Abstract
Ear length (EL) is a key trait that contributes greatly to grain yield in maize (Zea mays). While numerous quantitative trait loci for EL have been identified, few causal genes have been studied in detail. Here we report the characterization of ear apical degeneration1 (ead1) exhibiting strikingly shorter ears and the map-based cloning of the casual gene EAD1. EAD1 is preferentially expressed in the xylem of immature ears and encodes an aluminum-activated malate transporter localizing to the plasma membrane. We show that EAD1 is a malate efflux transporter and loss of EAD1 leads to lower malate contents in the apical part of developing inflorescences. Exogenous injections of malate rescued the shortened ears of ead1. These results demonstrate that EAD1 plays essential roles in regulating maize ear development by delivering malate through xylem vessels to the apical part of the immature ear. Overexpression of EAD1 led to greater EL and kernel number per row and the EAD1 genotype showed a positive association with EL in two different genetic segregating populations. Our work elucidates the critical role of EAD1 in malate-mediated female inflorescence development and provides a promising genetic resource for enhancing maize grain yield.
An ALMT transporter plays essential roles in regulating maize ear development by delivering malate through xylem vessels to the apical part of the immature ear.
Introduction
Maize (Zea mays) is one of the most important cereal crops worldwide. In addition to being a main source of food and feed, maize grains also currently provide massive raw materials for various industrial products. Thus, improvement of grain yield is always the primary target in maize breeding to meet an ever-increasing demand. Maize plants usually form a single main stem bearing one elite ear, making kernel number per ear a key determinant of yield potential. Large ears are thus preferred in maize breeding to maximize grain yield.
The great value of ears in maize production has driven the identification of genes involved in female inflorescence development, especially those regulating ear ontogeny. The genes BARREN STALK1 (BA1), BA2, BARREN INFLORESCENCE1 (BIF1), BIF2, BIF4, SPARSE INFLORESCENCE1 (SPI1), BARREN STALK FASTIGIATE1 (BAF1), and VANISHING TASSEL2 (VT2) are all required for the initiation of axillary meristems in maize (Gallavotti, 2004; McSteen et al., 2007; Gallavotti et al., 2008, 2011; Phillips et al., 2011; Galli et al., 2015; Yao et al., 2019). Single loss-of-function mutations in these genes are either unable to produce ears (ba1, ba2, and baf1), make fewer ear shoots (bif2), or bear shorter ears with fewer kernels (bif1, bif4, spi1, and vt2). These genes encode transcription factors (BA1, BA2, and BAF1), enzymes (BIF2, SPI1, and VT2), or Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) proteins (BIF1 and BIF4) involved in auxin biosynthesis or transport. RAMOSE1 (RA1), RA2, RA3, and RAMOSE ENHANCER LOCUS2 (REL2) are four classical loci regulating the identity and determinacy of spikelet-pair meristems (SPMs; Vollbrecht et al., 2005; Bortiri et al., 2006; Satoh-Nagasawa et al., 2006; Gallavotti et al., 2010). Mutants in these genes exhibit more branches in both tassels and ears. BRANCHED SILKLESS1, INDETERMINATE SPIKELET1 (IDS1), and SISTER OF IDS1 are three genes controlling the identity and determinacy of spikelet meristems (SMs; Chuck et al., 1998, 2002, 2008). Several genes were also reported to control the identity and determinacy of floral meristems (FMs) in maize, including ZEA AGAMOUS1 (ZAG1), ZAG3, DROOPING LEAF1 (DRL1), DRL2, INDETERMINATE FLORAL APEX1, and GROWTH REGULATING FACTOR–INTERACTOR FACTOR (Mena et al., 1996; Laudencia-Chingcuanco and Hake, 2002; Thompson et al., 2009; Zhang et al., 2018; Strable and Vollbrecht, 2019). These genes define a regulatory module that provides crucial control of floret patterning and FM determinacy in the development of grain-producing structures.
In addition to inflorescence morphogenesis, the number of florets determined by the inflorescence meristem (IM) proliferation is another critical factor that contributes to kernel number in maize. IM activity is maintained by the feedback signaling pathway of the stem cell promoting factor WUSCHEL (WUS) and its repressor CLAVATA (CLV), first discovered in Arabidopsis (Arabidopsis thaliana) but conserved in other species including maize (Schoof et al., 2000; Daum et al., 2014; Wu et al., 2018). ZmWUS1 is expressed in the organizing center of the meristem, and its overexpression causes stem cell overproliferation and IM rearrangement as well as severely affects productivity in maize (Chen et al., 2021). In addition, maize THICK TASSEL DWARF1, FASCIATED EAR2 (FEA2), and CLAVATA3/EMBRYO SURROUNDING REGION-RELATED7 (CLE7) are the respective orthologs of Arabidopsis CLV1, CLV2, and CLV3 (Taguchi-Shiobara et al., 2001; Bommert et al., 2005, 2013b; Je et al., 2016, 2018; Rodriguez-Leal et al., 2019; Liu et al., 2021), and function similarly in restricting IM proliferation. The corresponding mutants show enlarged IM and produce fasciated ears with increased disorganized kernel row number (KRN). Several other mutants that display similar features have also been characterized and the casual genes have been functionally studied (Bommert et al., 2013a; Chuck et al., 2014; Je et al., 2016, 2018). In contrast to these genes regulating ear size through improving IM width, KERNEL NUMBER PER ROW6 (KNR6), ZmACO2, and YIGE1 were recently shown to regulate ear size by improving EL (Jia et al., 2020; Luo et al., 2022; Ning et al., 2021). KNR6 encodes a serine/threonine protein kinase that phosphorylates Arf GTPase-activating protein and enhances grain yield by raising EL and KNR (Jia et al., 2020). ZmACO2 encodes 1-aminocyclopropane-1-carboxylate oxidase 2 regulating ethylene biosynthesis, and a loss-of-function mutation in ZmAOC2 leads to lower ethylene production in developing ears, resulting in higher grain yield (Ning et al., 2021). YIGE1 regulates EL by positively affecting pistillate floret number which may be involved in sugar and auxin signal pathways (Luo et al., 2022). These studies highlight the potential to manipulate maize inflorescences architecture and yield enhancement.
Plant malnutrition affects vegetative and reproductive growth. In maize, it usually causes a severe drop in grain yield by disturbing normal ear development. Three plant metabolism genes have been studied in maize with a link to ear development. THIAMINE BIOSYNTHESIS2 (THI2) encodes a putative enzyme involved in thiamine formation (Woodward et al., 2010). Plants from the corresponding thi2 mutant produce severely truncated ears and barren tassel branches, but these phenotypes can be rescued by supplementation with exogenous thiamine. ROTTEN EAR (RTE) and TASSEL-LESS1 (TLS1) are two boron transporters and their loss-of-function mutants share very similar phenotypes, characterized by smaller and sterile ears and tassels (Chatterjee et al., 2014; Durbak et al., 2014). tls1/TLS1 rte/RTE double heterozygous plants exhibit a dosage-dependent defect under boron-limited conditions, which indicate they may function together to maintain boron homeostasis in maize (Durbak et al., 2014).
Malate is an intermediate metabolite of the tricarboxylic acid cycle, the glyoxylate cycle, the C4 and crassulacean acid metabolism pathway, and exhibits multiple important functions including providing NADH for the assimilation of carbon (C), nitrogen (N), and sulfur (S) as indirect hydrogen carriers, regulating reactive oxygen species (ROS) production and scavenging (Igamberdiev and Eprintsev, 2016; Igamberdiev and Bykova, 2018; Zhao et al., 2018; Selinski and Scheibe, 2019). The metabolic enzymes fumarase (FUM), malate synthase (MS), malate dehydrogenase (MDH), and NAD(P)-dependent malic enzyme (ME; NAD(P)-ME) participate in malate metabolism (Maurino and Engqvist, 2015). FUM catalyzes the reversible hydration/dehydration of fumarate to/from malate. MS catalyzes the irreversible production of malate from acetyl-CoA and glyoxylate, and the encoding gene is highly expressed in senescing organs (Graham et al., 1992). MDH catalyzes the reversible reduction of oxaloacetate (OAA) to malate with different coenzyme specificity and subcellular localizations. Malate/OAA shuttles, also termed malate valves, are powerful systems to balance metabolic fluxes by opening an indirect transfer route of reducing equivalents (Selinski and Scheibe, 2019). There are two types of MDH enzymes: NAD-MDH is widely distributed in the cytosol, mitochondria, and peroxisomes, while NADP-MDH accumulates exclusively in plastids. Cytosolic MDH, together with OAA/malate antiporters and plastid-, mitochondrial-, and peroxisomal-localized MDH, mediates the transport of reducing equivalent NAD(P)+/NAD(P)H between the cytosol and organelles (Pastore et al., 2003). NAD(P)-ME catalyzes the oxidative decarboxylation of malate, generating pyruvate, NAD(P)H, and CO2 used for the production of carbohydrates (Brown et al., 2010). These enzymes coordinately regulate the malate metabolism, which plays essential roles in mediating the metabolic status and redox homeostasis of plant cells in response to changing environmental conditions and developmental requirements (Maurino and Engqvist, 2015; Heng et al., 2018; Zhao et al., 2018; Luo et al., 2019).
In this study, we characterized the maize mutant ear apical degeneration1 (ead1) with lower EL and KNR. We show that the casual gene EAD1 encodes an aluminum (Al)-activated malate transporter (ALMT) localizing to the plasma membrane (PM). Dysfunction of EAD1 resulted in lower malate contents in the apical parts of immature ears and severely degenerated apical IMs. Exogenous malate injection rescued the phenotypic defects of ead1, indicating that the shorter ears of ead1 are caused by a drop in malate contents. Collectively, we demonstrate that EAD1 regulates maize ear development by mediating the delivery of malate to the apical parts of immature ears. The identification of EAD1 will expand our knowledge of the molecular mechanism of maize ear development.
