Abstract
WRINKLED1 (WRI1) is an important transcription factor that regulates seed oil biosynthesis. However, how WRI1 regulates gene expression during this process remains poorly understood. Here, we found that BLISTER (BLI) is expressed in maturing Arabidopsis thaliana seeds and acts as an interacting partner of WRI1. bli mutant seeds showed delayed maturation, a wrinkled seed phenotype, and reduced oil content, similar to the phenotypes of wri1. In contrast, BLI overexpression resulted in enlarged seeds and increased oil content. Gene expression and genetic analyses revealed that BLI plays a role in promoting the expression of WRI1 targets involved in fatty acid biosynthesis and regulates seed maturation together with WRI1. BLI is recruited by WRI1 to the AW boxes in the promoters of fatty acid biosynthesis genes. BLI shows a mutually exclusive interaction with the Polycomb-group protein CURLY LEAF (CLF) or the chromatin remodeling factor SWITCH/SUCROSE NONFERMENTING 3B (SWI3B), which facilitates gene expression by modifying nucleosomal occupancy and histone modifications. Together, these data suggest that BLI promotes the expression of fatty acid biosynthesis genes by interacting with WRI1 to regulate chromatin dynamics, leading to increased fatty acid production. These findings provide insights into the roles of the WRI1–BLI–CLF–SWI3B module in mediating seed maturation and gene expression.
LISTER promotes seed maturation and activates fatty acid biosynthesis genes by linking WRINKLED1 to chromatin remodeling factors and by regulating nucleosomal occupancy and histone modifications.
IN A NUTSHELL.
Background: Storage substances such as starch, protein, and oil in crop seeds are major sources of food for humans. These substances mainly accumulate in seeds during the maturation stage. The transcription factor WRINKLED1 (WRI1) is a master regulator of plant oil biosynthesis in Arabidopsis thaliana. However, the underlying mechanism by which WRI1 regulates gene expression remains unclear.
Question: How does WRI1 regulate the expression of genes involved in fatty acid biosynthesis during seed maturation?
Findings: We found that BLISTER (BLI) plays a role in promoting the expression of WRI1 target genes involved in fatty acid biosynthesis and co-regulates seed maturation in Arabidopsis together with WRI1. BLI is recruited by WRI1 to the AW boxes in the upstream regions of fatty acid biosynthesis genes. In turn, BLI functions in the repression of CURLY LEAF (CLF, repressor of gene expression) and the recruitment of SWITCH/SUCROSE NONFERMENTING 3B (promoter of gene expression) to facilitate gene expression by modifying the chromatin state.
Next steps: Our work identified a WRI1-interacting seed maturation regulator and provided insights into the roles of the BLI–WRI1 module in controlling seed maturation and gene expression. Next, we will continue to analyze the structure of BLI in detail and the roles of the different regions of this protein in plant growth and development. We also want to explore which proteins are recruited by the BLI–WRI1 module, and how they work together with BLI–WRI1 to regulate the accumulation of various storage substances in seeds.
Introduction
Seed development is a crucial process in the lifecycle of flowering plants that can be roughly divided into two distinct stages: embryogenesis and seed maturation (Baud et al., 2002; Vicentecarbajosa and Carbonero, 2005). In Arabidopsis thaliana, embryogenesis begins with a single embryogenic cell formed after sexual fertilization and ends at the heart stage of embryo development (6-day after pollination [DAP]). During this process, the activities of specific genes are required to generate the apical–basal axis (Vicentecarbajosa and Carbonero, 2005), and seeds are white or pale yellow due to high water content (>80%) and relatively low fatty acid content (Baud et al., 2008). The transition from embryogenesis to embryo maturation is characterized by the cessation of cell division and specific gene expression. During the maturation phase (7–20 DAP), developing seeds initially accumulate starch, which is subsequently degraded to generate energy and supply carbon for the synthesis of lipids and seed storage proteins (Baud et al., 2002; Ruuska et al., 2002). Triacylglycerol (TAG), an ester derived from glycerol and fatty acids, is the major storage form of lipids in Arabidopsis embryos.
Regulatory networks of highly conserved transcription factors (TFs) play a major role in regulating early embryogenesis and seed maturation in plants (Braybrook et al., 2006). In Arabidopsis, three members of the B3 domain TF family, LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID INSENSITIVE3 (ABI3), and FUSCA3 (FUS3), as well as NF-YB TFs LEC1 and LEC1-like (L1L) are involved in storage protein synthesis and seed maturation (Lotan et al., 1998; Gutierrez et al., 2007; Suzuki and Mccarty, 2008; Boulard et al., 2017); these five TFs are collectively referred to as LAFL proteins. LAFL regulatory factors play important roles in the transition from embryogenesis to seed maturation (Yamamoto et al., 2009) and are required for the accumulation of storage materials such as storage proteins and lipids during seed maturation (To et al., 2006; Roscoe et al., 2015). Overexpression of LEC1 can cause an increase in the expression of all genes involved in fatty acid biosynthesis in plants, which in turn significantly promotes the accumulation of fatty acids (Mu et al., 2008). L1L, a homologous protein of LEC1, restored the phenotype of the lec1 mutant when it was expressed under the control of the LEC1 promoter (Kwong et al., 2003) L1L also plays an important regulatory role in the biosynthesis of seed fatty acids and seed storage oils (Mu et al., 2008). Also LEC2 regulates fatty acid biosynthesis (Baud et al., 2007). The ectopic expression of Arabidopsis LEC2 activated the expression of genes encoding seed storage proteins, enzymes required for oil biosynthesis, and lipid droplet-associated proteins, thereby promoting the accumulation of oils in rosette leaves (Braybrook et al., 2006). ABI3 can form a regulatory complex with the TFs bZIP10, bZIP25, and bZIP53 to deactivate the expression of genes encoding late embryonic enriched proteins, cruciferins, and oleosins (Mönke et al., 2012). FUS3 promotes the accumulation of oils in plants by promoting the expression of photosynthesis and fatty acid biosynthesis-related genes (Zhang et al., 2016).
On the other hand, WRINKLED1 (WRI1), an important regulator of seed maturation, regulates the expression of genes involved in late stages of glycolysis and fatty acid biosynthesis (Focks and Benning, 1998; Cernac and Benning, 2004; Baud et al., 2007; Maeo et al., 2009; Ma et al., 2013). WRI1 is a plant-specific TF of the APETALA2 (AP2)/ethylene-responsive element-binding protein family. The Arabidopsis wri1 mutant produces wrinkled seeds with only 20% of wild-type (WT) TAG contents and increased starch levels (Focks and Benning, 1998) and exhibits higher GH3.3 and lower PIN gene expression than the WT, which affects auxin homeostasis in roots (Kong et al., 2017). In addition, wri1 seeds contain an altered fatty acid composition, with higher levels of linolenic acid (C18:3) and erucic acid (C22:1) but lower levels of oleic (C18:1), linoleic (C18:2), and eicosenoic (C20:1) acids (Focks and Benning, 1998; Baud et al., 2007). In contrast, overexpressing WRI1 in Arabidopsis upregulated a number of glycolytic and fatty acid biosynthetic genes in seedlings and increased the overall seed oil content without altering the seed fatty acid composition (Cernac and Benning, 2004; Sanjaya et al., 2011). Similar results for WRI1 overexpression were obtained in oilseed rape (Brassica napus) and maize (Zea mays; Liu et al., 2010; Pouvreau et al., 2011), indicating functional conservation of WRI1 in both monocots and dicots.
In the lipid biosynthesis pathway in Arabidopsis seeds, WRI1 is positively regulated by LEC1, LEC2, and FUS3 and is negatively regulated by MYB89 (Casson and Lindsey, 2006; Baud et al., 2007; Mu et al., 2008; Santos-Mendoza et al., 2008; Marchive et al., 2014; Li et al., 2017a). The mechanism by which LEC1/LEC2 regulates WRI1 expression appears to be conserved in soybean (Glycine max) and maize as well (Shen et al., 2010; Zhang et al., 2017). Moreover, FUS3 is thought to activate the expression of WRI1, similar to LEC2 (Yamamoto et al., 2009). Although chromatin immunoprecipitation (ChIP) followed by DNA microarray hybridization (ChIP-chip) assays confirmed that WRI1 is a direct target of LEC1 and FUS3 (Wang and Perry, 2013; Pelletier et al., 2017), the DNA motifs for LEC1/LEC2/FUS3 binding to the promoter of WRI1 have not yet been identified (Santos-Mendoza et al., 2008; Marchive et al., 2014). In addition to these TFs, oil palm (Elaeis guineensis) EgNF-YA3, EgNF-YC2, and EgABI5 can bind to the EgWRI1 promoter and activate its expression (Yeap et al., 2017). Recent studies have begun to shed light on WRI1 function at the protein level. The CULLIN3-based E3 ligase adaptor BTB/POZMATH proteins (Chen et al., 2013), 14-3-3 proteins (Ma et al., 2016), and the protein kinase KIN10 (Zhai et al., 2017, 2018) were recently identified as WRI1-interacting proteins that modulate WRI1 stability. WRI1 possesses three intrinsically disordered regions (IDRs) and a PEST motif (a peptide signal for proteolysis) located in the third IDR (IDR3) (Ma et al., 2015). Modified WRI1 proteins with either deletions of IDR3-PEST or mutations at possible phosphorylation sites in IDR3-PEST result in increased protein stability and enhanced oil production (Ma et al., 2015).
WRI1 binds to the AW box sequence [CnTnG](n)7[CG], which is enriched in promoters of fatty acid biosynthetic genes (Baud et al., 2007; Maeo et al., 2009). Most AW boxes are located near the transcription start site (TSS) and in the 5′-untranslated regions of target genes (Maeo et al., 2009). The Arabidopsis mediator complex subunit MED15 physically interacts with WRI1 and promotes WRI1 target gene expression by directly associating with their promoter regions (Kim et al., 2016). However, the detailed mechanism underlying how WRI1 regulates target gene transcription and its impact on chromatin structure and accessibility are not well understood.
LAFL and WRI1 gene expression is repressed during late seed development and after germination by the highly conserved Polycomb-repressive complexes 1 and 2 (PRC1/2) proteins; these proteins are key epigenetic regulators of phase transitions and histone methylation (Makarevich et al., 2006; Liu et al., 2016; Merini et al., 2017). The plant-specific protein BLISTER (BLI) was previously identified as an interactor of CLF, a component of the PRC2 complex. BLI regulates cell differentiation, the expression of a subset of Polycomb group (PcG) target genes, and seed development (Schatlowski et al., 2010). However, the mechanisms underlying how BLI regulates seed development and gene expression are largely unknown.
In this study, we identified BLI as an interactor of WRI1 that play important role in seed maturation and fatty acid biosynthesis by regulating the expression of WRI1 target genes. BLI is involved in CLF repression and SWITCH/SUCROSE NONFERMENTING 3B (SWI3B) recruitment via mutually exclusive interactions with CLF or SWI3B to remodel the chromatin structures of WRI1 target genes by modifying nucleosomal occupancy and histone modifications. This study characterizes a WRI1-interacting co-regulator of seed maturation and fatty acid production and elucidates the role of WRI1 in chromatin regulation.
Results
BLI interacts with WRI1 in vitro and in vivo
To gain further insight into the seed maturation mechanism, we generated an Arabidopsis cDNA library from polyadenylated mRNA from ecotype Columbia-0 (Col-0) seeds of 5–20 DAP. We used the full-length WRI1 protein as a bait to identify its putative partners through yeast two-hybrid (YTH) library screening, and ∼120 clones were retrieved (Supplemental Table S1). Among these, BLI, which is involved in gene regulation (Schatlowski et al., 2010), was found 9 times and was chosen for further studies of its connection with WRI1 and its role in seed maturation.
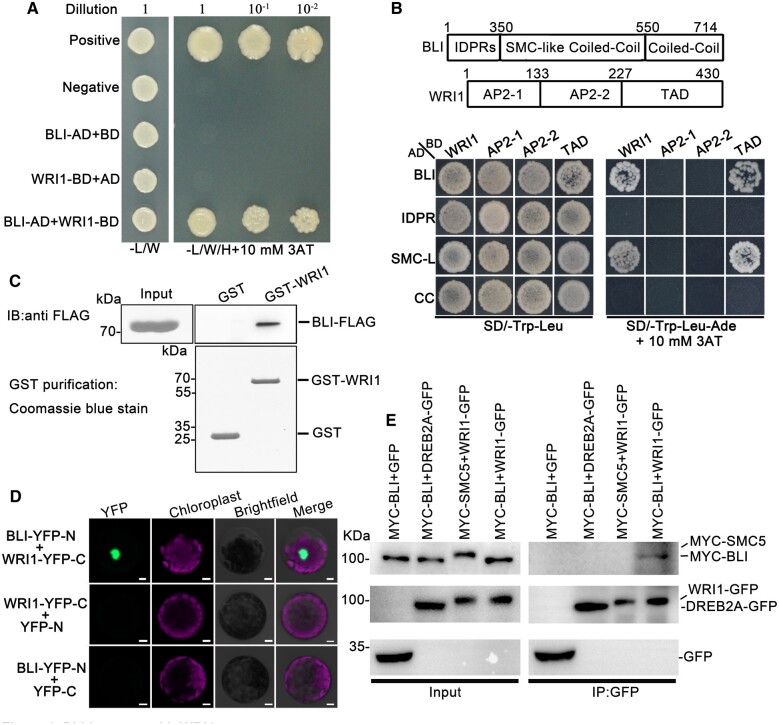
First, we confirmed the BLI–WRI1 interaction through YTH using WRI1 as bait and BLI as prey (i.e. as fusions to the GAL4-BD and GAL4-AD, respectively; Figure 1A). As additional controls, we fused another coiled-coil (CC) domain protein (STRUCTURAL MAINTENANCE OF CHROMOSOME 5 [SMC5]) to GAL4-AD and another AP2 transcription factor (DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A [DREB2A]) to GAL4-BD and tested their interaction with WRI1-BD and BLI-AD, respectively. Although these proteins share similar domains with WRI1 and BLI, they did not show any interaction with BLI or WRI1. Only when WRI1-BD was combined with clones of BLI-AD was growth on selective medium observed, suggesting that the interaction is specific (Figure 1A; Supplemental Figure S1).
Figure 1.
