Abstract
We examined the relative roles of acetogenic and sulfate-reducing bacteria in H2 consumption in a previously characterized subsurface sandstone ecosystem. Enrichment cultures originally inoculated with ground sandstone material obtained from a Cretaceous formation in central New Mexico were grown with hydrogen in a mineral medium supplemented with 0.02% yeast extract. Sulfate reduction and acetogenesis occurred in these cultures, and the two most abundant organisms carrying out the reactions were isolated. Based on 16S rRNA analysis data and on substrate utilization patterns, these organisms were named Desulfomicrobium hypogeium sp. nov. and Acetobacterium psammolithicum sp. nov. The steady-state H2 concentrations measured in sandstone-sediment slurries (threshold concentration, 5 nM), in pure cultures of sulfate reducers (threshold concentration, 2 nM), and in pure cultures of acetogens (threshold concentrations 195 to 414 nM) suggest that sulfate reduction is the dominant terminal electron-accepting process in the ecosystem examined. In an experiment in which direct competition for H2 between D. hypogeium and A. psammolithicum was examined, sulfate reduction was the dominant process.
In recent years, there have been many studies describing competition among microorganisms carrying out terminal electron-accepting processes in anaerobic ecosystems. Thermodynamically favored processes typically dominate when competition occurs among microorganisms that use respiratory electron acceptors (7). Acetogenic bacteria are common inhabitants of almost all anaerobic ecosystems and are known to grow rapidly with H2 and CO2. However, thermodynamic calculations (4) and the results of studies with pure cultures (7) suggest that both sulfate-reducing bacteria and methanogenic bacteria outcompete autotrophic acetogens for available hydrogen or formate. Nevertheless, acetogenesis is favored in certain ecosystems. Of note is the termite hindgut; in the hindguts of several termite species acetogenesis is known to preclude methanogenesis (3). The results of studies performed with tundra soils suggest that acetogenesis has a greater temperature tolerance than methanogenesis and can therefore compete effectively for electrons under psychrophilic conditions (27). It has also been suggested that acidic or carbon-limited environments may also favor acetogenesis (17, 28). Acetogenesis has been shown to occur and may be responsible for the accumulation of acetate in fine-grained subsurface Atlantic Coastal Plain sediments deposited during the Cretaceous period (5). Competition was not examined in the latter study.
Only one acetogen, strain SS1, has been isolated from the terrestrial subsurface (22). Many acetogens, including strain SS1, can grow mixotrophically with H2 and aryl O-methyl groups, and thermodynamic calculations suggest that acetogens could compete for H2 with sulfate-reducing bacteria (22) under these conditions. However, experiments have not been carried out to test this hypothesis.
Our previous studies with subsurface sandstones and shales deposited during the Cretaceous period (about 100 million years ago) revealed that both sulfate reduction and acetate accumulation occurred in batch incubation experiments (20). The source of the acetate was not clear, but it is possible that this compound may have originated from CO2 reduction. To investigate this phenomenon, we established enrichment cultures in which sulfate was included as an electron acceptor. In this paper, we present data obtained with these enrichment cultures, and our data demonstrate that both sulfate-reducing and acetogenic bacteria are present in the sandstones examined. We also describe the isolation and characterization of these organisms and their abilities to compete for H2 as a substrate.
MATERIALS AND METHODS
Batch incubation of sandstones and shales.
Rock cores (diameter, 6 cm) were obtained from the Cerro Negro drilling site in central New Mexico as previously described (11, 12). Ground rock material was aseptically removed from the interiors of the cores by either drilling from freshly fractured core faces or by grinding pared core material with a mortar and pestle (20). This ground rock material (0.45 to 1.0 g) was incubated in serum tubes with 1 ml of filter-sterilized groundwater from the site. The groundwater contained 3.9 mM endogenous sulfate, was supplemented with 10 mg of Na2HPO4 per liter and 20 mg of NH4Cl per liter, and was reduced with 1 mM Na2S · 9H2O. Groundwater that originally was maintained under an N2 atmosphere was adjusted to pH 7.8 by adding N2-CO2 (4:1) to the headspace. The headspaces of the serum tubes (headspace-to-liquid ratio, 2.6:1) were sampled by withdrawing 0.3-ml gas samples with a syringe. The H2 contents were then determined by injecting the samples into a reduction-gas analyzer (model RGA3; Trace Analytical, Menlo Park, Calif.).
Similar batch incubation preparations were used to measure sulfate reduction activity linked to the consumption of H2. Ground rock material and groundwater (0.75 g of sandstone from a depth of 213 m or 1.5 g of sandstone from a depth of 247 m and 5 ml of amended water), as well as H2 (39 kPa) and 10 μCi of Na235SO4 (20), were included in these incubation mixtures. Samples (0.5 ml of slurry) were removed periodically, and the total reduced inorganic sulfur was extracted and quantitated by the procedure of Ulrich et al. (35).
In the experiment described above, acetate and sulfate contents in culture supernatants were determined by ion chromatography (Dionex Instruments, Sunnyvale, Calif.) with an AS-11 column and a 0.25 to 22.5 mM NaOH gradient mobile phase. In subsequent experiments, acetate contents were determined by gas chromatography (32).
Enrichment cultures.
