Abstract
The inheritance of neurophysiologic and neuropsychologic complex diseases can only partly be explained by the Mendelian concept of genetic inheritance. Previous research showed that both psychological disorders like post-traumatic stress disorder and metabolic diseases are more prevalent in the progeny of affected parents. This could suggest an epigenetic mode of transmission. Human studies give first insight into the scope of intergenerational influence of stressors but are limited in exploring the underlying mechanisms. Animal models have elucidated the mechanistic underpinnings of epigenetic transmission. In this review, we summarize progress on the mechanisms of paternal intergenerational transmission by means of sperm RNA in mouse models. We discuss relevant details for the modelling of RNA-mediated transmission, point towards currently unanswered questions and propose experimental considerations for tackling these questions.
Keywords: sperm, epigenetic inheritance, RNA
Intergenerational Effects at Odds with Classic Heredity
The Mendelian concept of genetic inheritance can only partly explain the inheritance of complex multifactorial neurophysiologic and neuropsychologic diseases. Pioneering research showed that post-traumatic stress disorder (PTSD) had a higher prevalence in offspring of parents who were Holocaust victims with PTSD independently of the known genetic predispositions [1, 2]. Similarly, the Dutch Famine study and its successors showed multiple metabolic diseases being prevalent in offspring and grand-offspring of mothers starving during the period of the Dutch Famine [3–5]. This suggests an impact of the environment psychologically and physiologically, not only limited on the generation directly exposed but also on their descendants, and thus a potential epigenetic constituent in the inheritance of complex diseases. The human studies mentioned above allude to an influence of environmental stressors across generations. Due to their descriptive nature and the complexity of the human environment that has to account for the socio-economic effects on the individual, they are limited in examining the mechanisms of epigenetic transmission. Animal studies allow a mechanistic interrogation since they can strictly control the environment throughout the entire life span of the individuals at stake. They can exclude genetic confounds by using inbred strains. The influence of psychological or metabolic stress has been previously studied extensively using rodent studies, building a foundation for research on the epigenetic inheritance of stress.
A variety of stressors can be effectively modelled by psychological or metabolic challenges in the environment. Exposures that have been used to model psychological and metabolic stress include acute social defeat, predators, immobilization, foot shocks or acute food restriction [6]. In addition, stress can be induced by pharmaceutical interventions like injections or inhalation of GABA antagonists, opioids, inflammation-promoting agents, components of the HPA axis and stress response like CRH or ACTH and their mimics, as nicely summarized in Patchevs’ review [7]. While corticosterone-mediated stress typically boosts physical performance [8], it can also have negative impacts [9–11], especially when chronic [12–14]. Not only does chronic stress disrupt specific psychological and metabolic functions in the subject experiencing it, but also in its offspring. Long-lasting alterations in gene expression and protein abundance caused by chronic stress have been found to be also transmitted to the progeny, resulting in impaired stress resilience as extensively described by Safi-Stibler and Gabory [15].
While effects of chronic stress on the offspring have been investigated extensively, researchers just recently started to look into the possible effects of acute stress not only on the individual but also on the offspring. In mice, males receiving a foot shock were mated to non-exposed females a few weeks later. The resulting offspring displayed altered body weight and glucose metabolism [16]. These results were confirmed to rely on germline-transmission by artificial insemination [17]. Acute paternal glucocorticoid receptor challenge influenced offspring molecular profile and altered glucose metabolism in mice [18]. In rats, males exposed to predator odours influenced not only maternal investment, affecting licking, grooming, retrieval and feeding, but also led to the development of anxiety-like behaviour in offspring [19]. In tree swallows (Tachycineta bicolor), brief corticosterone treatment simulating acute stress responses led to progeny being smaller [20].
Using both chronically and acutely stressed animal models, several mechanisms potentially underlying epigenetic transmission have been investigated. DNA methylation, histone modifications and RNA in the germline have primarily been explored as targets for parental signals as they have been proven malleable by the environment and experiences [21, 22]. The concept of epigenetic germline inheritance or meiotic epigenetic inheritance determines that the transmission of altered regulating epigenetic marks like DNA methylation, histone modifications and RNAs occurs through gametes, from one generation to their offspring [23]. Noteworthy, once induced, those altered regulators need to maintain their altered state in the gametes in order to deliver information about environment and experiences to the offspring. DNA methylation and histone modifications undergo epigenetic reprogramming during gametogenesis and in the pre-implantation embryo [24].
RNAs in the male germline are exempt from further reprogramming, making them an interesting target to investigate the mechanism of intergenerational epigenetic transmission from parental generation to their direct offspring. Obviously, intergenerational transmission is not limited to father-offspring effects but occurs conceivably even to a bigger extent between mother and progeny. This is because the oocyte contributes a larger amount of RNA to the embryo [25] and gestational signalling holds great potential to convey additional information to the growing embryo. Post-gestational maternal care, comprising not only nutrition but also licking and grooming, further represents a layer of potential influence on offspring health [26]. This richness of potential effectors of offspring health together with the very limited number of oocytes available per mouse complicates the study of female line effects. Regardless, while not the focus of our review, extensive research has also been conducted on epigenetic modifications and their function in epigenetic heredity in the oocyte, which is reviewed elsewhere [27, 28].
In cases where effects trespass not only the parent–offspring generation, but are further perpetuated to the offspring, they are often labelled as transgenerational, especially when the affected individuals are born from gametes that were not directly exposed to an environmental insult.
Here, we will summarize findings on intergenerational transmission by means of sperm RNA, pointing out relevant subgroups of sperm RNA, (i) small non-coding RNAs, including (a) microRNAs (miRNAs), (b) transfer RNA (tRNA)-derived RNA fragments (tRFs) and (c) P-element Induced WImpy testis (PIWI)-interacting RNAs, and (ii) long linear RNAs and (iii) circular RNAs (circRNAs) (Fig. 1). We then explore key questions with regard to RNA-mediated inheritance. Finally, we suggest experimental considerations when testing these questions.
Figure 1:
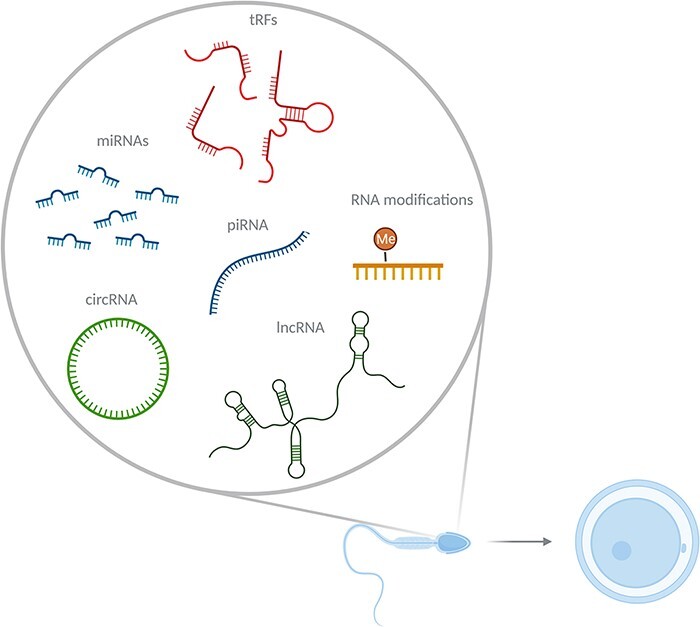
Sperm delivers a host of RNA to the oocyte during fertilization
Sperm RNA as a Vertical Information Vector
Historically, the main experimental approach to causally test the effects of altered sperm RNAs on the offspring was sperm RNA injection into non-exposed fertilized oocytes [21] and examining offspring phenotypes in comparison to controls. In this crucial method, a superovulated female is mated with a wild-type male, and fertilized oocytes are extracted. At the 1-cell stage, when still both pronuclei are visible, RNA isolated from sperm of experimental or control males is microinjected into the male pronucleus. The embryos are then implanted into and fostered by pseudopregnant females to generate offspring. This offspring generated can then be investigated for its phenotype (Fig. 2). While it is a fundamental experiment in the field, the protocols used by different groups vary slightly, which can make direct comparisons between results difficult. Most groups inject RNA directly into the male pronucleus [29], but RNA can also be injected into the cytoplasm, and this is sometimes not specified [30, 31]. Furthermore, the amounts of RNA injected vary between labs, usually between 1 and 2 pl of a 0.5 [29, 32], 1 [31, 33], 2 [32, 34, 35] and 10  g/ml [30] RNA solution. When injecting total RNA, RNA sizes might vary based on whether groups used TRIzol or column-based methods to purify RNA. Lastly, injections of synthesized RNA will by default lack potentially important RNA modifications [32, 34].
