Abstract
Since April 2014 all presumptive Salmonella isolates received by Public Health England (PHE) have been characterised using whole genome sequencing (WGS) and the genomic data generated used to identify clusters of infection. To inform the implementation and development of a national gastrointestinal infection surveillance system based on WGS we have retrospectively identified genetically related clusters of Salmonella Enteritidis and Salmonella Typhimurium infection over a one year period and determined the distribution of these clusters by PHE operational levels. Using a constrained WGS cluster definition based on single nucleotide polymorphism distance, case frequency and temporal spread we demonstrate that the majority of clusters spread to multiple PHE operational levels. The greatest investigative burden is on national level staff investigating small, geographically dispersed clusters. We also demonstrate that WGS identifies long-running, slowly developing clusters that may previously have remained undetected. This analysis also indicates likely increased workload for local health protection teams and will require an operational strategy to balance limited human resources with the public health importance of investigating small, geographically contained clusters of highly related cases. While there are operational challenges to its implementation, integrated cluster detection based on WGS from local to international level will provide further improvements in the identification of, response to and control of clusters of Salmonella spp. with public health significance.
Key words: Epidemiology, outbreaks, Salmonella, surveillance, whole genome sequencing
Background
Traditional automated cluster detection for Salmonella spp. in England and Wales using serological typing-based surveillance has relied on monitoring weekly reports that exceed an expected threshold based on a rolling average of data from similar weeks in previous years. This system has a number of drawbacks: it is prone to artefacts from reporting delays, exhibits a low level of strain discrimination and it is not designed to readily identify long-term clusters of common serovars where low numbers of cases are detected each week. This can result in public health teams investigating epidemiologically unlinked cases of Salmonella infection while other linked cases remain undetected.
Whole genome sequencing (WGS) is a molecular method for characterising organisms and has improved power to discriminate between closely related strains compared to previously deployed methods. It has proven to be a rapid, cost-effective method for infectious disease surveillance. Since April 2014 all presumptive Salmonella isolates received by Public Health England (PHE) from England have been sequenced while the application of parallel traditional phenotyping methods on these isolates has been greatly reduced. Clusters can be detected using analysis of WGS data by comparing isolates based on their single nucleotide polymorphism (SNP) ‘address’ [1]. Investigations into the feasibility and methodology for the implementation of a national surveillance system based on WGS mooted action thresholds for counts of isolates at different levels of genetic relatedness [2].
It is unclear how best to use these data operationally and their implications for the workload of public health teams investigating clusters identified at different thresholds. To inform the implementation and development of a national gastrointestinal infection surveillance system based on WGS, we retrospectively identified genetically related clusters of Salmonella enterica subsp. enterica serovars Enteritidis and Typhimurium – the first and second most commonly reported Salmonella serovars in humans in England, respectively – over a 1-year period using differing SNP detection thresholds and determined the distribution of these clusters by PHE operational levels.
Methods
As of April 2016, the operational hierarchy within PHE for following up notifications of Salmonella spp. cases from the PHE national Gastrointestinal Bacteria Reference Unit (GBRU) in England involved 23 health protection teams (HPTs) responsible for local investigation (‘local’ level). HPTs are aggregated into nine administrative PHE Centres (PHECs; ‘regional’ level). Both local and regional level investigations might be supported by Field Epidemiology Service (FES) teams. The population within HPT boundaries range from 1 365 847 to 4 637 413 persons for South Yorkshire HPT and East Midlands HPT, respectively, and within PHEC boundaries range from 2 618 710 to 8 614 573 persons for North East PHEC and South East PHEC, respectively [3]. GBRU in collaboration with the Gastrointestinal Infections Department are responsible for national surveillance of gastrointestinal infections and might either lead on or support FES teams leading on national level outbreak investigations, as well as contribute to regional and local investigations.
Data on S. Enteritidis (e-burst group (EBG) 4) and S. Typhimurium (EBG 1) [4] isolates received and processed by the GBRU were extracted from PHE's Gastrointestinal Data Warehouse (GDW) – a central web-based repository of reference laboratory data associated with a unique sample identifier – for the study period, 01 April 2014–31 March 2015. Isolates were mapped to HPT, first according to residential postcode, then originating sample hospital postcode or originating laboratory postcode, as available. Once an HPT was defined, each isolate was then assigned a corresponding PHEC based on existing geographical hierarchies. Quality assurance, non-clinical and duplicated samples and isolates with no SNP address were excluded.
