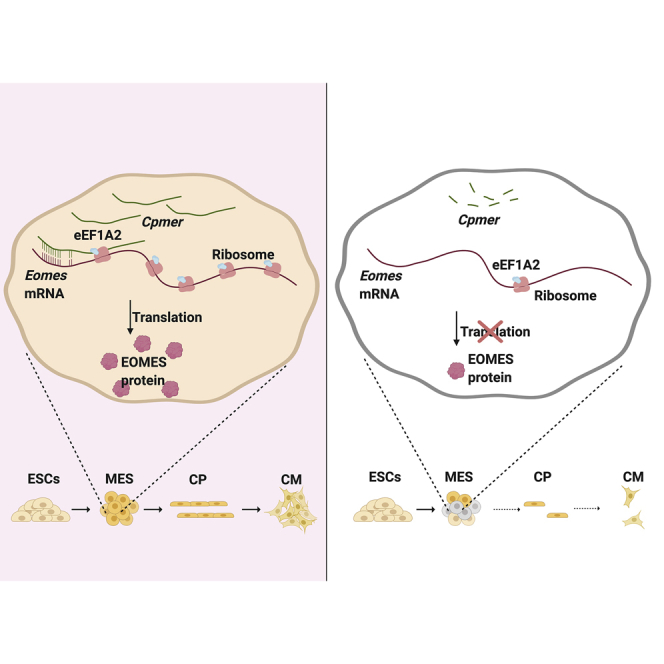
Summary
Previous studies have shown that eukaryotic elongation factor 1A2 (eEF1A2) serves as an essential heart-specific translation elongation element and that its mutation or knockout delays heart development and causes congenital heart disease and death among species. However, the function and regulatory mechanisms of eEF1A2 in mammalian heart development remain largely unknown. Here we identified the long noncoding RNA (lncRNA) Cpmer (cytoplasmic mesoderm regulator), which interacted with eEF1A2 to co-regulate differentiation of mouse and human embryonic stem cell-derived cardiomyocytes. Mechanistically, Cpmer specifically recognized Eomes mRNA by RNA-RNA pairing and facilitated binding of eEF1A2 with Eomes mRNA, guaranteeing Eomes mRNA translation and cardiomyocyte differentiation. Our data reveal a novel functionally conserved lncRNA that can specifically regulate Eomes translation and cardiomyocyte differentiation, which broadens our understanding of the mechanism of lncRNA involvement in the subtle translational regulation of eEF1A2 during mammalian heart development.
Keywords: cardiomyocyte differentiation, eEF1A2, lncRNA, mRNA translation, Eomes
Graphical abstract
Highlights
-
•
Cpmer promotes cardiomyocyte differentiation in mouse and human
-
•
Cytoplasmic Cpmer binds to heart-specific translation elongation factor eEF1A2
-
•
Cpmer specifically recognizes the mRNAs of Eomes and promote its translation
-
•
Cpmer plays dual roles of mRNA pairing and protein binding for Eomes translation
Lyu et al. demonstrated a novel translation regulation mechanism of the cytoplastic lncRNA Cpmer, which promotes cardiomyocyte differentiation in mouse and human. Cpmer specifically recognized the mRNAs of the key mesoderm gene Eomes to promote its translation by partnering with eEF1A2. These findings provide a new dimension of epigenetic regulation specificity in the research field of mRNA translation.
Introduction
Mammalian heart development is a multistep process, with multiple factors and pathways acting in concert (Murry and Keller, 2008). Proper expression of key protein-coding genes at the indicated stages is a prerequisite for heart development (Chang and Bruneau, 2012; Moore-Morris et al., 2018). Previous studies have revealed that the process of transcription of key protein-coding genes is strictly regulated during heart development. Post-transcriptional regulation of key genes, which is also essential for proteins to perform their biological functions, has not been fully investigated, especially in mammalian heart development (Simpson et al., 2020). eEF1A (consisting of eEF1A1 and eEF1A2) plays an important role in regulating translation elongation. The eEF1A1 paralog is widely expressed, but the eEF1A2 paralog is specifically expressed in the heart and brain (Liu et al., 2019). Mutation or knockout of Eef1a2 has been shown to cause delayed heart development, congenital heart disease, and death in zebrafish, suggesting an important role of eEF1A2 in early heart development of zebrafish (Cao et al., 2017). eEF1A2 is highly conserved among species, but so far, the function and specific regulatory mechanisms of eEF1A2 in mammalian heart development remain largely unknown.
Long noncoding RNAs (lncRNAs), as tissue- and stage-specific epigenetic regulators, have attracted extensive attention for their roles in various biological processes, including heart development (Gibb et al., 2011; Wilusz et al., 2009). Previous studies have shown that Uph2 binds directly to the enhancer of Hand2 to control its transcription and right ventricular development in mice (Anderson et al., 2016). yylncT promotes human embryonic stem cells (hESCs) to differentiate into mesoderm by inhibiting DNMT3B and methylation at the T gene promoter (Frank et al., 2019). Recent studies have also found that a cytoplasmically distributed lncRNA, Arin, regulates translation of Igf2bp2 mRNA, affecting cardiomyocyte (CM) survival (Hosen et al., 2018). However, it is still necessary to investigate how lncRNAs function in translational regulation of mRNAs. Whether the heart-expressed translation elongation factor eEF1A2 acts together with specific lncRNAs and precisely regulates translation of key genes in CM differentiation has not been reported.
Eomesodermin (Eomes) is an important T-box transcription factor that is expressed in the primitive streak (embryonic day 6.5 [E6.5]–E7.5) of mammalian development and serves as a marker of mesoderm formation (Tosic et al., 2019). Deletion or insufficient expression of Eomes causes defects in mesoderm formation and heart development, eventually resulting in death. EOMES activates transcription of Mesp1, a key step in promoting specification of cardiac mesoderm and cardiac lineage commitment (Costello et al., 2011). Previous studies have found that Wnt (wingless/integrated) signaling can directly activate Eomes transcription to promote mesendodermal differentiation, and Eomes can also be regulated by Nodal/Smad signaling to coactivate downstream cardiac mesoderm genes (Costello et al., 2011; Teo et al., 2011; Tosic et al., 2019). lncRNAs also regulate CM differentiation in concert with EOMES. Meteor, transcribed from the enhancer region of the Eomes gene, regulates transcription of Eomes in CM differentiation (Alexanian et al., 2017). Linc1405 (large intergenic non-coding RNA 1405) binds with EOMES and guides specific activation of EOMES at the enhancer region of the Mesp1 gene (Guo et al., 2018). However, the specific mechanism that regulates mRNA translation of Eomes has not been reported.
In this study, we identified a new lncRNA (cytoplasmic mesoderm regulator [Cpmer]) that could interact with the translation elongation factor eEF1A2 and specifically recognize Eomes mRNA sequences, contributing to proper translation of Eomes mRNA and CM differentiation. We observed that the human ortholog CPMER had consistent effects on eEF1A2 binding and translational regulation of human EOMES mRNA.
