Summary
RNA-sequencing and quantitative proteomic profiling simultaneously measure thousands of molecules and provide opportunities to decipher the transcriptomic and proteomic landscapes of cohort specimens for basic and health research. We present a protocol for the analysis of paired transcriptome and proteome data to identify and compare molecular subgroups among cohort specimens. We demonstrate a streamlined analysis workflow, applicable for both transcriptome and proteome data, which allows the comparison of two data types for RNA-protein variations and for derivation of biological implications.
For complete details on the use and execution of this protocol, please refer to Yang et al. (2021).
Subject areas: Bioinformatics, Cancer, RNAseq, Gene Expression, Proteomics
Graphical abstract

Highlights
-
•
Identify and compare subgroups using transcriptome and/or proteome data
-
•
Streamlined workflow from data preprocessing to cluster and differential analysis
-
•
Examine mRNA-protein correlation from paired transcriptome and proteome data
-
•
Discover subgroups and identify distinguishing features from any groups of interest
RNA-sequencing and quantitative proteomic profiling simultaneously measure thousands of molecules and provide opportunities to decipher the transcriptomic and proteomic landscapes of cohort specimens for basic and health research. We present a protocol for the analysis of paired transcriptome and proteome data to identify and compare molecular subgroups among cohort specimens. We demonstrate a streamlined analysis workflow, applicable for both transcriptome and proteome data, which allows the comparison of two data types for RNA-protein variations and for derivation of biological implications.
Before you begin
Protocol overview
Technological advances in high-throughput profiling technologies have enabled the molecular characterization of biological samples to identify fundamental and translational insights. RNA-sequencing (RNA-seq) and quantitative global proteomics are two approaches that have been exploited to enumerate thousands of RNA or protein molecules within a sample, respectively (Ellis et al., 2013; Weinstein et al., 2013). While either of these two methods has been widely used in omics studies, few studies have adapted both approaches to simultaneously profile the paired transcriptomes and proteomes of clinical specimens. In our previous study, we applied RNA-seq and quantitative proteomic mass spectrometry to pancreatic neuroendocrine neoplasm specimens and identified biologically distinct subgroups with potential therapeutic vulnerabilities (Yang et al., 2021). This protocol presents a generic version of the analysis workflow used in our previous study and details the preparation and analysis of RNA-seq and quantitative global proteomic data for subgroup identification and comparison. The identified subgroups and their molecular differences may be further investigated for clinical significance and biological differences. While we illustrate the workflow using cancer specimen data, this protocol or parts of the protocol can be adapted for RNA-seq and/or quantitative proteomic data from any sample or species to discover molecular subgroups based on global gene expression or protein abundance patterns, compare RNA or protein -level differences between conditions, and examine RNA-protein correlations. Combining paired RNA and protein -level data enables cross-referencing between the two independent measurements that substantiates the analysis results obtained from individual datasets.
Note: Throughout this protocol, each line of executable code is preceded by a greater than sign (>), while text outputs are italicized.
Pause point: Given that this protocol is exclusively bioinformatics-based, the users may pause at any time.
Institutional permissions
This protocol uses the molecular datasets from our previous study (Yang et al., 2021) to demonstrate the detailed steps and potential outcomes. These datasets were used in accordance with the ethical approval by the UBC Clinical Research Ethics Board (H12-03484) and the University of British Columbia BC Cancer Research Ethics Board (H16-01577). Users adapting this protocol are reminded to acquire the necessary permissions to use their datasets from the relevant institutions, if applicable.
Software setup
Timing: < 1 h
All the analysis steps described in this protocol are performed using R, a free software environment for statistical computing and visualization (R Core Team, 2020), which can be run on most operating systems including UNIX, Windows and MacOS. The current protocol was developed using R version 3.6.3 running on a Linus system (CentOS 7). In addition to R, a list of R packages is required as they provide the necessary functions for the data processing, analysis or visualization steps covered in this protocol. The list of packages and their versions used are included in the key resources table.
-
1.
Download and install R from https://cran.r-project.org/ (R Core Team, 2020).
Note: By default, the most recent release is downloaded, but older releases (eg. version 3.6.3) can also be found from the website. To install R, follow the installation steps specific to the operating system described on the website.
Optional: Download and install RStudio from https://www.rstudio.com/products/rstudio/ (RStudio Team, 2018). RStudio is an integrated development environment for R that facilitates code writing and execution with plotting capabilities. While RStudio is not required to follow this protocol, its usage provides convenient data visualization and is highly recommended.
-
2.
Download and install required R packages from CRAN or Bioconductor (https://bioconductor.org). We recommend using BiocManager to install R packages from either source, which automatically identifies the most updated versions of the packages for the R version used.
>if (!requireNamespace(“BiocManager”, quietly = T)) install.packages(“BiocManager”)
>required.packages <- c(“tidyverse”, “reshape2”, “biomaRt”, “org.Hs.eg.db”, “edgeR”, “limma”, “NMF”, “sva”, “vsn”, “ggpubr”, “ggfortify”, “GSEABase”)
>BiocManager::install(required.packages, ask = F)
-
3.
Attach the installed packages.
>sapply(c(required.packages, “magrittr”), require, character.only = T)
Note: Packages must be reattached/reloaded at each restart of R.
Note: The magrittr package is included in the tidyverse suite download but not automatically loaded with the tidyverse suite and must be explicitly loaded.
Load datasets
Timing: < 30 min
For the purpose of this protocol, we assume that the users start with data of a rectangular structure, with features (RNAs or proteins) in rows, and samples in columns. Additional columns with feature information such as gene identifiers or feature subtypes may be present. Depending on the source and prior workflow, the starting datasets may be of different structures or formats, and we leave it to the users’ discretion to reformat their data to that similar to our input data shown below.
In this protocol, we use the RNA-seq count and transcript-per-million (TPM) data and proteomic peptide spectral match (PSM) data from our previous study (Yang et al., 2021). In brief, samples from a specimen cohort were subjected to RNA-seq and proteomic profiling. The RNA-seq count data were generated using STAR aligner (Dobin et al., 2012) and featureCounts (part of Subread) (Liao et al., 2013). The proteomic PSM data were generated from five tandem mass tag (TMT) 10-plex mass-spectrometry runs using Orbitrap Fusion and Proteome Discoverer (Thermo Fisher). This protocol is also applicable to RNA expression data from other technologies (e.g., gene expression microarray), provided it is normalized according to the standard practices for the particular technologies.
-
4.
Read in transcriptomic data.
>COUNT <- read_tsv(“COUNT.tsv”)
>dim(COUNT)
[1] 57905 41
>head(COUNT)
# A tibble: 6 × 44
EnsemblGeneID Start End Strand Length S1 S2 S3
<chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 ENSG000002239… 1186… 1222… +;+;+… 1756 0 1 0
2 ENSG000002272… 1436… 1482… -;-;-… 2073 104 82 61
3 ENSG000002434… 2955… 3003… +;+;+… 1021 0 0 0
4 ENSG000002376… 3455… 3517… -;-;-… 1219 0 0 0
5 ENSG000002680… 5247… 5331… +;+;+ 947 0 0 0
6 ENSG000002403… 62948 63887 + 940 1 0 0
# … with 36 more variables: S4 <dbl>, S5 <dbl>, S6 <dbl>,
# S7 <dbl>, S8 <dbl>, S9 <dbl>, S10 <dbl>, S11 <dbl>,
# S12 <dbl>, S13 <dbl>, S14 <dbl>, S15 <dbl>, S16 <dbl>,
# S17 <dbl>, S19 <dbl>, S21 <dbl>, S22 <dbl>, S23 <dbl>,
# S24 <dbl>, S25 <dbl>, S26 <dbl>, S27 <dbl>, S28 <dbl>,
# S29 <dbl>, S30 <dbl>, S31 <dbl>, S32 <dbl>, S33 <dbl>,
# S37 <dbl>, S38 <dbl>, S39 <dbl>, S40 <dbl>, …
>TPM <- read_tsv(“TPM.tsv”)
>head(TPM)
# A tibble: 6 × 44
EnsemblGeneID Start End Strand Length S1 S2 S3
<chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 ENSG000002239… 1186… 1222… +;+;+… 1756 0 0.114 0
2 ENSG000002272… 1436… 1482… -;-;-… 2073 9.68 7.90 5.23
3 ENSG000002434… 2955… 3003… +;+;+… 1021 0 0 0
4 ENSG000002376… 3455… 3517… -;-;-… 1219 0 0 0
5 ENSG000002680… 5247… 5331… +;+;+ 947 0 0 0
6 ENSG000002403… 62948 63887 + 940 0.205 0 0
# … with 36 more variables: S4 <dbl>, S5 <dbl>, S6 <dbl>,
# S7 <dbl>, S8 <dbl>, S9 <dbl>, S10 <dbl>, S11 <dbl>,
# S12 <dbl>, S13 <dbl>, S14 <dbl>, S15 <dbl>, S16 <dbl>,
# S17 <dbl>, S19 <dbl>, S21 <dbl>, S22 <dbl>, S23 <dbl>,
# S24 <dbl>, S25 <dbl>, S26 <dbl>, S27 <dbl>, S28 <dbl>,
# S29 <dbl>, S30 <dbl>, S31 <dbl>, S32 <dbl>, S33 <dbl>,
# S37 <dbl>, S38 <dbl>, S39 <dbl>, S40 <dbl>, …
Note: Here, the “COUNT.tsv” and “TPM.tsv” files imported are tab-delimited files previously generated and saved to the working directory. These files contain RNA-seq count and TPM data, respectively, for our cohort of specimens. The first five columns of each dataset contain aggregated gene-level information such as the start and end positions of the reads mapped to each gene.