BLI Interacts with WRI1. A, The interaction between BLI and WRI1 confirmed in YTH assay. Positive control: pGADT7-T + pGBKT7-53; negative control: pGADT7-T + pGBKT7-lam. WRI1-BD, WRI1 fused to GAL4-BD; AD, GAL4-AD vectors without insertion; BLI-AD, BLI fused to GAL4-AD; BD, GAL4-BD vectors without insertion; –L/W, selective medium Trp and Leu. –L/W/H, selective medium lacking Trp, Leu, and Ade. B, Identification of the domains in BLI and WRI1 required for their interaction by YTH analysis. IDPRs, the intrinsically disordered protein regions in BLI. SMC-like CC/SMC-L, the region similar to the CC domain of the chromosomal structural maintenance protein in BLI. CC: the CC domain in BLI. AP2-1/2-2: AP2 domains in WRI1. TAD: Region that contains a transcriptional activation domain in WRI1. C, Interaction between BLI and WRI1 confirmed by in vitro pull-down assay. The BLI-FLAG proteins were incubated with immobilized GST or GST-WRI1, and proteins immunoprecipitated with glutathione Sepharose were detected using anti-FLAG antibody. The amounts of GST and GST-WRI1 are shown in the bottom. D, The interaction between BLI and WRI1 in an in vivo BiFC assay in Arabidopsis protoplasts. Bars = 5 µm. E, The interaction between BLI and WRI1 in an in vivo Co-IP assay. Total protein extracts from transgenic plants carrying both 35S:MYC-BLI and 35S:GFP or both 35S:MYC-BLI and 35S:YFP-WRI1 were immunoprecipitated with the immobilized anti-GFP antibody. The interactions between WRI1 and SMC5 (containing a CC domain), or BLI and the AP2 transcription factor DREB2A were chosen as negative controls. The proteins from crude lysates (left) and IPs (right) were detected using anti-MYC or anti-GFP antibody.
As both BLI and WRI1 are large proteins containing several domains, we generated deletion derivatives of the two proteins and used them to localize their interacting regions. We tested several truncations of WRI1 as bait and BLI as prey (Figure 1B). Yeast carrying WRI1 truncations fused to GAL4-BD and the GAL4-AD vectors without insertion did not grow on selective medium. When the WRI1 version carrying the highly conserved transcription activation domain (TAD) was combined with clones of BLI-AD, growth on selective medium was observed, indicating an interaction. For BLI GAL4-AD fusions, growth on selective medium and therefore an interaction with WRI1 was only obtained with the BLI truncations harboring the highly conserved SMC-like CC domain (Figure 1B).
To obtain additional evidence for the interaction of BLI and WRI1, we performed in vitro pull-down analysis. Both the FLAG-BLI and glutathione S-transferase (GST)-WRI1 fusion proteins were produced in and purified from Escherichia coli extracts. Subsequently, the cultures were mixed and GST fusion proteins and the associated proteins were purified. Whereas no copurification of BLI and GST was observed, FLAG-BLI specifically copurified with GST-WRI1 (Figure 1C). Therefore, BLI and WRI1 directly interact in vitro.
To determine whether the in vitro interaction of the two proteins could be recapitulated in vivo, we performed a bimolecular fluorescence complementation (BiFC) assay. BLI was fused to an yellow fluorescent protein N-terminal (YFP-N) fragment, whereas WRI1 was fused to the C-terminal fragment (YFP-C) of enhanced YFP. YFP-N and YFP-C were used as negative controls. YFP fluorescence was reconstituted when BLI and WRI1 were co-expressed in Arabidopsis protoplasts, and the BLI/WRI1 complex localized to the nucleus (Figure 1D). Fluorescence was not detected in protoplasts harboring BLI-YFP-N and YFP-C or YFP-N and WRI1-YFP-C. To confirm the BLI–WRI1 interaction in vivo, BLI-GFP and mCherry-WRI1 were co-transformed with the nuclear marker CFP-H3 (histone 3) into Arabidopsis leaf protoplasts and transiently expressed, revealing their co-localization in the nucleus (Supplemental Figure S2). Furthermore, MYC-BLI was co-immunoprecipitated by WRI1-GFP in an in vivo co-immunoprecipitation (Co-IP) assay but not by GFP or DREB2A-GFP, while MYC-SMC5 was not co-immunoprecipitated by WRI1-GFP, providing further evidence for a specific BLI–WRI1 interaction (Figure 1E).
Loss of BLI disturbs seed maturation
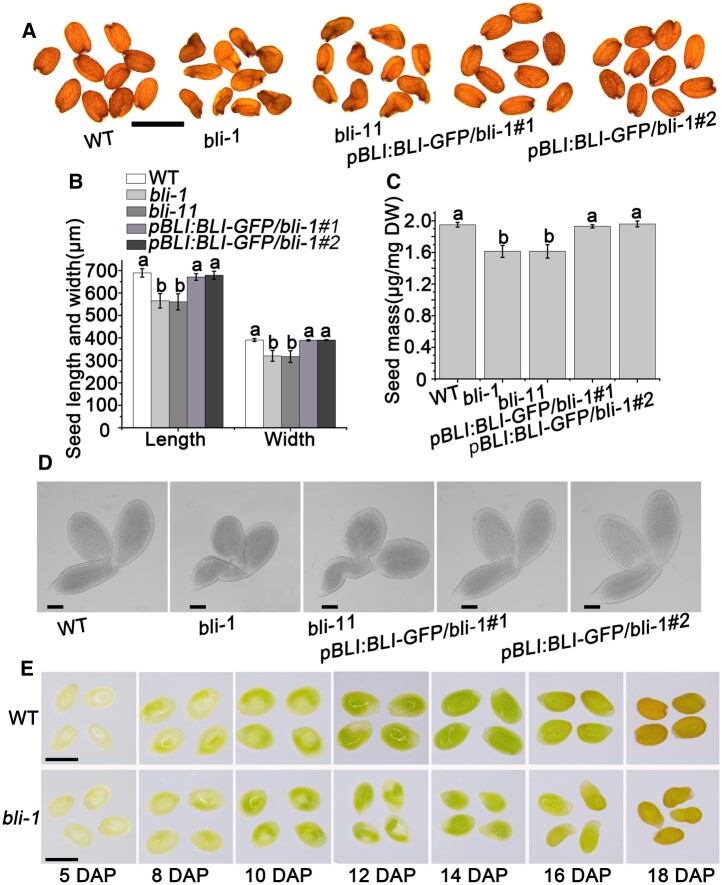
To investigate the role of BLI in seed development, we obtained two T-DNA insertion alleles with severely reduced BLI levels due to strongly reduced BLI transcript levels (SAIL_107_D04 and GABI-Kat_663H12), termed bli-1 and bli-11, in the Col-0 background from the Arabidopsis Biological Resources Center (ABRC) and the Nottingham Arabidopsis Stock Centre, respectively (Supplemental Figure S3; Schatlowski et al., 2010; Kleinmanns et al., 2017). Interestingly, besides the partially unfertilized ovules described previously (Schatlowski et al., 2010), the fertilized bli-1 and bli-11 ovules produced wrinkled and slightly smaller seeds (Figure 2, A and B), and the bli-1 and bli-11 seeds weighed less than WT seeds (Figure 2C), suggesting that bli-1 and bli-11 are defective in seed development, resulting in similar phenotypes. We, therefore, concentrated our analysis on one allele, bli-1.
Figure 2.
Roles of BLI in seed maturation. A, Mature seed phenotypes of WT, bli mutants and bli transgenic complementation plants. bli-1 and bli-11 are two independent BLI loss-of-function mutant lines. pBLI:BLI/bli-1#1 and pBLI:BLI/bli-1#2 indicate two independent bli-1 complementation plant lines. Bar = 1 mm. B, Seed sizes of lines with different BLI expression levels. Values are means ± SD (N = 3); each of the three assays for each biological replicate contained 50 seeds. Seed size does not significantly differ when labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05). C, 100-grain weight of plants with different BLI genomic constructs. Data shown are means ± standard deviation (sd) (N = 3). Seed mass does not significantly differ when labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05). D, Mature embryo phenotypes of different BLI complementation plants. Bars = 100 μm. E, Observation of the maturation process of bli seeds. Bars = 1 mm.
Seed development consists of two major processes, embryonic morphogenesis, and seed maturation. We analyzed bli-1 embryos from the early globular to the torpedo stage and identified no obvious difference between bli-1 and WT embryos (Supplemental Figure S4). We also characterized the seed maturation stages of bli-1. Intriguingly, the bli-1 mutant showed smaller and wrinkled embryos compared to WT plants at the maturation stage (Figure 2D). The wrinkled embryo phenotype was first detectable at 16 DAP in bli-1 but was fully penetrant at later stages of seed maturation (Supplemental Figure S5). Altered endosperm development is associated with wrinkled seed development (Garcia et al., 2005). We, therefore, analyzed endosperm development of bli-1 ovules from 2 to 6 DAP using the ovule autofluorescence technique. Compared to WT seeds, endosperm development was normal in bli-1 seeds (Supplemental Figure S6A). Overall, these results suggest that BLI might be an important regulator of seed maturation, but not of early embryonic or endosperm development. To verify this hypothesis, we studied embryonic development from 5 to 18 DAP using light microscopy and paraffin sectioning. Seed maturation in the bli-1 mutant was clearly delayed compared to WT plants (Figure 2E; Supplemental Figure S6B). The wrinkled seed phenotype could be complemented by the introduction of a transgene carrying BLI cDNA fused to the native promoter (30 independent transgenic lines; Figure 2A; Supplemental Figure S7).
Importantly, BLI overexpression resulted in plants with bigger seeds and increased seed weight in comparison to WT plants, but no obvious difference in seed number per silique (Supplemental Figure S8, A–E). As expected, BLI overexpression caused larger mature embryos compared to the WT (Supplemental Figure S8F). Overall, these results suggest that BLI plays an important role in regulating seed development at the maturation stage.
BLI is expressed in maturing seeds
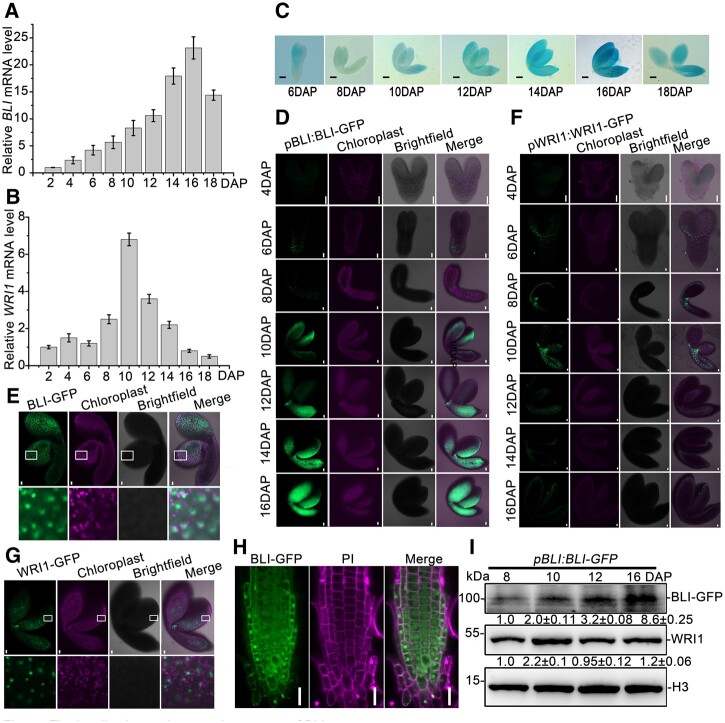
Given that BLI might be involved in seed maturation, we analyzed BLI steady-state mRNA levels during seed maturation using reverse transcription quantitative polymerase chain reaction (RT-qPCR) in WT plants. BLI was expressed at low levels at early embryonic stages from 2 to 6 DAP but its expression progressively increased during seed maturation beginning at 8 DAP, peaked at 16 DAP, and slightly decreased at 18 DAP (Figure 3A). WRI1 expression peaked at 10 DAP and progressively decreased thereafter (Figure 3B). Therefore, the expression patterns of BLI and WRI1 overlap only during a specific developmental window, with the strongest expression of both genes between 10 and 14 DAP. To monitor the detailed expression pattern of BLI, we generated the pBLI:β-glucuronidase (GUS) reporter construct and used it to transform WT plants. All 15 independent pBLI:GUS lines showed similar GUS staining patterns in developing seeds, which were highly consistent with the results of RT-qPCR analysis (Figure 3C). GUS staining was also observed in other tissues, which is consistent with the finding that cotyledons, leaves, and flowers are affected in the bli mutant (Schatlowski et al., 2010). This dynamic expression regulation of BLI might be relevant for the accumulation of seed storage materials, which mainly occurs at the seed maturation stage.
Figure 3.
The localization and expression pattern of BLI. A, BLI expression levels in Arabidopsis developing seeds detected by RT-qPCR. B, WRI1 expression levels in Arabidopsis developing seeds detected by RT-qPCR. C, BLI expression pattern in Arabidopsis developing seeds detected by GUS staining. Bars = 100 μm. D, Accumulation pattern of BLI protein during embryonic development. GFP fluorescence (green) was observed in pBLI:BLI-GFP/bli-1 embryos. Chloroplast, chloroplast fluorescence (magenta), Merge, overlap of BLI-GFP and chloroplast fluorescence by brightfield microscopy. Bars = 20 μm. E, Enlarged view of BLI protein localization in embryonic cells at 10 DAP. Bars = 20 μm. F, Accumulation pattern of WRI1 protein during embryo development. GFP fluorescence (green) was observed in pWRI1:WRI1-GFP/wri1–4 embryos. Chloroplast, chloroplast fluorescence (magenta), Merge, overlap of BLI-GFP and chloroplast fluorescence revealed by brightfield microscopy. Bars = 20 μm. G, Enlarged view of WRI1 protein localization in embryonic cells of at 10 DAP. Bars = 20 μm. H, Subcellular localization analysis of BLI protein in the roots of 35S:BLI-GFP transgenic plants. BLI-GFP, BLI fusion green fluorescent protein. Merge, overlap of BLI-GFP and PI iodide staining. Bars = 50 μm. I, BLI and WRI1 protein levels detected in pBLI:BLI-GFP seeds. BLI or WRI1 protein was detected using antibodies against GFP and WRI1, respectively.
To explore whether BLI and WRI1 proteins overlap during seed development, we observed the protein accumulation patterns of BLI and WRI1. We constructed pBLI:BLI-GFP and pWRI1:WRI1-GFP and analyzed GFP signals, revealing an accumulation of WRI1 and BLI in the nuclei of embryonic cells from 6 to 16 DAP (Figure 3, D–G). The nuclear localization of BLI in embryo cells was similar to that in root cells (Figure 3, E and H; Supplemental Figure S9). Consistent with BLI promoter activity, BLI protein levels gradually increased from 6 DAP to a maximum level at 16 DAP, and WRI1 expression peaked at 10 DAP and decreased thereafter (Figure 3, D, F, and I). Importantly, both protein were present at the 6–16 DAP stages.