Results
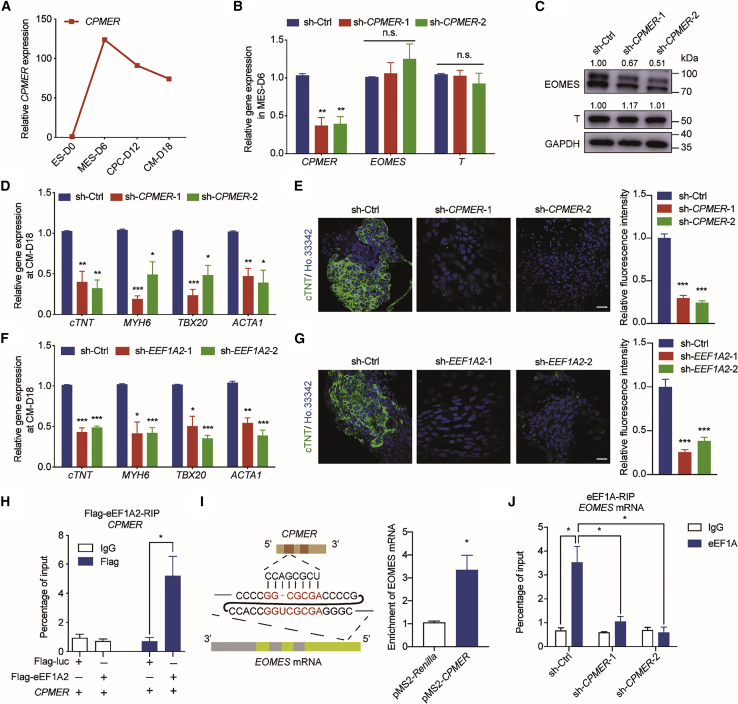
Cpmer is a potentially important cytoplasmic lncRNA in CM differentiation
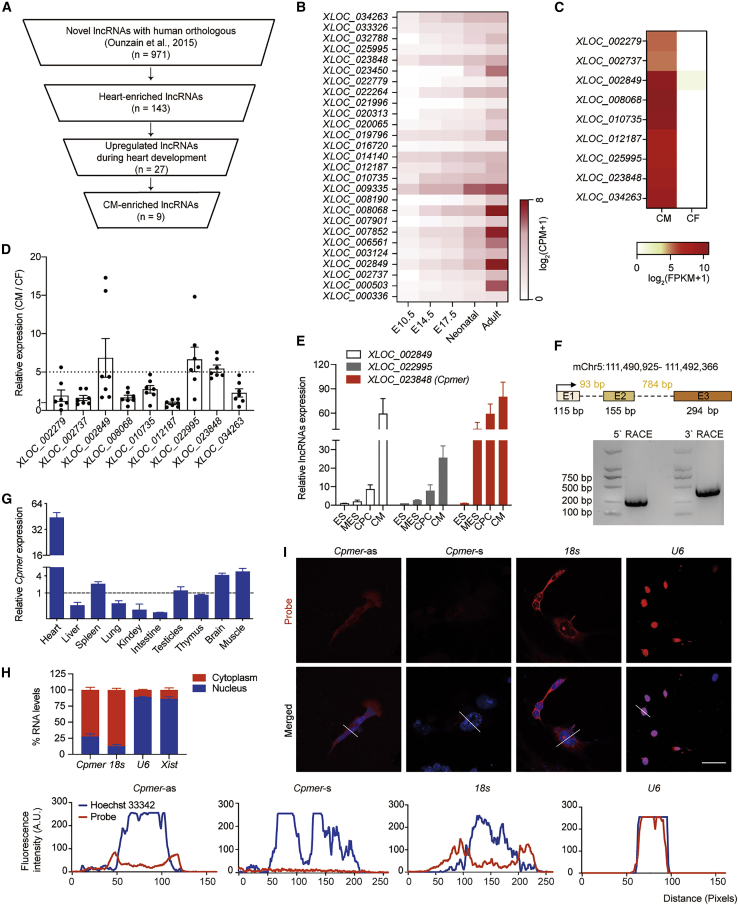
To investigate the function of unknown lncRNAs in regulating CM differentiation, we analyzed RNA sequencing (RNA-seq) data that had been used to identify a set of novel lncRNAs in the murine heart and focused on novel transcripts with putative human orthologs; we narrowed it down to 971 transcripts (Ounzain et al., 2015). Then we analyzed differentially expressed transcripts on the indicated days (E10.5, E14.5, E17.5, neonatal, and adult) of mouse heart development (GSE158202) and identified 143 transcripts specifically enriched in the heart, 27 of which were positively correlated with the heart development process (Figures 1A and 1B; Table S1). 9 of the candidate lncRNAs were found to be enriched in CMs (Mullin et al., 2017; Figure 1C), and qRT-PCR results confirmed that 3 (XLOC_002489, XLOC_022995, and XLOC_023848) of these 9 lncRNAs were expressed at 5-fold higher levels in CMs than in cardiac fibroblasts (CFs) (Figure 1D). The expression levels of these 3 lncRNAs were quantified during CM differentiation, and we found that expression of XLOC_023848, hereafter called Cpmer (GenBank: OL365371), was significantly upregulated at the mesoderm stage, suggesting that Cpmer might be important in regulating CM differentiation (Figure 1E). Cpmer was located at mm10 chromosome 5 (111,490,925–111,492,366) and consisted of three exons near the protein-coding gene Mn1. Rapid amplification of cDNA ends (RACE) was employed to amplify the full-length Cpmer transcript (Figure 1F). Cpmer was specifically expressed in the heart of mice compared with other fetal tissue (Figure 1G). Coding potential analysis of Cpmer indicated that it had low coding potential, similar to Xist (Figure S1A). We also analyzed the ribosome profiling sequencing (Ribo-seq) data of mesoderm cells (GSE86467; Fujii et al., 2017) and found that there were no mapping reads at the genomic location of Cpmer, suggesting that the ribosome was not occupied on Cpmer. These results confirmed that Cpmer did not have the potential for protein coding (Figure S1B). Nuclear/cytoplasmic fractionation analysis and RNA fluorescence in situ hybridization (FISH) confirmed the cytoplasm distribution of Cpmer in mesoderm cells (Figures 1H and 1I). These data suggest that Cpmer may play a functional role in regulating cardiac differentiation in the cytoplasm.
Figure 1.
Cpmer is a potentially important cytoplasmic lncRNA in CM differentiation
(A) Strategy for selecting novel lncRNAs related to heart development.
(B) The expression pattern of selected lncRNAs on the indicated days of mouse development. CPM, counts per million.
(C) Heatmap of the lncRNAs significantly enriched in CMs compared with cardiac fibroblasts (CFs). FPKM, fragments per kilobase of exon model per million mapped fragments.
(D) Validation of lncRNA expression in CMs versus CFs. Data shown are mean ± SEM, n = 7 mice.
(E) The expression pattern of candidate lncRNAs during CM differentiation. MES, mesoderm cells. Data shown are mean ± SEM, n = 3 independent experiments.
(F) Detection of 5′ and 3′ termini of Cpmer by RACE.
(G) Histograms showing the relative expression levels of Cpmer in neonatal mouse tissues. The mean of expression levels of Cpmer in the listed tissues was taken as 1 (dashed line). Data represent mean ± SEM, n = 3 mice.
(H) qRT-PCR analysis of the RNAs (Cpmer, 18s, U6, and Xist) derived from the cytosolic or nuclear fractions of MESs. Data shown are mean ± SEM, n = 3 independent experiments.
(I) RNA FISH assay showing the representative images of the Cpmer-antisense (Cpmer-as) probe targeting Cpmer in MESs (top). The Cpmer-sense (Cpmer-s) probe served as a negative control. U6, positive control for nuclear RNA; 18s, positive control for cytoplasmic RNA. Nuclei were stained with Hoechst 33342 (blue). Scale bar, 20 μm. The variation patterns of the RNA probe and nuclear signals were analyzed (bottom).
