Abstract
The properties of the pediocin AcH precursor, prepediocin AcH, have been studied to gain insight into how producer cells may protect themselves from the activity of intracellular prebacteriocins. The native 62-amino-acid precursor and the 44-amino-acid mature species were expressed in Escherichia coli host strains that lack the leader peptide processing enzyme, PapD. Both forms inhibited the growth of the test bacterium Listeria innocua Lin11, indicating that the native precursor is biologically active. The two species also were synthesized in the context of maltose-binding protein chimeric proteins to facilitate the measurement of their relative specific activities. The chimeric form of the precursor was ∼80% as active as the chimeric mature species. Of relevance to cell protection and pediocin AcH production, it was determined that the precursor is strongly susceptible to inactivation by reducing agents and to degradation by chymotrypsin and endogenous E. coli proteases. Taken together, the results indicate that the activity of prepediocin AcH may have to be controlled prior to secretion to prevent toxicity to the host. Perhaps producer cells avoid membrane damage by maintaining the precursor in a reduced inactive state or by degrading molecules whose secretion is delayed.
The bacteriocins produced by gram-positive bacteria are gene-encoded antimicrobial peptides that kill target cells by forming pores in the cytoplasmic membrane (1, 3, 4, 14). Nearly all bacteriocins contain N-terminal leader peptides that direct precursors to dedicated, ABC-export system proteins that translocate the active C-terminal prodomain across the cytoplasmic membrane and remove the leader peptide (2, 6, 29, 31). Currently, it is unclear why specialized export systems are used for secretion, and it has been shown that pediocin AcH (which is the same as pediocin PA-1 [17]) can be secreted via the Escherichia coli sec machinery when targeted to it via linkage to a standard secretory protein (18, 19). The only bacteriocin that normally is secreted by the general sec machinery of the cell is divergicin A (32).
Leader peptides are structurally and functionally distinct from signal sequences of sec-dependent secretory proteins (14). They contain 23 to 30 amino acids for nisin-like prebacteriocins and 18 to 24 amino acids for pediocin-like prebacteriocins. Although leader peptide sequences differ considerably between these two families, the members of each group show a great degree of sequence identity, suggesting that conserved residues have functional significance (6, 14). For example, amino acids in the −2 and −1 positions relative to the processing site are highly conserved, probably because they comprise the recognition site for processing enzymes (6, 12, 14, 28). Leader peptides generally are relatively hydrophilic and may assume an amphipathic α-helical structure in lipophilic environments (6, 9, 14, 26, 27). Currently, the features of the leader peptide that are recognized by ABC-export system proteins are unknown. At least some of these features may be in common because a given prebacteriocin sometimes can be secreted by an unrelated ABC-export system (28). After their removal, leader peptides may be secreted or degraded, as occurs for signal sequences of sec-dependent secretory proteins (21).
Leader peptides may perform other functions besides directing the interaction of prebacteriocins with ABC-export systems. These could include guiding and maintaining conformation during posttranslational modification of lantibiotics such as nisin, stabilizing prebacteriocin molecules to degradation prior to translocation, and/or keeping molecules inactive against the cytoplasmic membranes of producer cells prior to secretion (6, 14, 15, 30). With regard to the latter point, it has been shown that the fully modified nisin precursor (29) and a pediocin-like prebacteriocin, preleucocin A (10), are inactive, and that another pediocin-like prebacteriocin, precarnobacteriocin B2, is 125-fold less active than its mature form (24). In contrast, the prepediocin PA-1 precursor appears to display activity (31), but its activity relative to that of mature pediocin PA-1 has not been measured. In cases where precursors are inactive, leader peptides may alter the conformations of propeptide domains or interfere with their interaction with the cytoplasmic membrane of target as well as producer cells (6, 10, 12, 24).
