment for upregulation upon AGO1&2 loss (Figures 2E and S2F). However, only 14% of the upregulated genes and 11% of the downregulated genes belong to cluster_1 (Figures 2E, S2D, and S2E, and Table S4). These results suggest that the loss of H3K27me3 has a minor impact on the expression of Agos_sDEGs. Thus, other pathways might be specifically affected in the absence of the AGO proteins to explain Agos_sDEGs.
Loss of AGO1&2 affects chromatin accessibility in mESCs
To determine how AGOs affect gene expression, we measured chromatin accessibility by Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) (Buenrostro et al., 2013). We identified 3,137 regions exhibiting significant differential accessibility (DA) in Ago2&1_KO versus WT (2,290 with increased accessibility, 847 with decreased accessibility) (Figures 3A and S3A, and Table S2). In contrast, only minor differences in chromatin accessibility were observed in single AGO mutants, suggesting that only the combined loss of AGO1&2 can affect chromatin structure of mESCs (Figures S3B and S3C). These observations are in parallel with the changes observed at the transcriptomic level in these mutant cell lines, where Ago2&1_KO, but not the single KO mutants, exhibited a strongly perturbed transcriptome (Figures S1A and S1B), suggesting that Agos_sDEGs might at least be partially explained by changes in chromatin accessibility.
Figure 3.
Integration of the Ago2&1_KO transcriptome with chromatin accessibility and TF binding
(A) Heatmap and profile plots for WT and Ago2&1_KO chromatin accessibility as assessed by ATAC-seq. TSS to transcription end sites (TES) with 3-kbp margins are shown for the full set of annotated transcripts. Representative samples of biological duplicates are shown.
(B and C) Average signal of chromatin accessibility at TSS/promoter regions (B) and enhancer regions (C), as annotated by (González-Ramírez et al., 2021) for WT and Ago2&1_KO samples as assessed by ATAC-seq.
(D) Volcano plot of differential chromatin accessibility (DA) at TF binding sites (BS) for 88 expressed TFs as computed by diffTF (Berest et al., 2019) (Table S3). The x axis shows the difference in chromatin accessibility between Ago2&1_KO and WT ATAC-seq samples, where the red area denotes an increase in chromatin accessibility in the Ago2&1_KO samples and the blue area a decrease. TFs are annotated as activators (green) or repressors (red) according to the DA at their binding sites and their expression levels, based on RNA-seq data.
(E and F) Scatterplots showing differential expression (RNA-seq) versus DA of potential target genes associated with TFBS from (D) for the five TFs with most significant DA binding sites from (D). TFBS were associated with genes by promoter proximity (E, <1 kbp distance to TSS) or enhancer proximity (F) (González-Ramírez et al., 2021). Genes are denoted in orange if they are Agos_sDEGs. See also Figure S3.
Next, we retrieved genes that were associated with significant DA regions (Table S2) at their promoter regions and studied their DE in Ago2&1_KO mESCs. The DE of genes with increased chromatin accessibility showed a strong enrichment for increased expression, while genes with decreased chromatin accessibility showed a tendency for decreased expression levels (Figure S3D). The difference between the DE distributions of the two groups showed statistical significance (t test p < 1.8e-5). Nevertheless, out of the 384 genes that showed increased chromatin accessibility, only 21 were Agos_sDEGs and only 14 of them were upregulated (Figures S3D and S3E, Tables S2 and S4). Further, none of the downregulated Agos_sDEGs showed significant decrease in promoter accessibility (Figures S3E and S3G). Thus, chromatin opening at gene promoter regions alone is not sufficient to explain the Agos_sDEGs.
