Abstract
Production of phenazine antibiotics by the biological control bacterium Pseudomonas aureofaciens 30-84 is regulated in part by the PhzI/PhzR N-acyl-homoserine lactone (AHL) response system (L. S. Pierson III, V. D. Keppenne, and D. W. Wood, J. Bacteriol. 176:3966–3974, 1994; D. W. Wood and L. S. Pierson III, Gene 168:49–53, 1996). Two mutants, 30-84W and 30-84.A2, were isolated and were found to be deficient in the production of phenazine, protease, hydrogen cyanide (HCN), and the AHL signal N-hexanoyl-homoserine lactone. These mutants were not complemented by phzI, phzR, or the phenazine biosynthetic genes (phzFABCD) (L. S. Pierson III, T. Gaffney, S. Lam, and F. Gong, FEMS Microbiol. Lett. 134:299–307, 1995). A 2.2-kb region of the 30-84 chromosome which fully restored production of all of these compounds in strain 30-84W was identified. Nucleotide sequence analysis of this region revealed a single open reading frame encoding a predicted 213-amino-acid protein which is very similar to the global response regulator GacA. Strain 30-84.A2 was not complemented by gacA or any cosmid from a genomic library of strain 30-84 but was complemented by gacS (formerly lemA) homologs from Pseudomonas fluorescens Pf-5 (N. Corbel and J. E. Loper, J. Bacteriol. 177:6230–6236, 1995) and Pseudomonas syringae pv. syringae B728a (E. M. Hrabek and D. K. Willis, J. Bacteriol. 174:3011–3020, 1992). Transcription of phzR was not altered in either mutant; however, phzI transcription was eliminated in strains 30-84W and 30-84.A2. These results indicated that the GacS/GacA two-component signal transduction system of P. aureofaciens 30-84 controls the production of AHL required for phenazine production by mediating the transcription of phzI. Addition of exogenous AHL did not complement either mutant for phenazine production, indicating that the GacS/GacA global regulatory system controls phenazine production at multiple levels. Our results reveal for the first time a mechanism by which a two-component regulatory system and an AHL-mediated regulatory system interact.
Precise regulation of gene expression in response to changing environmental conditions is essential for the survival of bacterial populations. Regulation is coordinated in response to a variety of environmental cues, including self-produced diffusible molecules that allow cell-to-cell communication within a population. One class of signal molecules, known as N-acyl-homoserine lactones (AHLs), is utilized by many gram-negative bacteria to coordinate gene expression. AHLs accumulate in the extracellular environment and allow gene expression to be regulated in response to population density. In addition, AHL-mediated signal exchange between nonisogenic co-occurring bacterial populations (cross-communication) has been shown to affect gene expression in situ (13). AHL-mediated gene regulation in these bacteria is controlled by proteins belonging to the LuxI/LuxR family of quorum-sensing regulators (3, 5).
Regulation of bacterial gene expression in response to external environmental conditions is also often facilitated by two-component signal transduction systems (12). These systems consist of a sensor kinase (SK) and a cytoplasmic response regulator (RR). In a typical two-component system, the SK is capable of autophosphorylation and, in response to a specific environmental signal(s), transfers the phosphate to the RR. Upon phosphorylation, the RR activates transcription of its target genes. A two-component signal transduction system that has been found in many plant-associated pseudomonads is the GacS/GacA (Global antibiotics and cyanide control) regulon. The SK GacS (formerly LemA) was first identified in the plant pathogen Pseudomonas syringae pv. syringae B728a, in which it is required for lesion formation on bean plants (24). The RR GacA was first identified as a mediator of antibiotic production in the biological control bacterium Pseudomonas fluorescens CHA0 (9). GacS and GacA regulate the expression of multiple phenotypes, and therefore this system is known as a global regulatory system.
It is increasingly evident that two-component regulatory systems and AHL-mediated regulatory systems rarely function independently; instead, they are components of complex regulatory signal cascade mechanisms (19). A hierarchical cascade that regulates elastase production in Pseudomonas aeruginosa PAO1 includes the LasR/LasI and RhlR/RhlI AHL-mediated response systems, as well as the alternate sigma factor RpoS (8). Recently, it was reported that production of N-butyryl-homoserine lactone (BHL), the cognate signal of the RhlR/RhlI system, is reduced or delayed in gacA mutants of strain PAO1 (21). However, a mechanism by which this two-component system affects BHL production was not defined. In this paper we describe the mechanism responsible for the linkage between a two-component signal transduction system and an AHL-mediated response system.
