Abstract
CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), a well-characterized E3 ubiquitin ligase, is a central repressor of seedling photomorphogenic development in darkness. However, whether COP1 is involved in modulating abscisic acid (ABA) signaling in darkness remains largely obscure. Here, we report that COP1 is a positive regulator of ABA signaling during Arabidopsis seedling growth in the dark. COP1 mediates ABA-induced accumulation of ABI5, a transcription factor playing a key role in ABA signaling, through transcriptional and post-translational regulatory mechanisms. We further show that COP1 physically interacts with ABA-hypersensitive DCAF1 (ABD1), a substrate receptor of the CUL4-DDB1 E3 ligase targeting ABI5 for degradation. Accordingly, COP1 directly ubiquitinates ABD1 in vitro, and negatively regulates ABD1 protein abundance in vivo in the dark but not in the light. Therefore, COP1 promotes ABI5 protein stability post-translationally in darkness by destabilizing ABD1 in response to ABA. Interestingly, we reveal that ABA induces the nuclear accumulation of COP1 in darkness, thus enhancing its activity in propagating the ABA signal. Together, our study uncovers that COP1 modulates ABA signaling during seedling growth in darkness by mediating ABA-induced ABI5 accumulation, demonstrating that plants adjust their ABA signaling mechanisms according to their light environment.
COP1 positively regulates ABA signal transduction during Arabidopsis seedling growth in darkness by targeting ABD1 for 26S proteasome-mediated degradation to facilitate ABA-induced ABI5 accumulation.
IN A NUTSHELL.
Background: The phytohormone abscisic acid (ABA) rapidly accumulates in response to environmental stresses such as drought, and plays important roles in plant adaptation to these stresses. Binding of ABA by ABA receptors triggers a signaling cascade that modulates the expression of ABA-responsive genes, and the transcription factor ABA INSENSITIVE5 (ABI5) plays an important role in mediating ABA-regulated gene expression. Although plants are subjected to darkness at night under natural conditions and their roots mostly grow in the dark, the mechanisms behind ABA signaling in the dark remain largely obscure. CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) is a well-characterized E3 ubiquitin ligase, and cop1 mutant seedlings grown in the dark develop a phenotype resembling wild-type seedlings grown in the light, indicating that COP1 is a central repressor of seedling photomorphogenic development in darkness.
Question: Is COP1 involved in regulating ABA signaling in the dark?
Findings: We discovered that COP1 is a positive regulator of ABA signaling during Arabidopsis seedling growth in the dark. ABI5 protein accumulates to lower levels in dark-grown cop1 seedlings, while overexpression of ABI5 rescued the ABA-hyposensitive phenotype of cop1 seedlings, indicating that COP1 positively regulates ABA signaling in darkness by mediating ABA-induced ABI5 accumulation. We further show that COP1 physically interacts with ABA-hypersensitive DCAF1 (ABD1), a substrate receptor of the CUL4-DDB1 E3 ligase targeting ABI5 for degradation. In addition, COP1 directly ubiquitinates ABD1 in vitro, and negatively regulates ABD1 protein abundance in vivo in the dark but not in the light. Therefore, COP1 promotes ABI5 protein stability post-translationally in darkness by destabilizing ABD1 in response to ABA. Interestingly, we reveal that ABA induces the nuclear accumulation of COP1 in darkness, thus enhancing its activity in propagating the ABA signal. Finally, we conclude that wild-type seedlings grown in the light are more resistant to ABA than those grown in the dark, indicating that plants adjust their ABA signaling mechanisms according to their light environment.
Next steps: We aim to elucidate the molecular mechanism underlying ABA induction of COP1 nuclear accumulation in the dark. In addition, we are interested in investigating the roles of photoreceptors in regulating ABA signaling in response to light.
Introduction
Plants have evolved a remarkable ability to survive abiotic stresses. The phytohormone abscisic acid (ABA) rapidly accumulates in response to environmental stresses such as drought, cold, or salinity, and plays important roles in plant adaptation to these stresses (Cutler et al., 2010; Finkelstein, 2013; Li et al., 2017; Chen et al., 2020). Therefore, ABA has long been regarded as a “stress hormone”. However, ABA also clearly regulates additional aspects of plant growth and development, including seed maturation and dormancy, seed germination, seedling establishment, root growth, stomatal movement, flowering, and senescence (Cutler et al., 2010; Finkelstein, 2013; Li et al., 2017; Chen et al., 2020).
Over the past four decades, an array of components involved in the ABA signaling network has been identified, among which, ABA INSENSITIVE1-3 (ABI1-ABI3) were the first ABA-responsive loci identified in Arabidopsis (Arabidopsis thaliana; Koornneef et al., 1984). The abi4 and abi5 mutations were isolated later by forward genetic screening for Arabidopsis mutants showing ABA insensitivity (Finkelstein, 1994). Molecular cloning of ABI1 to ABI5 genes revealed that ABI1 and ABI2 proteins are group A protein phosphatase 2Cs (PP2Cs; Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998), whereas ABI3, ABI4, and ABI5 proteins are transcription factors of the B3-domain, APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR, and basic leucine zipper (bZIP) families, respectively (Giraudat et al., 1992; Finkelstein, 1994, 1998; Finkelstein and Lynch, 2000). In 2009, two groups independently reported the molecular characterization of PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTOR (RCAR) proteins as ABA receptors (Ma et al., 2009; Park et al., 2009). This breakthrough led to the current model of the ABA signaling pathway, which includes a central signaling module consisting of three protein classes, i.e. ABA receptors (PYR1/PYL/RCARs), coreceptors (group A PP2Cs, including ABI1 and ABI2), and sucrose nonfermenting1 (SNF1)-related protein kinase 2s (SnRK2s). In the absence of ABA, PP2Cs directly interact with and inhibit the kinase activity of SnRK2s, thus repressing downstream signaling events of the ABA signaling pathway. In the presence of ABA, however, ABA-bound PYR1/PYL/RCAR proteins form coreceptor complexes with PP2Cs and inhibit their phosphatase activity, thus releasing the kinase activity of SnRK2s to subsequently phosphorylate downstream signaling targets such as ABA-RESPONSIVE ELEMENT-BINDING FACTORS (abbreviated as either AREBs or ABFs) or ABI5 (Cutler et al., 2010; Finkelstein, 2013; Li et al., 2017; Chen et al., 2020).
ABI5 was shown to play fundamental roles in transducing ABA signals during seed germination and early seedling growth (Finkelstein, 1994; Finkelstein and Lynch, 2000). Interestingly, ABI5 belongs to the same clade as ABFs, and they share several conserved functional motifs in addition to the bZIP domain (Jakoby et al., 2002; Corrêa et al., 2008). However, only ABI5 in this clade was identified by forward genetic screens, whereas ABFs were isolated in yeast one-hybrid screening by using ABA-responsive elements (ABREs) as bait (Choi et al., 2000; Uno et al., 2000). ABI5 and ABFs share multiple conserved RXXS/T sites, and SnRK2 kinases have been shown to play a major role in phosphorylating the Ser/Thr residues of these sites in an ABA-dependent manner (Uno et al., 2000; Furihata et al., 2006; Fujii et al., 2007, 2009; Nakashima et al., 2009). Moreover, both ABI5 transcript and ABI5 protein levels are induced by ABA, and ABA can stabilize ABI5 by preventing its turnover (Lopez-Molina et al., 2001). The fact that ABI5 proteins accumulate to higher levels in seedlings treated with 26S proteasome inhibitors and in mutant seedlings lacking a subunit of the 26S proteasome suggests that ABI5 turnover is mediated through the 26S proteasome pathway (Lopez-Molina et al., 2001; Smalle et al., 2003). Consistent with these results, several types of E3 ubiquitin ligases have been identified that participate in ABI5 ubiquitination and degradation. KEEP ON GOING (KEG), a RING-type E3 ligase, directly interacts with and targets ABI5 for ubiquitination in vitro (Stone et al., 2006; Liu and Stone, 2010). Interestingly, KEG appears to target ABI5 for degradation mainly in the cytoplasm in the absence of ABA (Liu and Stone, 2013), whereas in response to ABA treatment, ABI5 accumulates in the nucleus and its stability may be controlled by several CULLIN4 (CUL4)-based E3 ligases specified by the substrate receptors DWD HYPERSENSITIVE TO ABA1 (DWA1), DWA2, and ABA-HYPERSENSITIVE DCAF1 (ABD1) (Lee et al., 2010; Seo et al., 2014). DWA1, DWA2, and ABD1 interact with ABI5 and negatively regulate ABI5 protein stability in response to ABA, thus acting as negative regulators of ABA signaling (Lee et al., 2010; Seo et al., 2014).
CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) is an evolutionarily conserved E3 ubiquitin ligase involved in multiple processes in many different organisms, including plant development and mammalian metabolism, tumorigenesis, and neuron development (Yi and Deng, 2005; Li et al., 2011; Marine, 2012; Han et al., 2020). The cop1 mutant was first isolated in a genetic screen for mutants displaying constitutive photomorphogenic phenotypes in the dark (Deng et al., 1991). COP1 protein is composed of three domains: an N-terminal RING-finger domain (RING), a coiled-coil domain (Coil) in the middle, and a C-terminal WD40 domain (Deng et al., 1992; Han et al., 2020). COP1 interacts with SUPPRESSOR OF phyA-105 (SPA) proteins to form E3 ubiquitin ligase complexes that target multiple photomorphogenesis-promoting factors for ubiquitination and degradation, such as ELONGATED HYPOCOTYL5 (HY5; Osterlund et al., 2000), HY5 HOMOLOG (HYH; Holm et al., 2002), LONG AFTER FAR-RED LIGHT1 (LAF1; Seo et al., 2003), LONG HYPOCOTYL IN FAR-RED1 (HFR1; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005), and a series of B-BOX (BBX) proteins (Xu et al., 2016; Lin et al., 2018; Song et al., 2020). COP1 localizes to the nucleus in darkness; upon light exposure, COP1 migrates from the nucleus to the cytosol, thus allowing the nuclear accumulation of photomorphogenesis-promoting factors (such as HY5) and the initiation of photomorphogenesis (von Arnim and Deng, 1994; Pacín et al., 2014; Podolec and Ulm, 2018). In addition to its pivotal role in regulating seedling photomorphogenesis, COP1 is also involved in mediating a variety of plant responses to developmental and environmental cues, such as flowering, stomatal opening and development, responses to environmental stresses, and crosstalk between light and phytohormonal signaling pathways (reviewed in Yi and Deng, 2005; Lau and Deng, 2012; Huang et al., 2014; Han et al., 2020).
We recently reported that PHYTOCHROME-INTERACTING FACTORs (PIFs) regulate ABA signaling specifically in darkness by binding directly to the ABI5 promoter and mediating ABA-induced expression of ABI5 (Qi et al., 2020). Interestingly, the pifq mutant (the pif1 pif3 pif4 pif5 quadruple mutant) seedlings exhibited a cop phenotype (Leivar et al., 2008). The prominent cop phenotype of cop1 mutants prompted us to investigate whether COP1 is involved in mediating ABA signaling in the dark as well. In this study, we report that COP1 positively regulates ABA signaling during Arabidopsis seedling growth in the dark by mediating ABA-induced accumulation of ABI5 through transcriptional and post-translational regulatory mechanisms. We further show that COP1 physically interacts with ABD1, a substrate receptor of the CUL4-based E3 ligase targeting ABI5 for 26S proteasome-mediated degradation, ubiquitinating ABD1 in vitro and negatively regulating ABD1 protein abundance in vivo. Interestingly, we observed that ABA treatment promotes nuclear enrichment of COP1 in darkness, thus promoting COP1-mediated ABD1 degradation and ABI5 protein accumulation. Together, our data demonstrate that COP1 plays a critical role in regulating ABA signaling in the dark.
Results
COP1 regulates ABA signaling in darkness
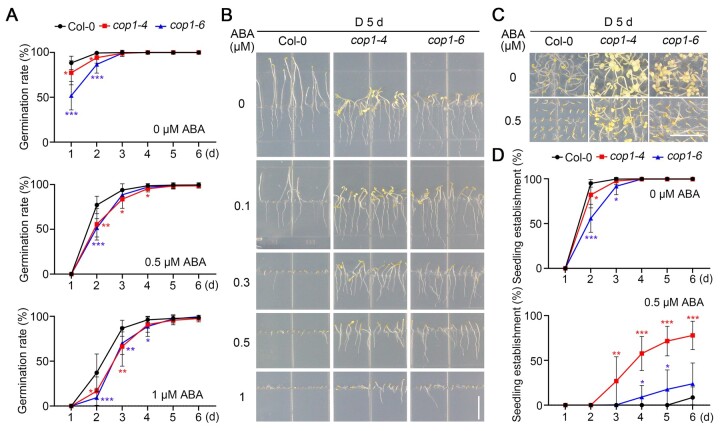
To investigate whether COP1 plays a role in mediating ABA signaling in the dark, we compared the seed germination rates of wild-type (Col-0) and two weak cop1 mutant alleles (cop1-4 and cop1-6) sown on Murashige and Skoog (MS) medium or MS medium containing 0.5- or 1-µM ABA in darkness. We observed that the germination rates of both cop1 mutants are significantly lower than those of Col-0 in all tested conditions (Figure 1A), indicating that COP1 promotes seed germination in the dark in the presence or absence of ABA.