Results
ead1 is a shortened ear mutant with degenerated female inflorescence
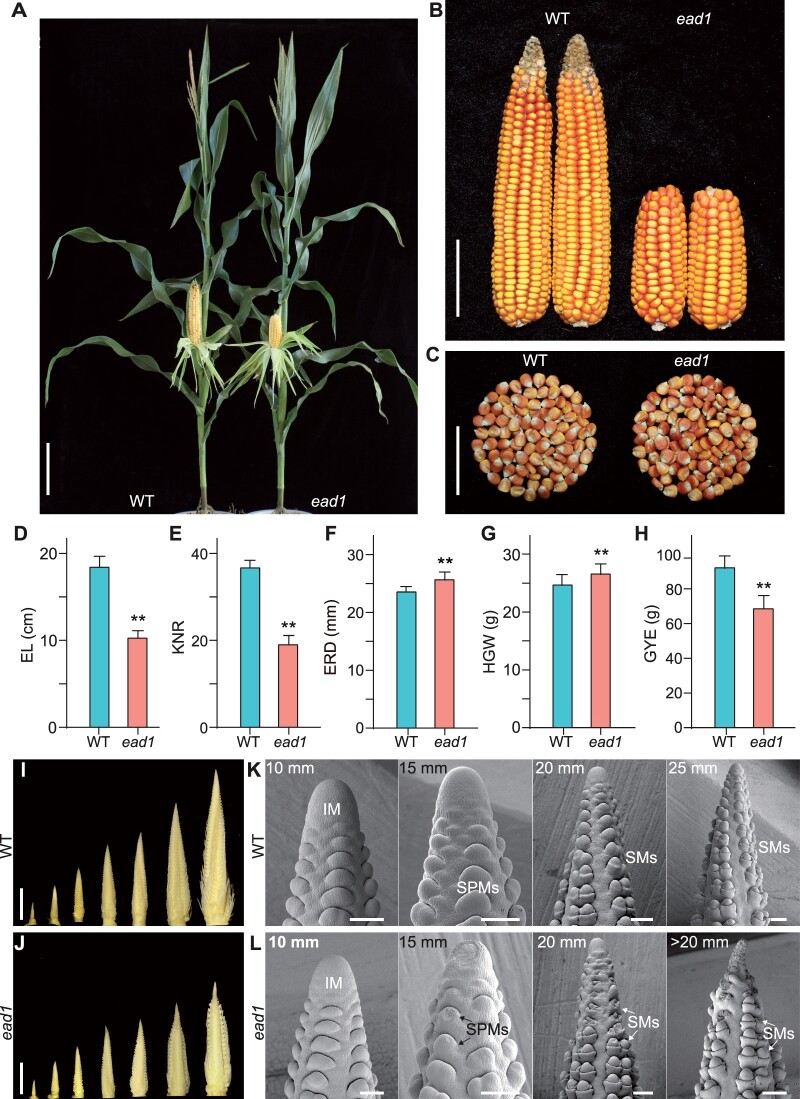
Maize ear length (EL) is an important agronomic trait highly proportional to overall grain yield (Li et al., 2018). To elucidate the genetic and molecular mechanisms underlying EL development, we isolated a natural mutant, designated ead1, exhibiting shorter ears from the elite inbred line HN321 widely used in maize breeding programs in China for its long ears and high specific combining ability in crosses. ead1 mutant plants exhibited no significant differences relative to its wild-type (WT) siblings during vegetative growth but developed shorter ears at maturity (Figure 1, A and B; Supplemental Figure S1). Compared to the WT siblings, ead1 displayed a significant decrease of EL by 44% (10.26 ± 0.86 cm in ead1; 18.42 ± 1.26 cm in WT; P < 0.01; Figure 1D) and of KNR by 48% (19.00 ± 2.12 in ead1 versus 36.67 ± 1.76 in WT; P < 0.01; Figure 1E), leading to a loss of 25.7% in grain yield per ear (GYE; 68.46 ± 7.71 g in ead1 versus 92.19 ± 7.51 g in WT; P < 0.01; Figure 1H), while ear rachis diameter (ERD) and hundred-grain weight (HGW) of ead1 were higher than those of the WT (P < 0.01; Figure 1, C, F, and G).
Figure 1.
Phenotypic comparisons of WT and ead1. A–C, Corn plants at the milk-ripe stage (A), ears (B), and kernels (C) at the full-ripe stage. WT HN321, WT. Scale bar = 20 cm in (A), 5 cm in (B) and (C). D–H, Comparison of ear traits between WT and ead1.Values are means, error bars represent standard deviation (sd) (n = 15). **P < 0.01 as determined by two-tailed Student’s t test between WT and ead1. I and J, Developing immature ears at 5, 10, 15, 20, 25, 30, and 40 mm in length in WT (I) and at 5, 10, 15, 20, and >20 mm length in ead1 (J). Scale bars = 10 mm. K and L, SEM analysis of the apical parts of immature ears in WT (K) and ead1 (L). Scale bars = 200 μm.
We followed the development of female inflorescences from WT and ead1 to determine the stage of ear degeneration in ead1. The ear developmental rates of ead1 were comparable to those of the WT before vegetative stage 11 (V11), but ear development became visibly delayed in ead1 from V12 onward when the EL is ∼20 mm (Supplemental Figure S2). Indeed, we observed no obvious changes in ear development until the length of the inflorescence reached 20 mm, at which point the ead1 apex started to shrink (Figure 1, I and J). We obtained close-up views of the developing inflorescences by scanning electron microscopy (SEM; Figure 1, K and L; Supplemental Figure S3). Again, we detected no obvious differences between WT and ead1 in the apical IM or lateral primordia at the 10-mm inflorescence stage. However, we noticed significant alterations, with shrunken apical IM and collapsed SPMs/SMs around the apex in ead1 at the 15-mm inflorescence stage. The observed apical IM degeneration and SPMs/SMs collapse extended and deteriorated in ≥20-mm inflorescences, and ultimately led to the abortion of SMs and FMs in ead1. By contrast, the lateral primordia at the middle and basal parts of ead1 inflorescences developed normally and produced fertile florets similar to those in WT (Figure 1, K and L; Supplemental Figure S3). We further compared the main axis length and branch number of mature tassels and observed no differences between ead1 and WT (Supplemental Figures S1, B, and C and S4, G). We confirmed these results by anatomical studies of the mature male florets in the apex of tassel main axes in ead1, which produced fertile pollen like the WT (Supplemental Figure S4). Collectively, our results indicate that ead1 causes apical degeneration of the female inflorescence, leading to shorter ear, while the mutation has no effects on vegetative growth or male inflorescence formation, suggesting the specificity of EAD1 function in modulating ear development.
ROS burst and programmed cell death occur in ead1 apical ears
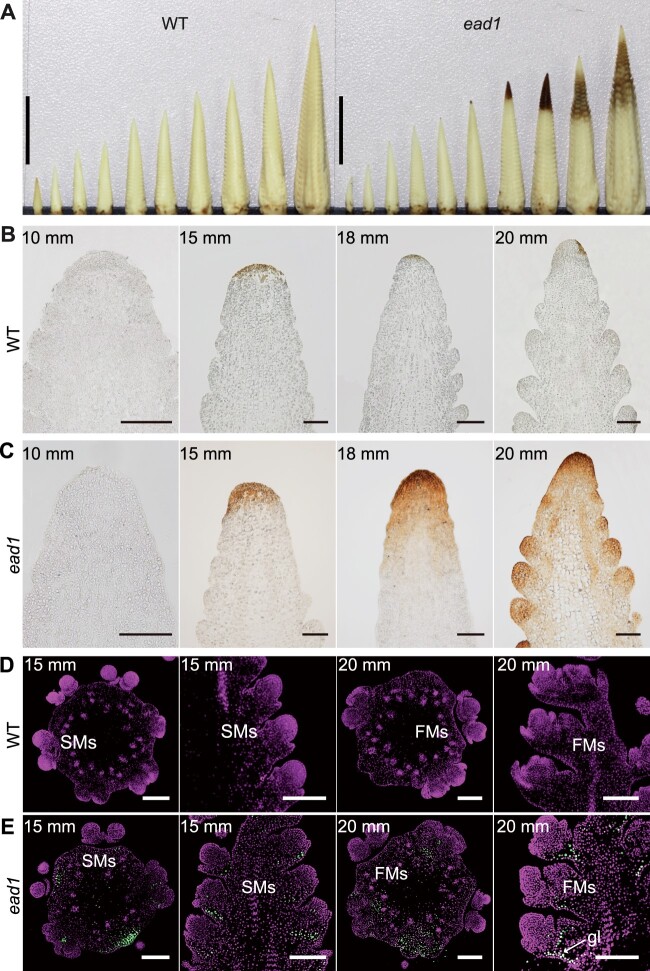
We performed 3,3′-diaminobenzidine (DAB) staining of developing inflorescences from WT and ead1 to investigate the changes in H2O2 levels. We detected H2O2 deposition, as indicated by the rotten head, at the 15-mm ear-length stage and became aggravated with the elongation of the ead1 inflorescence (Figure 2A). We also inspected paraffin sections of apical IM to obtain close-up views. Consistently, we initially detected strong signals for H2O2 accumulation in the apex of 15-mm inflorescences; the signal gradually spread to the adjacent lateral primordia with the development of inflorescence in ead1 (Figure 2, B and C). The terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay is widely used to examine programmed cell death (PCD) due to its high sensitivity (Wang et al., 2018; Zafar et al., 2020). Here, we conducted a TUNEL assay to ask whether PCD occurred in degenerating ead1 ears. In both longitudinal and transverse sections of inflorescences, we observed strong positive signals at the base of apical SMs and FMs in ead1 (Figure 2E), while we detected no signal in WT (Figure 2D). The results indicated that the abortion of the inflorescence apex in ead1 is caused by PCD of the apical SM and FM cells. To take a closer look, we performed transmission electron microscopy (TEM) analysis on the apex of the 20-mm inflorescence in both WT and ead1. Compared to WT, destructive events occurred in ead1 including the vacuolization of the cytoplasm, nuclear atrophy as well as the collapse of the cell wall. We also observed numerous abnormal mitochondria in ead1 (Supplemental Figure S5). Together, we conclude that the excessive accumulation of ROS in the apical IM triggers PCD, resulting in ear apical degeneration in ead1.
Figure 2.
ROS burst and PCD occur in ead1 apical ears. A, DAB-stained immature ears at 5, 7.5, 10, 12.5, 15, 17.5, 20, 22.5, 25, and 30-mm length in WT (left) and at 5, 7.5, 10, 12.5, 15, 17.5, 20, and >20-mm length in ead1 (right). Scale bars = 10 mm. B and C Paraffin sections of DAB-stained ear tips of WT (B) and ead1 (C) at different ELs. Scale bars = 100 μm. D and E TUNEL analysis of apical ears in WT (D) and ead1 (E). The first and third columns are from transverse sections and the second and fourth columns are from longitudinal sections. Sections were counterstained with propidium iodide. Signal was detected in ead1 (E), but not in the WT (D). gl, glume. Scale bars = 200 μm.
The causal gene underlying ead1
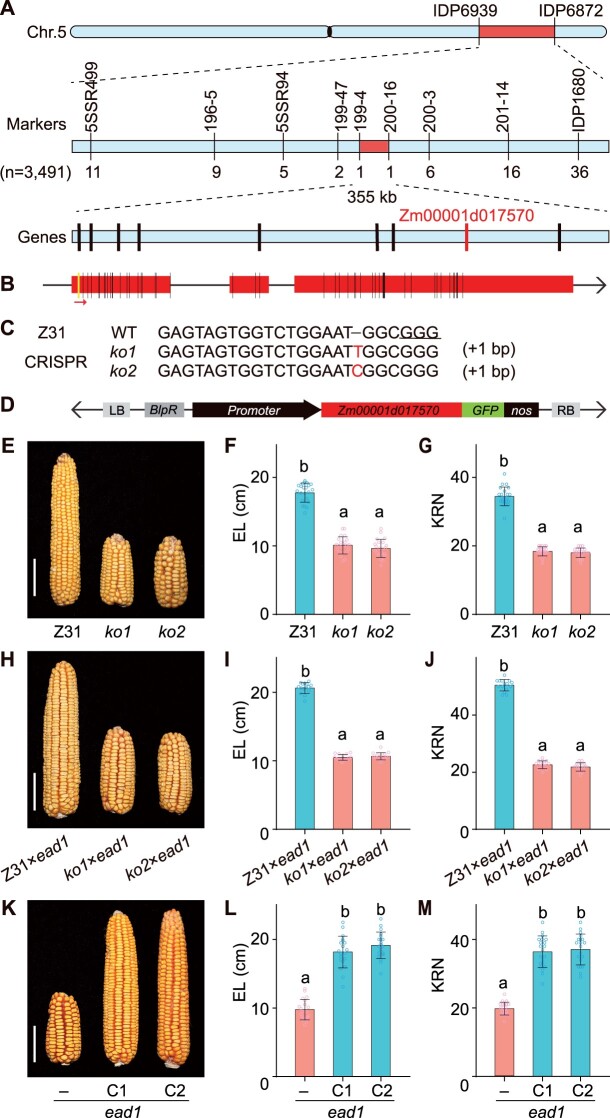
To identify the genetic nature of the ead1 mutation, we crossed the mutant to inbred line B73. All F1 progenies produced well-developed ears without apical degeneration. We backcrossed the F1 plants to the ead1 mutant; the resulting 862 BC1F1 (ead1/B73//ead1) individuals segregated in a 1:1 ratio (445 long ears versus 417 degenerated ears, χ2 = 0.85, P > 0.05; Supplemental Figure S6; Supplemental Table S1), indicating that ead1 is caused by a single recessive mutation. We used the above segregating population for map-based cloning of EAD1, which we initially located to bin 5.06 on the long arm of chromosome 5 between markers IDP6939 and IDP6872. We screened a total of 3,491 individuals for recombinants and narrowed down the mapping region to a 355-kb interval between newly developed markers 199-4 and 200-16. The mapping interval harbored nine putative protein-coding genes based on the B73 reference genome (B73_RefGen_v4, http://www.maizegdb.org), with only Zm00001d017570 exhibiting differences between WT and ead1 (Figure 3A). The predicted gene structure and amino acid sequence of Zm00001d017570 revealed a 3-bp deletion in the first exon and 51 single-nucleotide polymorphisms, resulting in the deletion of one amino acid and 25 amino acid alterations in ead1 (Figure 3B; Supplemental Figures S7 and S8B).
