Abstract
Diarrhea is a major health problem in neonatal and young calves worldwide. It can be caused by a variety of infectious agents, including the bacteria Salmonella enterica serovar Dublin (S. Dublin), enterotoxigenic Escherichia coli (ETEC), and Clostridium perfringens. Preventive alternatives to antibiotic treatment should be identified. As a first step toward this, the aim of the current study was to examine whether cell-free supernatants from cow milk fermented by lactic acid bacteria affects virulence-gene expression in strains of S. Dublin, ETEC E. coli F5 and C. perfringens. pH-neutralized, cell-free, spent medium of milk (nCFSM) fermented by 61 different lactic acid bacteria (LAB) and non-LAB starter cultures belonging to 17 genera was assayed for their effect on expression of important virulence factors (S. Dublin hilA, ssrB, ssaG, flhD, prgI, fliC; ETEC E. coli F5 fanC, estA, fim41a; C. perfringens cpa), when the bacteria were grown in the nCFSM. Screening was done using either a promoter-reporter expression system or RT-qPCR. nCFSM from Bifidobacterium longum BL-15955 and Limosilactobacillus reuteri LR-33016 downregulated the expression of fanC, fim41a and estA genes in the four tested ETEC E. coli F5 strains without affecting their growth, while mainly B. longum BL-15955 downregulated expression of cpa in the four tested strains of C. perfringens. nCFSM from the mixed cultures; NU-TRISH® BY-Mild (Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus and Bifidobacterium BL-15954) and COMBO4 (Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus), as well as Lactobacillus helveticus CNRZ32 downregulated the tested virulence genes in the three tested strains of S. Dublin. To enable possible downregulation of the expression of virulence genes in all three target bacteria simultaneously, nCFSM was prepared from NU-TRISH® By-Mild in combination with B. longum BL-15955 (i.e. a four-strain combination). The nCFSM from this combination downregulated the virulence genes expression in all the three species. In the future, NU-TRISH® By-Mild and B. longum BL-15955 in combination could potentially be used for prevention of neonatal calf diarrhea caused by S. Dublin, E. coli F5, and C. perfringens, reducing the need for antimicrobial treatment, however, field studies are needed to prove that.
Keywords: calf diarrhea, Salmonella Dublin, Escherichia coli F5, Clostridium perfringens, virulence inhibition
Introduction
Diarrhea is common in young calves causing severe welfare problems and economic losses to cattle producers worldwide. Ten different enteric pathogens are recognized as either major [(bovine rotavirus (BRV), bovine coronavirus (BCoV), bovine viral diarrhea virus (BVDV), Salmonella enterica, Escherichia coli, Clostridium perfringens, Cryptosporidium parvum and Eimeria] or emerging pathogens [(bovine norovirus (BNoV) and bovine torovirus (BToV)] of diarrhea (Smith, 2009; Cho and Yoon, 2014). Antimicrobial treatment may lead to development of antimicrobial resistance, which threatens public health as well as future treatment possibilities against the pathogens of calf diarrhea. Alternatives to antimicrobial treatment for control of the disease are highly needed. Inhibition of the virulence factors which enables the bacteria to cause diarrhea have been suggested as a possibility (Rasko and Sperandio, 2010).
Among the bacterial pathogens, strains with particular traits dominate. In E. coli, strains that produce the K99 (F5) fimbriae and the heat-stable enterotoxin, STa, are traditionally associated with the disease and are known to cause more severe diarrhea than other types. Among Salmonella types, the cattle-adapted serovar, S. Dublin is the most commonly reported and economically important one (Nataro and Kaper, 1998; Acha et al., 2004; Cho and Yoon, 2014). S. Dublin, unlike most other Salmonella serovars, tends to cause systemic disease. It invades in the intestine using a Type Three Secretion system (TTSS) encoded from Salmonella pathogenicity island 1. From there it spreads to systemic sites, where it can survive intracellularly due to genes from another TTSS, encoded from Salmonella pathogenicity island 2. Pathology of the third bacterial species, C. perfringens, depends on toxin production. Calf diarrhea is in particular associated with strains of toxin types B and C, but type A can also be associated with the diseases in rare cases. The α toxin is the main lethal toxin shared between these three types. It promotes cell lysis through the hydrolysis of membrane phospholipids (Songer, 1997; Perez et al., 1998).
Lactic acid bacteria (LAB) are a group of Gram-positive, lactic acid producing Firmicutes. They have been used for centuries as starter or adjunct cultures in dairy fermentations. Milk proteins are a major source of bioactive peptides and an increasing number of bioactive peptides have been identified in milk protein hydrolysates and fermented dairy products (Nagpal et al., 2011). The breakdown of milk proteins by LAB plays an important role in generating peptides and amino acids for bacterial growth and in the formation of metabolites that contribute to flavor formation of fermented products. Recent studies indicate that degradation products from certain LAB, as well as some Bifidobacterium strains affect expression of virulence-associated genes in specific pathogens. For instance, probiotic Lactobacillus acidophilus La-5 and Bifidobacterium longum NCC2705 strains have the ability to downregulate virulence genes (ciaB and flaA) expression in Campylobacter jejuni (Mundi et al., 2013). Likewise, B. bifidum ATCC25921, B. bifidum BBA1, B. crudilactis FR/62/B/3, and L. acidophilus La-5 produce metabolites inhibiting virulence gene expression of enterohemorrhagic E. coli O157:H7 (Medellin-Pena et al., 2007; Medellin-Pena and Griffiths, 2009; Zeinhom et al., 2012) and S. Typhimurium (Zeinhom et al., 2012; Bondue et al., 2016). L. acidophilus GP1B has been shown to cause downregulation of virulence genes in Clostridium difficile (Yun et al., 2014), and L. bulgaricus NRRL B548, Lacticaseibacillus rhamnosus NRRL B442, L. paracasei DUP-13076, and L. helveticus LH-2 affect attachment and invasion of Salmonella in vitro and in vivo through significant inhibition of the virulence genes expression (Tellez et al., 2011; Bayoumi and Griffiths, 2012; Muyyarikkandy and Amalaradjou, 2017; Ali et al., 2019).
Based on this we hypothesized that factors produced during fermentation of cow-milk by LAB and non-LAB starter cultures could inhibit virulence factors in S. Dublin, ETEC E. coli F5 and C. perfringens of importance for induction of diarrhea in young calves. The aim of the study was to determine the effects of spent milk fermented by different probiotic strains on virulence gene expression in these bacteria, and to identify one or more combinations of probiotic strains, which when pooled could prevent expression of the virulence genes in all three bacteria simultaneously.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains of pathogenic bacteria used (Table 1) were stored at −80 °C in 15% glycerol. Strains were whole genome sequenced (WGS) and analyzed for presence of virulence genes as previously described (Thomas et al., 2017). S. Dublin and E. coli F5 strains were cultivated aerobically in Luria-Bertani (LB) (Sigma, Copenhagen, Denmark) broth overnight at 37°C with shaking at 180 rpm. C. perfringens strains were cultivated anaerobically in Brain Heart Infusion (BHI) broth (Sigma, Copenhagen, Denmark) overnight at 37°C in an anaerobic jar plus Oxoid™ AnaeroGen™ 2.5L Sachets (ThermoFisher Scientific, Waltham, MA, USA) without shaking. A bioluminescent reporter strain of Salmonella Typhimurium enterica serovar LT2 containing a hilA::luxCDABE construct (Bayoumi and Griffiths, 2010) was grown overnight at 37 °C in LB broth supplemented with 50 μg/mL of ampicillin (Amp) (Sigma, Copenhagen, Denmark) to an approximate cell density of 1 × 109 cells mL−1.
Table 1.
Strains of pathogenic bacteria used in this study.
| Strain or construct | Serotype | Relevant virulence genes/genotypea | References |
|---|---|---|---|
| E. coli E21-79 | H10:O9 | fanC, fim41a, estA | This study |
| E. coli E38-72 | H37:O101 | fanC, fim41a, estA | This study |
| E. coli E242-3 | H9:O101 | fanC, fim41a, estA | This study |
| E. coli LG3 | H10:O101 | fanC, fim41a, estA | This study |
| S. Dublin MS14334 | S. Dublin | hilA, prgI, ssrB, ssaG, flhD, fliC | (Knudsen et al., 2011) |
| S. Dublin MS17265 | S. Dublin | hilA, prgI, ssrB, ssaG, flhD, fliC | (Knudsen et al., 2011) |
| S. Dublin MS17266 | S. Dublin | hilA, prgI, ssrB, ssaG, flhD, fliC | (Knudsen et al., 2011) |
| S. Dublin JEO3665 | S. Dublin | hilA, prgI, ssrB, ssaG, flhD, fliC | (Olsen et al., 2012) |
| C. perfringens C4-5 | C. perfringens Type A | Cpa | This study |
| C. perfringens C9-3 | C. perfringens Type A | Cpa | This study |
| C. perfringens C16-3 | C. perfringens Type A | Cpa | This study |
| C. perfringens C17-3 | C. perfringens Type A | Cpa | This study |
| S. Typhimurium hilA::lux | LT-2 | PhilA, luxCDABE, AmpR | (Bayoumi and Griffiths, 2010) |
aWhole genome sequences of the strains are deposited at the European Nucleotide Archive under study accession number PRJEB46410 with individual strain accession numbers.