In this study, we have examined possible auxiliary roles played by the pediocin AcH leader peptide during secretion. It has been determined that the precursor is nearly as active as the mature species and is highly susceptible to protease degradation and disulfide bond reduction. The implications of the results for cell protection and pediocin AcH production are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli host strain BL21(DE3) was used for expression of the mature and precursor forms of pediocin AcH (Table 1). This strain contains the T7 bacteriophage RNA polymerase gene cloned on the chromosome under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lac promoter. The papA and prepapA genes, which encode the respective mature and precursor species, were cloned into a T7 RNA polymerase promoter plasmid, pET3c, and the resulting plasmids were introduced into BL21(DE3) for expression. Transformed BL21(DE3) strains were grown at 37°C in Luria-Bertani (LB) broth containing 100 μg of ampicillin per ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Enterococcus faecalis M1 | Pediocin AcH sensitive | This study |
| Escherichia coli BL21(DE3) | T7 RNA polymerase host | 27a |
| Escherichia coli E609L | E609 lpp::Tn10, Tcr | Henry C. Wu |
| Lactococcus plantarum NCDO955 | Pediocin AcH sensitive | This study |
| Leuconostoc mesenteroides Ly | Pediocin AcH sensitive | This study |
| Listeria innocua Lin11 | Nonpathogenic, pediocin AcH sensitive | Jean Richard |
| Pediococcus acidilactici LB42 | Pediocin AcH sensitive | This study |
| Pediococcus acidilactici LB42-923 | Pediocin AcH producer | 33 |
| Plasmids | ||
| pMBR1.0 | papABCD operon in pHPS9, Cmr Emr | 2 |
| pET3c | T7 RNA polymerase promoter plasmid, Apr | 27a |
| pT71 | pET3c derivative with papA gene, Apr | This study |
| pT72 | pET3c derivative with prepapA gene, Apr | This study |
| pPR682 | malE plasmid, Apr | New England Biolabs |
| pPR6821 | malE-papA plasmid, Apr | 18 |
| pPR6823 | malE-prepapA plasmid, Apr | This study |
The E. coli strain E609L, which releases about half of its periplasmic proteins into the culture medium (19), served as host for expression of maltose-binding protein (MBP) chimeric proteins. Plasmid pPR6821 contains a malE-papA fusion gene that directs secretion of an MBP-pediocin AcH chimeric protein (18), and pPR6823 contains a malE-prepapA fusion gene that directs secretion of an MBP-prepediocin AcH chimeric protein. The two plasmids were constructed from the malE expression plasmid pPR682 and plasmid pMBR1.0 which contains a DNA insert encoding the papABCD operon (2, 20). The transcription of fusion genes is controlled by the IPTG-inducible tac promoter. Transformed E609L strains were grown at 37°C in LB broth containing 100 μg of ampicillin and 12.5 μg of tetracycline per ml unless otherwise indicated.
The pediocin AcH-sensitive strains listed in Table 1 were used as indicator bacteria in activity testing. All of these strains were grown at 30°C in tryptone-glucose-yeast extract (TGE) medium. Pediococcus acidilactici LB42-923 was used for partial purification of native pediocin AcH (33).
Construction of expression plasmids.
The method used for the construction of plasmid pPR6823 was similar to that used for the construction of pPR6821 (18). The prepapA coding region of plasmid pMBR1.0 was amplified by PCR by using a 5′ PCR primer (5′-AAAAAAATTGAAAAATTAACTGAAAAAGAAATG-3′) that begins with the Lys−17 codon of the prepapA DNA sequence and a 3′ PCR primer (5′-GGGTCGACCTAGCATTTATGATTACCT-3′) that contains a SalI restriction endonuclease site located downstream of the prepapA termination codon. The conditions used for DNA synthesis and the methods used for DNA fragment manipulation and cloning have been described in detail previously (18). After synthesis of the DNA fragment, it was treated with SalI restriction endonuclease and ligated between the StuI (blunt end) and SalI restriction endonuclease sites of pPR682 to construct pPR6823. As a result of the cloning steps, the Lys−17 codon of the leader peptide coding sequence was fused in frame to a 14-amino-acid linker peptide-coding sequence located between the malE and prepapA genes. The sequence of the prepapA region was confirmed by double-stranded DNA sequencing by using Sequenase DNA polymerase (U.S. Biochemicals).
T7 RNA polymerase expression plasmids were constructed for synthesis of the precursor and mature forms of pediocin AcH. PCR-amplified DNA fragments encoding the prepapA and papA sequences were prepared by using pMBR1.0 as a template and were ligated into the NdeI-to-BamHI restriction endonuclease sites of the pET3c T7 RNA polymerase promoter plasmid (27a). The 5′ PCR primer (5′-GGCATATGAAATACTACGGTAATGGG-3′) that was used for construction of the pediocin AcH expression plasmid (designated pT71) begins with a methionine preceding Lys+1 of the mature sequence region and contains an NdeI restriction endonuclease site for ligation to the vector. The 5′ PCR primer (5′-GGAGATTGGGCATATGAAAAAAATTG-3′) that was used for construction of the prepediocin AcH expression plasmid (designated pT72) begins with Met−18 of the leader peptide region and also contains an NdeI restriction endonuclease site for ligation to the vector. A 3′ PCR primer (5′-CTCGGATCCCTAGCATTTATGATTAC-3′) that contains a BamHI restriction endonuclease site was used for amplification of both DNA fragments. After their ligation into pET3c, both PCR inserts were sequenced in their entirety by double-stranded DNA sequencing.
Measurement of chimeric protein activity levels.