Pseudomonas aureofaciens 30-84 is a biological control bacterium that inhibits the fungal pathogen Gaeumannomyces graminis var. tritici, the causal agent of take-all disease of wheat. Strain 30-84 produces three phenazine antibiotics, which are primarily responsible for the control of G. graminis var. tritici (17). Phenazine production is regulated by the PhzI/PhzR quorum-sensing system (16, 27). PhzI is responsible for the synthesis of a specific AHL, N-hexanoyl-homoserine lactone (HHL), which accumulates as the bacterial population increases. As the concentration of HHL increases, PhzR is activated, which results in transcription of the phenazine biosynthetic operon (phzFABCD) (15). In this paper we describe an additional level of regulation of phenazine production that involves transcriptional control of AHL production by a GacS/GacA two-component regulatory system. Preliminary findings concerning this linkage have been reported previously (14, 19).
MATERIALS AND METHODS
Bacteria and plasmids.
The bacterial strains and plasmids used in this study are described in Table 1. A spontaneous rifampin-resistant derivative of P. aureofaciens 30-84 (17) was used in all experiments. The media, conditions for growth, and antibiotic concentrations used have been described previously (16).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Pseudomonas aureofaciens strains | ||
| 30-84 | Phz+ Rifr wild type | W. W. Bockus |
| 30-84Z | Phz− RifrphzB::lacZ genomic fusion | 16 |
| 30-84R | Phz− RifrphzR::Tn5lacZ genomic fusion | 16 |
| 30-84Ice | Phz− RifrphzB::inaZ genomic fusion | 26 |
| 30-84I | Phz− RifrphzI::Kmr | 27 |
| 30-84Z/I | Phz− RifrphzB::lacZ and phzI::Kmr genomic fusions | 27 |
| 30-84W | Phz− Rifr spontaneous gacA mutant | This study |
| 30-84.A2 | Phz− Rifr Kmr spontaneous gacS mutant | This study |
| 30-84.gacA | Phz− RifrgacA::Kmr | This study |
| Escherichia coli strains | ||
| DH5α | F− recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(argF-lacZYA)I169 φ80lacZ ΔM15 | GIBCO-BRL |
| HB101 | F− hsdS20 (rB−mB−) supE44 recA1 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-5 | GIBCO-BRL |
| Plasmids | ||
| pDW7311 | pLAFR3 containing ExoIII-truncated phzI of strain 30-84 | This study |
| pDW7311uidA | pLAFR3 containing a phzI::uidA transcriptional reporter fusion | This study |
| pEMH97 | pLAFR3 containing gacS from P. syringae pv. syringae B728a | 24 |
| pHP45Ω-Kmr | pUC18 containing Kmr cassette | 4 |
| pJEL5771 | pLAFR3 containing the gacS homolog apdA of P. fluorescens Pf-5 | 1 |
| pLAFR3 | IncP1 Tetr cos+ rlx+ | 23 |
| pLSP20H-2.7#7 | pIC20H containing 2.7-kb PstI fragment of pLSP259 with phzI/phzR | 16 |
| pLSP259 | pLAFR3 containing phzI, phzR, and the phenazine biosynthetic locus on a 20.4-kb EcoRI fragment of strain 30-84 chromosomal DNA | 17 |
| pLSP259Tn5lac#42 | pLAFR3 containing Tn5lacZ insertion within phzR in pLSP259 | 16 |
| pLSP619 | pLAFR3 containing 33 kb of strain 30-84 chromosomal DNA as multiple EcoRI fragments | This study |
| pME3066 | pLAFR3 containing gacA from P. fluorescens CHA0 | 9 |
| pRK2013 | IncP1 tra oriE1 Kmr | 2 |
| pSTC110 | pLAFR3 containing 15-kb EcoRI fragment of pLSP619 | This study |
| pSTC121 | pLAFR3 containing 4.5-kb EcoRI-PstI gacA fragment of pSTC110 | This study |
| pSTC122 | pLAFR3 containing 2.2-kb KpnI-PstI gacA fragment of pSTC121 | This study |
| pSTC140 | pUC18 containing 2.2-kb KpnI-PstI gacA fragment of pSTC121 | This study |
| pSTC161 | pLAFR3 containing gacA disrupted by the Kmr cartridge of pHP45Ω-Kmr | This study |
| pUC18 | ColE1 Apr | 28 |
| pWM6 | ColE1 vector containing uidA transcriptional fusion cassette | 10 |
DNA manipulations.
DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation were all performed as described previously (17).
Quantitation of phenazine.