Figure 3.
Map-based cloning and functional validation of EAD1. A, EAD1 was initially mapped to bin 5.06 between markers IDP6939 and IDP6872 on the long arm of chromosome 5. Fine mapping narrowed the region to a 355-kb interval between markers 199-4 and 200-16, covering nine putative genes based on the B73 version 4 reference genome sequence. Genes were indicated by black vertical lines. Only Zm00001d17570 (in red) has mutations between WT and ead1. B, Putative gene structure of Zm00001d17570. Red boxes represent exons. Vertical lines indicate nucleotide variations in the exons of ead1 consisting of a 3-bp deletion (yellow line) and 51 single-nucleotide polymorphisms (black lines). The red arrow indicates the sgRNA target site. C, Genotype of CRISPR/Cas9-mediated ko mutants (ko1 and ko2) of Zm00001d17570. The protospacer adjacent motif is underlined, and nucleotide insertions are shown in red. D, Schematic diagram of the Zm00001d17570 complementation vector. E–G, Functional validation of CRISPR/Cas9 alleles ko1 and ko2 (n = 22). Compared to a nontransgenic line (Z31), the ko mutants produce shorter ears. H–J, Allelism test between the ko mutants ko1 and ko2 and ead1 (n = 15). K–M, Functional validation of genetic complementation. C1 and C2 represent two separate positive transformation events. K, Representative ears from ead1 and complementation lines. Average EL and KNR are shown in L and M (n = 20). Scale bar = 5 cm in (E, H, and K). All values are means ± sd. Different letters indicate significant differences at P < 0.05 determined by analysis of variance (ANOVA) with Tukey’s honestly significant difference (HSD) test.
To determine whether the polymorphisms detected in Zm00001d017570 are responsible for apical degeneration in ead1, we introduced a construct for clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing with a single-guide RNA (sgRNA) targeting the first exon of Zm00001d017570 into maize inbred line Z31 to create mutant alleles. We obtained six independent T0 transgenic events with two types of mutations leading to frameshifts and premature termination of translation of the predicted protein encoded by Zm00001d017570 (Figure 3C; Supplemental Figure S9). Importantly, we observed apical degeneration, H2O2 accumulation and shorter ears, typical phenotypes of ead1, in T2 homozygous knockout (ko) individuals (Figure 3, E–G; Supplemental Figures S10 and S11). Crosses between these ko plants and ead1 failed to rescue the ead1 phenotype, confirming that these new mutants are allelic to ead1 (Figure 3, H–J). We also generated complementation lines by introducing a 4,463-bp genomic fragment encompassing 2,996 bp of upstream promoter region and 1,467 bp of the Zm00001d017570 coding region into Z31 (Figure 3D). We obtained 20 positive transgenic events, of which we randomly selected six for further testing. When introduced into the ead1 background by crossing/selfing and marker-assisted selection, all six transgenic events rescued the ead1 phenotype. We characterized two representative events further, as shown in Figure 3, K–M. These results demonstrate that Zm00001d017570 is the causal gene responsible for the apical degeneration in ead1.
EAD1 is preferentially expressed in vascular tissues of ears and EAD1 localizes to the PM
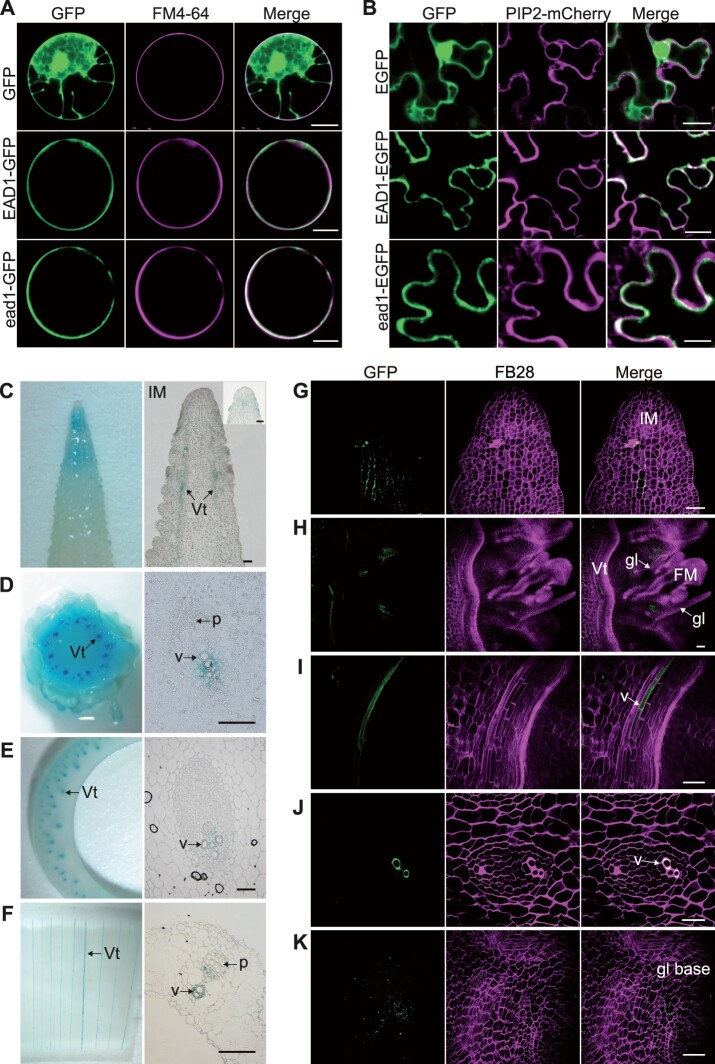
EAD1 was predicted to encode an ALMT containing six potential transmembrane helices close to its N terminus and a cytoplasmic tail at the C-terminus (Supplemental Figure S8). To reveal the subcellular localization of EAD1, we cloned the full-length coding sequences of EAD1 and ead1 in-frame and upstream of the sequence for the enhanced green fluorescent protein (EGFP) to generate the Ubipro:EAD1-GFP and Ubipro:ead1-GFP constructs. While we detected free GFP signal throughout the cell, both EAD1-GFP and ead1-GFP specifically colocalized with the PM-specific dye FM4-64 (Figure 4A; Supplemental Figure S12A). We also generated the constructs 35Spro:EAD1–EGFP and 35Spro:ead1-EGFP (driven by the cauliflower mosaic virus 35S promoter) and co-infiltrated them with the PM marker construct PIP2-mCherry (encoding a fusion between mCherry and PLASMA MEMBRANE INTRINSIC PROTEIN 2 [PIP2]) into Nicotiana benthamiana epidermal cells. Again, both EAD1–EGFP and ead1–EGFP signals overlapped with the mCherry signal (Figure 4B; Supplemental Figure S12B). These results indicated that EAD1 protein localizes to the PM and that the ead1 mutations do not alter the PM localization of the ead1 protein.
Figure 4.
Gene expression of EAD1 and protein tissue-specific localization of EAD1. A, Subcellular localization analysis in maize mesophyll protoplasts. The constructs Ubipro:GFP, Ubipro:EAD1-GFP, and Ubipro:ead1-GFP were transfected in maize protoplasts; the PM was stained with the dye FM4–64. Scale bars = 10 μm. B, Subcellular localization analysis in N. benthamiana epidermal cells. The constructs 35Spro:EGFP, 35Spro:EAD1–EGFP, and 35Spro:ead1–EGFP were separately co-infiltrated with the PM marker PIP2-mCherry. Scale bars = 20 μm. C–F, GUS staining results of ear related tissues in EAD1pro:GUS transgenic lines. C, Ear apical part at the 15-mm stage. D, Ear basal part at the 30-mm stage. E, Sheath of ear-leaf. F, Immature ear bract. Left: entire tissue; right, paraffin sections of GUS-stained tissues. Scale bars = 100 μm. G–K, Tissue-specific accumulation of the EAD1–GFP fusion protein in EAD1pro:EAD1–GFP transgenic maize ears. Positive signals were detected below the apical IM (G), the basal parts of FM glumes (H and K), and xylem vessels of the immature ears (H, I, and J). G, H, I, and K, the longitudinal sections and (J) indicates the transverse section from immature ears. K, the close-up view of the basal part of glume. Vt, vascular tissue; v, xylem vessel; p, phloem; gl, glume; BF, brightfield. FB28 is cell wall dye. Scale bars = 50 μm.
We then turned to an analysis of the EAD1 expression pattern in various tissues including immature ears, tassels, leaves, stems, and roots. Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that EAD1 is expressed in all tissues examined in the WT and showed preferential expression in the apical parts of immature ears and roots; notably, expression levels were much lower decreased in all ead1 mutant tissues examined (Supplemental Figure S13). We also examined the transgenic plants harboring a EAD1pro:GUS reporter construct, whereby the β-glucuronidase (GUS) reporter gene was placed under the control of the EAD1 promoter. We detected GUS activity in both the apical parts and the vascular tissues of ears (left parts of Figure 4, C and D), as well as a weak signal in leaf-sheaths and bracts (left parts of Figure 4, E and F). Detailed observations through longitudinal and transverse sections demonstrated the localization of strong GUS signal in xylem parenchyma cells (right parts of Figure 4, C–F). We also investigated the tissue-specific localization of the EAD1–EGFP fusion protein in EAD1pro:EAD1–EGFP transgenic plants. We detected strong EGFP signals in the xylem vessels of immature ears (Figure 4, H–J) and relatively weak signals in vascular cambium cells below the IM central zone (Figure 4G) and the base of the outer glume of FMs (Figure 4, H and K). Together, these results demonstrated that EAD1 is preferentially expressed in xylem parenchyma cells and that EAD1 protein tissue specifically localizes to the xylem vessels of immature ears.