-
5.
Read in proteomic data.
>PSM <- paste0("PSM_", 1:5) %>%
setNames(., paste0("TMT", 1:5)) %>%
as.list %>%
lapply(., function(x) paste0(x, ".tsv") %>% read_tsv)
>lapply(PSM, dim)
$TMT1
[1] 150236 25
$TMT2
[1] 139264 25
$TMT3
[1] 153412 25
$TMT4
[1] 121423 25
$TMT5
[1] 128379 25
>head(PSM$TMT1)
# A tibble: 6 × 25
Confidence `PSM Ambiguity` `Annotated Sequence… Contaminant
<chr> <chr> <chr> <lgl>
1 High Unambiguous [R].nPDNLEELLLNETA… FALSE
2 High Unambiguous [R].lALDDVAALHGPVV… FALSE
3 High Unambiguous [K].gkPAAPGGAGNTGT… FALSE
4 High Unambiguous [K].dLYANNVLSGGTTm… FALSE
5 High Unambiguous [R].vAPEEHPVLLTEAP… FALSE
6 High Unambiguous [K].mGGMEGPFGGGMEN… FALSE
# … with 21 more variables: Number of Protein Groups <dbl>,
# Number of Proteins <dbl>,
# Master Protein Accessions <chr>,
# Protein Accessions <chr>, Rank <dbl>,
# Search Engine Rank <dbl>, mz in Da <dbl>,
# Percolator q-Value <dbl>, Percolator PEP <dbl>,
# Peptide Quan Usage <chr>, Quan Info <chr>, S8 <dbl>, …
Note: Here, the PSM_[1–5].tsv files contain PSM abundance data from the five runs. Due to the random fractionation nature of the approach, each run identified and quantitated a slightly different set of peptides and are thus kept separate as objects in a list for processing prior to analysis. Each run contains 8 specimens plus two technical control samples: a pooled standard (P1∼5) consisting of fractions of the included specimens and a cell line supermix (SM1∼5) made up with a panel of cell lines. The first 15 columns of each dataset contain information on each PSM, including its confidence, its matching peptide and the protein(s) in which the peptide can be found.
Note: Due to a few specimens being excluded because of inferior RNA quality or failure in RNA-seq library construction, the RNA-seq count dataset contains fewer samples compared to the proteomic datasets as shown below.
>PSM %>%
sapply(., function(x) {
x %>%
dplyr::select(matches("S[0-9]")) %>%
colnames
}) %>% c %>%
intersect(colnames(COUNT)) %>% {
print(paste("Number of overlapping samples:",
length(.)))
}
[1] "Number of overlapping samples: 35"
Key resources table
Step-by-step method details
Preparation of RNA-seq data for cluster analysis
Timing: < 30 min
Both RNA-seq and proteomic data require transformation and/or normalization prior to analysis. For analysis of RNA-seq data, the TPM values are used for cluster analysis as they account for gene length and sequencing depth, and the RNA-seq count data is used for differential analysis. To prepare these two datasets for analyses, RNA annotations are added. To prepare the TPM data for cluster analysis, protein-coding mRNAs are log-transformed, and low level mRNAs are removed as background noise.
-
1.
Add annotations to RNA features for the TPM and COUNT data loaded in above.
Note: Additional annotations of RNA features are necessary for downstream analysis, particularly when comparing to proteomic data. For the purpose of this protocol, it is critical to add two types of annotations to the RNA features: the gene biotype and the HGNC symbol. The former allows filtering for protein-coding mRNAs, while the latter serves as a common identifier type in analyses involving both mRNA and protein data.
>GRCh37.87 <- useMart(biomart = "ENSEMBL_MART_ENSEMBL",
host = "dec2016.archive.ensembl.org",
dataset = "hsapiens_gene_ensembl")
>RNA.anno <- getBM(attributes = c("ensembl_gene_id", “hgnc_symbol", "entrezgene", "gene_biotype"), filters = "ensembl_gene_id", values = COUNT$EnsemblGeneID, mart = GRCh37.87, useCache = F) %>%
set_colnames(c("EnsemblGeneID", "HGNCSymbol", "EntrezID", "Biotype")) %>%
dplyr::mutate(EntrezID = as.character(EntrezID)) %>%
group_by(EnsemblGeneID, Biotype) %>%
dplyr::summarise(HGNCSymbol = ifelse(length(unique(HGNCSymbol)) > 1, paste(unique(HGNCSymbol), collapse = ", "), unique(HGNCSymbol)), EntrezID = ifelse(length(unique(EntrezID)) > 1, paste(unique(EntrezID), collapse = ", "), unique(EntrezID))) %>%
ungroup %>%
mutate_all(function(x) na_if(x, "NA")) %>%
mutate_all(function(x) na_if(x, ""))
>head(RNA.anno)
# A tibble: 6 × 4
EnsemblGeneID Biotype HGNCSymbol EntrezID
<chr> <chr> <chr> <chr>
1 ENSG00000000003 protein_coding TSPAN6 7105
2 ENSG00000000005 protein_coding TNMD 64102
3 ENSG00000000419 protein_coding DPM1 8813
4 ENSG00000000457 protein_coding SCYL3 57147
5 ENSG00000000460 protein_coding C1orf112 55732
6 ENSG00000000938 protein_coding FGR 2268
>COUNT <- left_join(COUNT, RNA.anno, by = "EnsemblGeneID")
>TPM <- left_join(TPM, RNA.anno, by = "EnsemblGeneID")
>COUNT %>%
dplyr::select(where(is.character), where(is.numeric)) %>%
head
# A tibble: 6 × 44
EnsemblGeneID Start En Strand Biotype HGNCSymbol
<chr> <chr> <chr> <chr> <chr> <chr>
1 ENSG00000223972 11869;11… 1222… +;+;+… transc… DDX11L1
2 ENSG00000227232 14363;14… 1482… -;-;-… unproc… WASH7P
3 ENSG00000243485 29554;30… 3003… +;+;+… lincRNA MIR1302-2
4 ENSG00000237613 34554;35… 3517… -;-;-… lincRNA FAM138A
5 ENSG00000268020 52473;53… 5331… +;+;+ unproc… OR4G4P
6 ENSG00000240361 62948 63887 + unproc… OR4G11P
# … with 38 more variables: EntrezID <chr>, Length <dbl>,
# S1 <dbl>, S2 <dbl>, S3 <dbl>, S4 <dbl>, S5 <dbl>,
# S6 <dbl>, S7 <dbl>, S8 <dbl>, S9 <dbl>, S10 <dbl>,
# S11 <dbl>, S12 <dbl>, S13 <dbl>, S14 <dbl>, S15 <dbl>,
# S16 <dbl>, S17 <dbl>, S19 <dbl>, S21 <dbl>, S22 <dbl>,
# S23 <dbl>, S24 <dbl>, S25 <dbl>, S26 <dbl>, S27 <dbl>,
# S28 <dbl>, S29 <dbl>, S30 <dbl>, S31 <dbl>, …
Note: Three feature annotation columns are added to the COUNT and TPM data: Biotype, HGNCSymbol and EntrezID, corresponding to the biotype, HGNC symbol and Entrez ID of each gene. The addition of Entrez IDs is optional but sometimes necessary depending on downstream analysis requirements.
Note: Users wishing to use a different genome assembly may do so by changing the “host” and “dataset” arguments to obtain gene annotations from a different version or species, respectively.
-
2.
Prepare log-transformed TPM matrix
Note: For cluster analysis, a matrix of log2-transformed TPM values is needed. Here, a numeric matrix is generated from the TPM values of protein-coding genes. Lowly expressed mRNAs, defined here as those expressed below 1 TPM in more than 50% of the cohort, are removed before being log2-transformed.