The SNP analysis pipeline has been described previously [1] and generates a seven number ‘SNP address’ that groups isolate into clusters of increasing levels of genetic similarity. Three independent analyses were conducted using different SNP thresholds to define single linkage clusters from available isolates: 0-SNP (zero SNP differences between isolates in cluster), 5-SNP (all isolates have no more than five SNP differences from at least one other isolate in the cluster) and 10-SNP (all isolates have no more than ten SNP differences from at least one other isolate in the cluster). In addition, clustering at each SNP threshold had to satisfy the following temporal and frequency criteria. Regardless of SNP threshold, each isolate within a cluster must have had a receipt date at the GBRU within seven days of at least one other case in a given cluster. Two or more cases were required for a cluster using a 0-SNP threshold, five or more for a cluster using a 5-SNP threshold and ten or more using a 10-SNP threshold. Clusters defined using each SNP threshold were stratified into mutually exclusive geographical levels: local (all cases within the cluster were within a single HPT area), regional (all cases within a cluster were within one PHEC area but multiple HPTs) and national (cases within a cluster were within more than one PHEC). Of all identified clusters, 10% were manually validated by cross-referencing summary level outputs with raw, case-level data.
Clusters within each serovar, as determined using the aforementioned SNP thresholds, were described by geographical level, duration, number of cases, whether the SNP address was common to any other clusters identified over the study period, the proportion with at least two cases with a common postcode suggesting possible household transmission, where available, and the proportion for which at least 75% of intra-cluster cases reported recent foreign travel. Data were summarised as medians and ranges, as necessary. Clusters identified using each SNP threshold at individual local and regional levels were summarised including minimum, the range of cases and duration of clusters. Counts of cases that were part of any cluster across the three geographic levels but resident within a given local or regional boundary were also presented.
A secondary clustering analysis was run on the dataset using the same criteria as the primary analysis but without the temporal requirement of 7 days between one case and any other in a given cluster. Clusters derived from this approach were summarised by SNP-threshold-geographical level strata identified and percentage difference comparison made with findings from the primary analysis.
Clustering analyses and postcode mapping were conducted using R v3.3.1 statistical programming language (R Development Core 2008). Deduplication was conducted using STATA v13 (StataCorp, Texas). Trends in proportions were investigated using a χ2 test for trend and differences in proportions assessed using a χ2 or Fishers exact test, as appropriate, where underlying assumptions were met, using OpenEpi v3.01 [5].
Investigation of associations between population size of the geographical area, month of cluster detection and count or presence of clusters, as appropriate, for 0-, 5- and 10-SNP clusters were conducted separately using regression methods. Table 1 indicates the type of generalised linear model that was used for each outcome. Further details of decision making while constructing these models can be found in the Supplementary Technical Description S1 (all supplementary material are available on the Cambridge Core website).
Table 1.
Choice of generalised linear models used to investigate associations between serovar-specific SNP threshold cluster outcomes and population and month of cluster identifications
| Geographical level | Outcome | Model type |
|---|---|---|
| Local | S. Enteritidis 0 SNP 0 clusters | Logistic |
| S. Typhimurium 0 SNP 0 clusters | Logistic | |
| Regional | S. Enteritidis SNP 0 clusters | Logistic |
| National | S. Enteritidis SNP 0 clusters | Poisson |
| S. Enteritidis SNP 5 clusters | Poisson | |
| S. Enteritidis SNP 10 clusters | Logistic | |
| S. Typhimurium SNP 0 clusters | Poisson | |
| S. Typhimurium SNP 5 clusters | Logistic | |
| S. Typhimurium SNP 10 clusters | Logistic |
Records of clusters of S. Enteritidis and S. Typhimurium identified using traditional typing and cluster detection methods and investigated by HPTs over the study period, according to the PHE electronic foodborne and non-foodborne gastrointestinal outbreak surveillance system (eFOSS) [6], and the national team was summarised.
Results
Primary analyses
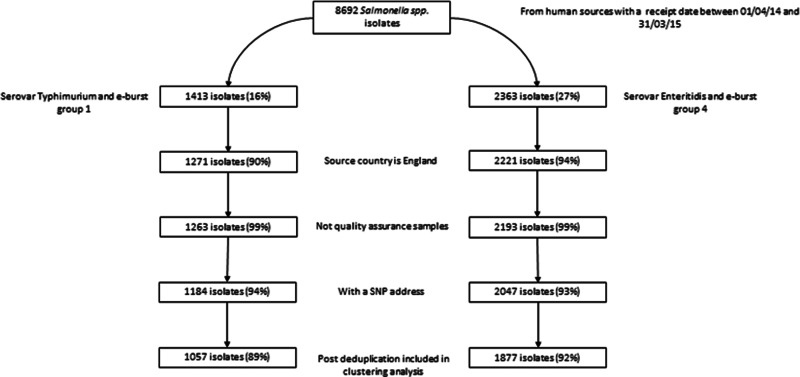
Of 8692 clinical isolates of Salmonella received by GBRU for analysis between April 2014 and March 2015, 2362 (27%) were S. Enteritidis (and belonged to EBG 4) and 1413 (16%) were S. Typhimurium (and belonged to EBG 1) (Fig. 1). Post-exclusion (including 7% and 6% of valid isolates without an SNP address for S. Enteritidis and S. Typhimurium, respectively) and deduplication, there were 1877 and 1057 case-isolates (hereafter referred to as cases) of S. Enteritidis and S. Typhimurium, respectively, for inclusion in clustering analyses.