KLF4 regulates the majority of Agos_sDEGs
While chromatin accessibility can influence TF binding (Spitz and Furlong, 2012), TF binding has conversely been suggested to modulate chromatin accessibility in many cases (Baek et al., 2017), potentially affecting the expression of associated genes. To assess whether TFs might mediate differential chromatin accessibility, we analyzed the DA specifically at promoter and enhancer regions (González-Ramírez et al., 2021). We observed an increased chromatin accessibility at these regulatory elements in Ago2&1_KO mESCs (Figures 3B and 3C), suggesting increased TF activity. Next, we integrated our chromatin accessibility data with motif-based TF binding site (TFBS) predictions using quantification of differential transcription factor activity and multiomics-based classification into activators and repressors (diffTF) (Figure 3D and Table S3) (Berest et al., 2019). Notably, five TFs (CTCF, KLF4, ERR2, REST, and MYC) showed highly significant differential binding (Figure 3D and Table S3), which might impact gene expression of their downstream targets in Ago2&1_KO mESCs. To assess this impact, we further associated the motif-based TFBS predictions from diffTF with genes, based on promoter- (transcription start site [TSS]-distance <1 kbp) and enhancer-proximity (González-Ramírez et al., 2021), and compared DA at TFBS with DEG (Figures 3E and 3F, Tables S3 and S4). For most TFs, a notable positive correlation between DA and DE was observed, indicating that differential binding of TFs can affect the expression of Agos_sDEGs. Combined, the five identified TFs positively correlate with around 17% of Agos_sDEGs (152 up- and 137 downregulated), from which CTCF and KLF4 binding sites correspond to the largest portion of them (149 and 147 genes respectively, Figures 3E and 3F, Tables S3 and S4). CTCF is well known for its role in chromatin looping and organization (Oudelaar and Higgs, 2021). KLF4 has also been linked to chromatin organization during the reprogramming of mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells (iPSCs) by conferring enhancer-promoter contacts (Campigli et al., 2019). Thus, altering CTCF and KLF4 levels or their binding site accessibility might affect the interaction of regulatory elements and the underlying gene expression. We did not observe a change in CTCF expression at RNA or protein levels by qPCR and by WB in Ago2&1_KO mESCs (Figures S4A and S4B). In contrast, KLF4 was significantly downregulated in Ago2&1_KO compared with WT mESCs (Figures 4A, S4C and Table S1), suggesting that the decrease in KLF4 levels might affect gene expression. To determine whether CTCF and KLF4 are associated with the promoter and enhancers of Agos_sDEGs, we analyzed published CTCF- and KLF4-ChIP-seq performed in mESCs (Campigli et al., 2019; Nora et al., 2017). Consistent with the increase in chromatin accessibility at CTCF-TFBS in Ago2&1_KO cells (Figure 3D), we observed that the number of CTCF-bound promoters and enhancers was higher for the upregulated Agos_sDEGs (35%, 260) than for the downregulated ones (19%, 201) (Figure 4B and Table S4). Remarkably, the analysis of KLF4-ChIP-seq data revealed a much larger fraction of the promoter and enhancer regions of Agos_sDEGs that were bound by KLF4 (upregulated genes, 497, 73%; downregulated genes, 550, 47%), suggesting a major role in the regulation of Agos_sDEGs (Figures 4C–4E and Table S4).
Figure 4.
Identification of KLF4 and CTCF targets and complete integration
(A) Representative WB (top) for KLF4 in WT and Ago2&1_KO mESCs and quantification (bottom) out of n = 3 independent experiments, ∗∗∗p < 0.001, unpaired t test.
(B and C) Venn diagrams of genes identified by CTCF (B) and KLF4 (C) ChIP-seq peak analysis with Agos_sDEGs. ChIP-seq peaks were associated with genes by promoter proximity (TSS <1,000 bp, green circle) and by enhancer-proximity (overlap with annotated enhancers by González-Ramírez et al. [2021]). Different Venn diagrams are shown for upregulated (top) and downregulated (bottom) gene sets and only statistically significant Ago2&1_KO DE-Gs are considered.
(D and E) UpSet plots of upregulated (D) and downregulated (E) Agos_sDEGs explained by one or multiple combined regulatory mechanisms as studied in this paper (Table S4). The dot-connected lines indicate which gene explanation sets were combined and the bar above denotes the total number of genes explained by that combination. The bars on the right indicate the total number of explained genes explained by each individual analysis (these are thus redundant with the columns representing a single dot). Notably, KLF4 ChIP-seq analysis (red box, and top-most bar on the right) alone already explained the majority of all explained genes. See also Figure S4.