Figure 1.
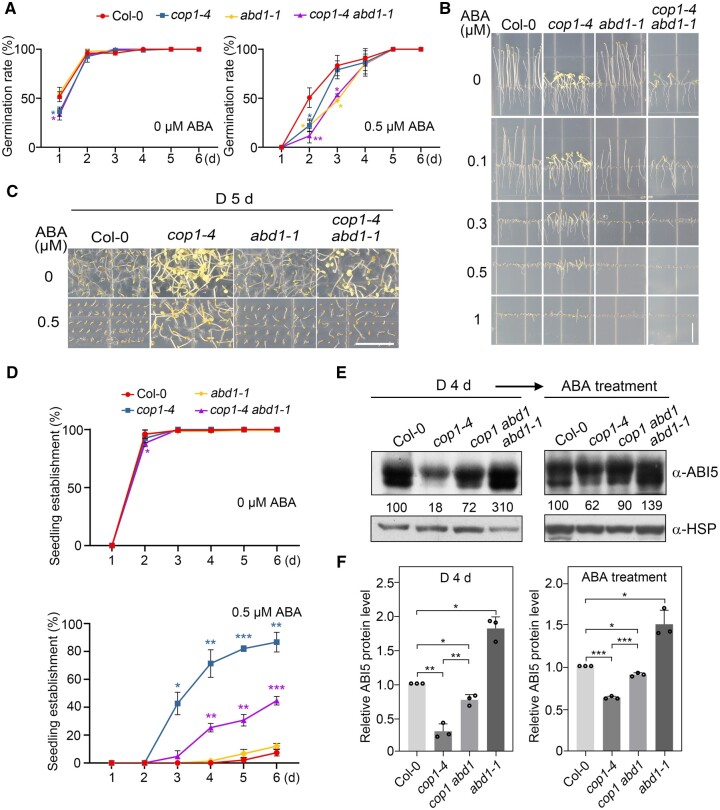
cop1 mutants are less sensitive to ABA-inhibited seedling growth in darkness. A, Germination rate measurements. Hydrated seeds for Col-0 and the cop1 mutants were sown on MS medium containing various concentrations of ABA, and then grown in darkness. Seed germination was defined as the first sign of radicle tip emergence and scored daily until day 6. Error bars represent the standard deviation (sd) of three independent sets of seeds, each set containing 50 seeds. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seeds. B, Growth of cop1 seedlings is insensitive to ABA. Col-0 and cop1 seedlings were grown vertically on MS medium with the indicated ABA concentrations in darkness (D) for 5 days. Bar = 1 cm. C, Seedling establishment of cop1 mutants is less sensitive to ABA. Col-0 and cop1 seedlings were grown horizontally on MS medium or MS medium containing 0.5-µM ABA in darkness for 5 days. Bar = 1 cm. D, Seedling establishment rate measurements. Col-0 and cop1 seedlings were grown horizontally on MS medium or MS medium containing 0.5-µM ABA in darkness. Error bars represent sd of three independent pools of seedlings, each pool containing 50 seedlings. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seedlings.
Seedling growth is another well-characterized developmental process regulated by ABA (reviewed in Finkelstein, 2013; Li et al., 2017; Chen et al., 2020). To examine whether COP1 was involved in mediating ABA regulation of seedling growth, we allowed Col-0 and cop1 mutant seedlings to grow vertically on various concentrations of ABA (ranging from 0 to 1 µM) in darkness. Notably, we determined that seedling growth of both cop1 alleles is more resistant to ABA than Col-0 in darkness (Figure 1B). We also grew Col-0 and the cop1 mutants horizontally on MS medium containing no ABA or 0.5-µM ABA in darkness for various days, and measured the rates of seedling establishment based on the criteria recently used by Yadukrishnan et al. (2020) that hypocotyls and cotyledons should emerge completely in established seedlings. Our data indicated that both Col-0 and the cop1 mutant seedlings are well established after growing for 4 days in darkness in the absence of ABA; however, in the presence of 0.5-µM ABA, the seedling establishment rates of both cop mutant alleles were remarkably higher than those of Col-0 in the dark (Figure 1, C and D). Together, these data demonstrate that COP1 mediates ABA inhibition of seedling growth in the dark.
Notably, we observed that cop1-4 and cop1-6 mutant seedlings display different levels of ABA resistance in the dark, which may be explained by different mutations in these two cop1 alleles (in cop1-4, a C-to-T mutation results in a truncated COP1 protein that contains only the N-terminal 282 amino acids [aa], whereas in cop1-6, an A-to-G substitution at the 3′-end of the fourth intron leads to four cryptically spliced products; McNellis et al., 1994). Collectively, our data demonstrate that COP1 negatively regulates ABA inhibition of seed germination but positively regulates ABA inhibition of seedling growth in darkness.
COP1 positively regulates ABA signaling during seedling growth in darkness through ABI5
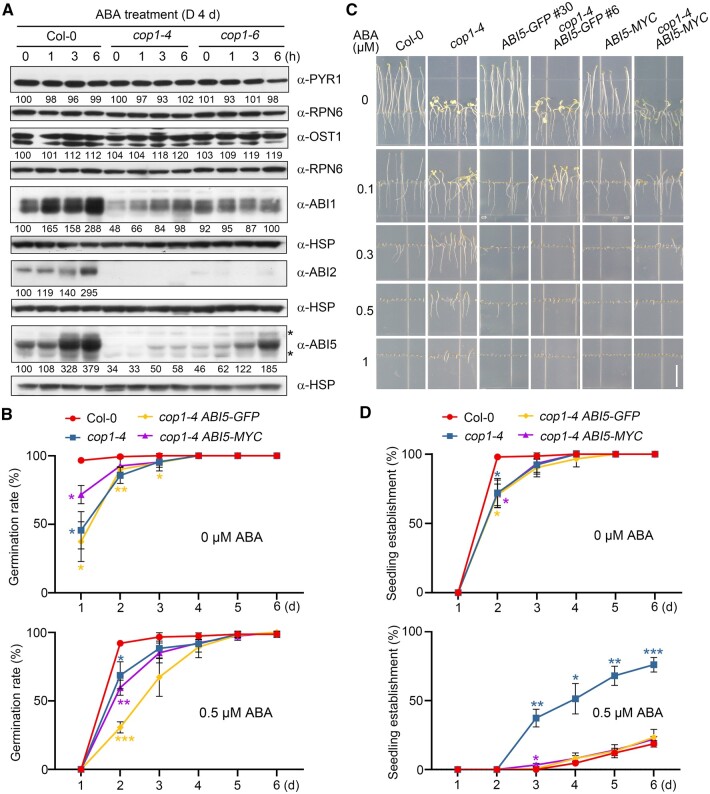
Next, we explored the molecular mechanism underlying COP1 regulation of ABA signaling during seedling growth. We recently generated antibodies against ABI1 (Kong et al., 2015), ABI2 (Wang et al., 2019b), and ABF1 to ABF4 transcription factors (Wang et al., 2019b), and in this study we successfully generated new antibodies against the ABA receptor PYR1 and the kinase OPEN STOMATA1 (OST1; Supplemental Figure S1). We then asked whether COP1 might modulate the abundance of these key proteins of the ABA signaling pathway in darkness. We first grew Col-0 and the cop1 mutant seedlings in darkness for 4 days without ABA, and then treated them with 50-µM ABA for 1, 3, or 6 h, respectively. Our immunoblot data indicated that PYR1 and OST1 abundance is not substantially altered after ABA treatment in either Col-0 or cop1 mutant seedlings in darkness (Figure 2A). However, whereas the levels of both ABI1 and ABI2 proteins were highly induced by ABA in dark-grown Col-0 seedlings, ABA-induced ABI1 and ABI2 protein accumulation was greatly impaired in cop1 mutants (Figure 2A). We recently reported that ABA rapidly and dramatically induces the transcription of ABI1 and ABI2 and thus the accumulation of ABI1 and ABI2 proteins in the light, which is due to a feedback regulatory mechanism mediated by ABI5-clade transcription factors (likely including ABI5 itself) to desensitize ABA signaling (Wang et al., 2019b). In this study, our data showed that addition of the protein synthesis inhibitor cycloheximide (CHX) efficiently inhibits the ABA-mediated induction of ABI1/ABI2 protein accumulation in the dark, indicating that ABA-induced ABI1/ABI2 protein accumulation in dark-grown seedlings is also due to the upregulation of the expression of their encoding genes (Supplemental Figure S2). Consistently, the expression of ABI1 and ABI2 was notably lower in cop1 mutants before and after ABA treatment (Supplemental Figure S3), demonstrating that COP1 positively regulates ABI1/ABI2 transcription and thus ABI1/ABI2 protein accumulation in the presence or absence of ABA.
Figure 2.
COP1 positively regulates ABA signaling during seedling growth in darkness through ABI5. A, Immunoblots showing the abundance of PYR1, OST1, ABI1, ABI2, and ABI5 in Col-0 and cop1 mutant seedlings before and after ABA treatment. Four-day-old Col-0, cop1-4, and cop1-6 mutant seedlings grown in darkness were first treated with 50-μM ABA for the indicated times, and then harvested and subjected to immunoblotting using antibodies against PYR1, OST1, ABI1, ABI2, and ABI5 proteins. Anti-RPN6 or anti-HSP was used as sample loading control. Numbers below the immunoblots indicate the relative band intensities of the respective proteins normalized to those of loading control for each panel. The ratio of the first band was set to 100 for each blot. The asterisks indicate nonspecific bands. B, Germination rate measurements. Hydrated seeds for Col-0, cop1-4, cop1-4 ABI5-GFP, and cop1-4 ABI5-MYC were sown on MS medium or MS medium containing 0.5-µM ABA. Error bars represent sd of three independent sets of seeds, each set containing 50 seeds. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seeds. C, Phenotypic analyses of 5-day-old dark-grown Col-0, cop1-4, ABI5-GFP, cop1-4 ABI5-GFP, ABI5-MYC, and cop1-4 ABI5-MYC seedlings. The seedlings were grown vertically on MS medium or MS medium with various ABA concentrations for 5 days in darkness. Bar = 1 cm. D, Seedling establishment rate measurements. Col-0, cop1-4, cop1-4 ABI5-GFP, and cop1-4 ABI5-MYC seedlings were grown horizontally on MS medium or MS medium containing 0.5-µM ABA in darkness. Error bars represent sd of three independent pools of seedlings, each pool containing 50 seedlings. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seedlings.
As reported recently (Qi et al., 2020), ABA dramatically induces ABI5 protein accumulation in dark-grown Col-0 seedlings (Figure 2A). Interestingly, although ABA still induced ABI5 accumulation in the two cop1 alleles, the levels of ABI5 protein drastically decreased in cop1-6, and further decreased in cop1-4 mutant seedlings (Figure 2A). Consistent with these results, the seedling establishment rates of cop1-4 mutants were higher than those of cop1-6 mutants in the dark in response to ABA (Figure 1, C and D), suggesting that the different levels of ABI5 protein in cop1-4 and cop1-6 mutant seedlings may account for their ABA-hyposensitive phenotypes in darkness (Figure 1, B–D). To further investigate whether COP1 specifically mediates ABA regulation of ABI5, we examined the levels of ABF1 and ABF4, two other ABI5-clade transcription factors, in dark-grown cop1 mutants in response to ABA treatment. Our immunoblot data showed that the levels of ABF1 protein are higher in both cop1 mutant alleles before and after ABA treatment, which contradicts the ABA-hyposensitive phenotype of cop1 mutants (Supplemental Figure S4A). In contrast, ABF4 protein levels decreased slightly in both cop1 alleles; however, ABF4 did not accumulate to lower levels in the stronger ABA-hyposensitive cop1-4 mutant relative to cop1-6 (Supplemental Figure S4B). Together, these data suggest that COP1 differentially mediates ABA regulation of ABI5-clade transcription factors in the dark.
To determine the genetic relationship between COP1 and ABI5, we generated homozygous cop1-4 abi5-8 double mutant (Supplemental Figure S5, A and B) by genetic crossing. Notably, dark-grown cop1-4 abi5-8 double mutant seedlings displayed similar ABA insensitivity as the abi5 and cop1-4 single mutants in terms of root growth (Supplemental Figure S5C). To further investigate whether COP1 regulates ABA signaling in darkness through ABI5, we transformed Super:ABI5-GFP (encoding a fusion protein of ABI5 and the green fluorescent protein [GFP], under the control of the Super promoter) separately into Col-0 and the cop1-4 mutant and obtained multiple homozygous lines for each transgene (Supplemental Figure S6). We also generated homozygous cop1-4 35S:ABI5-MYC seedlings by genetic crossing. Notably, cop1-4 Super:ABI5-GFP seeds exhibited enhanced ABA inhibition of germination in comparison with cop1-4 mutants in the dark (Figure 2B). In contrast, overexpression of ABI5-GFP or ABI5-MYC efficiently rescued the ABA-hyposensitive phenotype of the cop1-4 mutant during seedling growth in darkness (Figure 2, C and D; Supplemental Figure S6). Collectively, our data demonstrate that COP1 positively regulates ABA signaling during seedling growth in darkness by mediating ABA-induced ABI5 accumulation.