>logTPM <- TPM %>%
filter(., Biotype == "protein_coding") %>%
column_to_rownames("EnsemblGeneID") %>%
dplyr::select(matches("S\\d{1,2}")) %>%
as.matrix %>% {
mat = .
mat %>%
is_greater_than(1) %>%
rowSums %>%
is_weakly_greater_than(ncol(mat)*0.5) %>%
mat[.,] %>% return
} %>%
add(1) %>%
log2
>logTPM[1:5, 1:8]
S1 S2 S3 S4
ENSG00000187634 6.121744 5.120782 7.141506 1.659699
ENSG00000188976 5.596282 5.030179 5.147660 4.232472
ENSG00000187961 4.295906 2.720688 3.048589 2.280037
ENSG00000188290 4.405557 4.465438 4.933465 4.386167
ENSG00000187608 3.753190 3.237547 3.458798 3.622116
S5 S6 S7 S8
ENSG00000187634 3.180383 3.709654 2.282866 3.597135
ENSG00000188976 5.044459 4.852175 4.906102 5.338667
ENSG00000187961 3.991705 3.105126 3.108855 4.104080
ENSG00000188290 4.704014 3.947958 3.864484 5.879325
ENSG00000187608 2.700303 2.703028 2.438892 4.112869
Note: The gene expression threshold is arbitrarily set but should take into consideration the research question and cohort composition. Users wishing to adjust the gene expression threshold may do so by changing the 8th and 10th lines in the code chunk above. The is_greater_than(1) line identifies all TPM measurements above 1 while the is_weakly_greater_than(ncol(mat)∗0.5) identifies features with expression above 1 TPM in 50% or more of the cohort.
CRITICAL: A small constant (in this case 1) must be added to all TPM values prior to log2-transformation to avoid errors arising from taking the logarithm of zeroes. We prefer to use a constant of 1 because log2 of 1 returns zero.
Preparation of proteomic data for cluster analysis
Timing: < 1 h
Similar to RNA-seq data, the proteomic data should be transformed and normalized. Additionally, the PSM data should be converted into peptide and eventually protein -level data for analysis. Since the proteomic data were generated based on relative abundance, all identified proteins with quantitative values are used.
Note: Measurements obtained from RNA-seq and quantitative proteomic mass spectrometry are distinct in nature due to the differences in the corresponding technology. While RNA-seq can provide absolute counts, our TMT-based proteomic method estimated the abundances of PSMs in each sample relative to other samples within the same run. The two datasets therefore require different preparation steps for transformation and normalization to account for differences such as their data distributions. For further details on RNA-seq and quantitative proteomic mass spectrometry comparisons, see references (Nesvizhskii, 2010; Ning et al., 2012; Wang et al., 2014).
-
3.
Normalize and aggregate PSMs into proteins.
Note: The PSM abundance measurements loaded in above are aggregated into peptide and then into protein abundance using the median values as the representation. Variance stabilizing normalization (VSN) is applied to normalize and transform peptide abundances prior to collapsing into proteins. The measurements from ambiguously mapped peptides (ie. present in more than one protein) are excluded.
>PRO <- PSM %>%
lapply(., function(x) {
pep <- x %>%
filter(., `Number of Protein Groups` == 1) %>%
dplyr::select(`Master Protein Accessions`, `Annotated Sequence`, 16:25) %>%
dplyr::rename(ProteinAccession = `Master Protein Accessions`, Sequence = `Annotated Sequence`) %>%
dplyr::mutate(Sequence =
toupper(str_extract(Sequence,
"(?<=\\.).*(?=\\.)"))) %>%
na.omit %>%
filter(., !grepl("sp", ProteinAccession)) %>%
group_by(ProteinAccession, Sequence) %>%
summarise_all(median) %>%
ungroup
pep <- pep %>%
dplyr::select(-ProteinAccession, -Sequence) %>%
as.matrix %>%
justvsn %>%
data.frame(ProteinAccession = pep$ProteinAccession, .)
pro <- pep %>%
group_by(ProteinAccession) %>%
summarise_all(median) %>%
ungroup
return(pro)
})
>head(PRO$TMT1)
# A tibble: 6 × 11
ProteinAccession S8 S9 S14 S21 S20 S30 S40
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A0AUZ9 5.57 6.20 6.84 5.88 6.29 7.00 5.89
2 A0AV96 3.22 2.22 3.20 2.61 4.01 2.37 2.20
3 A0AVF1 1.12 1.32 0.785 0.878 1.05 0.944 1.17
4 A0AVT1 4.84 4.70 5.20 5.08 4.85 5.77 5.11
5 A0FGR8 3.95 4.28 4.64 4.05 4.70 4.48 4.35
6 A0JNW5 1.12 2.16 2.06 1.65 1.65 1.38 2.08
# … with 3 more variables: S22 <dbl>, P1 <dbl>, SM1 <dbl>
>sapply(PRO, nrow)
TMT1 TMT2 TMT3 TMT4 TMT5
8183 7882 8058 7498 7847
Note: Users may opt for their preferred normalization method by changing the justvsn function on the 18th line in the above code to their desired function. For example, the normalizeBetweenArrays() function from the limma package can be used to perform median or quantile -normalization. We use VSN in our protocol as it was shown to be superior in reducing variation between technical replicates (Välikangas et al., 2018). After variance stabilizing normalization, the peptide abundance values are in log2-scale and no further transformation is necessary.
-
4.
Compile protein abundance from separate TMT runs.
Note: As mentioned earlier, each proteomic run may identify a different set of proteins. For proteomic analysis of the entire cohort of specimens, only proteins identified and quantitated across all specimens are used.
>TMT <- sapply(PRO, function(x) x[,-1] %>% colnames) %>%
melt %>%
extract(,2:3) %>%
set_colnames(c("Batch", "SampleID"))
>head(TMT)
Batch SampleID
1 TMT1 S8
2 TMT1 S9
3 TMT1 S14
4 TMT1 S21
5 TMT1 S20
6 TMT1 S30
>PRO <- PRO %>%
purrr::reduce(inner_join, by = "ProteinAccession")
>PRO
# A tibble: 6,037 × 51
ProteinAccession S8 S9 S14 S21 S20 S30 S40
<fct <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 A0AV96 3.22 2.22 3.20 2.61 4.01 2.37 2.20
2 A0AVF1 1.12 1.32 0.785 0.878 1.05 0.944 1.17
3 A0AVT1 4.84 4.70 5.20 5.08 4.85 5.77 5.11
4 A0FGR8 3.95 4.28 4.64 4.05 4.70 4.48 4.35
5 A0MZ66 5.22 4.77 4.38 4.71 4.72 4.34 3.90
6 A1IGU5 5.39 7.23 6.18 6.54 4.71 7.04 5.61
7 A1KXE4 2.41 3.26 2.34 2.97 2.49 3.01 3.45
8 A1L0T0 6.17 5.81 7.36 5.45 6.97 5.71 4.92
9 A1L188 3.09 2.84 3.20 3.44 3.02 2.57 3.56
10 A1L390 5.23 4.21 4.46 4.03 2.38 2.71 4.17
# … with 6,027 more rows, and 43 more variables: S22 <dbl>,
# P1 <dbl>, SM1 <dbl>, S12 <dbl>, S5 <dbl>, S4 <dbl>,
# S28 <dbl>, S25 <dbl>, S2 <dbl>, S7 <dbl>, S29 <dbl>,
# P2 <dbl>, SM2 <dbl>, S27 <dbl>, S39 <dbl>, S36 <dbl>,
# S1 <dbl>, S13 <dbl>, S19 <dbl>, S35 <dbl>, S34 <dbl>,
# P3 <dbl>, SM3 <dbl>, S15 <dbl>, S18 <dbl>, S17 <dbl>,
# S26 <dbl>, S10 <dbl>, S24 <dbl>, S32 <dbl>, S38 <dbl>, …
-
5.
Add annotations to protein features to enable downstream analysis and mRNA-protein comparison.