BLI facilitates seed oil accumulation in Arabidopsis
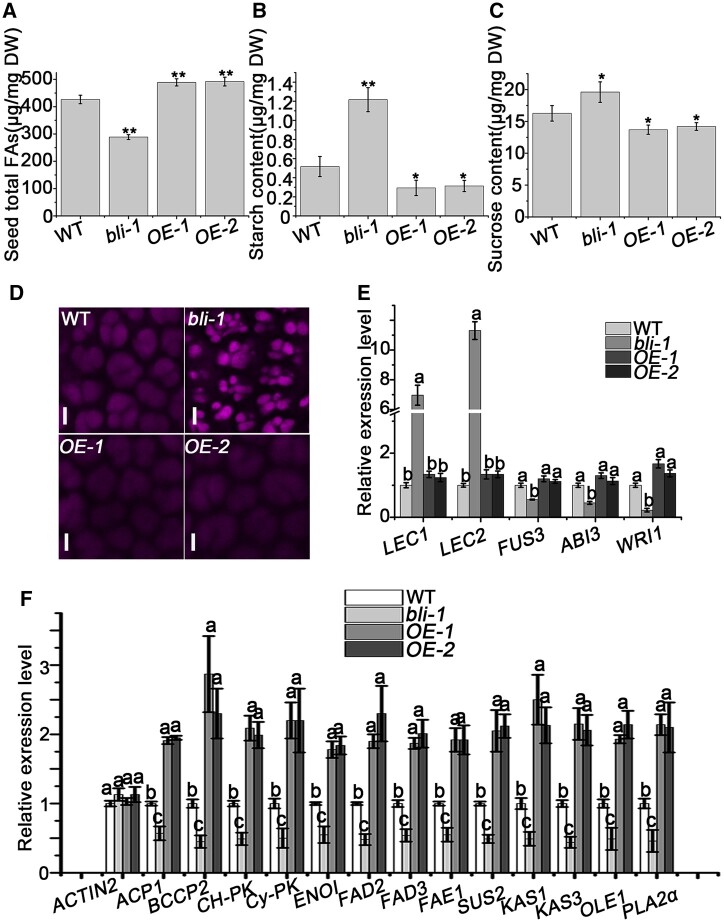
To test whether BLI affects the accumulation of seed oil during seed maturation, we measured the total fatty acid contents in mature dry seeds of bli-1, BLI overexpression, and WT plants. Whereas total fatty acid contents in bli-1 seeds showed a significant reduction of ∼33% compared to WT seeds, BLI overexpression resulted in an increase of 15%, suggesting that BLI might function as a dosage-dependent positive regulator of seed oil accumulation (Figure 4A).
Figure 4.
BLI affects fatty acid accumulation in seeds. A, Fatty acid contents in WT, bli, and BLI overexpression seeds. Data shown are means ± sd (N = 3). Asterisks denote significant differences compared with WT, as determined by Student’s t test (*P < 0.05; **P < 0.01). B, Starch contents in WT, bli and BLI overexpression seeds. Data shown are means ± sd (N = 3). Asterisks denote significant differences compared with WT, as determined by Student’s t test (*P < 0.05; **P < 0.01). C, Sugar contents in WT, bli and BLI overexpression seeds. Data shown are means ± sd (N = 3). Asterisks denote significant differences compared with WT, as determined by Student’s t test (*P < 0.05; **P < 0.01). D, Auto-fluorescence of seed PSVs in mature embryo cotyledons from dry seeds of WT, bli mutants and BLI overexpression plants examined by confocal microscopy. Bar = 5 µm. E, RT-qPCR analysis of the expression of key regulatory genes of seed maturation in 12 DAP seeds of WT, bli mutants and BLI overexpression plants. Data shown are means ± sd (N = 3). UBQ10 was used as a reference gene. The relative expression levels do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05). F, RT-qPCR analysis of genes involved in glycolysis, fatty acid biosynthesis and modification, and TAG accumulation in 12 DAP seeds of WT, bli mutants and BLI overexpression plants. Data shown are means ± sd (N = 3). UBQ10 was used as a reference gene, and ACTIN2 was used as a negative control. The relative expression levels do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05).
To reveal whether the altered levels of total fatty acids in bli-1 mutants/BLI overexpressors were compensated for by a change in other seed storage components, we measured the starch and sugar contents in dry seeds of lines with different BLI levels. Starch quantification revealed up to a 2.5-fold increase in bli-1 dry seeds and an approximately two-fold decrease in dry seeds of BLI overexpressing lines compared to WT dry seeds (Figure 4B). Similarly, free sucrose levels increased by 25% in bli-1 dry seeds and decreased by 18% in BLI overexpression seeds compared to the WT (Figure 4C).
We analyzed the influence of BLI on the quality and quantity of seed storage proteins by running crude seed protein extracts on 20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting (IB) using anti-2S albumin antibodies. Whereas bli-1 seeds accumulated more 2S albumins, seeds of the BLI overexpression lines had less 2S albumins compared to WT seeds (Supplemental Figure S10). These reciprocal changes in protein accumulation may be specific to storage proteins that are transported into seed protein storage vacuoles (PSVs), as PSV autofluorescence was stronger in bli-1 embryos than in WT or BLI overexpression embryos (Figure 4D). Consistently, the 2S albumins genes 2S1, 2S2, 2S3, 2S4, and 2S5 were upregulated in the bli-1 mutant compared with WT plants (Supplemental Figure S11).
To gain insight into the regulation of genes functioning in oil accumulation, we measured the expression levels of the TF gene WRI1 and LAFL genes ABI3, LEC1, LEC2, and FUS3 in 12 DAP developing seeds of plants with different BLI levels. While the B3 domain TF genes ABI3 and FUS3 showed decreased expression, LEC1 and LEC2 were expressed 10- and 7-fold higher, respectively, in bli-1 seeds than the WT (Figure 4E). Consistent with the decreased fatty acid content in bli-1, WRI1, encoding a transcriptional activator of the fatty acid biosynthesis pathway, was downregulated in bli-1 compared to WT seeds (Figure 4E). However, no significant expression changes in any of the analyzed TF genes was observed in the BLI overexpression lines (Figure 4E).
We analyzed the transcript levels of 13 key genes involved in glycolysis, fatty acid biosynthesis and modification, and TAG accumulation in 12 DAP developing seeds of lines with different BLI levels. The transcript levels of BIOTIN CARBOXYL CARRIER PROTEIN 2 (BCCP2), ACYL CARRIER PROTEIN 1 (ACP1), KETOACYL ACP SYNTHASE 1 (KAS1), KAS3, FATTY ACID ELONGASE 1 (FAE1), OLEOSIN 1 (OLE1), SUCROSE SYNTHASE 2 (SUS2), CHLOROPLAST PYRUVATE KINASE α (Ch-PKα), CYTOSOL PYRUVATE KINASE β (Cy-PKβ), PHOSPHOLIPASE A2α (PLA2α), ENOLASE 1 (ENO1), FATTY ACID DESATURASE 2 (FAD2), and FAD3 decreased by two- to three-fold in bli-1 compared to WT seeds (Figure 4F). All of these genes were significantly upregulated in seeds of the BLI overexpression lines (Figure 4F). Taken together, these results suggest that BLI promotes fatty acid accumulation during seed maturation by regulating the expression of genes involved in glycolysis, fatty acid biosynthesis and modification, and TAG accumulation.
Genetic interaction of BLI and WRI1 in regulating seed maturation and fatty acid biosynthesis
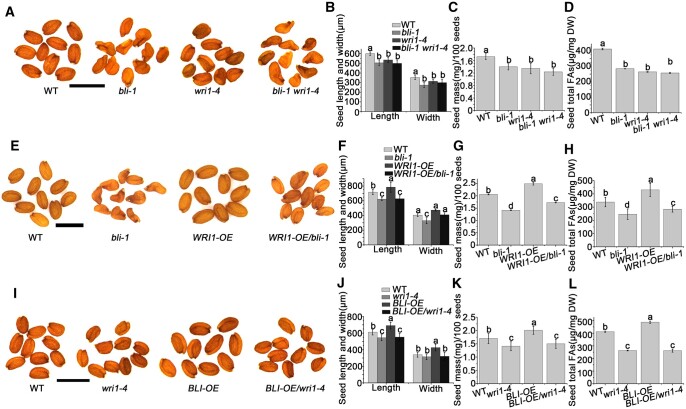
To investigate whether BLI and WRI1 function together in seed maturation, we generated BLI overexpression lines (BLI-OE) in the WT and wri1–4 backgrounds and WRI1 overexpression lines (WRI1-OE) in the WT and bli-1 backgrounds and constructed wri1–4 bli-1 double mutant plants. bli-1 wri1–4 double mutant seeds were wrinkled, like bli-1 seeds (Figure 5, A and B). However, the seed mass and total fatty acid content of bli-1 wri1–4 seeds were similar to those of wri1–4 seeds (Figure 5, C and D). Importantly, WRI1 overexpression in the bli-1 background partially rescued the wrinkled phenotype and decreased fatty acid content of bli-1 seeds (Figure 5, E–H; Supplemental Figure S12), whereas BLI overexpression did not rescue the seed phenotype and decreased FA content of wri1–4 seeds (Figure 5, I–L). Thus, BLI regulates fatty acid content in a WRI1-dependent manner.
Figure 5.
The genetic relationship between BLI and WRI1. A, Mature seed phenotypes of WT, bli-1, wri1–4, and bli wri1–4 plants determined by microscopy. Bars = 1 mm. B, Seed size (length and width) of WT, bli-1, wri1–4, and bli-1 wri1–4. C, Seed mass of WT, bli-1, wri1–4, and bli-1 wri1–4. D, Fatty acid contents in WT, bli-1, wri1–1, and bli-1 wri1–4 seeds. E, Mature seed phenotypes of WT, bli-1, WRI1-OE/WT, and WRI1-OE/bli-1 plants determined by microscopy. Bars = 1 mm. F, Seed size (length and width) of WT, bli-1, WRI1-OE/WT, and WRI1-OE/bli-1. G, Seed mass of WT, bli-1, WRI1-OE/WT, and WRI1-OE/bli-1. H, Fatty acid contents in WT, bli-1, WRI1-OE/WT, and WRI1-OE/bli-1 seeds. I, Mature seed phenotypes of WT, wri1–4, BLI-OE/WT, and BLI-OE/wri1–4 plants determined by microscopy. Bars = 1 mm. J, Seed size (length and width) of WT, wri1–4, BLI-OE/WT, and BLI-OE/wri1–4. K, Seed mass of WT, wri1–4, BLI-OE/WT, and BLI-OE/wri1–4. L, Fatty acid contents in WT, wri1–4, BLI-OE/WT, and BLI-OE/wri1–4 seeds. For seed size measurements, values are means ± sd (N = 3); each of the three assays for each biological replicate contained 50 seeds. For seed mass, the data shown are means ± SD (N = 3); each of the three assays for each biological replicate contained 100 seeds. Seed size or seed mass do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05). For fatty acid contents, data shown are means ± sd (N = 3). The contents do not differ significantly if they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05).
We analyzed the fatty acid composition in the seeds of WT, wri1–4, bli-1, wri1–4 bli-1, and transgenic plants with altered BLI and/or WRI1 level. The C18:3, C20:1, C20:2, and C22:1 levels relative to total fatty acid levels were significantly lower in bli-1 compared to the WT, whereas the relative amounts of C16:0 and C18:1 were higher (Table 1). However, there were no significant changes in fatty acid composition in BLI-OE seeds (Table 1). Consistent with previous results (Baud et al., 2007), the total fatty acid content of wri1–4 seeds was reduced by ∼31% compared to the WT (Figure 5D). This reduction was accompanied by increases in C18:3, C20:0, C20:2, and C22:1 levels and decreases in C16:0, C18:0, C18:1, and C18:2 levels (Table 1). Moreover, WRI1 overexpression increased the amount of total fatty acids by ∼20% without altering fatty acid composition compared to WT seeds (Figure 5H; Table 1).
Table 1.
BLI regulates the fatty acid composition in seeds in conjunction with WRI1
| Fatty Acid Composition | WT (mol%) | bli-1 (mol%) | BLI-OE (mol%) | wri1–4 (mol%) | WRI1-OE (mol%) | bli-1 wri1–4 (mol%) | WRI1-OE/bli-1 (mol%) | BLI-OE/wri1–4 (mol%) |
|---|---|---|---|---|---|---|---|---|
| C16:0 | 8.59 ± 0.10a | 10.02 ± 0.04b | 8.44 ± 0.04a | 5.87 ± 0.03c | 8.12 ± 0.11a | 9.95 ± 0.07b | 9.75 ± 0.21b | 6.17 ± 0.08c |
| C18:0 | 3.27 ± 0.01a | 3.66 ± 0.01b | 3.36 ± 0.03a | 2.06 ± 0.03c | 2.92 ± 0.01a | 3.37 ± 0.01a | 3.68 ± 0.05b | 2.27 ± 0.14c |
| C18:1 | 12.89 ± 0.17a | 19.83 ± 0.08c | 11.72 ± 0.11b | 10.56 ± 0.10b | 13.18 ± 0.20a | 21.60 ± 0.12d | 20.21 ± 0.17c | 9.90 ± 0.45b |
| C18:2 | 31.63 ± 0.13a | 32.47 ± 0.02a,b | 30.90 ± 0.11a | 24.63 ± 0.01c | 30.73 ± 0.11a | 32.13 ± 0.38a | 31.96 ± 0.02a | 26.38 ± 0.33c |
| C18:3 | 18.65 ± 0.22a | 16.23 ± 0.04c | 18.89 ± 0.24a | 20.19 ± 0.28b | 18.60 ± 0.02a | 14.78 ± 0.06d | 16.20 ± 0.30c | 20.52 ± 0.44b |
| C20:0 | 2.01 ± 0.11a | 1.85 ± 0.01a,b | 2.43 ± 0.01a,c | 3.65 ± 0.02c | 2.15 ± 0.01a | 1.53 ± 0.02b | 1.77 ± 0.02a,b | 3.56 ± 0.18c |
| C20:1 | 19.22 ± 0.52a | 14.12 ± 0.05c | 20.35 ± 0.17b | 20.52 ± 0.15b | 20.32 ± 0.04b | 14.84 ± 0.33c | 14.51 ± 0.09c | 19.21 ± 0.02a |
| C20:2 | 2.03 ± 0.06a | 1.02 ± 0.01b | 2.08 ± 0.01a | 3.84 ± 0.02c | 2.13 ± 0.02a | 0.94 ± 0.03b | 1.05 ± 0.03b | 3.60 ± 0.25c |
| C22:1 | 1.71 ± 0.07a | 0.80 ± 0.01b | 1.84 ± 0.04a | 8.68 ± 0.05c | 1.85 ± 0.01a | 0.87 ± 0.01b | 0.87 ± 0.02b | 8.39 ± 0.05c |
The contents of different fatty acids were detected in bli-1, BLI-OE, wri1–4, WRI1-OE, bli-1 wri1–4, WRI1-OE/bli-1, and BLI-OE/wri1–4 seeds compared to WT seeds. Data are means ± se. Three technical replicates were performed for each of three biological replicates. The same letter indicates that the mean contents do not differ significantly according to Tukey’s HSD test (P = 0.05).