In conclusion, the intersection of transcriptomic, chromatin accessibility, TF binding predictions, and ChIP-seq data identified AGO1&2 as regulators of gene expression in mESCs through the regulation of KLF4, a master regulator of pluripotency.
Discussion
Given that DGCR8, DROSHA, DICER, and AGO1&2 are involved in the canonical miRNA biogenesis pathway, one might assume that their KOs would lead to similar transcriptomic perturbations. Consistent with previous results (Schäfer et al., 2021), DEGs in Ago2&1_KO showed strong similarities to Dicer_KO, but not to Dgcr8_KO or Drosha_KO. This might be partially attributable to noncanonical miRNA pathways, which function independently of the Microprocessor (DGCR8/DROSHA), but still require DICER and the AGO proteins (Bodak et al., 2017a). Furthermore, AGO2 protein levels are strongly reduced in miRNA_KO mESCs due to its targeted proteasome degradation in the absence of miRNAs (Bodak et al., 2017b; Smibert et al., 2013). Some miRNA-independent AGO-mediated functions might therefore also be partially affected in miRNA_KO mESC lines. Indeed, we also observed a downregulation of Klf4 in Dicer_KO mESCs (Table S1). Surprisingly, despite these similarities, comparing DEGs between Ago2&1_KO and other miRNA_KO mESCs revealed a large number of DEGsthat were specific to individual mutants and especially to Ago2&1_KO (Figure 1A), indicating potential miRNA-independent AGO-specific functions. AGO-specific functions have already been described in cancer cell lines and more recently also in mESCs (Meister, 2013; Müller et al., 2022; Sarshad et al., 2018). Interestingly, mutation of a single Ago gene had very little impact on gene expression (Figure S1A) (Müller et al., 2022). Given the strong observed perturbation in the Ago2&1_KO transcriptome, it might indicate that AGO1&2 have global compensatory functions in mESCs that can be identified through the deletion of both.
The integrative analyses of differential chromatin accessibility at predicted TFBS revealed a strong regulatory potential of two TFs, CTCF, and, in particular, the pluripotency factor KLF4 that can display both an activating and a repressive function (Bialkowska et al., 2017). We found that KLF4 binds the promoter of about 50% to 70% of up- and downregulated Agos_sDEGs, respectively. Importantly, we also observed that the expression of KLF4 is strongly reduced in Ago2&1_KO mESCs. KLF4 is a pluripotency factor, which has been reported to occupy promoters of other key pluripotent TFs, thereby affecting their expression and vice versa (Bialkowska et al., 2017; Jiang et al., 2008).
KLF4 has also been implicated in genome reorganization in mESCs and important functions in conferring enhancer connectivity. There, the disruption of KLF4 and its binding sites in mESCs showed an abrogation of enhancer contacts, which consequently decreased expression levels of associated genes (Campigli et al., 2019). The loss of KLF4, along with many AGO1&2-regulated genes in our study, potentially may lead to alterations in chromatin conformation, which might be linked directly or indirectly to the AGO proteins. Indeed, previous studies already pointed toward potential alterations of the chromatin conformation linked to the AGO proteins (Moshkovich et al., 2011; Shuaib et al., 2019). Our study further supports a potential role for the AGO proteins in the modulation of chromatin conformation. Thus, mESCs might provide a unique opportunity to further investigate chromatin-related functions of the AGOs in mammals.
In conclusion, our study revealed noncanonical functions of AGO1&2 in mESCs that do not overlap with the canonical miRNA pathway and revealed a novel axis of gene regulation through the transcription factor KLF4.
Experimental procedures
Detailed methods are provided in the supplemental experimental procedures.
Mouse ESC lines
WT and miRNA_KO mESC lines were all generated in the Ciaudo lab in the E14 129/Ola background and were cultured as described in Schäfer et al. (2021).