Phenazine antibiotics were extracted from P. aureofaciens 30-84 and quantitated by UV-visible light spectroscopy as described previously (17), with the following modifications. Briefly, cultures were grown in PPMD medium amended, when appropriate, with tetracycline (50 μg/ml) at 28°C for 24 h. Five milliliters of each culture was centrifuged (3,000 × g), and the supernatant was acidified (pH <2) with concentrated HCl. Following addition of 5 ml of benzene, samples were mixed for 1 h and centrifuged. Four milliliters of the benzene layer was decanted and dried under air. Samples were resuspended in 1 ml of 0.1 N NaOH, and the absorbance at 367 nm was determined.
AHL extraction and biological assays.
AHL preparations were isolated from cell-free supernatants as described previously (13). Briefly, 5-ml cultures of the Pseudomonas strains were grown overnight at 28°C with shaking in PPMD broth. Supernatants were collected following centrifugation (3,000 × g) of the cultures. The supernatants were mixed for 1 h with a volume of acidified ethyl acetate (0.1 ml of acetic acid per liter) that was equal to the original volume of culture. After centrifugation (3,000 × g), the ethyl acetate was decanted and evaporated under nitrogen. Extracts were resuspended in volumes of PPMD broth equal to the volumes of ethyl acetate decanted.
To assay for the production of AHL, 5-ml overnight cultures of Pseudomonas test strains were extracted as described above. The extracts were resuspended in PPMD broth amended with kanamycin (50 μg/ml). Each sample was then inoculated with the AHL-specific P. aureofaciens reporter strain 30-84Z/I (phzI− phzB::lacZ) and allowed to grow with shaking at 28°C. β-Galactosidase activity was determined after 24 h as described by Miller (11).
Effect of exogenous AHL on phenazine production.
AHL was extracted from a 200-ml overnight culture of AHL+ strain 30-84Ice as described above. The dried extract was resuspended in 200 ml of PPMD broth and filter sterilized. The PPMD broth containing AHL was divided into 3-ml aliquots, and each aliquot was inoculated with one of the 30-84 derivative strains that were tested. Cultures were grown with shaking overnight at 28°C, and phenazine production was quantified as described above.
Assays for secondary metabolites.
Assays to determine the production of hydrogen cyanide and extracellular protease were performed as described previously (13, 17). Briefly, extracellular protease production was measured qualitatively by spotting 5-μl portions of overnight cultures of each test strain on skim milk agar (Difco Laboratories). Formation of a zone of clearing around a colony indicated that extracellular protease was produced. Production of hydrogen cyanide was determined qualitatively by using Cyantesmo paper (Machery-Nagel GmbH & Co.) as recommended by the manufacturer. The relative fluorescence of each Pseudomonas strain was determined by spotting 5-μl portions from overnight cultures onto King’s medium B (26) and comparing the relative intensities of fluorescence under UV light.
Complementation of strain 30-84W.
Introduction of a partial EcoRI-generated pLAFR3 genomic library of strain 30-84 into strain 30-84W resulted in identification of a single cosmid (pLSP619) based on its ability to restore phenazine production in the mutant. A 15-kb EcoRI fragment of the 33-kb fragment of 30-84 chromosomal DNA contained on pLSP619 was subcloned into pLAFR3 to generate pSTC110, which retained the ability to complement strain 30-84W for phenazine production. The complementing region was localized to a 4.5-kb EcoRI-PstI fragment of pSTC110, which was subcloned into pSTC121. Finally, a 2.2-kb KpnI-PstI fragment of pSTC121 was subcloned into pSTC122, which retained the ability to restore phenazine production to strain 30-84W. The KpnI-PstI fragment was subcloned further into pUC18 to generate pSTC140, which was used for DNA sequence determination.
DNA sequencing.
The DNA sequence of the 2.2-kb KpnI-PstI fragment was determined at the University of Arizona Biotechnology Center by using an Applied Biosystems automatic DNA sequencer (model 373A, version 1.2.1). The primers used included M13 forward and reverse primers. Additional primers were designed by using sequence data and Oligo 4.05 Primer Analysis Software (National Biosciences, Inc., Plymouth, Minn.) and were synthesized by Gibco-BRL (Gaithersburg, Md.). The DNA sequence analysis was performed with the University of Wisconsin Genetics Computer Group software packages (version 9.1).
Construction of a gacA mutant.
The P. aureofaciens 30-84 gacA gene contained on pSTC121 was disrupted by replacing an internal 50-base SmaI fragment with the kanamycin resistance cartridge from pHP45Ω-Kmr. The resulting plasmid, pSTC161, was introduced into strain 30-84 by triparental mating. The disrupted gacA gene was marker exchanged into the 30-84 chromosome by homologous recombination. A kanamycin-resistant, tetracycline-sensitive, phenazine-defective recombinant was identified, and disruption of gacA was verified by Southern blot analysis (data not shown). This mutant was designated 30-84.gacA.