Batch incubation mixtures containing sediments from a depth of 213 m and groundwater from the same Cubero sandstone (20) along with H2 (4 ml per 25-ml serum bottle) as a substrate were incubated for approximately 6 months. These cultures produced 4.1 mM acetate. Subsamples of these cultures were transferred (10%) into a mineral medium prepared as described by Balch et al. (2) and modified (18, 21) so that it contained vitamins, trace metals, 0.02% yeast extract, 2% rumen fluid, 10 mM Na2SO4, 40 mM HCO3−, 20% CO2, and 50 KPa H2 (pH 6.8). After 1 week of incubation, the cultures reached the stationary phase, as determined by measuring optical density. The acetate in culture supernatants was quantified, and sulfide contents were then determined by a colorimetric procedure (6). Another transfer was made into the same medium and also into a similar medium at pH 7.9 to determine the influence of pH on the growth of the organisms in the enrichment culture. The latter medium was prepared under N2 rather than under N2-CO2 (as was the case at pH 6.8). After the medium was autoclaved, NaHCO3 (7% solution equilibrated under N2-CO2) was added to a final concentration of 0.7%.
Isolation of cultures.
The pH 6.8 enrichment culture was transferred (10%) into a mineral medium containing sulfate, yeast extract, and rumen fluid along with H2 as an electron donor (20% CO2 and 40 mM HCO3−). After the culture had reached the stationary phase (about 1 week), it was transferred again and grown with H2 as the electron donor. This enrichment culture was serially diluted and inoculated into roll tubes containing the medium described above supplemented with 2% agar (16). After a 3-week incubation period, several colonies were picked from the agar-containing roll tubes originally inoculated with the 10−7 dilution of the enrichment culture. These colonies were transferred to the same medium lacking agar. Isolates were then categorized based on whether sulfide or acetate was the major product. One acetogenic strain, strain CN-E, and one sulfate-reducing strain, strain CN-A, were chosen for further study. Both of these isolates grew more rapidly at 30°C than at 23 or 37°C.
Pure-culture studies.
In general, both the acetogenic isolate and the sulfate-reducing isolate grew in a defined mineral medium (similar to the medium described above but lacking yeast extract and rumen fluid), but the growth rates were considerably slower in the absence of yeast extract. The sulfate reducer required acetate (10 mM) as a carbon source when yeast extract was not included. Utilization of substrates by both organisms was determined in the same media containing 0.02% yeast extract. The growth rate and H2 concentrations for the acetogenic isolate were determined in the mineral medium lacking yeast extract.
Hydrogen competition experiments.
Competition for H2 between strain CN-E and strain CN-A was studied in mixed cultures containing both organisms. The bacteria were grown separately in a mineral medium containing 0.01% yeast extract, 3 mM syringate, 10 mM Na2SO4, 2 atm of H2-CO2 (4:1), and 40 mM HCO3−. Mid-log-phase cultures were used as inocula for the competition experiment described below. Triplicate incubation mixtures were prepared by using the same medium used to grow the inocula and either a low H2 concentration (1% of the serum tube headspace) or a high H2 concentration (80% of the serum tube headspace). For each experiment, the following four incubation mixtures were prepared: (i) uninoculated medium, (ii) medium containing strain CN-A, (iii) medium containing strain CN-E, and (iv) medium containing both organisms. Inoculation mixtures were prepared such that the two cultures were present at the same optical density at 600 nm at the start of incubation. The preparations were incubated at 30°C. Acetate and sulfide production were monitored over time in an effort to determine the fate of the available reducing equivalents for each treatment.
16S ribosomal DNA sequencing.
Genomic DNAs were extracted from 500-ml cultures of the two strains by using a standard miniprep procedure, as described previously (1). The nearly full-length 16S rRNA gene was amplified from genomic DNA by PCR using forward primer Eubac27F and reverse primer 1492R (8). PCR mixtures were prepared and thermal cycling was carried out as described previously (8). In addition, acetamide (5%, wt/vol) was added to the reaction mixtures to enhance the specificity of the amplification reactions (29). All reactions were performed in triplicate, and the reaction mixtures were overlaid with mineral oil. Following amplification, each DNA mixture was purified by using a Wizard PCR Prep purification kit (Promega Corp., Madison, Wis.) and was resuspended in sterile H2O. Both strands of the purified PCR products were sequenced by automated dye dideoxy terminator sequencing at the Michigan State University Sequencing Facility with a model 373A DNA sequencing system (Applied Biosystems, Foster City, Calif.). Oligonucleotides complementary to the conserved regions of the eubacterial 16S rRNA were used to prime the sequencing reactions and were synthesized with either a model 394 DNA-RNA synthesizer or a model 380B DNA synthesizer (Applied Biosystems). The sequencing reaction mixtures contained 12 pmol of sequencing primer and 50 to 250 ng of PCR template in 20 μl (total volume) of sterile H2O.
Phylogenetic analysis.