g/ml [30] RNA solution. When injecting total RNA, RNA sizes might vary based on whether groups used TRIzol or column-based methods to purify RNA. Lastly, injections of synthesized RNA will by default lack potentially important RNA modifications [32, 34].
Figure 2:
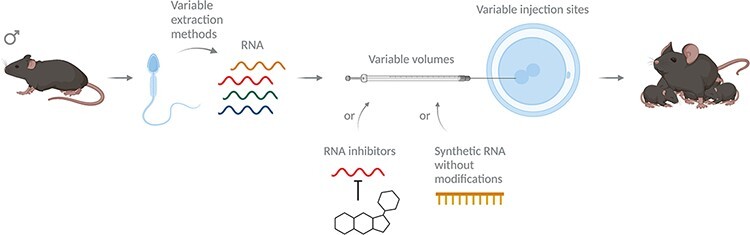
Methodological considerations of RNA injections into fertilized oocytes
Interestingly, the first work exploring sperm RNA-mediated inheritance as an epigenetic mechanism using this method reaches back as far as 2006. In this study by the group of Minoo Rassoulzadegan, the offspring of animals heterozygous for a mutation in the Kit gene displayed white tail tips similar to their parents despite being of wild-type genotype, which could be traced back to altered Kit RNA levels and sizes in parental testis. When injecting total sperm or testis RNA of Kit mutant mice into non-mutant zygotes, the offspring displayed similar white tail tips as the mutant [30]. This provided the first evidence for a mechanistic involvement of sperm RNA in epigenetic germline inheritance. Since then, studies using that injection method and studies showing that specific sperm RNA species are delivered to the oocyte [36, 37], laid the foundation for a sperm to oocyte RNA signalling system to be postulated. Similar to the histone code, the makeup of the sperm RNA signal is suggested to determine the health of the offspring [38]. Sperm RNA differs substantially in its composition from RNA found in somatic cells and can be affected qualitatively or quantitatively by environmental influences [39–41]. How the sperm RNA signal is regulated by the environment in the first place and delivered to the offspring, and what challenges these questions are facing are discussed elsewhere in detail [22, 38, 42]. This review focuses on the findings on the specific subtypes of RNAs making up the sperm RNA signal in mice, small non-coding RNAs and their subtypes and long non-coding RNAs (lncRNAs), how these could influence the offspring and how to test their involvement in the transmission of phenotypes in a further refined way.
Small Non-coding RNAs
MicroRNAs
miRNAs are small non-coding RNAs, 21–25 nucleotides (nt) in size, which are involved in gene expression regulation in somatic cells of non-vertebrae and vertebrae. By engaging the RNA-induced silencing complex and the three prime UTR of mRNA, they lead to inhibition or degradation of mRNA [43]. miRNAs have been reported to direct DNA methylation in plants [44], yet there is no evidence for this in mammals yet. In sperm, it might seem surprising to find miRNAs given their transcriptionally rather silent state and low amounts of mRNAs.
Two controversial studies reported the necessity of sperm miRNAs for early embryonic development [45, 46]. In the context of environmental exposures, Rodgers et al. reported for the first time that males receiving 42 days of chronic variable stress throughout puberty or adulthood altered sperm small RNA composition measured with quantitative real-time polymerase chain reaction (qRT-PCR), where expression levels of nine miRNAs were altered [40].
Soon thereafter Gapp et al. [29] showed that total RNA purified from mouse sperm of fathers exposed to early life stress in form of unpredictable maternal separation and maternal stress (MSUS) in the first 2 weeks of life persistently affected behaviour and metabolism in offspring generated from RNA injections into fertilized non-exposed oocytes. Using RNA-seq, said RNA showed alterations in a payload of 43 miRNAs in sperm, 5 of which were confirmed by quantitative PCR (qPCR) in sperm, brain and serum [29]. One of the confirmed miRNAs, miR 375, was also found to be altered in a study using a chronic stress model by Rodgers et al. [40]. Gapp and colleagues concluded that the altered sperm miRNA effectively transmitted the phenotype to the offspring [29]. Around the same time, injection of nine miRNAs previously identified to be elevated in sperm of mice exposed to chronic variable stress [40] into the zygote led to dysregulated stress responses in the resulting offspring, with reduced corticosterone response and reduced gene expression levels of a set of genes important for the response HPA axis [47].
A relation between sperm miRNA and transmission of phenotype in the offspring could not only be made for traumatic stress-induced behavioural changes, but also for dietary stress–induced metabolic changes. High-fat diet is commonly used to introduce metabolic stress. In comparison to standard chow, animals are fed a diet with significantly higher digestible energy by providing butterfat in the diet. Grandjean et al. used such a high-fat diet, feeding it to males 3 weeks of age for 4 months. At an age of 16 weeks, 13 miRNAs, as of RNA-seq, were elevated in sperm and testis in animals displaying the metabolic phenotype, with higher body weight and blood glucose levels. They could confirm five of those miRNAs by qRT-PCR, none of which overlapped with the miRNAs altered in the studies of Rodgers et al. [40] and Gapp et al. [29]. Microinjection of those miRNAs into a wild-type zygote led to similar metabolic alterations in the resulting offspring [31]. It had previously been shown that a high-fat diet, using same amounts of fat as Grandjean and colleagues, caused alterations in testes and sperm miRNA payload in microarray, confirming with qPCR 11 miRNAs of those to be altered in testes, 4 of which in sperm. Progeny sperm RNA content was not examined, although F1 and F2 progeny showed the paternal obese phenotype and insulin resistance while being fed a standard diet [39]. One of the paternal miRNAs demonstrated to be altered, miRNA 340-5p, has been reported to be altered in the high-fat study by Grandjean and colleagues [31].
Further confirming a causal relation between sperm miRNA and epigenetic transmission to the offspring, a study showed that the effects of transmitted miRNA could be reversed with a specific miRNA inhibitor. Adult male mice were exposed to environmental enrichment, enhancing their synaptic plasticity and cognition tested for contextual fear-conditioning and in the Morris water maze paradigm. Their sperm displayed altered levels of miR212 and miR132 as of qPCR, none of which overlapped with the studies by Rodgers et al. [40], Fullston et al. [39], Gapp et al. [29] or Grandjean et al. [31]. The phenotype was also apparent in their offspring generated with natural breeding and could be counteracted by co-injection of an inhibitor to miR212/132 [48]. While the transmission of beneficial effects of enriched environment to the offspring had already been suggested before [49, 50], this was the first time this could be conclusively related to alterations in miRNAs in sperm.