A total of 61 LAB and non-LAB starter cultures, including strains belonging to 17 different genera, were used in this study (Table 2). Despite including a few non-LAB starter cultures, we will use the term LAB strains for this collection throughout the study. Fifty-one of the cultures consisted of single strains, while 10 were multi-strain cultures (RD-1, NU-TRISH® By-Mild, COMBO4, COMBO1, COMBO2, MIX-1, MIX-2, MIX-3, LD20 and COMBO3). The LAB strains were cultivated under anaerobic conditions overnight at appropriate temperature (30–42°C) in modified De Man, Rogosa and Sharpe medium (mMRS; 10 g peptone from casein, 8 g beef extract, 4 g yeast extract, 5 g sucrose, 1 mL tween 80, 2 g dipotassium hydrogen phosphate, 0.5 g L-cysteine HCL, 2 g diammonium hydrogen citrate, 5 g sodium acetate, 0.2 g magnesium sulfate, and 0.04 g manganese sulfate in 1 liter distilled water) (Medellin-Pena et al., 2007). The LAB strains were enumerated using MRS Agar (ThermoFisher Scientific) after 48–72 h incubation. Bifidobacterium spp. were enumerated by pour plating using MRS agar (Oxoid LTD, Hampshire, England) with 0.5 g L-cysteine (Sigma Aldrich, St. Louis, MO, USA). A schematic overview of the general experimental design is presented in Supplementary Figure S1.
Table 2.
Lactic acid bacteria strains used in this study.
| Strainsa | Abbr. | Incubation temperature (°C) | Source |
|---|---|---|---|
| (+) Bifidobacterium lactis, Lactobacillu delbrueckii subsp. bulgaricus, Streptococcus thermophilus |
NU-TRISH® By-Mild | 42 | Chr. Hansen A/S |
| Bifidobacterium animalis subsp. lactis | BLC1 | 37 | SACCO |
| Bifidobacterium animalis subsp. lactis | BL-15954 | 37 | Chr. Hansen A/S (DSM15954) |
| Bifidobacterium longum subsp. infantis | BI-33361 | 37 | Chr. Hansen A/S (DSM33361) |
| Bifidobacterium longum subsp. longum | BL-15955 | 37 | Chr. Hansen A/S (DSM15955) |
| Enterococcus faecium-669 | EF-669 | 37 | Chr. Hansen A/S |
| Enterococcus faecium-202 | EF-202 | 37 | Chr. Hansen A/S |
| Enterococcus faecium-339 | EF-339 | 37 | Chr. Hansen A/S |
| Kocuria varians | KV | 37 | This study |
| (+) Kocuria varians, Latilactobacillus cuvatus, Staphylococcus carnosus | RD-1 | 37 | Frutarom |
| Lactobacillus acidophilus | LA-3 | 37 | SACCO |
| Lactobacillus acidophilus | LA-13241 | 37 | Chr. Hansen A/S (DSM13241) |
| Lactobacillus acidophilus | LA-20079 | 37 | DSM20079 |
| Ligilactobacillus animalis-506 | LA-506 | 37 | Chr. Hansen A/S |
| Lentilactobacillus buchneri-881 | LB-881 | 37 | Chr. Hansen A/S |
| Lactobacillus delbrueckii subsp. bulgaricus | SP5 | 42 | SACCO |
| (+) Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus | COMBO4 | 37 | Chr. Hansen A/S |
| Levilactobacillus brevis | LB | 30 | Chr. Hansen A/S |
| Lacticaseibacillus paracasei subsp. paracasei | LP-33451 | 37 | Chr. Hansen A/S (DSM33451) |
| Lacticaseibacillus casei | BGP93 | 37 | SACCO |
| Latilactobacillus curvatus | LC | 37 | This study |
| Lactobacillus delbrueckii | LB-20074 | 37 | DSM20074 |
| (+) Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus |
COMBO1 | 42 | Chr. Hansen A/S |
| (+) Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophiles |
COMBO2 | 37 | Chr. Hansen A/S |
| Limosilactobacillus fermentum | LF | 37 | Chr. Hansen A/S |
| Lactobacillus helveticus | LH521 | 42 | ATTC-521 |
| Lactobacillus helveticus | CNRZ32 | 42 | (Jensen et al., 2009) |
| Lactobacillus helveticus-02 | LH-02 | 42 | Chr. Hansen A/S |
| Lactobacillus johnsonii | LJ-10533 | 37 | DSM10533 |
| Lacticaseibacillus paracasei | BGP1 | 37 | SACCO |
| Lacticaseibacillus paracasei | BGP2 | 37 | SACCO |
| Lacticaseibacillus paracasei | LMG P-17806 | 37 | Chr. Hansen A/S |
| Lacticaseibacillus paracasei | LP-20006 | 37 | DSM20006 |
| Lactiplantibacillus plantarum | LPAL | 37 | SACCO |
| Lactiplantibacillus plantarum-672 | LP-672 | 37 | Chr. Hansen A/S |
| Lactiplantibacillus plantarum-673 | LP-673 | 37 | Chr. Hansen A/S |
| Lactiplantibacillus plantarum-072 | LP-072 | 37 | Chr. Hansen A/S |
| Lactiplantibacillus plantarum | LP-20174 | 30 | DSM20174 |
| Limosilactobacillus reuteri | LR-33016 | 37 | Chr. Hansen A/S (DSM33016) |
| Limosilactobacillus reuteri | LR-20016 | 30 | DSM 20016 |
| Lacticaseibacillus rhamnosus | IMC 501 | 37 | SACCO |
| Lacticaseibacillus rhamnosus | LR-33156 | 37 | Chr. Hansen A/S (DMS33156) |
| Lacticaseibacillus rhamnosus | LR-20021 | 37 | DSM20021 |
| Lacticaseibacillus rhamnosus | SP1 | 37 | SACCO |
| Lactococcus lactis-955 | LL-995 | 30 | Chr. Hansen A/S |
| Lactococcus lactis-671 | LL-671 | 30 | Chr. Hansen A/S |
| (+) Lactococcus cremoris,Leuconostoc, Lactococcus lactis subsp. lactis, Lactococcus lactis subsp. Lactis biovar diacetylactis | MIX-1 | 30 | Chr. Hansen A/S |
| (+) Lactococcus cremoris, Leuconostoc, Lactococcus lactis subsp. lactis, Lactococcus lactis subsp. Lactis biovar diacetylactis | MIX-2 | 30 | Chr. Hansen A/S |
| (+) Lactococcus cremoris, Lactococcus lactis subsp. lactis, Lactococcus lactis subsp. Lactis biovar diacetylactis, Leuconostoc spp. | MIX-3 | 30 | Chr. Hansen A/S |
| Lactococcus lactis spp. lactis biovar diacetylactis (L. diacetylactic) | LD | 37 | Chr. Hansen A/S |
| (+) Latilactobacillus sakei subsp. sakei, Staphylococcus carnosus | LD-20 | 37 | BiTEC |
| (+) Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophiles | COMBO3 | 37 | Chr. Hansen A/S |
| Pediococcus acidilactici-839 | PA-839 | 37 | Chr. Hansen A/S |
| Pediococcus pentosaceus-354 | PP-354 | 30 | Chr. Hansen A/S |
| Pediococcus pentosaceus-670 | PP-670 | 30 | Chr. Hansen A/S |
| Pediococcus pentosaceus-674 | PP-674 | 30 | Chr. Hansen A/S |
| Propionibacterium freudenreichii subsp. shermanii | PF7 | 30 | SACCO |
| Propionibacterium freudenreichii subsp. shermanii | PF8 | 30 | SACCO |
| Propionibacterium freudenreichii subsp. shermanii | PB-1 | 30 | SACCO |
| Propionobacterium freudenreichii-507 | PF-507 | 30 | Chr. Hansen A/S |
| Staphylococcus carnosus | SC | 37 | This study |
aMulti-strain cultures indicated with (+).
Fermentation of Cow Milk
Neutralized-cell-free spent medium (nCFSM) was prepared as previously described (Medellin-Pena et al., 2007) with minor modifications. Briefly, the LAB strains were seeded from agar media and grown anaerobically in milk for 16 h at appropriate temperature and then subcultured in sterilized 3.5% whole milk (Thise Dairy, Denmark) using 10% inoculum and 18 h fermentation according to the results of preliminary tests described below. The sterilization of milk was carried out by heating in a water bath at 95°C for 30 min. The fermented milk samples were first centrifuged at 18,500 × g for 3 min at 20°C, and the coagulated fat layer at the top of the tube was removed with a sterile spoon. Then the cells were precipitated by centrifugation at the same conditions for 10 min at 20 °C, the supernatants were harvested and neutralized to pH 7.2 ± 0.2 with 6 M NaOH in order to avoid effects caused merely by acidification. The pH adjusted supernatants were further centrifuged for 5 min to remove all coagulated fat layer to enable easy filter-sterilization through a 0.2-um-pore-size filter (Millipore) by vacuum to remove live bacteria. As a control, nCFSM from unfermented milk was prepared as described above including pH adjustment to 7.2. nCFSM preparations were stored at−20 °C prior to use.
The potential of LAB fermented milk to elicit inhibition of virulence factors in pathogenic bacteria may depend on the fermentation conditions. Therefore, optimal fermentation time and percent inoculation for this was determined in preliminary experiments using whole milk 3.5% fat (Thise Dairy, Denmark) or reconstituted milk powder (Thise Dairy, Denmark). The milk/reconstituted milk powder were fermented with four different fermentation protocols with 1% and 10% v/v inoculation level of the starter culture combined with either 6 h or 18 h fermentation time using three different starter cultures (L. helveticus CNRZ32, L. acidophilus LA-13241 and L. rhamnosus LR-33156).