Transcriptional analysis.
Transcriptional analyses of phzI were performed by utilizing a plasmid-borne phzI::uidA transcriptional fusion. To generate this construct, 410 bp was deleted from the 3′ end of phzI on pLSP20H-2.7#7 by exonuclease III digestion. The truncated phzI was blunt ended by using S1 nuclease and was cloned into the EcoRI site of pLAFR3, which was also treated with S1 nuclease, in order to generate pDW7311. The 3.6-kb BamHI fragment of pWM6 containing the uidA-bla cassette was inserted into the BamHI site in pDW7311. The resultant plasmid, pDW7311uidA, was introduced into strain 30-84 and its derivatives. β-Glucuronidase (GUS) activity was assayed as described by Wilson et al. (25) after growth with shaking in PPMD medium for 24 h at 28°C. Transcriptional analyses of phzR were performed by utilizing a phzR::lacZ transcriptional fusion on plasmid pLSP259Tn5lac#42 (16). β-Galactosidase was assayed as described by Miller (11).
Statistical analysis.
Treatment effects were determined by analysis of variance by using SAS software (version 6.12 for UNIX, 1993; SAS Institute Inc., Cary, N.C.). Means were compared by performing an analysis of variance after least significant difference multiple comparisons were performed.
Nucleotide sequence accession number.
The nucleotide sequence of P. aureofaciens 30-84 gacA has been deposited in the GenBank database under accession no. AF115381.
RESULTS
Isolation of two novel phenazine mutants.
Two spontaneous mutants of P. aureofaciens 30-84, 30-84W and 30-84.A2, were selected based on their failure to produce the orange phenazines characteristic of strain 30-84. Strain 30-84W was isolated as a spontaneously occurring white colony on a PPMD agar plate. Strain 30-84.A2 was isolated as a single white colony on a PPMD agar plate following Tn5 mutagenesis of P. aureofaciens 30-84. However, sequence analysis of the DNA regions flanking the Tn5 insertion in strain 30-84.A2 did not reveal an open reading frame or extensive similarity to any other gene in the database, suggesting that a second, spontaneous mutation is responsible for the mutant phenotype (data not shown). UV-visible light spectroscopy of culture extracts of strains 30-84W and 30-84.A2 revealed that the ability to produce phenazine was completely lost by the mutants (Table 2). Both of the mutants had a characteristic fluorescent green appearance and, when plated onto King’s medium B agar, produced more intense fluorescent halos than wild-type strain 30-84 produced. Cosmid pLSP259 containing phzI, phzR, and the phenazine biosynthetic locus (phzFABCD) of strain 30-84 failed to restore phenazine production when it was introduced into strain 30-84W or 30-84.A2, indicating that each mutant was mutated in a gene outside the immediate phenazine regulon. Analysis of a phzB::lacZ genomic fusion indicated that the level of phenazine gene expression in a gacA mutant was <1% of the level of expression in strain 30-84Z. Introduction of gacA in trans fully restored phzB expression, indicating that the effect of the gacA mutation on phenazine production occurred at the level of transcription of the phenazine biosynthetic genes (data not shown).
TABLE 2.