Strains E609L/pPR6821 and E609L/pPR6823 were grown overnight at 37°C in LB broth containing ampicillin and tetracycline. On the next day, cells were pelleted by centrifugation, washed with LB broth, and diluted in LB broth lacking antibiotics. Cultures were grown until the optical density at 600 nm (OD600) reached ∼0.5, and then chimeric protein synthesis was induced by adding IPTG to 1 mM final concentration. Samples of the culture broths (cells plus supernatant), or the cell pellet and supernatant fractions isolated by 5-min centrifugation and filtration, were collected 3 h after IPTG induction. One portion of the samples was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis as described below to determine the relative synthesis levels of chimeric proteins. Another portion was boiled 10 min to inactivate the proteases, and aliquots were spotted onto TGE agar plates spread with Listeria innocua Lin11 to calculate the activity units (AU) per milliliter for each broth. Plates were incubated overnight at 30°C and examined to determine the smallest aliquot that produced a zone of growth inhibition in the lawn. The number of AU per milliliter of each broth was calculated based on the size of the smallest aliquot that formed a zone of inhibition (33). These values for the two broths were corrected for variations in the OD600 readings of the samples and the levels of chimeric proteins present. The MBP-prepediocin AcH chimeric protein also was tested for activity against the other pediocin AcH-sensitive strains listed in Table 1. In this case, activity titrations were not performed.
Effects of reducing agents on chimeric protein activity.
Samples of E609L/pPR6821 and E609L/pPR6823 induced by a 3-h treatment with IPTG were prepared from cultures grown at 37°C in the absence of antibiotics. Culture broths were split into three portions. One portion served as a control, dithiothreitol (40 mM) was added to the second portion, and 2-mercaptoethanol (50 mM) was added to the third. The samples then were boiled for 10 min to reduce the pediocin AcH domains of the chimeric proteins and to inactivate proteases present in the broths. Control samples boiled with and without reducing agents were made by using LB medium alone and LB medium containing dissolved pediocin AcH from P. acidilactici LB42-923.
To determine the effects of reducing agents on activity, 1-ml aliquots of the samples were incubated for 1 h at 25°C with ∼550 L. innocua Lin11 cells. After incubation, the mixtures were centrifuged, the supernatant fractions were removed, and the cells were resuspended and plated on TGE agar to determine the number of viable cells present. In addition, the supernatant fractions were boiled and tested for activity by being spotted onto a lawn of L. innocua Lin11. The numbers of AU per milliliter of the supernatant fractions were calculated and corrected for variations in culture ODs and chimeric protein synthesis levels, as described above.
Chymotrypsin digestion of chimeric proteins.
Boiled culture broths from strains E609L/pPR6821 and E609L/pPR6823 induced by a 3-h treatment with IPTG and grown at 37°C in the absence of antibiotics were incubated for up to 30 min at 25°C with 10 μg of chymotrypsin per ml. Aliquots of the reactions were withdrawn at specific intervals over the 30-min incubation period and were boiled to inactivate the enzyme. The digested samples and samples of the unincubated culture broths were spotted onto a lawn of L. innocua Lin11 to determine their AU values. These values were corrected based on the original OD600 readings of the broths and the levels of chimeric proteins present. The samples also were examined by SDS-PAGE and Western immunoblotting to assess the degradation of the chimeric proteins.
SDS-PAGE analysis of protein synthesis levels and activities.
The synthesis levels of MBP chimeric proteins were determined by SDS-PAGE with 10% acrylamide-bisacrylamide–SDS gels (16). Gels were stained with Coomassie brilliant blue G-250 dye and scanned with a Bio-Rad Gel Dock laser densitometer to determine the relative levels of chimeric proteins present in samples. The same gel system was used for Western immunoblotting. In this case, chimeric proteins were electroblotted onto nitrocellulose membranes and were detected by staining with rabbit anti-MBP primary antiserum and goat anti-rabbit immunoglobulin G-alkaline phosphatase secondary-antibody complex.
The activities of MBP chimeric proteins and of the precursor and mature forms of pediocin AcH were analyzed by SDS-PAGE–gel overlay screening (18). Samples were obtained from chimeric protein expression strains grown at 37°C in LB-ampicillin-tetracycline medium and from T7 RNA polymerase expression strains grown at 37°C in LB-ampicillin medium. Proteins were separated on 20% acrylamide-bisacrylamide–SDS gels (25), and subsequently the gels were soaked in sterile water to remove SDS, placed on TGE agar plates, and overlaid with TGE top agar containing L. innocua Lin11 cells. Plates were incubated overnight at 25°C and examined for zones of growth inhibition caused by active proteins.
RESULTS
Properties of native and chimeric proteins.
The sequences of the native mature and precursor forms of pediocin AcH are shown in Fig. 1A. The two species were produced in the respective E. coli T7 RNA polymerase expression strains BL21(DE3)/pT71 and BL21(DE3)/pT72. The BL21(DE3) strain lacks the leader peptide processing enzyme, PapD, and therefore can be used to obtain the unprocessed precursor. Both the precursor and mature forms of the bacteriocin accumulate in the cytoplasm of the strains.