In order to understand the relation between growth of LAB and the final pH in the fermented milk, 10 selected LABs were seeded to pasteurized whole milk at 106-107 CFU/mL and incubated at the optimal growth temperature for 18 hours. At this time, CFU and pH was determined as described above. Determinations were done in triplicate. The strains were selected to include the strains used in the experiment described above (CNRZ32, LA-13241 and LR-33156), five of the 10 strains used later in the more detailed characterization of the inhibitory effect on virulence gene expression (BL-15955, NU-TRISH® By-Mild, COMBO4, CNRZ32, LR-33016) and three additional strains (BL-15954, LP-673 and LMG P-17806).
Effect of NCFSM on HilA Gene Expression in S. Typhimurium LT2 Reporter Construct
The putative anti-virulence effects was first assayed using the bioluminescent reporter strain S. Typhimurium LT2 hilA::luxCDABE (Bayoumi and Griffiths, 2010), where the expression of the lux genes is controlled by the hilA promoter, so that light emits when the hilA gene is expressed. S. Typhimurium LT2 hilA::luxCDABE was grown in nCFSM, and expression of hilA was measured as previously reported (Muyyarikkandy and Amalaradjou, 2017; Ali et al., 2019). Briefly, an overnight culture (19 h) of the reporter strain with an approximate cell density of 1 × 109 cells mL−1 was diluted 1:100 in nCFSM supplemented with 10% (v/v) 10x LB medium. Then 200 μl of each inoculated nCFSM was distributed into triplicate wells of a sterile opaque corning 96-well plate (Sigma-Aldrich), which was placed in a Victor multi-lable counter (Wallac, PerkinElmer Life Sciences Canada, Woodbridge, ON, Canada) for incubation at 37 °C. The growth of the reporter strain expressed by cell density (OD490) and hilA gene expression measured as bioluminescence were measured hourly for 24 h. The bioluminescence was expressed as relative light units (RLU) defined as bioluminescence counts min−1 and adjusted by OD490 (RLU/OD490). Accumulated gene expression over 12 h was calculated by summarizing bioluminescence signal for the first 12 h of measuring and termed Sum (12 h). The proportional change in expression of hilA in S. Typhimurium LT2 hilA::luxCDABE construct has been termed change in virulence activity (%VA) and was calculated as 100 – [(expression in nCFSM / expression in the non-fermented milk (control)] * 100. Since this assay was only used for initial screening and results were confirmed by RT-PCR, only one biological assay with two technical replicates was performed.
QPCR Measurement of the Effect of NCFSM on Transcription of Virulence Genes in S. Dublin, E. Coli F5 and C. Perfringens
Culture of Pathogens in NCFSM
Single colonies of E. coli and S. Dublin were grown in LB broth aerobically with shaking (180 rpm), and C. perfringens were grown in BHI broth anaerobically in anaerobic jars that utilized Oxoid™ AnaeroGen™ 2.5 L Sachets (ThermoFisher Scientific, Waltham, MA, USA) without shaking overnight at 37°C. The overnight cultures were diluted to a final OD600 = 0.05 by mixing with 4.5 mL nCFSM supplemented with 0.5 mL of 10x appropriate broth (LB or BHI) and then grown at 37°C to early stationary phase (OD600 =1.0) as recommended (Bayoumi and Griffiths, 2012). To serve as controls, the overnight cultures were similarly diluted into unfermented milk supplemented with LB or BHI and grown to OD600 =1.0.
RNA Extraction
RNA was isolated using a FastPrep cell disrupter system (Qbiogene, Illkirch, France) and RNeasy Mini Kit (Qiagen, Sollentuna, Sweden) for total RNA isolation by mechanical disruption, according to the manufacturer's instructions. Quantity of the extracted RNA was determined by A260 measurements and purity by A260/A280 ratio measurements using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Hvidovre, Denmark). RNA samples were purified by DNA digestion using TURBOTM DNase kit (2 U/μl) (Ambion, Life Technologies, Nærum, Denmark) to remove contaminating DNA.
Reverse-Transcribed-Quantitative Real-Time Polymerase Chain Reaction (RT-QPCR)
RNA was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Nærum, Denmark) under the following conditions: 25°C for 10 min, 37°C for 120 min, 85 °C for 5 min, and a cooling step to 4°C. RT-qPCR was performed using FastStart Essential DNA Green Master (Roche, Hvidovre, Denmark). For E. coli and C. perfringens RT-qPCR were performed using a LightCycler 96 (Roche, Hvidovre, Denmark), where as an Applied Biosystems QuantStudio 5 (Thermo Fisher Scientific, Roskilde, Denmark) were used for S. Dublin. The standard qPCR cycling program was as following: initial activation denaturation (95 °C for 10 min), and followed by 40 cycles (95°C for 15 s, 60°C for 30 s, 72°C for 30 s).
Expression levels of the virulence factors listed in Table 1 were measured. Primers are listed in Supplementary Table S1. Real-time PCR amplification efficiencies were calculated from the slope of the standard curve, which was constructed using the data collected from serial dilutions of the template DNA for each gene according to the equation: E = 10 [−1/slope] (Pfaffl, 2001). The transcript levels were normalized to validated reference genes, gapA, nusG, 16S rRNA and GAPDH (Wise and Siragusa, 2005; Moller et al., 2017; Shi et al., 2019). RT-qPCR was performed on three biological replicates with two technical replicates each, and the relative changes in gene expression were calculated by using the formula: ΔCT = CT (target) – CT (normalizer), and ΔΔCT was calculated by subtracting the ΔCT of the untreated sample (pathogens grown in non-fermented milk) from the treated one (pathogen grown in nCFSM): ΔΔCT = ΔCT (treated) - ΔCT (untreated). The relative gene expression was calculated as 2−ΔΔCT. Finally, a fold change was calculated as −1/2−ΔΔCT (Pfaffl, 2004).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software) version 7.03. Comparison of gene expression was analyzed using Student's t-test with Welch's correction. P ≤ 0.05 were considered statistically significant.
Results
Optimal Fermentation Protocol for Inhibition of Virulence Genes
In order to choose optimal fermentation conditions for maximizing the effect on virulence gene expression, cell free spent medium (CFSM) was prepared from whole milk and reconstituted milk powder fermented with the strains L. helveticus CNRZ32, L. acidophilus LA-13241 or L. rhamnosus LR-33156 based on four different fermentation protocols with variation in seeding quantity and fermentation time. After pH neutralization, the nCFSM obtained from fermented reconstituted milk powder resulted in less inhibition of the hilA gene expression in S. Typhimurium compared with nCFSM from whole milk-based fermentation (data not shown). Thus, whole milk was used in the following experiments.
The pH in CFSM from L. helveticus CNRZ32 was 4.5 (1% inoculation) and 4.0 (10% inoculation) after 6 h fermentation, which decreased to 3.5 (1% inoculation) and 3.0 (10% inoculation) after 18 h fermentation, respectively. CFSM from L. acidophilus LA-13241 showed a pH of 6.5 (1% inoculation) and 5.0 (10% inoculation) after 6 h fermentation while 18 h of fermentation decreased pH to 5.0 (1% inoculation) and 4.5 (10% inoculation). CFSM of L. rhamnosus LR-33156 was 6.5 (1% inoculation) and 6.3 (10% inoculation) after 6 h and was 6.0 at both inoculation levels after 18 h. For these three strains, and seven other strains tested for comparison, the final pH generally correlated inversely with the Log CFU/ml at the end of the 18 hours of fermentation (R = −0.69). In this testing the end cell density of L. helveticus CNRZ32 (9.1±0.1 CFU/mL) exceeded that of the other strains, and the resulting pH was below 4, while L. rhamnosus LR33156 had a more than one log lower end CFU/ml and a higher end pH close to 6.0 (Supplementary Figure S2). However, there were exceptions to the general correlation, for example both B. lactis BL-15954 and B. longum BL15955 has a final pH >5 with approximate 8 Log CFU/ml, while L. plantarum LP-673 had a pH ≈5, with a final cell concentrations at 7 Log CFU/mL, as seen in Supplementary Figure S2. To rule out that the effect of different pH could influence down-stream results, all CFSM were neutralized (nCFSM) before they were used as growth medium for pathogenic bacteria.
In general, nCFSM from L. helveticus CNRZ32 showed the highest inhibition of hilA expression in S. Typhimurium irrespective of fermentation protocols. The trend was that 10% inoculation level resulted in a higher inhibition of hilA expression than 1% inoculum, and that 18 h fermentation was superior to 6 h fermentation, however this was not uniform among the strains (Table 3).
Table 3.
The proportional change (VA%) in expression of the virulence gene hilA in S. Typhimurium after growth in nCFSM from L. helveticus CNRZ32, L. acidophilus LA-13241, and L. rhamnosus LR-33156.
| 6 h fermentation (VA%) | 18 h fermentation (VA%) | |||||||
|---|---|---|---|---|---|---|---|---|
| LAB strains | 1% a | 10% a | 1% a | 10% a | ||||
| 12 hb | Sum (12 h)b | 12 hb | Sum (12 h)b | 12 hb | Sum (12 h)b | 12 hb | Sum (12 h)b | |
| L. helveticus CNRZ32 | −45 ± 2 | −40 ± 1 | −69 ± 7 | −65 ± 4 | −30 ± 11 | −15 ± 11 | −96 ± 1 | −57 ± 15 |
| L. acidophilus LA-13241 | −3 ± 10 | 13 ± 8 | −27 ± 1 | −7 ± 2 | −41 ± 14 | −1 ± 5 | −55 ± 4 | −20 ± 3 |
| L. rhamnosus LR-33156 | −26 ± 10 | −12 ± 10 | −19 ± 11 | −17 ± 13 | −41 ± 15 | −1 ± 10 | −18 ± 11 | −4 ± 13 |
aInoculation level.
bInhibition of hilA expression in the hilA::luxCDABE bioluminescence reporter estimated as nCFSM relative to the non-fermented milk control at time point 12 h and as the area under the accumulated expression curve during 12 h [sum(12h)].