Phenotypic characteristics of strain 30-84 and derivatives of 30-84
| Strain | Relevant phenotype and/or genotype | Phenazine absorbanceab | β-Galactosidase activity (Miller units)bc | Presence ofd:
|
||
|---|---|---|---|---|---|---|
| Protease | HCN | Fluorescence | ||||
| 30-84(pLAFR3) | Wild type (+ vector) | 1.03 ± 0.03 A | NDe | + | + | + |
| 30-84(pSTC121) | Wild type (+ gacA) | 0.99 ± 0.04 A | ND | + | + | + |
| 30-84(pME3066) | Wild type (+ gacA) | 1.05 ± 0.02 A | ND | + | + | + |
| 30-84(pEMH97) | Wild type (+ gacS) | 1.02 ± 0.01 A | ND | + | + | + |
| 30-84Ice(pLAFR3)f | phzB::inaZ (+ vector) | ND | 1,486 ± 168 C | + | + | + |
| 30-84Ice(pSTC121) | phzB::inaZ (+ gacA) | ND | 1,275 ± 42 C | + | + | + |
| 30-84Ice(pME3066) | phzB::inaZ (+ gacA) | ND | 1,284 ± 194 C | + | + | + |
| 30-84Ice(pEMH97) | phzB::inaZ (+ gacS) | ND | 1,258 ± 153 C | + | + | + |
| 30-84.gacA(pLAFR3) | gacA::Kmr (+ vector) | 0.05 ± 0.03 B | 140 ± 71 D | − | − | ++ |
| 30-84.gacA(pSTC121) | gacA::Kmr (+ gacA) | 0.98 ± 0.06 A | 1,486 ± 46 C | + | + | + |
| 30-84.gacA(pME3066) | gacA::Kmr (+ gacA) | 1.01 ± 0.00 A | 1,229 ± 29 C | + | + | + |
| 30-84.gacA(pEMH97) | gacA::Kmr (+ gacS) | 0.05 ± 0.06 B | 53 ± 42 D | − | − | ++ |
| 30-84W(pLAFR3) | gacA (+ vector) | 0.13 ± 0.04 B | 105 ± 72 D | − | − | ++ |
| 30-84W(pSTC121) | gacA (+ gacA) | 1.01 ± 0.02 A | 1,426 ± 150 C | + | + | + |
| 30-84W(pME3066) | gacA (+ gacA) | 0.99 ± 0.02 A | 1,225 ± 139 C | + | + | + |
| 30-84W(pEMH97) | gacA (+ gacS) | 0.11 ± 0.03 B | 84 ± 69 D | − | − | ++ |
| 30-84.A2(pLAFR3) | gacS (+ vector) | 0.13 ± 0.02 B | 89 ± 63 D | − | − | ++ |
| 30-84.A2(pSTC121) | gacS (+ gacA) | 0.14 ± 0.04 B | 150 ± 111 D | − | − | ++ |
| 30-84.A2(pME3066) | gacS (+ gacA) | 0.10 ± 0.01 B | 132 ± 108 D | − | − | ++ |
| 30-84.A2(pEMH97) | gacS (+ gacS) | 0.98 ± 0.01 A | 1,225 ± 188 C | + | + | + |
Absorbance at 367 nm of phenazine extracted from supernatants of overnight cultures resuspended in 0.1 N NaOH and diluted 10−1.
The values are means ± standard errors based on three replicates per treatment. Means followed by the same letter are not significantly different.
β-Galactosidase activity produced by AHL reporter strain 30-84Z/I grown in PPMD medium amended with AHL extracted from culture supernatants of selected test strains (11).
Determined qualitatively as described in Materials and Methods.
ND, not determined.
30-84Ice was used as a positive control for AHL production due to coextraction of phenazines with AHL from strain 30-84.
Characterization of strains 30-84W and 30-84.A2.
To identify the region(s) of the 30-84 chromosome responsible for the observed phenotypes, a genomic library of strain 30-84 was introduced into strains 30-84W and 30-84.A2 by triparental mating. Transconjugants were screened for the restoration of phenazine production. We identified a single cosmid, pLSP619, containing a 33-kb fragment of the 30-84 chromosome, which complemented strain 30-84W to wild-type levels of phenazine production (data not shown). A complementation analysis identified a 2.2-kb KpnI-PstI fragment that was present in subclone pSTC122 and was sufficient to restore phenazine production to strain 30-84W. Sequence analysis of this fragment revealed a single 213-amino-acid open reading frame whose predicted product was very similar to the products of gacA of P. fluorescens CHA0 (98%) and gacA of P. syringae pv. syringae B728a (93%). In order to verify that mutation of gacA in the spontaneous mutant strain 30-84W was responsible for these phenotypes, a gacA disruption mutant, strain 30-84.gacA, was constructed. The phenotype of strain 30-84.gacA was identical to the phenotype of strain 30-84W, and each mutant phenotype was completely restored by pSTC122. Because GacA in other pseudomonads is known to regulate a variety of secondary metabolites, including protease and hydrogen cyanide (HCN) (9, 22), strains 30-84W and 30-84.gacA were assayed to determine whether they produced these compounds. Neither strain produced protease or HCN. Complementation of the mutants with pSTC122 restored production of both compounds to the mutants. Strains 30-84W and 30-84.gacA were also complemented by gacA of P. fluorescens CHA0 contained on pME3066.
Strain 30-84.A2 had a phenotype similar to that of mutants 30-84W and 30-84.gacA (Phz− protease− HCN−). Due to this similarity and due to the fact that this mutant was not complemented by gacA, we predicted that the mutation in strain 30-84.A2 may reside in the cognate GacA SK encoded by gacS in other pseudomonads. Southern hybridization of strain 30-84 genomic DNA probed with gacS from P. syringae pv. syringae B728a detected a single hybridizing EcoRI fragment (data not shown). However, we identified no cosmid from the 30-84 genomic library that complemented 30-84.A2. Nevertheless, complementation with the heterologous gacS (lemA) gene from P. syringae pv. syringae B728a (6) (Table 2) or the gacS (apdA) gene from P. fluorescens Pf-5 (1) (data not shown) restored all phenotypes in strain 30-84.A2. These data are consistent with the hypothesis that strain 30-84.A2 is a gacS mutant.