Cpmer is critically required for proper CM differentiation
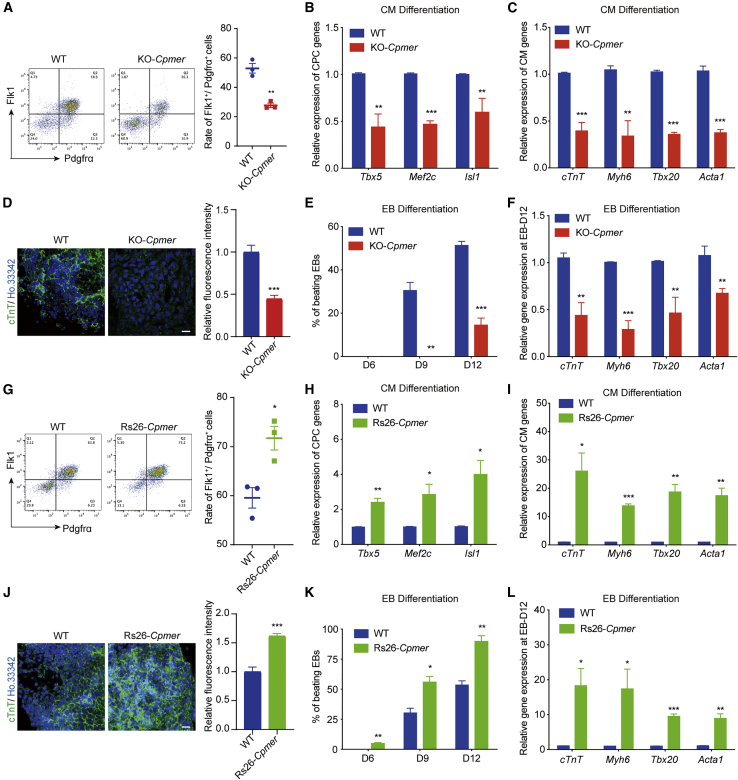
Next, to explore the role of Cpmer in CM differentiation, we constructed Cpmer knockout (KO-Cpmer) mouse embryonic stem cell (mESC) 46C cell line using CRISPR-Cas9 technology (Figure S2A). Our data showed that KO of Cpmer did not affect the expression level of pluripotency genes or of its neighbor gene Mn1 (Figures S2B–S2D) but significantly reduced the percentage of Flk1+/Pdgfrα+ cells that appeared at the mesoderm stage (Figure 2A). Moreover, cardiac progenitor cell (CPC) marker genes (Tbx5, Gata4, and Mef2c) and CM marker genes (cTnT, Myh6, Tbx20, and Acta1) exhibited lower expression levels in KO-Cpmer cells (Figures 2B and 2C). KO of Cpmer resulted in a lower proportion of cTnT+ CMs than wild-type (WT) cells (Figure 2D). Similarly, KO of Cpmer led to a delay in spontaneous contraction compared with WT cells (spontaneous contraction appeared on day 8, and the contraction rate was about 30% on day 9 and 60%–70% on day 12) (Figure 2E). Expression of cardiac genes was significantly decreased in KO-Cpmer cells on day 12 of embryoid body (EB) differentiation (Figure 2F). KO of Cpmer did not affect the neuroectoderm and definitive endoderm differentiation but resulted in upregulated expression of marker genes of CFs and endothelial cells and decreased expression of smooth-muscle-related genes (Figures S2E and S2F).
Figure 2.
Cpmer is critically required for proper CM differentiation
(A) FACS analysis and statistics for the percentage of Flk1+Pdgfrα+ cells. Data shown are the mean ± SEM, n = 3 independent experiments.
(B and C) qRT-PCR analysis of CPC (B) and CM (C) marker genes. Data shown are the mean ± SEM, n = 3 independent experiments.
(D) cTnT immunostaining (green) at the CM stage (left) and statistics for relative fluorescence intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM, n = 6 fields from 3 independent experiments.
(E) Percentage of spontaneously contracting EBs determined on days 6, 9, and 12 of EB differentiation. Data shown are the mean ± SEM, n = 3 independent experiments.
(F) qRT-PCR analysis of CM marker genes on day 12 of EB differentiation (EB-day 12). Data shown are the mean ± SEM, n = 3 independent experiments.
(G) FACS analysis and statistics of the percentage of Flk1+Pdgfrα+ cells. Data shown are the mean ± SEM, n = 3 independent experiments.
(H and I) qRT-PCR analysis of CPC (H) and CM (I) marker genes. Data shown are the mean ± SEM, n = 3 independent experiments.
(J) cTnT immunostaining (green) at the CM stage (left) and statistics for relative fluorescence intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM, n = 6 fields from 3 independent experiments.
(K) Percentage of spontaneously contracting EBs of EB differentiation. Data shown are the mean ± SEM, n = 3 independent experiments.
(L) qRT-PCR analysis of CM marker genes at EB-day 12. Data shown are the mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (versus WT); Student’s t test.
See also Figure S2.
We then inserted the full-length Cpmer sequence into the ROSA26 locus (used for constitutive gene expression) to increase Cpmer expression (Rs26-Cpmer) (Figure S2G). Overexpression of Cpmer had no effect on expression of pluripotency genes and the neighbor gene Mn1 (Figures S2H–S2J). Fluorescence-activated cell sorting (FACS) assays showed that Cpmer overexpression significantly increased the percentage of Flk1+/Pdgfrα+ mesoderm cells (Figure 2G), along with increased expression of CPC and CM marker genes (Figures 2H and 2I) and ratio of cTnT+ cells in Rs26-Cpmer cells (Figure 2J). Spontaneously contracted EBs appeared earlier in Cpmer-overexpressing cells, on day 6, along with an increased beating ratio and cardiac marker expression (Figures 2K and 2L). In contrast, the expression levels of CF and endothelial cell markers were attenuated in Rs26-Cpmer cells compared with WT cells (Figure S2K). These results indicate that Cpmer plays a critical role during proper CM differentiation.
eEF1A2 is a Cpmer-binding protein and similarly regulates CM differentiation
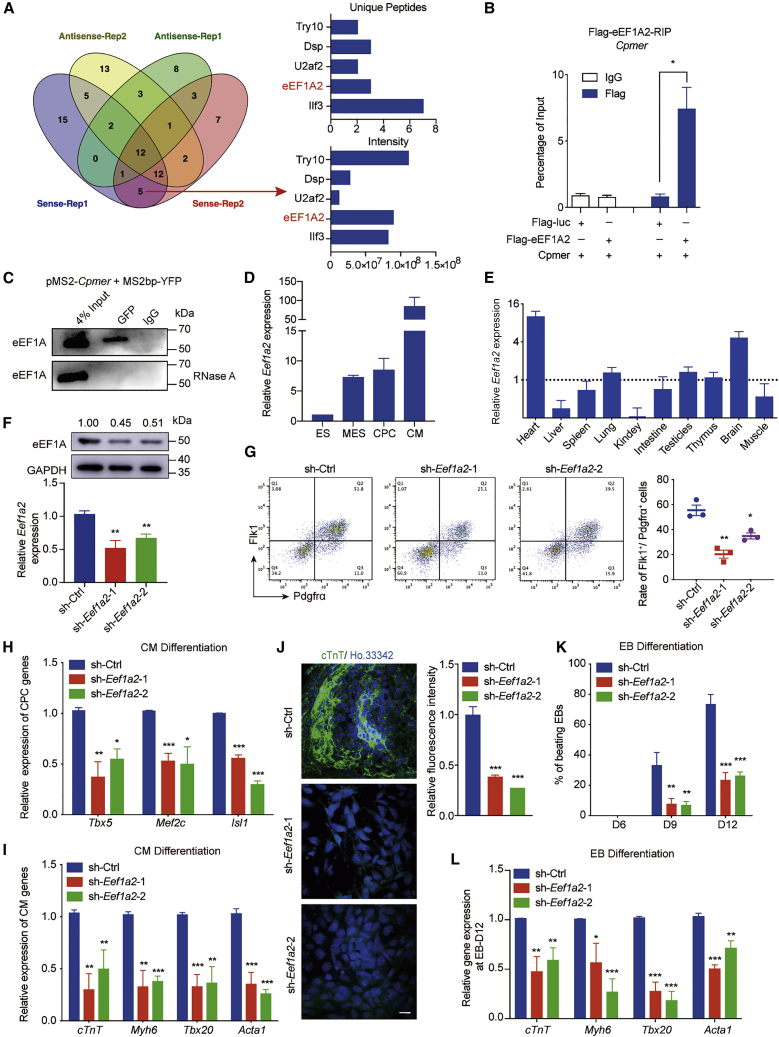
Because cytoplasmic lncRNAs usually interact with RNA-binding proteins to execute their cellular functions, we performed an RNA pull-down assay followed by mass spectrometry analysis to identify the interactome of Cpmer (Figure 3A; Table S2). Among the potential interaction proteins of Cpmer, eEF1A2, usually expressed in terminally differentiated cells (i.e., CMs and nerve cells), exhibited a remarkable interaction intensity with Cpmer (Figure 3A). Binding of Cpmer and FLAG-eEF1A2 was confirmed by exogenous RNA immunoprecipitation (RIP) experiments (Figure 3B). An endogenous RNA pull-down assay mediated by MS2bp-YFP (yellow fluorescent protein) also confirmed that Cpmer interacted with eEF1A protein in mesoderm cells (unable to distinguish eEF1A2 from eEF1A1 with an anti-eEF1A antibody) (Figure 3C). Similar to Cpmer expression, Eef1a2 expression was significantly increased during CM differentiation and was specifically detected in the heart of fetal mice (Figures 3D and 3E).
Figure 3.
eEF1A2 is a Cpmer-binding protein and similarly regulates CM differentiation
(A) MS to identify the potential Cpmer-binding proteins (left). The unique peptides and intensities of 5 candidate proteins are shown (right).