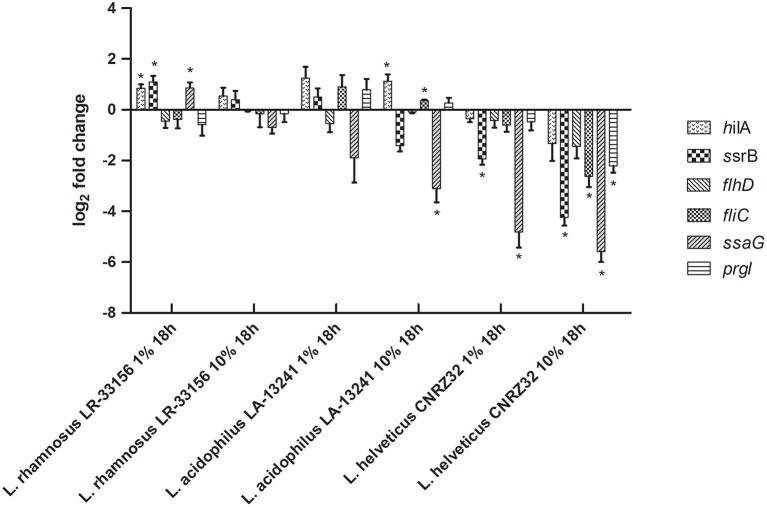
S. Typhimurium may cause calf diarrhea, but it is not the most common and most important serovar of Salmonella in relation to this disease, which is S. Dublin. Therefore, although the S. Typhimurium bioluminescence assay provides a relatively easy screening approach, RT-qPCR was applied to assess expression of six relevant virulence genes in S. Dublin after growth in nCFSM from each of the three LAB for 18 h with 10% inoculation vs. 1% inoculation. This showed variation with both up- and downregulations of virulence genes with the exception of L. helveticus CNRZ32, which showed consistent downregulation for all tested virulence genes of S. Dublin (Figure 1). L. helveticus CNRZ32 showed significant downregulation of ssrB (18.9-fold), flhD (2.73-fold), fliC (6.14-fold), ssaG (48.0-fold) and prgI (4.63-fold) at 10% inoculation level, while expression of hilA (2.52-fold) and flhD (2.73-fold) was lowered, but not significantly different from the control. Based on the results on hilA expression in the S. Typhimurium reporter strain and virulence genes in S. Dublin, a fermentation protocol with a 10% inoculum level of the starter cultures and a fermentation time of 18 h was applied for the onward testing of effects of nCFSMs.
Figure 1.
Effect of nCFSM of L. helveticus CNRZ32, L. acidophilus LA-13241 and L. rhamnosus LR-33156 on the expression of hilA, ssrB, flhD, fliC, ssaG and prgI in S. Dublin analyzed by RT-qPCR. The CFSM were prepared from milk fermented 18 h with an inoculation level of 1% and 10%. The change in expression of genes is relative to the non-fermented milk control. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The stars indicate statistical significance at different levels: *P ≤ 0.05.
Effect of NCFSM on HilA Gene Expression in S. Typhimurium LT2 Reporter Construct
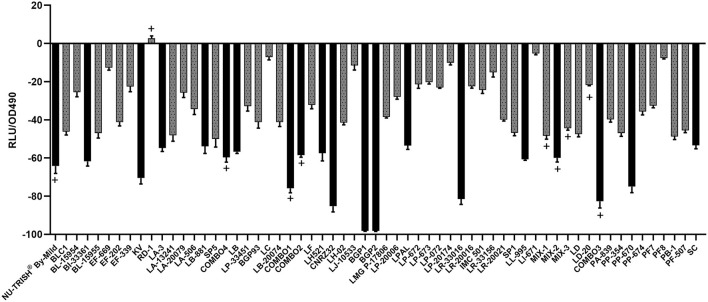
The bioluminescent reporter strain, S. Typhimurium LT2 hilA::luxCDABE, was used for initial screening of the effects of nCFSM obtained from milk fermented with 61 different LAB strains/combinations of LAB strains. No effects were observed on growth phenotypes of S. Typhimurium LT2 hilA::luxCDABE during 24 h incubation at 37°C in presence of 10% nCFSM from 59 LAB, while nCFSM from L. paracasei BGP1, BGP2 and P. pentosaceus PP-670 did not sustain good growth (OD490 <0.5 after 24 h).
Bioluminescence of the S. Typhimurium LT2 hilA::luxCDABE after 12 h of growth in nCFSM compared to growth in non-fermented milk (control) was reduced for all tested LAB strains except RD1 (Figure 2). nCFSM from L. paracasei BGP1 and BGP2 elicited the highest downregulation of hilA expression by 98.2%, however, due to the effect on growth phenotype, these results were dis-regarded. Other single strain starter cultures eliciting more than 50% downregulation of hilA expression were L. helveticus CNRZ32 (85.3%), L. reuteri LR-33016 (81.5%), P. pentosaceus PP-670 (74.7%), K. varians (70.5%), B. infantis BI-33361 (61.8%), L. lactis LL-995 (60.7%), L. helveticus LH521 (57.5%), L. brevis (57%), L. acidophilus LA-3 (54.8%), L. buchneri LB-881 (53.9%), L. plantarum LPAL (53.3%), and S. carnosus (53%) (Figure 2). For seven out of ten multi-strain starter cultures, downregulation of hilA expression was more than 50% [(COMBO3 (82.7%), COMBO1 (75.9%), NU-TRISH® By-Mild (64.2%), MIX-2 (60.0%), COMBO4 (59.7%), COMBO2 (58.5%)] (Figure 2).
Figure 2.
Change in bioluminescence (%VA) of S. Typhimurium LT2 hilA::luxCDABE after 12 h incubation in nCFSM from 61 different LAB strain cultures. Strains where growth in the nCFSM resulted in more than 50 % reduction in bioluminescence compared to growth in unfermented milk are shown as black bars. The hilA measurement was based on one biological replicate of nCFSM with technical triplicates. The multi-strain starter cultures are marked with (+).
Effect of NCFSM on Transcription of Virulence Genes in S. Dublin, E. Coli F5 and C. Perfringens
Even if bioluminescent reporter genes are considered a valuable tool for a first screening of gene expression, bioluminescence may be influenced by several factors, including components of the medium (Ali et al., 2019). Therefore, RT-qPCR as a highly sensitive and more specific method is necessary to complement the bioluminescence results. A two-step procedure was used. First, the effects of nCFSM from the 61 LAB strains on virulence gene expression was measured in one strain of each of E. coli F5 (E21-79), S. Dublin (JEO3665) and C. perfringens (C4-5) after incubation to OD600 = 1.0. Based on the combined results of bioluminescent and RT-qPCR analysis (Supplementary Table S2), the 10 most promising nCFSMs were selected to make sure all pathogens were covered sufficiently. Among the 10 selected nCFSMs, NU-TRISH® By-Mild, L. casei BGP93, L. reuteri LR-33016, P. acidilactici PA-839, P. pentosaceus PP-674, Pr. freudenreichii PF-507 and L. helveticus CNRZ32 inhibited all tested virulence genes in S. Dublin and E. coli. B. longum BL-15955 and Lentilactobacillus buchneri LB-881 inhibited all tested virulence genes in E. coli and C. perfringens, while COMBO4 inhibited all tested virulence genes in S. Dublin and C. perfringens.