GacA is required for AHL production.
To determine whether complementation of phenazine production in strains 30-84.gacA and 30-84.A2 by gacA and gacS, respectively, was correlated with the ability to produce AHL, the effects of mutations in gacA or gacS on AHL production were determined with strain 30-84Z/I (phzB::lacZ phzI::Kmr) as a reporter (Table 2). Plasmids pSTC121, pME3066, pEMH97, and pLAFR3 were conjugated into strains 30-84.gacA, 30-84W, 30-84.A2, and 30-84Ice. 30-84Ice is a Phz− AHL+ derivative of 30-84 and was used in this study as a positive control for AHL production because coextraction of phenazines with AHL made assaying wild-type strain 30-84 for AHL difficult. The level of production of AHL in the gacA and gacS mutants was ca. 10% of the level of production in the strain 30-84Ice control (Table 2). The levels of production of AHL by strains 30-84.gacA and 30-84W were restored to wild-type levels by the presence in trans of either gacA from strain 30-84 on pSTC121 or gacA from P. fluorescens CHA0 on pME3066. Introduction of gacS from P. syringae pv. syringae on pEMH97 had no effect on the expression of AHL in either strain 30-84.gacA or strain 30-84W. In contrast, strain 30-84.A2 complemented by the presence of gacS from P. syringae pv. syringae on pEMH97 produced wild-type AHL levels (Table 2), but addition of gacA in trans had no effect. Additional copies of gacA or gacS had no effect on the production of AHL in the positive control strain 30-84Ice.
Exogenous AHL does not restore phenazine production to mutants.
Strain 30-84I does not produce phenazines in the absence of exogenous AHL (27). To determine if the inability of strains 30-84.gacA, 30-84W, and 30-84.A2 to produce phenazine is due solely to the inability of the organisms to produce AHL, AHL extracted from strain 30-84Ice (AHL+ Phz−) culture supernatants was added to cultures of strains 30-84.gacA, 30-84W, and 30-84.A2. No significant phenazine production was detected in strains 30-84.gacA, 30-84W, and 30-84.A2 grown in the presence or absence of exogenous AHL (Table 3). The activity of the extracted AHL was confirmed by its ability to restore phenazine production to the AHL− strain 30-84I.
TABLE 3.
Effect of exogenous AHL on phenazine production in gacA and gacS mutants
| Prepn | Phenazine absorbancea |
|---|---|
| 30-84I (AHL− control) | 0.035 ± 0.002 A |
| 30-84I + AHL | 0.563 ± 0.040 B |
| 30-84.gacA | 0.040 ± 0.007 A |
| 30-84.gacA + AHL | 0.040 ± 0.005 A |
| 30-84W | 0.034 ± 0.003 A |
| 30-84W + AHL | 0.038 ± 0.004 A |
| 30-84.A2 | 0.042 ± 0.002 A |
| 30-84.A2 + AHL | 0.048 ± 0.007 A |
Absorbance at 367 nm of phenazine extracted from culture supernatants and resuspended in 0.1 N NaOH. Exogenous AHL was extracted from 30-84Ice and resuspended in PPMD medium. PPMD medium containing AHL was filter sterilized and inoculated with each test strain. Strain 30-84I was included as the AHL-negative control. The values are means ± standard errors based on three replicates per treatment. Means followed by the same letter are not significantly different.
GacA is required for phzI transcription.
To determine whether the loss of AHL production in the gacA or gacS mutants was due to a direct effect on the PhzI/PhzR system, a phzI::uidA transcriptional fusion was utilized to determine if GacA regulates AHL production at the level of transcription of phzI. The phzI::uidA construct in pDW7311uidA measures phzI transcription as GUS activity. The GUS activities from the phzI::uidA fusion in all of the gacA and gacS mutants were less than 8% of the GUS activity in wild-type AHL strain 30-84Z (Table 4). The expression of phzI in the phzR mutant 30-84R was about one-half the expression in the PhzR+ strain 30-84Z, which is consistent with the supposition that phzI transcription is increased by AHL-activated PhzR (27). To test if GacS/GacA affected the transcription of phzR, expression of a plasmid-borne phzR::lacZ transcriptional fusion was measured. There was no significant difference in phzR transcription between positive control strain 30-84Ice and mutant strains 30-84.gacA, 30-84W, and 30-84.A2, as judged by β-galactosidase activity (Table 4).