As EAD1 was predicted to encode an ALMT, we extracted the protein sequences for all nine putative ALMT members from the maize reference proteome. We then constructed a maximum-likelihood (ML) tree with ALMT members from maize, rice (Oryza sativa), and Arabidopsis, as well as determined the chromosomal distribution of ALMT members in the maize and rice genomes (Supplemental Figures S14 and S15; Sharma et al., 2016). Notably, maize EAD1, Zm00001d017571, and rice OsAMLT7 belonged to a monophyletic lineage, suggesting that maize EAD1 and rice OsAMLT7 are possible orthologs. Zm00001d017571 appeared to encode an additional copy of EAD1, while OsAMLT7 was present as a single copy in rice. Zm00001d017571 and EAD1 located ∼70-kb apart on chromosome 5, and their encoded proteins shared 94.1% identity, suggesting that Zm00001d017571 and EAD1 are derived from a tandem duplication event specifically during maize genome evolution (Supplemental Figure S15).
EAD1 functions as a malate transporter responsible for malate transport into apical ears
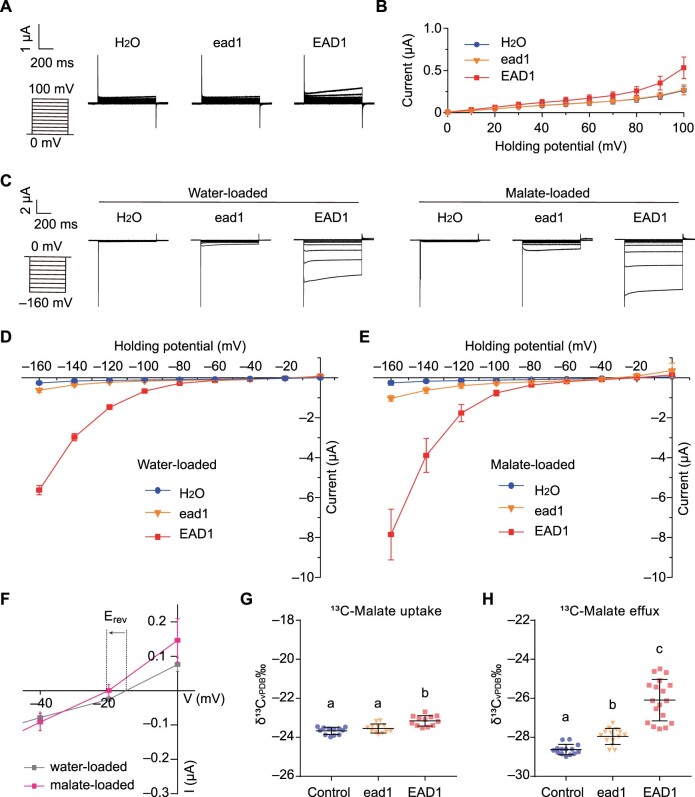
To address the role of EAD1 in maize ear development, we examined its transport properties for malate in the Xenopus laevis oocyte system. Accordingly, we separately injected full-length purified complementary RNA (cRNA) for EAD1 and ead1 into oocytes and analyzed transport using the two-electrode voltage clamp (TEVC) technique and stable isotope labeling. In TEVC experiments, with malate supplemented in the bathing solution, we observed slight outward currents (anion influx) in the oocytes injected with the EAD1-cRNA compared to control oocytes injected with water or oocytes injected with ead1-cRNA (Figure 5, A and B). We also assessed the malate efflux properties of EAD1. We thus preloaded oocytes injected with water, or with EAD1 or ead1 cRNA, with H2O or malate, before measuring their elicited currents in ND96 buffer solution (pH 4.5). EAD1-accumulating oocytes mediated larger inward currents than those recorded in oocytes accumulating ead1 or in oocytes injected with water only (Figure 5, C and D). Increasing the intracellular malate concentration (i.e. malate loaded) resulted in an increase in EAD1-mediated inward currents (Figure 5, C–E), which was accompanied by a negative shift in Erev, the holding potential at which the current reverses sign (Figure 5F). Consistently, when testing malate influx with a 10-mM 13C-labeled malate solution, we measured a slightly greater amount of 13C-labeled malate uptake in EAD1-accumulating oocytes than those with ead1 or water-injected control (Figure 5G). To assess oocytes for malate efflux, we preloaded EAD1-injected oocytes and ead1-injected oocytes with 13C-labeled malate and measured the efflux 2 h after incubation in ND96 bath solution (pH 4.5). The total 13C amount exuded from EAD1-injected oocytes was significantly higher than those with ead1 or water-injected control (Figure 5H). Taken together, these results confirmed that EAD1 mainly mediates malate efflux across the membrane while we barely detected a malate efflux function for ead1.
Figure 5.
Malate transport activity of EAD1. A and B, Outward current recordings in X. laevis oocytes injected with EAD1 or ead1 cRNA. A, Whole-cell currents recorded in oocytes injected with water control or with different cRNAs: EAD1, ead1. The outside solution contained malate at a final concentration of 20 mM. The holding potentials ranged from 0 to 100 mV (in 10-mV increments). The time and current scale bars for the recordings are shown. B, Mean I/V curves in oocytes constructed from steady-state currents recordings in (A). Values are means ± standard error (n = 10). C–E, Inward current recordings in X. laevis oocytes injected with EAD1 or ead1 cRNA. C, The oocytes were preloaded with either water (left) or malate (right). The holding potentials ranged from –160 to 20 mV (in 20-mV increments). The time and current scale bars for the recordings are shown. D and E, Mean I/V curves of the steady state currents in oocytes preloaded with water (D) or malate (E). F, Difference in the magnitude of EAD1-mediated current and shift in the holding potential (Erev) at which the current reverses sign between water-loaded and malate-loaded oocytes accumulating EAD1. G, Malate uptake activity of EAD1 cRNA- and ead1 cRNA-injected oocytes detected in a 10-mM 13C-labeled malate solution. Water-injected oocytes were used as negative control (n = 12). H, Malate efflux activity of EAD1- and ead1-accumulating oocytes. Oocytes preloaded with 36 nL of 100-mM 13C-labeled malate were incubated in the bath solution (ND96, pH 4.5) for 2 h. The total 13C amount of efflux was measured (n = 18). The scatterplots indicate the value of each sample and the black line indicates the means ± sd. Different letters in (G and H) indicate significant difference at P < 0.05 determined by Tukey’s HSD test.
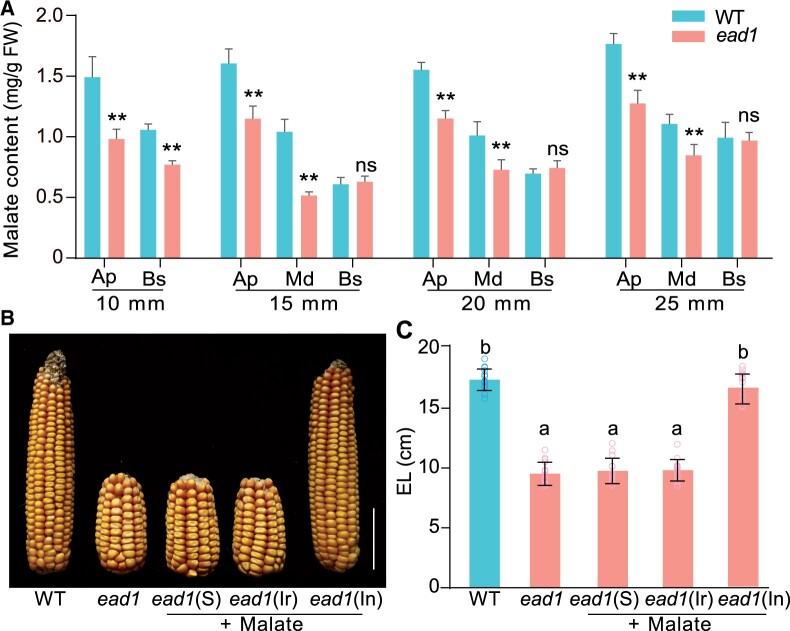
If EAD1 facilitates the transport of malate into the apical parts of immature ears, then malate contents should be altered in different parts of ead1 ears. We thus compared the malate contents in apical, middle, and basal parts of developing ears from WT and ead1. Indeed, we observed a gradual decrease pattern in malate contents from the apical to the basal parts of WT immature ears (Figure 6A). Compared to WT, the malate contents were lower in both the apical and the middle parts of ead1 immature ears when they were 10, 15, 20, and 25 mm in length. We detected significantly lower malate contents in the basal part only in 10-mm immature ears (P < 0.01; Figure 6A). These findings confirm that EAD1 is responsible for the heterogeneous malate distribution in different parts of immature ears and plays an essential role in maintaining malate supply for normal immature ear development.
Figure 6.
Malate content analysis and rescue of the ead1 phenotype in maize. A, Malate contents in the apical, middle and basal parts of developing ears in WT and ead1. About 10-mm ears were divided into two parts (apical and basal parts); the remaining ears were divided into three parts (apical, middle, and basal parts). Pairwise comparisons were conducted between WT and ead1 in each part. Ap, apical; Md, middle; Bs, basal. Values are means, error bars indicate the sd of three biological replicates; **P < 0.01 as determined by two-tailed Student’s t test between WT and ead1. B and C Rescue of the ead1 phenotype by exogenous malate supply. Representative ear phenotype (B) and corresponding average EL (C) of ead1 after exogenous malate supplementation by spraying (indicated by S), irrigation of roots (indicated by Ir) or injection of the stem-adhered ear (indicated by In) with 5-mM malate. Scale bar = 5 cm. Values are means, error bars indicate sd (n = 15). Different letters indicate significant difference at P < 0.05 determined by Tukey’s HSD test.
To further study the role of malate in ear development, we tested whether exogenous applications of malate might rescue the shorter ears of ead1. To this end, we applied exogenous malate solutions following three different methods (spraying plants; root irrigation and injection into ear-adhered stems) when the immature ear was ∼5 mm in length. Only exogenous malate injections resulted in longer mature EL, with a complete rescue of the ead1 mutant phenotype (Figure 6, B and C). The above results validate the notion that the shorter ears in ead1 are caused by endogenous malate deficiency that can be rescue by exogenous malate injection at an early stage of ear development.