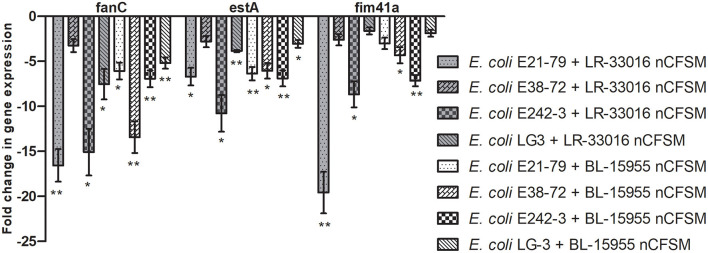
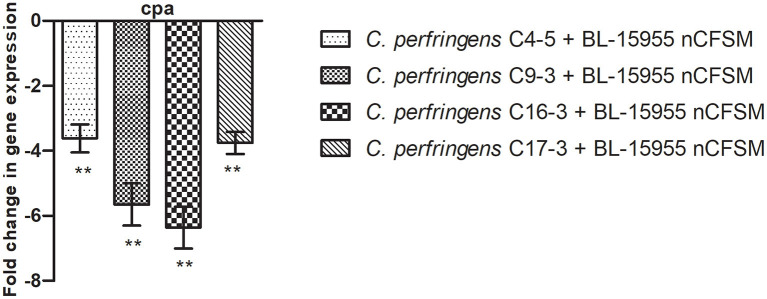
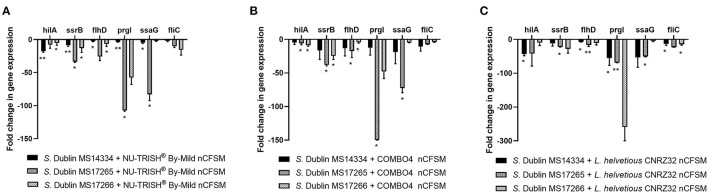
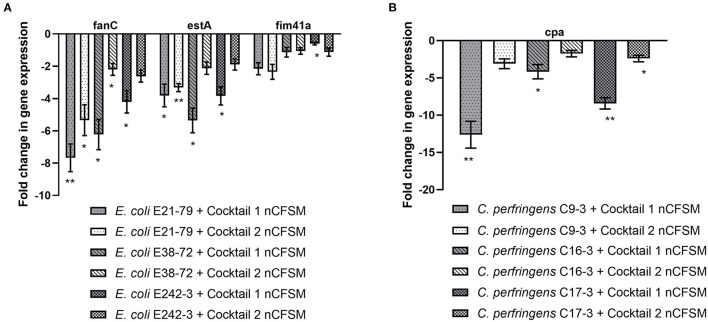
As a second step, four strains of E. coli F5, 3 strains of S. Dublin and 4 strains of C. perfringens were then grown in the 10 selected nCFSMs, and virulence gene expression was tested on by RT-qPCR to verify pathogen strain coverage. It was found that fanC, fim41a and estA genes were significantly downregulated in the four E. coli strains after growth in nCFSM from L. reuteri LR-33016 and B. longum BL-15955 (Figure 3). nCFSM from B. longum BL-15955 significantly downregulated fanC expression with 5.19 to 13.43 fold, estA expression with 3.06 to 6.90 fold, and fim41a with 1.88 to 7.15 fold. L. reuteri LR-33016 significantly downregulated fanC expression with 7.53 to 15.09 fold, estA expression with 3.86 to 10.78 fold, and fim41a with 2.62 to 19.58 fold, except in E. coli E38-72, where it caused a non-significant 2.62 to 3.26 fold decrease in expression (Figure 3). B. longum BL-15955 also significantly downregulated cpa expression in all four tested C. perfringens strains with 3.65 to 6.24 fold (Figure 4), while little effect of other nCFSMs was observed (Supplementary Dataset S1). In the 3 tested S. Dublin strains, two multi-strain cultures, NU-TRISH® By-Mild (L. delbrueckii subsp. bulgaricus, S. thermophilus and B. lactis BL-15954) and COMBO4 (L. delbrueckii subsp. bulgaricus and S. thermophilus), as well as L. helveticus CNRZ32 significantly downregulated hilA expression with 4.6 to 43 fold, ssrB expression with 8.7 to 38.1 fold, flhD with 2.7 to 25.7 fold, prgL with 3.7 to 258 fold, ssaG with 2.0 to 82.8 fold, and fliC with 2.2 to 22.8 fold expression (Figure 5).
Figure 3.
Effect of L. reuteri LR-33016 and B. longum BL-15955 nCFSM on virulence gene expression of 4 E. coli F5 strains presented as fold change of expression levels in nCFSM relative to non-fermented milk. The expression data were normalized to two validated reference genes, gapA and nusG. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The stars indicate statistical significance at different levels: *P ≤ 0.05, ** P ≤ 0.01.
Figure 4.
Effect of B. longum BL-15955 nCFSM on expression of cpa in 4 C. perfringens strains presented as fold change of expression levels in nCFSM relative to non-fermented milk. The expression data were normalized to two validated reference genes, 16sRNA and GAPDH. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The stars indicate statistical significance at different levels: ** P ≤ 0.01.
Figure 5.
Effect of nCFSM from (A) NU-TRISH® By-Mild (L. delbrueckii subsp. bulgaricus, S. thermophilus and B. lactis BL-15954, (B) COMBO4 (L. delbrueckii subsp.bulgaricus and S. thermophilus) and (C) L. helveticus CNRZ32 on virulence gene expression in 3 S. Dublin strains. The expression data were normalized to validated reference gene gapA and presented as fold change of expression levels in nCFSM relative to a control (non-fermented milk). Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The stars indicate statistical significance at different levels: *P ≤ 0.05, ** P ≤ 0.01.
Activity of NCFSM Cocktails on Expression of Virulence Genes in Bacterial Pathogens of Calf Diarrhea
None of the nCFSM made from fermentations with each of the 61 individual LAB strains served to affect virulence gene expression in all three pathogens equally well. We therefore tested the possibility to obtain a nCFSM affecting the virulence genes in all the E. coli F5, S. Dublin and C. perfringens strains based on fermentations with mixtures of LAB strains. As the ability of LAB strains to grow and degrade milk may be affected by the presence of competing strains, we could not assume that the outcome from mixtures would simply be the sum of the results obtained from the individual nCFSMs. Hence, two cocktails were tested: Cocktail 1 consisted of a mix of NU-TRISH® By-Mild and B. longum BL-15955 and cocktail 2 of a mix of COMBO4 with B. longum BL-15955. After growth in nCFSM from cocktail 1, virulence genes expression was reduced in the three tested E. coli F5 strains. The effect was particularly notable for fanC with 4.21 to 7.67 fold downregulation and estA with 3.83 to 5.36 fold regulation. Cocktail 2 also significantly decreased expression of fanC with 2.20 to 5.33 fold, and estA with 1.89 to 3.32 fold (Figure 6A). As seen in Figure 6B, nCFSM from cocktail 1 also significantly downregulated cpa in the 3 C. perfringens strains with 4.17 to 12.63 fold, while no significantly downregulation was found for this gene after growth in cocktail 2 (1.74 to 3.10 fold).
Figure 6.
Effect of nCFSM from two cocktails of LAB strains on expression of fanC, estA and fim41a in 3 E. coli strains (A), and cpa in 3 C. perfringens strains (B), and the data are presented as fold change of expression levels in nCFSM relative to non-fermented milk. The expression data of virulence genes for E. coli were normalized to gapA and nusG, and C. perfringens were normalized to 16sRNA and GAPDH. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The stars indicate statistical significance at different levels: *P ≤ 0.05, ** P ≤ 0.01.
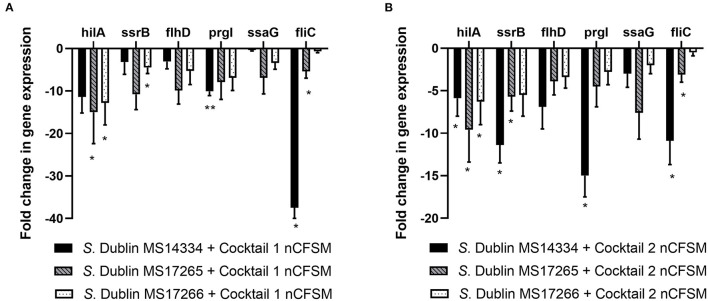
For S. Dublin, similar effects with significant downregulation of virulence genes were observed for the two cocktails. Cocktail 1 showed significant downregulation of hilA expression with 5.9 to 15.0 fold, ssrB expression with 3.2 to 10.8 fold, flhD with 3.1 to 9.9 fold, prgI with 7.9 to 15.0 fold, ssaG with 0.4 to 6.9 fold, and fliC with 0.7 to 37.5 fold in 3 different S. Dublin strains. The corresponding results for cocktail 2 was 5.9 to 9.6 fold (hilA), 5.5 to 11.4 fold (ssrB), 3.4 to 6.9 fold (flhD), 2.8 to 15.0 fold (prgL), 2.0 to 7.6 fold (ssaG), and 0.5 to 10.9 fold (fliC) (Figure 7).
Figure 7.
Effect of nCFSM from (A) cocktail 1 and (B) cocktail 2 on virulence gene expression in 3 S. Dublin strain. The expression data were normalized to validated reference gene gapA and presented as fold change of expression levels in nCFSM relative to non-fermented milk. Three independent replicates including two technical replicates each were performed. The data shown represents the mean and the error bars represent standard deviations. The stars indicate statistical significance at different levels: *P ≤ 0.05, ** P ≤ 0.01.
Discussion
It has previously been shown that certain LAB affects expression of virulence genes in enteropathogenic bacteria (Brovko et al., 2003; Medellin-Pena et al., 2007; Vinderola et al., 2007), and the use of this ability of LAB has been suggested as an effective measure to control entero-pathogens (Fooks and Gibson, 2002; Gusils et al., 2002). Based on this, the current study was concerned with the possible use of fermentation products from LAB strains to prevent expression of virulence genes necessary for induction of diarrhea in calves. This disease is caused by a multitude of pathogens, and focus was on infections caused by Salmonella, E. coli and C. perfringens, since they are the drivers of use of antimicrobials for this disease. Because anti-virulence approaches give rise to a milder evolutionary pressure and does not affect the ability of the pathogen to grow, it is less likely that resistance will be developed toward substances that affects virulence genes than toward conventional antibiotics (Rasko and Sperandio, 2010).
LAB are used in food processing because of their characteristic flavor formation and ability to lower the pH and to produce antimicrobial substances (Verluyten et al., 2003). The effect with certain LAB is seen even when using pH-neutral, cell-free supernatants of the strains (Bayoumi and Griffiths, 2010), suggesting that the effect on expression of virulence genes associated to substances produced by the LAB strains during growth, either products exported from the bacteria or degradation products of the growth medium. In the current study, we investigated the effect of pH neutralized, spent cow milk from 61 food grade starter cultures on virulence gene expression in the three bacteria associated with calf diarrhea. pH neutralization was done because there were measurable differences in the final pH after growth of LABs, and we wanted to rule out this factor as a contributor to the virulence inhibition. Milk is readily accessible in dairy farms where calf diarrhea is a disease problem, and fermented milk is already used to feed calves (Maldonado et al., 2018). In addition to the nutritional and specific antibacterial effect, milk components are known to have a variety of health benefits in humans, including opioid, antimutagenic, antimicrobial, and immunomodulating activity, in addition to their basic nutritional properties (Smacchi and Gobbetti, 2000). It has been reported that the consumption of milk fermented by lactic acid bacteria produces a specific humoral immune response, and may have effects on the digestive, cardiovascular and nervous systems (Matar et al., 2001; Korhonen, 2009). Whether similar effects can be seen in calves remains to be shown, but could be an additional benefit of the approach.