>PRO <- getBM(attributes = c("uniprot_swissprot", "hgnc_symbol", "entrezgene"), filters = "uniprot_swissprot",values = PRO$ProteinAccession, mart = GRCh37.87, useCache = F) %>%
set_colnames(c("ProteinAccession", "HGNCSymbol", "EntrezID")) %>%
dplyr::mutate(EntrezID = as.character(EntrezID)) %>%
group_by(ProteinAccession) %>%
dplyr::summarise(HGNCSymbol =
ifelse(length(unique(HGNCSymbol)) > 1,
paste(unique(HGNCSymbol), collapse = ", "),
unique(HGNCSymbol)), EntrezID =
ifelse(length(unique(EntrezID)) > 1,
paste(unique(EntrezID), collapse = ", "),
unique(EntrezID))) %>%
ungroup %>%
mutate_all(function(x) na_if(x, "NA")) %>%
mutate_all(function(x) na_if(x, "")) %>%
left_join(PRO, ., by = "ProteinAccession") %>%
dplyr::select(ProteinAccession, HGNCSymbol, EntrezID, everything())
>PRO
# A tibble: 6,037 × 53
ProteinAccession HGNCSymbol EntrezID S8 S9 S14
<chr> <chr> <chr> <dbl> <dbl> <dbl>
1 A0AV96 RBM47 54502 3.22 2.22 3.20
2 A0AVF1 TTC26 79989 1.12 1.32 0.785
3 A0AVT1 UBA6 55236 4.84 4.70 5.20
4 A0FGR8 ESYT2 57488 3.95 4.28 4.64
5 A0MZ66 SHTN1 57698 5.22 4.77 4.38
6 A1IGU5 ARHGEF37 389337 5.39 7.23 6.18
7 A1KXE4 FAM168B 130074 2.41 3.26 2.34
8 A1L0T0 ILVBL 10994 6.17 5.81 7.36
9 A1L188 NDUFAF8 284184 3.09 2.84 3.20
10 A1L390 PLEKHG3 26030 5.23 4.21 4.46
# … with 6,027 more rows, and 47 more variables: S21 <dbl>,
# S20 <dbl>, S30 <dbl>, S40 <dbl>, S22 <dbl>, P1 <dbl>,
# SM1 <dbl>, S12 <dbl>, S5 <dbl>, S4 <dbl>, S28 <dbl>,
# S25 <dbl>, S2 <dbl>, S7 <dbl>, S29 <dbl>, P2 <dbl>,
# SM2 <dbl>, S27 <dbl>, S39 <dbl>, S36 <dbl>, S1 <dbl>,
# S13 <dbl>, S19 <dbl>, S35 <dbl>, S34 <dbl>, P3 <dbl>,
# SM3 <dbl>, S15 <dbl>, S18 <dbl>, S17 <dbl>, S26 <dbl>, …
Note: HGNC symbols are added to the protein data as with RNA-seq data. To facilitate subsequent analyses that involve both mRNAs and proteins, the same column name, HGNCSymbol, is used here.
-
6.
Check for batch effects.
Note: It is recognized that multiplex proteomic profiling, particularly using TMT-based approaches, suffers from batch-to-batch variations (Brenes et al., 2019). These batch effects can jeopardize comparability between runs, so it is critical to ascertain whether batch effects are present in the dataset. The following code generates a quick principal component analysis (PCA) plot from the normalized protein data where the samples are color-coded according to their TMT run batch. As shown in Figure 1A, batch-related sample variations are evident in our proteomic dataset where the technical replicates of the pooled internal standard (P1∼5; solid circles) are dispersed yet cluster with other samples from the same runs. This result suggests the batch effects related to the TMT runs are pronounced and conceal the proteomic similarities expected between the technical replicates.
Figure 1.
Principal component analysis of the proteomic profiles before and after applying batch correction
(A) Principal component analysis (PCA) plot of samples based on their proteomic profiles. The samples are color-coded according to tandem mass tag (TMT) run. The patient specimen samples are shown as hollowed circles. Technical replicates of the pooled internal standard sample (P1∼5) and cell line supermix (SM1∼5) are shown as solid circles and solid triangles, respectively.
(B) PCA plot of the same samples after applying ComBat to mitigate the observed batch effects. The samples are color-coded and shown as in (A).
>PRO %>%
column_to_rownames("ProteinAccession") %>%
dplyr::select(-HGNCSymbol, -EntrezID) %>%
as.matrix %>%
t %>%
prcomp %>% {
mat = .
dat <- TMT %>%
dplyr::mutate(Type = ifelse(SampleID %in% paste0("SM", 1:5), 17, ifelse(SampleID %in% paste0("P", 1:5), 19, 1)))
autoplot(., data = TMT, colour = "Batch", label = F, shape = dat$Type, size = 2) +
theme_pubr(base_size = 10)
}
Optional: For subgroup identification, it is critical to mitigate prominent batch effects (if present) to prevent them from confounding the true biological variations (Leek et al., 2010). Several batch effect correction methods exist (Nygaard et al., 2016). For our analysis, we use ComBat to correct for the batch effects in our proteomic data prior to subgroup identification. ComBat uses an empirical Bayes approach for batch correction which avoids over-correction in datasets with small batch sizes (Johnson et al., 2007). The code below applies ComBat to correct for the TMT run-associated batch effects and uses PCA plot to check for post-correction sample dispersion. As shown in Figure 1B, ComBat effectively reduces batch-associated clustering and results in comparability between the technical replicates of the pooled internal standard (P1∼5; solid circles) while leaving the technical replicates of the cell line supermix (SM1∼5; solid triangles) in a distinct cluster.
>PRO.CB <- PRO %>%
dplyr::select(-HGNCSymbol, -EntrezID) %>%
column_to_rownames("ProteinAccession") %>%
as.matrix %>%
ComBat(dat = ., batch = TMT$Batch)
>PRO.CB[1:5, 1:5]
S8 S9 S14 S21 S20
A0AV96 3.951024 3.063534 3.935030 3.408637 4.662523
A0AVF1 3.518845 3.706400 3.208412 3.294879 3.455464
A0AVT1 4.931518 4.804685 5.255306 5.151811 4.937922
A0FGR8 4.458965 4.772773 5.112917 4.558818 5.171048
A0MZ66 5.275175 4.840055 4.461929 4.784848 4.793794
>PRO.CB %>%
t %>%
prcomp %>% {
mat = .
dat <- TMT %>%
dplyr::mutate(Type = ifelse(SampleID %in% paste0("SM", 1:5), 17, ifelse(SampleID %in% paste0("P", 1:5), 19, 1)))
autoplot(., data = TMT, colour = "Batch", label = F, shape = dat$Type, size = 2) +
theme_pubr(base_size = 10)
}
Note: Correction of batch effects should be limited to data on which exploratory analysis is performed. For differential analysis of data with known batch effects, the batch information can be included as a covariate in the statistical model to accommodate batch-related differences. This approach is demonstrated in the differential analysis section below.
Cluster analysis of RNA-seq and proteomic data
Timing: 1 h ∼ days (depending on the sample size and computing power)
Non-negative matrix factorization (NMF) is used to perform cluster analysis for the identification of subgroups. To ensure the identification of robustly segregated subgroups, a consensus approach is utilized to aggregate results from multiple iterations. Running consensus NMF can become time-consuming as the sample size increases. We therefore first perform a rank survey using a few iterations to identify the optimal number of clusters (rank), then increase the number of iterations to derive robust subgroup assignments at the optimal rank. In addition, only the most variable features (mRNAs or proteins) are used in the cluster analysis.
-
7.
Create wrapper function to perform NMF.
Note: Since NMF will be run for four separate instances (ie. rank survey and final clustering using mRNA or protein), a wrapper function to perform NMF can be utilized to minimize code repetition. Below, a wrapper function is created to 1) select the top 25% most variably expressed mRNAs or variably abundant proteins based on coefficient of variation ranking, 2) median-center feature expression/abundance, and 3) perform NMF using a specified rank(s) and number of iterations.
>doNMF <- function(mat, rank, nrun, save = T, save.name = NULL) {
if(save & is.null(save.name)) {
stop("A file name (save.name) must be provided for the result to be saved")
}
newmat <- apply(mat, 1, sd) %>%
divide_by(rowMeans(mat)) %>%
order(decreasing = T) %>%
mat[., ] %>%
extract(1:round(nrow(mat)*0.25), )
newmat <- apply(newmat, 1, median) %>%
sweep(newmat, 1, ., "-") %>%
as.matrix
res <- posneg(newmat) %>%
subset(., rowSums(.) > 0) %>%
nmf(x = ., rank = rank, nrun = nrun, seed = 123456)
if (save) {
saveRDS(res, save.name)
}
return(res)
}
CRITICAL: As with most clustering approaches, a random seed is used to initialize each NMF run. To ensure reproducible results, a 6-digit random seed number must be provided and is used to initialize the random number generator when running NMF. In the above wrapper function, the number 123456 is used.
CRITICAL: NMF requires an input matrix of non-negative values, but standardization may result in negative values. In the above code, the posneg() function is used to split the positive and negative values, where the latter are transformed into non-negative values and added as additional features that maintain variations across samples.
Note: Since running NMF can become very time-consuming, we recommend saving the result as soon as the computation is complete to avoid its accidental loss. The above wrapper function includes a parameter to save a copy of the result in .rds format that can be easily read back into the workspace using the function readRDS().
-
8.
Perform rank surveys by assuming a range of ranks and running NMF for 50 iterations at each possible rank. The quality measures of the result from each rank can then be evaluated (next step) to choose the optimal rank at which the samples are best segregated into clusters.