RNA-seq, ChIP-seq, and ATAC-seq
All procedures, from library preparation, sequencing, and analysis, are detailed in the supplemental information.
Data and software availability
All data and software used in this paper are listed in the supplemental information. Data visualization has been performed with the tools described in the supplemental information. If not mentioned otherwise, graphs have been generated by using PRIMS.
All omics datasets generated in this study have been deposited in the NCBI Gene Expression Omnibus under the accession number GSE185410 (H3K27me3 ChIP-seq and ATAC-seq).
Author contributions
Conceptualization, M.M., M.S., and C.C.; laboratory experiments, M.M., T.F., V.H., R.P.N.; ChiP-seq libraries preparation, M.M., R.P.H.; computational analysis, M.S. and D.S.; writing original draft preparation, M.S., M.M., and C.C.; writing, review and editing, C.C.; expertise and editing, R.S.; visualization, M.S., M.M., and C.C.; supervision, C.C.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Conflicts of interests
The authors declare no competing interests.
Acknowledgments
We thank the members of the Ciaudo lab and Dr. Tobias Beyer for fruitful discussions and the critical reading of this manuscript. This work was supported by the Swiss National Science Foundation (grants 31003A_173120 and 310030_196861) to C.C., and by the Swiss National Science Foundation (31003A_173056 and 201268), and ERC grant (ERC-AdG-787074-NucleolusChromatin) to R.S. In addition, C.C., M.M., and R.S. were supported by the NCCR RNA and Disease, a National Center of Excellence in Research, funded by the Swiss National Science Foundation (grant number 182880). We also thank the Functional Genomics Center Zurich (FGCZ) for their support with the preparation of ChIP-seq libraries and sequencing. We also thank Quick Biology for their help with the ATAC-seq library preparation, sequencing, and computational support.
Published: April 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.03.014.
Supplemental information
References
- Asenjo H.G., Gallardo A., López-Onieva L., Tejada I., Martorell-Marugán J., Carmona-Sáez P., Landeira D. Polycomb regulation is coupled to cell cycle transition in pluripotent stem cells. Sci. Adv. 2020;6:eaay4768. doi: 10.1126/sciadv.aay4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S., Goldstein I., Hager G.L. Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 2017;19:1710–1722. doi: 10.1016/j.celrep.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berest I., Arnold C., Reyes-Palomares A., Palla G., Rasmussen K.D., Giles H., Bruch P.M., Huber W., Dietrich S., Helin K., et al. Quantification of differential transcription factor activity and multiomics-based classification into activators and repressors: diffTF. Cell Rep. 2019;29:3147–3159.e12. doi: 10.1016/j.celrep.2019.10.106. [DOI] [PubMed] [Google Scholar]
- Bialkowska A.B., Yang V.W., Mallipattu S.K. Krüppel-like factors in mammalian stem cells and development. Development. 2017;144:737–754. doi: 10.1242/dev.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodak M., Cirera-Salinas D., Luitz J., Ciaudo C. The role of RNA interference in stem cell biology: beyond the mutant phenotypes. JMB. 2017;429:1532–1543. doi: 10.1016/j.jmb.2017.01.014. [DOI] [PubMed] [Google Scholar]
- Bodak M., Cirera-Salinas D., Yu J., Ngondo R.P., Ciaudo C. Dicer , a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells. FEBS Open Bio. 2017;7:204–220. doi: 10.1002/2211-5463.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campigli D., Giammartino D., Kloetgen A., Polyzos A., Liu Y., Kim D., Murphy D., Abuhashem A., Cavaliere P., Aronson B., et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat. Cell Biol. 2019;21:1179–1190. doi: 10.1038/s41556-019-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K., et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ramírez M., Ballaré C., Mugianesi F., Beringer M., Santanach A., Blanco E., Di Croce L. Differential contribution to gene expression prediction of histone modifications at enhancers or promoters. PLOS Comput. Biol. 2021;17:e1009368. doi: 10.1371/journal.pcbi.1009368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B., Marcais A., Dharmalingam G., Carroll T., Kanellopoulou C., Graumann J., Nesterova T.B., Bermange A., Brazauskas P., Xella B., et al. MicroRNAs of the miR-290-295 family maintain bivalency in mouse embryonic stem cells. Stem Cell Rep. 2016;6:635–642. doi: 10.1016/j.stemcr.