Fig. 1.
Exclusions of available S. Enteritidis and S. Typhimurium isolates prior to clustering analysesa. aPercentages calculated using isolates prior to a given step as the denominator.
A total of 147 0-SNP, 40 5-SNP and 16 10-SNP clusters, defined using temporal criteria, were identified from S. Enteritidis cases and 64 0-SNP, 11 5-SNP and 4 10-SNP clusters from S. Typhimurium cases (Table 2). Regardless of the SNP threshold used for defining a cluster, the majority were nationally distributed (58%, 95% and 100% of 0-, 5- and 10-SNP clusters, respectively, of S. Enteritidis and 65%, 96% and 100% of 0-, 5- and 10-SNP clusters, respectively, of S. Typhimurium). Local level clusters of both serovars were only identified when a 0 SNP threshold (two or more cases) was used and accounted for 36% and 31% of all 0 SNP S. Enteritidis and S. Typhimurium clusters, respectively. Regional level clusters of either serovar were identified at 0- and 5-SNP thresholds and accounted for no more than 9% of the total number of clusters. No errors were detected among the 10% of manually reviewed clusters in the finalised dataset.
Table 2.
Summary of identified 0-, 5- and 10-SNP clusters of S. Enteritidis and S. Typhimurium by HPT (local), PHEC (regional) and national levels in England, April 2014–March 2015
| Serovar | SNP level | Geographical level | Total n of clusters (%)a | Total n of cases in clusters (%)a | Median no. of cases (range)a | Median duration of cluster in days (range) | N (%) of clusters with a SNP address common to another cluster | N (%) of clusters with household contactsb | N (%) of clusters with 75% known foreign travelc |
|---|---|---|---|---|---|---|---|---|---|
| Enteritidis | 0 | Local | 53/147 (36%) | 164/522 (31%) | 2 (2–25) | 3 (1–27) | 10/53 (19%) | 13/53 (25%) | 7/53 (13%) |
| Regional | 9/147 (6%) | 28/522 (5%) | 2 (2–8) | 4 (1–16) | 0 (–) | 1/9 (11%) | 2/9 (22%) | ||
| National | 85/147 (58%) | 330/522 (63%) | 2 (2–62) | 6 (1–30) | 28/85 (33%) | 4/85 (5%) | 22/85 (26%) | ||
| 5 | Local | 0 (–) | 0 (–) | – | – | – | – | – | |
| Regional | 2/40 (5%) | 14/602 (2%) | 7 (6–8) | 11.5 (9–14) | 0 (–) | 2/2 (100%) | 0 (–) | ||
| National | 38/40 (95%) | 588/602 (98%) | 7 (5–239) | 18.5 (4–134) | 31/38 (82%) | 9/38 (24%) | 3/38 (8%) | ||
| 10 | Local | 0 (–) | 0 (–) | – | – | – | – | – | |
| Regional | 0 (–) | 0 (–) | – | – | – | – | – | ||
| National | 16 (100%) | 505 (100%) | 14.5 (10–239) | 33.5 (16–134) | 11/16 (69%) | 6/16 (38%) | 0 (–) | ||
| Typhimurium | 0 | Local | 27/64 (42%) | 64/207 (31%) | 2 (2–11) | 3 (1–15) | 2/27 (7%) | 9/27 (33%) | 1/27 (4%) |
| Regional | 4/64 (6%) | 9/207 (4%) | 2 (2–3) | 7 (6–8) | 0 (–) | 0 (–) | 0 (–) | ||
| National | 33/64 (52%) | 134/207 (65%) | 2 (2–31) | 6 (1–27) | 4/33 (12%) | 2/33 (6%) | 4/33 (12%) | ||
| 5 | Local | 0 (–) | 0 (–) | – | – | – | – | – | |
| Regional | 1/11 (9%) | 5/117 (4%) | 5 (5–5) | 15 (15–15) | 0 (–) | 1/1 (100%) | 0 (–) | ||
| National | 10/11 (91%) | 112/117 (96%) | 7 (5–36) | 16 (1–29) | 0 (–) | 3/10 (30%) | 1/10 (10%) | ||
| 10 | Local | 0 (–) | 0 (–) | – | – | – | – | – | |
| Regional | 0 (–) | 0 (–) | – | – | – | – | – | ||
| National | 4/4 (100%) | 111 (100%) | 26.5 (15–43) | 32 (16–61) | 3/4 (75%) | 2/4 (50%) | 0 (–) |
SNP level column percentages.
Two or more of cases in a cluster had the same postcode.
75% or more of cases in a cluster had known travel history.