>rank.r <- doNMF(logTPM, 2:7, 50, save.name = "RankSurvey_mRNA.rds")
>rank.p <- colnames(PRO.CB) %>%
str_detect(., "S\\d{1,2}") %>%
PRO.CB[, .] %>%
doNMF(., 2:7, 50, save.name = "RankSurvey_protein.rds")
Note: For the proteomic data, the technical control samples are excluded from the cluster analysis to prevent their inclusion from confounding the identification of potential subgroups from the specimens. The sample exclusion was done using line 4 of the above code chunk which selects columns based on their names containing “S” followed by a one-to-two digit number.
Note: Considering that our cohort contains 35–36 samples with proteome and transcriptome data, respectively, we only test a range of ranks up to 7. Users with larger sample size may increase the range of ranks by changing the second parameter (“2:7” in the above code chunk) of the doNMF function.
-
9.
Examine cophenetic and silhouette coefficients to determine the optimal ranks.
Note: The cophenetic correlation and silhouette width of a cluster result reflect how well the cluster result recapitulates the inter-sample distances in the original data and how well each sample is grouped in its cluster compared to neighboring clusters, respectively. These two coefficients from the consensus NMF at each rank tested can be evaluated to determine the optimal rank at which samples fall into distinct clusters. The code below plots the cophenetic and silhouette coefficients at the rank of 2–7 for mRNA and protein -based rank surveys. As shown in Figures 2A and 2B, the consensus cophenetic correlation and silhouette coefficients peak at the rank of 2 or 4 in both mRNA and protein -based analyses suggesting 2 or 4 is likely the optimal rank for NMF analysis using either data type.
Figure 2.
Rank surveys of consensus non-negative matrix factorization on mRNA or protein measurements
(A) Cophenetic correlation and silhouette coefficients (y-axis) at the rank 2–7 (x-axis) from consensus non-negative matrix factorization (NMF) using the top 25% variably expressed mRNAs.
(B) Cophenetic correlation and silhouette coefficients (y-axis) at the rank 2–7 (x-axis) from consensus NMF using the top 25% variably abundant proteins.
>rank.r$measures %>% {
dat = .
c <- ggline(dat, x = "rank", y = "cophenetic", xlab = "Rank", ylab = "Cophenetic coefficient", ggtheme = theme_pubr(base_size = 10))
s <- ggline(dat, x = "rank", y = "silhouette.consensus", xlab = "Rank", ylab = "Silhouette coefficient", ggtheme = theme_pubr(base_size = 10))
ggarrange(c, s, ncol = 2)
}
>rank.p$measures %>% {
dat = .
c <- ggline(dat, x = "rank", y = "cophenetic", xlab = "Rank", ylab = "Cophenetic coefficient", ggtheme = theme_pubr(base_size = 10))
s <- ggline(dat, x = "rank", y = "silhouette.consensus", xlab = "Rank", ylab = "Silhouette coefficient", ggtheme = theme_pubr(base_size = 10))
ggarrange(c, s, ncol = 2)
}
Note: While the cophenetic correlation and silhouette coefficients are helpful for choosing the optimal rank, the ultimate decision of the optimal rank should also take into consideration the research question and the cohort examined. For our analysis, while the coefficients appear the highest at the rank of 2, we had performed auxiliary NMF analyses which indicated the cluster solutions at the rank of 4 align more with clusters of biological differences. We therefore determined the rank of 4 to be the optimal rank.
-
10.
Perform NMF at the optimal ranks to obtain subgroup assignments.
Note: Since one of the goals of this protocol is to draw comparisons between paired transcriptome and proteome, only the samples with both RNA and protein information are used to determine the subgroup assignments and for further analyses. To determine the subgroup assignment of each sample, the consensus cluster solution at the rank of 4 is obtained by running consensus NMF for 200 iterations using either mRNA or protein data. Hierarchical cluster analysis is then performed using the resultant consensus matrix to obtain the subgroup assignments.
>samp <- intersect(colnames(logTPM), colnames(PRO.CB))
>nmf.r <- logTPM[, samp] %>%
doNMF(., 4, 200, save.name = "NMF_mRNA.rds")
>nmf.p <- PRO.CB[, samp] %>%
doNMF(., 4, 200, save.name = "NMF_protein.rds")
>assignment.r <- cutree(hclust(as.dist(1-nmf.r@consensus)), 4)
>assignment.p <- cutree(hclust(as.dist(1-nmf.p@consensus)), 4)
-
11.
Compare mRNA and protein -derived subgroup assignments.
>rbind(mRNA = assignment.r, Protein = assignment.p)
S1 S2 S3 S4 S5 S6 S7 S8 S9 S10 S11 S12 S13 S14 S15
mRNA 1 1 2 2 3 3 3 3 2 3 1 1 1 3 1
Protein 1 1 2 2 3 3 3 3 2 3 1 1 2 3 2
S16 S17 S19 S21 S22 S23 S24 S25 S26 S27 S28 S29 S30
mRNA 4 2 3 2 1 3 3 4 2 3 2 2 4
Protein 4 3 2 2 1 3 1 4 2 3 2 2 2
S31 S32 S33 S37 S38 S39 S40
mRNA 3 1 2 1 2 4 4
Protein 3 2 2 1 2 4 4
Note: Comparison of subgroup assignments from different analyses can be done using various approaches. The simplest and most intuitive is to tabulate the subgroup assignments for visual comparison as shown above. Evidently, the subgroup assignments derived from mRNA or protein -based clustering are highly concordant.
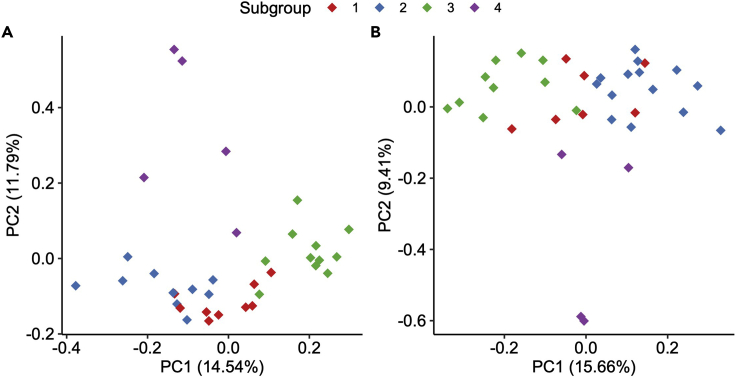
Optional: The NMF-derived subgroup assignments can be superimposed onto PCA plot to provide a visualization of the spatial separation between samples of the different subgroups. In the below code, PCA is performed using log-transformed TPM or batch-corrected abundance from mRNAs or proteins, respectively, and plotted with the samples color-coded according to their mRNA or protein -derived subgroup assignments. As shown in Figure 3A and 3B, PCA plots using the first two principal components largely recapitulate NMF-derived subgroups.
Figure 3.
Consensus NMF-derived subgroups illustrated using PCA plots
(A and B) Analysis and visualization strategies adapted from Yang et al. (Yang et al., 2021). (A) PCA plot using all expressed mRNAs. The samples are color-coded according to their NMF-derived subgroups. (B) PCA plot using all proteins. The samples are color-coded according to their NMF-derived subgroups.
>assignment.r[colnames(logTPM)] %>%
na.omit %>%
as.data.frame %>%
set_colnames("Subgroup") %>%
dplyr::mutate(Subgroup = as.character(Subgroup)) %>% {
sg = .
logTPM[, rownames(sg)] %>%
t %>%
prcomp %>%
autoplot(., data = sg, colour = "Subgroup", shape = 19, label = F) +
scale_color_brewer(palette = "Set1") +
theme_pubr(base_size = 12)
}
>assignment.p[colnames(PRO.CB)] %>%
na.omit %>%
as.data.frame %>%
set_colnames("Subgroup") %>%
dplyr::mutate(Subgroup = as.character(Subgroup)) %>% {
sg = .
PRO.CB[, rownames(sg)] %>%
t %>%
prcomp %>%
autoplot(., data = sg, colour = "Subgroup", shape = 19, label = F) +
scale_color_brewer(palette = "Set1") +
theme_pubr(base_size = 12)
}
Comparison of mRNA and protein variation
Timing: < 30 min
Given availability of paired transcriptome and proteome data, it can be informative to compare the transcriptome and proteome profiles of each sample, especially if the correlation between transcriptome and proteome is not known for the particular disease/condition of interest. Here, correlation analysis is used to compare the correlation between individual mRNAs and proteins in each specimen and across the cohort.
-
12.
Identify genes with paired mRNA and protein data.
Note: As mentioned earlier, HGNC gene symbols are used as the identifiers when cross-referencing RNA and protein. However, Ensembl gene IDs and protein accessions, the primary identifiers used in RNA and protein datasets respectively, should be preserved to prevent unnecessary loss of data.
>genes <- intersect(COUNT$HGNCSymbol, PRO$HGNCSymbol) %>%
extract(!is.na(.))