Finally, the contribution of miRNAs in transmitting effects of chronic stress to the offspring has been corroborated by Wang et al. [51] in a study that blocked the transmission of the stress-induced phenotype by injecting a pool of miRNA inhibitors. Males aged 8 weeks were exposed to unpredictable mild stresses daily for 5 weeks, resulting in a depression-like model as tested with a forced swim test and a sugar preference test. RNA-seq and qPCR revealed altered levels for 17 miRNAs in their sperm. Directly after the stress paradigm, sperm small RNA was isolated and injected into non-exposed zygotes to generate offspring. At 2 months of age, the offspring was susceptible to depression-like symptoms but did not display altered small RNA levels in sperm. Injecting zygotes that had been generated with sperm of males that underwent the unpredictable mild stress with antisense strands to neutralize miRNA rescued the phenotype [51]. Thereby, the role of miRNAs in transmitting the effects of chronic stress can now be judged as fairly established.
While the above-mentioned studies all point towards a classical miRNA-induced regulation of mRNA targets in the early embryo, it cannot be excluded that miRNA changes in sperm primarily affect mRNAs in sperm cells, a class that will be discussed further down.
tRNA-Derived RNA Fragments
Importantly, the analysis of total sperm RNA showed that miRNAs do not constitute the largest portion of small non-coding RNAs, but instead tRFs [52]. TRFs in sperm are derived from the 5ʹ and 3ʹ ends of tRNAs and are distinctly bigger in size than miRNAs with 29–34 nt. In somatic mammalian cells, tRFs are involved with Argonaute proteins to promote cleavage of sequence matched targets [53]. Sperm tRFs are proposed to regulate genes linked to retroelements in the early embryo [32]. Sharma et al. [32] measured the abundance of tRFs in epididymal sperm with small RNA-seq, five of which were upregulated in adult males that were fed a low protein diet consisting of 10% instead of 19% protein. Their in vitro fertilization-derived offspring displayed upregulated expression for biosynthesis-related genes. They found tRFs in paternal testes not to be affected, yet epididymal tissue also displayed the same changes in tRF population, from which they concluded that altered tRFs in sperm were not derived during spermatogenesis. Based on coincubation experiments of sperm cells and epidydimal epithelial exosomes followed by comparative small RNA sequencing, they reasoned that at least a large proportion of them are provided to spermatozoa by epididymal epithelial cells via extracellular vesicles during epididymal transit from caput to cauda. They further provided evidence for tRFs functioning in sperm as regulators for genes linked to the endogenous retroelement MERVL, a murine endogenous retrovirus [32].
Corroborating tRF uptake during epididymal transit, Gapp et al. [18] demonstrated that one tRF was unaltered in the caput epididymis of adult males 3 h after injection of dexamethasone, a glucocorticoid receptor agonist, yet altered in cauda epididymis using a combination of small RNA-seq and qRT-PCR. They further proposed that upregulation of tRF Arg-CCT-2 in cauda epididymal sperm was potentially derived from serum-circulating exosomes, which also showed tRF Arg-CCT-2 upregulation [18].
Also using a high-fat diet mouse model like Grandjean et al. [31] for their study on miRNAs, Chen et al. showed even earlier that such a diet in male mice altered tRFs abundance and RNA methylation levels (see in the section on RNA modifications). When injecting total sperm RNA or only sperm tRFs from males fed this diet into a wild-type zygote, the resulting offspring developed metabolic disorders similar to their fathers while injection of miRNA or long RNA fractions resulted in no effect [34]. This study by Chen and colleagues together with the one by Sharma et al. [32] provided the first causal evidence of sperm tRFs transmitting acquired information to the offspring.
Furthermore, a study by Cropley et al. described altered sperm RNA—especially tRFs—in F1 offspring of prediabetic males. When mated with wild-type females, the resulting F2 offspring showed similar changes in glucose metabolism as their F1 fathers with the phenotype only wearing off in the F3 generation [54]. Another study confirmed the role of tRFs in epigenetic transmission, by injecting total RNA or tRFs, but not larger RNAs (sized 40–90 nt) isolated from sperm of males with a high-fat diet-induced hedonic and metabolic phenotype into fertilized egg cells with the resulting progeny displaying the same phenotype. Based on their results together with the findings of Sharma et al. [32], they speculated that tRFs transmit acquired information via the regulation of MERVL elements in the early embryo [32, 35].
Overall, the above studies clearly demonstrate the responsiveness of tRFs to environmental, in their majority dietary, challenges and their involvement in the transmission of non-genetically transmitted phenotypes.
PIWI-Interacting RNAs
Besides alterations in miRNA, Gapp et al. [29] also described changes in one cluster of PIWI-interacting RNAs (piRNA) in sperm of males exposed to early life stress in form of MSUS in the first 2 weeks of life. piRNAs are small non-coding RNA (26–31 nt) involved in the regulation of gene expression by interacting with the PIWI subfamily of Argonaute proteins [55]. In the brain, they facilitate the methylation of promoter regions to enhance synaptic plasticity [56]. In the germline, prepachytene piRNAs are similarly involved in the methylation of genomic sequences harbouring transposable elements to stabilize the germline genome [55], whereas pachytene piRNAs and post-meiotic spermatid piRNAs contribute to post-translational mRNA cleavage [57, 58].
A concomitant change in piRNA and tRFs requires consideration with interpreting RNA injection results, since sperm RNA injections using size selected fractions of total sperm RNA cannot distinguish piRNAs from tRFs. Hence, Gapp et al. [29] could not exclude a contribution of piRNA alterations to the transmission of effects of MSUS to the resulting offspring [29]. While Sharma et al. and Chen et al. did not comment on piRNA changes [32, 34], Grandjean et al. also noted changes in piRNA expression in their study [31]. Males that were fed a high-fat diet had elevated piRNA levels in their spermatozoa for 63 different piRNA clusters [31]. While environmental influences seem to regulate piRNA payload, the role of piRNAs in epigenetic germline inheritance remains to be elucidated. piRNAs are crucial to chromatin remodelling [59] and retrotransposon silencing [60]. A recent study demonstrated that fertilization of wild-type oocytes with sperm harbouring a homozygous deletion of a specific piRNA cluster on chromosome 6 resulted in abnormal heterozygous embryos with reduced embryonic survival [61]. This was attributed to regulatory functions of piRNAs on mRNAs prior fertilization but could well have also a component relevant post-fertilization. On the other hand, a study by Yuan et al. found that paternal pachytene piRNAs are not required for persistent fertility in mice lacking MIWI in round spermatids [62]. In non-vertebrae, piRNAs have been clearly involved in epigenetic inheritance as is discussed elsewhere [22].
Convincing proof for the involvement of piRNAs in epigenetic inheritance needs yet to be established in mammalian systems.
Long Linear RNAs
In contrast to the small RNA species discussed so far, we define long linear RNAs as RNAs >200 nt. It comprises both coding mRNAs as well as lncRNA. The latter can be subclassified as sense, antisense, bidirectional, intronic, and intergenic lncRNAs. They are involved in a host of functions of gene regulation concerning chromatin structure and factor recruitment in somatic cells, as summarized elsewhere [63]. However, they are equally involved in regulating gene expression in sperm where they modulate spermatogenesis [64]. As described above, studies on the effect of diet-induced transmission, using a high-fat diet, were unable to phenocopy effects in the offspring by sperm lncRNA injections [34, 35]. Whether the long RNA fraction was affected in those models had not been assessed. Different so for a model of early trauma, consisting of exposing males to maternal separation and unpredictable maternal stress in the first two weeks of life: these males displayed considerable changes in their sperm long RNA payload, assessed by next-generation sequencing, including alterations in transposable element abundance [65]. Although not specifically investigated, this could be related to changes in sperm piRNAs observed in the same model. Typical behavioural changes in increased risk-taking and behavioural despair and metabolic changes in glucose response were partially mimicked by injection of sperm long RNA fraction. Interestingly, injection of small RNA fraction was sufficient to copy increased behavioural despair. Gapp and colleagues interpreted that both lncRNAs and small RNAs were necessary to transmit all behavioural and metabolic alterations to the offspring yet concluded that sperm long RNAs are functional [65].