The 16S ribosomal DNA secondary structures were determined manually by using templates in the Ribosomal Database Project database (26) to aid in identification of homologous sequence positions. Sequences were aligned manually in the Genetic Data Environment (31). All of the reference sequences and the basic alignment to which the sequences of the new strains were added were obtained from the Ribosomal Database Project. Only homologous sites at which the 16S rRNA sequences aligned unambiguously were included in further analyses. Phylogenetic and bootstrap analyses were performed by using the DNADIST program with the Jukes-Cantor correction, the SEQBOOT program with 100 replicates, and the FITCH program with input randomization and global rearrangement, all of which were contained in the PHYLIP 3.5 package (10) implemented through the Genetic Data Environment. Bootstrap values are given if they were greater than 50%.
Nucleotide sequence accession numbers.
The 16S rRNA sequences of strains CN-A and CN-E have been deposited in the GenBank database under accession no. AF132738 and AF132739, respectively.
RESULTS
Enrichment cultures.
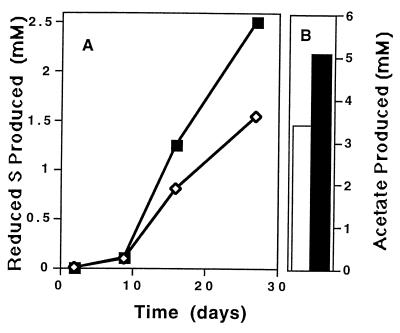
Sandstone-groundwater slurries incubated with actively sulfidogenic (20) rock material from the Cerro Negro site initially produced H2 reaching a concentration of about 500 nM. With time, the H2 was consumed, and the H2 concentration reached a steady state level near 5 nM. Similar preparations to which H2 was added produced both sulfide and acetate (Fig. 1) at levels well above the levels observed when there was no exogenous electron donor (20). These results indicated that both acetogenesis and sulfate reduction were stimulated when H2 was available as an electron donor.
FIG. 1.
Microbial activity in ground sandstone-groundwater mixtures incubated with added H2. (A) Sulfate reduction over time, expressed as the total amount of reduced inorganic sulfur formed. (B) Concentrations of acetate after incubation for 3 months. Sandstones were obtained from depths of 213 m (◊ and open bar) and 247 m (■ and solid bar).
Competition within the enrichment culture.
After subsequent transfers of the cultures, both sulfate and acetate continued to be produced. Because groundwater from the site had a pH of 8.3 and pH is thought to influence competition among acetogens and other hydrogen-utilizing bacteria (28), an attempt was made to test the influence of pH on the use of H2 in the two competing processes. Both acetate production and sulfide production were measured in parallel enrichment cultures at pH 6.8 and 7.9. The results showed that there was virtually no difference between the fates of electrons at the two pH values, suggesting that pH is not the major factor controlling competition in this environment. Under the conditions of the enrichment culture, sulfate reduction consumed the lesser amount of electrons (23 to 30%) available from H2, which suggested that laboratory culture may favor acetogenesis over sulfate reduction.
Characterization of the sulfate reducer.
To further understand the competing processes in the subsurface system studied, we isolated pure cultures of bacteria for additional study. Strain CN-A was isolated as a gray-black colony that grew well with H2 and sulfate in the medium used for enrichment and isolation. It did not grow under these conditions in the absence of sulfate. Originally, the cultures were flocculent, and the majority of the biomass apparently was present as 0.5- to 1.0-mm flocs. These flocs were not broken up when a culture was shaken or subjected to mild sonication. Therefore, in the initial studies we could not measure optical density. After repeated transfers, the cultures lost the ability to produce large flocs, and later transfers produced turbid cultures with finer, almost undetectable flocs.
Cells of CN-A were short rods that were 0.6 to 0.8 μm wide by 1.4 to 1.9 μm long. A single polar flagellum was observed on each phosphotungstic acid-stained cell, as determined with a JEOL model 2000FX transmission electron microscope (data not shown) This organism grew in a mineral medium containing 10 mM acetate as a carbon source but grew more rapidly and abundantly in a medium containing 0.02% yeast extract. The substrates tested which supported growth when sulfate (10 mM) was the electron acceptor included ethanol (10 mM), pyruvate (10 mM), lactate (10 mM), formate (20 mM), and H2 (1 atm). Acetate was produced during growth on lactate. The substrates which did not support growth in the presence of sulfate included fructose (2 mM), acetate (10 mM), fumarate (10 mM), methanol (10 mM), malate (10 mM), and choline (10 mM). Pyruvate (10 mM) supported growth in the presence of sulfate but not fermentatively. Growth occurred when H2 was the electron donor and thiosulfate (10 mM) or sulfite (10 mM) was the electron acceptor. Growth was not detected when nitrate or fumarate was the electron acceptor. Desulfoviridin was not detected.
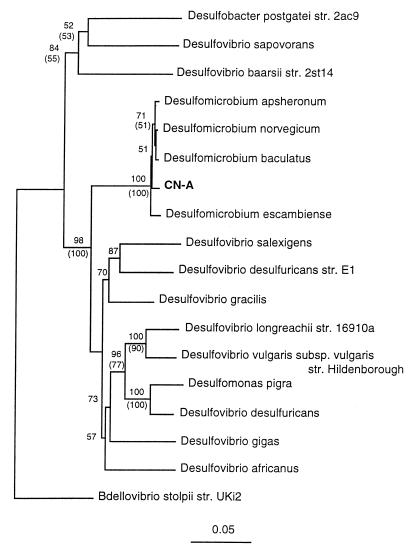
Based on an analysis of the 16S rRNA sequence (Fig. 2), strain CN-A is related to the tightly clustered members of the genus Desulfomicrobium. This new strain exhibited 98.4 to 99.0% homology with the previously described strains of this genus. On the other hand, the levels of homology with Desulfovibrio species were less than 90%. Desulfovibrio desulfuricans Norway4 (to which CN-A is closely related phylogentically) has been recently renamed Desulfomicrobium norvegicum (13).