COP1 stabilizes ABI5 protein by inhibiting 26S proteasome pathway-mediated ABI5 degradation
We hypothesized that COP1 may mediate ABA induction of ABI5 protein accumulation through transcriptional or post-translational regulatory mechanisms. To test whether COP1 may transcriptionally regulate ABI5, we compared ABI5 transcript levels in Col-0 and cop1 mutants in response to exogenous ABA treatment. Our reverse transcription quantitative polymerase chain reaction (RT-qPCR) assays showed that, indeed, ABI5 transcript levels accumulate to lower levels in response to ABA in both cop1 mutants relative to the wild-type (Supplemental Figure S7). However, the decreased ABA induction of ABI5 expression in cop1 mutants may be explained in part by the previous findings that fewer PIF proteins accumulate in dark-grown cop1 mutants (Bauer et al., 2004; Ling et al., 2017; Pham et al., 2018), since PIFs mediate ABA induction of ABI5 expression in darkness by directly binding to the ABI5 promoter (Qi et al., 2020). In addition, since ABI5 was shown to directly activate the transcription of its own gene (Xu et al., 2014), ABI5 transcript levels should also be reduced in cop1 mutants if COP1 promotes ABI5 protein stability.
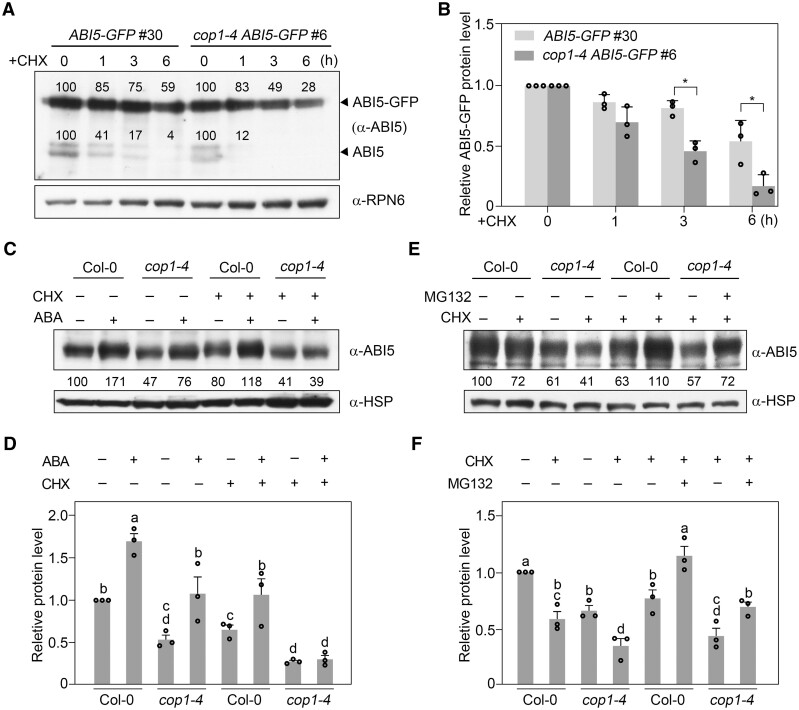
To investigate whether COP1 may post-translationally regulate ABI5 abundance, we grew Super:ABI5-GFP and cop1-4 Super:ABI5-GFP seedlings in darkness for 4 days, before being treated with 100-µM CHX for 1–6 h. Our immunoblot data showed that the already translated ABI5-GFP proteins are steadily degraded in Col-0 seedlings after CHX treatment; however, this degradation occurred much faster in cop1-4 than in Col-0 (Figure 3, A and B). Moreover, endogenous ABI5 protein levels displayed a surprisingly faster degradation in the cop1-4 mutant than in Col-0 after CHX treatment (Figure 3A; Supplemental Figure S8). Together, these data indicate that COP1 promotes the stability of ABI5 protein in the dark.
Figure 3.
COP1 stabilizes ABI5 protein by inhibiting 26S proteasome pathway-mediated ABI5 degradation. A, B, Immunoblots showing the abundance of ABI5-GFP and endogenous ABI5 in ABI5-GFP and cop1-4 ABI5-GFP seedlings in darkness. Four-day-old Super:ABI5-GFP and cop1-4 Super:ABI5-GFP seedlings grown in darkness were first treated with 100-µM CHX for the indicated times, and then harvested and subjected to immunoblotting using antibodies against ABI5. Anti-RPN6 was used as a sample loading control. Representative pictures are shown in (A) and the relative levels of ABI5-GFP are shown in (B). Numbers in the immunoblots in (A) indicate the relative band intensities of ABI5-GFP and endogenous ABI5 normalized to the loading control. The ratio was set to 100 for the 0 h samples. Error bars in (B) represent sd from three independent assays. *P < 0.05 (Student’s t test; Supplemental Data Set S1) for the indicated pair of samples. C, D, COP1 is essential for ABA-induced stabilization of ABI5 proteins. Four-day-old Col-0 and cop1-4 mutant seedlings grown in darkness were first treated with mock (ethanol), 100-μM CHX, 50-μM ABA, or 100-μM CHX together with 50-μM ABA for 3 h, and then harvested and subjected to immunoblotting using antibodies against ABI5. Anti-HSP was used as a sample loading control. Representative pictures are shown in (C) and the relative ABI5 levels are shown in (D). Numbers below the immunoblots in (C) indicate the relative band intensities of ABI5 normalized to the loading control. The ratio of the first band was set to 100. Error bars in (D) represent the standard error (se) from three independent assays. Different letters represent significant differences by one-way ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set S1). E, F, COP1 inhibits 26S proteasome pathway-mediated ABI5 degradation. Four-day-old Col-0 and cop1-4 mutant seedlings grown in darkness were first treated with 100-μM CHX, or 100-μM CHX together with 50-μM MG132 for 3 h, and then harvested and subjected to immunoblotting using antibodies against ABI5. Anti-HSP was used as a sample loading control. Representative pictures are shown in (E) and the relative ABI5 levels are shown in (F). Numbers below the immunoblots in (E) indicate the relative band intensities of ABI5 normalized to the loading control. The ratio of the first band was set to 100. Error bars in (F) represent se from three independent assays. Different letters represent significant differences by one-way ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set S1).
To further explore the regulatory mechanism underlying COP1 regulation of ABI5 stability, we grew Col-0 and cop1-4 mutant seedlings in darkness for 4 days, and then treated them with CHX, ABA, or CHX together with ABA. Our immunoblot data indicated that the already translated ABI5 protein is stabilized by exogenous ABA treatment (Figure 3, C and D), consistent with a previous study (Lopez-Molina et al., 2001); however, ABA-enhanced ABI5 stability was abolished in the absence of COP1 (Figure 3, C and D). We also treated Col-0 and cop1-4 mutant seedlings with CHX, or CHX together with MG132, an inhibitor of the 26S proteasome. Notably, addition of MG132 efficiently blocked the degradation of ABI5 in cop1-4 mutants (Figure 3, E and F), indicating that COP1 stabilizes ABI5 by inhibiting its degradation via the 26S proteasome pathway. Collectively, our data demonstrate that COP1 mediates ABA induction of ABI5 protein accumulation by upregulating ABI5 transcription, and by inhibiting ABI5 degradation through the 26S proteasome pathway.
COP1 physically interacts with ABD1
As a well-known E3 ubiquitin ligase, COP1 has been shown to ubiquitinate and target a wide range of substrates for degradation (Yi and Deng, 2005; Marine, 2012; Han et al., 2020). We therefore explored how COP1 promotes the stability of ABI5. We first performed yeast two-hybrid assays to investigate whether COP1 directly interacts with ABI5. Our results showed that there is no obvious interaction between ABI5 and COP1 in yeast cells, although we did detect an interaction between COP1 and HY5 as positive control (Supplemental Figure S9, A and B). We also performed co-immunoprecipitation (co-IP) assays using 4-day-old 35S:ABI5-GFP and 35S:HY5-GFP seedlings grown in the dark, which revealed that HY5-GFP, but not ABI5-GFP, coimmunoprecipitates COP1 (Supplemental Figure S9C). Collectively, our data suggest that ABI5 may not physically interact with COP1 in yeast cells or in vivo.
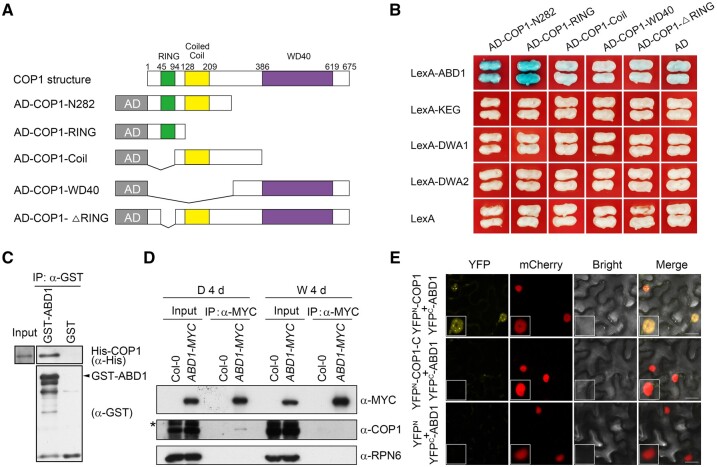
We then hypothesized that COP1 may target certain E3 ubiquitin ligase(s) normally acting on ABI5 for their 26S proteasome-mediated degradation, thus leading to enhanced stability of ABI5. ABI5 was previously shown to be targeted by several types of E3 ubiquitin ligases, including KEG, a RING-type E3 ligase, as well as DWA1, DWA2, and ABD1, three substrate receptors for CUL4-based E3 ligases (Stone et al., 2006; Lee et al., 2010; Seo et al., 2014). To test our hypothesis, we performed yeast two-hybrid assays to investigate whether COP1 physically interacts with any of these ABI5 E3 ligases. Bait vectors encoding the full-length KEG, DWA1, DWA2, or ABD1, and prey vectors encoding various fragments of COP1 (Figure 4A), were used in the assays. Notably, we observed that of these four ABI5 E3 ligases, only ABD1 directly associated with COP1 in yeast cells (Figure 4B). Pull-down assays showed that, indeed, recombinant glutathione S-transferase (GST)-tagged ABD1, but not GST alone, was able to pull down His-tagged COP1 in vitro (Figure 4C). Specifically, the RING-finger domain alone, or the N-terminal 282-aa region of COP1 that contains the RING-finger domain, physically interacted with ABD1, whereas deletion of the RING-finger domain abolished COP1 interaction with ABD1, demonstrating that the RING-finger domain of COP1 is responsible for interacting with ABD1 (Figure 4B).
Figure 4.
COP1 physically interacts with ABD1. A, Schematic diagram of prey proteins (COP1-N282, COP1-RING, COP1-Coil, COP1-WD40, and COP1-ΔRING fused with AD domains). B, Yeast two-hybrid assays showing that the RING-finger domain of COP1 is responsible for the interaction of COP1 with ABD1. Empty vectors were used as negative controls. C, Pull-down assays showing that GST-tagged ABD1, but not GST alone, can pull down His-tagged COP1 in vitro. His-tagged COP1 was incubated with immobilized GST or GST-tagged ABD1, and then the precipitated proteins were analyzed with anti-GST and anti-His antibodies, respectively. D, Co-IP assays showing that COP1 associates with ABD1 in vivo in the dark. Col-0 and 35S:ABD1-MYC seedlings were first grown in darkness or continuous white light for 4 days, and then the total proteins were extracted and incubated with Myc-Trap beads (ChromoTek). The total and precipitated proteins were subjected to immunoblotting with antibodies against MYC, COP1 and RPN6, respectively. The asterisks indicate nonspecific bands. E, BiFC assays in N. benthamiana leaves. The indicated combinations of YFPC-ABD1, YFPN-COP1, YFPN-COP1-C, and YFPN constructs were co-infiltrated into N. benthamiana leaves. YFP signals were observed 2 days after infiltration using confocal fluorescence microscopy. Bars = 20 μm.
To substantiate the physical interaction between COP1 and ABD1 in vivo, we conducted co-IP assays using 4-day-old Col-0 and 35S:ABD1-MYC seedlings grown in darkness or continuous white light. Our data showed that COP1 was coimmunoprecipitated by anti-MYC antibodies from 35S:ABD1-MYC protein extracts, but not from Col-0 (Figure 4D). Moreover, we observed that COP1 associates with ABD1 in vivo in the dark, but not in the light (Figure 4D). We also performed bimolecular fluorescence complementation (BiFC) assays (Waadt et al., 2008) by transiently infiltrating constructs encoding YFPN-COP1 (the N-terminal fragment of yellow fluorescent protein [YFP] fused with COP1) and YFPC-ABD1 (the C-terminal fragment of YFP fused with ABD1) fusions in Nicotiana benthamiana leaf cells. We determined that co-infiltration of YFPN-COP1 and YFPC-ABD1 constructs leads to strong YFP fluorescence, whereas YFPC-ABD1 co-infiltrated with YFPN-COP1-C (aa 283–675 of COP1 fused to the N-terminal fragment of YFP) or the empty vector showed no detectable YFP fluorescence (Figure 4E). Importantly, COP1 interacted with ABD1 in the nucleus, clearly forming nuclear bodies (Figure 4E). Collectively, our data demonstrate that COP1 physically interacts with ABD1.