A recent study addressed a long-standing controversy around sperm long RNA, that is whether cloned mRNA fragments might represent remnants and would not reflect full-length functional RNAs. Previous analysis used bioinformatic tools to establish whether the read fragments at least generally cover the full-length RNAs in question. A new study uses single-molecule long-read sequencing to unambiguously determine the presence of intact RNA molecules in mature sperm [66]. Such sequencing will in the future certainly prove useful also in the context of the assessment of changes following an exposure.
In conclusion, surprisingly little research has been conducted on environmental effects on long linear RNAs in sperm despite their potential of transposable elements to translate into genomic changes and thereby inducing long-term more stable consequences. It will be interesting to see whether this will change given the new technical possibilities to determine intact long RNA and the growing certainty that those are indeed present in mature sperm.
Circular RNAs
To add yet another interesting class of sperm RNAs to the long list of mobile molecules, Gapp and colleagues recently reported for the first time changes in circRNA in response to a strong stress mimic in the form of a dexamethasone injection [18]. circRNAs are circular long RNAs, 500–4000 nt in size, that are produced by a form of alternative splicing called back splicing. During this event, the 5ʹ and 3ʹ end of a pre-mRNA are covalently linked. They are particularly interesting since they are highly stable in comparison to linear counterparts [67, 68] and could thus be protected during sperm to oocyte delivery and affect offspring embryonic phenotype. Many regulatory functions have been described for circRNAs in somatic cells, ranging from regulation of pluripotency and differentiation to control of proliferation [69]. CircRNAs have been demonstrated to be very highly expressed in testis [70]. In the context of epigenetic inheritance, three specific mechanistic properties stand out. First, circRNAs have been shown to be stored in the maturing germline when transcription ceases as templates for the translation of peptides during later stages of spermatogenesis [71]. Upon delivery to the oocyte, they could also potentially engage in peptide translation. Second, one very hotly debated role concerns the sponging of miRNAs. Such was shown conclusively for Circ SRY that has 16 mir138 binding sites [72]. Hence, if transmitted to the oocyte, circRNAs could act as miRNA sponges, thereby regulating targets of miRNAs in the offspring embryo and amplifying effects on gene expression. In the study by Gapp et al. [18] data from single 2-cell embryo expression indeed point towards such regulation [18]. Sat1, a mRNA target of the miRNA that has predicted binding sites on both circRNAs, that are upregulated in sperm, was shown to be upregulated in the 2-cell embryos, suggesting a sponging of the regulatory miRNA. Lastly, circRNAs were suggested to function as protein decoys, which prevent or promote the interaction of proteins with binding partners [73] and may support or repress their mobility between cytoplasm and nucleus [74]. This last mechanism could influence epigenetically transmitted proteins in their function or target proteins translated in the early embryo.
Overall, circRNAs show great potential to amplify intergenerational signals in the early embryo and are therefore a target worthwhile considering.
RNA Modifications
The transmission of diet-induced effects was suggested to be reliant on the changes in RNA modifications [75], building on earlier results from the group of Minoo Rassoulzadegan. Her results had shown that a knockout of the tRNA methyltransferase Dnmt2, which engages tRFs as its substrate, prevented the Kit mutant phenotype from being transmitted to the offspring. Microinjection of sperm RNA from Dnmt2−/− males into non-mutant zygotes did not result in the Kit mutant phenotype in the resulting progeny [33]. Chen’s findings corroborated this, in that a deletion in Dnmt2 prevented the transmission of a high-fat diet-induced metabolic phenotype by sperm RNA and was associated with altered sperm RNA expression, especially tRFs, and prohibited the rise in RNA modifications they observed in high-fat diet mice. They concluded that those modifications and altered levels were necessary to compose the sperm signal for the transmission of paternal acquired information [75]. This stands in contrast to the results obtained by Sharma et al. [32] when injecting synthesized tRFs mimicking those tRFs that showed changes upon altered diet. They were able to copy the paternal low protein diet-induced gene expression changes in the early embryo [32]. This could indicate that each study was capturing distinct aspects of a complex phenotype and demonstrates the potential of plural interpretations.
A new preprint by Jung et al. [76] on the effects of bisphenol A (BPA) exposure on intergenerational obesity also emphasizes the involvement of chromatin changes at the Fto gene, encoding a N6-methyladenosine (m6A) methyltransferase, leading to decreased m6A levels. Genetic deletion of a transcription factor binding site at the Fto gene abolishes the transmission of obesity [76]. This again points towards a critical involvement of m6A. RNA hypomethylation has been implicated in increased recruitment of enhancer RNAs to DNA thereby altering chromatin accessibility and transcription [77], but simultaneously the binding might confer a stability advantage during fertilization as to influence early embryonic transcription. Furthermore, m6A levels have been shown crucial for circRNA biogenesis during sperm maturation [71], potentially indicating a complex interplay of transcription factor-mediated chromatin looping, non-coding RNAs and RNA modifications in the transmission of regulatory signals of gene expression that perpetuate the effects of BPA across generations.
Altered RNA modifications have also been reported for other types of exposures apart from diet. Depressive patients challenged with glucocorticoids and mice exposed to stress show a tight regulation of m6A and N6,2ʹ-O-dimethyladenosine in the brain [78]. That m6A levels of mRNAs might be also affected in the germline of stress-exposed males is to be expected but remains to be determined.
RNA modifications are also relevant for technical reasons. Certain modified bases can lead to lower cloning efficiencies and bias the assessment in genome-wide sequencing approaches. A recent preprint by tRF experts from the same group that first reported the importance of tRFs in transmission benchmarked different sequencing methods and concluded conventional small RNA sequencing library preparation methods to be inaccurate for tRF profiling due to their high degree of modifications [79]. This prompts reconsideration of prior studies on the contribution of tRFs. Luckily, novel technologies embrace the potential discrimination of certain modified RNAs [80] and will hopefully accelerate the elucidation of the effects of environmental exposures on RNA modifications and their contribution to intergenerational phenotypes.
Taken together, it is clear that several RNA classes and their modifications are contributing to epigenetic germline inheritance. The complexity of the sperm RNA payload calls for investigation on the origin of sperm RNA. Furthermore, it will be crucial to determine which of the presented RNA classes are indeed delivered to the oocyte, and whether this changes in response to environmental exposures. The mechanisms underlying such transmission might bear some clues on how a given RNA excerpts its function.
Where Does Sperm RNA Come from?
The transcriptionally silent state of mature sperm poses an obvious question. Where or when is mature sperm RNA transcribed? Early studies explored the possibility of mobile RNA. Sperm RNAs might originate from somatic cells elsewhere in the body and get taken up by sperm cells. This was tested by xenografting tumour cells to germline distal sites. Exogenous tumour RNA, being technically more distinguishable from endogenous sperm RNA, could be detected in sperm and thereby provided the first proof of principle [81]. Later studies by Sharma et al. [32] revisited the idea of exosomal delivery of tRNA fragments in a more natural setting focusing on epididymal exosomes, then termed epididymosomes [32]. With the advent of novel RNA-labelling techniques such as SlamITseq [82], Sharma provided data on the presence of metabolically labelled epididymal RNA in sperm [83]. This RNA was presumably taken up during the transit from caput to cauda epididymis and focused on miRNAs as opposed to tRNA fragments. A disputed follow-up study claimed that these acquired miRNAs were required upon delivery to the oocyte for embryonic development [84]. The importance of epididymal miRNA contributions for the transmission of the effects of chronic stress has been proposed by a study from the Bale lab [85]. They mimicked the transmission of effects to the offspring by injections of sperm incubated with epididymosomes from stressed males, suggesting their importance in a natural mating setting of chronically stressed fathers. The sperm RNA payload however likely also contains remnants of prior transcription as has been shown for circRNAs [71] and tRNA fragments [18] and suggested for long RNAs [86].