Previous studies have showed that metabolites produced by L. helveticus LH-2 and L. acidophilus LA-5 affected expression of hilA, hilD, ssrA, and ssrB in Salmonella, and tir, ler, eaeA, fliC, and hlyB in E. coli O157:H7 after growth in chemically defined media and milk (Medellin-Pena and Griffiths, 2009; Tellez et al., 2010; Bayoumi and Griffiths, 2012; Zeinhom et al., 2012). Here, we investigated the effects of neutralized cell-free spent milk (nCFSM) on a wider range of virulence genes in Salmonella and E. coli, and we included the main virulence gene shared between all types of C. perfringens. The most commonly observed fimbriae on ETEC from calves with diarrhea are F5 and F41 (Nagy and Fekete, 1999), and most bovine ETEC produce STa (Guth, 2000). In Salmonella, the gene hilA directly controls and activates all the genes of SPI1 for invasion (Lostroh et al., 2000), and ssrB is the main regulator of SPI2, which is responsible for systemic infection and replication inside macrophages and epithelial cells (Feng et al., 2004). Meanwhile, flagella are also involved in adhesion and host invasion, which is controlled by the regulator flhD/C (Kirov, 2003). Besides these three key regulator genes, three structural genes (prgL, ssaG and fliC) of each apparatus were also included in this study. Among C. perfringens, type A is the most common of all the C. perfringens types. The alpha-toxin encoded by cpa gene is the main lethal toxin of type A, and it is also produced by all other genotypes of C. perfringens. Type B and C which produce beta-toxin have been frequently reported in conjunction with calf diarrhea (Rings, 2004), but those types infection are not very common in calves. Though all types have the ability to produce the alpha-toxin, type A usually produces a greater amount (Katayama et al., 1993). In young calves, severe abomasitis associated with C. perfringens type A is characterized by sudden onset of disease, abomasal tympany, abdominal pain, and hemorrhagic diarrhea (Glenn Songer and Miskimins, 2005; Schlegel et al., 2012). Success in suppression these key virulence factors would putatively enable use of feeding with fermented milk as a way of preventing calf diarrhea caused by S. Dublin, ETEC F5, and C. perfringens. This would be a sustainable prevention measure, especially with the emerging problem of antibiotic-resistant strains.
In previous experiments, nCFSMs obtained from reconstituted milk powder have shown to be capable of modulating macrophage activity, as well as inhibit virulence genes expression (Brovko et al., 2003; Ding et al., 2005; Vinderola et al., 2007; Tellez et al., 2010, 2011), suggesting that reconstituted milk could be used as an alternative to fresh milk. In the current study, we compared the anti-virulence activity of nCFSMs made based on whole and reconstituted milk, and interestingly nCFSMs from whole milk showed better inhibition of virulence gene expression than reconstituted milk. The reason for this remains unknown. There was no observations to suggest that the LAB strains grew any different in the fresh compared to when they grew in re-constituted milk and the pathogens also grew equally well in nCFSM from the two sources (data not shown). It may be that some components of the milk, which are important for formation of the antibacterial principle is affected by the drying procedure used to make milk powder, as the chemical composition of reconstituted milk and fresh milk differ (Mehta, 2015). Further studies are needed to understand this difference.
Most previous studies on the effect of fermentation products from LAB on virulence gene expression have used 1% LAB inoculation level for fermentation (Brovko et al., 2003; Casey et al., 2007; Medellin-Pena et al., 2007; Medellin-Pena and Griffiths, 2009; Bayoumi and Griffiths, 2012), 2% (Matar et al., 2001; Vinderola et al., 2007), 4% (Ding et al., 2005), while 10% inoculation is less frequent (Tellez et al., 2010). We found that 10% inoculation level lead to nCFSMs with better anti-virulence activity. This may be because 10% inoculation resulted in more effective fermentation and thus lower pH and probably more production of the anti-virulence substances.
Different methodological strategies were used to screen for the ability of nCFSM to repress the virulence gene. A broad screen was carried out using bioluminescence activity of a plasmid borne promoter-reporter fusions hilA::luxCDABE. Bioluminescent reporter genes are considered a rapid tool for initial monitoring of gene expression; however, bioluminescence may be influenced by several factors, including other components of the medium. Previous observations using bioluminescent constructs suggest the occurrence of false positives and negatives with respect to virulence expression (Medellin-Pena et al., 2007; Delcenserie et al., 2012; Zeinhom et al., 2012; Guri et al., 2016). Therefore, in the current study, the samples were further analyzed using RT-qPCR to confirm the results and to have the opportunity to analyze the virulence expression of more genes in several pathogens. In this way it was shown that nCFSM from B. infantis BI-33361, MIX-2, and COMBO3 did not result in significant downregulation of hilA when measured by qPCR opposed to bioluminescent assay. Two L. paracasei strains, BGP1 and BGP2, together with P. pentosaceus PP-670 did not sustain growth of Salmonella. For this reason, these strains showed the highest inhibition of hilA expression in the bioluminescent assay, underlining the importance of controlling for growth, when analyzing effect of virulence gene expression. hilA expression varies with growth phase and is maximally expressed in late logarithmic phase (Miao et al., 1999; Mouslim and Hughes, 2014), thus comparison of effect of a substance on expression of hilA can only be done if the strains are in the same growth phase.
The 61 nCFSMs were made using LAB strains belonging to 17 genera, enabling analysis of whether the ability to affect virulence gene expression was a trait of strains from a particular genus. From the initial screening of the 61 strains, it seems that nCFSM of the Enterococcus genus had no effects on virulence gene expression. A previous study reported that Enterococcus faecalis Symbioflor® downregulated virulence genes of E. coli O157:H7 through direct bacteria-bacteria interactions in a Caenorhabditis elegans model (Neuhaus et al., 2017), however, in line with our results, nCFSM with anti-virulence activity from Enterococcus genus has not been reported. Only three strains of Enterococcus were included in the current study, and care should be taken not to over-conclude. The observation may help in future to identify the active substances produced during fermentation, by comparing nCFSM from Enterococcus to nCFSM from the other genera.
A previous study showed that fractions extracted from L. acidophilus LA-5 cell-free spent medium were able to downregulate several virulence genes in verotoxigenic E. coli O157:H7 and that this could be used to decrease the severity of diarrhea in mice (Zeinhom et al., 2012). In the current study, virulence factors of enterotoxigenic E. coli (ETEC), namely fanC and fim41a genes encoding major subunits of the F5 and F41 fimbriae, and estA encoding STa enterotoxins, were investigated. nCFSM from the mixed culture NU-TRISH® By-Mild and from B. infantis BI-33361, B. longum BL-15955, L. buchneri LB-881, L. delbrueckii subsp. bulgaricus LB20074, L. reuteri LR-33016, P. acidilactici PA-839 and P. pentosaceus PP-674, and Pr. freudenreichii PF-507 showed anti-virulence activity when only one E. coli strain was assayed, however, when extra strains were included, only nCFSM from B. longum BL-15955 and L. reuteri LR-33016 stably reduced the expression of the three genes in all tested E. coli strains. These LAB strains were thus indicated as the best candidates to control calf diarrhea caused by ETEC F5 and F41. L. johnsonii F19785 has been reported to reduce the colonization and shedding of C. perfringens in poultry as a defined competitive exclusion agent (La Ragione et al., 2004). In the current study, nCFSM from B. longum BL-15955, L. buchneri LB-881, L. johnsonii LJ-10533, L. rhamnosus LR-20021, L. lactis LL-995, and the multi-strain cultures COMBO3 and COMBO4 showed potent downregulation of cpa expression in one C. perfringens strain. Due to an inhibitory effect on virulence genes in E. coli, B. longum BL-15955, L. buchneri LB-881 and COMBO4 were selected for testing with more C. perfringens strains. Only B. longum BL-15955 showed significant downregulatory effect on the cpa gene of all tested C. perfringens strains.
With respect to Salmonella, previous studies have included effect on hilA and ssrB encoding key regulators of TTSSs on SPI-1 and SPI-2, and for these virulence factors, there is evidence that the downregulation affects virulence in cell cultures and mice. For example, nCFSM from B. bifidum ATCC 29521 and L. helveticus LH-2 affected attachment and invasion of Salmonella in vitro and in vivo through inhibition of these virulence genes (Tellez et al., 2011; Bayoumi and Griffiths, 2012). In the current study, we revisited these key regulator genes, and in addition we included the main regulator of the flagella operon, flhD. For each of these important virulence associated systems, we further included a structural genes (prgI of T3SS-1, ssaG of T3SS-2 and fliC of the flagella) to have a stronger argument for a possible effect. Previous studies have been concerned with S. Typhimurium, while we have focused on S. Dublin, the main serovar involved in calf diarrhea. Many LAB strains showed inhibition of these genes, suggesting that the factors which affect virulence gene expression in Salmonella are relatively commonly formed during lactic acid bacteria fermentation of milk. Characteristically, when downregulation was seen in regulator genes, this was always followed by at least the same trend in the corresponding structural gene (Supplementary Dataset S1). We were primarily concerned with finding LAB strains which inhibit a broad selection of virulence genes and in as many strains as possible. nCFSM of two multi-strain cultures, NU-TRISH® By-Mild and COMBO4, and the single strain culture, L. helveticus CNRZ32, showed significant downregulation in all tested Salmonella strains.