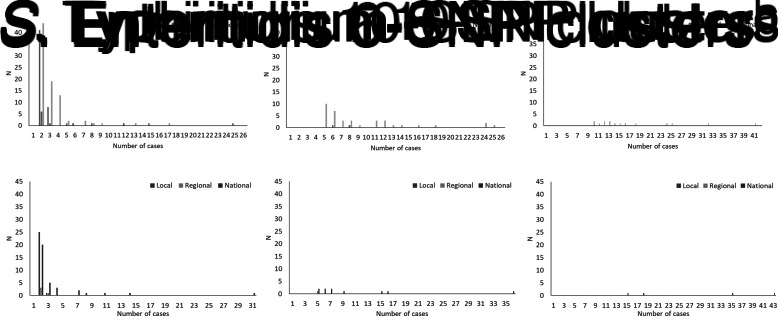
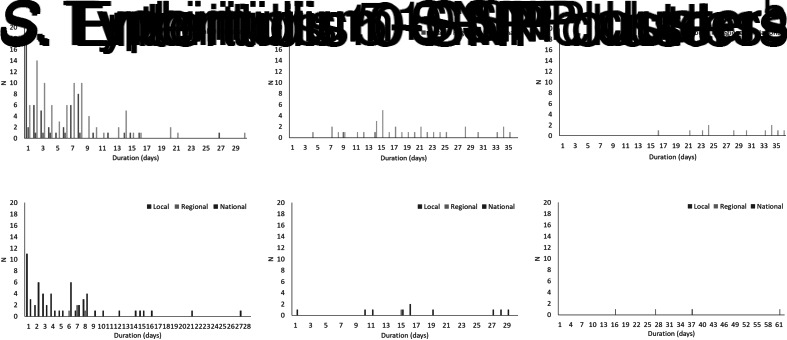
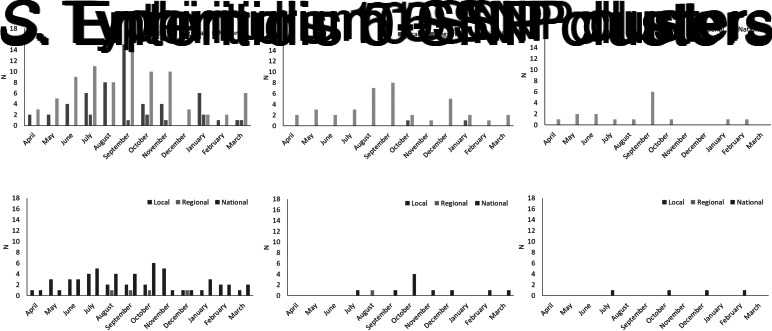
The total number of cases of S. Enteritidis within identified 0-, 5- and 10-SNP clusters was 522, 602 and 505, respectively, (Table 2) which is 28%, 32% and 27% of cases included in the clustering analysis (n = 1877; Fig. 1). Across local, regional and national geographical levels, the medians of number of cases and duration of 0-SNP clusters were less than or equal to two cases (all geographical levels) and 6 days (national clusters), respectively, and the maximum values within any 0-SNP cluster were less than or equal to 62 cases and 30 days, respectively, for national clusters (Table 2; Figs 2 and 3). The medians of number of cases and duration of 5-SNP clusters across the geographical levels for which clusters were identified was less than or equal to seven cases (regional and national clusters) and 18.5 days (for national clusters), respectively, and the maxima were less than or equal to 239 cases and 134 days, respectively, for national clusters. The medians of a number of cases and duration of national 10-SNP clusters were 14.5 cases and 33.5 days, respectively, and the maxima were 239 cases and 134 days, respectively. The national cluster with the maximum number and duration of cases at the 5-SNP level was comprised the same 239 cases as that at the 10-SNP level. The peak month of local and/or national level S. Enteritidis clusters, regardless of SNP threshold, was September (Fig. 4).
Fig. 2.
Distribution of size of clusters of S. Enteritidis and S. Typhimurium using the 0-, 5- and 10-SNP level thresholds in England, April 2014–March 2015. aOne national cluster, not shown, had 62 cases. bOne national cluster, not shown, had 239 cases. cOne national cluster, not shown, had 239 cases.
Fig. 3.
Distribution of duration of clusters of S. Enteritidis and S. Typhimurium using the 0-, 5- and 10-SNP level thresholds in England, April 2014–March 2015. aIndividual national clusters, not shown, had durations of 57, 58, 66 and 134 days, respectively. bIndividual national clusters, not shown, had durations of 57, 58, 66 and 134 days, respectively.
Fig. 4.
Distribution of detection of clusters of S. Enteritidis and S. Typhimurium using the 0-, 5- and 10-SNP levels thresholds in England, April 2014–March 2015.