(B) RIP analysis for exogenous expression of Cpmer and FLAG-eEF1A2 in 293T cells, determined by an anti-FLAG antibody. Data shown are the mean ± SEM, n = 3 independent experiments.
(C) MS2bp-YFP RNA pull-down analysis for the co-transfected pMS2-Cpmer and MS2bp-YFP in MESs with or without RNase A treatment. An anti-GFP antibody was used to recognize the YFP protein.
(D) The expression pattern of Eef1a2. Data shown are the mean ± SEM, n = 3 independent experiments.
(E) The relative expression levels of Eef1a2 in neonatal mouse tissues. Data shown are the mean ± SEM, n = 3 mice.
(F) Detection of mRNA and protein expression levels of Eef1a2 at the MES stage after Eef1a2 knockdown. Data shown are the mean ± SEM, n = 3 independent experiments.
(G) FACS analysis and statistics of the percentage of Flk1+Pdgfrα+ cells. Data shown are the mean ± SEM, n = 3 independent experiments.
(H and I) qRT-PCR analysis of CPC (H) and CM (I) marker genes. Data shown are the mean ± SEM, n = 3 independent experiments.
(J) cTnT immunostaining (green) at the CM stage (left) and statistics for relative fluorescence intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM, n = 6 fields from 3 independent experiments.
(K) Percentage of spontaneously contracting EBs of EB differentiation. Data shown are the mean ± SEM, n = 3 independent experiments.
(L) qRT-PCR analysis of the CM marker genes. Data shown are the mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (versus sh-Ctrl); Student’s t test.
See also Table S2.
We constructed Eef1a2 knockdown mESC lines by short hairpin RNA (shRNA) viruses, which were confirmed at the mRNA and protein levels (Figure 3F). Our findings showed that Eef1a2 knockdown led to a significant decrease in the percentage of Flk1+/Pdgfrα+ cells and the mRNA expression levels of CPC and CM marker genes compared with control group (sh-Ctrl) (Figures 3G–3I). Immunofluorescence analysis confirmed a significant reduction in cTnT+ cells after Eef1a2 knockdown (Figure 3J). The results of EB differentiation confirmed that Eef1a2 knockdown resulted in a decreased percentage of beating EBs and expression of CM marker genes (Figures 3K and 3L). These results indicate that the translation elongation factor eEF1A2 serves as a potential partner of the lncRNA Cpmer in CM differentiation.
Cpmer/eEF1A2 recognizes the mRNA of the mesoderm gene Eomes and regulates its translation
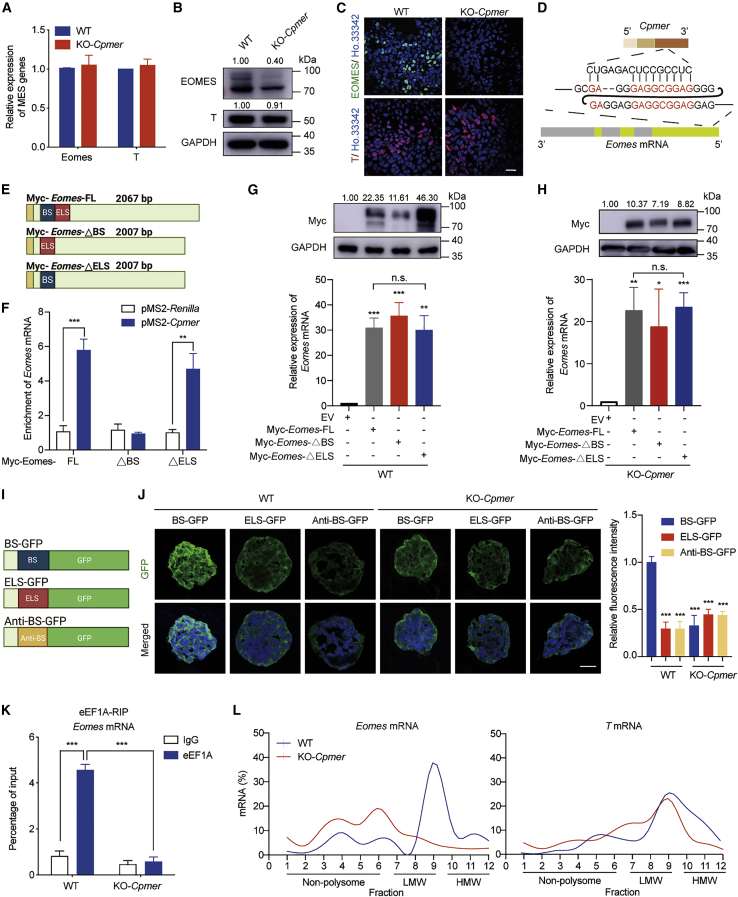
We sought to investigate the mechanism by which Cpmer/eEF1A2 regulates CM differentiation and analyzed the potential mRNAs (791 genes) that theoretically interacted with Cpmer via RNA pairing by RIblast (Table S3; Fukunaga and Hamada, 2017). 3 genes (Eomes, Bmp4, and Lef1) were found when overlapping with genes associated with mesoderm formation (GO_ID: 00010707) (Figure S3A). Among these genes, Eomes was thought to be a well-known transcription factor in the mesoderm and significantly increased at the mesoderm stage (Figure S3B). We confirmed that knockdown of Eomes significantly reduced the percentage of spontaneously beating EBs and decreased the expression levels of CM markers on day 12 of EB differentiation (Figures S3C–S3E). Bone morphogenetic protein 4 (Bmp4), as a secreted ligand of the transforming growth factor β (TGF-β) superfamily, mainly functions in differentiation of pluripotent stem cells (PSCs) into the mesodermal instead of the ectodermal lineage (Johansson and Wiles, 1995; Monteiro et al., 2004). Lef1, as a response molecule of Wnt/β-catenin signaling, has been reported to be associated with neurogenesis and T cell differentiation, whereas deletion of Lef1 did not affect cardiogenesis (Galceran et al., 1999; Oosterwegel et al., 1993; Xing et al., 2019). Previous studies have reported that BMP4 treatment can upregulate expression of Eomes and T (another key mesoderm transcription factor), whereas deficiency of Bmp4 resulted in significant downregulation of Eomes and T in embryos (Amita et al., 2013; Soares et al., 2005), suggesting that BMP4 might serve as the upstream regulator of the Eomes and T genes. Our findings showed that the mRNA expression levels of Eomes and T were not influenced by Cpmer (Figure 4A), indicating that Bmp4 was not a target of Cpmer. The protein level of EOMES, but not T, was significantly decreased in KO-Cpmer mesoderm cells (Figures 4B and 4C). Thus, we speculated that Cpmer might affect the protein level of EOMES at the mesoderm stage.
Figure 4.
Cpmer/eEF1A2 recognizes mRNA of the mesoderm gene Eomes and regulates its translation
(A–C) qRT-PCR (A), western blot (B), and immunofluorescence (C) detection of the expression of representative mesodermal marker genes (Eomes and T). Data shown are the mean ± SEM, n = 3 independent experiments. Scale bar, 20 μm.
(D) Regions of potential recognition between Cpmer and mouse Eomes mRNA.
(E) Schematic of the FL and deletion mutants of Eomes mRNA. BS, blue; ELS, red; Myc tag, yellow.
(F) MS2bp-YFP RNA pull-down analysis for exogenous expression of Cpmer with FL or deletion mutant Eomes mRNA in 293T cells. pMS2-Renilla RNA was used as a negative control. The fold enrichment relative to the immunoglobulin G (IgG) control is shown. Data shown are the mean ± SEM, n = 3 independent experiments.
(G and H) qRT-PCR and western blot analysis of Eomes mRNA and protein levels in WT (G) or KO-Cpmer (H) cells transfected with FL or deletion mutant Eomes. Data shown are the mean ± SEM, n = 3 independent experiments.
(I) Schematic of the GFP reporters. BS, blue; ELS, red; anti-BS, yellow.
(J) GFP signal analysis at EB-day 4 in cells transfected with distinct GFP reporters (left) and statistics for relative GFP signal intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM (n = 6 fields from 3 independent experiments).
(K) RIP analysis of the interaction of eEF1A with Eomes mRNA. Data shown are the mean ± SEM, n = 3 independent experiments.