>paired.TPM <- TPM %>%
filter(., HGNCSymbol %in% genes) %>%
dplyr::select(HGNCSymbol, any_of(samp)) %>%
group_by(HGNCSymbol) %>%
summarise_all(median) %>%
ungroup %>%
column_to_rownames("HGNCSymbol") %>%
add(1) %>%
log2
>paired.PRO <- PRO[,c("ProteinAccession", "HGNCSymbol")] %>%
filter(., HGNCSymbol %in% genes) %>%
left_join(., PRO.CB %>% as.data.frame %>% rownames_to_column("ProteinAccession"),
by = "ProteinAccession") %>%
dplyr::select(HGNCSymbol, any_of(samp)) %>%
group_by(HGNCSymbol) %>%
summarise_all(median) %>%
ungroup %>%
column_to_rownames("HGNCSymbol")
-
13.
Compute mRNA-protein correlation for each sample.
Note: The correlation between paired transcriptome and proteome can be computed for each sample to evaluate how well the transcriptome and proteome landscapes are correlated with respect to mRNA expression and protein abundance in each sample on a global scale. The correlations can then be plotted (as shown in Figure 4A) for visual inspection. The code below computes and plots the mRNA-protein correlation for each sample.
Figure 4.
Correlations between mRNA expression and protein abundance
(A and B) Analysis and visualization strategies adapted from Yang et al. (2021). (A) Sample-wise mRNA-protein correlation computed as Spearman’s Rho (y-axis). Samples are ordered along the x-axis based on increasing correlation. (B) The distribution of gene-wise mRNA-protein correlations computed as Spearman’s Rho (x-axis). A histogram of 20 bins is shown with height of each bar proportional to the number of genes in each bin. The median correlation is depicted by a red vertical line.
>cor(paired.TPM, paired.PRO, method = "spearman") %>%
as.data.frame %>%
set_colnames("Rho") %>%
rownames_to_column("Sample") %>%
ggdotchart(., x = "Sample", y = "Rho", ggtheme = theme_pubr(base_size = 10), add = "segment", ylim = c(0, 1), ylab = "mRNA-protein correlation (rho)")
-
14.
Compute correlation between mRNA and protein variation for each gene to evaluate how well mRNA and protein variations are correlated in the sample cohort.
>cor(t(paired.TPM), t(paired.PRO), method = "spearman") %>%
diag %>%
gghistogram(., bins = 20, theme = theme_pubr(base_size = 10), fill = "gray",
xlab = "mRNA-protein correlation (rho)",
ylab = "Number of genes")
Note: As shown in Figure 4B, based on the list of genes with paired mRNA and protein information, diverse correlations between mRNA and protein variations are evident in our cohort specimens.
Differential analysis
Timing: < 30 min
Differential analysis of gene expression or protein abundance can be performed using the same analysis workflow, with slight differences in their input structures. Here, we illustrate our workflow for differential analysis of gene expression and protein abundance between the four subgroups identified earlier. For simplicity, only the mRNA-derived subgroup assignments are used. Differential analysis through the limma r package uses linear models and an empirical Bayes approach that allow analysis of data as a whole and permit inclusion of covariates (e.g., batch effects) that may confound with analysis objectives in the model design (Ritchie et al., 2015). In addition to differential analysis, a gene set test is performed to showcase added functionalities of the limma package in identifying biological differences from molecular comparisons.
DGEList objects from the edgeR package (Robinson et al., 2010) and ExpressionSet objects from the Biobase package (Huber et al., 2015) are two input formats accepted by limma. Both DGEList and ExpressionSet objects are of the S4 class that are designed to store expression, phenotype and feature data. For our purpose of analyzing transcriptome and proteome data, the transcriptome data are converted into a DGEList object while the proteome data are converted into an ExpressionSet object for analysis using limma.
Note: The utility of a DGEList class object is to facilitate normalization and transformation of count data and is not applicable to the proteomic data showcased in this protocol. Users working with microarray data may wish to proceed with an ExpressionSet object followed by appropriate normalization.
-
15.
Construct a DGEList object from the RNA-seq data.
Note: As mentioned earlier, the raw counts are used in the differential analysis, as compared to the TPMs in the cluster analyses. Since differential analysis compares gene-level measurements between groups, gene-length normalization is irrelevant. Instead, raw counts can be normalized to scale for library size and composition differences using the trimmed mean of M values (TMM) method from edgeR (Robinson and Oshlack, 2010).
>dge <- COUNT %>% {
dat = .
p <- assignment.r %>%
as.data.frame %>%
set_colnames("Subgroup") %>%
dplyr::mutate(Subgroup = paste0("Subgroup", Subgroup))
m <- dat %>%
dplyr::select(EnsemblGeneID, one_of(rownames(p))) %>%
column_to_rownames("EnsemblGeneID") %>%
as.matrix
f <- dat %>%
dplyr::select(-matches("S\\d{1,2}")) %>%
column_to_rownames("EnsemblGeneID")
res <- DGEList(counts = m,
samples = p,
genes = f)
fkeep <- rowSums(cpm(res) > 1) >= round(nrow(p)∗0.1)
res <- res[fkeep, keep.lib.sizes = F] %>%
calcNormFactors(., method = "TMM")
res
}
>dim(dge)
[1] 25373 35
Note: As done in the preparatory steps of cluster analysis, noise-level RNAs are removed from the constructed DGEList object. Here, only RNAs with expression above 1 count-per-million (CPM) in at least 10% of the cohort are retained for further analyses.
Note: The DGEList is analogous to a list of three objects: a gene matrix (counts), a feature annotation dataframe (genes) and a sample annotation dataframe (samples).
-
16.
Construct an ExpressionSet object from the proteomic data.
Note: For differential abundance analysis of proteomic data, the normalized protein abundance values are used. To account for the TMT-associated batch effects in the linear modeling, the batch covariate (eg. TMT run) is added to the sample annotations.
>eset <- PRO %>% {
dat = .
p <- assignment.r %>%
as.data.frame %>%
set_colnames("Subgroup") %>%
merge(., TMT, by.x = "row.names", by.y = "SampleID") %>%
column_to_rownames("Row.names") %>%
dplyr::mutate(Subgroup = paste0("Subgroup", Subgroup)) %>%
extract(samp,) %>%
new("AnnotatedDataFrame", .)
m <- dat %>%
dplyr::select(ProteinAccession, one_of(samp)) %>%
column_to_rownames("ProteinAccession") %>%
as.matrix
f <- dat %>%
dplyr::select(ProteinAccession, HGNCSymbol, EntrezID) %>%
column_to_rownames("ProteinAccession") %>%
new("AnnotatedDataFrame", .)
ExpressionSet(assayData = m,
phenoData = p,
featureData = f)
}
>eset
ExpressionSet (storageMode: lockedEnvironment)
assayData: 6037 features, 35 samples
element names: exprs
protocolData: none
phenoData
sampleNames: S1 S2 … S40 (35 total)
varLabels: Subgroup Batch
varMetadata: labelDescription
featureData
featureNames: A0AV96 A0AVF1 … Q9Y6Y8 (6037 total)
fvarLabels: HGNCSymbol EntrezID
fvarMetadata: labelDescription
experimentData: use 'experimentData(object)'
Annotation:
Note: Similar to a DGEList, the ExpressionSet contains a protein abundance matrix (assayData), a feature annotation dataframe (featureData) and a sample annotation dataframe (phenoData).
-
17.
Create a wrapper function to perform limma.
Note: Differential analysis using limma involves several steps that are mostly identical regardless of the input. As such, a wrapper function can be created to reduce code repetition, as previously done for performing NMF. In addition to differential analysis capabilities, limma is equipped with a function camera() that performs a competitive gene set test against any reference gene panel of interest (Wu and Smyth, 2012). This added capability can be leveraged to develop a streamlined workflow combining differential analysis and gene set tests.
>doLimma <- function(dat, type, model.m, contr.m, gs = NULL)
{
if (type == "rna") {
v <- voom(dat, design = model.m)
if (!is.null(gs)) {
gset <- gs %>%
ids2indices(., id = dat$genes$HGNCSymbol)
}
} else if (type == "protein") {
v <- dat
if (!is.null(gs)) {
gset <- gs %>%
ids2indices(., id = fData(dat)$HGNCSymbol)
}
} else {
stop("Data type must be specified.")
}
vfit <- lmFit(v, model.m) %>%
contrasts.fit(., contrasts = contr.m)
efit <- eBayes(vfit)
if (!is.null(gs)) {
groups = colnames(contr.m)
cam <- groups %>%
setNames(.,.) %>%
as.list %>%
lapply(., function(x) {
camera(v, gset, model.m, contrast = contr.m[,x])
})
return(list(v=v, efit = efit, camera = cam))
} else {
list(v = v, efit = efit)
}
}
Note: Since the transcriptome data was only subjected to normalization, the wrapper function transforms the transcriptome data using voom prior to performing the analysis. The proteome data, on the other hand, was subjected to transformation during variance-stabilizing normalization and no further processing is required.