COP1 mediates the degradation of ABD1 via the 26S proteasome pathway
Next, we investigated whether COP1 regulates the degradation of ABD1. Accordingly, we conducted cell-free degradation assays using recombinant His-ABD1 proteins and total protein extracts from 4-day-old Col-0 and cop1-4 mutant seedlings grown in darkness. We added ATP to enhance the protein degradation rate in cell-free degradation assays, as previously described (Kong et al., 2015). Immunoblot analysis showed that His-ABD1 is degraded when incubated with total protein extracts from both Col-0 and cop1-4 mutant seedlings; however, the degradation rate of His-ABD1 was much slower in samples from the cop1-4 mutant than from Col-0 (Figure 5, A and B). Notably, addition of MG132 efficiently blocked the degradation of His-ABD1 incubated with total protein extracts from Col-0 seedlings (Figure 5A). We also transiently co-transfected Arabidopsis protoplasts with the same amount of Super:ABD1-GFP plasmid DNA and various amounts of 35S:HF-COP1 plasmid. We observed that the levels of ABD1-GFP proteins decreased substantially with increasing levels of HF-COP1; however, COP1-mediated degradation of ABD1-GFP was fully inhibited by the addition of MG132 (Figure 5, C and D). These data indicate that COP1 mediates the degradation of ABD1 via the 26S proteasome pathway.
Figure 5.
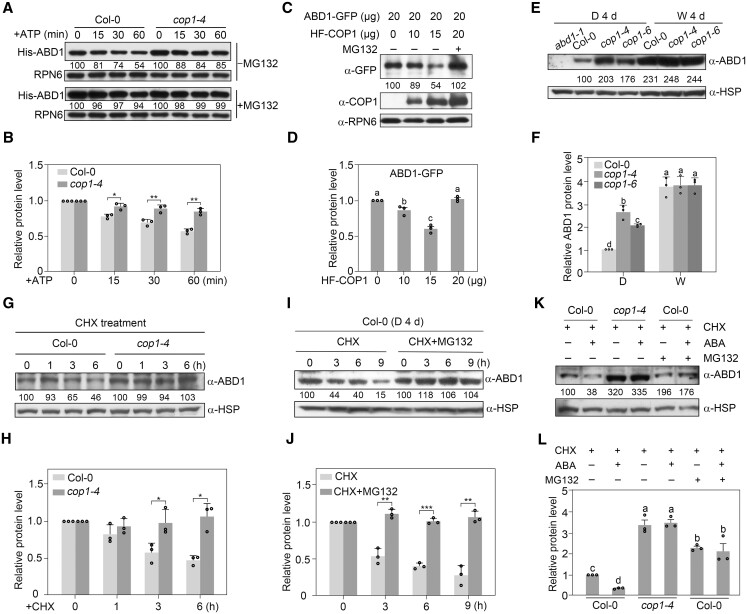
COP1 mediates ABD1 degradation via the 26S proteasome pathway. A, B, COP1 promotes the degradation of ABD1 in cell-free degradation assays. Recombinant purified His-ABD1 was incubated with equal amounts of total protein extracts from 4-day-old dark-grown Col-0 or cop1-4 seedlings in the presence of ATP, and then His-ABD1 was detected by immunoblotting using anti-His antibodies. Anti-RPN6 was used as a sample loading control. Representative pictures are shown in (A) and the relative levels of His-ABD1 are shown in (B). In (A), numbers below the immunoblots indicate the relative band intensities of His-ABD1 normalized to those of loading control. The ratio was set to 100 for the 0 h samples for each genotype. In (B), error bars represent sd from three independent assays. *P < 0.05 and **P < 0.01 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of samples. C, D, ABD1 is destabilized by COP1 in Arabidopsis protoplasts. Super:ABD1-GFP and different amounts of 35S:HF-COP1 plasmids were cotransfected into Arabidopsis (Col-0) protoplasts. After incubation for 18 h in darkness, ABD1-GFP and HF-COP1 proteins were detected with anti-GFP and anti-COP1 antibodies, respectively. Anti-RPN6 was used as a sample loading control. Representative pictures are shown in (C) and the relative levels of ABD1-GFP are shown in (D). Numbers below the immunoblots in (C) indicate the relative band intensities of ABD1-GFP normalized to the loading control. The ratio of the first ABD1-GFP band was set to 100. Error bars in (D) represent se from three independent assays. Different letters represent significant differences by one-way ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set S1). E, F, Immunoblots showing the levels of ABD1 proteins in Col-0 and two cop1 mutants. Four-day-old seedlings grown in darkness (D) or continuous white (W) light were subjected to immunoblotting. Anti-HSP was used as a sample loading control. Representative pictures are shown in (E) and the relative levels of ABD1 are shown in (F). Numbers below the immunoblots in (E) indicate the relative band intensities of ABD1 normalized to the loading control. The ratio of the first ABD1 band was set to 100. Error bars in (F) represent sd from three independent assays. Different letters represent significant differences by one-way ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set S1). G, H, COP1 regulates the stability of ABD1 proteins in vivo. Four-day-old Col-0 and cop1-4 mutant seedlings grown in darkness were treated with 300-μM CHX for the indicated times, and then the total proteins were extracted and subjected to immunoblotting with anti-ABD1 antibodies. Anti-HSP was used as a sample loading control. Representative pictures are shown in (G) and the relative levels of ABD1 are shown in (H). Numbers below the immunoblots in (G) indicate the relative band intensities of ABD1 normalized to the loading control. The ratio was set to 100 for the first ABD1 band of each genotype before CHX treatment. Error bars in (H) represent sd from three independent assays. *P < 0.05 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of samples. I, J, ABD1 is degraded via the 26S proteasome pathway. Four-day-old Col-0 seedlings grown in darkness were treated with 300-μM CHX or 300-μM CHX together with 50-μM MG132 for the indicated times, then the total proteins were extracted and subjected to immunoblotting with anti-ABD1 antibodies. Anti-HSP was used as a sample loading control. Representative pictures are shown in (I) and the relative levels of ABD1 are shown in (J). Numbers below the immunoblots in (I) indicate the relative band intensities of ABD1 proteins normalized to the loading control. The ratio was set to 100 for the first ABD1 band before each treatment. Error bars in (J) represent sd from three independent assays. **P < 0.01 and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of samples. K, L, COP1 mediates ABA-induced ABD1 turnover via the 26S proteasome pathway. Four-day-old Col-0 and cop1-4 mutant seedlings grown in darkness were treated for 3 h with 300-μM CHX or 300-μM CHX together with 50-μM MG132 in the presence or absence of 50-μM ABA. The total proteins were subjected to immunoblotting with antibodies against ABD1. Anti-HSP was used as a sample loading control. Representative pictures are shown in (K) and the relative levels of ABD1 are shown in (L). Numbers below the immunoblots in (K) indicate the relative band intensities of ABD1 normalized to the loading control. The ratio of the first ABD1 band was set to 100. Error bars in (L) represent se from three independent assays. Different letters represent significant differences by one-way ANOVA with Duncan's post hoc test (P < 0.05; Supplemental Data Set S1).
To investigate whether COP1 regulates ABD1 protein accumulation in vivo, we generated monoclonal anti-ABD1 antibodies that specifically recognize endogenous ABD1 protein (Supplemental Figure S10). We then grew Col-0 and cop1 mutant seedlings in darkness or continuous white light for 4 days before determining ABD1 protein abundance by immunoblotting. Indeed, ABD1 accumulated to higher levels in the cop1 mutants in the dark compared to the wild-type (Figure 5, E and F). Notably, larger amounts of ABD1 proteins accumulated in cop1-4 than in cop1-6 in the dark (Figure 5, E and F), consistent with the observations that cop1-4 mutant seedlings accumulate less ABI5 and display enhanced ABA insensitivity compared to cop1-6 seedlings in darkness (Figures 1 and 2, A). ABD1 accumulated to higher levels in the light than in darkness in Col-0 seedlings, with COP1 not obviously regulating ABD1 protein abundance in the light, as evidenced by the comparable ABD1 levels in Col-0 and the cop1 mutants (Figure 5, E and F). These results indicate that COP1 specifically regulates ABD1 protein accumulation in darkness.
To further verify the role of COP1 in mediating ABA regulation of ABD1 protein stability in vivo, we treated 4-day-old etiolated Col-0 and cop1-4 mutant seedlings with CHX for various times. Our immunoblot data showed that the already translated ABD1 becomes steadily degraded in the Col-0 background after CHX treatment; in contrast, this degradation was abolished in the absence of COP1 or upon the addition of MG132 (Figure 5, G−J). We also treated 4-day-old etiolated Col-0 and cop1-4 mutant seedlings with CHX either alone or together with ABA. We observed that ABA treatment markedly induces the degradation of translated ABD1 protein in Col-0 seedlings; however, ABA-induced ABD1 degradation was abolished in cop1-4 mutant seedlings or by the addition of MG132 (Figure 5, K and L). Taking these data together, we conclude that COP1 mediates ABA-induced ABD1 turnover in vivo by targeting ABD1 for degradation via the 26S proteasome pathway.
COP1 mediates the ubiquitination of ABD1
Next, we asked whether ABD1 is a substrate of the COP1 E3 ligase. To this end, we conducted in vitro ubiquitination assays using recombinant GST-tagged ABD1 and maltose-binding protein (MBP)-tagged COP1 produced in and purified from Escherichia coli. We established that in the presence of recombinant E1, E2, and FLAG-tagged ubiquitin (FLAG-UBQ), GST-ABD1 was polyubiquitinated by MBP-COP1, but not by boiled MBP-COP1, as detected by anti-GST and anti-FLAG antibodies (Figure 6A). Thus, these data indicate that COP1 can ubiquitinate ABD1 in vitro.
Figure 6.
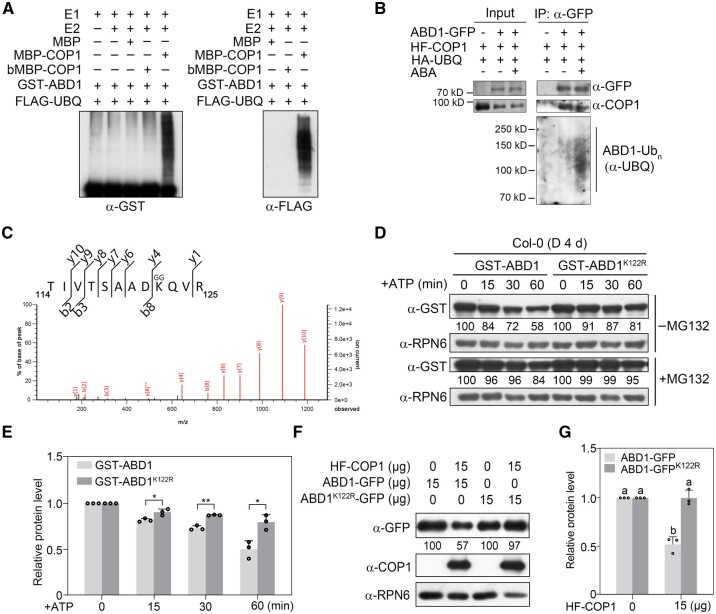
COP1 directly ubiquitinates ABD1 in vitro and in vivo. A, COP1 ubiquitinates ABD1 in vitro. Recombinant purified MBP, MBP-COP1, GST-ABD1, and denatured COP1 (bMBP-COP1) were used in the assays. The ubiquitinated ABD1 was analyzed by immunoblots using anti-GST and anti-FLAG antibodies. B, COP1 ubiquitinates ABD1 in vivo. Arabidopsis protoplasts expressing different combinations of HA-UBQ, HF-COP1 and ABD1-GFP were incubated in darkness for 18 h in the presence of 5-μM MG132 with or without 5-μM ABA. Total proteins were then extracted and incubated with GFP-Trap beads. The total and precipitated proteins were subjected to immunoblotting with antibodies against GFP, COP1, and ubiquitin, respectively. C, Peptides produced by trypsin proteolysis of ABD1 in the LC–MS/MS showed the fragment ion mass with intact diglycine modification at K122. D, E, Cell-free degradation assays showing that the K122R mutation decreases the degradation of ABD1 in total protein extracts from Col-0 seedlings. Recombinant purified GST-ABD1 or GST-ABD1K122R proteins were incubated with equal amounts of total proteins extracted from 4-day-old dark-grown Col-0 seedlings in the presence of ATP, and then detected by immunoblotting using anti-GST antibodies. Anti-RPN6 was used as a sample loading control. Representative pictures are shown in (D) and the relative levels of GST-ABD1 from three independent assays are shown in (E). In (D), numbers below the immunoblots indicate the relative band intensities normalized to the loading control. The ratio was set to 100 for the first band before ATP treatment. In (E), error bars represent sd from three independent assays. *P < 0.05 and **P < 0.01 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of samples. F, G, The K122R mutation improves the stability of ABD1 in vivo. The same amounts of Super:ABD1-GFP and Super:ABD1K122R-GFP were transfected with or without 35S:HF-COP1 plasmids into Arabidopsis (Col-0) protoplasts. After incubation for 18 h in darkness, ABD1-GFP and HF-COP1 were detected with anti-GFP and anti-COP1 antibodies, respectively. Anti-RPN6 was used as a sample loading control. Representative pictures are shown in (F) and the relative levels of ABD1-GFP proteins are shown in (G). Numbers below the immunoblots in (F) indicate the relative band intensities normalized to the loading control. The ratio was set to 100 in the absence of HF-COP1. Error bars in (G) represent sd from three independent assays. Different letters represent significant differences by one-way ANOVA with Duncan’s post hoc test (P < 0.05; Supplemental Data Set S1).