FIG. 1.
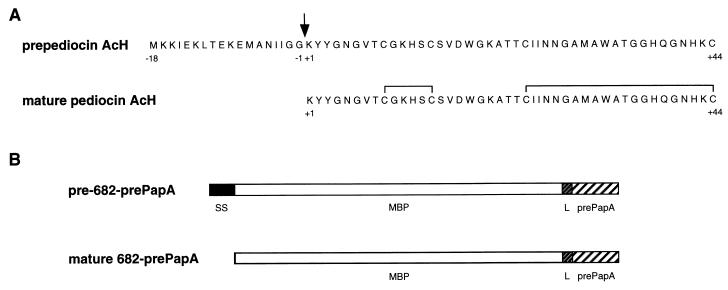
(A) Amino acid sequences of native prepediocin AcH and mature pediocin AcH that is formed after leader peptide processing (arrow) and disulfide bond oxidation. (B) Structure features of the MBP-prepediocin AcH chimeric protein. The pre(682-prePapA) translation product contains a wild-type MBP domain with a functional signal sequence (SS), a 14-amino-acid linker peptide (-Asn-Ser-Ser-Ser-Val-Pro-Gly-Arg-Gly-Ser-Ile-Asp-Gly-Arg-) (L), and the 61-amino-acid prepediocin AcH domain (prePapA) that begins with Lys−17. The mature 682-prePapA protein is generated from pre(682-prePapA) by processing of the MBP signal sequence.
The properties of the MBP-prepediocin AcH chimeric protein [pre(682-prePapA)] synthesized by strain E609L/pPR6823 are summarized in Fig. 1B. The pre(682-prePapA) protein contains a wild-type MBP domain fused to Lys−17 of the prepediocin AcH leader peptide. The design enables, after MBP targeting to the cellular sec machinery and signal sequence processing, the secretion of the mature 682-prePapA protein into the periplasm of the E. coli host (18). Due to disruption of the outer membrane protein structural gene lpp in this strain, about half of the secreted chimeric protein is released into the culture medium. The 682-PapA protein produced by strain E609L/pPR6821 is similar to 682-prePapA except that it lacks the pediocin AcH leader peptide sequence (18).
The native and chimeric forms of prepediocin AcH are biologically active.
The activities of both native and chimeric prepediocin AcH molecules were examined. It was necessary to study chimeric molecules because of uncertainties in quantitating the levels of the native species produced by the T7 RNA polymerase expression strains and because native species could be obtained only by growing these strains in the presence of ampicillin (data not shown). The latter issue made it impossible to directly test culture broths from these strains against the pediocin AcH (and ampicillin)-sensitive bacteria listed in Table 1. On the other hand, significant levels of chimeric proteins were obtained from E609L strains grown in the absence of ampicillin, and therefore, culture broths from chimeric protein expression strains could be used directly in activity testing.
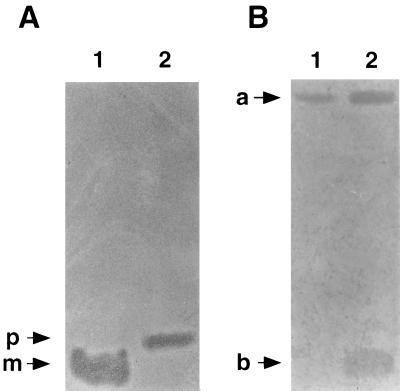
To verify that native prepediocin AcH is biologically active, SDS-PAGE–gel overlay screening analysis was performed. The presence of ampicillin in samples did not interfere with activity determination performed by this technique. BL21(DE3)/pT72 cells were grown in LB-ampicillin medium, and synthesis of prepediocin AcH was induced for 30 min by the addition of 1 mM IPTG to the culture. BL21(DE3)/pT71 cells were grown under identical conditions to obtain pediocin AcH as a positive control for activity. As shown in Fig. 2A, both samples formed zones of growth inhibition against L. innocua Lin11, indicating that prepediocin AcH is active. The zone of growth inhibition produced by the precursor appeared at a slightly higher molecular weight than the zone of growth inhibition produced by the mature form of the molecule.
FIG. 2.
SDS-PAGE–gel overlay screening analysis of native mature and precursor pediocin AcH (A) and MBP-prepediocin AcH (B). In panel A, equivalent amounts of cell pellet fractions were loaded in lane 1 [BL21(DE3)/pT71] and lane 2 [BL21(DE3)/pT72]. The migration positions of the precursor (p) and mature (m) species are marked with arrows. In panel B, equivalent amounts of cell pellet (lane 1) and culture supernatant (lane 2) fractions from E609L/pPR6823 were loaded onto the gel. Arrows a and b mark the respective positions of the high-molecular-weight MBP-prepediocin AcH chimeric protein and its low-molecular-weight degradation product. The activity was tested against L. innocua Lin11.