How and Where Are RNA Molecules Functional?
As we explore which of the sperm RNA subgroups is relevant for epigenetic germline inheritance, it is also important to investigate which of these are effectively transmitted to the oocyte. The standard method for exploring the effects of a certain sperm RNA type has been to isolate total sperm RNA and to inject it or RNA fractions into non-exposed fertilized oocytes. This provided information about the functionality of sperm RNA, but not the transfer from sperm to oocyte itself. So far, sperm RNA delivery has been inferred from comparisons of microarray or sequencing data from unfertilized and fertilized oocytes [37, 87, 88]. The lack of statistical comparisons makes them prone to artefacts introduced by for instance divergent sequencing depth or sample quality. Metabolic labelling methods such as mentioned above in the context of exosomal RNA delivery [82, 83] could instead be employed to unambiguously establish which RNA classes are transmitted.
The prime curiosity remains RNA fragility. How can RNA retain sufficient stability? As alluded to in the prior section, circRNAs would have a clear advantage over free linear RNAs and therefore make them a likely relevant candidate. To experimentally approach their potential ability to sponge miRNAs in the embryo, such miRNAs could be provided in excess to counterbalance the circRNA-induced reduction. This reversal would expectedly lead to a downregulation of mRNA targets of said miRNAs and thereby normalize the circRNA-induced upregulation of mRNAs.
DNA-Bound RNA
In opposition to inherently stable circRNAs, linear sperm RNA subgroups most likely rely on certain stabilization to transfer information to the offspring. It is well recognized that some RNA is bound to DNA [77]; however, experimental procedures discussed here so far with the exception of the study from Jung et al. [91] ignored this fraction.
Chromatin-associated RNAs have been described as highly stable structures [89], revealing that there are cis- and trans-interacting RNAs. Cis-interacting DNA-bound RNAs stay on site of their transcription, whereas trans-interacting DNA-bound RNAs leave their site of synthesis to interact with another genomic locus [90]. Western blot showed the presence of RNA polymerase II in mature sperm [91], a remainder of active transcription during spermatogenesis. As evidenced by findings of the group of Victor Corces, a small fraction of stalled nascent RNA appears bound to chromatin in spermatozoa. This RNA could possibly remain on site and thus form a cis-interaction with its DNA template. Given the evidence that spermatozoa receive a majority of their RNA content from the epididymis via epididymosomes [42], a trans-interacting mode is expected as to how RNAs can associate with sperm DNA. Trans-interactions can occur directly via DNA:RNA hybrids or indirectly via a mediating RNA binding protein where they are described to regulate transcription [92]. Trans-interacting RNA binds directly to DNA by forming triplex structures. By Hoogsteen base-pairing, RNA binds via hydrogen bonds to the major groove of the double-stranded DNA, winding around the double helix [93, 94]. This leads to stabilization of the RNA [95] and allows it to guide transcription regulators to distinct sites, best experimentally proven so far with lncRNAs [93].
A multitude of chromatin enriched lncRNAs have been found in adjacency to active genes [96] and have later been confirmed as transcriptional co-activators [97]. The insights on specific lncRNAs creating triplex formations and their implications for genomic regulation are reviewed elsewhere [93]. Interestingly, there has been recent evidence that precursor mRNA might also function as regulatory lncRNA. Skalska and colleagues proposed in 2017 that precursor mRNA functions similarly to non-coding RNAs by forming transcription hubs around their site of transcription to regulate gene expression [98]. Regarding the high abundance of protein-coding RNAs in the DNA-bound RNA fraction of the presented data, it might be possible that some of these are functioning as expression regulators, as suggested by Skalska et al. and Wei et al. [98, 99]. This possibility needs to be investigated further, as the understanding of DNA-bound RNAs in sperm is very rudimental. lncRNA content in sperm could be analysed by nanopore sequencing to account for the transcript lengths and compared with previous findings [96, 97]. Furthermore, to confirm these lncRNAs are bound to sperm DNA directly, they should be compared after mapping with results from a genome-wide characterization of DNA:RNA triplex structures [100]. Alternatively, chromatin isolation by RNA purification [101] or mapping RNA–genome interactions [102] might be employed to discover and examine DNA-bound RNA. In a follow-up, the question of whether this specific fraction bears any advantage during sperm to oocyte transmission could be explored. The characterization of lncRNA content and associated structures in the zygote after mating or in vitro fertilization, in comparison to the non-exposed oocyte, might prove useful to test such hypotheses.
Similar to lncRNAs, several miRNAs have been shown to bind double-stranded DNA by forming triplex structures. Bioinformatic analysis investigated triplex-forming binding sites for miRNA. The sites were enriched in genes that positively correlated their expression with miRNA expression of the miRNAs binding those sites [103]. Similarly, a recent in silico study compared genomic sites where miRNAs bind with transcription factor binding sites in the genome. It revealed conserved motifs in miRNA transcripts and predicted them to bind specific DNA sequences [104]. Other research so far covered the indirect binding of miRNAs with DNA via Ago2 protein [105] or triplex formation miRNA-mediated detection [106, 107]. Experimental evidence for miRNAs binding directly to genomic DNA by the formation of triplex structures is still lacking to confirm the in silico predictions. If indeed specific triplex-forming miRNAs can be proven experimentally, this adds to the idea that sperm RNAs—in this case miRNAs—might benefit from stabilization by the formation of triplex structures.
Two studies suggest tRF binding to DNA via Ago, with an impact on gene expression [108, 109]. While these studies indicate an indirect binding to the DNA, it is currently unclear whether tRFs also engage in direct DNA binding.
Similarly, piRNAs most likely bind directly or indirectly to DNA as to induce DNA methylation and gene silencing [110, 111].
The most widely studied DNA-binding RNA is telomeric repeat–containing RNA (TERRA). Interestingly, TERRAs bind to DNA by forming R-loops, opposing to the triplex formation of the RNAs described above. R-loops are formed by the DNA double helix opening up and the RNA attaching to one of the DNA strands. TERRAs are involved in telomere maintenance at chromosome ends [112, 113]. They form G-quadruplex structures, and in vitro studies indicate that co-binding of the human protein FUS to this structure and DNA quadruplexes at telomeres could recruit epigenetic modifiers and thereby regulate heterochromatin formation [114]. Furthermore, DNA G-quadruplexes in gene bodies have also been observed to help resume transcription [115], or opposingly to stall transcription by facilitating the stabilization of nascent transcripts into R-loops [116]. Alternatively, the formation of DNA–RNA quadruplexes might terminate transcription of that locus altogether [117, 118]. This rather resembles the cis-interacting mode of RNA binding DNA [90].
Not only the stabilization of RNA by the formation of triplex structures with the DNA double helix and its delivery have to be proven relevant for epigenetic inheritance. The mechanism of action post-fertilization also requires further investigation. Detailed intersection of sperm RNA data, DNA Pol II data and genome-wide characterization of DNA:RNA triplex structures [100] and DNA quadruplex structures will likely reveal interesting further indications on the role of DNA–RNA interactions in sperm.