The total number of cases of S. Typhimurium within identified 0-, 5- and 10-SNP clusters was 207, 117 and 111, respectively (Table 2), which is 20%, 11% and 11% of cases included in the clustering analysis (n = 1057; Fig. 1). Across local, regional and national geographical levels, the medians of number of cases within and duration of 0-SNP clusters across geographical levels were no more than two cases (all geographical levels) and 7 days (regional clusters), respectively and the maxima were less than or equal to 31 cases and 27 days, respectively, for national clusters (Table 2; Figs 2 and 3). The medians of the number of cases and duration of 5-SNP clusters across the geographical levels for which clusters were identified were less than or equal to seven cases and 16 days, respectively, for national clusters and the maxima were less than or equal to 36 cases and 29 days, respectively, also for national clusters. The medians of a number of cases and duration of national 10-SNP clusters were 26.5 cases and 32 days, respectively, and the maxima of the ranges were 43 cases and 61 days, respectively. Where observable, the peak month of S. Typhimurium clusters was November for local level and October for national level clusters (Fig. 4).
When counting repeat clusters over the study period only once, 49% of S. Enteritidis and 25% of S. Typhimurium 0-SNP clusters, were nested within clusters of more genetic diversity at the 5- or 10-SNP level and, of unique 5-SNP clusters, 53% of S. Enteritidis and 27% S. Typhimurium, respectively, were nested within 10-SNP clusters (Table 3). Of cases of S. Enteritidis and S. Typhimurium in all 0-SNP clusters, 66% and 46%, respectively, were nested within clusters of more genetic diversity at the 5- or 10-SNP level and, of cases within 5-SNP clusters, 75% and 57%, respectively, were nested within 10-SNP clusters (Table 3).
Table 3.
Nestedness of clusters and associated cases identified using 0-, 5- and 10-SNP threshold clustering
| SNP levels | S. Enteritidis (%) | S. Typhimurium (%) | ||
|---|---|---|---|---|
| Clustersa | Casesb | Clustersa | Casesb | |
| In 0-SNP and 5-SNP outputs | 52/121 (43) | 325/522 (62) | 13/60 (22) | 94/207 (45) |
| In 0-SNP and 10-SNP outputs | 42/121 (35) | 274/522 (52) | 6/60 (10) | 60/207 (29) |
| In 5-SNP and 10-SNP outputs | 10/19 (53) | 453/602 (75) | 3/11 (27) | 67/117 (57) |
| In 0-SNP and 5-SNP or 10-SNP outputs | 59/121 (49) | 342/522 (66) | 13/60 (25) | 96/207 (46) |
Denominator is the total number of unique clusters at SNP level with least genetic diversity.
Denominator is the total number of cases in all clusters at SNP level with least genetic diversity.
Across SNP threshold-geographical level strata with more than two clusters of S. Enteritidis or S. Typhimurium, a maximum of 82% and 75% of clusters, respectively, had an SNP addresses that was common to another cluster over the study period; 38% and 50% of clusters, respectively, had two or more cases which shared a common postcode; and 26% and 12% clusters, respectively, were composed of cases where at least 75% had a recorded history of recent travel (Table 2).
The maximum number of 0-SNP clusters and cases attributed to a single local team was seven clusters of S. Enteritidis and 30 cases and three clusters of S. Typhimurium and 11 cases (Supplementary Table S1, Fig. 5). The maximum number of 0-SNP clusters attributed to a single regional team was two for both S. Enteritidis and S. Typhimurium clusters. The maximum number of cases in a 0-SNP cluster in any single regional team was eight cases and five cases for S. Enteritidis and S. Typhimurium clusters, respectively. There were no 5-SNP clusters attributed to a local team and, in total, two small clusters of S. Enteritidis (no more than eight cases) and one small cluster of S. Typhiumurium (five cases) attributed to regional teams at this SNP threshold (Supplementary Table S2, Fig. 5). There were no local or regional level clusters detected using a 10-SNP threshold (Supplementary Table S3, Fig. 5).
Fig. 5.
Map showing the number of cases of Salmonella Enteritidis and Salmonella Typhimurium associated with clusters using 0-, 5- and 10-SNP thresholds at local (a and c, for S. Enteritidis and S. Typhimurium, respectively) and regional (b and d) geographical levels in England, April 2014–March 2015.
A single local team had up to 75 S. Enteritidis cases (at 5-SNP threshold) and 46 S. Typhimurium cases (at 0-SNP threshold) associated with any cluster regardless of the geographical level to which clusters were assigned in this analysis (Supplementary Table S4).
Secondary analysis (omitting temporal criteria from cluster definitions)
Rerunning the clustering analysis without a temporal component resulted in an increase in identified clusters of both serovars in most SNP threshold-geographical level cluster strata for which there were clusters identified in the primary analysis (Table 4). Typically the median size of clusters was comparable with those identified when a temporal component was considered but the maximum sizes tended to increase and the median and maximum cluster durations were substantially greater, often by hundreds of days. A notable outlier to this pattern was with 10-SNP national clusters of S. Typhimurium, for which identification of additional smaller clusters resulted in the median cluster size being less than in the primary analysis. There were clusters of both S. Enteritidis and S. Typhimurium which, when the temporal criteria were removed, covered the full study period.