The MBP-prepediocin AcH chimeric protein also was found to be active by SDS-PAGE–gel overlay screening. E609L/pPR6823 was grown in LB-ampicillin-tetracycline medium, and synthesis of the 682-prePapA protein was induced for 3 h by the addition of 1 mM IPTG to the culture. As shown in Fig. 2B, zones of growth inhibition corresponding the high-molecular-weight 682-prePapA protein appeared in both the cell pellet (lane 1) and the culture supernatant (lane 2) fractions obtained from this strain. Activity due to proteolysis fragments with sizes similar to native pediocin AcH also was observed in these samples in the low-molecular-weight region of the gel. These fragments probably do not contain much or any of the MBP domain since they were not detectable by Western immunoblotting with anti-MBP antiserum. Taken together, the results indicate that the MBP-prepediocin AcH molecule is active and that it accounts for much of the activity associated with this expression strain.
Specific activities of chimeric proteins.
The specific activities of the 682-PapA and 682-prePapA chimeric proteins were calculated to permit estimation of the relative activity of the precursor. Cultures of E609L/pPR6821 and E609L/pPR6823 were grown in LB medium in the absence of antibiotics, chimeric protein synthesis was induced for 3 h with IPTG, and aliquots of the culture broths were boiled and spotted onto lawns of L. innocua Lin11. The numbers of AU per milliliter of the two broths were calculated and corrected for variations in culture ODs and chimeric protein synthesis levels determined by SDS-PAGE. As shown in Table 2, the corrected AU value of the E609L/pPR6823 culture broth was 78% of that of the E609L/pPR6821 broth. Although breakdown products contributed to the calculated AU values of the broths, the fractions of the total activities in the strains attributable to the full-length chimeric proteins were about the same (data not shown). Thus, the corrected activity levels calculated for the broths are reflective of the specific activities of the two chimeric proteins.
TABLE 2.
Relative bactericidal activities of the 682-PapA and 682-prePapA proteins
| Protein | OD600a | AU/ml of culture brothb | Protein levelc | Corrected AU/mld |
|---|---|---|---|---|
| 682-PapA | 2.1 | 4,000 | 1 | 4,000 |
| 682-prePapA | 2.0 | 2,000 | 0.67 | 3,134 (78)e |
Culture broth ODs were measured after a 3-h IPTG induction period.
Calculated by determining the minimum volume of culture broth required to create a zone of growth inhibition on a lawn of L. innocua Lin11 (see Materials and Methods).
Relative protein synthesis levels determined by laser scanning densitometry of Coomassie brilliant blue-stained gel samples.
Corrected for differences in OD600 values and relative levels of chimeric proteins in culture broths.
The percentage of 682-PapA activity is given in parentheses.
Activity spectra of chimeric proteins.
Culture broths from E609L/pPR6821 and E609L/pPR6823 induced for 3 h with IPTG and grown in the absence of ampicillin were tested against several pediocin AcH-sensitive strains. As has been reported for the 682-PapA protein (18, 19), the 682-prePapA protein was active against all five pediocin AcH-sensitive strains listed in Table 1 (data not shown). This suggests that the two molecules may have the same primary mode of action. In addition, both proteins were inactive against eight pediocin AcH producer strains that have been isolated in our laboratories (data not shown). All of the latter strains probably synthesize the PapB immunity protein (2), and therefore the results suggest that the activity of the precursor is blocked by the standard immunity mechanism.
Susceptibility of chimeric proteins to inactivation by reducing agents.
The pediocin AcH precursor normally is found only in the cytoplasm of producer cells (14). Experiments were performed to determine whether the activity of the precursor could be decreased by exposing it to reducing conditions analogous to those that exist in the cytoplasm. A culture broth of strain E609L/pPR6823 induced for 3 h with IPTG was prepared and treated with or without dithiothreitol or 2-mercaptoethanol to reduce the 682-prePapA protein. An E609L/pPR6821 culture broth grown under identical conditions also was prepared, as was a sample of native pediocin AcH from P. acidilactici LB42-923. The effects of the reducing agents on activity were tested in two ways. Aliquots of treated samples were incubated with L. innocua Lin11 cells, the cells were pelleted by centrifugation, and the number of cells surviving the incubation was determined by plating. In addition, the supernatant fractions from the centrifuged incubation mixtures were analyzed by spot test assays to determine their AU values. As shown in Table 3, the activities of all three proteins were substantially lowered by treatment with the reducing agents. The number of cells that survived incubation increased for samples exposed to reducing agents, and the numbers of AU per milliliter of treated supernatants were lower than for untreated samples. Control experiments performed with LB broth containing the reducing agents showed that these compounds have no effects per se on the viability of the indicator bacterium, at least at the concentrations used in the assays. The results indicate that the pediocin AcH precursor, like the mature bacteriocin (14), is sensitive to reducing agents.