(L) Percentage of Eomes and T mRNAs in the gradient total RNA, as measured by qRT-PCR in each fraction collected from the polysome profiling assay. LMW, low molecular weight; HMW, high molecular weight. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., not significant; Student’s t test.
See also Figures S3 and S4 and Table S3.
Analysis of the potential interaction of Cpmer and Eomes mRNA indicated RNA pairing between the third exon of Cpmer and the coding sequence (CDS) region of Eomes (95–125 nt) (Figure 4D). To demonstrate the potential binding of Cpmer and the Eomes mRNA, we constructed Eomes full-length (Myc-Eomes-FL), Myc-Eomes-ΔBS (deletion of the predicted binding sequences [BSs]), and Myc-Eomes-ΔELS (deletion of equal-length sequences [ELSs]) overexpression vectors (Figure 4E). The exogenous RNA pull-down assay showed that the Eomes mRNA interacted with Cpmer via the predicted BS (Figure 4F). We then studied whether the interaction between Cpmer and the Eomes mRNA affected expression of the EOMES protein. When transfecting FL and deletion mutant Eomes into WT cells, our results showed that the protein level of exogenous Eomes-ΔBS was significantly lower than that of FL Eomes and Eomes-ΔELS (Figure 4G). Similarly, deficiency of Cpmer impaired the protein expression level of FL Eomes or its deletion mutants but not the mRNA level (Figure 4H). The expression level of EOMES protein remained quite low in KO-Cpmer cells compared with WT cells with or without transfection of FL Eomes (Figure S3F). We constructed GFP reporters by separately inserting the BS of Eomes mRNA and Cpmer (BS), the ELS (as a scramble sequence), and the antisense BS (anti-BS) upstream of the GFP gene and then introduced these GFP reporters into the WT and KO-Cpmer cell lines (Figure 4I). The fluorescence intensity of BS-GFP was significantly stronger than that of ELS-GFP or anti-BS-GFP in WT mesoderm cells, suggesting that the BS of Eomes mRNA with Cpmer facilitated GFP mRNA translation. The fluorescence intensity of all GFP reporters was extremely low in KO-Cpmer mesoderm cells, indicating that Cpmer was required for proper GFP mRNA translation (Figure 4J). Considering that the effect of Cpmer KO occurred at the post-transcriptional level, we then treated mesoderm cells with actinomycin D or MG132 to detect mRNA stability or protein degradation. KO of Cpmer did not affect the stability of Eomes and T mRNAs or degradation of EOMES or T protein (Figures S3G and S3H). These results indicated that the direct RNA-RNA pairing of Cpmer and Eomes greatly affects Eomes mRNA translation.
The RIP assay showed that eEF1A interacted with Eomes mRNA in WT cells but not in KO-Cpmer cells (Figure 4K). A previous study has shown that eEF1A2 directly interacts with the Utrn mRNA (Miura et al., 2010). Enrichment of eEF1A2 on Utrn mRNAs was not affected by Cpmer KO, suggesting that Cpmer mediated the specific binding of eEF1A2 and the Eomes mRNA (Figure S3I). KO of Cpmer did not affect expression of Eef1a2 or other translation-related elements (Eef1a1, Eif2a, Rpl3, and Rpsa) (Figure S3J), suggesting that Cpmer might act as a scaffold for proper binding of eEF1A2 and the Eomes RNA. A polysome profile assay followed by qRT-PCR showed that loss of Cpmer resulted in low enrichment of the polysome fraction (fractions 7–12) on Eomes mRNA but not T (Figures 4L and S3K), suggesting specificity of Cpmer-mediated Eomes translation. Distribution of eEF1A in the polysome fractions showed no significant change in WT and KO-Cpmer cells, which suggested that eEF1A recruitment to ribosomal complexes was independent of Cpmer (Figure S3L). Consistent with the finding that eEF1A2 was mainly involved in translational regulation of genes, we concluded that Cpmer recruited eEF1A2 on Eomes mRNA by RNA-RNA recognition, regulating its translation.
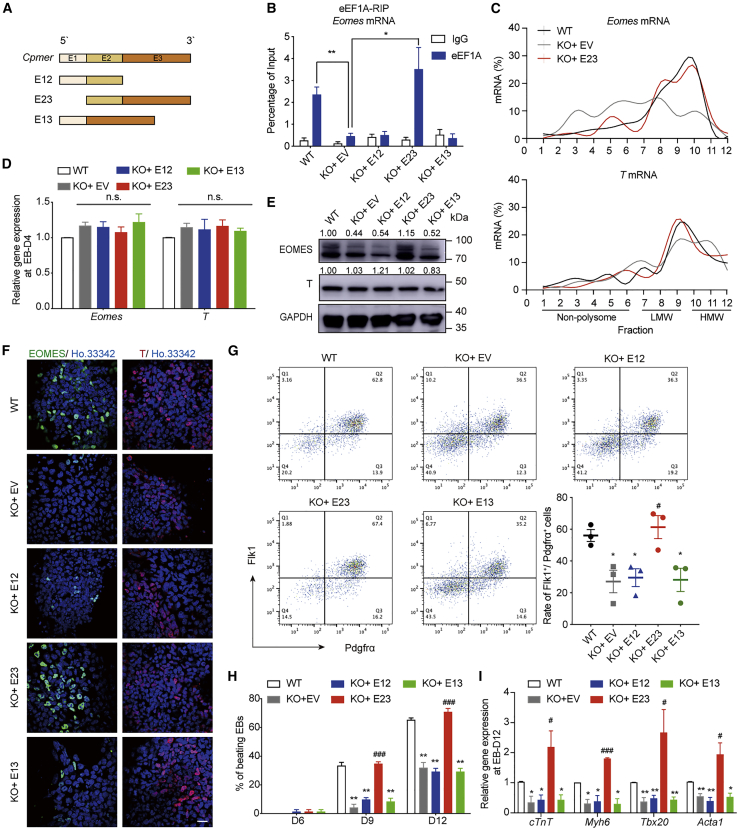
E23 of Cpmer is responsible for Eomes translation and CM differentiation
We then constructed plasmids overexpressing Cpmer fragments (E1, E2, and E3) (Figure S4A), and only the second fragment of Cpmer (E2) interacted with the eEF1A2 protein (Figures S4B and S4C). Overexpression of an individual Cpmer fragment neither affected the mRNA levels of Eomes and T at the mesoderm stage (Figure S4D) nor rescued the protein level of EOMES (Figure S5E) or the ratio of spontaneously beating EBs (Figure S4F). We then constructed overexpression plasmids containing two exons of Cpmer (E12, E23, and E13) (Figure 5A) and found that only the Cpmer mutant E23 could recover enrichment of the eEF1A2 protein on Eomes mRNA (Figure 5B). Accordingly, overexpression of E23 rescued polysome enrichment and EOMES protein expression in KO-Cpmer cells, whereas the other two Cpmer mutants (E12 and E13) were not able to rescue expression of the EOMES protein (Figures 5C–5F). FACS assays also showed that the ratio of Flk1+/Pdgfrα+ cells, the ratio of beating EBs, and CM marker expression could be rescued only by E23 overexpression (Figures 5G–5I). We also analyzed the RNA structure of Cpmer by RNAfold, and these results showed that FL and E23 of Cpmer contained unique stem-loop structures with low positional entropy (Figures S5G and S5H). Neither E1 nor E13 of Cpmer contained a similar structure (Figures S5I and S5J), which suggested that the stem-loop structure of Cpmer might be considered its protein-binding characteristic. Combined with the finding that Cpmer bound to the eEF1A protein and Eomes mRNA through the distinct RNA fragments E2 and E3, respectively, these data indicate that Cpmer establishes a link between the eEF1A2 protein and Eomes mRNA, which ensures Eomes translation and proper cardiac differentiation.
Figure 5.
E23 of Cpmer is responsible for Eomes translation and CM differentiation
(A) Schematic of Cpmer mutants.
(B) RIP analysis of eEF1A binding with Eomes mRNA in KO-Cpmer cells transfected with Cpmer mutants. Data shown are the mean ± SEM, n = 3 independent experiments.
(C) Polysome profile analysis of Eomes and T mRNAs.