Malate transport and metabolism play essential roles in cell proliferation and inflorescence development
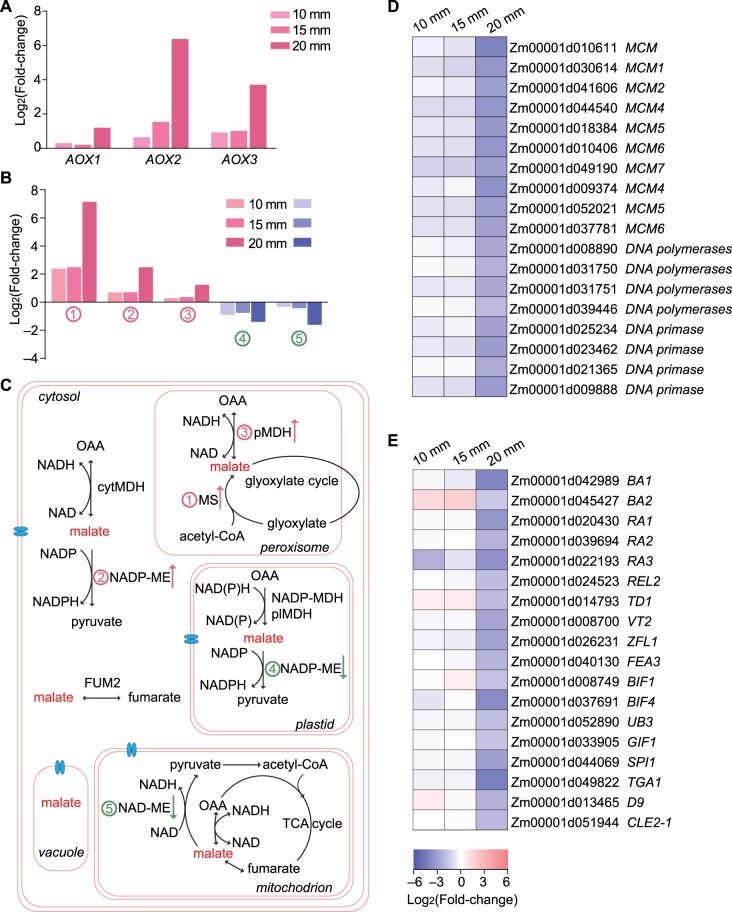
To better understand the gene regulatory network of EAD1 involved in maize female inflorescence development, we extracted total RNA from the apical parts of 10-mm, 15-mm, and 20-mm immature ears of WT and ead1 plants and performed transcriptome deep sequencing (RNA-seq). We confirmed the quality of the data by principal component analysis (Supplemental Figure S16) and generated corresponding transcriptome profiles. We identified 639 (491 upregulated and 148 downregulated, ead1 versus WT) and 609 (389 upregulated and 220 downregulated) differentially expressed genes (DEGs) in 10-mm and 15-mm ears. We also detected 7,411 (2,909 upregulated and 4,502 downregulated) DEGs between the two genotypes at the 20-mm ear stage, when clear apical shrinking of ead1 ear can be observed. The Venn diagram showing the overlapping DEG sets and a heatmap representation of DEG expression levels between WT and ead1 mutant ears are presented in Supplemental Figure S17. Gene ontology (GO) analysis revealed that the most significant enriched DEGs in the cell component category is related to membrane integrity at all three EL stages (Supplemental Figure S18), indicating that EAD1 dysfunction may disrupt the membrane system in ead1. For ears of 10 and 15 mm in length, DEGs were mainly enriched in the biological processes (BPs) RNA metabolism, DNA binding, protein modification (hydrolase activity, catalytic activity, and peptidase activity), and primary metabolism (Supplemental Figure S18, A and B). For ears of 20 mm in length, the upregulated DEGs were enriched in BPs linked protein ubiquitination, H2O2, and ROS response as well as organic acid catabolism and transport (Supplemental Figure S18C). The three well-known ALTERNATIVE OXIDASE (AOX) genes (AOX1, AOX2, and AOX3) with critical roles in counteracting excess ROS (Liu et al., 2019) were upregulated in ead1 (Figure 7A). The DEGs involved in malate metabolism including MS (Zm00001d003247) and pMDH (Zm00001d041243) whose encoding proteins localize to peroxisomes as well as NADP-ME (Zm00001d043601) with NADP-ME accumulating in the cytosol were upregulated, while NADP-ME (Zm00001d003090, encoding a plastid-localized protein) and NAD-ME (Zm00001d013911, encoding a mitochondrion-localized protein) were downregulated across all three stages (Figure 7, B and C). These results may reflect the disorder of malate metabolism across different organelles when EAD1 is not functional. The enriched molecular functions (MFs) were mainly involved in glutathione transferase activity, hydrogen symporter activity, and transmembrane transporter activity (Supplemental Figure S18C). By contrast, the downregulated DEGs were enriched in BP related to the cell cycle, chromosome organization, nucleic acid metabolism, DNA replication, and cell proliferation (Figure 7D; Supplemental Figure S18D). The enriched MFs were mainly related to RNA, DNA, and protein binding activities (Supplemental Figure S18D). In addition, genes involved in inflorescence development were globally downregulated in ead1 (Figure 7E).
Figure 7.
Malate transport and metabolism play essential roles in cell proliferation and inflorescence development in developing ears. A, DEGs involved in the AOX pathway at all three stages. B, Differential expression of malate metabolism-related genes. (1) MS (Zm00001d003247); (2) NADP-ME (Zm00001d043601); (3) pMDH (Zm00001d041243); (4) NADP-ME (Zm00001d003090); (5) NAD-ME (Zm00001d013911). C, Malate metabolism is involved in multiple processes and malate can be easily transported across cell membranes separating various compartments through its transporter (indicated as a blue transmembrane structure). The numbers are the same as in (B). D and E, DEGs involved in DNA replication (D) and inflorescence development (E) are broadly expressed at lower levels in ead1 relative to WT at all three stages.
Overexpression of EAD1 produces longer ears
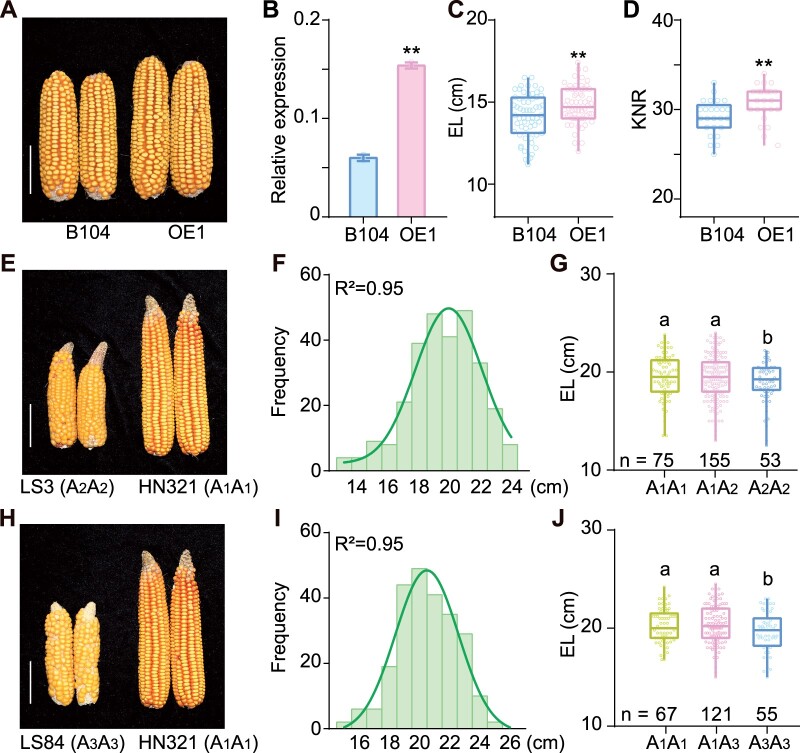
Given that EAD1-mediated malate transport plays a substantial role in immature ear development and elongation, we wondered if EAD1 overexpression might enhance EL and KNR. We thus generated a construct with the EAD1 coding sequence driven by the Ubiquitin (Ubi) promoter, transformed it into the inbred line B104 and obtained the two EAD1-overexpressing lines OE1 and OE2 (Figure 8A). OE1 led to a three-fold increase in EAD1 transcript levels, a 4.9% increase in EL (0.7 ± 0.23 cm; Figure 8, B and C) and a 5.6% increase in KNR (1.64 ± 0.56; Figure 8D) relative to the nontransgenic siblings. We obtained similar results in OE2, with a 7.31% increase seen for EL (Supplemental Figure S19A). We also compared EL between the F1 progeny of the crosses Z31 × B104 and Z31 × B104OE. The ELs of Z31 × B104OE1 F1 and Z31 × B104OE2 F1 progeny were 7.7% and 8.1% longer than those of Z31 × B104 F1 progeny, respectively (Supplemental Figure S19B). These results confirmed that higher EAD1 expression positively regulates EL and KNR in maize.
Figure 8.
Contribution of EAD1 to EL. A–D, EAD1-overexpression line (OE1) produces longer ears than a nontransgenic line (B104). A, Typical ears at the full-ripe stage. B, Relative EAD1 transcript levels, as analyzed by RT-qPCR. Values are means ± sd (n = 3). C and D, Average EL (C) (n = 60) and KNR (D) (n = 25) in OE1 line and B104. **P < 0.01 as determined by two-tailed Student’s t test. E–J, Effect of different EAD1 alleles on EL. E, Representative ears in two inbred lines with the A2A2 (LS3) or A1A1 (HN321) alleles of EAD1. F, Distribution of EL in the F2 segregating population derived from the HN321 × LS3 cross. G, Average EL of the A1A1 and A1A2 genotypes is significantly higher than that of the A2A2 genotype. H, Ears from two inbred lines with the A3A3 (LS84) or A1A1 (HN321) alleles of EAD1. I, Distribution of EL in the F2 segregating population derived from the HN321 × LS84 cross. J, Average EL of the A1A1 and A1A3 genotypes is significantly higher than that of the A3A3 genotype. Boxplots represent the range between first and third quartiles (25%–75%); whiskers, full data range; horizontal center line, median. Different letters in (G and J) indicate significant difference at P < 0.05 determined by Tukey’s HSD test. Scale bars = 5 cm in (A, E, and H).
Furthermore, to study the effect of EAD1 on EL in different genetic backgrounds, we developed two F2 segregating populations derived from HN321 × LS3 and HN321 × LS84 crosses. We determined the genotype of the three EAD1 alleles in HN321, LS3 and LS84 inbred lines (Supplemental Table S4), which we designated here as A1, A2, and A3, respectively (Figure 8, E and H). We also developed allele-specific DNA markers (Supplemental Table S2). EL in both F2 populations followed a normal distribution (R2 = 0.95; Figure 8, F and I). The EL of plants carrying the A1 allele from HN321 (A1A1, A1A2, and A1A3) was significantly higher than that of plants harboring the other alleles from LS3 and/or LS84 (A2A2 and A3A3). Indeed, the A1 allele increased EL by 4.8%–5.2% (0.89–0.97 cm) and 3.5%–3.7% (0.68–0.73 cm) compared to the A2 and A3 alleles, respectively (Figure 8, G and J). These results demonstrate that diverse EAD1 alleles exist in maize inbred lines and different alleles have different effects on EL.
Discussion
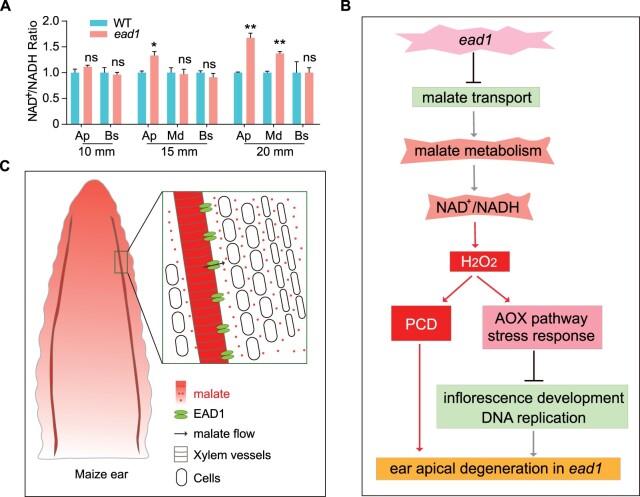
Malate is a vital plant organic acid that participates in the transfer of redox equivalents between cell compartments (Maurino and Engqvist, 2015; Selinski and Scheibe, 2019). Given that malate is always accompanied by transfer of reducing equivalent, we investigated the contents of NAD+ and NADH from different parts of immature ears and determined that these two parameters are less abundant in the apical parts of ead1 immature ears (Supplemental Figure S20, A and B). However, the ratio of NAD+/NADH, typically considered a pivotal determinant of cellular redox status (Noctor et al., 2006; Ying, 2008), had increased by 1.11- to 1.68-fold in ead1 relative to the WT (Figure 9A). As ammonium is the product of nitrate reduction (Tischner, 2000), we also measured ammonium contents to assess the N assimilative capacity of immature ears. The results showed that the ammonium contents of the apical and middle parts of ead1 immature ears start to decrease at the 15-mm stage and was more pronounced at the 20-mm stage (Supplemental Figure S20C). Together, we conclude that transcriptional regulation, redox potential, and metabolism status are disturbed in ead1 apical immature ears. We propose that low malate contents, disturbed malate metabolism, unbalanced cell redox state (NAD+/NADH) together with high H2O2 levels, trigger PCD in ead1 inflorescences. The activated AOX pathway, upregulated stress responses, and the downregulation of genes related to DNA replication and inflorescence development lead to the developmental arrest of ead1 apical ears (Figure 9B). Based on the facts that (1) EAD1 is highly expressed in xylem parenchyma cells and the protein mainly localizes in xylem vessels of immature ears; (2) EAD1 is responsible for the efflux of malate; and (3) EAD1 dysfunction results in lower malate contents in the apical parts of developing inflorescences, we conclude that EAD1 plays essential roles in the delivery of malate through xylem vessels to the apical parts of the immature ear. A working model of EAD1 involved in maize immature ear development is shown in Figure 9C.