TABLE 4.
Effect of the loss of gacA or gacS on transcription of phzI and phzR
| Strain | phzI expression (GUS units)ab | phzR expression (Miller units)bc |
|---|---|---|
| 30-84Z (control) | 364 ± 45 A | NDd |
| 30-84Ice (control) | ND | 1,470 ± 214 D |
| 30-84.gacA | 24 ± 2 B | 1,753 ± 132 D |
| 30-84.W | 23 ± 2 B | 1,446 ± 76 D |
| 30-84.A2 | 23 ± 2 B | 1,789 ± 57 D |
| 30-84R | 163 ± 6 C | ND |
GUS activity produced from a plasmid-borne phzI::uidA transcriptional fusion (25).
Strains 30-84Z and 30-84Ice were included as controls for phzI and phzR expression, respectively. The values are means ± standard errors based on three replicates per treatment. Means followed by the same letter are not significantly different.
β-Galactosidase activity produced by a plasmid-borne phzR::lacZ transcriptional fusion (11).
ND, not determined.
DISCUSSION
This study is the first to describe a mechanism for direct linkage between a two-component sensory transduction system and an AHL-mediated regulatory system to control gene expression. Previous work showed that phenazine production in P. aureofaciens 30-84 is regulated by the AHL-mediated PhzI/PhzR response system and its cognate signal HHL (16, 18, 27). In this study, we demonstrated that GacS/GacA controls AHL production via transcriptional regulation of phzI. The level of expression of phzI in a gacA or gacS mutant was less than 8% of the level of expression in the wild type, and expression was fully restored by complementation of the mutations with the appropriate wild-type gacA or gacS gene. Since AHL is required for phenazine gene expression, GacS/GacA mutants of 30-84 are not able to produce phenazine antibiotics.
Our data suggest that in addition to control of phzI transcription, GacS/GacA also controls phenazine production in an AHL-independent manner. Since GacS/GacA controls AHL production and AHL is required for phenazine expression, we determined whether exogenous AHL functionally complemented gacA and gacS mutants. Surprisingly, addition of AHL extracted from AHL-producing derivatives of strain 30-84 failed to restore phenazine production to the mutants (Table 3). The simplest explanation for this is that GacS/GacA regulates transcription of phzR as well. However, phzR transcription was not altered in the GacS/GacA mutants (Table 4).
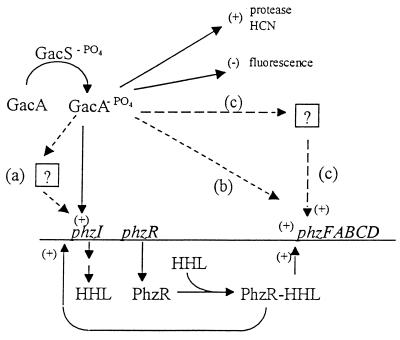
One hypothesis to explain the second level of phenazine regulation by GacA is that multiple regulatory proteins bind to the phenazine promoter region to activate phenazine biosynthesis. According to current models for AHL-mediated gene activation, PhzR binding to a consensus lux box is required (5). A potential lux box is located upstream of both phzI (27) and the phenazine biosynthetic locus (phzFABCD) (20). Extra copies in trans of the region containing this lux box result in a reduction in phenazine gene expression (16). However, introduction in trans of a 0.3-kb subclone of this region in which one-half of the lux box was deleted also caused a reduction in phenazine gene expression. This is consistent with the hypothesis that the concentration of a second transcriptional activator of phenazine expression may be reduced by titration. The second transcriptional activator may be GacA itself or another protein under regulation of GacA (Fig. 1). The hypothesis that a second regulatory protein is involved is supported by the recent identification of SalA, a regulator of syringomycin production in P. syringae pv. syringae B728a, which is under GacS/GacA control (7). The requirement for two activators, although unusual, would explain the inability of exogenous AHL to complement GacS/GacA mutants of strain 30-84.
FIG. 1.
Model for control of phenazine production in P. aureofaciens 30-84. GacS responds to the presence of an unknown signal by transphosphorylating GacA. GacA controls phenazine production by regulating AHL synthesis through transcriptional control of phzI either directly or indirectly (reaction a). GacA also controls phenazine production at a second level, possibly by direct binding of GacA (reaction b) or by some unidentified regulatory protein controlled by GacA (reactions c). The solid arrows indicate known regulatory controls, and the dashed arrows indicate possible regulatory controls. Places where positive and negative regulation occur are indicated by plus and minus signs, respectively.