Note: A model matrix and a contrast matrix are required to perform linear modeling and to specify group comparisons. These matrices are left for the users to supply to the wrapper function to allow flexibility to add or modify as appropriate. Examples of how these matrices can be constructed are shown in steps 19 and 20.
Note: The above wrapper function outputs a list containing the input (v), the result of fitted gene-wise linear model as a MArrayLM object (efit), and a gene set test result if applicable (camera).
-
18.
Import reference gene sets for gene set testing.
>Hallmark <- getGmt("h.all.v7.4.symbols.gmt") %>%
geneIds
>length(Hallmark)
[1] 50
>Hallmark$HALLMARK_PANCREAS_BETA_CELLS
[1] "PAX6" "NEUROD1" "ISL1" "NKX2-2" "PCSK1"
[6] "NKX6-1" "SLC2A2" "SEC11A" "DCX" "SPCS1"
[11] "FOXA2" "GCK" "MAFB" "INS" "PDX1"
[16] "ABCC8" "IAPP" "SRP9" "NEUROG3" "FOXO1"
[21] "AKT3" "GCG" "DPP4" "PAX4" "SYT13"
[26] "SCGN" "HNF1A" "STXBP1" "CHGA" "VDR"
[31] "PCSK2" "INSM1" "SST" "ELP4" "SRPRB"
[36] "PAK3" "G6PC2" "PKLR" "LMO2" "SRP14"
Note: For illustrative purpose, here we only use the Hallmark gene sets from MSigDB, which consist of 50 curated gene sets with cancer relevance (Liberzon et al., 2015) downloaded from www.gsea-msigdb.org/gsea/downloads.jsp - msigdb and saved to the working directory. This same code is applicable to any other gene sets from MSigDB.
Note: In theory, any reference gene panels of the same data structure (a list of named objects, each containing a vector of genes) can be used in placed of the “Hallmark” gene set generated from the above code.
CRITICAL: The type of gene identifiers used in the reference gene panels must also be present in the data to be analyzed. In our example, our transcriptome and proteome data contain HGNC symbols, thus the MSigDB gene sets consisting of HGNC symbols were used.
-
19.
Perform differential analysis using transcriptomic data.
Note: Here, differential expression analysis is performed to compare RNA expression between each subgroup to the average of the other subgroups. Since one subgroup was very different from the others (Subgroup 4, purple in Figure 3), it was not included in the analysis of the other three subgroups as reflected in the contrast matrix design.
>lm.r <- dge %>% {
dat = .
model.m <- model.matrix(∼0 + factor(Subgroup), dat$samples) %>%
set_colnames(paste0("Subgroup", 1:4))
contr.m <- makeContrasts(Subgroup1 = Subgroup1 - (Subgroup2 + Subgroup3)/2,
Subgroup2 = Subgroup2 - (Subgroup1 + Subgroup3)/2,
Subgroup3 = Subgroup3 - (Subgroup1 + Subgroup2)/2,
Subgroup4 = Subgroup4 - (Subgroup1 + Subgroup2 + Subgroup3)/3,
levels = colnames(model.m))
doLimma(dat, "rna", model.m = model.m, contr.m = contr.m, Hallmark)
}
Note: The model (model.m) and contrast (contr.m) matrices can be changed to suit users’ needs, depending on the experimental context and analysis objectives, such as if a different comparison is desired. A model matrix can be constructed using the model.matrix function where a formula and phenotype data should be supplied. In the above code chunk, the sample annotation dataframe from the RNA DGEList is used as the phenotype data and the “Subgroup” column from it used to define the groups of interest. A contrast matrix specifies the desired comparisons and can be constructed using the makeContrasts function. In the above example, four comparisons are made, one for each evaluation between a subgroup (Subgroup1∼4 preceding the minus sign) and others (Subgroups following the minus sign). Users interested in other comparisons may alter the formula accordingly. For instance, to compare Subgroup 4 to all other subgroups, one can use: “Subgroup4 = Subgroup4 – (Subgroup1 + Subgroup2 + Subgroup3)/3”.
-
20.
Perform differential analysis using proteomic data.
>lm.p <- eset %>% {
dat = .
model.m <- model.matrix(∼0 + factor(Subgroup) + factor(Batch),
pData(dat)) %>%
set_colnames(c(paste0("Subgroup", 1:4), paste0("TMT", 2:5)))
contr.m <- makeContrasts(Subgroup1 = Subgroup1 - (Subgroup2 + Subgroup3)/2,
Subgroup2 = Subgroup2 - (Subgroup1 + Subgroup3)/2,
Subgroup3 = Subgroup3 - (Subgroup1 + Subgroup2)/2,
Subgroup4 = Subgroup4 - (Subgroup1 + Subgroup2 + Subgroup3)/3,
levels = colnames(model.m))
doLimma(dat, "protein", model.m = model.m, contr.m = contr.m, Hallmark)
}
Note: The code for running differential protein abundance analysis is nearly identical to that used for differential mRNA expression analysis, with the addition of the batch covariate (TMT run grouping) in the design matrix to account for the batch effects associated with the different TMT runs. In experiments where confounding variables are known, users may replace the “Batch” covariate or add to the formula to account for more than one covariate that can adversely impact the analysis.
-
21.
Extract differential analysis results.
Note: Differential mRNA expression or protein abundance analysis results can be easily extracted using the topTable() function on the MArrayLM object outputted by limma. A set of significantly differential genes for each subgroup is derived by imposing expression/abundance and/or significance thresholds. The code below illustrates how to extract the significantly differential genes for each subgroup from the transcriptomic and proteomic data. As in our previous work (Yang et al., 2021), a significance threshold is set at adjusted p-value < 0.05, and log-fold change thresholds of 2 and 1 are set for mRNA and protein, respectively.
>deg <- paste0("Subgroup", 1:4) %>%
setNames(., .) %>%
as.list %>%
lapply(., function(x) topTable(lm.r$efit, coef = x, p.value = 0.05, lfc = 2, n = Inf))
>dap <- paste0("Subgroup", 1:4) %>%
setNames(., .) %>%
as.list %>%
lapply(., function(x) topTable(lm.p$efit, coef = x, p.value = 0.05, lfc = 1, n = Inf))
Note: The output from the above code is a Iist of data frames, each containing significantly differential genes for a subgroup from the transcriptomic (deg) or proteomic (dap) data. The number of significantly differential genes varies between subgroups and data as shown below.
>sapply(deg, nrow)
Subgroup1 Subgroup2 Subgroup3 Subgroup4
332 527 905 1124
>sapply(dap, nrow)
Subgroup1 Subgroup2 Subgroup3 Subgroup4
34 75 203 112
Note: The significance threshold here is imposed on the adjusted p-value, after correction for multiple comparisons. The default p-value adjustment method is the Benjamini-Hochberg procedure.
Note: To obtain the entirety of the differential analysis results, one can drop the p.value and lfc parameter.
Note: The gene set test results are structured as data frames with each row corresponding to a gene set and test statistics in columns. Below is a demonstration of the gene set test result from one of the subgroups.
>head(lm.r$camera$Subgroup4)
NGenes Direction
HALLMARK_E2F_TARGETS 197 Up
HALLMARK_G2M_CHECKPOINT 190 Up
HALLMARK_MYC_TARGETS_V1 195 Up
HALLMARK_INTERFERON_GAMMA_RESPONSE 193 Down
HALLMARK_MYC_TARGETS_V2 58 Up
HALLMARK_INTERFERON_ALPHA_RESPONSE 94 Down
PValue FDR
HALLMARK_E2F_TARGETS 1.979853e-47 9.899264e-46
HALLMARK_G2M_CHECKPOINT 2.665669e-33 6.664172e-32
HALLMARK_MYC_TARGETS_V1 2.409791e-17 4.016318e-16
HALLMARK_INTERFERON_GAMMA_RESPONSE 1.850506e-07 2.313132e-06
HALLMARK_MYC_TARGETS_V2 2.334552e-07 2.334552e-06
HALLMARK_INTERFERON_ALPHA_RESPONSE 5.432535e-07 4.527113e-06
Expected outcomes
Given paired transcriptomic and proteomic data, this protocol is designed to achieve three goals: identify molecular subgroups, compare variations between mRNA expression and protein abundance, and perform differential analysis between the identified subgroups. Steps are provided to apply appropriate normalization and transformation to RNA-seq or proteomic quantitative data (steps 1–6) for cluster and differential analyses. Consensus NMF analysis (steps 7–11) identifies robust clusters using either mRNA or protein data (Figures 3A and 3B). Using a common gene identifier, the correlations between mRNA expression and protein abundance are computed (steps 12–14) for each sample or for each gene (Figures 4A and 4B) to inform the degree of independence and interdependence between transcriptomic and proteomic landscapes. Differential analysis through limma (steps 15–21) identifies differentially expressed mRNAs and differentially abundant proteins for further investigation.