FIG. 2.
Phylogenetic distance tree for Desulfomicrobium hypogeium CN-A based on 16S rDNA sequences. Bootstrap values greater than 50% are shown at the nodes. Bootstrap values are shown for both distance (numbers without parentheses) and parsimony (numbers within parentheses) analyses. Bar = five nucleotide differences per 100 nucleotide positions. str., strain.
Characterization of the acetogenic culture.
Strain CN-E was isolated from a colony after 2 weeks of incubation at 24°C. It grew well on H2 and CO2 in the medium used for enrichment and isolation. It grew slowly in a mineral medium in the absence of yeast extract or rumen fluid, with turbidity appearing only after more than 1 month when a 5% inoculum was used. However, addition of yeast extract (0.02%) or rumen fluid (5%) increased both the growth rate and the yield.
Cells of CN-E were rods that were 1.1 μm wide by 1.7 to 3.3 μm long. Flagella were not detected when phosphotungstic acid-stained cells were examined.
The substrates tested that supported growth in the presence of yeast extract (0.02%) included H2-CO2 (1 atm of H2), formate (30 mM), methanol (10 mM), glucose (2 mM), syringate (5 mM), pyruvate (10 mM), lactate (10 mM), betaine (5 mM), ethanol (10 mM), propanol (10 mM), glycerol (10 mM), and acetoin (5 mM). The substrates which did not support growth included fructose (2 mM), choline (5 mM), and ethylene glycol (5 mM).
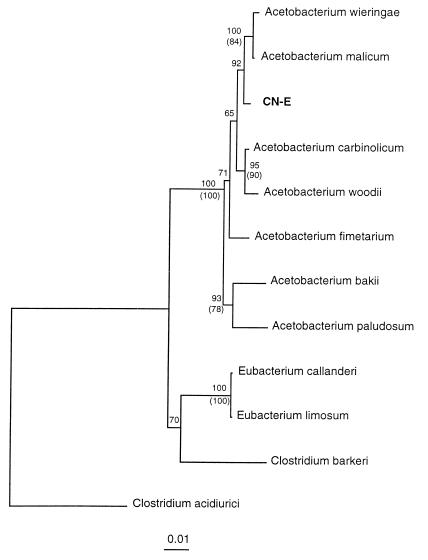
Analysis of the 16S rRNA sequence indicated that strain CN-E is related to the tightly clustered members of the genus Acetobacterium (Fig. 3). This new strain exhibited 97.0 to 99.3% homology with the previously described strains of this genus. The closest relatives based on phylogenetic analysis data are Acetobacterium wieringae and Acetobacterium malicum.
FIG. 3.
Phylogenetic distance tree for Acetobacterium psammolithicum CN-E based on 16S rDNA sequences. Bootstrap values greater than 50% are shown at the nodes. Bootstrap values are shown for both distance (numbers without parentheses) and parsimony (numbers within parentheses) analyses. Bar = one nucleotide difference per 100 nucleotide positions.
Studies were carried out to determine some of the factors that promote the growth of this organism (Table 1). When the organism was grown on formate alone or on syringate alone in a mineral medium, growth was slow and unreliable. We grew this strain on a combination of C1 substrates (formate and syringate), which resulted in more rapid and reliable growth, and then added glucose at different concentrations. This experiment allowed us to determine the influence of glucose on the growth of this organism. Glucose stimulated growth of strain CN-E. The final optical densities obtained all three substrates were greater than the sums of the individual optical densities (Table 1). This effect was not observed when we used the combination of glucose and formate (data not shown). The latter combination of substrates produced the same growth yield as glucose alone produced. Formate alone resulted in only a low growth yield which did not detectably change the optical density of the culture.
TABLE 1.
Final optical densities and doubling times for strain CN-E grown with different concentrations of glucose, 4 mM syringate, and 20 mM formate
| Glucose concn (mM) | Presence of syringate and formate | Final optical density at 600 nm | Doubling time (days) |
|---|---|---|---|
| 0 | + | 0.16 | 1.1 |
| − | NMIa | NDb | |
| 0.5 | + | 0.23 | ND |
| − | 0.015 | ND | |
| 1.0 | + | 0.32 | ND |
| − | 0.03 | ND | |
| 2.0 | + | 0.39 | 0.92 |
| − | 0.07 | 1.53 |
NMI, no measurable increase in growth.
ND, not determined.
Determination of H2 threshold values.
To characterize the abilities of the isolates to metabolize H2, we grew cultures and measured the H2 concentrations at the end of the exponential phase of growth. Cultures (10 ml) were grown as described above with 2.4 ml of H2 added to the 18-ml headspace. Strain CN-A removed H2 so that the final H2 concentration was 2.1 nM when it was grown in a medium containing 10 mM Na2SO4 and 0.02% yeast extract. When strain CN-E was grown in mineral media containing combinations of growth substrates, the following H2 threshold concentrations (mean ± standard deviation) were observed: 414 ± 104 nM for H2 alone; 278 ± 46 nM for H2 plus glucose; 204 ± 18 nM for H2 plus syringate; and 195 ± 13 nM for H2 plus syringate plus glucose. In these experiments, the glucose and syringate concentrations were 2 and 4 mM, respectively.