Figure 9.
Proposed working model of EAD1 function in maize female inflorescence development. A, Average NAD+/NADH ratio in WT and ead1 in different parts of immature ears. Ap, apical; Md, middle; Bs, basal. Values are means, error bars indicate sd of three biological replicates. *P < 0.05; **P < 0.01 as determined by two-tailed Student’s t test between WT and ead1. B, A working model of impaired maize ear development in the ead1 mutant. C, A proposed working model of functional EAD1 in WT maize immature ear development.
The ALMT protein family comprise a group of anion channels participating in diverse physiological processes including Al resistance, phosphate starvation response, stomatal aperture, panicle development, and plant–microbial interactions in different species (Sasaki et al., 2004; Takanashi et al., 2016; Heng et al., 2018; Medeiros et al., 2018; Zhou et al., 2020). Recently, the rice ALMT gene OsALMT7 was shown to be essential for rice panicle development (Heng et al., 2018). In this study, we discovered that EAD1 shares 73.8% identity with rice OsALMT7. Despite being possible orthologs and having similar biological functions during inflorescence development, major differences also exist between the two proteins. First, maize ead1 mutant plants show apical degeneration of the female inflorescence during IM differentiation, while Osalmt7 mutant plants exhibit apical degeneration after the successful differentiation and determination of the IM (Figure 1, K and L; Heng et al., 2018). These distinct phenotypes may reflect the specific functional stages of maize and rice inflorescence development. Second, our results demonstrated a gradual decrease in malate contents from the apical to the basal parts of WT immature ears, while malate contents of rice panicles are homogeneous. These different distribution patterns may also be attributed to the different inflorescence architecture in maize and rice. Third, EAD1 specifically modulates female ear development but has no effect on the male tassel, which is unique to maize and absent in rice. Fourth, we detected a second copy of EAD1 (Zm00001d017571), likely derived from a tandem duplication event in the maize genome, while OsAMLT7 was present as a single copy in the rice genome, which provides a clue for the evolutionary functional divergence between EAD1 and OsAMLT7, or sub-functionalization of EAD1 and Zm00001d017571. Finally, we showed that EAD1 is highly expressed in xylem parenchyma cells and that the protein mainly localizes to xylem vessels of immature ears. OsALMT7 was reported to be predominantly expressed in the phloem and phloem parenchyma cells of vascular tissues of the spikelet hull and rachis, supporting a role for OsALMT7 in phloem loading of malate. This apparent discrepancy for the cell type expression pattern between maize and rice may have inadvertently been caused by our focus on expression levels mainly in the key stages of EAD1 function, which does not rule out the possibility that EAD1 is expressed in phloem of mature ears. In maize, there are nine putative ALMT members and none of their physiological roles have been fully deciphered (Pineros et al., 2008; Ligaba et al., 2012). The map-based cloning and functional demonstration of EAD1 will lay a solid foundation for further functional studies of the ALMT gene family.
Given that large ears generally produce more kernels to maximize grain yield, researchers have poured much efforts into identifying functional genes to improve ear size. These genes mainly include modulators of IM width through the WUS-CLV signaling pathway. However, complete loss-of-function of these genes often causes dramatic IM fasciation and eventually shorter and disorganized ears, possibly as a consequence of the competition for resources between the massive overproliferated IM and developing kernels. Thus, weak alleles of UB3, FEA2, FEA3, and CLE7 have been tentatively mined to reach a balance between IM proliferation and kernel development, resulting in greater KRN while producing normal ears with well-developed kernels (Bommert et al., 2013b; Liu et al., 2015; Je et al., 2016; Liu et al., 2021). Compared to these genes modulating ear width, few genes regulating EL have been identified (Xiao et al., 2016; Li et al., 2018), largely because EL is a typical quantitative trait controlled by multiple loci. Three QTLs, KNR6, ZmACO2, and YIGE1, have recently been cloned that regulate ear size by improving KNR and EL, providing new insights into ear development and will ultimately contribute to maize grain yield improvement (Jia et al., 2020; Ning et al., 2021; Luo et al., 2022). Our studies reveal that EAD1 indeed plays an essential role in regulating maize EL by maintaining adequate malate supply to apical inflorescences. In contrast to the other malnutrition-related genes THI2, RTE, and TLS1, which are involved in both reproductive and vegetative processes (Woodward et al., 2010; Chatterjee et al., 2014; Durbak et al., 2014), EAD1 specifically modulates ear development but has no effect on tassel or vegetative growth. The ead1 mutant is characterized by shorter ears but increased ERD and HGW, which may be attributed to the balance between “source” and “sink” organs. When EAD1 was nonfunctional, the transport of malate to the apical inflorescence was blocked, thus mimicking the destruction of an apical “sink.” However, the “source” would retain normal function, as vegetative growth is not affected. Nutrients and energy would thus accumulate at the basal part of the degenerated ear, leading to larger seeds and increased ERD and HGW. EAD1 showed very weak expression in the tassel, which may explain the lack of phenotype in mutant tassels. Our results demonstrate a gradual decrease in malate contents from the apical portion to the basal part of WT immature ears. This pattern may mean that the apical part of the immature ear is the most developmentally active region that needs more malate to sustain proper structure and growth. This greater need for malate in the apical region may also explain why lines overexpressing EAD1 produced longer ears than their untransformed siblings. We also found that different alleles of EAD1 have different effects on EL, as the HN321 allele of EAD1 exhibited a positive association with EL in two different segregating populations. Possible reasons include higher expression of the HN321 allele or that the encoded protein displays a higher affinity toward malate. Further study is needed to address the mechanistic difference between EAD1 alleles in maize ear formation. Our work provides a potential gene resource for future yield improvement in maize.
Materials and methods
Plant materials and growth conditions
The maize lines and populations used in this study were planted in the experimental fields either in Beijing (39°N, 116°E) from the end of April to the start of May, or in Sanya (18°N, 108°E), Hainan province from the end of October to the start of December. The transgenic plants were grown in the greenhouse with a 16-h light (25°C)/8-h dark (18°C) cycle. Nicotiana benthamiana plants used for subcellular localization experiments were grown in a light incubator with a 16-h light/8-h dark photoperiod at 22°C.
Phenotypic characterization of the ead1 mutant
A Canon digital camera (EOS 550D) was used to take all photographs. Air-dried ears from the WT (HN321) and ead1 were used to measure (cm), KNR, ERD (mm), HGW (g), GYE (g), and KRN. Tassel main axis length (cm), tassel branch number as well as other vegetative traits were all measured from mature plants after flowering. Pollen grains were stained with 1% (w/v) I2-KI and photographed with a BX53 microscope (Olympus, Tokyo, Japan). For cryo-SEM analysis, fresh immature ears of WT and ead1 were fixed to the loading table and rapidly frozen in liquid N. The samples were then freeze-dried, sputter-coated with gold, and analyzed by cryo-SEM with a HITACHI S-3000N Scanning Electron Microscope carrying a Quorum refrigeration system. For TEM analysis, apical parts of fresh immature ears were vacuum infiltrated and fixed in 4% (w/v) glutaraldehyde, followed by rinsing, replacement, dehydration, embedding, and sectioning as described previously (Huo et al., 2020). Images were obtained with a HITACHI H-7500 transmission electron microscope.
DAB staining and TUNEL assay
Fresh immature ears were immersed in freshly prepared DAB staining solution (1 mg mL−1, pH 3.8) and infiltrated under vacuum for 10 min. Stained ears were cleared of pigments in 100% ethanol for 60 min with three changes at room temperature, and then stored in 75% (v/v) ethanol. Apical parts of the stained ears were then dehydrated with a graded ethanol series and embedded in paraffin. Eight-micrometer thick sections were prepared using a microtome (RM2235; Leica, Wetzlar, Germany) and photographed with a BX53 microscope. The TUNEL assay was performed using the DeadEnd Fluorometric TUNEL System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Samples were analyzed under a laser scanning confocal microscope (LSM 710 NLO; Zeiss, Oberkochen, Germany).
Fine mapping of EAD1 and generation of maize transgenic plants
F1 plants were generated by crossing ead1 with the maize reference cultivar B73, and a BC1F1 segregating population was obtained by backcrossing. The locus was initially mapped to chromosome 5. Additional insertion/deletion and simple sequence repeat markers were developed for fine mapping. The primers for mapping are listed in Supplemental Table S2.
For functional complementation, the coding sequence of EAD1 (without stop codon) was cloned in-frame and upstream of the GFP sequence, placed under the control of a 2,996-bp EAD1 promoter fragment, and terminated by the Nos terminator. The entire cassette was inserted into pCAMBIA3300 to generate the complementation construct EAD1pro:EAD1-GFP. Positive transgenic plants were selected based on BASTA resistance and backcrossed to ead1; F2 individuals were assayed. A 20-bp sgRNA (5′-GAGTAGTGGTCTGGAATGGC-3′) targeting the first exon of Zm00001d017570 was inserted into the pBUN411 vector to generate CRISPR/Cas9-mediated mutants. Positive transgenic plants carrying the CRISPR/Cas9 construct were genotyped to test for mutations. The 2,996-bp EAD1 promoter fragment from HN321 was cloned into pCAMBIA1301 to generate the construct EAD1pro:GUS. The coding sequence of EAD1 was inserted into pCAMBIA2300-EGFP between the maize Ubi promoter and GFP to generate the overexpression construct Ubipro:EAD1–EGFP. All individual constructs were introduced into maize via Agrobacterium (Agrobacterium tumefaciens)-mediated infection (strain EHA105; Frame et al., 2002, 2011), with EAD1pro:EAD1-GFP, the CRISPR/Cas9 construct and EAD1pro:GUS transformed into inbred line Z31, and Ubipro:EAD1–EGFP into inbred line B104. The primers used for cloning are listed in Supplemental Table S3.
Subcellular localization of EAD1
The TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict putative transmembrane domains in EAD1. The full-length coding sequence of EAD1 and ead1 were cloned in-frame with GFP to generate the Ubipro:EAD1-GFP and Ubipro:ead1-GFP constructs using pJIT-163-GFP vector as backbone and the 35S:EAD1–EGFP and 35S:ead1–EGFP constructs using the pCAMBIA1300-EGFP vector. The resulting plasmids were transfected into maize mesophyll protoplasts using polyethylene glycol-mediated transient transfection or individually co-transfected with the PM marker PIP2-mCherry into N. benthamiana mesophyll cells (Yoo et al., 2007). The fluorescence signals were recorded with a laser scanning confocal microscope. The indicated numbers of randomly selected protoplasts or N. benthamiana mesophyll cells with mCherry and GFP accumulation were quantified for fluorescence colocalization analyses using Fiji (Schindelin et al., 2012). The primers used are listed in Supplemental Table S3.