To investigate whether COP1 mediates the ubiquitination of ABD1 in vivo, we transiently transfected Arabidopsis protoplasts with constructs encoding HA- and FLAG-tagged COP1 (HF-COP1) and HA-tagged ubiquitin (HA-UBQ) with or without ABD1-GFP in the presence of 5-μM MG132. We extracted total proteins and incubated these extracts with anti-GFP beads, followed by detection of the precipitated proteins using anti-UBQ antibodies. Our data showed that IP-enriched ABD1-GFP is clearly polyubiquitinated; moreover, ABA treatment drastically induced the polyubiquitination levels of ABD1-GFP (Figure 6B). Together, our data demonstrate that COP1 mediates ABD1 ubiquitination in vitro and in vivo.
Then, we sought to identify the possible ubiquitination sites on ABD1. We employed a recently described cell-free assay system (Yu and Xie, 2019) using recombinant GST-ABD1 proteins and total proteins extracted from 4-day-old Col-0 seedlings grown in darkness. After incubation at 4°C for 4 h in the presence of 50-μM MG132, we separated the samples on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel, excised the gel sample containing the ubiquitinated target proteins, in-gel digested the slice, and subjected the digests to liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis. These efforts led to the identification of a modified peptide (114-TIVTSAADKQVR-125) ubiquitinated at the Lys122 residue (Figure 6C), suggesting that the K122 residue of ABD1 is a ubiquitination site.
To investigate whether this K122 residue plays an essential role in modulating ABD1 stability, we performed cell-free degradation assays using recombinant GST-tagged wild-type ABD1 protein and a K122R ABD1 variant, and total proteins extracted from 4-day-old etiolated Col-0 seedlings. The K122R variant of ABD1 exhibited a much lower degradation when incubated with total protein extracts from Col-0 seedlings (Figure 6, D and E). In addition, when we transiently transfected Arabidopsis protoplasts with equal amounts of Super:ABD1-GFP or Super:ABD1K122R-GFP alone, or together with 35S:HF-COP1 plasmid, we observed that the presence of HF-COP1 substantially decreased the abundance of ABD1-GFP, but not the abundance of ABD1K122R-GFP (Figure 6, F and G). Together, our data demonstrate that the K122R mutation improved the stability of ABD1, suggesting that the K122 residue of ABD1 is an active ubiquitination site in vivo.
COP1 partially promotes ABI5 protein stability in darkness by degrading ABD1
To investigate whether COP1 modulates ABA signaling in darkness through ABD1, we introduced the abd1-1 mutation into the cop1-4 background by genetic crossing and identified homozygous cop1-4 abd1-1 double mutant (Supplemental Figure S11). We observed that the delayed germination phenotype of the cop1-4 mutant in the dark is not rescued by the loss of ABD1 function (Figure 7A). However, the abd1-1 mutation largely suppressed the ABA-hyposensitive growth of cop1-4 seedlings (Figure 7B), and partially counteracted the higher seedling establishment rates of cop1-4 mutants when grown on MS medium containing ABA (Figure 7, C and D). These observations suggest that COP1 positively regulates ABA signaling, at least in part, by degrading ABD1 during the seedling stage in darkness.
Figure 7.
COP1 partially promotes ABI5 protein stability in darkness by degrading ABD1. A, Germination rate measurements. Hydrated seeds for Col-0, cop1-4, abd1-1, and cop1-4 abd1-1 mutants were sown on MS medium or MS medium containing 0.5-µM ABA, and then grown in darkness. Error bars represent sd of three independent sets of seeds, each set containing 50 seeds. *P < 0.05, and **P < 0.01 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seeds. B, Phenotypic analyses of 5-day-old dark-grown Col-0, cop1-4, abd1-1, and cop1-4 abd1-1 seedlings. The seedlings were grown vertically on MS medium with the indicated concentrations of ABA in darkness for 5 days. Bar = 1 cm. C, Phenotypes of Col-0, cop1-4, abd1-1, and cop1-4 abd1-1 mutants grown on MS medium or MS medium containing 0.5-µM ABA in darkness for 5 days. Bar = 1 cm. D, Seedling establishment rates for Col-0, cop1-4, abd1-1, and cop1-4 abd1-1 mutants. The seedlings were grown horizontally on MS medium or MS medium containing 0.5-µM ABA. Error bars represent sd of three independent pools of seedlings, each pool containing 50 seedlings. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seedlings. E, F, Immunoblots showing the levels of ABI5 protein in Col-0, cop1-4, abd1-1, and cop1-4 abd1-1 mutant seedlings. Four-day-old seedlings grown in darkness were treated with 50-μM ABA for 3 h, and then subjected to immunoblotting with anti-ABI5 antibodies. Anti-HSP was used as a sample loading control. Representative pictures are shown in (E) and the relative levels of ABI5 are shown in (F). Numbers below the immunoblots in (E) indicate the relative band intensities of ABI5 normalized to the loading control. The ratio of the first ABI5 band was set to 100 for each blot. Error bars in (F) represent sd from three independent assays. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of samples.
We further compared ABI5 protein abundance in cop1-4 abd1-1 double mutant seedlings with that of the respective single mutants by immunoblot analysis. While ABI5 accumulated to lower levels in cop1-4 mutant seedlings than in Col-0 seedlings when grown in the dark for 4 days, loss of ABD1 function led to an increase in ABI5 protein abundance in the cop1-4 abd1-1 double mutant, although not to the same level as in Col-0 seedlings (Figure 7, E and F). However, after 3 h of ABA treatment, the level of ABI5 in the cop1-4 abd1-1 double mutant was close to that in Col-0 seedlings (Figure 7, E and F). Collectively, these data demonstrate that COP1 partially promotes ABI5 protein accumulation during the seedling stage in darkness by degrading ABD1 in response to ABA.
ABA signaling induces the nuclear enrichment of COP1 in darkness
Finally, we asked how COP1 might be regulated by ABA. To this end, we treated 4-day-old etiolated Col-0 seedlings with 50-µM ABA or mock (equal volume of ethanol) for various times. Our RT-qPCR assays indicated that COP1 transcript levels are not significantly affected by ABA treatment (Supplemental Figure S12A). In addition, our immunoblot data showed that the abundance of COP1 protein remains largely unchanged in etiolated Col-0 seedlings treated with ABA (Supplemental Figure S12, B and C). Together, these data indicate that COP1 transcript and COP1 protein levels are not significantly regulated by ABA.
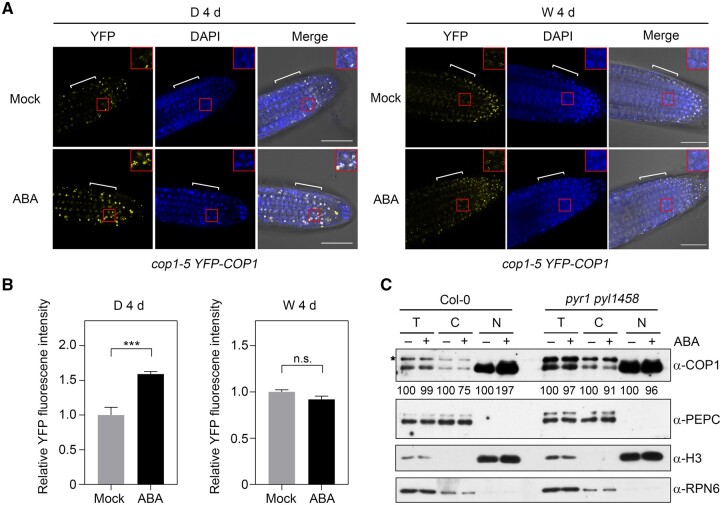
Light tightly controls the nucleo-cytoplasmic partitioning of COP1 by inducing its rapid translocation from the nucleus to the cytosol (von Arnim and Deng, 1994; Pacín et al., 2014). To investigate whether the nucleo-cytoplasmic distribution of COP1 is regulated by ABA, we grew cop1-5 35S:YFP-COP1 seedlings in darkness or continuous white light for 4 days, and then treated them with mock (ethanol only) or ABA for 1 h. Confocal microscopy analyses showed that compared to mock-treated seedlings, ABA treatment induced the nuclear enrichment of YFP-COP1 in the dark (Figure 8, A and B). In contrast, ABA treatment did not obviously affect the nuclear accumulation of YFP-COP1 in the light (Figure 8, A and B). To further verify the ABA regulation of COP1 nuclear accumulation in the dark, we treated 4-day-old etiolated Col-0 and pyr1 pyl1458 seedlings with mock (ethanol only) or ABA for 1 h, and then subjected their protein extracts to nuclear-cytoplasmic fractionation assays. We determined that ABA treatment indeed induces a drastic increase in the abundance of nuclear COP1 protein in dark-grown Col-0 seedlings (Figure 8C). However, this induction was abolished in pyr1 pyl1458 mutant seedlings grown in the dark (Figure 8C), indicating that ABA induces the nuclear enrichment of COP1 through the ABA signaling pathway. Taken together, our data demonstrate that ABA signaling induces the nuclear enrichment of COP1 in darkness.
Figure 8.
ABA induces the nuclear accumulation of COP1 in darkness. A, ABA treatment induces the nuclear accumulation of YFP-COP1 in darkness. cop1-5 35S:YFP-COP1 seedlings were first grown in darkness (D) or continuous white (W) light for 4 days, and were treated with mock (ethanol) or 60-μM ABA for 1 h. YFP signals were observed under a confocal laser scanning microscope. Bar = 50 μm. 4′,6-Diamidino-2-phenylindole staining shows the nuclei of the cells. Red boxed regions are enlarged and shown as insets. The regions indicated by white brackets were used for the quantification of YFP fluorescence signals using Image J shown in (B). B, Relative intensities of YFP fluorescence signals of cop1-5 35S:YFP-COP1 seedlings after 1 h of mock or ABA treatment. For each treatment, at least 10 seedlings were observed, each seedling being taken three confocal images at different imaging depths, and error bars represent se from at least 30 confocal images. ***P < 0.001 (Student’s t test; Supplemental Data Set S1) for the indicated pairs of seedlings. n.s., not significant. C, Nuclear/cytoplasmic fractionation assays showing that ABA signaling induces the nuclear enrichment of COP1 in darkness. Four-day-old Col-0 and pyr1 pyl1458 mutant seedlings grown in darkness were treated with mock (ethanol) or 60-μM ABA for 1 h, and then the cytoplasmic and nuclear fractions were separated and subjected to immunoblotting. PEPC was used as a cytosolic marker, and histone H3 served as a nuclear marker. RPN6 was used as a total protein loading control. Numbers below the immunoblots indicate the relative band intensities of COP1 normalized to RPN6 for total, PEPC for cytosolic, and H3 for nuclear fractions, respectively. The ratio of the COP1 band treated with mock was set to 100 for each fraction. The asterisk indicates a nonspecific band.
Discussion
COP1 and ABI5 were shown to colocalize in nuclear bodies long ago (Lopez-Molina et al., 2003); however, the relationship between COP1 and ABI5 remains largely obscure. Recently, COP1 was suggested to act genetically downstream of ABI5 in mediating ABA inhibition of seedling establishment in long-day conditions (16-h light/8-h dark; Yadukrishnan et al., 2020). In addition, COP1 may not influence ABI5 transcript or ABI5 protein levels under long-day conditions, but facilitates ABI5 binding to its target promoters (Yadukrishnan et al., 2020). In this study, we demonstrated that COP1 positively regulates the ABA signaling pathway during seedling growth in darkness by mediating ABA-induced accumulation of ABI5. Notably, overexpression of ABI5 was unable to rescue the ABA-hypersensitive seed germination of the cop1-4 mutant (Figure 2B), indicating that COP1 mediates ABA regulation of seed germination through different mechanisms, which should be investigated in future studies.
Our RT-qPCR analyses showed that COP1 mediates ABA-induced ABI5 expression in dark-grown seedlings (Supplemental Figure S7). This result may be explained in part by the fact that COP1 is required for the accumulation of PIF proteins in darkness (Bauer et al., 2004; Ling et al., 2017; Pham et al., 2018), while our recent study showed that PIFs mediate ABA-induced ABI5 expression in darkness by directly binding to the ABI5 promoter (Qi et al., 2020). In addition, as ABI5 was shown to directly activate its own transcription (Xu et al., 2014), a lower stability of ABI5 may also contribute to the decreased expression levels of ABI5. Indeed, our data demonstrated that COP1 directly ubiquitinates ABD1 and negatively regulates ABD1 protein stability in response to ABA (Figures 5 and 6), thus playing an essential role in mediating ABA-enhanced ABI5 stability in dark-grown seedlings (Figure 3). Collectively, our data established that COP1 positively regulates ABA signaling during seedling growth in darkness by mediating ABA-induced ABI5 accumulation through transcriptional and post-translational regulatory mechanisms (Figure 9).