TABLE 3.
Effects of reducing agents on the bactericidal activities of pediocin AcH, 682-PapA, and 682-prePapA
| Samplea | No. (%) of surviving cellsb | Corrected AU/mlc (%) |
|---|---|---|
| LBd | 550 (100) | 0 |
| LB + DTT | 510 (93) | 0 |
| LB + 2-ME | 580 (105) | 0 |
| PapAe | <10 (<2) | 2,500 (100) |
| PapA + DTT | 500 (91) | 1,000 (40) |
| PapA + 2-ME | 450 (82) | 1,000 (40) |
| 682-PapAf | <10 (<2) | 2,500 (100) |
| 682-PapA + DTT | 400 (73) | 1,000 (40) |
| 682-PapA + 2-ME | 500 (91) | 1,000 (40) |
| 682-prePapAg | 140 (25) | 2,000 (100) |
| 682-prePapA + DTT | 430 (78) | 500 (25) |
| 682-prePapA + 2-ME | 380 (69) | 500 (25) |
The indicated samples contained 40 mM dithiothreitol (DTT) or 50 mM 2-mercaptoethanol (2-ME).
The number of surviving L. innocua Lin11 cells remaining after incubation with the indicated samples, as determined by a cell viability assay, is given. The percentage of L. innocua Lin11 cells that survived incubation is given in parentheses. The percentage is calculated based on 550 cells per incubated sample.
Corrected values were determined only for supernatant fractions of the 682-PapA and 682-prePapA samples after their incubation with L. innocua Lin11, as explained in Materials and Methods. The percentage of activity observed in the absence of reducing agents is shown in parentheses.
Boiled LB medium.
Boiled LB medium containing partially purified pediocin AcH (PapA) from strain P. acidilactici LB42-923.
Boiled culture broth of strain E609L/pPR6821.
Boiled culture broth of strain E609L/pPR6823.
Susceptibility of chimeric proteins to protease inactivation.
Experiments were performed to compare the susceptibilities of the chimeric proteins to proteolytic attack. To assess protease sensitivity, culture broths of E609L/pPR6821 and E609L/pPR6823 induced for 3 h with IPTG were incubated with chymotrypsin, and the numbers of AU remaining in the samples after incubation were determined (Table 4). Although the AU values of both culture broths declined during the incubations, the number of AU per milliliter for E609L/pPR6823 declined more rapidly and was nearly eliminated after 15 min of incubation. In contrast, significant activity remained in the E609L/pPR6821 broth even after 30 min of incubation. It was confirmed by SDS-PAGE and Western immunoblotting that both chimeric proteins were degraded by chymotrypsin and that the 682-prePapA protein was degraded more rapidly than 682-PapA (data not shown). It also has been noted that the 682-prePapA protein is more susceptible to degradation in unboiled culture broths than is the 682-PapA protein (data not shown). Taken together, the data suggest that prepediocin AcH is more susceptible to both in vitro and in vivo proteolysis than is mature pediocin AcH.
TABLE 4.
Relative sensitivities of the 682-PapA and 682-prePapA proteins to chymotrypsin digestion
| Samplea | Corrected AU/ml at time (min)b:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | 30 | |
| 682-PapA | 4,000 | 2,000 | 1,000 | 1,000 | 500 | 500 |
| 682-prePapA | 3,100 | <150c | <150 | <30d | <30 | <30 |
Boiled culture broths of strain E609L/pPR6821 (682-PapA protein) and E609L/pPR6823 (682-prePapA protein) after a 3-h induction with IPTG.
The AU values against L. innocua Lin11 were corrected for differences in the OD600 readings and chimeric protein synthesis levels in the unincubated culture broths. The time refers to minutes of incubation at 25°C with 10 μg of chymotrypsin per ml.
No activity was detected in 10 μl of boiled culture broth.
No activity was detected in 50 μl of boiled culture broth.
DISCUSSION
Limited available information indicates that many of the prebacteriocins produced by gram-positive bacteria are virtually inactive. Such observations suggest that in many cases producer cells are protected from the action of precursors mainly because these molecules display little or no activity. It seems possible that leader peptides could neutralize the activities of prebacteriocins in several different ways (24, 26, 29). For example, leader peptides, which generally are polar, may increase the solubility of prebacteriocins in water (26) and therefore may cause them to partition more into the aqueous phase than into the membrane. In addition, leader peptides could interact directly with mature domains to reduce their affinity for membranes (24). In this regard, both leader peptides and mature regions are moderately amphipathic, and their nonpolar regions may interact in water and shield nonpolar membrane interaction sequences from contact with lipids.