H2 competition experiments.
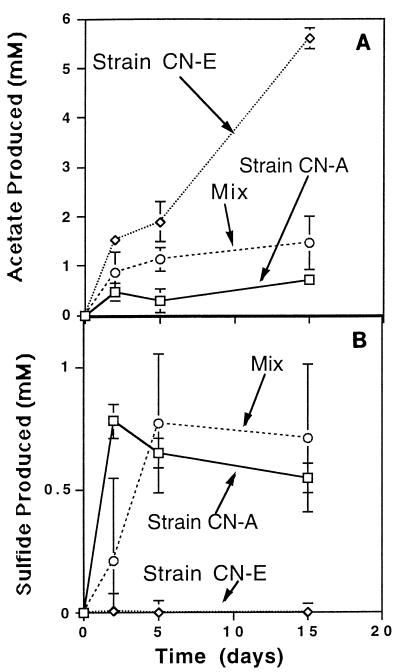
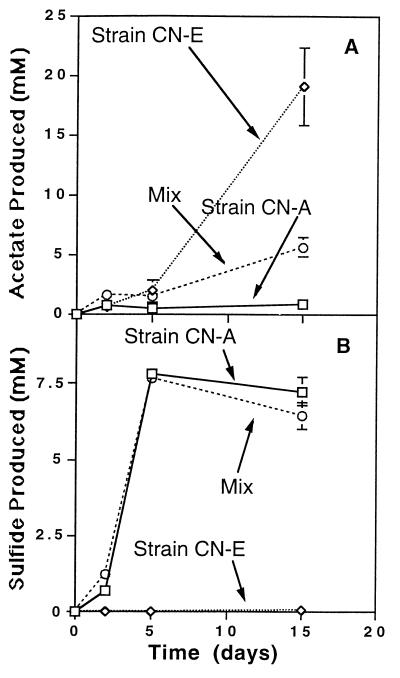
In H2 competition experiments, pure cultures were grown separately, mixed, and incubated. The fates of reducing equivalents in cocultures and pure cultures were then compared. Strain CN-E accumulated 6.5 and 19.8 mM acetate (Fig. 4A and 5A) under headspaces containing limiting and saturating H2 concentrations, respectively. The pattern of end product formation for strain CN-A was similar in that more sulfide was produced under saturating H2 conditions (Fig. 4B and 5B). However, under limiting H2 conditions, the presence of strain CN-A in a coculture with strain CN-E resulted in a 75% decrease in the total amount of acetate produced (Fig. 4A). Similarly, the presence of strain CN-A dramatically reduced the level of acetogenesis when H2 was included at saturating levels (Fig. 5A). In contrast, the levels of sulfide produced, used as a measure of H2 consumption by the sulfate reducer, were virtually unaffected by the presence of the acetogen (Fig. 4B and 5B). The results of these experiments suggest that the acetogen was not able to compete effectively for reducing equivalents under either of the incubation conditions used.
FIG. 4.
End product formation in a competition experiment performed with a low H2 concentration (1% H2, 1 atm).
FIG. 5.
End product formation in a competition experiment performed with a high H2 concentration (80% H2, 2.2 atm).
Pure cultures of strain CN-E did not accumulate sulfide, and strain CN-A did not accumulate acetate during the incubation period (data not shown). In addition, uninoculated controls did not produce acetate or sulfide during the experiment.
DISCUSSION
The terminal steps of the microbial food chain in anaerobic subsurface communities are generally thought to function as they do in surficial anaerobic communities; that is, consumption of H2 and consumption of acetate occur by mechanisms similar to the mechanisms in the surface counterparts. It was with this in mind that we began to examine H2 consumption in active sandstones from the Cerro Negro Site in central New Mexico.
Our initial observations indicated that sulfate reduction could occur in ground sandstones, but methanogenesis was not detectable (20). Acetate also accumulated during incubation. H2 concentration is thought to be an indicator of the terminal electron-accepting microbial process (24). We measured the concentrations of H2 in groundwater-ground rock slurries and found that the equilibrium concentration was about 5 nM, suggesting that sulfate reduction was the dominant terminal electron-accepting microbial process (24). However, when H2 was added to the incubation mixtures, acetate was also produced (Fig. 1) at levels (3 to 5 mM) which suggested that autotrophic acetogenesis was occurring. In the absence of added H2, the acetate levels remained less than 1.5 mM. We also observed relatively rapid sulfate reduction under these conditions (Fig. 1).
In order to produce the 3 to 5 mM acetate observed in our experiments, acetogenic bacteria would have had to compete effectively with the sulfate reducers for hydrogen during the early part of the incubation (when hydrogen levels were still high). This suggested to us that acetogenic bacteria were relatively abundant in the rock material and therefore important members of the subsurface microbial community. It then became imperative to determine the roles of both sulfate-reducing bacteria and acetogenic bacteria in hydrogen consumption in this subsurface community.