Expression analysis of EAD1 and promoter GUS assay
Maize tissues were harvested for total RNA isolation using the TRIzol reagent (Invitrogen, Waltham, MA, USA). For RT-qPCR analysis, total RNA samples were digested with RNase-free DNaseI (Transgen, Beijing, China) and cDNA was produced using total RNA and oligo(dT)18 primers (Thermo RevertAid First-Strand cDNA Synthesis Kit). qPCR was performed with SYBR Premix (Roche LightCycler 480 SYBR Green I Master) using a MyiQ5 single color Real-Time PCR Detection System (Roche, Basel, Switzerland). The comparative threshold cycle method was used for determining relative transcript levels using GAPDH as an internal control. Three biological and three technical replicates were performed. The primer sequences used are listed in Supplemental Table S3.
For GUS staining, tissues from EAD1pro:GUS transgenic T2 lines were immersed in 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid/cyclohexyl ammonium salt (X-gluc) staining solution (1× phosphate-buffered saline, pH 7.0; 10-mM EDTA, pH 7.0; 0.5-mM potassium ferricyanide; 0.5-mM potassium ferrocyanide; 0.1% [v/v] Triton X-100, and 0.1% [v/v] X-gluc), vacuum infiltrated for 15 min and incubated at 37°C overnight. Chlorophyll and pigments were removed from the stained tissues in 70% (v/v) ethanol and examined under a SZX7 stereomicroscope (Olympus). The samples were then dehydrated with a graded ethanol series and embedded in paraffin. Eight-micrometer thick sections were prepared using a microtome (RM2235; Leica) and photographed with a BX53 microscope.
Confocal analysis of EAD1–GFP fusion protein
Fresh immature ears from EAD1pro:EAD1–GFP transgenic lines were collected and immediately placed in 2.5% (w/v) paraformaldehyde (Sigma-Aldrich, St Louis, MO, USA) at pH 7.0 at 4°C, and then vacuum infiltrated for 30 min before being stored overnight at 4°C. Sample preparation for confocal analysis was conducted as previously described (Wang et al., 2014). Fifty-micrometer thick sections were prepared using a Leica VT1000S vibratome. Samples were stained with 50-μgmL−1 cell wall dye FB28 (Sigma-Aldrich) for 20 min. Images were acquired with a two-photon fluorescence microscope (LSM 780 NLO; Zeiss).
Phylogenetic analysis
The amino acid sequences of the nine ALMT members from maize were retrieved from EnsemblPlants (http://plants.ensembl.org/index.html) and tested against the HMM profiles in Pfam (http://pfam.xfam.org/). The sequence alignment was performed with ClustalW (https://www.genome.jp/tools-bin/clustalw) using default parameters. An unrooted maximum likelihood (ML) tree was constructed with the program MEGA version 6.0 (https://www.megasoftware.net/) using the best model LG with Rates among Sites as Gamma Distributed (G), partial deletion, and the 1,000-bootstrap method. The graphs of chromosomal distribution of ALMT members were retrieved from TBtools (Chen et al., 2020).
Electrophysiological measurements in X. laevis oocytes
The coding sequences of EAD1 and ead1 were amplified and cloned into the plasmid pGHME2 (Chen et al., 2010). The resulting constructs were then linearized and transcribed into cRNA using T7 polymerase. Oocytes were injected with 36 ng of cRNA and cultured in ND96 solution (96-mM NaCl, 2-mM KCl, 1-mM MgCl2, 1.8-mM CaCl2, 5-mM HEPES, pH 7.4) at 18°C for 2 days. The two-electrode voltage clamp technique was used for electrophysiological studies. The outward currents were recorded in a bath solution containing malate (20-mM Na-malate, 60-mM Na-gluconate, 1-mM Ca-gluconate, 1-mM Mg-gluconate, 1-mM K-gluconate, and 10-mM MES, pH 5.6). Oocytes were clamped with 800 ms pulses from 0 to 100 mV with 10 mV increments and resting for 1.2 s between two pulses. The inward currents were recorded in oocytes preloaded with 36-nL nuclease-free water or 100-mM Na-malate. The recording solution (96-mM NaCl, 2-mM KCl, 1-mM MgCl2, 1.8-mM CaCl2, 5-mM HEPES, 0.1-mM LaCl3, pH 4.5) was used. Oocytes were clamped with 800-ms pulses from –160 to 0 mV with 20-mV increments and resting for 1.2 s between two pulses. The mean current–voltage (I/V) curves were preliminarily processed by Clampfit (Axon, American) and plotted in Origin2020b. Reversal potential was calculated through cross points of the I–V curves and x axes (I = 0). Experiments were performed at least 3 times independently; representative data from one experiment were shown.
13C-malate uptake and efflux activity analysis in X. laevis oocytes
Stable isotope labeling was used to measured EAD1 transport activity. For malate uptake assays, X. laevis oocytes were transferred into 13C-labeled (99% atom; Sigma-Aldrich) malate solution (ND96 supplemented with 10-mM 13C-malate, pH 5.6). After an incubation of 2 h, oocytes were washed and dried at 80°C. For malate efflux experiments, oocytes were preloaded with 36-nL 13C-malate (100 mM) and separately incubated in 110-μL ND96 solution (pH 4.5) for 2 h. The effluxes were respectively collected and dried in tin capsules at 80°C. Finally, samples were ground and the δ13CvPDB‰ was determined using an isotope ratio mass spectrometer with an elemental analyzer (Thermo Finnigan DeltaPlus XP; Flash EA 1112).
Analysis of malate contents
Developing ears (EL = 10, 15, 20, and 25 mm) from WT and ead1 were collected. The 10-mm ears were divided into two parts (apical and basal), while the other ears were divided into three parts (apical, middle, and basal) to measure their malate contents according to the previously described enzymatic method (Delhaize, 1993; Heng et al., 2018). Briefly, samples were ground to a fine powder in liquid N, and ∼50 mg powder was extracted with 1-mL water and incubated at 95°C for 20 min. The supernatant was then collected after centrifugation (more than 10,000 rcf) for 15 min at room temperature. A 0.1-mL aliquot of each supernatant or standards (ranging from 0 to 0.5 mM) was taken and mixed with buffer (1 mL of 0.05-M glycine, pH 10; 0.2 mL of 30-mg/mL NAD; 2 μL of 4 U/μL glutamic-oxaloacetic transaminase) and 0.9-mL water. The mix was preincubated for 10 min at room temperature to obtain a stable absorbance reading (SYNERGY HTX, BioTek) at 340 nm, defined as R1, followed by the addition of 4-μL 25 U/μL MDH to activate the reaction for 10 min. The absorbance readings were recorded at 340 nm as R2. The values of (R1–R2) were used to plot a standard curve and the malate contents were determined. Experiments were performed three times and representative data from one experiment were shown.
Exogenous malate supplementation to ead1
ead1 plants growing in the field were used for the malate supplementation tests. A volume of 0.5-mL malate solution (5 mM, pH 5.6) was injected into the ear-adhered stem of ead1 at the 5-mm ear-length stage with a 1-mL plastic syringe. Additional groups including ead1 plants without exogenous malate supplementation and ead1 plants sprayed with exogenous malate (50 mL, 5 mM, pH 5.6) or irrigated (50 mL, 5 mM, pH 5.6) were set as controls. An additional malate supplementation was performed 3 days later. At least 15 individuals were used for each group. EL was evaluated at the mature stage. The supplementation experiments were performed three times over three successive years.
Transcriptome analysis
Transcriptome profiling was carried out on the apical parts of 10-, 15-, and 20-mm immature ears from WT and ead1. To improve the representativeness of transcriptome data of the samples, 10 immature ears were collected as one sample pool, with four biological replicates for each stage group, corresponding to a total of 24 immature ear groups. All samples were processed into cDNA libraries using the Illumina TruSeq Stranded Total RNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) and were sequenced on an Illumina NovaSeq instrument (Illumina) as 150-nt paired-end reads. Clean data after adaptor and quality trimming were mapped to the B73 reference genome (B73_RefGen_v4) with the program STAR (Dobin et al., 2013). Read counts per gene were used for differential expression analysis with the tool DESeq2 (Love et al., 2014) with 1% false discovery rate as the threshold to declare significant DEGs (P < 0.05, fold-change > 2) between WT and ead1. GO analysis was performed using agriGO version 2.0 (Tian et al., 2017).
Analysis of NAD+, NADH, and ammonium contents
The BioAssay Systems EnzyChrom NAD+/NADH assay kit (E2ND-100) was used to determine the contents for NAD+ and NADH according to the manual. Indophenol blue colorimetry was performed at 550 nm to measure ammonium concentration (Santoni et al., 2001). The content analyses were performed three times and representative data from one experiment were shown.
Accession numbers
The RNA-seq datasets are available from the National Center for Biotechnology Information, under BioProject number PRJNA718682 and SRA accession numbers SAMN18558475–SAMN18558486, SAMN22518025–SAMN22518036.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparisons of ear, tassel, and vegetative traits between WT and ead1.
Supplemental Figure S2. Quantitative analyses of ear development rate in WT and ead1.
Supplemental Figure S3. SEM analysis of lateral meristems in apical ears.
Supplemental Figure S4. Comparisons of mature male florets at the flowering stage and mature tassels after powdering in WT and ead1.
Supplemental Figure S5. Transmission electron microscopy analysis of apical ears at the 20-mm stage in WT and ead1.
Supplemental Figure S6. Representative ear phenotypes of the BC1F1 (ead1/B73//ead1) segregating population.
Supplemental Figure S7 . Coding sequence alignment of EAD1 in WT and ead1.
Supplemental Figure S8. Amino acid sequence analysis of EAD1.
Supplemental Figure S9. CRISPR/Cas9-mediated mutations in Zm00001d17570.
Supplemental Figure S10. Developing immature ears in Z31 and Zm00001d017570 knockout line.
Supplemental Figure S11. Pairwise comparison of DAB-stained immature ears of ead1 ko mutants and Z31.
Supplemental Figure S12. Quantitative analyses of fluorescence co-localization.
Supplemental Figure S13. RT-qPCR analysis of EAD1 expression in different tissues of WT and ead1.
Supplemental Figure S14. Maximum-likehood (ML) phylogenetic tree of the ALMT protein family in maize, rice and Arabidopsis.
Supplemental Figure S15. Chromosomal distribution of ALMT members in maize and rice.
Supplemental Figure S16. Quality control analysis of the transcriptome data.
Supplemental Figure S17. EAD1 dysfunction leads to transcriptional reprogramming in maize immature ears.
Supplemental Figure S18. Gene ontology (GO) analysis of DEGs between WT and ead1 in three ear length stages.
Supplemental Figure S19. Contribution of EAD1 overexpression to EL.
Supplemental Figure S20. Contents of NAD+, NADH, and ammonium in different parts of immature ears.
Supplemental Table S1. Chi-square fitness test of the BC1F1 segregating population (ead1/B73//ead1).
Supplemental Table S2. List of primers used for map-based cloning of EAD1.
Supplemental Table S3. List of primers used for functional analysis.
Supplemental Table S4. Polymorphic amino acids in the three EAD1 alleles.
Supplemental File S1. Amino acid alignment used for the phylogenetic tree shown in Supplemental Figure S14.
Supplemental File S2. Newick format of the phylogenetic tree shown in Supplemental Figure S14.