Table 4.
Summary of identified 0-, 5- and 10-SNP clusters of S. Enteritidis and S. Typhimurium derived using no temporal consideration by HPT (local), PHEC (regional) and national levels in England, April 2014–March 2015
| Serovar | SNP level | Geographical level | Total n of clusters (% differencea) | Total n of cases in clusters | Median no. of cases (range) | Median duration of cluster in days (range) |
|---|---|---|---|---|---|---|
| Enteritidis | 0 | Local | 45 (−15%) | 160 | 2 (2–29) | 8 (1–317) |
| Regional | 20 (122%) | 60 | 2 (2–9) | 28 (1–182) | ||
| National | 125 (47%) | 551 | 3 (2–63) | 28 (1–307) | ||
| 5 | Local | 1 (–b) | 5 | 5 (5–5) | 20 (20–20) | |
| Regional | 1 (−50%) | 12 | 12 (12–12) | 197 (197–197) | ||
| National | 62 (63%) | 1025 | 7 (5–254) | 167.5 (30–365) | ||
| 10 | Local | 0 (0%) | – | – | – | |
| Regional | 0 (0%) | – | – | – | ||
| National | 33 (106%) | 1041 | 15 (10–254) | 243 (57–365) | ||
| Typhimurium | 0 | Local | 34 (26%) | 84 | 2 (2–11) | 9 (1–120) |
| Regional | 4 (0%) | 9 | 2 (2–3) | 41 (7–172) | ||
| National | 57 (73%) | 206 | 2 (2–31) | 21 (1–176) | ||
| 5 | Local | 0 (0%) | – | – | – | |
| Regional | 1 (0%) | 6 | 6 (6–6) | 191 (191–191) | ||
| National | 29 (190%) | 277 | 7 (5–37) | 120 (9–338) | ||
| 10 | Local | 0 (0%) | – | – | – | |
| Regional | 0 (0%) | – | – | – | ||
| National | 14 (250%) | 332 | 15.5 (11–131) | 253 (35–365) |
Compared with the summary results of the primary analysis in which clusters were defined using a temporal component.
A percentage could not be calculated; 0 clusters were identified in the primary analysis.
Regression analyses
In multivariable regression analyses, there was evidence of an independent association, having controlled for population size, between month and identification of 0-SNP S. Enteritidis clusters at local level (odds ratio (OR) 5.13, 95% confidence interval (CI) 1.18–22.3 for September vs. April, P = 0.001) (Supplementary Table S5). There was evidence of an association between season and count of 0-SNP and 5-SNP S. Enteritidis clusters at the national level (June–November vs. April/May, P < 0.001; and OR 3.00, 95% CI 1.09–8.25 for August/September vs. April/May, P = 0.02, respectively). Having controlled for the month of identification of 0-SNP S. Typhimurium clusters, there was evidence of an independent association between HPT size of population and identification of clusters (OR 1.74 per 100 000 persons, 95% CI 1.05–2.89, P = 0.04).
Outbreaks identified using traditional approaches
In total, 13 outbreaks of either serovar identified using traditional methods and investigated were reported during the study period (Table 5). Local teams reported investigating seven S. Enteritidis outbreaks, with a median number of 17 cases, and two S. Typhimurium outbreaks, with a median number of 23 cases. Five of seven reported outbreaks of S. Enteritidis were the same phage type (14b) and the same phage type was also reported as one investigated national outbreak. In total, the national team reported investigating four S. Enteritidis clusters, with a median number of 30.5 cases, and no S. Typhimurium outbreaks over the study period.
Table 5.
Reported number of clusters and cases identified by traditional means and investigated by local and national teams in England, April 2014–March 2015
| Outbreak lead |
S. Enteritidis clusters (median; range of cases) |
S. Typhimurium clusters (median; range of cases) |
|---|---|---|
| Local team | 7 (17; 9–173) | 2 (23; 2–44) |
| National team | 4 (30.5; 23–60) | 0 (–) |
Discussion
This retrospective clustering analysis of S. Enteritidis and S. Typhimurium cases in England between April 2014 and March 2015 based on WGS surveillance data provides insight to inform the use of routine cluster detection as part of routine surveillance activities. Between a quarter and one-third of the cases of S. Enteritidis reported to PHE were assigned to clusters, as per the varying SNP, temporal and frequency criteria thresholds, while for S. Typhimurium fewer cases identified through surveillance were assigned to clusters (between 10% and 20% depending on SNP level).
The greatest burden of cluster notifications from the use of WGS data would be at the national level and all large, longer running clusters at the 10-SNP level were nationally distributed. There were relatively few clusters identified at the regional level using the 0- and 5-SNP thresholds for either serovar (<10% of total clusters) and none using 10-SNP thresholds indicating that these clusters were relatively small, brief and without substantial genetic diversity. While the number of 0-SNP cluster investigations (and associated cases) of S. Enteritidis or S. Typhimurium that would be led by any local team is low (no more than seven and three, respectively), this would represent an increase in workload compared with the number of reported outbreaks actually investigated by these teams during the study period. Local level clusters were all relatively small, brief and closely related genetically (identified using the 0 SNP threshold only). Local teams would also be involved in the investigation of cases within their boundary that are part of outbreaks led by the EES or the national team, thereby further adding to their workload.