Therefore, we isolated both the most abundant sulfate reducer and the most abundant acetogen in our enrichment cultures and studied their activities. Strain CN-A was characterized and was found to be similar in terms of substrate utilization pattern to other species of the genus Desulfomicrobium (14). It differs from Desulfomicrobium escambiense in that it does not ferment pyruvate, from Desulfomicrobium baculatus in that it oxidizes ethanol but not malate, and from Desulfomicrobium apsheronum in that it does not oxidize fumarate or malate or ferment pyruvate or fumarate. It also differs in its 16S rRNA sequence (Fig. 2). The differences between the 16S rRNA sequence of CN-A and the 16S rRNA sequences of the other Desulfomicrobium strains are greater than the differences among the 16S rRNA sequences of D. apsheronum, D. baculatus, and D. norvegicum. However, differentiation of D. apsheronum, D. baculatus, and D. norvegicum has recently been justified based on DNA-DNA similarities, restriction fragment length analysis results, and phenotypic characteristics (14). For these reasons, we suggest that strain CN-A should be placed in a new species Desulfomicrobium hypogeium.
Strain CN-E was also characterized. Based on rRNA sequence analysis data, this organism is most closely related to A. malicum and A. wieringae. However, CN-E appears to catabolize a broader range of substrates (33). The A. malicum strains examined did not use methanol, ethanol, propanol, and glucose, which were used for growth by strain CN-E, but one strain used choline and the other used ethylene glycol, two compounds which were not used by the new isolate. A. wieringae differs from strain CN-E in that it does not use methanol, ethanol, propanol, pyruvate, glucose, or O-methylated aromatic acids. It does use fructose, ethylene glycol, and choline. Since strain CN-E was isolated from a different habitat, exhibits different physiological features, and has a different rRNA sequence, we placed this strain in a new species, Acetobacterium psammolithicum.
Each of the two new isolates differs somewhat from previously isolated species, and this is not a surprise because of the unique habitat of these organisms. However, strains CN-A and CN-E are similar to surface-dwelling isolates in many more characteristics, which supports our belief that subsurface organisms do not differ dramatically from their surface counterparts.
Previous results have shown that acetogenic bacteria have complex and different growth requirements. For example, Syntrophococcus sucromutans requires an electron donor and an electron acceptor, as well as phospholipids as growth factors (9, 19). Similar patterns have been observed with Acetobacterium woodii cultures, in which a small amount of yeast extract stimulates growth (34). Lui and Suflita (22, 23) have reported that the combination of H2 and O-methyl groups of aromatic acids provides the requirements for growth of acetogenic bacteria and have contended that H2 is needed for utilization of aryl OCH3 groups. Our data for the acetogen described here indicate that neither substrate individually provides ideal growth conditions, but either formate or syringate supports growth to a limited extent. When both formate and syringate are present, growth is relatively abundant and is further enhanced by the addition of glucose. On its own, glucose supports only a low level of growth. Yeast extract is rich in sugars (19), and both yeast extract and glucose stimulate growth of this organism. Therefore, it is possible that some of the growth-stimulating response due to yeast extract observed with this organism and other Acetobacterium strains may be due to utilization of sugars in the yeast extract. These sugars may provide carbon necessary for growth, which ultimately results in higher growth yields.
Sulfate-reducing bacteria are thought to compete with acetogenic bacteria for H2 in a variety of environments. Competition may be regulated by thermodynamic, kinetic, or other factors (25). The kinetic parameters of individual cells that have been studied include Km, the maximal rate (Vmax), and the threshold value. Each parameter individually or all of the parameters together could influence the ability of the organisms to compete for H2. When sulfate-reducing bacteria and acetogenic bacteria are compared, the Km values are typically lower (about 1 μM for the sulfate reducers and 5 μM for the acetogens) and the Vmax is typically higher (by a factor of 10) for sulfate reducers (15, 30), which gives them a significant kinetic advantage.
Thermodynamic calculations for acetogenesis also suggest that sulfate reducers may have an advantage (22). However, when the calculations for growth of the acetogen under mixotrophic conditions are done, the results suggest that acetogenesis might compete with sulfate reduction as a terminal electron-accepting process (22). In this study, we found that the acetogenic isolate had a H2 threshold value that was on the order of 100-fold higher than the H2 threshold value observed with the sulfate reducer. Our H2 threshold results are similar to those reported for other acetogens (7) grown under similar conditions. However, when Cord-Ruwisch et al. (7) incubated A. woodii in the presence of caffeate, the H2 threshold value was reduced to levels comparable to those obtained with sulfate-reducing bacteria. We determined the threshold values with several different substrate combinations for the acetogen (strain CN-E). Addition of syringate or syringate and glucose had a significant effect on the threshold value and resulted in a decrease of about 50% compared to cells grown on H2 alone. The threshold value was still much greater than the threshold value observed during sulfate reduction, whereas the thermodynamics of mixotrophic acetogenesis and sulfate reduction are comparable (22, 23). These results suggest that one of the following hypotheses is true: (i) thermodynamics is not the major factor that controls H2 threshold values in acetogens or (ii) consumption of H2, consumption of glucose, and consumption of O-methyl groups by these organisms are not sufficiently coupled to allow a highly favorable reaction to provide energy that drives a less favorable reaction forward.