(D–F) qRT-PCR (D), western blot (E), and immunofluorescence (F) detection of expression of Eomes and T. Data shown are the mean ± SEM, n = 3 independent experiments. Scale bar, 20 μm.
(G) Representative FACS results and the statistics of percentage of Flk1+Pdgfrα+ cells. Data shown are the mean ± SEM, n = 3 independent experiments.
(H) Percentage of spontaneously contracting EBs. Data shown are the mean ± SEM, n = 3 independent experiments.
(I) qRT-PCR analysis of the CM marker genes. Data shown are the mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01 (versus the WT); #p < 0.05 and ###p < 0.001 (versus KO + EV [empty vector]); Student’s t test.
See also Figure S5.
Because Cpmer levels kept rising during CM differentiation and sustained high expression in CMs, but the expression level of Eomes was significantly increased at the mesoderm stage and decreased rapidly after mesoderm differentiation, we were curious about the roles of Cpmer after mesoderm differentiation. Surprisingly, it was found that there were 33 mRNAs overlapped with potentially recognized mRNAs and genes associated with heart development (GO_ID: 0007507), including Tbx18 and Tbx20 (CPC transcription factors) and cTnT (a CM marker) (Figure S5A). We effectively knocked down Cpmer by shRNA virus after the mesoderm stage, and our results showed that knockdown of Cpmer after the mesoderm stage also resulted in lower expression levels of CM marker genes compared with the control (Figures S5B and S5C), as well as the lower proportion of cTnT+ CMs (Figure S5D). Hence, we believed that Cpmer might target distinct mRNAs during CM differentiation and that its recognition and regulation of Eomes mRNA at the mesoderm stage was critical for proper CM differentiation.
CPMER/eEF1A2 conservatively functions in EOMES translation and human CM differentiation
CPMER (ENST00000440255.1) was identified as a putative ortholog of Cpmer in the human genome (hg38 chromosome 22: 27,676,559–27,714,970). We first detected the stage-specific makers of hESC-derived CM differentiation (Figure S6A) and found that expression of CPMER was similarly upregulated at the mesoderm stage (Figure 6A). Next, two CPMER knockdown cell lines were constructed, and knockdown of CPMER significantly inhibited expression of the EOMES protein but not EOMES mRNA (Figures 6B and 6C). CPMER deficiency significantly reduced the differentiation efficiency of hESCs toward CMs (Figures 6D and 6E). Expression of EEF1A2 was similarly induced at the mesoderm stage and increased along with human CM differentiation (Figure S6B). Knockdown of EEF1A2 in hESCs significantly reduced the expression levels of key CM marker genes and the percentage of cTNT+ cells compared with the control group (Figures 6F and 6G). Mechanistically, an exogenous RIP assay was employed to confirm the interaction of human CPMER and eEF1A2 (Figure 6H). We considered that the RNA pairing was similar between CPMER and EOMES mRNA (94–118 nt) (Figure 6I, left), which was verified by exogenous RNA pull-down assay (Figure 6I, right). Endogenous RIP results showed that enrichment of eEF1A on EOMES mRNAs was significantly reduced after CPMER knockdown (Figure 6J). Our data highlight that CPMER and eEF1A2 play a conserved role in regulation of EOMES mRNA translation and human CM differentiation.
Figure 6.
CPMER/eEF1A2 conservatively functions in Eomes translation and human CM differentiation
(A) Expression pattern of CPMER during hESC-derived CM differentiation.
(B) qRT-PCR analysis of expression of CPMER and mesodermal marker genes (EOMES and T) in CPMER knockdown cells. Data shown are the mean ± SEM, n = 3 independent experiments.
(C) Western blot analysis of EOMES and T protein levels.
(D) qRT-PCR analysis of CM marker gene expression. Data shown are the mean ± SEM, n = 3 independent experiments.
(E) cTnT immunostaining (green) on day 18 (left) and statistics for relative fluorescence intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM, n = 6 fields from 3 independent experiments.
(F) qRT-PCR analysis of CM marker gene expression. Data shown are the mean ± SEM, n = 3 independent experiments.
(G) cTnT immunostaining (green) on day 18 (left) and statistics for relative fluorescence intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM, n = 6 fields from 3 independent experiments.
(H) RIP analysis for the exogenous expression of CPMER and FLAG-eEF1A2 in 293T cells, determined by an anti-FLAG antibody. Data shown are the mean ± SEM, n = 3 independent experiments.
(I) Regions of potential recognition between CPMER and EOMES mRNA (left) and MS2bp-YFP RNA pull-down analysis for exogenous expression of pMS2-CPMER and human EOMES mRNA in 293T cells (right). Data shown are the mean ± SEM, n = 3 independent experiments.
(J) Endogenous RIP analysis for the interaction of eEF1A with EOMES mRNA. Data shown are the mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (versus sh-Ctrl); Student’s t test.
See also Figure S6.
Discussion
Subtle regulation of mRNA translation is critical for controlling the expression of pivotal proteins (Roux and Topisirovic, 2018; Simpson et al., 2020). A few studies have reported that translational regulation of specific genes is closely related to protein function and heart disease. The translation efficiency of Pabpc1 mRNA varies significantly at different stages of mouse heart development and is closely related to pathological cardiac hypertrophy (Chorghade et al., 2017). In contrast, the RNA-binding protein RBM24 prevents excessive activation of the P53 protein and diminishes heart defects by specifically inhibiting assembly of the translation initiation complex on P53 mRNA (Zhang et al., 2018). However, the precise regulation of mRNA translation in cardiac development is not completely understood. eEF1A2, the key element of the translation complex, is specifically expressed in the heart and muscle. Previous studies have found that knockdown of Eef1a2 causes abnormal heart development and heart failure in zebrafish, whereas deletion or mutation of Eef1a2 leads to ataxia, muscle atrophy, and premature death in 3-week-old mice. Mutation of Eef1a2 has also been found in individuals with dilated cardiomyopathy and growth retardation (Cao et al., 2017). These studies suggest a correlation between eEF1A2 and abnormal heart development, but the rationale is still unclear. Our results indicated that knockdown of Eef1a2 significantly inhibits differentiation of ESCs into CMs in mice and humans by decreasing the translation efficiency of the key Eomes mRNA. Our study revealed, for the first time, that eEF1A2 is a conserved translation regulator in cardiac lineage commitment.
Previous studies have reported that lncRNAs often exert their exquisite regulatory functions by partnering with RNA-binding proteins. The lncRNA UCA1 binds to PTBP1 to regulate the stability of ALAS2 mRNA and, thus, affects heme synthesis (Liu et al., 2018). Lnc-DC promotes dendritic cell differentiation by binding STAT3 and protecting it from dephosphorylation (Pfeiffer et al., 2018). The eEF1A2 protein has RNA-binding capability and functions as an mRNA translation regulator, but its synergistic molecules and regulatory mechanisms remain unknown. We found that a heart-specific lncRNA, Cpmer, localized in the cytoplasm, interacted with eEF1A2 and significantly affected the efficiency of CM differentiation. More importantly, we also determined that the human homolog CPMER bound with human eEF1A2 to regulate the process of human CM differentiation. eEF1A1 and eEF1A2 proteins have nearly identical amino acid sequences, which makes it difficult to distinguish eEF1A2 from eEF1A1. So the possibility remains that Cpmer endogenously binds eEF1A1 and eEF1A2 to execute its regulatory function, but the precise binding specificity of Cpmer with eEF1A2 rather than eEF1A1 need to be explored. Our study revealed a novel function of the lncRNA Cpmer in regulating CM differentiation by partnering with eEF1A2 and a conserved mechanism by which the eEF1A2/Cpmer interaction regulates cardiac differentiation.