These results suggest that BLI and WRI1 have opposite roles in regulating long-chain fatty acid biosynthesis. Thus, we analyzed the fatty acid composition of bli-1 wri1–4 double mutant seeds, revealing an identical long-chain fatty acid composition in this mutant and the bli-1 single mutant (Table 1), suggesting that BLI acts epistatically to WRI1 in regulating long-chain fatty acid biosynthesis. To investigate possible roles of BLI and WRI1 in cooperatively regulating fatty acid composition, we examined the fatty acid compositions in BLI-OE/wri1–4 and WRI1-OE/bli-1 seeds. Overexpression of BLI or WRI1 did not rescue the altered total fatty acid composition of wri1–4 or bli-1, respectively (Table 1).
BLI associates with the promoters of WRI1 target genes and requires WRI1 to promote transcriptional activation
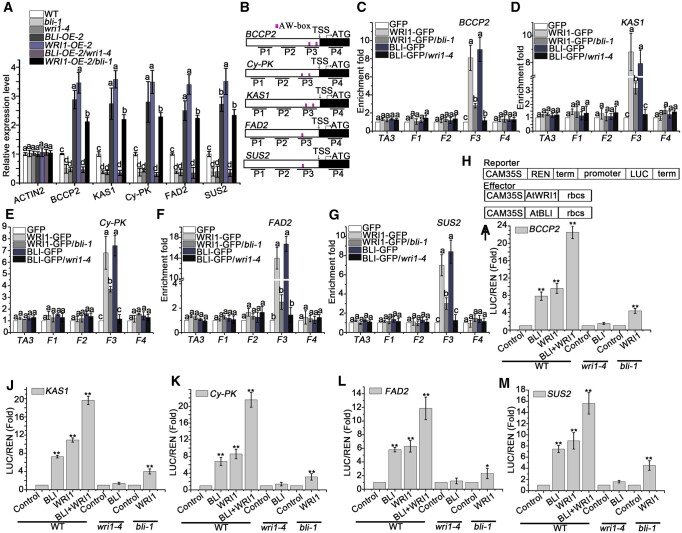
To further understand how BLI and WRI1 regulate fatty acid biosynthesis at the molecular level, we analyzed the transcript levels of 13 genes involved in glycolysis, fatty acid biosynthesis, and modification, and TAG accumulation (see also above) in early-stage developing seeds (3–18 DAP) of different genetic backgrounds (Figure 6A). Overexpression of BLI or WRI1 in the WT background (BL1-OE-2 and WRI1-OE-2) increased the transcript levels of WRI1 target genes BCCP2, KAS1, Cy-PKβ, FAD2, and SUS2 by approximately three-fold compared to WT plants (Figure 6A) and upregulated ACP1, Ch-PKα, ENO1, FAD3, FAE1, KAS3, PLA2α, and OLE1 (Supplemental Figure S13). These genes were downregulated in wri1–4 and bli-1. The reduced expression of these genes could be rescued by WRI1 overexpression in the WRI1-OE/bli-1 line, whereas none of the genes were significantly upregulated by BLI overexpression in the BLI-OE/wri1–4 line. Intriguingly, the expression levels of these genes in the bli-1 wri1–4 double mutant were similar to those in the wri1–4 or BLI-OE/wri1–4 lines (Figure 6A). These results suggest that BLI1 acts as a transcriptional activator and depends on WRI1 function to regulate the expression of glycolysis-related and fatty acid biosynthetic genes.
Figure 6.
BLI regulates the expression of WRI1 target genes associated with WRI1 and the transcriptional activity of WRI1. A, RT-qPCR analysis of WRI1 target genes in 12 DAP bli-1, BLI-OE, wri1–4, WRI1-OE, WRI1-OE/bli-1, and BLI-OE/wri1–4 seeds compared to the WT. Data shown are means ± sd (N = 3). UBQ10 was used as a reference gene, and ACTIN2 was used as a negative control. The mean relative expression levels do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05). B, Schematic diagram of ChIP-qPCR-detected sequences in the WRI1 target genes. White box represents promoters; Magenta vertical line indicates AW-box or AW-box like cis-element sites; Black line indicates ChIP-qPCR-detected sequence; F1–F4 indicate different DNA fragments. C–G, Occupancy of BLI and WRI1 on the promoters of fatty acid biosynthesis genes BCCP2 (C), KAS1 (D), Cy-PK (E), FAD2 (F), and SUS2 (G) in pBLI:BLI-GFP/bli-1 (BLI-GFP), BLI-GFP/wri1–4, pWRI1:WRI1-GFP/wri1-4 (WRI1-GFP), and WRI1-GFP/bli-1 seed compared to GFP control seeds. Data shown are means ± sd (N = 3). The means of enrichment folds do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05). H, Scheme of the constructs used for dual-LUC assay. The reporter construct consists of the CaMV 35S promoter, five repeats of the GAL4 binding sequence (5xGAL4BS), NOS terminator (NOSter), and firefly LUC coding sequence. Effector constructs express the GAL4 DNA-binding domain (GAL4DB)-fused protein under the control of the CaMV 35S promoter. I–M, Effects of BLI or/and WRI1 on BCCP2 (I), KAS1 (J), Cy-PK (K), FAD2 (L), and SUS2 (M) transcriptional regulation in the WT, bli-1 and wri1–4 Arabidopsis protoplast transient expression assay. Data are means ± sd (n = 3 experiments). Asterisks denote significant difference compared to the control, as determined by Student’s t test (*P < 0.05; **P < 0.01).
One possible scenario is that BLI is recruited to the promoters of WRI1 target genes by WRI1. To explore this possibility, we performed ChIP experiments to address whether BLI binds to the WRI1 target gene loci using the pBLI:BLI-GFP/bli-1 (BLI-GFP) transgenic line in which the bli-1 morphological defects were rescued by the functional transgene. WRI1 binds to the AW box-containing regions [CnTnG](n)7[CG] of fatty acid biosynthesis genes to promote their expression (Maeo et al., 2009). In developing seeds, the AW motif-containing regions of the promoters of WRI1 target loci BCCP2, KAS1, Cy-PKβ, FAD2, and SUS2 were enriched in the anti-GFP immunoprecipitant from BLI-GFP relative to the GFP controls (Figure 6, B–G). The association of BLI to these promoters is dependent on WRI1, as BLI promoter occupancy was lost in BLI-GFP/wri1–4 seeds (Figure 6, C–G). WRI1 occupancy at AW motif-containing regions was significantly reduced, but not completely lost, in WRI1-GFP/bli-1 compared to WRI1-GFP plants (Figure 6, C–G). Therefore, BLI might regulate the association of WRI1 to fatty acid biosynthesis genes in vivo by directly affecting WRI1 recruitment, protein level, or activity or indirectly by BLI’s effect on WRI1 target gene expression. These results indicate that BLI is recruited to and associates with the promoters of WRI1 target genes to regulate their transcription.
To investigate whether BLI possesses transcriptional activity in vivo, we performed a dual-luciferase (LUC) assay in Arabidopsis protoplasts. The dual-LUC reporter harbored five copies of the GAL4 DNA-binding element and CaMV 35S fused to the firefly LUC reporter, whereas a Renilla (REN) LUC reporter under the control of the 35S promoter was used as an internal control. The protein-coding regions of BLI or WRI1 were fused to the yeast GAL4 DNA-binding domain (GAL4-BLI or GAL4-WRI; Figure 6H). A reporter construct harboring the firefly LUC driven by the BCCP2 promoter was constructed to assay the transcriptional activity of BLI and/or WRI1 to regulate BCCP2 expression.
Following co-transformation of the BLI or WRI1 effector and reporter constructs, the relative LUC/REN activity increased by ∼6.5- or 8.5-fold, respectively, compared to transformation of the reporter only (Figure 6I). Intriguingly, the relative LUC activity increased by ∼21-fold after co-transformation of BLI and WRI1 effectors at the same time (Figure 6I). Similar experiments revealed that BLI enhanced the transcription of KAS1, Cy-PKβ, FAD2, and SUS2 (Figure 6, J–M). Whereas BLI-induced reporter gene activation was lost in the wri1–4 mutant, WRI1 still activated the reporters in the bli-1 mutant, albeit with reduced efficiency (Figure 6, I–M). Thus, BLI-dependent transcriptional activation is dependent on WRI1, which is consistent with the reduced expression of fatty acid biosynthesis genes in the bli-1 mutant (Figures 4, E and 6, A). To determine whether BLI promotes the transcriptional activity of WRI1 in stable transgenic lines, we examined the seed phenotypes and total fatty acid contents of seeds of the co-overexpression lines (BLI-OE/WRI1-OE), revealing slightly larger seeds and higher fatty acid contents than either BLI-OE or WRI1-OE seeds (Supplemental Figure S14).
BLI regulates nucleosomal occupancy and histone modification at WRI1 target loci
To gain insight into the mechanism by which BLI promotes WRI1 target gene expression during the seed maturation progress, we examined the nucleosome positioning and occupancy at the WRI1 target gene promoters using high-resolution micrococcal nuclease (MNase) mapping (Chodavarapu et al., 2010; Rafati et al., 2011). We identified one well-positioned nucleosome in the WRI1 target gene promoter region (-1 nucleosome) upstream of a 100-bp nucleosome-depleted region (−100 to 0 bp) in BCCP2, KAS1, Cy-PKβ, FAD2, and SUS2 (Figure 7, A–E). Moreover, the −1 or −2 nucleosome at the WRI1 target loci protected ∼150 bp of genomic DNA from MNase digestion (Figure 7, A–E), which is consistent with the typical protection of 147 bp by a nucleosome (Yen et al., 2012). Intriguingly, the −1 nucleosome positions at WRI1 target genes overlapped with the WRI1-binding DNA elements (Figure 7, A–E), suggesting that WRI1 or WRI1-interacting proteins may be important for nucleosome positioning or the maintenance of occupancy. Thus, we examined the nucleosome positioning and occupancy of the WRI1 target genes in wri1–4, bli1-1, BLI-OE, and BLI-OE/wri1–4 compared to WT plants. In bli-1 and wri1–4 plants, we observed decreased WRI target gene expression (Figure 6A) and a consistent increase in nucleosomal occupancy at the -1 nucleosome positions of KAS1, Cy-PKβ, FAD2, and SUS2 and at the -2 nucleosome of BCCP2 (Figure 7, A–E). In contrast, the nucleosome occupancy in BLI-OE plants was strongly reduced (Figure 7, A–E). Intriguingly, the decreased nucleosome occupancy in BLI-OE plants was reversed and increased in BLI-OE/wri1–4 plants (Figure 7, A–E). However, no BLI-dependent alteration in nucleosome positioning was observed at the -2 or -1 nucleosome at the WRI1 target gene loci (Figure 7, A–E). These results suggest that BLI is required to inhibit high occupancy at the −1 or −2 nucleosome at WRI1 target loci and that this function requires the presence of WRI1.
Figure 7.
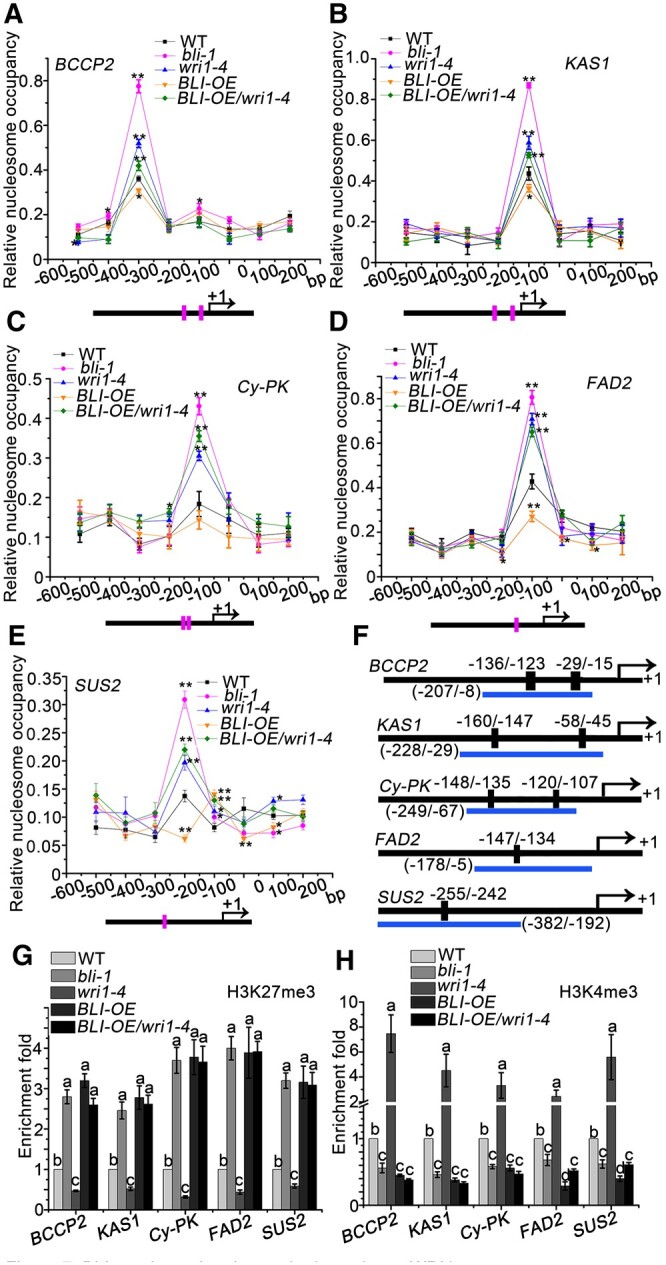
BLI regulates the chromatin dynamics at WRI1 target genes. A–E, MNase digestion followed by tiled primer qPCR to monitor nucleosome positioning and occupancy at the BCCP2 (A), KAS1 (B), Cy-PK (C), FAD2 (D), and SUS2 (E) locus in WT, bli-1, wri1–4, BLI-OE, and BLI-OE/wri1–4 plants. The number on the x-axis denotes distance (bp) from the TSS (0 bp). Horizontal black lines below the scheme represent the gene loci; Magenta vertical line indicates AW-box or AW-box like cis-element sites. Values are mean ± se (standard error) of three technical replicates from one representative experiment. Asterisks denote significant difference compared to the control, as determined by Student’s t test (*P < 0.05; **P < 0.01). F, Diagram of the fatty acid biosynthesis gene loci tested for histone modifications. Horizontal blue lines below the scheme, regions amplified by qPCR; vertical black rectangles, AW-motifs; black box before arrows, promoter regions; arrows, the TSSs. G, qPCR after anti-H3K27me3 ChIP in plants with different BLI genotypes. H, qPCR after anti-H3K4me3 ChIP in plants with different BLI genotypes. Relative enrichment is the percentage of input fold change after the percentage of input of the WT was set to 1. Data shown are means ± sd (N = 3). Means of enrichment folds do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05).