Analyses using regression modelling indicated evidence of an association between early autumn and counts or identification of any clusters, which confirms what has been observed in the descriptive analysis and elsewhere [7, 8]. There was also some evidence of an association between increasing population size within local level boundaries and occurrence of 0-SNP clusters of S. Typhimurium. However, with only 1 year of data and a small number of observations, these analyses were limited. Further analyses with multiple years of data would allow for investigation of seasonality while consideration of other covariates such as population density and measure of rurality, and cluster size as an outcome might also be informative.
During the study period, there was a large outbreak of S. Enteritidis phage type 14b with 287 cases (many in Wessex HPT) linked to eggs from a common international supplier [9]. A corresponding cluster was identified in our clustering analyses at the 5- and 10-SNP national levels and represented the longest and largest of these clusters; these clusters were made of the same cases and contributed more than 50% of the nestedness of 5-SNP cases among those in 10-SNP clusters. Large outbreaks of S. Enteritidis and S. Typhimurium are relatively frequent [10–12] and the frequencies of both serovars over the study period were in the range of annual data reported in England between 2006 and 2015 [8]. We therefore consider these data to be representative in terms of clustering activity for these serovars and valid for informing the prospective application of this method. Furthermore, for either serovar, no more than 7% of isolates reported to PHE in the study period and meeting inclusion criteria could not be assigned an SNP address, which minimises the possible introduction of any ascertainment bias.
The application of WGS increases the sensitivity of cluster detection by identifying additional genetically related isolates that are geographically dispersed which might have previously been considered to be sporadic even if temporally linked. Furthermore, the improved specificity of cluster detection using WGS would reduce the amount of resources allocated to investigating groups of isolates that might have been considered a cluster using traditional approaches but are genetically distant and unlikely to be epidemiologically linked.
We have modelled the service implications for PHE teams using various genetic thresholds for cluster detection using WGS, which is particularly important in a resource-constrained environment. It has been suggested by previous work that the use of 5-SNP thresholds as the default to detect clusters might be appropriate given that such isolates would likely originate from a common source [2] but there were no 5-SNP clusters restricted to the local level over the study period. In practice, it is likely that all three SNP threshold levels would need to be reviewed on a regular basis at each geographical level in order to identify a range of cluster types, from small clusters made up of closely related cases – useful when investigating geographically contained, transient point or locally distributed source outbreaks – to large, geographically dispersed clusters with more genetic variation among cases, which may indicate a longstanding source of infection in which genetic variation exists as a result of a large infected source population.
The temporal window of the cluster definition was intended to better limit the clusters to cases that were more likely to be involved with a common source or vehicle of infection, which is most pertinent for the least genetically related clusters, and this has been shown to be a robust, systematic approach for identifying long-term clusters. Removing this temporal parameter resulted in increases in the number of clusters for most SNP thresholds and geographical level strata as additional clusters of temporally spaced cases, which previously were not included in the temporal window (>7 days between cases), were identified. These clusters were typically similarly sized but additional cases for some resulted in substantially longer cluster duration, up to a full year, indicating that some clusters might have run beyond the duration of the study period. There were instances of a reduction in the number of clusters identified as recurring events would only be counted once.
Since the implementation of industry-led controls, including vaccination of egg layers in the UK against S. Enteritidis, clinical infection in humans with this serovar has fallen [13, 14] and is now mainly attributed to foreign-sourced chicken and or eggs which are typically nationally distributed produce. Given the likely national distribution of such goods, the identification of predominantly national clusters of S. Enteritidis might be expected. S. Typhimurium outbreaks in England are attributed to a diverse range of sources or vehicles of infection [10, 11]. S. Enteritidis and S. Typhimurium are the two most commonly reported serovars of Salmonella in humans in England and in total accounted for 43% of all Salmonella isolates received by GBRU over the study period. We consider these serovars to be good exemplars for understanding the distribution of Salmonella clusters within PHE operational boundaries and the comparative resource implications for different teams as a result of routine WGS surveillance but findings of these analyses likely under-represents the burden on PHE teams investigating all Salmonella serovars. It is also important to consider that WGS is used for the routine surveillance of other gastrointestinal pathogens in England and there are increasing demands on teams to investigate clusters of other organisms.