Supplemental Data Set 1. Summary of statistical analyses.
Supplementary Material
Acknowledgments
We thank Prof. Chengcai Chu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), Prof. Baichen Wang (Institute of Botany, Chinese Academy of Sciences) and Prof. Mingliang Xu (China Agricultural University) for their critical discussions.
Funding
This work was supported by the National Key Research and Development Programs of China (2016YFD0101803) and the National Natural Science Foundation of China (31671699).
Conflict of interest statement. None declared.
Contributor Information
Yuanrong Pei, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Yanan Deng, College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China; State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Huairen Zhang, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Zhaogui Zhang, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Jie Liu, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Zhibin Chen, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Darun Cai, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Kai Li, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Yimo Du, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Jie Zang, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China.
Peiyong Xin, National Center for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Jinfang Chu, National Center for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Yuhang Chen, College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing 100039, China; State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Li Zhao, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Juan Liu, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Huabang Chen, State Key Laboratory of Plant Cell and Chromosome Engineering, Innovative Academy of Seed Design, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
H.C. designed, supervised, and revised the project. Y.P. performed most of the experiments and field works. Y.Deng designed and performed electrophysiological experiments and stable isotope labeling experiments. Juan L. and H.Z. analyzed the transcriptome data. L.Z., Z.Z., Jie L., Z.C., D.C., K.L., Y.Du., and J.Z. conducted field works and experiments. Y.C., J.C., and P.X. provided technical assistance. Y.P. and Juan L. wrote the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plcell) is Juan Liu (juanliu@genetics.ac.cn).
References
- Bommert P, Je BI, Goldshmidt A, Jackson D (2013a) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr W (2005) thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bommert P, Nagasawa NS, Jackson D (2013b) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet 45: 334–337 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Palmer BG, Stanley S, Hajaji H, Janacek SH, Astley HM, Parsley K, Kajala K, Quick WP, Trenkamp S, et al. (2010) C4 acid decarboxylases required for C4 photosynthesis are active in the mid-vein of the C3 species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. Plant J 61: 122–133 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Tabi Z, Galli M, Malcomber S, Buck A, Muszynski M, Gallavotti A (2014) The boron efflux transporter ROTTEN EAR is required for maize inflorescence development and fertility. Plant Cell 26: 2962–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative Toolkit developed for interactive analyses of big biological data. Mol Plant 13: 1194–1202 [DOI] [PubMed] [Google Scholar]
- Chen YH, Hu L, Punta M, Bruni R, Hillerich B, Kloss B, Rost B, Love J, Siegelbaum SA, Hendrickson WA (2010) Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature 467: 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Li W, Gaines C, Buck A, Galli M, Gallavotti A (2021) Structural variation at the maize WUSCHEL1 locus alters stem cell organization in inflorescences. Nat Commun 12: 2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Gene Dev 12: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Chuck GS, Brown PJ, Meeley R, Hake S (2014) Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci USA 111: 18775–18780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck GS, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum aestivum L.) II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbak AR, Phillips KA, Pike S, O'Neill MA, Mares J, Gallavotti A, Malcomber ST, Gassmann W, McSteen P (2014) Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. Plant Cell 26: 2978–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame B, Main M, Schick R, Wang K (2011) Genetic transformation using maize immature zygotic embryos. Methods Mol Bio 710: 327–341 [DOI] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SE, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P (2008) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Long JA, Stanfield S, Yang X, Jackson D, Vollbrecht E, Schmidt RJ (2010) The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Malcomber S, Gaines C, Stanfield S, Whipple C, Kellogg E, Schmidt RJ (2011) BARREN STALK FASTIGIATE1 is an AT-hook protein required for the formation of maize ears. Plant Cell 23: 1756–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pè ME, Schmidt RJ (2004) The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, et al. (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci USA 112: 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA, Leaver CJ, Smith SM (1992) Induction of malate synthase gene expression in senescent and detached organs of cucumber. Plant Cell 4: 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y, Wu C, Long Y, Luo S, Ma J, Chen J, Liu J, Zhang H, Ren Y, Wang M, et al. (2018) OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell 30: 889–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Pei Y, Tian Y, Zhang Z, Li K, Liu J, Xiao S, Chen H, Liu J (2020) IRREGULAR POLLEN EXINE2 encodes a GDSL lipase essential for male fertility in maize. Plant Physiol 184: 1438–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Bykova NV (2018) Role of organic acids in the integration of cellular redox metabolism and mediation of redox signalling in photosynthetic tissues of higher plants. Free Radic Bio Med 122: 74–85 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Eprintsev AT (2016) Organic acids: the pools of fixed carbon involved in redox regulation and energy balance in higher plants. Fron Plant Sci 7: 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Arevalo ED, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, et al. (2016) Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet 48: 785–791 [DOI] [PubMed] [Google Scholar]
- Je BI, Xu F, Wu Q, Liu L, Meeley R, Gallagher PJ, Corcilius L, Payne RJ, Bartlett ME, Jackson D (2018) The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. eLife 7: e35673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Li M, Li W, Liu L, Jian Y, Yang Z, Shen X, Ning Q, Du Y, Zhao R, et al. (2020) A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat Commun 11: 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco D, Hake S (2002) The indeterminate floral apex1 gene regulates meristem determinacy and identity in the maize inflorescence. Development 129: 2629–2638 [DOI] [PubMed] [Google Scholar]
- Li M, Zhong W, Yang F, Zhang Z (2018) Genetic and molecular mechanisms of quantitative trait loci controlling maize inflorescence architecture. Plant Cell Physiol 59: 448–457 [DOI] [PubMed] [Google Scholar]
- Ligaba A, Maron L, Shaff J, Kochian L, Pineros M (2012) Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ 35: 1185–1200 [DOI] [PubMed] [Google Scholar]
- Liu L, Du YF, Shen XM, Li MF, Sun W, Huang J, Liu ZJ, Tao YS, Zheng YL, Yan JB, et al. (2015) KRN4 controls quantitative variation in maize kernel row number. Plos Genet 11: e1005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu Q, Bartlett M, Jackson D (2021) Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat Plants 7: 287–294 [DOI] [PubMed] [Google Scholar]
- Liu Q, Galli M, Liu X, Federici S, Buck A, Cody J, Labra M, Gallavotti A (2019) NEEDLE1 encodes a mitochondria localized ATP-dependent metalloprotease required for thermotolerant maize growth. Proc Natl Acad Sci USA 116: 19736–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, He Y, Zhao Y, Xu Q, Wu J, Ma H, Guo H, Bai L, Zuo J, Zhou JM, et al. (2019) Regulation of mitochondrial NAD pool via NAD+ transporter 2 is essential for matrix NADH homeostasis and ROS production in Arabidopsis. Sci China Life Sci 62: 991–1002 [DOI] [PubMed] [Google Scholar]
- Luo Y, Zhang M, Liu Y, Liu J, Li W, Chen G, Peng Y, Jin M, Wei W, Jian L, et al. (2022) Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New phytol 234: 513–526 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Engqvist MK (2015) 2-Hydroxy acids in plant metabolism. Arabidopsis Book 13: e0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S (2007) barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros DB, Fernie AR, Araujo WL (2018) Discriminating the function(s) of guard cell ALMT channels. Trends Plant Sci 23: 649–651 [DOI] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ (1996) Diversification of C-function activity in maize flower development. Science 274: 1537–1540 [DOI] [PubMed] [Google Scholar]
- Ning Q, Jian Y, Du Y, Li Y, Shen X, Jia H, Zhao R, Zhan J, Yang F, Jackson D, et al. (2021) An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield. Nat Commun 12: 5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Queval G, Gakière B (2006) NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 57: 1603–1620 [DOI] [PubMed] [Google Scholar]
- Pastore D, Di Pede S, Passarella S (2003) Isolated durum wheat and potato cell mitochondria oxidize externally added NADH mostly via the malate/oxaloacetate shuttle with a rate that depends on the carrier-mediated transport. Plant Physiol 133: 2029–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineros MA, Cancado GM, Maron LG, Lyi SM, Menossi M, Kochian LV (2008) Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: the case of ZmALMT1 - an anion-selective transporter. Plant J 53: 352–367 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Xu C, Kwon CT, Soyars C, Demesa-Arevalo E, Man J, Liu L, Lemmon ZH, Jones DS, Van Eck J, et al. (2019) Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat Genet 51: 786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni S, Bonifacio E, Zanini E (2001) Indophenol blue colorimetric method for measuring cation exchange capacity in sandy soils. Commun Soil Sci Plan 32: 2519–2530 [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ju Ahn S, R. Ryan P, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Selinski J, Scheibe R (2019) Malate valves: old shuttles with new perspectives. Plant Biol 21: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T, Dreyer I, Kochian L, Pineros MA (2016) The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front Plant Sci 7: 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable J, Vollbrecht E (2019) Maize YABBY genes drooping leaf1 and drooping leaf2 regulate floret development and floral meristem determinacy. Development 146: 171181. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Gene Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi K, Sasaki T, Kan T, Saida Y, Sugiyama A, Yamamoto Y, Yazaki K (2016) A Dicarboxylate transporter, LjALMT4, mainly expressed in nodules of Lotus japonicus. Mol Plant Microbe In 29: 584–592 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S (2009) bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45: W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischner R (2000) Nitrate uptake and reduction in higher and lower plants. Plant Cell Environ 23: 1005–1024 [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES, Martienssen R (2005) Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz E, Jiao Y (2014) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QL, Sun AZ, Chen ST, Chen LS, Guo FQ (2018) SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat Plants 4: 280–288 [DOI] [PubMed] [Google Scholar]
- Woodward JB, Abeydeera ND, Paul D, Phillips K, Rapala-Kozik M, Freeling M, Begley TP, Ealick SE, McSteen P, Scanlon MJ (2010) A maize thiamine auxotroph is defective in shoot meristem maintenance. Plant Cell 22: 3305–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Xu F, Jackson D (2018) All together now, a magical mystery tour of the maize shoot meristem. Curr Opin Plant Biol 45: 26–35 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Tong H, Yang X, Xu S, Pan Q, Qiao F, Raihan MS, Luo Y, Liu H, Zhang X, et al. (2016) Genome-wide dissection of the maize ear genetic architecture using multiple populations. New Phytol 210: 1095–1106 [DOI] [PubMed] [Google Scholar]
- Yao H, Skirpan A, Wardell B, Matthes MS, Best NB, McCubbin T, Durbak A, Smith T, Malcomber S, McSteen P (2019) The barren stalk2 gene is required for axillary meristem development in maize. Mol Plant 12: 374–389 [DOI] [PubMed] [Google Scholar]
- Ying W (2008) NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Sign 10: 179–206 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zafar SA, Patil SB, Uzair M, Fang J, Zhao J, Guo T, Yuan S, Uzair M, Luo Q, Shi J, et al. (2020) DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine beta-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol 225: 356–375 [DOI] [PubMed] [Google Scholar]
- Zhang D, Sun W, Singh R, Zheng Y, Cao Z, Li M, Lunde C, Hake S, Zhang Z (2018) GRF-interacting factor1 (gif1) regulates shoot architecture and meristem determinacy in maize. Plant Cell 30: 360–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Luo L, Xu J, Xin P, Guo H, Wu J, Bai L, Wang G, Chu J, Zuo J, et al. (2018) Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res 28: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Neuhauser B, Neumann G, Ludewig U (2020) LaALMT1 mediates malate release from phosphorus-deficient white lupin root tips and metal root to shoot translocation. Plant Cell Environ 43: 1691–1706 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.