BLI interacts with CLF to control the expression of PcG target genes and cellular differentiation (Schatlowski et al., 2010). PcG proteins are involved in the establishment and maintenance of a repressed chromatin state by setting the H3K27me3 mark. PcG silencing is counteracted by the activity of Trithorax group (TrxG) proteins, which catalyze H3K4me3. We, therefore, examined whether BLI and WRI1 are required for H3K4me3 and/or H3K27me3 deposition at WRI1 target gene loci in seeds by performing chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR). The levels of H3K27me3 at the AW box-containing regions of WRI1 target gene loci were higher in bli-1 and wri1–4 compared to WT plants (Figure 7, F and G), but other regions lacking the AW box were not enriched by the H3K27me3 antibody in bli-1 (Supplemental Figure S15). Moreover, H3K27me3 levels were reduced at the AW motif-containing regions in BLI-OE compared to WT plants (Figure 7G), but this was reversed and increased in BLI-OE/wri1–4 plants (Figure 7G), suggesting that the BLI-dependent H3K27me3 changes are dependent on WRI1. Reciprocal observations were made for H3K4me3, with a decrease of H3K4me3 at WRI1 target genes in bli-1 and wri1–4 seeds and a WRI1-dependent increase of H3K4me3 in BLI-OE seeds (Figure 7H). These results suggest that BLI interacts with multiple chromatin regulatory proteins and mediates WRI1 target recognition and chromatin remodeling.
BLI functions as adaptor protein linking WRI1 to chromatin remodeling factors
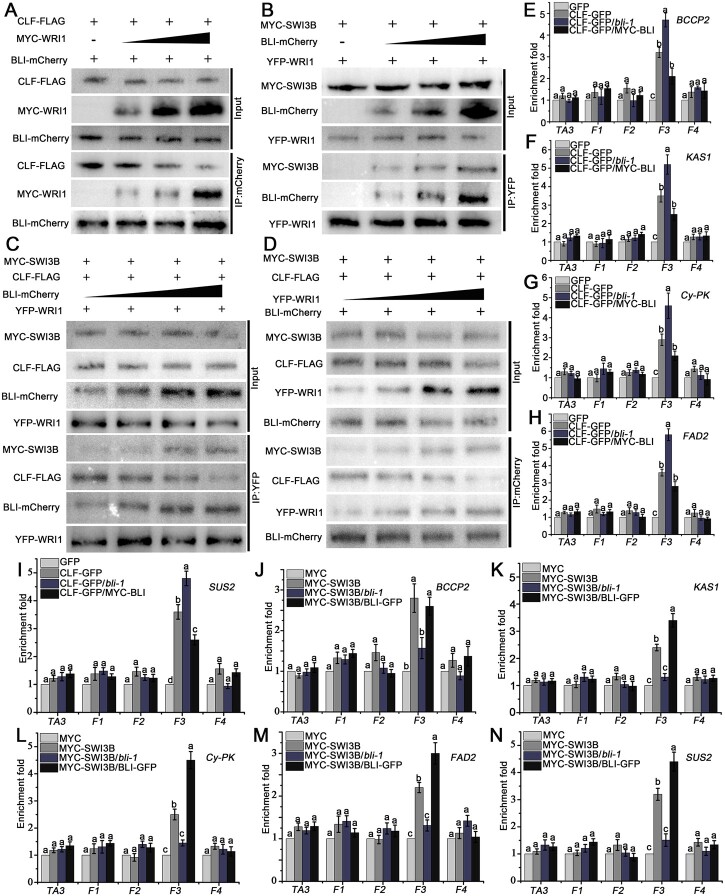
Besides CLF, BLI interacted with SWI3B, a core subunit of the Arabidopsis SWI2/SNF2 chromatin remodeling complex, using YTH and BiFC analyses (Supplemental Figure S16). To further explore the mechanism by which overexpression of BLI or WRI1 causes changes in nucleosome occupancy and histone modification at WRI1 target genes, we analyzed the interactions between WRI1, BLI, and CLF or SWI3B using Co-IP. Interestingly, in the absence of WRI1, BLI strongly interacted with CLF, which is consistent with previous studies. However, an increase in WRI1 protein levels interfered with the interaction between BLI and CLF in a dosage-dependent manner (Figure 8A). These results indicate that WRI can competitively inhibit the interaction between CLF and BLI by interacting with BLI.
Figure 8.
BLI associates with WRI1 to repress CLF and recruit SWI3B during regulating WRI1 target genes. A, WRI1 and CLF competitively interact with BLI, as determined in an in vivo Co-IP. Total protein extracts from transformed WT protoplasts carrying both 35S:BLI-mCherry and 35S:CLF-FLAG without or with different amounts of 35S:MYC-WRI1 were immunoprecipitated with the immobilized anti-mCherry antibody. B, WRI1 and SWI3B synergistically interact with BLI, as determined in an in vivo Co-IP assay. Total protein extracts from transformed WT protoplasts carrying both 35S: YFP-WRI1 and 35S:MYC-SWI3B without or with different amounts of 35S:BLI-mCherry were immunoprecipitated with the immobilized anti-GFP antibody. C and D, CLF and SWI3B competitively interact with the BLI–WRI1 complex, as determined in an in vivo Co-IP assay. Total protein extracts from transformed 35S:YFP-WRI1 (C) or 35S:BLI-mCherry (D) protoplasts carrying both 35S:MYC-SWI3B and 35S:CLF-FLAG with different amounts of 35S:BLI-mCherry (C) or 35S:YFP-WRI1 (D) were immunoprecipitated with the immobilized anti-GFP (C) or anti-mCherry (D) antibody. The variable amounts of tagged BLI and WRI1 protein were achieved by adding 1, 2, 3, or 4 times the amount of expression vector, and the proteins from crude lysates (Input) and IPs were detected using different antibodies (A–D). E–I, Occupancy of CLF on the promoters of fatty acid biosynthesis genes BCCP2 (E), KAS1 (F), Cy-PK (G), FAD2 (H), and SUS2 (I) in CLF-GFP, CLF-GFP/bli-1, and CLF-GFP/MYC-BLI seeds compared to GFP control seeds. J–N, Occupancy of SWI3B on the promoters of fatty acid biosynthesis genes BCCP2 (J), KAS1 (K), Cy-PK (L), FAD2 (M), and SUS2 (N) in SWI3B-MYC, SWI3B-MYC/bli-1, and SWI3B-MYC/BLI-GFP seed compared to MYC control seeds. F1–F4 indicate different DNA fragments, as shown in Figure 6B; the TA3 locus was used as a negative control. Data shown are means ± sd (N = 3). Means of enrichment folds do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05).
We also tested the interactions of BLI, WRI1, and SWI3B. WRI1 and SWI3B did not interact when BLI was missing; however, with increasing BLI protein levels, the interaction between WRI1 and SWI3B was gradually enhanced (Figure 8B). Thus, BLI may function as an adaptor protein by simultaneously interacting with WRI1 and SWI3B to promote the indirect interaction between WRI1 and SWI3B to form a complex and recruit chromatin remodeling activity.
Furthermore, we analyzed the interactions of WRI1, BLI, CLF, and SWI3B by Co-IP when all proteins were present at the same time. An increase in BLI protein levels resulted in the enhanced co-precipitation of SWI3B when WRI1 was immunoprecipitated, whereas less CLF was precipitated. When WRI1 was immunoprecipitated, the interaction between BLI and SWI3B was enhanced with increasing amounts of BLI, while the interaction with CLF was weakened (Figure 8C). Moreover, an increase in WRI levels resulted in weakened co-precipitation of CLF when BLI was immunoprecipitated, whereas more SWI3B was precipitated. When BLI was immunoprecipitated, the interaction between WRI1 and SWI3B was enhanced with increasing amounts of WRI1, while the interaction with CLF was weakened (Figure 8D). Thus, the presence of WRI1 may switch BLI’s function in CLF interaction and PcG-mediated repression to activate genes by recruiting SWI3B and chromatin remodeling.
To obtain further evidence for these interactions in planta, we used ChIP-qPCR to detect the binding of CLF and SWI3B to the promoter regions of WRI1 target genes in seeds of with different BLI levels. CLF binding to the promoter regions of WRI1 target genes increased in bli-1 seeds, whereas less CLF protein bound to the promoter regions of WRI1 target genes in the seeds of BLI overexpression plants (Figure 8, E–I). In contrast, the ability of the SWI3B protein to bind to the promoters of WRI1 target genes was lost in the bli-1 mutant, while it was significantly enhanced in BLI overexpression plants (Figure 8, J–N). The SWI3B expression level was maintained during seed maturation, whereas CLF expression gradually decreased (Supplemental Figure S17).
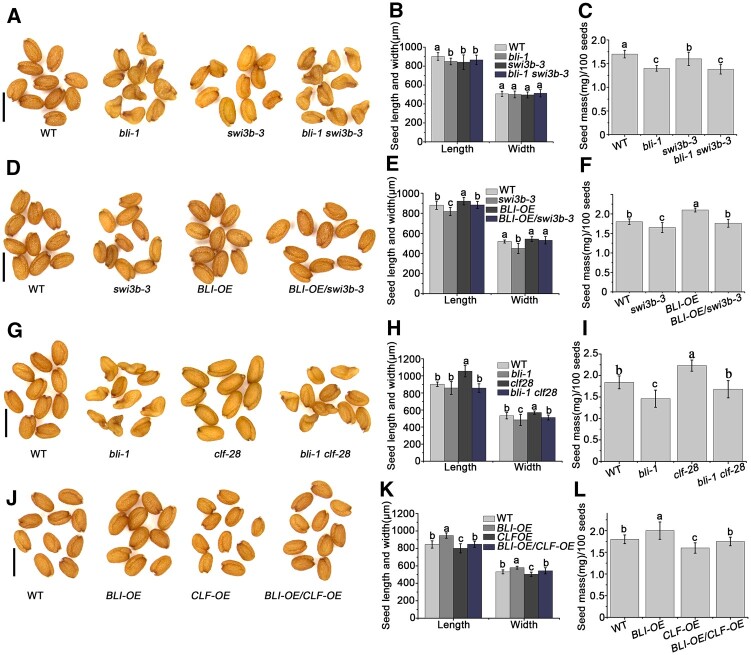
To investigate whether SWI3B is involved in seed maturation, we analyzed the seed phenotype of a weak allele of SWI3B (swi3b-3, carrying one single amino acid substitution), since the knockout alleles are known to be embryo-lethal (Saez et al., 2008). The seed size and seed mass of swi3b-3 were reduced compared to WT seeds (Figure 9, A–F). We introduced BLI-OE in the swi3b-3 background, and as expected, the increased seed size and mass of BLI-OE were reversed by the SWI3B mutation in BLI-OE/swi3b-3 (Figure 9, D–F). Furthermore, bli-1 swi3b-3 and SWI3B-OE/bli-1 seeds were wrinkled, like bli-1 seeds (Figure 9, A–C; Supplemental Figure S18). These results indicate that BLI and SWI3B function together in seed maturation and that SWI3B is required for BLI function. On the other hand, the seed phenotype of bli-1 was partially rescued by clf-28, and that of BLI-OE was reversed by CLF-OE. This suggests that BLI-promoted seed maturation is dependent on the inhibition of CLF (Figure 9, G–L; Supplemental Figure S16). These results indicate that BLI inhibits or promotes the regulation of WRI1 target genes by interacting with CLF or SWI3B, respectively.
Figure 9.
The genetic relationship between BLI and SWI3B or CLF in regulating seed maturation. A, Mature seed phenotypes of WT, bli-1, swi3b-3, and bli-1 swi3b-3 plants determined by microscopy. Bars = 1 mm. B, Seed size (length and width) of WT, bli-1, swi3b-3, and bli-1 swi3b-3. C, Seed mass of WT, bli-1, swi3b-3 and bli-1, swi3b-3. D, Mature seed phenotypes of WT, swi3b-3, BLI-OE, and BLI-OE/swi3b-3 plants determined by microscopy. Bars = 1 mm. E, Seed size (length and width) of WT, swi3b-3, BLI-OE, and BLI-OE/swi3b-3. F, Seed mass of WT, swi3b-3, BLI-OE, and BLI-OE/swi3b-3. G, Mature seed phenotypes of WT, bli-1, clf-28, and bli-1 clf-28 plants determined by microscopy. Bars = 1 mm. H, Seed size (length and width) of WT, bli-1, clf-28, and bli-1 clf-28. I, Seed mass of WT, bli-1, clf-28, and bli-1 clf-28. J, Mature seed phenotypes of WT, BLI-OE, CLF-OE, and BLI-OE/CLF-OE plants determined by microscopy. Bars = 1 mm. K, Seed size (length and width) of WT, BLI-OE, CLF-OE, and BLI-OE/CLF-OE. L, Seed mass of WT, BLI-OE, CLF-OE, and BLI-OE/CLF-OE. For seed size measurements, values are means ± sd (N = 3); each of the three assays for each biological replicate contained 50 seeds. For seed mass, data shown are means ± sd (N = 3); each of the three assays for each biological replicate contained 100 seeds. Seed size or seed mass do not significantly differ when they are labeled with the same letter, as determined by Tukey’s HSD test (P < 0.05).
Discussion
Here, we dissected the function of BLI in seed maturation and fatty acid accumulation. We demonstrated that BLI physically interacts with WRI1 and binds to the promoters of fatty acid biosynthesis genes to regulate seed maturation. Moreover, we showed that BLI regulates nucleosomal occupancy and histone modifications to regulate gene expression.
BLI regulates seed maturation
Seed maturation is an important stage of seed development during which embryos gradually accumulate seed storage proteins and oils, both of which account for 30%–40% of dry seed matter (Mansfield et al., 1992; Raz et al., 2001; Baud et al., 2002). Oil accumulate in lipid droplets in the form of TAG, which occupy ∼60% of the volume of cotyledon cells in the mature embryo. In the bli mutant, the maturation process is delayed and disturbed, accompanied by a significant decrease in seed oil content (Figure 2). Gene expression analysis revealed reduced transcript levels of FUS3, ABI3, and WRI1, whereas LEC1 and LEC2 are upregulated in the bli mutant (Figure 4E).