Limitations
This protocol relies on available transcriptomic and proteomic data from the samples of interest. While there are virtually no limitations to the types of research questions or samples for this protocol to work, the quality of the input data directly affects the quality of the result outputs. For data likely affected by confounding variables such as batch effects, a thoughtful experimental design is crucial to assess and mitigate such unwanted effects during the analysis and to thereby minimizing the impacts of the confounding variables on the results. For example, the inclusion of technical control samples in our proteomic profiling experiment enabled the discovery, assessment and mitigation of batch effects associated with TMT runs (Figures 1A and 1B). Failure to account for potential confounding variables in the experimental design may limit the reliability of the analysis results and the extent to which correction methods may aid with mitigating their effects. We therefore strongly recommend spending considerable effort on experimental design prior to commencing data generation and acquisition.
Troubleshooting
Problem 1
Encountering error messages when downloading or installing packages (Before you begin steps 1–3).
Potential solution
Error messages can often be encountered during package download and installation. Often, these errors are due to incompatibility between the R and package versions or missing package dependencies. While the most appropriate solution would depend on the actual errors encountered, updating R and/or the packages in questions while also enabling dependency installation/update can often resolve the errors. This approach, however, may sometimes trigger additional incompatibilities due to partially updated dependencies. The most straightforward solution for this problem may be to use the renv package (Ushey, 2021) to create a project-specific environment and use the BiocManager package (Morgan, 2021) to facilitate all package downloads and installations as described in step 2 of before you begin.
Problem 2
Encountering error messages when running executable codes throughout this protocol.
Potential solution
If steps from this protocol are followed properly, error messages encountered would likely be due to either inaccurate syntax or deviations in the input data structure(s). For the former, it is crucial to confirm proper and accurate placement of commas, brackets, parentheses, and quotation marks as well as correct spelling. For errors due to input data structure(s), users are advised to reformat their data to closely match those used in this protocol. Snapshots of input data are shown throughout the protocol.
Problem 3
More than one identifier is mapped to a feature when adding annotation(s) (steps 1 and 5).
Potential solution
Conversion of gene identifiers from omic data can almost always lead to multiple identifiers mapped to a feature. For example, an Ensembl gene identifier may have more than one HGNC symbol and Entrez ID due to various reasons such as gene splicing. A potential solution, as demonstrated in steps 1, 3, and 13 of the step-by-step method details, is to either treat the additional identifiers as new features and duplicate feature measurements or to simply use only one of the identifiers. Advantages and disadvantages of either approach would depend on the exact applications and downstream analysis, but such simplification is rarely detrimental to omic-scale analysis as only a marginal fraction of the feature pool is affected.
Problem 4
Too many or too few significantly differential genes (step 21).
Potential solution
The thresholds used to determine significantly differential genes are typically defined per experiment and should be set according to the particular context. While the significance threshold is most commonly set at 0.05 to allow a maximum of 5% error rate, the fold-change thresholds often vary between studies, and it is at the users’ discretion to decide on a reasonable threshold given their study objectives and the samples examined. Nevertheless, identification of significantly differential genes relies on the assumption that the groups compared are biologically different, and the number of significantly differential genes identified reflect the degree to which the compared groups differ.
Problem 5
Uncertainties surrounding the details and/or uses of a package or function throughout this protocol.
Potential solution
A well- developed and maintained package typically contains a vignette with elaborate information including usage examples. Users wishing to learn more about a specific package used in this protocol are encouraged to refer to the package vignette for additional information and package usage variations. The vignette for a package can be accessed through the package’s download link (listed in the key resources table) or within R using the function vignette(). For information on a particular function used, the help() function can be used within R to bring up the information page on the function.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sharon Gorski (sgorski@bcgsc.ca).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This study was supported by the 2017 Neuroendocrine Tumor Research Foundation-AACR Grant, Grant Number 17-60-33-GORS, the Pancreas Centre BC IDEAS Grant (2013–2015), and the Canadian Institutes of Health Research (CIHR) PJT-159536. The R logo used in the graphical abstract was adapted from https://www.r-project.org/logo/ under the CC-BY-SA 4.0 license.
Author contributions
Conceptualization, K.C.Y. and S.M.G.; Methodology, K.C.Y. and S.M.G.; Software, K.C.Y.; Validation, K.C.Y.; Formal Analysis, K.C.Y.; Investigation, K.C.Y.; Data Curation, K.C.Y.; Writing – Original Draft, K.C.Y. and S.M.G.; Writing – Review & Editing, K.C.Y., S.M.G.; Visualization, K.C.Y.; Supervision, S.M.G.; Funding Acquisition, S.M.G.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Kevin C. Yang, Email: kevyang@bcgsc.ca.
Sharon M. Gorski, Email: sgorski@bcgsc.ca.
Data and code availability
This study did not generate any new datasets or software. The datasets used for demonstration in this protocol were previously generated (Yang et al., 2021) and are available at the European Genome-phenome Archive (EGAS00001005024) and the Proteomics Identification Database (PXD024175). The codes generated and used in this study are presented throughout the text.
References
- Brenes A., Hukelmann J., Bensaddek D., Lamond A.I. Multibatch TMT reveals false positives, batch effects and missing values ∗. Mol. Cell Proteomics. 2019;18:1967–1980. doi: 10.1074/mcp.RA119.001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. [Software]; 2019. org.Hs.eg.db: Genome Wide Annotation for Human. [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M.J., Gillette M., Carr S.A., Paulovich A.G., Smith R.D., Rodland K.K., Townsend R.R., Kinsinger C., Mesri M., Rodriguez H., et al. Connecting genomic alterations to cancer biology with proteomics: the NCI clinical proteomic tumor analysis consortium. Cancer Discov. 2013;3:1108–1112. doi: 10.1158/2159-8290.CD-13-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R., Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M., Tang Y. [Software]; 2020. ggfortify: Data Visualization Tools for Statistical Analysis Results. [Google Scholar]
- Huber W., von Heydebreck A., Sueltmann H., Poustka A., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. {O}rchestrating high-throughput genomic analysis with {B}ioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kassambara A. [Software]; 2020. ggpubr: “ggplot2” Based Publication Ready Plots. [Google Scholar]
- Leek J.T., Scharpf R.B., Bravo H.C., Simcha D., Langmead B., Johnson W.E., Geman D., Baggerly K., Irizarry R.A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Fertig E.J., Jaffe A.E., Storey J.D., Zhang Y., Torres L.C. [Software]; 2019. sva: Surrogate Variable Analysis. [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures Database Hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. [Software]; 2021. BiocManager:: Access the Bioconductor Project Package Repository. [Google Scholar]
- Morgan M., Falcon S., Gentleman R. [Software]; 2019. GSEABase: Gene Set Enrichment Data Structures and Methods. [Google Scholar]
- Nesvizhskii A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning K., Fermin D., Nesvizhskii A.I. Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-Seq gene expression data. J. Proteome Res. 2012;11:2261–2271. doi: 10.1021/pr201052x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard V., Rødland E.A., Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17:29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . [Software]; 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . [Software]; 2018. RStudio: Integrated Development for R. [Google Scholar]
- Tang Y., Horikoshi M., Li W. ggfortify: unified interface to visualize statistical result of popular R packages. R. J. 2016;8:474. [Google Scholar]
- Ushey K. [Software]; 2021. renv: Project Environments. [Google Scholar]
- Välikangas T., Suomi T., Elo L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2018;19:1–11. doi: 10.1093/bib/bbw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu Q., Zhang B. Leveraging the complementary nature of RNA-Seq and shotgun proteomics data. Proteomics. 2014;14:2676–2687. doi: 10.1002/pmic.201400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M., Cancer Genome Atlas Research Network The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Reshaping data with the {reshape} package. J. Stat. Softw. 2007;21:1–20. [Google Scholar]
- Wickham H., Averick M., Bryan J., Chang W., McGowan L.D., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the {tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.C., Kalloger S.E., Aird J.J., Lee M.K., Rushton C., Mungall K.L., Mungall A.J., Gao D., Xu J., Karasinska J.M., et al. Proteotranscriptomic classification and characterization of pancreatic neuroendocrine neoplasms. Cell Rep. 2021;37:109817. doi: 10.1016/j.celrep.2021.109817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any new datasets or software. The datasets used for demonstration in this protocol were previously generated (Yang et al., 2021) and are available at the European Genome-phenome Archive (EGAS00001005024) and the Proteomics Identification Database (PXD024175). The codes generated and used in this study are presented throughout the text.