Experimental Considerations
Assuming that paternal sperm RNA bound to DNA is favoured in the transmission, two mechanistic scenarios are to be expected post-fertilization. (i) The RNA is released to find complementary specific sites on the maternal chromosomes and/or to regulate RNA transcripts post-transcriptionally. (ii) The RNA stays bound to the paternal allele and continues to regulate gene expression monoallelically in an imprinting-like fashion [119, 120].
It is important to mention that sperm RNA injections of exposed males into fertilized non-exposed oocytes are not suitable to assess the potential contributions of DNA-bound RNA. When RNA is harvested for such injection, its potential DNA binding is either disrupted or if maintained the fraction of DNA-bound RNA is left behind. Hence, injected RNA consists of either previously bound DNA that lacks potential stability or other functionality conferred by DNA binding or is deprived of the RNA fraction usually bound to DNA. This is a clear limitation of prior studies, including our own, and might explain at least in part why a full mimic of phenotypes has seldom been achieved. A concern that adds to this is the often highly complex secondary structure of RNAs, which might be key for stability and function yet could get altered in such experimental approaches. tRFs have complex secondary structures similar to tRNAs and bind with their 5ʹ fragments [121]. Similarly, lncRNAs rely on their secondary structure to bind their targets [122]. The cis- and trans-acting of lncRNAs in DNA:RNA hybrid triplex formations and their function in gene regulation has been summarized by Li and colleagues. They suggest lncRNAs bind the DNA double helix based on triplex-forming motifs [93].
To circumvent a potential loss of (i) secondary structures and (ii) stability, inhibition of DNA:RNA hybrids by complementary oligonucleotides could provide an elegant approach of reverse paternal phenotypes. A reversal by antisense oligos has been achieved so far by the group of Chen and by Benito et al. for miRNAs hence targeting post-transcriptional gene regulation [34, 48]. It remains to be seen whether approaches targeting DNA-bound RNA can be effective too. Targeting bound RNA is inevitably more challenging. Instead of using oligonucleotides, aptamers could be applied to bind on the DNA:RNA triplex. Aptamers are synthetic protein or deoxy-/ribonucleic acid-based small molecules that can bind biological structures by their three-dimensional conformation [123]. They have been established already as small interfering RNA (siRNA) chimeras [124]. Specific aptamers could be synthesized targeting DNA:RNA hybrids or specific DNA sites that usually form triplex structures, prohibiting the attachment of RNA to the DNA. A prevention of behavioural and metabolic phenotypes in the offspring of environmentally exposed males by aptamers would be an unequivocal indication that DNA-bound RNAs indeed are needed for epigenetic germline transmission.
In an alternative approach, recently employed by Jung et al. [76], a deletion in the region encoding m6A demethylase, where additionally a specific enhancer RNA (eRNA) would bind, achieved the correction of a BPA-induced phenotype [76].
Overall, the mechanism of how paternal RNA is contributing to embryonic expression in terms of how it is delivered from sperm to oocyte and how it interacts with the embryonic genome is not understood yet. We proposed experiments to dissect the chromatin–RNA interaction as a possible means to understand the mechanism of RNA-mediated epigenetic inheritance.
Conclusion
The field of epigenetic germline inheritance is vastly expanding, and new studies on paternal influence in the transmission of environmental influences emerge continuously. A so-called “sperm RNA code” was postulated [30] based on many studies demonstrating the influence of distinct RNA classes. It seems clear that all these RNA classes are involved in one way or the other in the transmission of environmental effects to influence the offspring. RNAs are altered through environmental challenges in sperm, either during spermatogenesis or in the epididymis, and delivered to the oocyte during fertilization. Nevertheless, the detailed mechanism remains largely unclear. It is unknown whether paternal RNA indirectly affects embryonic expression by regulation leading to a persistently altered expressive state or whether they persist until later embryonal stages where they directly modulate embryonic gene expression. Furthermore, it is unclear how a given metabolic or psychological stressor leads to changes in RNA payload or RNA modifications mechanistically and whether there are converging RNA signatures of distinctively different exposures. As outlined, several technically highly challenging experimental approaches might help to elucidate how a particular sperm RNA signal is established under specific environmental influences, how it is propagated and how it brings about phenotypic changes in the offspring.
Lastly, to come back to the notion of transgenerational effects, RNA arguably would require a self-perpetuating signal reminiscent potentially of that of the piRNA ping-pong cycle [125]. Alternatively, and potentially more likely, however, the sperm RNA signal is embedded in a complex interplay with other epigenetic modifications, such as seen for Fto enhancer and chromatin states [85]. Such interactions could (i) induce a translation of RNA signals into other more stable modifications in the offspring, (ii) induce another cycle of RNA-mediated inheritance or (iii) even lead to a genetic consolidation by altering the propensity for mutations via RNA-directed methylation [44] or transposable element activity [48, 126].
Lastly, we predict that a more holistic assessment of the interplay of different sperm RNA classes but also other epigenetic marks in combination with gene expression analysis and experimentation targeting reversal as opposed to mimics will substantially contribute to clarifying the mechanistic contribution of sperm RNA to intergenerational and potentially transgenerational effects.
Acknowledgements
We thank Vincent Fischer for critical reading and comments. Figures were created with https://BioRender.com.
Contributor Information
Miriam Kretschmer, Department of Health Sciences and Technology, ETH Zurich, Laboratory of Molecular and Behavioral Neuroscience, Institute for Neuroscience, Winterthurerstrasse 190, Zurich 8057, Switzerland; Neuroscience Centre Zurich, ETH Zurich and University of Zurich, Winterthurerstrasse 190, Zurich 8057, Switzerland.
Katharina Gapp, Department of Health Sciences and Technology, ETH Zurich, Laboratory of Molecular and Behavioral Neuroscience, Institute for Neuroscience, Winterthurerstrasse 190, Zurich 8057, Switzerland; Neuroscience Centre Zurich, ETH Zurich and University of Zurich, Winterthurerstrasse 190, Zurich 8057, Switzerland.
Data availability
Not applicable.
Funding
The lab of Katharina Gapp is currently supported by funding from the Swiss National Science foundation Prima fellowship (PR00P3_201543), an Olga Mayenfisch foundation grant and an ETH project grant (ETH-41 20-1).
Conflict of interest statement
None declared.
Author contributions
M.K. and K.G. conceived the idea for this review and wrote the manuscript.