LAFL genes form highly complex regulatory networks that participate in seed development and the accumulation of storage materials. For example, LEC1 and LEC2 are positive upstream regulators of WRI1, ABI3, and FUS3 and together with WRI1, ABI3, and FUS3, control the accumulation of lipids and storage proteins in seeds by regulating gene expression (Mu et al., 2008). In addition, ABI3 and FUS3 are subject to feedback regulation by regulating their own or each other’s expression (To et al., 2006). Thus, LAFL and WRI1 expression homeostasis may be altered in the bli mutant. For example, LEC1 and LEC2 may be upregulated to compensate for the adverse effects resulting from the downregulation of ABI3, FUS3, and WRI1 due to the loss of function of BLI. During seed maturation, BLI regulates WRI1 expression and is an important factor that regulates the accumulation of storage materials by maintaining expression homeostasis among LAFL TFs. Whether LAFL genes and WRI1 are direct targets of BLI remains to be elucidated. BLI might associate with WRI1 to promote its capacity for auto-regulation via binding to the promoter of WRI1. Whether the regulation of WRI1 expression by BLI is conserved in other plant species requires further investigation.
The identification of positive regulators of fatty acid biosynthesis is very important for agricultural production. Transgenic Arabidopsis, maize, Brachypodium distachyon, and soybean overexpressing AtWRI1 or WRI1 orthologs or WRI1 activators show elevated seed oil content (Cernac and Benning, 2004; Shen et al., 2010; Yang et al., 2015; Zhang et al., 2016, 2017). However, irregular growth or cell death was observed in some transgenic plants overexpressing WRI1 (Cernac and Benning, 2004; Yang et al., 2015). Thus, an increase in oil production in oil crops should avoid WRI1 overexpression. BLI promotes fatty acid contents by enhancing WRI1 activity without affecting WRI1 expression (Figures 4E and 6). The positive effect of WRI1 overexpression is abrogated when BLI is lacking, suggesting that BLI is important for fatty acid accumulation and seed maturation. Thus, enhancing BLI expression in oil crops may overcome undesirable effects associated with overexpression of WRI1 in transgenic plants. BLI is a plant-specific gene that is widely conserved in plants genomes (Schatlowski et al., 2010). Thus, identifying the roles of BLI in plant lipid metabolism in other plant species may permit the modification of fatty acid composition and levels.
BLI is required by WRI1 to regulate the expression of WRI1 target genes
Although WRI1 was cloned and characterized more than 15 years ago (Cernac and Benning, 2004), little is known about how its transcriptional activity is mediated. Here, we identified the PcG-associated protein BLI as an interacting partner of WRI1, suggesting a connection between target gene binding and chromatin regulation. BLI associates with the promoter regions of five WRI1 target genes that harbor a conserved AW sequence motif in their proximal upstream regions. As the reduced expression of WRI1 target genes in WRI1-OE/bli plants did not reach the level observed in WRI1-OE plants, WRI1 transcriptional activity is largely dependent on the function of BLI (Figure 6). BLI has transcription activation potential and may be a transcriptional co-regulator of fatty acid biosynthesis genes, as its transcription activity is dependent on WRI1 (Figure 6, H–M).
Overall, our data support a model in which WRI1/BLI function in a complex promoting the transcription of fatty acid biosynthesis genes. However, the relationship between BLI and WRI1 is complex. The bli seed phenotypes, including decreased fatty acid contents and elevated starch and sucrose contents, are similar to those of wri1 seeds (Focks and Bennings, 1998). bli seeds also showed obvious seedling establishment phenotypes similar to wri1 seeds when germinated in the absence of externally added sucrose (Supplemental Figure S19; Focks and Bennings, 1998), indicating that BLI and WRI1 function in the same pathway. Moreover, the partially rescued seed phenotypes, fatty acid content, and fatty acid biosynthesis gene expression by overexpressing WRI1 in bli mutants suggest that the role of BLI in regulating seed maturation is partially dependent on WRI1. Furthermore, the decreased WRI1 activity in bli mutants indicates that the function of WRI1 in regulating fatty acid biosynthesis is dependent on BLI. Thus, BLI and WRI1 can act in an interdependent manner to regulate fatty acid content and seed maturation.
On the other hand, bli seeds have a different fatty acid composition from wri1 seeds, resembling the fatty acid composition of WT seeds at the mid-maturation stage (Table 1). In addition, bli seeds showed some phenotypes, such as abnormal embryo hypocotyls and altered accumulation of seed storage proteins, which are not observed in wri1 mutants (Focks and Bennings, 1998). Thus, the oil accumulation defect observed in bli mutants may be partially due to the arrest in embryo maturation. These data suggest that BLI also has some unique functions in regulating seed maturation independent of WRI1. BLI is a multifunctional protein involved in the activation or repression of gene expression (Schatlowski et al., 2010; Kleinmanns et al., 2017). Thus, in addition to WRI1, BLI may recruit or be recruited by other interacting components to activate or repress seed maturation-related gene expression.
Intriguingly, the motifs that interact with BLI at the C-terminus of WRI1 include the TAD and PEST domains, both of which are very important for the function and stability of WRI1 (Ma et al., 2015; Figure 1). Thus, BLI may affect WRI1 transcriptional activity and stability. Consistent with this hypothesis, WRI1 activity is inhibited or reduced in the bli mutant (Figure 6, H–M). Potential phosphorylation residues have been identified in WRI1, and some have been experimentally shown to be vital for modulating the stability and function of WRI1 (Ma et al., 2015, 2016; Zhai et al., 2017). Interestingly, the phosphorylation level of WRI1 was clearly altered in the bli mutant (Supplemental Figure S20), indicating a mechanism by which BLI may modulate WRI1 activity. Thus, the identification and characterization of WRI1 kinases that interact with or are regulated by BLI would be quite interesting. The BLI protein possesses different motifs and domains, including the IDRs, an structural maintenance of chromosomes-like (SMC-like) domain, and CC domains (Schatlowski et al., 2010). However, the functions of these motifs and domains are currently unclear. Thus, it will be important to illuminate the functions of different BLI motifs and domains in more detail; future work should focus on investigating BLI dynamics regulated by protein–protein interactions and in response to cellular signals and environmental cues.
BLI is an epigenetic regulator of seed maturation
SMC proteins are involved in various processes of chromatin biology including chromosome condensation, sister chromatid cohesion, and DNA repair (Uhlmann et al., 2016). The SMC-like domain of BLI is essential for the interaction between BLI and WRI1 (Figure 1) or BLI and CLF (Schatlowski et al., 2010). We observed that WRI1 weakened the association of BLI with CLF, perhaps by competing with CLF for binding to the SMC-like domain (Figure 8A). CLF negatively regulates fatty acid biosynthesis by increasing H3K27me3 levels (Liu et al., 2016). Thus, BLI may inhibit CLF function through protein–protein interaction to moderate H3K27me3 levels. Consistent with this idea, BLI regulates the expression of several PcG target genes but likely also has PcG-independent functions (Schatlowski et al., 2010).
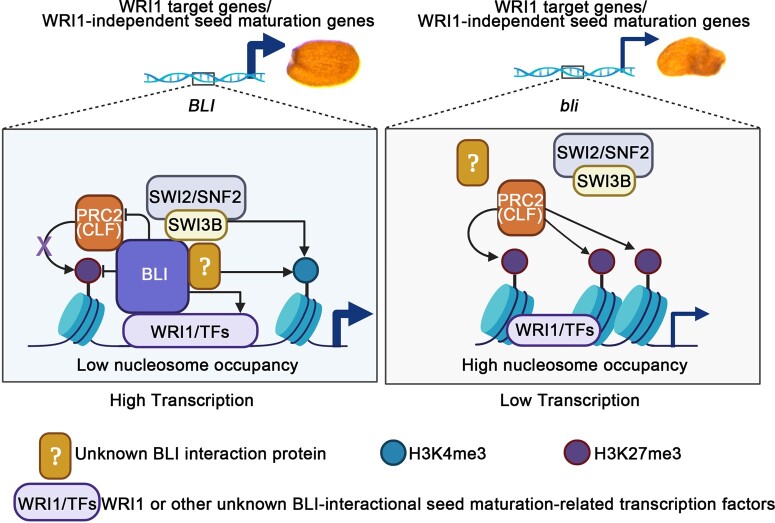
CLF mediates large-scale H3K27me3 programming/reprogramming during embryonic development, as ∼11.6% of genes of the Arabidopsis genome are repressed by CLF in various organs and ∼54% of these genes are preferentially repressed in siliques and embryos at the mature-green stage (Liu et al., 2016). clf-28 mutants produce larger and heavier seeds with higher oil content, larger oil bodies, and elevated expression levels of WRI1 in developing siliques compared to the WT. Consistently, the upstream TFs of WRI1 including AGAMOUS-Like 15, FUS3, and ABI3, and WRI1 downstream target genes that regulate fatty acid biosynthesis are derepressed in clf-28 siliques (Liu et al., 2016). In contrast, almost all of these genes were repressed in bli (Figure 4E). Furthermore, CLF occupancy on the promoters of WRI1 target genes is restrained by BLI (Figure 8, E–I), and the seed phenotype of bli-1 was partially rescued by clf-28, and that of BLI-OE was reversed by CLF-OE (Figure 9), suggesting that BLI may regulate seed maturation antagonistically to CLF. Hence, BLI and CLF may be recruited to the same locus by a cell-type-specific TF or other proteins to directly remodel chromatin status during the seed maturation stages. The increase in H3K27me3 levels in the bli-1 mutant could be explained by an increase in PcG occupancy, since the binding of BLI to the WRI1 target gene loci is required for the removal of PcG from WRI1 target genes.
The action of PcG proteins is counteracted by TrxG proteins, which set the activating H3K4me3 mark. The loss of PRC2 leads to increased H3K4me3 levels (Lafos et al., 2011). BLI is at least partially responsible for preventing a gain or increase of H3K4me3 at certain PcG target genes (Kleinmanns et al., 2017), suggesting that BLI might facilitate the recruitment of TrxG proteins to certain PcG target genes to regulate the switches between repressive and active chromatin states. BLI was found to interact with SWI3B (Supplemental Figure S16; Figure 8, B–D), a subunit of the Arabidopsis SWI2/SNF2 complex. This complex contains the chromatin remodeling ATPases BRAHMA and SPLAYED, which act as TrxG proteins to overcome polycomb repression (Wu et al., 2012; Li et al., 2016). Further analysis revealed that BLI recruits SWI3B to the promoter regions of WRI1 target genes (Figure 8, J–N) and that the swi3b-3 mutation reversed the effects of BLI overexpression on seed maturation (Figure 9). Thus, BLI may recruit the chromatin remodeling factor SWI3B to help WRI1 activate fatty acid biosynthesis genes. Therefore, BLI is involved in antagonizing CLF and SWI3B recruitment via a mutually exclusive interaction with CLF or SWI3B (Figure 8, C and D).
An as yet unanswered question is how the switch from the binding of BLI to CLF to its binding to SWI3B might occur. One likely scenario is that the plant SWI/SNF complex evicts Polycomb proteins, as does the mammalian SWI/SNF complex (Kadoch et al., 2017), and BLI plays a role in this process. Another possibility is that BLI functions like a GABA factor, switching from the PcG complex to the SWI/SNF complex with the help of other partners (Chetverina et al., 2021). Consistent with this notion, BLI could be also immunoprecipitated by the SWR1-C subunit ARP6 and its interacting partner, methyl-CpG binding domain 9, which interacts with the Imitation SWItch chromatin remodeling complex (Potok et al., 2019). Moreover, BLI was co-purified with SWC6, another core component of the Arabidopsis SWR1-C (Gomez-Zambrano et al., 2018). SWR1-C-mediated deposition of H2A.Z contributes to transcriptional activation by decreasing nucleosome occupancy (Nutzmann and Osbourn, 2015; Choi et al., 2016). Furthermore, SWR1-C mediated deposition of H2A.Z co-localizes with H3K4me3 near the TSSs of many genes (Choi et al., 2013) and promotes the deposition of H3K4me3 (Xu et al., 2018). Thus, BLI also has the potential to recruit SWR1-C to decrease nucleosome occupancy or deposit H3K4me3 to activate WRI1 target gene expression. Future work should be directed toward understanding how the BLI/WRI1 complex recruits other factors to alter the chromatin structure of target genes to activate transcription.
Here, we propose a working model for how BLI regulates seed maturation in Arabidopsis (Figure 10). For the BLI/WRI1 complex involved in fatty acid biosynthesis during seed maturation, WRI1 act as a TF to recruit BLI to the promoters of WRI1 target genes. BLI can remodel the chromatin, likely by recruiting other proteins such as SWI2/SNF2 chromatin remodeling complexes and preventing PRC2 action. Thus, the transcriptional activity of WRI1 is dependent on the function of BLI in chromatin remodeling. On the other hand, BLI may be required for the transcriptional activation of WRI1 via its post-translational (e.g. phosphorylation) regulation. Loss of BLI or WRI1 results in the repression of fatty acid biosynthesis genes and disturbed seed maturation. Moreover, BLI probably interacts with other unidentified TFs involved in distinct seed maturation-related processes. Therefore, BLI represents a promising seed maturation regulator that could be used to improve crop yield and quality.
Figure 10.
Proposed model for the role of BLI in regulating seed maturation and fatty acid biosynthesis. In WT and BLI-OE plants, WRI1 or other transcription factors recruit BLI to the promoter regions of WRI1 target genes or other WRI1-independent seed maturation genes. BLI may be required for the transcriptional activation of WRI1 or other TFs and affect their transcriptional activity by post-translational (e.g. phosphorylation) regulation. BLI affects chromatin by directly increasing H3K4me3 levels, decreasing H3K27me3 levels and nucleosome occupancy, or recruiting other proteins such as the SWI2/SNF2 complex and blocking PRC2 complex activity via a mutually exclusive interaction with CLF or SWI3B to help WRI1 or other TFs access and bind to the promoters of fatty acid biosynthesis or seed maturation genes. The H3K27me3 levels and nucleosome occupancy are enhanced at the promoters of fatty acid biosynthesis or other seed maturation genes in the bli loss-of-function mutant. This reduces the accessibility of AW-box cis-elements at the fatty acid biosynthesis genes or other cis-elements at the WRI1-independent seed maturation genes and reduces the binding of WRI1 or other TFs to the cis-elements. Black arrow indicates promotion; purple cross and black line indicates inhibition; thick blue arrow indicates high transcription; thin blue arrow indicates low transcription.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) ecotype Col-0 was used as the WT control. Seeds of bli-1 (SAIL_107_D04; Col background) and wri1–4 (SALK_ 008559) were obtained from the Arabidopsis Biological Resource Center. bli-11 (GABI-Kat_663H12) was provided by the Nottingham Arabidopsis Stock Centre. For analysis of genetic interactions with clf and swi3b, crosses were performed using clf-28 (Schatlowski et al., 2010) and swi3b-3 (Saez et al., 2008). All genotypes used in this study are in the Col-0 background. The pBLI:GUS transgenic lines were generated as follows. A 2.5-kb BLI promoter (pBLI) was cloned and fused with the GUS reporter in pCAMBIA-1300221(Huang et al., 2021), using the restriction endonuclease sites HindIII and BamHI, and the construct was transformed into WT to generate transgenic plants via the floral dip method.