Figure 9.
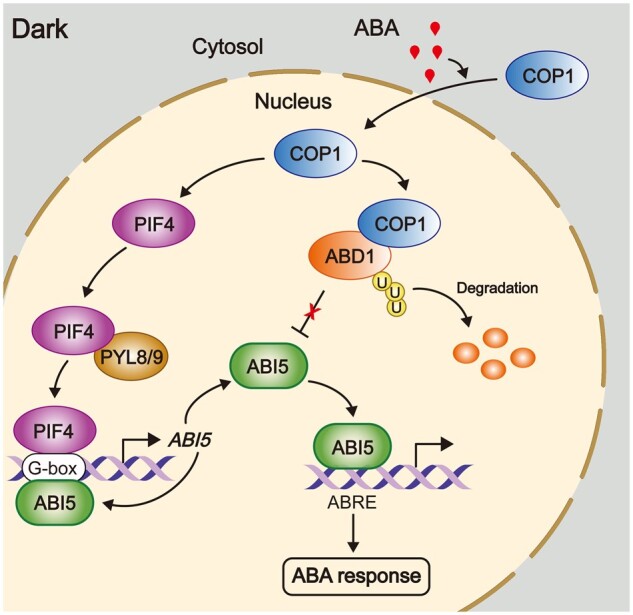
A working model depicting the role of COP1 in mediating ABA signaling during Arabidopsis seedling growth in darkness. During Arabidopsis seedling growth in darkness, COP1 promotes ABI5 stability by targeting ABD1 for 26S proteasome pathway-mediated degradation. In addition, COP1 also promotes the stability of PIF proteins in the dark. As PIFs and ABI5 itself can activate ABI5 transcription by directly binding to the G-box motifs in the ABI5 promoter (Xu et al., 2014; Qi et al., 2020), COP1 also mediates ABA-induced ABI5 transcription in the dark. Thus, COP1 mediates ABA-induced ABI5 accumulation through transcriptional and post-translational regulatory mechanisms, thereby playing a positive role in mediating ABA signaling during Arabidopsis seedling growth in darkness. ABA signaling induces the nuclear accumulation of COP1 in darkness, thus enhancing its activity in propagating the ABA signal.
Our data indicated that COP1 promotes ABI1/ABI2 protein accumulation in the dark before and after ABA treatment (Figure 2A). We reasoned that this was due, at least in part, to COP1-mediated upregulation of ABI1 and ABI2 transcription, as COP1 promotes the accumulation of ABI5 in the dark before and after ABA treatment (Figure 2A), while ABI5 mediates ABA-induced ABI1/ABI2 expression by directly binding to their promoters (Supplemental Figure S3; Wang et al., 2019b). Therefore, ABA rapidly induces ABI1 and ABI2 transcription and thus the accumulation of ABI1 and ABI2 both in the dark (this study) and in the light (Wang et al., 2019b), which likely serves as a feedback mechanism to desensitize ABA signaling when dramatically increased levels of ABA are synthesized under abiotic stress (Raghavendra et al., 2010; Wang et al., 2019b). Notably, a recent study reported that COP1 physically interacts with several group A PP2Cs (including ABI1 and ABI2) and degrades them through the 26S proteasome pathway under diurnal (12-h light/12-h dark) conditions to promote ABA-induced stomatal closure (Chen et al., 2021). Together with the findings that COP1 differentially regulates ABI5 accumulation in darkness (this study) and long-day conditions (Yadukrishnan et al., 2020), COP1 may thus act in ABA signaling via distinct mechanisms in different developmental processes and in response to various environmental cues.
Notably, our data showed that COP1 does not contribute to the regulation of ABD1 protein abundance in the light (Figure 5, E and F), suggesting that COP1-mediated ABI5 stabilization by degrading ABD1 only occurs in the dark. Consistent with this conclusion, our co-IP data showed that COP1 associates with ABD1 in vivo in the dark but not in the light (Figure 4D). A long-established mechanism for light inactivation of COP1 activity is its rapid translocation from the nucleus to the cytosol that is induced by photoactivated photoreceptors (von Arnim and Deng, 1994; Osterlund and Deng, 1998; Pacín et al., 2014). In addition, photoactivated photoreceptors also disrupt the interactions between COP1 and SPA proteins, thus inactivating the COP1/SPA E3 ubiquitin ligase complexes (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011; Lu et al., 2015; Sheerin et al., 2015). Since ABD1 physically interacts with and negatively regulates the stability of ABI5 in the nucleus (Seo et al., 2014), the inability of COP1 to degrade ABD1 in the light may be explained by the fact that photoactivated photoreceptors inactivate the COP1/SPA complexes by inducing the nuclear export of COP1 and by disrupting the COP1/SPA interactions. Interestingly, our data showed that ABA treatment induces the nuclear enrichment of COP1 in the dark but not in the light (Figure 8, A and B). That this induction was abolished in pyr1 pyl1458 mutant seedlings (Figure 8C) indicates that ABA signaling induces the nuclear accumulation of COP1, thus enhancing its activity in propagating the ABA signal in the dark. The underlying molecular mechanisms remain to be investigated in future studies.
Our genetic analyses indicated that the abd1-1 mutation only partially rescues the ABA-hyposensitivity of cop1-4 mutant seedlings (Figure 7, C and D) and the levels of ABI5 protein abundance (Figure 7, E and F). These observations might be explained as follows. First, as discussed above, decreased accumulation of PIFs in the absence of COP1 leads to reduced expression of ABI5, which may account for the partial restoration of ABI5 protein abundance and the ABA-hyposensitivity of cop1-4 mutant seedlings by the abd1-1 mutation. Second, although several types of E3 ubiquitin ligases involved in ABI5 degradation have been identified, there is still a strong possibility that some as-yet-unidentified E3 ubiquitin ligase(s) may participate in ABI5 ubiquitination and degradation. COP1 may target some of these unidentified E3 ubiquitin ligases for degradation as well, whereas these COP1-targeted E3 ubiquitin ligases can still degrade ABI5, thus leading to decreased ABI5 protein stability in the cop1-4 abd1-1 double mutant.
We recently reported that pifq mutant seedlings are hyposensitive to ABA in the dark (Qi et al., 2020). In this study, we observed that dark-grown cop1 mutant seedlings are also less sensitive to ABA than the wild-type (Figure 1, B–D). Interestingly, both cop1 and pifq mutants exhibit a cop phenotype, i.e. developing in darkness as wild-type seedlings would in the light (Deng et al., 1991; Leivar et al., 2008; Shin et al, 2009). These observations may suggest that light-grown seedlings are more resistant to ABA than dark-grown seedlings. Indeed, we did notice that wild-type (Col-0) seedlings exhibit a greater resistance to ABA treatment in continuous white light than in the dark (Supplemental Figure S13). In addition, a recent study showed that wild-type seedlings grown under long-day conditions are also more resistant to ABA than those grown in the dark (Yadukrishnan et al., 2020). This enhanced ABA resistance in the light is likely mediated by photoactivated photoreceptors, since our data in this study and earlier (Qi et al., 2020) demonstrated that both COP1 and PIFs positively regulate ABA signaling in dark-grown seedlings, whereas photoactivated phytochromes and cryptochromes inhibit the functions of these negative regulators of photomorphogenesis in the light (Li et al., 2011; Lau and Deng, 2012; Wang and Lin, 2020; Cheng et al., 2021). Photoactivated phytochromes (phys) and cryptochromes (Crys) have recently been shown to directly interact with the key components of several phytohormone pathways, including auxin/indole-3-acetic acid proteins (Aux/IAAs) and auxin response factors (ARFs) acting in auxin signaling (Xu et al., 2018; Yang et al., 2018; Mao et al., 2020), GIBBERELLIN-INSENSITIVE DWARF1 (GID1) and DELLAs for gibberellins (Xu et al., 2021; Yan et al., 2021; Zhong et al., 2021), and BRI1-EMS-SUPPRESSOR1 (BES1) for brassinosteroids (Wang et al., 2018b; Wu et al., 2019), to regulate their signaling. Thus, our data demonstrate that photoactivated photoreceptors employ a similar mechanism to modulate ABA signaling via the phy/Cry-COP1-ABD1-ABI5 pathway, since both phytochromes and cryptochromes directly interact with COP1 (Wang et al., 2001; Yang et al., 2001; Seo et al., 2004; Jang et al., 2010).
To summarize, our study provides further evidence supporting the notion that there are dark-specific ABA signaling components and pathways in plants. Therefore, further elucidation of ABA signaling mechanisms in darkness will shed more light on how plants adjust their responses to abiotic stresses under dynamic light environments.
Materials and methods
Plant materials and growth conditions
The Arabidopsis (A. thaliana) plants used in this study were in the Columbia-0 (Col-0) accession. The abd1-1 (SALK_051074; Seo et al., 2014), cop1-4 (McNellis et al., 1994), cop1-6 (McNellis et al., 1994), abi5-1 (Zhao et al., 2019), abi5-8 (SALK_013163; Zheng et al., 2012; Guan et al., 2014), ost1-3 (SALK_008068; Ding et al., 2015), pyr1 pyl124 (Gonzalez-Guzman et al., 2012), pyr1 pyl1458 (Antoni et al., 2013), cop1-5 35S:YFP-COP1 (Huang et al., 2013), 35S:ABD1-MYC (Seo et al., 2014), 35S:HY5-GFP (Li et al., 2022), and 35S:ABI5-MYC (Chen et al., 2012) mutants and transgenic lines have been described previously. The cop1-4 abd1-1, cop1-4 abi5-8 double mutants and cop1-4 35S:ABI5-MYC plants were generated by genetic crossing. The growth conditions were as described previously (Zhang et al., 2018). Germination was induced by incubation at 22°C under 50 µmol m−2 s−1 continuous white light (F17T8/TL841 bulb, Philips) for 12 h, and then the seeds were incubated at 22°C in complete darkness or in continuous white light for 4 days.
Plasmid construction and generation of transgenic Arabidopsis plants
To generate the LexA-ABD1, LexA-KEG, LexA-DWA1 and LexA-DWA2 constructs, the coding sequences of ABD1, KEG, DWA1 and DWA2 were individually amplified by polymerase chain reaction (PCR) with the primer pairs listed in Supplemental Table S1, and then cloned into the EcoRI–XhoI (for LexA-ABD1 and LexA-DWA1), XhoI (for LexA-DWA2), or MfeI-SalI (for LexA-KEG) sites of the pLexA vector (Clontech). The AD-COP1, AD-COP1-RING, AD-COP1-N282, AD-COP1-Coil, AD-COP1-ΔRING, AD-COP1-WD40, AD-COP1-ΔCoil constructs were described previously (Ang et al., 1998; Saijo et al., 2003). To generate the LexA-COP1, LexA-COP1-WD40 and LexA-COP1-Coil constructs, the corresponding PCR fragments of COP1 were amplified by PCR with the primer pairs listed in Supplemental Table S1, and then cloned into the EcoRI–XhoI sites of the pLexA vector (Clontech).
To generate the YFPC-ABD1 construct, the coding sequence of ABD1 was amplified by PCR with the primer pairs shown in Supplemental Table S1, and then cloned into the BamHI–XhoI sites of the pSPYCE (MR) vector (Waadt et al, 2008). To generate the YFPN-COP1 and YFPN-COP1-C constructs, the corresponding fragments of the COP1 coding sequence were individually cloned into the SpeI–SalI sites of the pSPYNE (R) 173 vector (Waadt et al, 2008).
The MBP-COP1 construct was described previously (Lian et al., 2017). To generate the His-ABD1 and His-COP1 constructs, the full-length coding sequences of ABD1 and COP1 were individually cloned into the EcoRI–XhoI sites of the pET-28a vector (Novagen). To generate the GST-ABD1 construct, the full-length coding sequence of ABD1 was cloned into the EcoRI–XhoI sites of the pGEX-4T-1 vector (Amersham Biosciences). To generate the GST-ABD1K122R construct, the fast mutagenesis kit (Vazyme) was employed using the GST-ABD1 construct as template and the primer pairs shown in Supplemental Table S1. To generate the His-PYR1 construct, the full-length coding sequence of PYR1 was cloned into the BamHI–EcoRI sites of the pET-28a vector. To generate the His-OST1 construct, the full-length coding sequence of OST1 was cloned into the EcoRI–XhoI sites of the pET-28a vector.
The pSuper1300 vector (the pCAMBIA1300 vector containing a Super promoter) was described previously (Liu et al., 2017). To generate the Super:ABD1-GFP construct, the coding sequence of ABD1 was amplified by PCR, and then cloned into the corresponding sites of the pSuper1300-GFP vector (Wang et al., 2019a). To generate the Super:ABD1-GFPK122R construct, the fast mutagenesis kit (Vazyme) was employed using the Super:ABD1-GFP construct as template and the primer pairs shown in Supplemental Table S1. To generate the 35S:HF-COP1 construct, the coding sequence of COP1 was cloned into the SalI–SpelI sites of the pCAM1307-HF vector (Wang et al., 2019a).