Recent evidence suggests that regulatory systems, such as two-component and AHL-mediated regulatory systems, may not function independently but may be components of integrated regulatory networks. A linkage between GacA and AHL-mediated regulation was demonstrated previously in P. aeruginosa PAO1 (21). Disruption of gacA in strain PAO1 resulted in reduced or delayed production of the BHL involved in regulation of pyocyanin, hydrogen cyanide, and lipase (21). However, no mechanism was proposed for the influence of GacA on BHL production in this strain. A linkage has also been demonstrated between an AHL regulatory system and the stationary-phase sigma factor RpoS in P. aeruginosa (8).
Integrated regulatory schemes may allow bacteria to regulate expression of a wide array of potentially unrelated genes in a coordinated manner in response to multiple environmental signals. In order to test this hypothesis, a better understanding of the mechanisms of interactions between regulatory systems is necessary. Future research will focus on identifying the second mechanism by which GacA is involved in the sensory transduction system to regulate phenazine biosynthesis in strain 30-84.
ACKNOWLEDGMENTS
We thank Françoise Blachere for technical assistance and Elizabeth Pierson for performing the statistical analysis. We also thank Uvini Gunawardena, Christina Kennedy, and Elizabeth Pierson for critical reviews of the manuscript.
This work was supported in part by USDA NRICGP grant 98-02129.
REFERENCES
- 1.Corbel N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunlap P V. N-Acyl-l-homoserine lactone autoinducers in bacteria: unity and diversity. In: Shapiro J A, Dworkin M, editors. Bacteria as multicellular organisms. London, United Kingdom: Oxford University Press; 1996. pp. 69–106. [Google Scholar]
- 4.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 5.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR/LuxI family of cell density responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 8.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 9.Laville J, Voisard C, Keel C, Maurhofer M, DeFago G, Haas D. Global control in P. fluorescens mediating antibiotic synthesis and suppression of black rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1567. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metcalf W M, Wanner B L. Construction of a new β-glucuronidase cassettes for making transcriptional fusions and their use with methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- 11.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 12.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling molecules. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 13.Pierson E A, Wood D W, Cannon J A, Blachere F M, Pierson L S., III Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol Plant Microbe Interact. 1998;11:1078–1084. [Google Scholar]
- 14.Pierson L S, III, Chancey S T, Wood D W. Phenazine antibiotic biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30-84 is regulated at multiple levels. In: Stacy G, Gresshoff P, editors. Advances in molecular genetics of plant-microbe interactions. Boston, Mass: Kluwer Academic Publishers; 1996. pp. 463–468. [Google Scholar]
- 15.Pierson L S, III, Gaffney T, Lam S, Gong F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30-84. FEMS Microbiol Lett. 1995;134:299–307. doi: 10.1111/j.1574-6968.1995.tb07954.x. [DOI] [PubMed] [Google Scholar]
- 16.Pierson L S, III, Keppenne V D, Wood D W. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J Bacteriol. 1994;176:3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierson L S, III, Thomashow L S. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol Plant Microbe Interact. 1992;5:330–339. doi: 10.1094/mpmi-5-330. [DOI] [PubMed] [Google Scholar]
- 18.Pierson L S, III, Wood D W, Pierson E A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu Rev Phytopathol. 1998;36:207–225. doi: 10.1146/annurev.phyto.36.1.207. [DOI] [PubMed] [Google Scholar]
- 19.Pierson L S, III, Wood D W, Pierson E A, Chancey S T. N-Acyl-homoserine lactone-mediated gene regulation in biological control by fluorescent pseudomonads: current knowledge and future work. Eur J Plant Pathol. 1998;104:1–9. [Google Scholar]
- 20.Pierson, L. S., III. Unpublished data.
- 21.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Hass D. The global activator GacA of Pseudomonas aeruginosa PAO1 positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 22.Rich J, Kinscherf T, Kitten T, Willis D. Genetic evidence that the gacA gene encodes the cognate response regulator for the gacS sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion forming ability on bean. Mol Plant Microbe Interact. 1990;3:149–156. [Google Scholar]
- 25.Wilson K J, Jefferson R A, Hughes S G. The Escherichia coli GUS operon: induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria. In: Gallagher R, editor. GUS protocols: using the GUS gene as a reporter of gene expression. San Diego, Calif: Academic Press Inc.; 1992. pp. 7–22. [Google Scholar]
- 26.Wood D W, Gong F, Aykin M M, Williams P, Pierson L S., III N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood D W, Pierson L S., III The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene. 1996;168:49–53. doi: 10.1016/0378-1119(95)00754-7. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]