Heart development starts with the cardiac mesoderm, which specializes from the mesoderm, and the mesoderm transcription factor EOMES plays a critical role in regulation of cardiac mesoderm specification (Costello et al., 2011; Pfeiffer et al., 2018). Deletion of Eomes causes significant impairment of differentiation of ESCs into CMs (van den Ameele et al., 2012). Previous studies have shown that the BMP and WNT signaling pathways activate Eomes transcription and that transcription factors such as NANOG (nanog homeobox), OCT4, and SOX2 also bind to the enhancer of the Eomes gene and activate its transcription (Arnold and Robertson, 2009; Teo et al., 2011; Tosic et al., 2019). Transcription of the Eomes gene is also regulated by the lncRNA Meteor, which is transcribed from the enhancer region of the Eomes gene (Alexanian et al., 2017). Only miR-29 and let-7 have been reported to target the 5′ UTR and open reading frame (ORF) region of Eomes mRNA at the post-transcriptional level, which leads to degradation of Eomes mRNA (Steiner et al., 2011; Wells et al., 2017). In this study, we found that Cpmer directly recognized Eomes mRNA sequences and affected the translation efficiency of Eomes mRNA. In-depth data revealed that Cpmer regulated translation of Eomes mainly by specifically affecting recruitment of eEF1A2 and ribosomes to Eomes mRNA. Our study of the precise mechanism by which Cpmer and eEF1A2 mediate translation regulation of Eomes mRNA improves our understanding of the upstream regulatory mechanisms of mRNA translation of Eomes in the context of cardiac development.
lncRNAs have been reported to perform their regulatory functions through RNA-RNA interactions. HBL1, for example, acts as a sponge of miR-1 and inhibits differentiation of human PSCs into CMs by directly competing with the interaction of miR-1 with its target genes (Liu et al., 2017). The lncRNA TINCR recognizes and affects the stability of mRNAs containing the TINCR box motif, regulating somatic tissue differentiation (Kretz et al., 2013). Recent studies have also revealed that lncRNAs can repress translation of specific genes through sequence recognition. By matching the long-length sequence of CTNNB1 and JUNB mRNAs, lincRNA-p21 leads to ribosome drop-off and inhibition of CTNNB1 and JUNB mRNA translation (Yoon et al., 2012). AS Uchl1 promotes Uchl1 mRNA translation by facilitating assembly of the translation initiation complex (Carrieri et al., 2012). SINEUP (lncRNAs contain a SINE element and up-regulate the translation of target mRNA)-like lncRNAs enhance translation initiation of target mRNAs by acting in conjunction with PTBP1/HNRNPK (heterogeneous nuclear ribonucleoprotein K), which can serve as a potentially exogenous intervention for diseases caused by insufficient protein expression through designed artificial SINEUP lncRNAs (Bon et al., 2019). In this study, we found that the specific recognition between Cpmer and the Eomes mRNA was indispensable for translation of the Eomes mRNA. Deleting the recognition sequence significantly diminished binding of eEF1A2 and Eomes mRNAs and mRNA translation efficiency. The short recognition sequences, located at the 5′ ORF region of the Eomes mRNA, perhaps have a conservative responsibility for proper translation elongation. Only the Cpmer mutant with the ability to recognize Eomes mRNA and bind eEF1A2 could restore the ribosome enrichment level, EOMES protein expression, and CM differentiation. Our study revealed the novel epigenetic mechanism by which Cpmer mediates eEF1A2 to specifically regulate translation of Eomes mRNA through its dual roles of sequence recognition and protein recruitment, providing a new dimension of epigenetic regulation specificity in the research field of mRNA translation.
Our study demonstrated that the functionally conserved Cpmer interacted with eEF1A2 and specifically regulated translation of Eomes mRNA and CM differentiation. Our research on Cpmer not only identified a novel lncRNA capable of regulating cardiac development but also discovered a new epigenetic mechanism of lncRNA-mediated specific gene translation, which improves our knowledge of the precise post-transcriptional regulation of key transcription factors during CM differentiation.
Experimental procedures
A detailed description of the experimental procedures is provided in the supplemental information.
Cell culture and differentiation
46C mESCs (Aubert et al., 2003) were maintained in DMEM (Gibco) supplemented with 15% fetal bovine serum (FBS; Gibco), 1× nonessential amino acids (NEAAs; Gibco), 1× GlutaMAX (Gibco), 1× sodium pyruvate (Gibco), β-mercaptoethanol (Gibco), and leukemia inhibitory factor (LIF) on feeder cells at 37°C and 5% CO2. Cultures were passaged every 2 days by adding 0.25% trypsin (Gibco). Mouse CM differentiation was performed as described previously (Kattman et al., 2011). H9 hESCs (WiCell) were maintained in DMEM/F12 (Gibco), 20% KO serum replacement (Gibco), 0.1 mM β-mercaptoethanol (Gibco), 1% NEAAs (Gibco), 0.5% GlutaMAX (Gibco), and 4 ng/mL hbFGF (human basic fibroblast growth factor) (R&D Systems) on a feeder layer. hESC-induced CM differentiation was performed as described previously (Protze et al., 2017).
Vectors
shRNAs targeting Eef1a2 (mouse and human), Eomes (mouse), and Cpmer (mouse and human) were designed and inserted into the pLKO.1 vector (Addgene). The cDNA fragments corresponding to the RACE findings on Cpmer were cloned into the donor vector (introduced into the Rosa26 locus of mESCs), pBSK vector, and pMS2 vector (Addgene), respectively. Cpmer E1, E2, E3, E12, E23, and E13 transcripts; the FL and deletion mutant Eomes (GenBank: NM_010136) sequences; Eomes sequences; and GFP fusion reporter sequences were cloned into the Fugw vector (Addgene). All constructed plasmids were verified by DNA sequencing. All primer sequences are listed in Table S4.
Biotin-RNA pull-down and liquid chromatography (LC)-mass spectrometry (MS)
Biotin-labeled Cpmer was obtained using RNA Labeling Mix (11685597910, Roche) with T7 (10881767001, Roche) or T3 RNA polymerase (11031171001, Roche). Total RNA was heated to 90°C for 2 min, held on ice for 2 min, and incubated in RNA structure buffer. Lysis was performed using streptavidin beads coated with biotin-labeled sense Cpmer or antisense Cpmer. The RNA-binding proteins were analyzed by LC-MS as described previously (Shevchenko et al., 2006).
RIP and MS2bp-YFP RNA pull-down assay
RIP and MS2bp-YFP-based RNA pull-down assays were performed as described previously (Ng et al., 2012). The eEF1A antibody was able to endogenously recognize eEF1A1 and eEF1A2 proteins.
Polysome profile analysis
Polysome profile analysis was performed as described previously (Panda et al., 2017). The eEF1A antibody was able to recognize eEF1A1 and eEF1A2 in the co-sedimenting protein of polysomes.
Statistical analysis
Statistical significance was analyzed with two-tailed Student’s t test from three independent experiments. Mean values ±SEM are shown.
Data and code availability
All data are available in the main text or the supplemental information. Additional data related to this paper may be requested from the lead contact. The MS proteomics data are available via ProteomeXchange with identifier ProteomeXchange: PXD031814. Datasets have been deposited in the Gene Expression Omnibus Database under accession number GSE158202.
Author contributions
Y.L., X.G., and J.K. designed the study and wrote the manuscript. Y.L., X.Z., and Y.X. performed the experiments and data analyses. W.J. and Y.W. contributed to the interpretation of data. All authors approved the final version of the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the Ministry of Science and Technology (2018YFA0800100), the National Natural Science Foundation of China (31830059, 31970599, 32170575, 31871298, 31721003, and 32000605), the Shanghai Rising-Star Program (20QA1409600), and the Fundamental Research Funds for the Central Universities (22120220104).
Published: April 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.03.006.
Contributor Information
Xudong Guo, Email: 19504@tongji.edu.cn.
Jiuhong Kang, Email: jhkang@tongji.edu.cn.