References
- 1. Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res 2008;167:121–35. [DOI] [PubMed] [Google Scholar]
- 2. Yehuda R, Daskalakis NP, Lehrner A. et al. Influences of maternal and paternal PTSD on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. Am J Psychiatry 2014;171:872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev 2006;82:485–91. [DOI] [PubMed] [Google Scholar]
- 4. Roseboom TJ, Painter RC, van Abeelen AF. et al. Hungry in the womb: what are the consequences? Lessons from the Dutch famine. Maturitas 2011;70:141–5. [DOI] [PubMed] [Google Scholar]
- 5. Veenendaal MV, Painter RC, de Rooij SR. et al. Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. Bjog 2013;120:548–53. [DOI] [PubMed] [Google Scholar]
- 6. Sutanto W, de Kloet ER. The use of various animal models in the study of stress and stress-related phenomena. Lab Anim 1994;28:293–306. [DOI] [PubMed] [Google Scholar]
- 7. Patchev VK, Patchev AV. Experimental models of stress. Dialogues Clin Neurosci 2006;8:417–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhabhar FS. The short-term stress response - mother nature’s mechanism for enhancing protection and performance under conditions of threat, challenge, and opportunity. Front Neuroendocrinol 2018;49:175–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim JW, Ko MJ, Gonzales EL. et al. Social support rescues acute stress-induced cognitive impairments by modulating ERK1/2 phosphorylation in adolescent mice. Sci Rep 2018;8:12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li S, Fan YX, Wang W. et al. Effects of acute restraint stress on different components of memory as assessed by object-recognition and object-location tasks in mice. Behav Brain Res 2012;227:199–207. [DOI] [PubMed] [Google Scholar]
- 11. Rashidy-Pour A, Sadeghi H, Taherain AA. et al. The effects of acute restraint stress and dexamethasone on retrieval of long-term memory in rats: an interaction with opiate system. Behav Brain Res 2004;154:193–8. [DOI] [PubMed] [Google Scholar]
- 12. Mudra Rakshasa A, Tong MT. Making "good" choices: social isolation in mice exacerbates the effects of chronic stress on decision making. Front Behav Neurosci 2020;14:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Sousa Rodrigues ME, Bekhbat M, Houser MC. et al. Chronic psychological stress and high-fat high-fructose diet disrupt metabolic and inflammatory gene networks in the brain, liver, and gut and promote behavioral deficits in mice. Brain Behav Immun 2017;59:158–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Kooij MA, Jene T, Treccani G. et al. Chronic social stress-induced hyperglycemia in mice couples individual stress susceptibility to impaired spatial memory. Proc Natl Acad Sci U S A 2018;115:E10187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Safi-Stibler S, Gabory A. Epigenetics and the developmental origins of health and disease: parental environment signalling to the epigenome, critical time windows and sculpting the adult phenotype. Semin Cell Dev Biol 2020;97:172–80. [DOI] [PubMed] [Google Scholar]
- 16. Hoyer C, Richter H, Brandwein C. et al. Preconceptional paternal exposure to a single traumatic event affects postnatal growth of female but not male offspring. Neuroreport 2013;24:856–60. [DOI] [PubMed] [Google Scholar]
- 17. Bohacek J, von Werdt S, Mansuy IM. Probing the germline-dependence of epigenetic inheritance using artificial insemination in mice. Environ Epigenet 2016;2:dvv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gapp K, Parada GE, Gross F. et al. Single paternal dexamethasone challenge programs offspring metabolism and reveals multiple candidates in RNA-mediated inheritance. iScience 2021;24:102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korgan AC, O’Leary E, Bauer J. et al. Effects of paternal predation risk and rearing environment on maternal investment and development of defensive responses in the offspring. eNeuro 2016;3:1–14. ENEURO.0231–16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vitousek MN, Taff CC, Ardia DR. et al. The lingering impact of stress: brief acute glucocorticoid exposure has sustained, dose-dependent effects on reproduction. Proc Biol Sci 2018;285:20180722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bohacek J, Mansuy IM. A guide to designing germline-dependent epigenetic inheritance experiments in mammals. Nat Methods 2017;14:243–9. [DOI] [PubMed] [Google Scholar]
- 22. Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 2018;19:774–90. [DOI] [PubMed] [Google Scholar]
- 23. Bohacek J, Mansuy IM. Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat Rev Genet 2015;16:641–52. [DOI] [PubMed] [Google Scholar]
- 24. Fraser R, Lin CJ. Epigenetic reprogramming of the zygote in mice and men: on your marks, get set, go! Reproduction 2016;152:R211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science 2007;316:407–8. [DOI] [PubMed] [Google Scholar]
- 26. Champagne FA, Meaney MJ. Stress during gestation alters postpartum maternal care and the development of the offspring in a rodent model. Biol Psychiatry 2006;59:1227–35. [DOI] [PubMed] [Google Scholar]
- 27. Stäubli A, Peters AH. Mechanisms of maternal intergenerational epigenetic inheritance. Curr Opin Genet Dev 2021;67:151–62. [DOI] [PubMed] [Google Scholar]
- 28. Clarke HJ, Vieux KF. Epigenetic inheritance through the female germ-line: the known, the unknown, and the possible. Semin Cell Dev Biol 2015;43:106–16. [DOI] [PubMed] [Google Scholar]
- 29. Gapp K, Jawaid A, Sarkies P. et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 2014;17:667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rassoulzadegan M, Grandjean V, Gounon P. et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006;441:469–74. [DOI] [PubMed] [Google Scholar]
- 31. Grandjean V, Fourré S, De Abreu DA. et al. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci Rep 2015;5:18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma U, Conine CC, Shea JM. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kiani J, Grandjean V, Liebers R. et al. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet 2013;9:e1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Q, Yan M, Cao Z. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397–400. [DOI] [PubMed] [Google Scholar]
- 35. Sarker G, Sun W, Rosenkranz D. et al. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc Natl Acad Sci U S A 2019;116:10547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet 2005;6:633–42. [DOI] [PubMed] [Google Scholar]
- 37. Ostermeier GC, Miller D, Huntriss JD. et al. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 2004;429:154. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Shi J, Rassoulzadegan M. et al. Sperm RNA code programmes the metabolic health of offspring. Nat Rev Endocrinol 2019;15:489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fullston T, Ohlsson Teague EM, Palmer NO. et al. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J 2013;27:4226–43. [DOI] [PubMed] [Google Scholar]
- 40. Rodgers AB, Morgan CP, Bronson SL. et al. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci 2013;33:9003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 2016;17:733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bohacek J, Rassoulzadegan M. Sperm RNA: quo vadis? Semin Cell Dev Biol 2020;97:123–30. [DOI] [PubMed] [Google Scholar]
- 43. Felden B, Paillard L. When eukaryotes and prokaryotes look alike: the case of regulatory RNAs. FEMS Microbiol Rev 2017;41:624–39. [DOI] [PubMed] [Google Scholar]
- 44. Erdmann RM, Picard CL. RNA-directed DNA methylation. PLoS Genet 2020;16:e1009034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu WM, Pang RT, Chiu PC. et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A 2012;109:490–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Conine CC, Sun F, Song L. et al. MicroRNAs absent in caput sperm are required for normal embryonic development. Dev Cell 2019;50:7–8. [DOI] [PubMed] [Google Scholar]
- 47. Rodgers AB, Morgan CP, Leu NA. et al. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A 2015;112:13699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benito E, Kerimoglu C, Ramachandran B. et al. RNA-dependent intergenerational inheritance of enhanced synaptic plasticity after environmental enrichment. Cell Rep 2018;23:546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arai JA, Li S, Hartley DM. et al. Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment. J Neurosci 2009;29:1496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gapp K, Bohacek J, Grossmann J. et al. Potential of environmental enrichment to prevent transgenerational effects of paternal trauma. Neuropsychopharmacology 2016;41:2749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Chen ZP, Hu H. et al. Sperm microRNAs confer depression susceptibility to offspring. Sci Adv 2021;7:eabd7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peng H, Shi J, Zhang Y. et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 2012;22:1609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Magee R, Rigoutsos I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res 2020;48:9433–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cropley JE, Eaton SA, Aiken A. et al. Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Mol Metab 2016;5:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chuma S, Nakano T. piRNA and spermatogenesis in mice. Philos Trans R Soc Lond B Biol Sci 2013;368:20110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rajasethupathy P, Antonov I, Sheridan R. et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012;149:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gou LT, Dai P, Yang JH. et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 2014;24:680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goh WS, Falciatori I, Tam OH. et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev 2015;29:1032–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 2015;16:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhu X, Zhi E, Li Z. MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis. Reproduction 2015;149:R229–35. [DOI] [PubMed] [Google Scholar]