In marked contrast, the pediocin AcH precursor appears to be ∼80% as active as the mature form. This suggests that the leader peptide has little effect on the function of the mature domain. It has been demonstrated here and previously (18, 19) that fusion of the large MBP molecule to the N terminus of the mature domain also does not block activity. Apparently, the tertiary structure and membrane-binding properties of the mature domain are relatively insensitive to what is attached to the N terminus. Until the properties of more prebacteriocins have been investigated, it is impossible to conclude whether the activity displayed by prepediocin AcH is exceptional or not.
Based on these findings, it becomes necessary to consider alternative ways for how producer cells overcome the potentially lethal action of prepediocin AcH in the time between its translation and its secretion. One possible way in which binding of the precursor to the cytoplasmic membrane may be limited is that the binding site on the putative pediocin AcH receptor (4) may not be accessible to prepediocin AcH from inside the cell. However, pediocin PA-1 has been shown to display membrane permeabilization activity against phospholipid vesicles in the absence of a receptor (3), and therefore some damage may be caused even though a receptor is inaccessible. In addition, the membrane insertion activity of the precursor may be limited due to the reverse orientation of the membrane electrochemical potential that it should encounter from inside the cell. In this regard, the membrane permeabilization activity of pediocin PA-1 is stimulated by a membrane potential difference that is trans-side negative (3, 4).
It also is possible that prepediocin AcH may be neutralized by the cytoplasmic PapB immunity protein (2, 31), for which the mechanism of action currently is unknown. In most cases, immunity proteins protect cells from the bacteriocins they have secreted by preventing their interaction with the cytoplasmic membrane (1, 6). For example, the specific immunity proteins for nisin, subtilin, and epidermin are secreted and anchored to the outer surface of the cytoplasmic membrane and are believed to bind these lantibiotics before they can interact with membrane lipids. Similarly, the lactococcin A immunity protein is membrane bound, but in this case it may bind to and shield the receptor for this bacteriocin (1). In contrast, the carnobacteriocin B2 immunity protein is located predominantly in the cytoplasm and may instead plug pores after they have been formed in the membrane (23). If PapB can inhibit the activity of prepediocin AcH, then it may need to do so by binding to it in solution. It seems unlikely that PapB could plug pores formed by prepediocin AcH because the molecules within these pore complexes might have the reverse orientation within the bilayer compared to molecules that have inserted from outside.
Another possible way by which producer cells could control the activity of intracellular prepediocin AcH molecules is by using the cytoplasmic redox potential to maintain cysteine thiol groups in a reduced state (7). It has been demonstrated that the four cysteines within the native pediocin AcH molecule must be oxidized to two disulfide bonds for the bacteriocin to be active (4, 14). The substitution of serines and other residues for cysteines completely inactivates native pediocin AcH and MBP-pediocin AcH (19). We have shown here that cysteines within MBP-prepediocin AcH also must be oxidized to achieve maximal activity. These findings suggest that the activity of prepediocin AcH, and perhaps the activities of the precursors of other “cystibiotics” that require disulfide bonds (8, 14), could be controlled within producer cells by the reducing state of the cytoplasm. However, there are indications that natural, but not synthetic, leucocin A and also carnobacteriocin B2 exhibit activity when their two cysteines are reduced (8, 11, 22).
The finding that prepediocin AcH is active raises the question of why its leader peptide is removed during secretion. One possible reason is that the leader peptide makes the molecule more susceptible to proteases in the environment. In this regard, MBP-prepediocin AcH was found to be more susceptible than MBP-pediocin AcH to chymotrypsin and proteases in the culture broths of the producer strains. This suggests that prepediocin AcH would be highly susceptible to proteases produced by target cells and would be a less-effective antimicrobial agent than pediocin AcH. Furthermore, it may be important for producer cells to export the precursor before it can be cleaved by intracellular proteases. Rapid export also could limit damage caused to producer cells by cytoplasmic prepediocin AcH molecules.
In conclusion, the results show that the pediocin AcH precursor has significant biological activity. In cases where a precursor is active, producer cells must employ alternative schemes to avoid cytoplasmic toxicity. It is suggested that these mechanisms may include rapid and efficient translocation of prebacteriocins out of cells and, in the case of most cystibiotics, maintenance of cysteines in the reduced state. In addition, precursors that are not immediately secreted may be destroyed by intracellular proteases. Poor translocation efficiency and proteolysis could be contributing factors in cases where hybrid bacteriocins are produced in relatively small amounts when expressed using swapped leader peptides (5, 13, 29).
ACKNOWLEDGMENTS
We thank the investigators for providing bacterial strains.
We acknowledge the financial support of the National Science Foundation, the State of Wyoming, and the Sustainable Directorate of the U.S. Army Natick Research, Development, and Engineering Center (contract number DAAK 60-97-R-9601).