Supplemental information
References
- Alexanian M., Maric D., Jenkinson S.P., Mina M., Friedman C.E., Ting C.C., Micheletti R., Plaisance I., Nemir M., Maison D., et al. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat. Commun. 2017;8:1806. doi: 10.1038/s41467-017-01804-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amita M., Adachi K., Alexenko A.P., Sinha S., Schust D.J., Schulz L.C., Roberts R.M., Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc. Natl. Acad. Sci. U S A. 2013;110:E1212–E1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.M., Anderson D.M., McAnally J.R., Shelton J.M., Bassel-Duby R., Olson E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S.J., Robertson E.J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Aubert J., Stavridis M.P., Tweedie S., O'Reilly M., Vierlinger K., Li M., Ghazal P., Pratt T., Mason J.O., Roy D., Smith A. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc. Natl. Acad. Sci. U S A. 2003;100(Suppl 1):11836–11841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon C., Luffarelli R., Russo R., Fortuni S., Pierattini B., Santulli C., Fimiani C., Persichetti F., Cotella D., Mallamaci A., et al. SINEUP non-coding RNAs rescue defective frataxin expression and activity in a cellular model of Friedreich's Ataxia. Nucleic Acids Res. 2019;47:10728–10743. doi: 10.1093/nar/gkz798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Smith L.L., Padilla-Lopez S.R., Guida B.S., Blume E., Shi J., Morton S.U., Brownstein C.A., Beggs A.H., Kruer M.C., Agrawal P.B. Homozygous EEF1A2 mutation causes dilated cardiomyopathy, failure to thrive, global developmental delay, epilepsy and early death. Hum. Mol. Genet. 2017;26:3545–3552. doi: 10.1093/hmg/ddx239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chang C.P., Bruneau B.G. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- Chorghade S., Seimetz J., Emmons R., Yang J., Bresson S.M., Lisio M., Parise G., Conrad N.K., Kalsotra A. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. Elife. 2017;6:e24139. doi: 10.7554/eLife.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I., Pimeisl I.-M., Dräger S., Bikoff E.K., Robertson E.J., Arnold S.J. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Ahuja G., Bartsch D., Russ N., Yao W., Kuo J.C.-C., Derks J.-P., Akhade V.S., Kargapolova Y., Georgomanolis T., et al. yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell. 2019;24:318–327.e8. doi: 10.1016/j.stem.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Fujii K., Shi Z., Zhulyn O., Denans N., Barna M. Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat. Commun. 2017;8:14443. doi: 10.1038/ncomms14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga T., Hamada M. RIblast: an ultrafast RNA-RNA interaction prediction system based on a seed-and-extension approach. Bioinformatics. 2017;33:2666–2674. doi: 10.1093/bioinformatics/btx287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J., Farinas I., Depew M.J., Clevers H., Grosschedl R. Wnt3a-/--like phenotype and limb deficiency in Lef1(-/-)Tcf1(-/-) mice. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Xu Y., Wang Z., Wu Y., Chen J., Wang G., Lu C., Jia W., Xi J., Zhu S., et al. A linc1405/Eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell. 2018;22:893–908.e6. doi: 10.1016/j.stem.2018.04.013. [DOI] [PubMed] [Google Scholar]
- Hosen M.R., Militello G., Weirick T., Ponomareva Y., Dassanayaka S., Moore J.B.t., Doring C., Wysoczynski M., Jones S.P., Dimmeler S., Uchida S. Airn regulates Igf2bp2 translation in cardiomyocytes. Circ. Res. 2018;122:1347–1353. doi: 10.1161/CIRCRESAHA.117.312215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B.M., Wiles M.V. Evidence for involvement of activin A and bone morphogenetic protein 4 in mammalian mesoderm and hematopoietic development. Mol. Cell. Biol. 1995;15:141–151. doi: 10.1128/mcb.15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J., et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Y., Lin B., Sheng Y., Yang L. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev. Cell. 2017;42:333–348.e5. doi: 10.1016/j.devcel.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Y., Tong J., Gao J., Guo Q., Zhang L., Wang B., Zhao H., Wang H., Jiang E., et al. Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development. Nat. Commun. 2018;9:4386. doi: 10.1038/s41467-018-06883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Hausmann S., Carlson S.M., Fuentes M.E., Francis J.W., Pillai R., Lofgren S.M., Hulea L., Tandoc K., Lu J., et al. METTL13 methylation of eEF1A increases translational output to promote tumorigenesis. Cell. 2019;176:491–504.e21. doi: 10.1016/j.cell.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P., Coriati A., Belanger G., De Repentigny Y., Lee J., Kothary R., Holcik M., Jasmin B.J. The utrophin A 5'-UTR drives cap-independent translation exclusively in skeletal muscles of transgenic mice and interacts with eEF1A2. Hum. Mol. Genet. 2010;19:1211–1220. doi: 10.1093/hmg/ddp591. [DOI] [PubMed] [Google Scholar]
- Monteiro R.M., de Sousa Lopes S.M., Korchynskyi O., ten Dijke P., Mummery C.L. Spatio-temporal activation of Smad1 and Smad5 in vivo: monitoring transcriptional activity of Smad proteins. J. Cell Sci. 2004;117:4653–4663. doi: 10.1242/jcs.01337. [DOI] [PubMed] [Google Scholar]
- Moore-Morris T., van Vliet P.P., Andelfinger G., Puceat M. Role of epigenetics in cardiac development and congenital diseases. Physiol. Rev. 2018;98:2453–2475. doi: 10.1152/physrev.00048.2017. [DOI] [PubMed] [Google Scholar]
- Mullin N.K., Mallipeddi N.V., Hamburg-Shields E., Ibarra B., Khalil A.M., Atit R.P. Wnt/beta-catenin signaling pathway regulates specific lncRNAs that impact dermal fibroblasts and skin fibrosis. Front. Genet. 2017;8:183. doi: 10.3389/fgene.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Ng S.Y., Johnson R., Stanton L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterwegel M., van de Wetering M., Timmerman J., Kruisbeek A., Destree O., Meijlink F., Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- Ounzain S., Micheletti R., Beckmann T., Schroen B., Alexanian M., Pezzuto I., Crippa S., Nemir M., Sarre A., Johnson R., et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36:353–368a. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A.C., Martindale J.L., Gorospe M. Polysome fractionation to analyze mRNA distribution profiles. Bio Protoc. 2017;7:e2126. doi: 10.21769/BioProtoc.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer M.J., Quaranta R., Piccini I., Fell J., Rao J., Ropke A., Seebohm G., Greber B. Cardiogenic programming of human pluripotent stem cells by dose-controlled activation of EOMES. Nat. Commun. 2018;9:440. doi: 10.1038/s41467-017-02812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., Keller G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- Roux P.P., Topisirovic I. Signaling pathways involved in the regulation of mRNA translation. Mol. Cell. Biol. 2018;38 doi: 10.1128/MCB.00070-18. e00070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Simpson L.J., Reader J.S., Tzima E. Mechanical regulation of protein translation in the cardiovascular system. Front. Cell Dev. Biol. 2020;8:34. doi: 10.3389/fcell.2020.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares M.L., Haraguchi S., Torres-Padilla M.E., Kalmar T., Carpenter L., Bell G., Morrison A., Ring C.J., Clarke N.J., Glover D.M., Zernicka-Goetz M. Functional studies of signaling pathways in peri-implantation development of the mouse embryo by RNAi. BMC Dev. Biol. 2005;5:28. doi: 10.1186/1471-213X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner D.F., Thomas M.F., Hu J.K., Yang Z., Babiarz J.E., Allen C.D., Matloubian M., Blelloch R., Ansel K.M. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K.K., Arnold S.J., Trotter M.W.B., Brown S., Ang L.T., Chng Z., Robertson E.J., Dunn N.R., Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosic J., Kim G.J., Pavlovic M., Schroder C.M., Mersiowsky S.L., Barg M., Hofherr A., Probst S., Kottgen M., Hein L., Arnold S.J. Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 2019;21:1518–1531. doi: 10.1038/s41556-019-0423-1. [DOI] [PubMed] [Google Scholar]
- van den Ameele J., Tiberi L., Bondue A., Paulissen C., Herpoel A., Iacovino M., Kyba M., Blanpain C., Vanderhaeghen P. Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep. 2012;13:355–362. doi: 10.1038/embor.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A.C., Daniels K.A., Angelou C.C., Fagerberg E., Burnside A.S., Markstein M., Alfandari D., Welsh R.M., Pobezinskaya E.L., Pobezinsky L.A. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife. 2017;6:e26398. doi: 10.7554/eLife.26398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Gai K., Li X., Shao P., Zeng Z., Zhao X., Zhao X., Chen X., Paradee W.J., Meyerholz D.K., et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 2019;216:847–866. doi: 10.1084/jem.20182010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang Y., Xu E., Mohibi S., de Anda D.M., Jiang Y., Zhang J., Chen X. Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 2018;25:1118–1130. doi: 10.1038/s41418-017-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplemental information. Additional data related to this paper may be requested from the lead contact. The MS proteomics data are available via ProteomeXchange with identifier ProteomeXchange: PXD031814. Datasets have been deposited in the Gene Expression Omnibus Database under accession number GSE158202.