- 61. Wu PH, Fu Y, Cecchini K. et al. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet 2020;52:728–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yuan S, Tang C, Schuster A. et al. Paternal pachytene piRNAs are not required for fertilization, embryonic development and sperm-mediated epigenetic inheritance in mice. Environ Epigenet 2016;2:dvw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qin T, Li J, Zhang KQ. Structure, regulation, and function of linear and circular long non-coding RNAs. Front Genet 2020;11:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Joshi M, Rajender S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod Biol Endocrinol 2020;18:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gapp K, van Steenwyk G, Germain PL. et al. Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol Psychiatry 2020;25:2162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sun YH, Wang A, Song C. et al. Single-molecule long-read sequencing reveals a conserved intact long RNA profile in sperm. Nat Commun 2021;12:1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol 2018;9:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci 2019;26:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ji P, Wu W, Chen S. et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep 2019;26:3444–60.e5. [DOI] [PubMed] [Google Scholar]
- 71. Tang C, Xie Y, Yu T. et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res 2020;30:211–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 73. Abdelmohsen K, Panda AC, Munk R. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14:361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Du WW, Yang W, Li X. et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018;37:5829–42. [DOI] [PubMed] [Google Scholar]
- 75. Zhang Y, Zhang X, Shi J. et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 2018;20:535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jung YH, Wang HV, Ruiz D. et al. Recruitment of CTCF to an Fto enhancer is responsible for transgenerational inheritance of obesity. Preprint In: bioRxiv 2020. 11.20.391672.doi: 10.1101/2020.11.20.391672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu J, Dou X, Chen C. et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020;367:580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Engel M, Eggert C, Kaplick PM. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 2018;99:389–403.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gustafsson T, Galan C, Yu T. et al. Deep sequencing of yeast and mouse tRNA and tRNA fragments using OTTR. Preprint In: bioRxiv 2022. 02.04.479139v1.doi: 10.1101/2022.02.04.479139. [DOI] [Google Scholar]
- 80. Kugelberg U, Nätt D, Skog S. et al. 5´XP sRNA-seq: efficient identification of transcripts with and without 5´ phosphorylation reveals evolutionary conserved small RNA. RNA Biol 2021;18:1588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cossetti C, Lugini L, Astrologo L. et al. Soma-to-germline transmission of RNA in mice xenografted with human tumour cells: possible transport by exosomes. PLoS One 2014;9:e101629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Matsushima W, Herzog VA, Neumann T. et al. Sequencing cell-type-specific transcriptomes with SLAM-ITseq. Nat Protoc 2019;14:2261–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sharma V, Hecker N, Roscito JG. et al. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat Commun 2018;9:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Conine CC, Sun F, Song L. et al. Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev Cell 2018;46:470–80.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chan JC, Morgan CP, Adrian Leu N. et al. Reproductive tract extracellular vesicles are sufficient to transmit intergenerational stress and program neurodevelopment. Nat Commun 2020;11:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gapp K, Bohacek J. Epigenetic germline inheritance in mammals: looking to the past to understand the future. Genes Brain Behav 2018;17:e12407. [DOI] [PubMed] [Google Scholar]
- 87. Dard-Dascot C, Naquin D, d’Aubenton-Carafa Y. et al. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 2018;19:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yeri A, Courtright A, Danielson K. et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics 2018;19:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 1992;258:1463–6. [DOI] [PubMed] [Google Scholar]
- 90. Li X, Fu XD. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet 2019;20:503–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jung YH, Kremsky I, Gold HB. et al. Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol Cell 2019;75:154–71.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xiao R, Chen JY, Liang Z. et al. Pervasive Chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 2019;178:107–21.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li Y, Syed J, Sugiyama H. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem Biol 2016;23:1325–33. [DOI] [PubMed] [Google Scholar]
- 94. Martianov I, Ramadass A, Serra Barros A. et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007;445:666–70. [DOI] [PubMed] [Google Scholar]
- 95. Kunkler CN, Hulewicz JP, Hickman SC. et al. Stability of an RNA•DNA-DNA triple helix depends on base triplet composition and length of the RNA third strand. Nucleic Acids Res 2019;47:7213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Werner MS, Ruthenburg AJ. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep 2015;12:1089–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Werner MS, Sullivan MA, Shah RN. et al. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat Struct Mol Biol 2017;24:596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Skalska L, Beltran-Nebot M, Ule J. et al. Regulatory feedback from nascent RNA to chromatin and transcription. Nat Rev Mol Cell Biol 2017;18:331–7. [DOI] [PubMed] [Google Scholar]
- 99. Wei C, Xiao R, Chen L. et al. RBFox2 binds nascent RNA to globally regulate polycomb complex 2 targeting in mammalian genomes. Mol Cell 2016;62:875–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sentürk Cetin N, Kuo CC, Ribarska T. et al. Isolation and genome-wide characterization of cellular DNA:RNA triplex structures. Nucleic Acids Res 2019;47:2306–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chu C, Qu K, Zhong FL. et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011;44:667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sridhar B, Rivas-Astroza M, Nguyen TC. et al. Systematic mapping of RNA-chromatin interactions in vivo. Curr Biol 2017;27:602–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Paugh SW, Coss DR, Bao J. et al. MicroRNAs form triplexes with double stranded DNA at sequence-specific binding sites; a eukaryotic mechanism via which microRNAs could directly alter gene expression. PLoS Comput Biol 2016;12:e1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chetta M, Di Pietro L, Bukvic N. et al. Rising roles of small noncoding RNAs in cotranscriptional regulation: in silico study of miRNA and piRNA regulatory network in humans. Genes (Basel) 2020;11:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. von Brandenstein M, Bernhart SH, Pansky A. et al. Beyond the 3ʹUTR binding-microRNA-induced protein truncation via DNA binding. Oncotarget 2018;9:32855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jin J, Vaud S, Zhelkovsky AM. et al. Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic Acids Res 2016;44:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang L, Arrabito G. Hybrid, multiplexed, functional DNA nanotechnology for bioanalysis. Analyst 2015;140:5821–48. [DOI] [PubMed] [Google Scholar]
- 108. Kuscu C, Kumar P, Kiran M. et al. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018;24:1093–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shigematsu M, Kirino Y. tRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul Syst Bio 2015;9:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem 2015;84:405–33. [DOI] [PubMed] [Google Scholar]
- 111. Ozata DM, Gainetdinov I, Zoch A. et al. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
- 112. Bettin N, Oss Pegorar C, Cusanelli E. The emerging roles of TERRA in telomere maintenance and genome stability. Cells 2019;8:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Koch L. Non-coding RNA: a protective role for TERRA at telomeres. Nat Rev Genet 2017;18:453. [DOI] [PubMed] [Google Scholar]
- 114. Takahama K, Takada A, Tada S. et al. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol 2013;20:341–50. [DOI] [PubMed] [Google Scholar]
- 115. Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res 2008;18:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Belotserkovskii BP, Soo Shin JH, Hanawalt PC. Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res 2017;45:6589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Varshney D, Spiegel J, Zyner K. et al. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol 2020;21:459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wanrooij PH, Uhler JP, Shi Y. et al. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res 2012;40:10334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 2014;6:a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Tucci V, Isles AR, Kelsey Get al. Erice Imprinting Group . Genomic imprinting and physiological processes in mammals. Cell 2019;176:952–65. [DOI] [PubMed] [Google Scholar]
- 121. Pliatsika V, Loher P, Telonis AG. et al. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics 2016;32:2481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zampetaki A, Albrecht A, Steinhofel K. Long non-coding RNA structure and function: is there a link? Front Physiol 2018;24:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lakhin AV, Tarantul VZ, Gening LV. Aptamers: problems, solutions and prospects. Acta Naturae 2013;5:34–43. [PMC free article] [PubMed] [Google Scholar]
- 124. Kruspe S, Giangrande PH. Aptamer-siRNA chimeras: discovery, progress, and future prospects. Biomedicines 2017;5:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Czech B, Hannon GJ. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 2016;41:324–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 2017;45:5142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