Under conditions where fermentation of organic matter supports the microbial community, as most likely occurs in the subsurface system we studied, the microbial population with the lowest H2 threshold value is expected to be the most successful population in competing for H2 (24). We therefore expect that acetogens would not be able to compete for H2 in this subsurface system. Pure-culture experiments showed that this was the case; the sulfate reducer dominated under all conditions (Fig. 4 and 5). However, our sediment cultures (Fig. 1) and enrichment cultures provided results which showed that acetogenesis was a significant process when saturating H2 concentrations were available. When H2 was present at threshold concentrations, all of our results suggested that the sulfate reducers were the major sink for the reducing equivalents present in H2. Sediment cultures initially produced hydrogen at a level greater than the threshold level for acetogens, possibly due to the perturbation associated with grinding sediments to create the slurry system. This may partially explain the high level of acetogenesis observed in our sediment incubation mixtures. It is also clear that Acetobacterium species are capable of using a wide range of compounds (33). The sandstones used in this study are continually being supplied with complex organic compounds that diffuse from adjacent shales (20). Therefore, it seems likely that the acetogens produce acetate both autotrophically and, more importantly, from the degradation of low-molecular-weight organic compounds which may be present as products of biodegradation.
Description of Desulfomicrobium hypogeium sp. nov.
Desulfomicrobium hypogeium (hy.po.gei′um. Gr. adj. hypo, under; Gr. fem. n. ge [or gaia], earth; M. L. neut. adj. hypogeium, under the earth).
Motile, gram-negative, short rods that are 0.6 to 0.8 μm wide by 1.4 to 1.9 μm long. A single polar flagellum is present. No spores are formed.
Strict anaerobe. Sulfate, sulfite, and thiosulfate are reduced. Ethanol, pyruvate, lactate, formate, and H2 serve as electron donors. Acetate is produced from lactate. This organism grows in mineral medium containing 10 mM acetate as a carbon source but grows more rapidly and abundantly in a medium containing 0.02% yeast extract. Substrates which do not support growth with sulfate include fructose (2 mM), acetate (10 mM), fumarate (10 mM), methanol (10 mM), malate (10 mM), and choline (10 mM). Pyruvate (10 mM) does not support fermentative growth. Nitrate and fumarate are not used as electron acceptors.
The type strain exhibits 98.4 to 99.0% homology with the previously described strains in the genus Desulfomicrobium based on an analysis of the 16S rRNA sequence.
Habitat: subsurface sandstone.
Type strain: CN-A, which has been deposited in the Subsurface Microbial Culture Collection as strain SMCC/W 750.
Description of Acetobacterium psammolithicum sp. nov.
Acetobacterium psammolithicum (psam.mo.li′thi.cum. Gr. fem. n. psammos, sand; Gr. masc. n. lithos, stone; M. L. neut. adj. lithicum, of stone; M. L. neut. adj. psammolithicum, of sandstone).
Gram-negative rods that are 1.1 μm wide by 1.7 to 3.3 μm long. Flagella are not observed. No spores are formed.
This organism grows in a mineral medium but grows more rapidly and abundantly in a medium containing 0.02% yeast extract. The substrates which support growth include H2, formate, methanol, glucose, syringate, pyruvate, lactate, betaine, ethanol, propanol, glycerol, and acetoin. Substrates which do not support growth include fructose, choline, and ethylene glycol.
The type strain exhibits 97.0 to 99.3% homology with the previously described strains in the genus Acetobacterium based on an analysis of the 16S rRNA sequence. The closest relatives based on phylogenetic analysis are A. wieringae and A. malicum.
Habitat: subsurface sandstone.
Type strain: CN-E, which has been deposited in the Subsurface Microbial Culture Collection as strain SMCC/W 751.
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of energy Subsurface Science Program.