The BLI-GFP (BLI-OE) and WRI1-GFP (WRI1-OE) transgenic lines were generated as follows. The BLI or WRI1 coding region was subcloned into the vector pCAMBIA-1300-GFP (driven by the UBQ10 promoter; Huang et al., 2021), using SmaI and BamHI. The resulting BLI-GFP and WRI1-GFP construct was transformed into WT to generate transgenic plants. After obtaining homozygous plants, the lines having the same seed phenotypes were selected for further analysis. The BLI-GFP plants were used for seed phenotype and GFP analysis and were crossed with wri1, clf, swi3b, or WRI1-OE plants for further analysis. The WRI1-GFP plants were used for seed phenotyping and were crossed with bli or BLI-OE plants for further study. The pBLI:BLI-GFP and pWRI:WRI-GFP transgenic lines were generated as follows. A 2.5-kb BLI promoter (pBLI) or WRI promoter region (pWRI1) was cloned into the vector BLI-GFP or WRI1-GFP by replacing the UBQ10 promoter using HindIII and BamHI, respectively. The constructs were transformed into WT to generate transgenic plants and were crossed with bli or wri1, respectively.
The MYC-BLI and MYC-WRI1 transgenic lines were generated as follows. The BLI or WRI1 coding region was subcloned into pCanG-MYC (driven by the 35S promoter; Huang et al., 2021), using BamHI and SpeI. The resulting 35S:MYC-BLI and 35S:MYC-WRI1 constructs were transformed into WT to generate transgenic plants. The WRI1 coding region was subcloned into the vector pZSC-YFP (driven by the 35S promoter) to generate YFP-WRI1 transgenic plants (Huang et al., 2021), using BglII and SalI. The 35S:MYC-WRI1 plants were used for crossing with BLI-GFP, and the 35S:MYC-BLI plants were used for crossing with YFP-WRI1 plants to observe seed phenotypes and for Co-IP analysis. The UBQ:CLF-GFP and 35S:SWI3B-GFP plants were constructed and used for crossing with bli or BLI-OE plants for seed phenotype observation.
Seeds were surface sterilized for 2 min in 75% ethanol followed by 5 min in 1% NaClO solution with 0.1% Triton X-100 and rinsed 5 times with double-distilled water. For germination, seeds were plated on Murashige and Skoog (MS) medium with 1.5% sucrose and 0.8% agar, vernalized at 4°C in the dark for 2 days, and transferred to a growth chamber at 22°C. Plants were grown under long-day conditions (16 h of light/8 h of dark) in a phytotron. Light was supplied by cool and warm white fluorescent bulbs, reaching an intensity of ∼100 µmol m−2 s−1 on the shelf surface.
Gene expression analysis
Total RNA was extracted from seeds dissected from siliques at different developmental stages using a Plant RNA Kit (PROMEGA, Madison, WI, USA) according to the manufacturer’s instructions. The total RNA was used to synthesize cDNA with oligo (dT) primer. Reverse transcription (RT) of total RNA was performed following the manufacturer’s protocol. Total RNA (1 μg) with oligo (dT) primer was heated at 70°C for 10 min, immediately cooled, and the mixture coupled with MMLV-RT SPCL reverse-transcriptase (Invitrogen, Waltham, MA, USA) was heated at 42°C for 1 h; UBQ10 (At4g05320) was used as a reference gene (Czechowski et al., 2005). The primers used for gene expression analysis by RT-PCR are listed in Supplemental Table S2. The qPCR was performed using a C1000 Thermal Cycler (CFX96 Real-Time System, Bio-Rad, Hercules, CA, USA). The expression levels were calculated and analyzed using the 2−ΔΔCq method with Bio-Rad CFX Manager software (version 2.1) and confirmed using the Pfaffl method (Pfaffl et al., 2002). Three technical replicates were performed using the same sample, and three biological replicates were performed using distinct samples at different times.
Transient gene expression assay
Rosette leaves of 4-week-old Arabidopsis were used for protoplast transformation following the protocol from Jen Sheen’s laboratory as described previously (Yoo et al., 2007). For the BiFC experiment, the BLI-YFP-N vector was constructed by cloning the BLI coding region into the pSPYNE plasmid, and WRI1/SWI3B-YFP-C was constructed by cloning the WRI1 coding region into the pSPYCE plasmid (Walter et al., 2004). The transformed protoplasts were cultured for 48 h at 23°C in the opaque background and the subcellular localization of GFP fusion protein was observed under a Zeiss LSM 710 laser scanning microscope with 514 nm for excitation and 530–600 nm for emission. The chlorophyll autofluorescence was also recorded. The photographs from the YFP, chlorophyll, and bright field channels were merged. For the LUC assay, 5-μg reporter plasmid and 5 μg of each effector plasmid were used for protoplast transformation. The control was transformed with 5-μg reporter plasmid and 5 μg of empty effector plasmid. The transformed protoplasts were cultured for 24 h at 23°C in the dark. Before the LUC activity was quantified, the transformed protoplasts were treated using Dual-Luciferase Reporter Assay reagent (Invitrogen) according to the manufacturer’s instructions. Three technical replicates were performed for each of three biological replicates.
Confocal microscopy
For confocal microscopy of homozygous transgenic plants, embryos at different developmental stages or 7-day-old seedlings grown in MS medium were used for fluorescence observation. Cell walls of roots were stained with 10 mg/mL propidium iodide (PI) for 5 min, washed once in distilled water, and mounted in water before confocal microscopy analysis as described (Truernit and Haseloff, 2008). Confocal images were taken using a Zeiss LSM 710 laser scanning microscope with the following excitation/emission wavelengths: 561/591 nm to 635 nm for PI, 488/505 nm to 530 nm for GFP, 514/530 nm to 600 nm for YFP. Three biological replicates were performed in each experiment. For each experiment, at least 10 embryos of each developmental stage or 10 roots were observed. A representative result is shown.
To observe PSVs in embryos, protein autofluorescence was imaged using the filter set for GFP. Confocal images were taken using a Zeiss LSM 710 laser scanning microscope. A representative result is shown. Three biological replicates were performed in each experiment. For each experiment, at least 20 embryos of each genotype were observed.
Embryo phenotype observation
Developing seeds dissected from siliques at various developmental stages were precleared in Hoyer’s solution for 1–12 h, depending on the developmental stage and experimental requirements, and observed under a differential interference contrast microscope (Leica, Wetzlar, Germany). Ovule autofluorescence was observed to analyze endosperm cellularization (Li et al., 2017b). A representative result is shown. Three biological replicates were performed in each experiment. For each experiment, at least 50 embryos or 15 ovules of each genotype were observed.
Histochemical staining
Histochemical staining for GUS activity in homozygous transgenic plants and developing embryos was performed as previously described (Jefferson et al., 1987) with some modifications. GUS stock solution [0.05-M NaPO4 buffer (pH 7.0), 5-mM K3Fe(CN)6, 5-mM K4Fe(CN)6, and 10-mM X-glucuronide] was made as described previously (Stahl et al., 2009). The tissues were stained in GUS staining solution, incubated at 37°C in the dark for 2–8 h (depending on experimental requirements), rinsed with 75% ethanol, and mounted in HCG solution before microscopy analysis (Huang et al., 2021). A representative result is shown. Three biological replicates were performed in each experiment. For each experiment, at least 20 embryos of each genotype were stained and observed.
YTH analysis
YTH analysis was performed as described previously (Matsui et al., 2004). In brief, yeast (Saccharomyces cerevisiae) strain L40 was transformed by the LiAc/DNA/PEG method (Gietz et al., 1992). The cDNAs were cloned into the pGBKT7 and pGADT7 vectors (Clontech, Mountain View, CA, USA), and plasmids of each pair were cotransformed into the yeast strain AH109. Transformants were selected on synthetic drop-out (SD) medium lacking Trp and Leu (−L/W), whereas the selection of interactions was conducted on SD medium lacking His, Trp, and Leu (−L/W/H) containing 0- to 15-mM 3-amino-1,2,4-triazole (3-AT). The experiments were performed at least 3 times independently, and similar results were obtained.
ChIP assays
The ChIP assay was carried out as described (Yamaguchi et al., 2014). Seeds of 12-DAP siliques from related plants were collected for ChIP assays. After fixation with formaldehyde, the chromatin was sheared to an average length of 500 bp by sonication and immunoprecipitated with GFP-Trap Agarose beads (ChromoTek, Planegg, Germany; gtma-20) or anti-MYC nanobody agarose beads (KT HEALTH, American Fork, UT, USA; KTSM1306) or H3K27me3 (Abcam, Cambridge, UK; ab6002) and H3K4me3 (Abcam, ab8580) antibodies with protein A agarose (Beyotime, Jiangsu, China; P2051), depending on the experimental requirements. After cross-linking was reversed, the amount of precipitated DNA fragments and input DNA was detected by quantitative RT-PCR using specific primers listed in Supplemental Table S3. The percentage of input was calculated by determining 2−ΔCt (=2−[Ct(ChIP)-Ct(Input)]). The exon region of retrotransposon TA3 was used as a negative control. Three technical replicates were performed using the same sample, and three biological replicates were performed using distinct samples at different times, and similar results were obtained.
MNase assay
Seeds of 12-DAP siliques were harvested in liquid nitrogen after cross-linking in 1% formaldehyde. Nuclei and chromatin were isolated as previously described (Chodavarapu et al., 2010) with the following modifications. The isolated nuclei were washed 3 times with Honda buffer (0.44-M Sucrose, 1.25% Ficoll, 2.5% Dextran T40, 20-mM HEPES-KOH pH 7.4, 10-mM MgCl2, 0.5% Triton X-100, 5-mM dithiothreitol (DTT), 1-mM Pefabloc, cOmplete), and the isolated chromatin was digested with 0.01–0.02 U μL−1 (final concentration) of Micrococcal Nuclease (NEB, Ipswich, MAs, USA) for 10 min in digestion buffer at 37°C. Subsequent steps were performed as previously described (Chodavarapu et al., 2010). DNA was purified by phenol/chloroform extraction and precipitated with salt and ethanol. Purified DNA was run on a 1.5% agarose gel, and the 150-bp bands corresponding to the mononucleosomal fraction were excised and extracted with a Gel Extraction Kit. The purified DNA was quantified using a NanoDrop ND-1000 spectrophotometer. Two nanograms of purified DNA were used for qPCR to monitor nucleosome occupancy. The fraction of input was calculated as 2−ΔCt (2−[Ct(mono)-Ct(gDNA)]) using undigested genomic DNA (Gévry et al., 2009). The primer used for RT-PCR is listed in Supplemental Table S2. Three technical replicates were performed using the same sample, and three biological replicates were performed using distinct samples at different times, and similar results were obtained.
In vitro pull-down assay
To produce recombinant BLI-FLAG and GST-WRI1 proteins, the corresponding open reading frames of BLI and WRI1 were cloned into the pCDF-Duet-1 and pGEX4T1 vector, respectively. GST or GST-WRI1 recombinant proteins were incubated with glutathione Sepharose (GE Healthcare, Chicago, IL, USA) in binding buffer (50 mM Tris, pH 7.4,120-mM NaCl, 5% glycerol, 0.5% Nonidet P-40, 1-mM PMSF, and 1-mM β-mercaptoethanol), for 2 h at 4°C, collected, mixed with supernatant containing 6XHis-BLI-FLAG, and incubated at room temperature for 60 min. After rinsing 5 times with washing buffer (50-mM Tris, pH 7.4, 120-mM NaCl, 5% glycerol, and 0.5% Nonidet P-40), the bound proteins were boiled in SDS sample buffer, subjected to SDS–PAGE, and immunoblotted and detected with anti-FLAG (TransGen, HT201-01). The experiments were performed in three biological replicates using distinct samples at different times, and similar results were obtained.
IB and Co-IP
To detect BLI and WRI1 protein levels during seed maturation, proteins were extracted from seeds of 6-, 10-, 12- or 16-DAP siliques of pBLI:BLI-GFP plants. The proteins were separated by SDS–PAGE and analyzed by IB with anti-GFP (TransGen, Beijing, China; HT801-01), anti-WRI1 (designed by GeneScript), or anti-H3 antibodies (Abcam; ab176842). To detect BLI and WRI1 interactions, the Co-IP assay was performed using transgenic plants harboring both 35S:MYC-BLI and 35S:YFP-WRI1. Proteins were extracted from the 12-DAP siliques of related transgenic plants in extraction buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2-mM MgCl2, 20% glycerol, and 0.1% Nonidet P-40) containing protease inhibitor cocktail (Roche, Basel, Switzerland). After centrifugation at 13,000 g for 12 min, the supernatant was incubated with GFP-Trap at 4°C for 2 h. The beads were centrifuged and washed 4 times with washing buffer (50-mM Tris–HCl, pH 7.4, 150-mM NaCl, 2-mM MgCl2, 10% glycerol, and 0.01% NP-40). Proteins were eluted with SDS sample buffer and analyzed by IB.
To detect the competitive interactions of WRI1 and CLF with BLI, total protein extracts from transformed WT protoplasts carrying both 35S:BLI-mCherry and 35S:CLF-FLAG without or with different amounts of 35S:MYC-WRI1 were immunoprecipitated with the immobilized anti-mCherry antibody. The detect synergistic interactions of WRI1 and SWI3B with BLI, total protein extracts from transformed protoplasts carrying both 35S:YFP-WRI1 and 35S:MYC-SWI3B without or with different amounts of 35S:BLI-mCherry were immunoprecipitated with the immobilized anti-GFP antibody. To detect competitive interactions of CLF and SWI3B with the BLI-WRI1 complex, total protein extracts from transformed 35S:YFP-WRI1 or 35S:BLI-mCherry protoplasts carrying both 35S:MYC-SWI3B and 35S:CLF-FLAG with different amounts of 35S:BLI-mCherry or 35S:YFP-WRI1 were immunoprecipitated with the immobilized anti-GFP or anti-mCherry antibody. Variable amounts of tagged BLI and WRI1 proteins were achieved by adding 1, 2, 3, or 4 times the amount of expression vector. All immunoprecipitated proteins (IPs) were detected with anti-FLAG (TransGen; HT201-01), anti-MYC (TransGen; HT101-01), anti-mcherry (Abcam; ab213511), or anti-GFP antibodies (TransGen; HT801-01) in IB experiments. The experiments were performed in three biological replicates using distinct samples at different times, and similar results were obtained.