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Chan Y.S., Loh Y.H., Cai J., Tong G.Q., Lim C.A., Robson P., Zhong S., Ng H.H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kennerdell J.R., Liu N., Bonini N.M. MiR-34 inhibits polycomb repressive complex 2 to modulate chaperone expression and promote healthy brain aging. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Moshkovich N., Nisha P., Boyle P.J., Thompson B.A., Dale R.K., Lei E.P. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Fazi F., Ciaudo C. Argonaute proteins : from structure to function in development and pathological cell fate determination. Front. Cell Dev. Biol. 2020;7:1–10. doi: 10.3389/fcell.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Fäh T., Schaefer M., Hermes V., Luitz J., Stalder P., Arora R., Ngondo R.P., Ciaudo C. AGO1 regulates pericentromeric regions in mouse embryonic stem cells. Life Sci. Alliance. 2022;5:1–14. doi: 10.26508/lsa.202101277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngondo R.P., Cirera-Salinas D., Yu J., Wischnewski H., Bodak M., Vandormael-Pournin S., Geiselmann A., Wettstein R., Luitz J., Cohen-Tannoudji M., et al. Argonaute 2 is required for extra-embryonic endoderm differentiation of mouse embryonic stem cells. Stem Cell Rep. 2018;10:1–16. doi: 10.1016/j.stemcr.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E.P., Goloborodko A., Valton A.L., Gibcus J.H., Uebersohn A., Abdennur N., Dekker J., Mirny L.A., Bruneau B.G. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudelaar A.M., Higgs D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021;22:154–168. doi: 10.1038/s41576-020-00303-x. [DOI] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarshad A.A., Juan A.H., Muler A.I.C., Anastasakis D.G., Wang X., Genzor P., Feng X., Tsai P.-F., Sun H.-W., Haase A.D., et al. Argonaute-miRNA complexes silence target mRNAs in the nucleus of mammalian stem cells. Mol. Cell. 2018;71:1040–1050.e8. doi: 10.1016/j.molcel.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M., Nabih A., Spies D., Bodak M., Stalder P., Ngondo R.P., Liechti L.A., Sajic T., Aebersold R., Gatfield D., et al. Integrative analysis allows a global and precise identification of functional miRNA target genes in mESCs. bioRxiv. 2021 doi: 10.1101/2021.09.24.461622. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivram H., Le S.V., Iyer V.R. MicroRNAs reinforce repression of PRC2 transcriptional targets independently and through a feed-forward regulatory network. Genome Res. 2019;29:184–192. doi: 10.1101/gr.238311.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M., Parsi K.M., Thimma M., Ravasi T., Carninci P., Orlando V., Shuaib M., Parsi K.M., Thimma M., Adroub S.A., et al. Nuclear AGO1 regulates gene expression by affecting chromatin architecture in human cells. Cell Syst. 2019;9:446–458.e6. doi: 10.1016/j.cels.2019.09.005. [DOI] [PubMed] [Google Scholar]
- Smibert P., Yang J.-S., Azzam G., Liu J.-L., Lai E.C. Homeostatic control of Argonaute stability by microRNA availability. NSMB. 2013;20:1–9. doi: 10.1038/nsmb.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.a., Doyle F., Epstein C.B., Frietze S., Harrow J., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2013;488:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and software used in this paper are listed in the supplemental information. Data visualization has been performed with the tools described in the supplemental information. If not mentioned otherwise, graphs have been generated by using PRIMS.
All omics datasets generated in this study have been deposited in the NCBI Gene Expression Omnibus under the accession number GSE185410 (H3K27me3 ChIP-seq and ATAC-seq).