In a previous study, administration of L. acidophilus alone showed no beneficial effects on lambs infected with E. coli O157:H7, while feeding the lambs with a mixture of L. acidophilus and E. faecium significantly lowered numbers of this pathogenic strain (Lema et al., 2001). The greatest reduction in numbers was seen with the use of a mixture of L. acidophilus, E. faecium, L. casei, L. fermentum, and L. plantarum (Lema et al., 2001). A five-strain probiotic combination consisted of two L. murinus strains and one strain of each L. salivarius, L. pentosus and P. pentosaceous also reduced pathogen shedding and alleviated diarrhea in pigs challenged with S. Typhimurium (Casey et al., 2007). In line with the beneficial effects of multi-strains probiotics, nCFSM made from multi-strains cultures generally showed good effect on virulence gene expression. Thus, NU-TRISH® By-Mild consisting of B. lactis BL-15954, L. delbrueckii subsp. bulgaricus and S. thermophilus, and COMBO4 consisting of L. delbrueckii subsp. bulgaricus and S. thermophilus both showed significant anti-virulence activity. The basis of the multifactorial mode of action of multi-strain probiotics remains largely un-clarified, but it may be that modulation of virulence is an important feature in their protection against enteric infections.
The experience from probiotics and the results of our initial screening indicated the possibility of using a multi-strain combination to control more than one pathogenic species. In order to identify the best candidate to reduce pathogens associated with calf diarrhea, cocktails were tested. A cocktail with NU-TRISH® By-Mild, which had excellent effect on genes in S. Dublin and B. longum BL-15955 with good effect on ETEC F5 and C. perfringens was shown to be very promising, and should be taken into further testing. This four-strain combination showed significant downregulation of virulence genes in all tested pathogens. However, the level of reduction was reduced for S. Dublin compared with NU-TRISH® By-Mild alone, underlying that combination of strains do not just have the added effects of each strain individually.
The observation that fermented milk with this combination down regulates virulence genes in the major bacterial pathogens associated with calf diarrheae opens the possibility that feeding with this milk may prevent the pathogens from causing disease in the calfes. However, there is a long way from showing this effect in the laboratory to concluding on the effect on field conditions. Not only will there be variation in intake of the fermented milk, but their effect on virulence genes may also be modified once they reach the active sites in the intestine. Further studies should thus be undertaken to investigate whether the principle will work in vivo by feeding calves with cow milk fermented with this four-strain combination.
Conclusions
In summary, the data presented in this study confirms that milk fermented by selected LAB strains may affect virulence genes in bacteria. The study performed a broad screening of single strains as well as multi-strains of LAB strains. In addition to previous studies, it included ETEC and C. perfringens virulence genes, and it focused on inhibition of virulence genes in bacteria relevant to induction of diarrhea in calves. A four-strain combination with B. lactis BL-15954, L. delbrueckii subsp. bulgaricus, S. thermophilus and B. longum BL-15955 was identified which gave stable anti-virulence effect in S. Dublin, ETEC F5 and C. perfringens, and these observations warrant further in vivo validation. Clearly, additional research must be conducted to demonstrate that this can lead to reduced incidence of diarrhea in calves and identify molecules responsible for the observed activity and determine the mechanisms whereby nCFSM inhibits the activity of the virulence genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: European Nucleotide Archive - PRJEB46410.
Author Contributions
GL, JO, MK, SA, and AJ have participated in the design of the study. GL and MK carried out the experiments. GL drafted the manuscript. All authors commented on and approved the final manuscript.
Funding
This work was funded by the GUDP Development and Demonstration Program through Grant No. 1167960.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to Resadije Idrizie for excellent technical skills in the laboratory. Lactic acid bacteria were kindly provided by Chr. Hansen (Denmark).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.828013/full#supplementary-material
References
- Acha S. J., Kuhn I., Jonsson P., Mbazima G., Katouli M., Mollby R. (2004). Studies on calf diarrhoea in Mozambique: prevalence of bacterial pathogens. Acta. Vet. Scand. 45, 27–36. 10.1186/1751-0147-45-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali E., Nielsen S. D., Abd-El Aal S., El-Leboudy A., Saleh E., Lapointe G. (2019). Use of Mass Spectrometry to Profile Peptides in Whey Protein Isolate Medium Fermented by Lactobacillus helveticus LH-2 and Lactobacillus acidophilus La-5. Front. Nutr. 6, 152. 10.3389/fnut.2019.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayoumi M. A., Griffiths M. W. (2010). Probiotics down-regulate genes in Salmonella enterica serovar typhimurium pathogenicity islands 1 and 2. J. Food. Prot. 73, 452–460. 10.4315/0362-028X-73.3.452 [DOI] [PubMed] [Google Scholar]
- Bayoumi M. A., Griffiths M. W. (2012). In vitro inhibition of expression of virulence genes responsible for colonization and systemic spread of enteric pathogens using Bifidobacterium bifidum secreted molecules. Int. J. Food Microbiol. 156, 255–263. 10.1016/j.ijfoodmicro.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Bondue P., Crevecoeur S., Brose F., Daube G., Seghaye M. C., Griffiths M. W., et al. (2016). Cell-Free Spent Media Obtained from Bifidobacterium bifidum and Bifidobacterium crudilactis Grown in Media Supplemented with 3'-Sialyllactose Modulate Virulence Gene Expression in Escherichia coli O157:H7 and Salmonella Typhimurium. Front. Microbiol. 7, 1460. 10.3389/fmicb.2016.01460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brovko L. Y., Vandenende C., Chu B., Ng K. Y., Brooks A., Griffiths M. W. (2003). In vivo assessment of effect of fermented milk diet on course of infection in mice with bioluminescent Salmonella. J. Food Prot. 66, 2160–2163. 10.4315/0362-028X-66.11.2160 [DOI] [PubMed] [Google Scholar]
- Casey P. G., Gardiner G. E., Casey G., Bradshaw B., Lawlor P. G., Lynch P. B., et al. (2007). A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 73, 1858–1863. 10.1128/AEM.01840-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. I., Yoon K. J. (2014). An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J. Vet. Sci. 15, 1–17. 10.4142/jvs.2014.15.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcenserie V., Lapointe G., Charaslertrangsi T., Rabalski A., Griffiths M. W. (2012). Glucose decreases virulence gene expression of Escherichia coli O157:H7. J. Food. Prot. 75, 748–752. 10.4315/0362-028X.JFP-11-384 [DOI] [PubMed] [Google Scholar]
- Ding W., Wang H., Griffiths M. W. (2005). Probiotics down-regulate flaA sigma28 promoter in Campylobacter jejuni. J. Food Prot. 68, 2295–2300. 10.4315/0362-028X-68.11.2295 [DOI] [PubMed] [Google Scholar]
- Feng X., Walthers D., Oropeza R., Kenney L. J. (2004). The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol. Microbiol. 54, 823–835. 10.1111/j.1365-2958.2004.04317.x [DOI] [PubMed] [Google Scholar]
- Fooks L. J., Gibson G. R. (2002). In vitro investigations of the effect of probiotics and prebiotics on selected human intestinal pathogens. FEMS Microbiol. Ecol. 39, 67–75. 10.1111/j.1574-6941.2002.tb00907.x [DOI] [PubMed] [Google Scholar]
- Glenn Songer J., Miskimins D. W. (2005). Clostridial abomasitis in calves: case report and review of the literature. Anaerobe. 11, 290–294. 10.1016/j.anaerobe.2004.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guri A., Paligot M., Crevecoeur S., Piedboeuf B., Claes J., Daube G., et al. (2016). In vitro screening of mare's milk antimicrobial effect and antiproliverative activity. FEMS Microbiol. Lett. 363, fnv234. 10.1093/femsle/fnv234 [DOI] [PubMed] [Google Scholar]
- Gusils C., Bujazha M., González S. (2002). Preliminary studies to design a probiotic foruse in swine feed. Interciencia. 27, 409–413. [Google Scholar]
- Guth B. E.. (2000). Enterotoxigenic Escherichia coli–an overview. Mem Inst Oswaldo Cruz. 95, 95–97. 10.1590/S0074-02762000000700017 [DOI] [PubMed] [Google Scholar]
- Jensen M. P., Vogensen F. K., Ardö Y. (2009). Variation in caseinolytic properties of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 19, 661–668. 10.1016/j.idairyj.2009.04.001 [DOI] [Google Scholar]
- Katayama S., Matsushita O., Minami J., Mizobuchi S., Okabe A. (1993). Comparison of the alpha-toxin genes of Clostridium perfringens type A and C strains: evidence for extragenic regulation of transcription. Infect Immun. 61, 457–463. 10.1128/iai.61.2.457-463.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov S. M.. (2003). Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol Lett. 224, 151–159. 10.1016/S0378-1097(03)00445-2 [DOI] [PubMed] [Google Scholar]