The Super:ABI5-GFP construct was described previously (Qi et al., 2020). To generate Super:ABI5-GFP and cop1-4 Super:ABI5-GFP transgenic plants, the Super:ABI5-GFP construct was transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101, and then into Col-0 and cop1-4 by the floral dip method (Clough and Bent, 1998).
All primers used to generate the above-mentioned constructs are listed in Supplemental Table S1, and all constructs were confirmed by sequencing prior to usage in various assays. Transgenic plants were selected on MS plates with the appropriate antibiotics, and homozygous transgenic plants were used in various assays.
ABA treatments and analyses of ABA responses
For gene expression and immunoblotting assays, 4-day-old Arabidopsis seedlings grown in darkness were first soaked in half-strength liquid MS medium containing 50-μM ABA (ABA treatment) or an equal volume of ethanol (mock) for the indicated times, and then harvested for various assays.
To examine the ABA phenotypes of various materials, all seedlings were grown vertically on MS medium alone or containing various concentrations of ABA for 5 days in darkness. For the germination assays, seeds (at least 50 seeds for each genotype at each time point) harvested at approximately the same time were surface sterilized and sown on MS medium or MS medium containing various concentrations of ABA in darkness. Germination was defined as the first sign of radicle tip emergence and scored daily under a green safety light until the sixth day in darkness, and the germination results of three independent experiments were calculated. The rates of seedling establishment were based on the criteria defined by Yadukrishnan et al. (2020) that hypocotyls and cotyledons should emerge completely in established seedlings; the results of three independent experiments were calculated.
RNA extraction and RT-qPCR analyses
Four-day-old seedlings grown in darkness were first treated with 50-μM ABA (ABA treatment) or an equal volume of ethanol (mock) for the indicated times and then harvested. Total RNA was extracted using the RNAprep Pure Plant Kit (TIANGEN), and the first-strand cDNAs were synthesized from 1 μg total RNA using ReverAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). qPCR assays were performed using gene-specific primers (listed in Supplemental Table S1) and PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) with a Step One Plus Real-Time PCR System (Applied Biosystems). Materials for RT-qPCR assays were collected from three independent pools of seedlings. PCR reactions were performed in triplicate for each sample, and the expression levels were normalized to those of TUBULIN3.
Yeast two-hybrid assays
The yeast two-hybrid assays were performed as described previously (Li et al., 2010). Briefly, the respective combinations of LexA- and AD-fusion plasmids were cotransformed into the yeast strain EGY48 (Estojak et al., 1995), which already contains the reporter plasmid p8op:LacZ. Yeast transformants were then streaked on synthetic dropout (SD) medium (SD–Ura–His–Trp) containing X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue color development. Yeast transformation was conducted as described in the Yeast Protocols Handbook (Clontech).
Co-IP and immunoblotting
For co-IP assays, 500 mg of Col-0 or the indicated transgenic seedlings were first grown in darkness or continuous white light for 4 days, and then homogenized in 1-mL of protein extraction buffer containing 50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 10-mM MgCl2, 1-mM EDTA, 0.1% (v/v) NP-40, 1-mM phenylmethanesulfonyl fluoride (PMSF), 40-μM MG132, and 1× complete protease inhibitor cocktail (Roche), and centrifuged twice for 15 min at 12,000g, 4°C. Of the 1-mL supernatant for each sample, 100 μL was set aside as total, and the remainder was incubated with Myc-Trap beads (ChromoTek) or GFP-Trap beads (ChromoTek) for 2 h at 4°C. The beads were then washed four times (10 min each time) with protein extraction buffer, and the immunoprecipitated proteins were eluted in 2× SDS loading buffer and analyzed by immunoblotting.
For anti-ABI1, ABI2, ABI5, ABD1, ABF1, and ABF4 immunoblots, total proteins were extracted as described previously (Qiu et al., 2017). Briefly, 4-day-old dark-grown seedlings (250 mg each sample) were ground in 750-µL extraction buffer (100-mM Tris–HCl, pH 7.5, 100-mM NaCl, 5-mM EDTA, pH 8.0, 5% [w/v] SDS, 20% [v/v] glycerol, 20-mM DTT, 40-mM β-mercaptoethanol, 2-mM PMSF, 1× EDTA-free protease inhibitor cocktail [Roche], 80-μM MG132, 80-μM MG115, 1× phosphatase inhibitor cocktail, and 10-mM N-ethylmaleimide) under dim green light. Samples were immediately boiled for 10 min and then centrifuged at 16,000g for 10 min at room temperature. Proteins from the supernatant were used in the subsequent immunoblot assays.
For all other immunoblots, total proteins were extracted by homogenizing 4-day-old seedlings in 100 μL of extraction buffer containing 50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 10-mM MgCl2, 0.1% (v/v) Tween-20, 1-mM PMSF, 40-μM MG132, and 1× EDTA-free protease inhibitor cocktail (Roche). Protein concentration was determined using Bradford Assay reagent (Bio-Rad), and then equal amounts of total proteins for each sample were boiled in 6× SDS loading buffer for 15 min.
Immunoblotting was performed as previously described (Yan et al., 2020). Primary antibodies used in this study include anti-GST (1:2,000 [v/v], catalog no. G7781; Sigma-Aldrich), anti-His (1:1,000 [v/v], catalog no. H1029; Sigma-Aldrich), anti-GFP (1:2,000 [v/v], catalog no. BE2002; EasyBio), anti-MYC (1:2,000 [v/v], catalog no. MF083; Mei5 Biotechnology), anti-FLAG (1:1,000 [v/v], catalog no. M20008H; Abmart), anti-Ubiquitin (1:2,000 [v/v], catalog no. 3936; Cell Signaling Technology), anti-HSP (1:5,000 [v/v], catalog no. AbM51099-31-PU; Beijing Protein Innovation), anti-H3 (1:2,000 [v/v], catalog no. ab1791; Abcam), anti-PEPC (1:1,000 [v/v], catalog no. AS09458; Agrisera), anti-COP1 (1:1,000 [v/v]; Saijo et al., 2008), anti-ABI5 (1:2,000 [v/v], catalog no. ab98831; Abcam), anti-ABI1 (1:2,000 [v/v]; Kong et al., 2015), anti-ABI2 (1:2,000 [v/v]; Wang et al., 2019b), anti-ABF1 (1:2,000 [v/v]; Wang et al., 2019b), anti-ABF4 (1:1,000 [v/v]; Wang et al., 2019b), anti-RPN6 (1:5,000 [v/v]; Zhou et al., 2018).
The anti-PYR1, anti-OST1, and anti-ABD1 antibodies were made by Beijing Protein Innovation. To generate the anti-PYR1 and anti-OST1 polyclonal antibodies, His-PYR1 and His-OST1 proteins were first produced in E. coli, and then purified and used as antigens to immunize rabbits for producing polyclonal antisera. Antigen affinity-purified antibodies were used in immunoblots (1:2,000 [v/v] for both His-PYR1 and His-OST1 antibodies). To generate anti-ABD1 monoclonal antibodies, His-ABD1 proteins produced in E. coli were used as antigens to immunize mice for generating the anti-ABD1 monoclonal antibodies. Purified ABD1 monoclonal antibodies were used in immunoblots (1:1,000 [v/v]).
Nuclear-cytoplasmic fractionation assays
Nuclear fractionation was conducted as described previously (Zhang et al., 2018). Briefly, 1 g of wild-type (Col-0) and pyr1 pyl1458 mutant seedlings grown in darkness for 4 days were first treated with mock (equal volume of ethanol) or 60-μM ABA for 1 h and ground in liquid nitrogen, thawed in 2 mL of pre-cooled (4°C) lysis buffer (20-mM Tris–HCl, pH 7.4, 25% [v/v] glycerol, 20-mM KCl, 2-mM EDTA, 2.5-mM MgCl2, 250-mM sucrose, 5-mM DTT) supplemented with 1× protease inhibitor cocktail (Roche), and filtered twice with two layers of Miracloth (Merck Millipore). The flow-through was centrifuged at 1,500g for 10 min at 4°C, and then the supernatant, consisting of the cytoplasmic solution, was centrifuged at 13,000g for 15 min at 4°C and collected. The pellet, containing the nuclear fraction, was washed four times with 2 mL of NRBT buffer (20-mM Tris–HCl, pH 7.4, 25% [v/v] glycerol, 2.5-mM MgCl2, and 0.2% [v/v] Triton X-100), and centrifuged for 2 min at 1,500g at 4°C. The pellet was resuspended in 300 μL of pre-cooled NRBT2 (250-mM sucrose, 10-mM Tris–HCl, pH 7.5, 10-mM MgCl2, 1% [v/v] Triton X-100, 0.035% [v/v] β-mercaptoethanol) containing 1× protease inhibitor cocktail (Roche), and carefully overlaid on top of 300 μL of NRBT3 (1.7-M sucrose, 10-mM Tris–HCl, pH 7.5, 2-mM MgCl2, 0.15% [v/v] Triton X-100, 0.035% [v/v] β-mercaptoethanol) containing 1× protease inhibitor cocktail (Roche). These samples were centrifuged for 45 min at 16,000g at 4°C, and the final nuclear pellet was resuspended in 2× SDS loading buffer. As quality control for the fractionation, PEPC protein was detected and used as a cytoplasmic marker, and histone H3 was probed and used as a nuclear marker.
In vitro pull-down assays
In vitro pull-down assays were performed as described previously (Dong et al., 2020). Briefly, 2.5 μg of purified recombinant bait proteins (GST-ABD1 and GST) and 2.5 μg of prey proteins (His-COP1) were added to 1 mL of binding buffer (50-mM Tris–HCl, pH 7.5, 100-mM NaCl, 0.2% [v/v] glycerol, and 0.6% [v/v] Triton X-100). After incubation for 2 h at 4°C, Glutathione Sepharose 4B beads (GE Healthcare) were added and incubated for another 2 h. After a gradient wash with washing buffer (50-mM Tris–HCl, pH 7.5, and 150- or 200- or 300-mM NaCl), the pulled-down proteins were eluted by boiling in 2× SDS loading buffer for 15 min. The input and eluted proteins were separated on 8% (w/v) SDS–PAGE gels, and then subjected to immunoblotting with anti-His (1:1,000 [v/v], catalog no. H1029; Sigma-Aldrich) or anti-GST antibodies (1:2000 [v/v], catalog no. G7781; Sigma-Aldrich).
BiFC assays
BiFC experiments were performed as described previously (Waadt et al., 2008). Briefly, the constructs were first individually transformed into Agrobacterium strain GV3101. The bacteria were then grown overnight, briefly pelleted by centrifugation and resuspended in infiltration buffer (10-mM MES, pH 5.7, 10-mM MgCl2, 150-µM acetosyringone). The cell suspensions for the appropriate constructs were mixed in equal volumes and infiltrated into young but fully expanded leaves of 7-week-old N. benthamiana leaves for transient expression using a needleless syringe. Two days after infiltration, the YFP fluorescence signal was detected by a confocal laser scanning microscope (ZEISS LSM 880).
In vitro and in vivo ubiquitination assays
In vitro ubiquitination assays were performed as described previously (Lian et al., 2017) with minor modifications. Briefly, ubiquitination reaction mixtures (60 μL) contained 100 ng of UBE1 (E1; Boston Biochem), 100 ng of UbcH5b (E2; Boston Biochem), 10 μg of FLAG-tagged ubiquitin (FLAG-Ub; Boston Biochem), 200 ng of GST-ABD1 or GST, and 0.5 μg of MBP-COP1 (incubated with 20 μM zinc acetate) or bMBP-COP1 (MBP-COP1 denatured by boiling for 10 min at 95°C before addition to the ubiquitination reaction system) in reaction buffer (50-mM Tris, pH 7.5, 10-mM MgCl2, 10-mM ATP). After 2 h of incubation at 30°C, the reactions were stopped by 5× SDS loading buffer. Thirty microliters of each mixture were then separated on 8% (w/v) SDS–PAGE gels, and the ubiquitinated GST-ABD1 proteins were detected using anti-GST (1:2,000 [v/v], catalog no. G7781; Sigma-Aldrich) and anti-FLAG antibodies (1:1,000 [v/v], catalog no. M20008H; Abmart).
In vivo ubiquitination assays were performed as described previously (Wang et al., 2019a) with minor modifications. Briefly, the indicated combinations of 35S:HF-COP1, Super:ABD1-GFP and 35S:HA-UBQ (Wang et al., 2018a) plasmids were cotransfected into Arabidopsis (Col-0) protoplasts and incubated for 18 h with 5-μM MG132 in the presence or absence of 5-μM ABA. Total proteins were then extracted with protein extraction buffer consisting of 50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 10-mM MgCl2, 1-mM EDTA, 0.1% (v/v) NP-40, 1-mM PMSF, 1× MG132, and 1× complete protease inhibitor cocktail (Roche), and centrifuged twice for 15 min at 12,000g at 4°C. Of the 1 mL of supernatant for each sample, 100 μL was set aside as total, and the remainder was incubated with GFP-Trap beads (ChromoTek) in the presence of 50-μM MG132. After 2 h of incubation at 4°C, the beads were washed four times (10 min each time) with protein extraction buffer, and the immunoprecipitated proteins were eluted in 2× SDS loading buffer at 100°C for 15 min, and analyzed by immunoblotting.