This cluster analysis is based on a typical year of reported isolate data and is therefore applicable to determining workload. However, it is limited in that it does not address the amount of resources needed to investigate clusters – contacting cases, completing surveillance questionnaires and reviewing reported exposures for the potential source or vehicle of infection – and not all clusters identified here would require a full investigation. Those clusters identified as having the same WGS profiles as others in the study period would likely be considered as part of the same outbreak, especially those derived using more restricted genetic relatedness criteria. In addition, clusters composed predominately of suspected household contacts, as determined by a common postcode, and those made up of cases of whom at least 75% had a reported recent travel history might not be investigated or require less resource than those without either of these characteristics. Considering the person-time required to investigate clusters in which there is weighting to reflect the heterogeneity in investigations given cluster size and intra-cluster case composition (e.g. travel-related cases and household contacts) might further inform how best to use WGS data to trigger cluster investigation.
England is one of the first countries to implement WGS as part of the routine surveillance of gastrointestinal infections; it is a powerful tool for cluster detection with an increased sensitivity over traditional methods, identifying additional clusters of previously unlinked, genetically related isolates at every geographical level. It has particularly added value as a robust means of identifying long-running, slowly developing clusters. While local surveillance and response are clearly important for point source clusters, this analysis indicates that the greatest burden of work generated through routine surveillance using WGS data for S. Enteritidis and S. Typhimurium will be on national level staff investigating small, geographically dispersed but temporally related clusters, regardless of the genetic threshold employed. Local public health teams might need support in prioritising response based on these data to balance limited available human resources with the public health importance of investigating small, geographically contained clusters of highly related cases. Guidance for prioritising cluster investigations, taking into account size, growth rate, travel history, demographic characteristics and geographical spread and potential public health benefit from the investigation is needed to best allocate resources for investigation. While there are operational challenges to its implementation, cluster detection based on WGS provides an integrated approach from local to national level in England – and potentially internationally – and should facilitate further improvements in the timeliness and capacity to respond and control clusters of public health significance. Other countries may find the experiences within PHE useful when planning to implement similar surveillance developments.
Acknowledgements
We thank the PHE health protection team staff responsible for investigating and reporting outbreaks of Salmonella to the Electronic Foodborne and Non-Foodborne Gastrointestinal Outbreak Surveillance System (eFOSS).
Financial support
The research was funded by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at the University of Liverpool in partnership with Public Health England (PHE), in collaboration with University of East Anglia, University of Oxford and the Institute of Food Research. Timothy J Dallman is based at the PHE. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or PHE.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818001589.
click here to view supplementary material
References
- 1.Ashton P et al. (2015) Revolutionising public health reference microbiology using whole genome sequencing: Salmonella as an exemplar. bioRxiv 033225. [Google Scholar]
- 2.Waldram A et al. Epidemiological analysis of Salmonella clusters identified by whole genome sequencing, England and Wales 2014. Food Microbiology 71, 39–45. [DOI] [PubMed] [Google Scholar]
- 3.Annual Mid-year Population Estimates – Office for National Statistics. Available at https://www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/bulletins/annualmidyearpopulationestimates/2015-06-25 (Accessed 1 May 2017).
- 4.Achtman M et al. (2012) Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathogens 8, e1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.OpenEpi Menu. Available at http://www.openepi.com/Menu/OE_Menu.htm (Accessed 1 May 2017).
- 6.Foodborne and non-foodborne gastrointestinal outbreaks surveillance – GOV.UK. Available at https://www.gov.uk/guidance/foodborne-and-non-foodborne-gastrointestinal-outbreaks-surveillance (Accessed 30 June 2017).
- 7.European Centre for Disease Prevention and Control (2016) Salmonellosis – Annual Epidemiological Report 2016 [2014 data]. Available at http://ecdc.europa.eu/en/publications-data/salmonellosis-annual-epidemiological-report-2016-2014-data (Accessed 12 December 2017).
- 8.Public Health England (2016) Salmonella data 2006 to 2015. Available at https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/598401/Salmonella_2016_Data.pdf (Accessed 28 June 2017).
- 9.Inns T et al. (2015) A multi-country Salmonella Enteritidis phage type 14b outbreak associated with eggs from a German producer: ‘near real-time’ application of whole genome sequencing and food chain investigations, United Kingdom, May to September 2014. Euro Surveillance: Bulletin Europeen Sur Les Maladies Transmissibles = European Communicable Disease Bulletin 20, pii: 21098. [DOI] [PubMed] [Google Scholar]
- 10.Harker KS et al. (2014) National outbreaks of Salmonella infection in the UK, 2000–2011. Epidemiology and Infection 142, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harker KS et al. (2011) An outbreak of Salmonella Typhimurium DT191a associated with reptile feeder mice. Epidemiology and Infection 139, 1254–1261. [DOI] [PubMed] [Google Scholar]
- 12.Inns T et al. (2017) Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiology and Infection 145, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Brien SJ (2013) The ‘decline and fall’ of nontyphoidal Salmonella in the United Kingdom. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 56, 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogan TA and Humphrey TJ (2003) The rise and fall of Salmonella Enteritidis in the UK. Journal of Applied Microbiology 94(suppl), 114S–119S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818001589.
click here to view supplementary material