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 2.Bukhitiyarova M, Yang R, Ray B. Analysis of the pediocin AcH gene cluster from plasmid pSMB74 and its expression in a pediocin-negative Pediococcus acidilactici strain. Appl Environ Microbiol. 1994;60:3405–3408. doi: 10.1128/aem.60.9.3405-3408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Shapira R, Eisenstein M, Montville T J. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl Environ Microbiol. 1997;63:524–531. doi: 10.1128/aem.63.2.524-531.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chikindas M L, Garcia-Garcera M J, Driessen A J M, Ledeboer A M, Nissen-Meyer J, Nes I F, Abee T, Konings W N, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC 1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chikindas M L, Venema K, Ledeboer A M, Venema G, Kok J. Expression of lactococcin A and pediocin PA-1 in heterologous hosts. Lett Appl Microbiol. 1995;21:183–189. doi: 10.1111/j.1472-765x.1995.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 6.de Vos W M, Kuipers O P, van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 7.Fahey R C, Brown W C, Adams W B, Worsham M B. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fimland G, Blingsmo O R, Sletten K, Jung G, Nes I F, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fremaux C, Ahn C, Klaenhammer T R. Molecular analysis of the lactacin F operon. Appl Environ Microbiol. 1993;59:3906–3915. doi: 10.1128/aem.59.11.3906-3915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher N L F, Sailer M, Niemczura W P, Nakashima T T, Stiles M E, Vederas J C. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry. 1997;36:15062–15072. doi: 10.1021/bi971263h. [DOI] [PubMed] [Google Scholar]
- 11.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M E. Characterization of leucocin A-UAL 187 and cloning of the bactericidal gene from Leuconostoc gelidum. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 13.Horn N, Martinez M I, Martinez J M, Hernandez P E, Gasson M J, Rodriguez J M, Dodd H M. Production of pediocin PA-1 by Lactococcus lactis using the lactococcin A secretory apparatus. Appl Environ Microbiol. 1998;64:818–823. doi: 10.1128/aem.64.3.818-823.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack W R, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung G. Lantibiotics—ribosomally synthesized biologically active polypeptides containing sulphide bridges and α,β-didehydro amino acids. Angew Chem Int Ed Engl. 1991;30:1051–1068. [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller K W, Schamber R, Chen Y, Ray B. Production of active chimeric pediocin AcH in Escherichia coli in the absence of processing and secretion genes from the Pediococcus pap operon. Appl Environ Microbiol. 1998;64:14–20. doi: 10.1128/aem.64.1.14-20.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller K W, Schamber R, Osmanagaoglu O, Ray B. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl Environ Microbiol. 1998;64:1997–2005. doi: 10.1128/aem.64.6.1997-2005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motlagh A M, Bukhtiyarova M, Ray B. Complete nucleotide sequence of pSMB74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol. 1994;18:305–312. doi: 10.1111/j.1472-765x.1994.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 21.Novak P, Dev I K. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol. 1988;170:5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 23.Quadri L E N, Sailer M, Terebiznik M R, Roy K L, Vederas J C, Stiles M E. Characterization of the protein conferring immunity to the antimicrobial peptide carnobacteriocin B2 and expression of carnobacteriocins B2 and BM1. J Bacteriol. 1995;177:1144–1151. doi: 10.1128/jb.177.5.1144-1151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quadri L E N, Yan L Z, Stiles M E, Vederas J C. Effect of amino acid substitutions on the activity of carnobacteriocin B2. J Biol Chem. 1997;272:3384–3388. doi: 10.1074/jbc.272.6.3384. [DOI] [PubMed] [Google Scholar]
- 25.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 26.Schnell N, Entian K-D, Schneider U, Götz F, Zähner H, Kellner R, Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 27.Schnell N, Entian K-D, Götz F, Hörner T, Kellner R, Jung G. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine-containing antibiotic. FEMS Microbiol Lett. 1989;58:263–268. doi: 10.1016/0378-1097(89)90050-5. [DOI] [PubMed] [Google Scholar]
- 27a.Studier F W, Rosenberg A H, Dunn J J, Budendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 28.van Belkum M J, Worobo R W, Stiles M E. Double-glycine-type leader peptides direct secretion of bacteriocins by ABC-transporters: colicin V secretion in Lactococcus lactis. Mol Microbiol. 1997;23:1293–1301. doi: 10.1046/j.1365-2958.1997.3111677.x. [DOI] [PubMed] [Google Scholar]
- 29.van der Meer J R, Polman J, Beerthuyzen M M, Siezen R J, Kuipers O P, de Vos W M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Meer J R, Rollema H S, Siezen R J, Beerthuyzen M M, Kuipers O P, de Vos W M. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J Biol Chem. 1994;269:3555–3562. [PubMed] [Google Scholar]
- 31.Venema K, Kok J, Marugg J D, Toonen M Y, Ledeboer A M, Venema G, Chikindas M L. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC 1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 32.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang R, Johnson M C, Ray B. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1992;58:3355–3359. doi: 10.1128/aem.58.10.3355-3359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]