- Knudsen G. M., Sommer H. M., Sorensen N. D., Olsen J. E., Aabo S. (2011). Survival of Salmonella on cuts of beef carcasses subjected to dry aging. J Appl Microbiol 111, 848–854. 10.1111/j.1365-2672.2011.05094.x [DOI] [PubMed] [Google Scholar]
- Korhonen H.. (2009). Milk-derived bioactive peptides: from science to applications. J. Funct. Foods. 1, 177–187. 10.1016/j.jff.2009.01.00730301185 [DOI] [Google Scholar]
- La Ragione R. M., Narbad A., Gasson M. J., Woodward M. J. (2004). In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett Appl Microbiol. 38, 197–205. 10.1111/j.1472-765X.2004.01474.x [DOI] [PubMed] [Google Scholar]
- Lema M., Williams L., Rao D. R. (2001). Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157:H7 in lambs by feeding microbial feed supplement. Small Rumin Res. 39, 31–39. 10.1016/S0921-4488(00)00168-1 [DOI] [PubMed] [Google Scholar]
- Lostroh C. P., Bajaj V., Lee C. A. (2000). The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37, 300–315. 10.1046/j.1365-2958.2000.01991.x [DOI] [PubMed] [Google Scholar]
- Maldonado N. C., Chiaraviglio J., Bru E., De Chazal L., Santos V., Nader-Macias M. E. F. (2018). Effect of milk fermented with lactic acid bacteria on diarrheal incidence, growth performance and microbiological and blood profiles of newborn dairy calves. Probiotics Antimicrob. Proteins. 10, 668–676. 10.1007/s12602-017-9308-4 [DOI] [PubMed] [Google Scholar]
- Matar C., Valdez J. C., Medina M., Rachid M., Perdigon G. (2001). Immunomodulating effects of milks fermented by Lactobacillus helveticus and its non-proteolytic variant. J. Dairy Res. 68, 601–609. 10.1017/S0022029901005143 [DOI] [PubMed] [Google Scholar]
- Medellin-Pena M. J., Griffiths M. W. (2009). Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Appl. Environ. Microbiol. 75, 1165–1172. 10.1128/AEM.01651-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medellin-Pena M. J., Wang H., Johnson R., Anand S., Griffiths M. W. (2007). Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl. Environ. Microbiol. 73, 4259–4267. 10.1128/AEM.00159-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta B. M.. (2015). “Chemical Composition of Milk and Milk Products,” in Handbook of Food Chemistry. p. 1–34. 10.1007/978-3-642-41609-5_31-1 [DOI] [Google Scholar]
- Miao E. A., Scherer C. A., Tsolis R. M., Kingsley R. A., Adams L. G., Baumler A. J., et al. (1999). Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34, 850–864. 10.1046/j.1365-2958.1999.01651.x [DOI] [PubMed] [Google Scholar]
- Moller T. S. B., Liu G., Boysen A., Thomsen L. E., Luthje F. L., Mortensen S., et al. (2017). Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an IncI1 plasmid in Escherichia coli. Front. Microbiol. 8, 2365. 10.3389/fmicb.2017.02365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouslim C., Hughes K. T. (2014). The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 10, e1003987. 10.1371/journal.ppat.1003987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundi A., Delcenserie V., Amiri-Jami M., Moorhead S., Griffiths M. W. (2013). Cell-free preparations of Lactobacillus acidophilus strain La-5 and Bifidobacterium longum strain NCC2705 affect virulence gene expression in Campylobacter jejuni. J. Food Prot. 76, 1740–1746. 10.4315/0362-028X.JFP-13-084 [DOI] [PubMed] [Google Scholar]
- Muyyarikkandy M. S., Amalaradjou M. A. (2017). Lactobacillus bulgaricus, Lactobacillus rhamnosus and Lactobacillus paracasei Attenuate Salmonella Enteritidis, Salmonella Heidelberg and Salmonella Typhimurium Colonization and Virulence Gene Expression In Vitro. Int. J. Mol. Sci. 18. 10.3390/ijms18112381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R., Behare P., Rana R., Kumar A., Kumar M., Arora S., et al. (2011). Bioactive peptides derived from milk proteins and their health beneficial potentials: an update. Food Funct. 2, 18–27. 10.1039/C0FO00016G [DOI] [PubMed] [Google Scholar]
- Nagy B., Fekete P. Z. (1999). Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 30, 259–284. [PubMed] [Google Scholar]
- Nataro J. P., Kaper J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201. 10.1128/CMR.11.1.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus K., Lamparter M. C., Zolch B., Landstorfer R., Simon S., Spanier B., et al. (2017). Probiotic Enterococcus faecalis Symbioflor((R)) down regulates virulence genes of EHEC in vitro and decrease pathogenicity in a Caenorhabditis elegans model. Arch. Microbiol. 199, 203–213. 10.1007/s00203-016-1291-8 [DOI] [PubMed] [Google Scholar]
- Olsen J. E., Hoegh-Andersen K. H., Casadesus J., Thomsen L. E. (2012). The importance of motility and chemotaxis for extra-animal survival of Salmonella enterica serovar Typhimurium and Dublin. J. Appl. Microbiol. 113, 560–568. 10.1111/j.1365-2672.2012.05363.x [DOI] [PubMed] [Google Scholar]
- Perez E., Kummeling A., Janssen M. M., Jimenez C., Alvarado R., Caballero M., et al. (1998). Infectious agents associated with diarrhoea of calves in the canton of Tilaran, Costa Rica. Prev. Vet. Med. 33, 195–205. 10.1016/S0167-5877(97)00038-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.. (2004). Quantification Strategies in Real-Time PCR, p 87–112. La Jolla, CA: AZ of quantitative PCR. International University Line. [Google Scholar]
- Pfaffl M. W.. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Sperandio V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. 10.1038/nrd3013 [DOI] [PubMed] [Google Scholar]
- Rings D. M.. (2004). Clostridial disease associated with neurologic signs: tetanus, botulism, and enterotoxemia. Vet. Clin. North Am. Food Anim. Pract. 20, 379–391. 10.1016/j.cvfa.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Schlegel B. J., Nowell V. J., Parreira V. R., Soltes G., Prescott J. F. (2012). Toxin-associated and other genes in Clostridium perfringens type A isolates from bovine clostridial abomasitis (BCA) and jejunal hemorrhage syndrome (JHS). Can. J. Vet. Res. 76, 248–254. [PMC free article] [PubMed] [Google Scholar]
- Shi H., Huang X., Yan Z., Yang Q., Wang P., Li S., et al. (2019). Effect of Clostridium perfringens type C on TLR4/MyD88/NF-kappaB signaling pathway in piglet small intestines. Microb. Pathog. 135, 103567. 10.1016/j.micpath.2019.103567 [DOI] [PubMed] [Google Scholar]
- Smacchi E., Gobbetti M. (2000). Bioactive peptides in dairy products: synthesis and interaction with proteolytic enzymes. Food Microbiol. 17, 129–141. 10.1006/fmic.1999.0302 [DOI] [Google Scholar]
- Smith G. W.. (2009). Treatment of calf diarrhea: oral fluid therapy. Vet. Clin. North Am. Food. Anim. Pract. 25, 55–72, vi. 10.1016/j.cvfa.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Songer J. G.. (1997). Bacterial phospholipases and their role in virulence. Trends Microbiol. 5, 156–161. 10.1016/S0966-842X(97)01005-6 [DOI] [PubMed] [Google Scholar]
- Tellez A., Corredig M., Brovko L. Y., Griffiths M. W. (2010). Characterization of immune-active peptides obtained from milk fermented by Lactobacillus helveticus. J. Dairy Res. 77, 129–136. 10.1017/S002202990999046X [DOI] [PubMed] [Google Scholar]
- Tellez A. C. M., Turner P. V., Morales R., Griffiths M. (2011). A peptidic fraction from milk fermented with Lactobacillus helveticus protects mice against Salmonella infection. Int. Dairy J. 21, 607–614. 10.1016/j.idairyj.2011.03.011 [DOI] [Google Scholar]
- Thomas M., Fenske G. J., Antony L., Ghimire S., Welsh R., Ramachandran A., et al. (2017). Whole genome sequencing-based detection of antimicrobial resistance and virulence in non-typhoidal Salmonella enterica isolated from wildlife. Gut Pathog. 9, 66. 10.1186/s13099-017-0213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verluyten J., Messens W., De Vuyst L. (2003). The curing agent sodium nitrite, used in the production of fermented sausages, is less inhibiting to the bacteriocin-producing meat starter culture Lactobacillus curvatus LTH 1174 under anaerobic conditions. Appl. Environ. Microbiol. 69, 3833–3839. 10.1128/AEM.69.7.3833-3839.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinderola G., Matar C., Perdigon G. (2007). Milk fermented by Lactobacillus helveticus R389 and its non-bacterial fraction confer enhanced protection against Salmonella enteritidis serovar Typhimurium infection in mice. Immunobiology. 212, 107–118. 10.1016/j.imbio.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Wise M. G., Siragusa G. R. (2005). Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. App.l Environ. Microbiol. 71, 3911–3916. 10.1128/AEM.71.7.3911-3916.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B., Oh S., Griffiths M. W. (2014). Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J. Dairy Sci. 97, 4745–4758. 10.3168/jds.2014-7921 [DOI] [PubMed] [Google Scholar]
- Zeinhom M., Tellez A. M., Delcenserie V., El-Kholy A. M., El-Shinawy S. H., Griffiths M. W. (2012). Yogurt containing bioactive molecules produced by Lactobacillus acidophilus La-5 exerts a protective effect against enterohemorrhagic Escherichia coli in mice. J. Food Prot. 75, 1796–1805. 10.4315/0362-028X.JFP-11-508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: European Nucleotide Archive - PRJEB46410.