Cell-free protein degradation assays
Cell-free protein degradation assays were performed as described previously (Kong et al., 2015). Briefly, total proteins were extracted from 4-day-old dark-grown wild-type (Col-0) and cop1-4 mutant seedlings with native protein extraction buffer (50-mM Tris–MES, pH 8.0, 0.5-M sucrose, 1-mM MgCl2, 10-mM EDTA, pH 8.0, 5-mM DTT). Then, 200 ng of recombinant His-ABD1 protein was incubated with equal amounts (500 μg) of total proteins extracted from Col-0 or cop1-4 seedlings, at 25°C for the indicated times. ATP (at a final concentration of 10 mM) was added as previously described. The reactions were stopped by 6× SDS loading buffer, and the His-ABD1 proteins were detected with the anti-His antibodies (1:1,000 [v/v], catalog no. H1029; Sigma-Aldrich).
Identification of ubiquitination sites
Cell-free assays using recombinant GST-ABD1 were performed as described previously (Yu and Xie, 2019). Briefly, total proteins were extracted from 4-day-old dark-grown wild-type (Col-0) seedlings with native protein extraction buffer (50-mM Tris–MES, pH 8.0, 0.5-M sucrose, 1-mM MgCl2, 10-mM EDTA, pH 8.0, 5-mM DTT, 1-mM PMSF, 1× complete protease inhibitor cocktail [Roche], 0.2% [v/v] NP40). GST beads (bound with ∼200 ng of GST-ABD1 protein) were added to 200 μL plant extracts (containing ∼500 μg of total proteins) and incubated for 30 min at room temperature in the presence of 50-μM MG132, and then incubated at 4°C for 4 h. GST beads were then washed six times (10 min each time) with modified GST-binding buffer (50-mM Tris–HCl, pH 8.0, 270-mM NaCl, 0.5% [v/v] NP40, 1-mM DTT, and 1-mM PMSF). Then, the immunoprecipitated proteins were eluted in 2× SDS loading buffer at 100°C for 5 min, and separated on an SDS–PAGE gel. The gel sample containing the ubiquitinated target proteins was excised, digested in-gel, and subjected to LC–MS/MS analysis.
Quantification and statistical analyses
Protein quantification was performed with ImageJ. Analyses of variance (ANOVAs) were performed with SPSS statistical software, and Student’s t tests were performed in Microsoft Excel. Different letters represent statistical significances, as determined by ANOVA (P < 0.05) for multiple comparisons, and levels that are not significantly different are indicated with the same letter.
Accession numbers
Sequence data from this article can be found in the Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: COP1 (At2g32950), ABD1 (At4g38480), ABI5 (At2g36270), KEG (At5g13530), DWA1 (At2g19430), DWA2 (At1g76260), PYR1 (At4g17870), and OST1 (At4g33950).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Specificity of anti-PYR1 and anti-OST1 antibodies.
Supplemental Figure S2. ABA promotes ABI1/ABI2 protein accumulation in darkness by rapidly inducing the expression of their encoding genes.
Supplemental Figure S3. RT-qPCR assays showing that COP1 and ABI5 mediate ABA-induced ABI1/ABI2 expression in darkness.
Supplemental Figure S4. Immunoblots showing the levels of ABF1 and ABF4 proteins in dark-grown Col-0 and cop1 mutant seedlings in response to ABA treatment.
Supplemental Figure S5. The cop1-4 abi5-8 double mutant is hyposensitive to ABA in the dark.
Supplemental Figure S6. Immunoblots showing the levels of transgenic or endogenous ABI5 proteins in various seedlings.
Supplemental Figure S7. RT-qPCR assays showing that COP1 mediates ABA-induced ABI5 expression in darkness.
Supplemental Figure S8. COP1 promotes the stability of endogenous ABI5 proteins in vivo.
Supplemental Figure S9. ABI5 does not directly interact with COP1 in yeast cells or in vivo.
Supplemental Figure S10. Specificity of anti-ABD1 antibodies.
Supplemental Figure S11. Genotyping of cop1-4 abd1-1 mutant.
Supplemental Figure S12. COP1 transcript and COP1 protein levels are not significantly regulated by ABA.
Supplemental Figure S13. Wild-type seedlings are more resistant to ABA in the light than in the dark.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set S1. Statistical analyses.
Supplementary Material
Acknowledgments
We thank Jae-Hoon Lee and Kyoung-In Seo (Pusan National University, Busan, Korea) for abd1-1 and 35S:ABD1-MYC, Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for 35S:ABI5-MYC, Haodong Chen (Tsinghua University and Tsinghua-Peking Center for Life Sciences, Beijing, China) for cop1-5 35S:YFP-COP1, Tonglin Mao (China Agricultural University, Beijing, China) for abi5-1, and Pedro L. Rodriguez (Consejo Superior de Investigaciones Cientificas-Universidad Politecnica de Valencia, Valencia, Spain) for pyr1 pyl124 and pyr1 pyl1458 seeds, and are grateful to Sourav Datta (Indian Institute of Science Education and Research, Bhopal, India) for helpful discussions.
Funding
This work was supported by grants from the Beijing Outstanding Young Scientist Program (BJJWZYJH01201910019026) to F.Q., the National Natural Science Foundation of China (31970262 and 31770321 to J.L., and 32000187 to C.L.), China National Postdoctoral Program for Innovative Talents (BX20200371 to L.Q.), China Postdoctoral Science Foundation (2020M670531 to C.L. and 2021M693432 to L.Q.), and Beijing Outstanding University Discipline Program.
Conflict of interest statement. The authors declare no conflict of interests.
Contributor Information
Jing Peng, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Meijiao Wang, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiaoji Wang, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Lijuan Qi, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Can Guo, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Hong Li, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Cong Li, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Yan Yan, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Yun Zhou, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, Henan University, Kaifeng 475004, China.
William Terzaghi, Department of Biology, Wilkes University, Wilkes-Barre, Pennsylvania 18766, USA.
Zhen Li, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Chun-Peng Song, State Key Laboratory of Crop Stress Adaptation and Improvement, Collaborative Innovation Center of Crop Stress Biology, Henan University, Kaifeng 475004, China.
Feng Qin, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Zhizhong Gong, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jigang Li, State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
J.L. and J.P. designed the research; J.P., M.W., X.W., L.Q., C.G., H.L., C.L., Y.Y., and Z.L. performed the research; J.L., Z.G., F.Q., C.-P.S. and Y.Z. analyzed the data; and J.L., J.P., and W.T. wrote the paper.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Jigang Li (jigangli@cau.edu.cn).
References
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. (2013) PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol 161: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E, et al. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020) Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62: 25–54 [DOI] [PubMed] [Google Scholar]
- Chen Q, Bai L, Wang W, Shi H, Ramón Botella J, Zhan Q, Liu K, Yang HQ, Song CP (2021) COP1 promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases. New Phytol 229: 2035–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W., Sun J, et al. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Kathare PK, Paik I, Huq E (2021) Phytochrome signaling networks. Annu Rev Plant Biol 72: 217–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M (2008) The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH (1991) cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Dong X, Yan Y, Jiang B, Shi Y, Jia Y, Cheng J, Shi Y, Kang J, Li H, Zhang D, et al. (2020). The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J 39: e103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Estojak J, Brent R, Golemis EA (1995) Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol 15: 5820–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (2013) Abscisic acid synthesis and response. Arabidopsis Book 11: e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C, Wang X, Feng J, Hong S, Liang Y, Ren B, Zuo J (2014) Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of abscisic acid insensitive 5 protein in Arabidopsis. Plant Physiol 164: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Huang X, Deng XW (2020) The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun 1: 100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol 21: 96–103 [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, Deng XW (2013) Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA 110: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, et al. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6: 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qi L, Zhang S, Dong X, Jing Y, Cheng J, Feng Z, Peng J, Li H, Zhou Y, et al. (2022) Mutual upregulation of HY5 and TZP in mediating phytochrome A signaling. Plant Cell 34: 633–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wu Y, Xie Q, Gong Z (2017) Abscisic acid. In Li J, Li C, Smith SM, eds, Hormone Metabolism and Signaling in Plants. Academic Press, Cambridge, MA, pp. 161–202 [Google Scholar]
- Li J, Li G, Wang H, Deng XW (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, et al. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian N, Liu X, Wang X, Zhou Y, Li H, Li J, Mao T (2017) COP1 mediates dark-specific degradation of microtubule-associated protein WDL3 in regulating Arabidopsis hypocotyl elongation. Proc Natl Acad Sci USA 114: 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Jiang Y, Li J, Yan T, Fan L, Liang J, Chen ZJ, Xu D, Deng XW (2018) B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30: 2006–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JJ, Li J, Zhu D, Deng XW (2017) Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci USA 114: 3539–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stone SL (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stone SL (2013) Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J Biol Chem 288: 20267–20279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017) Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66: 117–128 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S, et al. (2020). Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225: 848–865 [DOI] [PubMed] [Google Scholar]
- Marine JC (2012) Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer 12: 455–464 [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Deng XW (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ (2014) Rapid decline in nuclear CONSTITUTIVE PHOTOMORPHOGENESIS1 abundance anticipates the stabilization of its target ELONGATED HYPOCOTYL5 in the light. Plant Physiol 164: 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Xu X, Huq E (2018) Molecular bases for the constitutive photomorphogenic phenotypes in Arabidopsis. Development 145: dev.169870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Ulm R (2018) Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol 45: 18–25 [DOI] [PubMed] [Google Scholar]
- Qi L, Liu S, Li C, Fu J, Jing Y, Cheng J, Li H, Zhang D, Wang X, Dong X, et al. (2020) PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol Plant 13: 414–430 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Pasoreck EK, Reddy AK, Nagatani A, Ma W, Chory J, Chen M (2017) Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat Commun 8: 1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Zhu D, Li J, Rubio V, Zhou Z, Shen Y, Hoecker U, Wang H, Deng XW (2008) Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Seo KI, Lee JH, Nezames CD, Zhong S, Song E, Byun MO, Deng XW (2014) ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 26: 695–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Yan T, Liu J, Bian Y, Heng Y, Lin F, Jiang Y, Deng XW, Xu D (2020) BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J 104: 377–390 [DOI] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. (2018a) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69: 493–504 [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin C (2020) Mechanisms of cryptochrome-mediated photoresponses in Plants. Annu Rev Plant Biol 71: 103–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu X, Li L, Lian H, Mao Z, Xu P, Guo T, Xu F, Du S, Cao X, et al. (2018b). Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ding Y, Li Z, Shi Y, Wang J, Hua J, Gong Z, Zhou JM, Yang S (2019a) PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev Cell 51: 222–235 [DOI] [PubMed] [Google Scholar]
- Wang X, Guo C, Peng J, Li C, Wan F, Zhang S, Zhou Y, Yan Y, Qi L, Sun K, et al. (2019b). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol 221: 341–355 [DOI] [PubMed] [Google Scholar]
- Wu J, Wang W, Xu P, Pan J, Zhang T, Li Y, Li G, Yang H, Lian H (2019) phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol 60: 353–366 [DOI] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M (2014) Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet. 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11: 523–541 [DOI] [PubMed] [Google Scholar]
- Xu P, Chen H, Li T, Xu F, Mao Z, Cao X, Miao L, Du S, Hua J, Zhao J, et al. (2021). Blue light-dependent interactions of CRY1 with GID1 and DELLA proteins regulate gibberellin signaling and photomorphogenesis in Arabidopsis. Plant Cell 33: 2375–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadukrishnan P, Rahul PV, Ravindran N, Bursch K, Johansson H, Datta S (2020) CONSTITUTIVELY PHOTOMORPHOGENIC1 promotes ABA-mediated inhibition of post-germination seedling establishment. Plant J 103: 481–496 [DOI] [PubMed] [Google Scholar]
- Yan B, Yang Z, He G, Jing Y, Dong H, Ju L, Zhang Y, Zhu Y, Zhou Y, Sun J (2021) The blue light receptor CRY1 interacts with GID1 and DELLA proteins to repress gibberellin signaling and plant growth. Plant Commun 2: 100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Li C, Dong X, Li H, Zhang D, Zhou Y, Jiang B, Peng J, Qin X, Cheng J, et al. (2020) MYB30 is key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32: 2196–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L (2018) Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell 44: 29–41.e4. [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1-from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yu F, Xie Q (2019) Approaches to identify protein ubiquitination sites in plants. Methods Mol Biol 2026: 85–93 [DOI] [PubMed] [Google Scholar]
- Zhang S, Li C, Zhou Y, Wang X, Li H, Feng Z, Chen H, Qin G, Jin D, Terzaghi W, et al. (2018). TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell 30: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Dou L, Gong Z, Wang X, Mao T (2019) BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol 221: 908–918 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Zeng B, Tang D, Yang J, Qu L, Yan J, Wang X, Li X, Liu X, Zhao X (2021) The blue light receptor CRY1 interacts with GID1 and DELLA proteins to repress GA signaling during photomorphogenesis in Arabidopsis. Mol Plant 14: 1328–1342 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang L, Duan J, Cheng J, Shen Y, Wang X, Han R, Li H, Li Z, Wang L, et al. (2018) Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc Natl Acad Sci USA 115: e11864–e11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.