Biochemical analyses
Total fatty acids were extracted from dry seeds and quantified as previously described (Li et al., 2006). Starch and sugar contents in dry seeds were measured as previously described (Caspar et al., 1991). Proteins were extracted from 20 dry seeds per sample and suspended in Laemmli buffer (2×) with DTT. The crude seed protein extracts were incubated at 95°C for 10 min, followed by centrifugation at 15,000 g for 3 min, and the supernatants were transferred to a clean tube. The samples were separated in 15% SDS–PAGE, followed by Coomassie blue staining. The 2S protein was detected by IB with anti-2S antibody (Bioacademia, 81–121). Three technical replicates were performed for each of the three biological replicates.
Paraffin sections
Arabidopsis fruits or seeds at different developmental stages were placed in fixative solution (2.5 glutaraldehyde and 3% paraformaldehyde dissolved in 0.1-M phosphate buffer, pH 7.2) and fixed overnight at 4°C. The samples were washed 3 times with phosphate buffer, 20 min each time, dehydrated through an ethanol series (15%, 30%, and 50%), 20 min each time, and stained with 1% hematoxylin dye solution at room temperature for 24–48 h. After staining, the samples were washed 3 times with 50% ethanol, 20 min each time. The samples were dehydrated through an ethanol series (70%, 85%, 95%, and 100%), 20 min each time, incubated in pure xylene 2 times, 20 min each time, transferred to paraffin, and placed in a 40°C oven overnight. The material was transferred to a 60°C oven for 2 h, followed with changing the paraffin once and keep it in the 60°C oven for 2 h, and changing the paraffin again and keep it in the 60°C oven for 4 h; and embedded in paraffin. The embedded paraffin blocks were trimmed and sliced with a Reichert HistoSTAT 820 paraffin microtome to a thickness of 4 μm. Water was dripped on the glass slides, and the paraffin slice was placed on the drying table and stored in a 40°C oven. The prepared paraffin sections were deparaffinized with xylene for 3 times, the first for 10 min, the second for 5 min, and the third for 3 min. The slides were sealed with neutral gum and placed in an oven at 40°C for storage. The slides were observed under a Leica DM6 microscope and photographed.
Statistical analysis
Student’s t test was performed to evaluate statistically significant differences compared to the control (WT). One-way ANOVA followed by Tukey’s test was used to determine the genetic interaction of BLI and WRI1 in regulating seed maturation and fatty acid biosynthesis for a given variable. Differences between means were evaluated for significance using Tukey’s honest significant difference (HSD) test (Tukey’s HSD) (P < 0.05); means with the same letter are not significantly different. Statistical analyses were performed with SPSS version 22.0 statistical software (IBM Corp., Armonk, NY, USA). T test and ANOVA results are shown in Supplemental Data Set 1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: BLI (AT3G23980), WRI1 (AT3G54320), LEC1 (AT1G21970), LEC2 (AT1G28300), FUS3 (AT3G26790), ABI3 (AT3G24650), SUS2 (AT5G49190), ENO1 (AT1G74030), Cy-PKβ (AT5G52920), Ch-PKα (At3g22960), BCCP2 (AT5G15530), ACP1 (AT3G05020), KASI (AT5G46290), KASIII (AT1G62640), FAD2 (AT3G12120), FAD3 (AT2G29980), FAE1 (AT4G34520), PLA2α (AT2G06925), CLF (AT2G23380), and SWI3B (AT2G33610).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the interaction between BLI and DREB2A or WRI1 and SMC5 in YTH.
Supplemental Figure S2 . Subcellular localization of BLI and WRI1.
Supplemental Figure S3 . Schematic diagram of bli-1 and bli-11 and BLI expression levels in the bli-1 and bli-11 mutants.
Supplemental Figure S4. Embryo phenotypes at various stages of embryogenesis in the WT and bli-1 mutant.
Supplemental Figure S5. Observation of abnormal embryo hypocotyls in bli-1.
Supplemental Figure S6. Endosperm and embryo development of bli-1 during seed development.
Supplemental Figure S7 . BLI expression in bli complemented plants.
Supplemental Figure S8. The seed phenotypes and BLI expression levels in BLI-overexpression plants.
Supplemental Figure S9. Subcellular localization of BLI in 12 and 14 DAP embryonic cells.
Supplemental Figure S10. Seed reserve proteins identified by SDS–PAGE and IB using the anti-2S antibody.
Supplemental Figure S11 . The expression levels of 2S1, 2S2, 2S3, 2S4, and 2S5 in the bli mutant.
Supplemental Figure S12. The seed phenotypes and WRI1 expression levels in WRI1-OE/bli plants.
Supplemental Figure S13. RT-qPCR analysis of glycolysis, fatty acid biosynthesis, and modification, and TAG accumulation genes in seeds of different genotypes.
Supplemental Figure S14. The seed phenotypes of BLI and WRI1 co-overexpression plants.
Supplemental Figure S15 . RT-qPCR of the expression of the promoter regions of WRI1 target genes without the AW box after anti-H3K27me3 and anti-H3K4me3 ChIP in plants with different BLI genotypes.
Supplemental Figure S16. Interaction assay between BLI and SWI3B.
Supplemental Figure S17. The expression patterns of BLI and CLF during seed maturation.
Supplemental Figure S18. The genetic relationship between BLI and SWI3B or CLF in regulating seed maturation.
Supplemental Figure S19. The response of bli-1 seeds to added sucrose.
Supplemental Figure S20. The phosphorylation level of WRI1 in the bli mutant.
Supplemental Table S1. Putative WRI1-interacting proteins identified by cDNA library screening.
Supplemental Table S2. The primers used for gene expression analysis and the MNase assay.
Supplemental Table S3. The primers used for the ChIP assay and so on.
Supplemental Data Set 1. Statistical analysis using Student’s t test and ANOVA.
Supplementary Material
Acknowledgments
We thank Professor Andrzej Wierzbicki from the University of Michigan for technical support for the ChIP and MNase assays and the ABRC for kindly providing seeds used in this study.
Funding
This work was supported by the National Natural Science Foundation of China (31870301 and 31370350 for S.Z.), the Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2016 for S.Z.).
Conflict of interest statement. None declared.
Contributor Information
Ruihua Huang, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Mengling Liu, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Guanping Gong, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Pingzhi Wu, Key Laboratory of South Subtropical Fruit Biology and Genetic Resource Utilization, Ministry of Agriculture, Institution of Fruit Tree Research, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China.
Mei Bai, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou 510642, China.
Hongting Qin, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Guohe Wang, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Huimei Liao, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Xiaoxiu Wang, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Yanqun Li, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou 510642, China.
Hong Wu, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou 510642, China.
Xiaojing Wang, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Chengwei Yang, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
Daniel Schubert, Institute for Biology, Freie Universität Berlin, Berlin 14195, Germany.
Shengchun Zhang, Guangdong Key Laboratory of Biotechnology for Plant Development, College of Life Sciences, South China Normal University, Guangzhou 510631, China.
S.Z. conceived this project and designed all research. R.H. and M.L. performed most of the research. G.G., H.Q., X.W., G.W., and H.L. helped perform some experiments. P.W. helped measure the fatty acid contents. M.B., Y.L., H.W., and X.W. helped perform paraffin sectioning. S.Z. and R.H. analyzed the data. S.Z. and R.H. wrote the article. S.Z., C.Y., and D.S. revised the article.
The author for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Shengchun Zhang (sczhang@scnu.edu.cn).
References
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arabidopsis Book 6: e0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Boulard C, Fatihi A, Lepiniec L, Dubreucq B (2017) Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim Biophys Acta Gene Regul Mech 1860: 1069–1078 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Lindsey K (2006) The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol 142: 526–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chen L, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H (2013) Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D, Erokhin M, Schedl P (2021) GAGA factor: a multifunctional pioneering chromatin protein. Cell Mol Life Sci 78: 4125–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim J, Muller SY, Oh M, Underwood C, Henderson I, Lee I (2016) Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex. Plant Physiol 171: 1128–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC, et al. (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry N, Svotelis A, Larochelle M, Gaudreau L (2009) Nucleosome mapping. Methods Mol Biol 543: 281–291 [DOI] [PubMed] [Google Scholar]
- Gietz D, Jean AS, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Zambrano A, Crevillen P, Franco-Zorrilla JM, Lopez JA, Moreno-Romero J, Roszak P, Santos-Gonzalez J, Jurado S, Vazquez J, Kohler C, et al. (2018) Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes. Mol Plant 11: 815–832 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Wuytswinkel OV, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Plant Mol Biol 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Huang R, Liu M, Gong G, Wu P, Patra B, Yuan L, Qin H, Wang X, Wang G, Liao H, et al. (2021) The Pumilio RNA-binding protein APUM24 regulates seed maturation by fine-tuning the BPM-WRI1 module in Arabidopsis. J Integr Plant Biol 63: 1240–1259 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, Crabtree GR (2017) Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 49: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Jang IC, Chua NH (2016) The mediator complex MED15 subunit mediates activation of downstream lipid-related genes by the WRINKLED1 transcription factor. Plant Physiol 171: 1951–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinmanns JA, Schatlowski N, Heckmann D, Schubert D (2017) BLISTER regulates polycomb-target genes, represses stress-regulated genes and promotes stress responses in Arabidopsis thaliana. Front Plant Sci 8: 1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Ma W, Yang H, Ma G, Mantyla JJ, Benning C (2017) The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J Exp Bot 68: 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, Ai LF, et al. (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Jin C, Duan S, Zhu Y, Qi S, Liu K, Gao C, Ma H, Zhang M, Liao Y, et al. (2017a) MYB89 transcription factor represses seed oil accumulation. Plant Physiol 173: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zou W, Jian L, Qian J, Deng Y, Zhao J (2017b) Non-SMC elements 1 and 3 are required for early embryo and seedling development in Arabidopsis. J Exp Bot 68: 1039–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge JJP (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liu J, Deng S, Wang H, Ye J, Wu HW, Sun HX, Chua NH (2016) CURLY LEAF regulates gene sets coordinating seed size and lipid biosynthesis. Plant Physiol 171: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H (2010) Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem 48: 9–15 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto MA, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Arondel V, Kilaru A, Bates PD, Thrower NA, Benning C, Ohlrogge JB (2013) Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS One 8: e68887–e68887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Kong Q, Grix M, Mantyla JJ, Yang Y, Benning C, Ohlrogge JB, Ma W, Kong Q, Grix MJPJ (2015) Deletion of a C–terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J 83: 864–874 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Mantyla JJ, Yang Y, Ohlrogge JB, Benning C (2016) 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J 88: 228–235 [DOI] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Kohler C (2006) Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG, Erni S (1992) Early embryogenesis in Arabidopsis thaliana. Can J Bot 69: 447–460 [Google Scholar]
- Marchive C, Nikovics K, To A, Lepiniec L, Baud S (2014) Transcriptional regulation of fatty acid production in higher plants: molecular bases and biotechnological outcomes. Eur J Lipid Sci Technol 116: 1332–1343 [Google Scholar]
- Matsui K, Tanaka H, Ohmetakagi M (2004) Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol J 2: 487–493 [DOI] [PubMed] [Google Scholar]
- Merini W, Romero-Campero FJ, Gomez-Zambrano A, Zhou Y, Turck F, Calonje M (2017) The Arabidopsis polycomb repressive complex 1 (PRC1) components AtBMI1A, B, and C impact gene networks throughout all stages of plant development. Plant Physiol 173: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutzmann HW, Osbourn A (2015) Regulation of metabolic gene clusters in Arabidopsis thaliana. New Phytol 205: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier JM, Kwong RW, Park S, Le BH, Baden R, Cagliari A, Hashimoto M, Munoz MD, Fischer RL, Goldberg RB, et al. (2017) LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc Natl Acad Sci USA 114: E6710–E6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Wang Y, Xu L, Zhong Z, Liu W, Feng S, Naranbaatar B, Rayatpisheh S, Wang Z, Wohlschlegel JA, et al. (2019) Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat Commun 10: 3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau B, Baud S, Vernoud V, Morin V, Py C, Gendrot G, Pichon JP, Rouster J, Paul W, Rogowsky PM (2011) Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant physiol 156: 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T (2011) Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol 9:e1001206–e1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56: 1215–1228 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14:1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL (2008) HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya Durrett TP, Weise SE, Benning C (2011) Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol J 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schatlowski N, Stahl Y, Hohenstatt ML, Goodrich J, Schubert D (2010) The CURLY LEAF interacting protein BLISTER controls expression of polycomb-group target genes and cellular differentiation of Arabidopsis thaliana. Plant Cell 22: 2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Mccarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Haseloff J (2008) A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann J, Siemens N, Kai-Larsen Y, Fiedler T, Bergman P, Johansson L, Norrby-Teglund A (2016) Phosphoglycerate kinase—a novel streptococcal factor involved in neutrophil activation and degranulation. J Infect Dis 214: 1876–1883 [DOI] [PubMed] [Google Scholar]
- Vicentecarbajosa J, Carbonero P (2005) Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Leichty AR, Hu T, Poethig RS (2018) H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development 145: dev152868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kwon CS, William DA, Wagner D (2014) PROTOCOLS: chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Munz J, Cass C, Zienkiewicz A, Kong Q, Ma W, Sedbrook J, Benning C (2015) Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol 169: 1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap WC, Lee FC, Shabari Shan DK, Musa H, Appleton DR, Kulaveerasingam H (2017) WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J 91: 97–113 [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF (2012) Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Cho Y, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocol 2: 1565. [DOI] [PubMed] [Google Scholar]
- Zhai Z, Liu H, Shanklin J (2017) Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29: 871–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J (2018) Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhao M, Li S, Sun L, Wang W, Cai C, Dierking EC, Ma J (2017) Plasticity and innovation of regulatory mechanisms underlying seed oil content mediated by duplicated genes in the palaeopolyploid soybean. Plant J 90: 1120–1133 [DOI] [PubMed] [Google Scholar]
- Zhang M, Cao X, Jia Q, Ohlrogge J (2016) FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J 88: 95–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.