We thank Calvin Byre and Hans Trüper for help with nomenclature.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 2.Balch W E, Wolfe R S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brauman A, Kane M D, Labat M, Breznak J A. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science. 1992;257:1384–1387. doi: 10.1126/science.257.5075.1384. [DOI] [PubMed] [Google Scholar]
- 4.Breznak J A. Acetogenesis from carbon dioxide in termite guts. In: Drake H L, editor. Acetogenesis. London, United Kingdom: Chapman and Hall; 1994. pp. 303–330. [Google Scholar]
- 5.Chapelle F H, Bradley P M. Microbial acetogenesis as a source of organic acids in ancient Atlantic Coastal Plain sediments. Geology. 1996;24:925–928. [Google Scholar]
- 6.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 7.Cord-Ruwisch R, Seitz H-J, Conrad R. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch Microbiol. 1988;149:350–357. [Google Scholar]
- 8.DeLong E. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dore J, Bryant M P. Lipid growth requirement and influence of lipid supplement on fatty acid and aldehyde composition of Syntrophococcus sucromutans. Appl Environ Microbiol. 1989;55:927–933. doi: 10.1128/aem.55.4.927-933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Fredrickson J K, McKinley J P, Bjornstad B N, Long P E, Ringelberg D B, White D C, Krumholz L R, Suflita J M, Colwell F S, Lehman R M, Phelps T J. Pore-size constraints on the activity and survival of subsurface bacteria in a late Cretaceous shale-sandstone sequence, northwestern New Mexico. Geomicrobiol J. 1997;14:183–202. [Google Scholar]
- 12.Fredrickson J K, Phelps T J. Subsurface drilling and sampling. In: Knudsen G, Stetzenbach L, McInerney M M, Walter M, editors. Manual of Environmental Microbiology. Washington, D.C: American Society for Microbiology; 1996. pp. 526–540. [Google Scholar]
- 13.Genthner B R S, Friedman S D, Devereux R. Reclassification of Desulfovibrio desulfuricans Norway 4 as Desulfomicrobium norvegicum comb. nov. and confirmation of Desulfomicrobium escambiense (corrig., formerly “escambium”) as a new species in the genus Desulfomicrobium. Int J Syst Bacteriol. 1997;47:889–892. [Google Scholar]
- 14.Genthner B R S, Mundfrom G, Devereux R. Characterization of Desulfomicrobium escambium sp. nov. and proposal to assign Desulfovibrio desulfuricans strain Norway 4 to the genus Desulfomicrobium. Arch Microbiol. 1994;161:215–219. [Google Scholar]
- 15.Harris S H, Suflita J M. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Hydrogen consumption kinetics of acetogens and their abilities to compete with other hydrogenotrophs, abstr. N103. [Google Scholar]
- 16.Hungate R E. A roll tube method for cultivation of strict anaerobes. In: Norris J R, Ribbons D W, editors. Methods in microbiology. New York, N.Y: Academic Press, Inc.; 1969. pp. 117–132. [Google Scholar]
- 17.Jones J G, Simon B M. Interaction of acetogens and methanogens in anaerobic freshwater sediments. Appl Environ Microbiol. 1985;49:944–948. doi: 10.1128/aem.49.4.944-948.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumholz L R, Bryant M P. Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch Microbiol. 1986;144:8–14. [Google Scholar]
- 19.Krumholz L R, Bryant M P. Syntrophococcus sucromutans sp. nov. gen. nov. uses carbohydrates as electron donors and formate, methoxymonobenzenoids or Methanobrevibacter as electron acceptor systems. Arch Microbiol. 1986;143:313–318. [Google Scholar]
- 20.Krumholz L R, McKinley J P, Ulrich G A, Suflita J M. Confined subsurface microbial communities in Cretaceous rock. Nature. 1997;386:64–66. [Google Scholar]
- 21.Krumholz L R, Sharp R, Fishbain S S. A freshwater anaerobe coupling acetate oxidation to tetrachloroethylene dehalogenation. Appl Environ Microbiol. 1996;62:4108–4113. doi: 10.1128/aem.62.11.4108-4113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Suflita J M. H2-CO2-dependent anaerobic O-demethylation activity in subsurface sediments and an isolated bacterium. Appl Environ Microbiol. 1993;59:1325–1331. doi: 10.1128/aem.59.5.1325-1331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, Suflita J M. H2 as an energy source for mixotrophic acetogenesis from the reduction of CO2 and syringate by Acetobacterium woodii and Eubacterium limosum. Curr Microbiol. 1995;31:245–250. [Google Scholar]
- 24.Lovley D R, Goodwin S. Hydrogen concentration as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim Cosmochim Acta. 1988;52:2993–3003. [Google Scholar]
- 25.Lupton F S, Zeikus J G. Physiological basis for sulfate-dependent hydrogen competition between sulfidogens and methanogens. Curr Microbiol. 1984;11:7–12. [Google Scholar]
- 26.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nozhevnikova A N, Kotsyurbenko O R, Simankova M V. Acetogenesis at low temperature. In: Drake H L, editor. Acetogenesis. London, United Kingdom: Chapman and Hall; 1994. pp. 416–431. [Google Scholar]
- 28.Phelps T J, Zeikus J G. Influence of pH on terminal carbon metabolism in anoxic sediments from a mildly acidic lake. Appl Environ Microbiol. 1984;48:1088–1095. doi: 10.1128/aem.48.6.1088-1095.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson J A, Tiedje J M. Competition between sulfate reducing and methanogenic bacteria for H2 under resting conditions and growth conditions. Arch Microbiol. 1984;137:26–32. [Google Scholar]
- 31.Smith S W, Overbeck R, Olsen G, Woese C, Gillevet P M, Gilbert W. Genetic data environment and the Harvard genome database. In: Myers R M, Porteous D, Roberts R J, editors. Abstracts of papers presented at the 1992 Meeting on Genome Mapping and Sequencing. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. p. 190. [Google Scholar]
- 32.Supelco, Inc. GC separation of VFA C2-C5. Bulletin 749E. Bellafonte, Pa: Supelco Inc.; 1975. [Google Scholar]
- 33.Tanaka K, Pfennig N. Fermentation of 2-methoxyethanol by Acetobacterium malicum sp. nov. and Pelobacter venetianus. Arch Microbiol. 1988;149:181–187. [Google Scholar]
- 34.Tschech A, Pfennig N. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch Microbiol. 1984;137:163–167. [Google Scholar]
- 35.Ulrich G A, Krumholz L R, Suflita J M. A rapid and simple technique for measuring sulfate reduction activity and quantifying sulfides. Appl Environ Microbiol. 1997;63:1627–1630. doi: 10.1128/aem.63.4.1627-1630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]