Abstract
Despite the identification of temperature sensors and downstream components involved in promoting stem growth by warm temperatures, when and how previous temperatures affect current plant growth remain unclear. Here we show that hypocotyl growth in Arabidopsis thaliana during the night responds not only to the current temperature but also to preceding daytime temperatures, revealing a short-term memory of previous conditions. Daytime temperature affected the levels of PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and LONG HYPOCOTYL 5 (HY5) in the nucleus during the next night. These factors jointly accounted for the observed growth kinetics, whereas nighttime memory of prior daytime temperature was impaired in pif4 and hy5 mutants. PIF4 promoter activity largely accounted for the temperature-dependent changes in PIF4 protein levels. Notably, the decrease in PIF4 promoter activity triggered by cooling required a stronger temperature shift than the increase caused by warming, representing a typical hysteretic effect; this hysteretic pattern required EARLY-FLOWERING 3 (ELF3). Warm temperatures promoted the formation of nuclear condensates of ELF3 in hypocotyl cells during the afternoon but not in the morning. These nuclear speckles showed poor sensitivity to subsequent cooling. We conclude that ELF3 achieves hysteresis and drives the PIF4 promoter into the same behavior, enabling a short-term memory of daytime temperature conditions.
The transcriptional regulator ELF3 forms nuclear condensates in the warmth whose reversal shows poor sensitivity to cooler temperatures, thereby storing information about previous warm conditions.
IN A NUTSHELL.
Background: Plants increase their stature in response to warm temperatures by enhancing stem growth. One thermosensor is EARLY FLOWERING 3 (ELF3). ELF3 represses the expression of PHYTOCHROME INTERACTING FACTOR 4 (PIF4), encoding a protein that promotes stem growth. In turn, warmth decreases the activity of ELF3 via a process that involves transitions in the liquid phase where ELF3 is present in the nucleus, increasing the levels of PIF4 to enhance stem growth. Another thermosensor is phytochrome B, which represses the activity of CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1). In turn, COP1 represses the activity of ELONGATED HYPOCOTYL 5 (HY5), which represses growth; warm temperatures inhibit this pathway to promote growth.
Question: Since plants experience fluctuating temperatures, we wanted to investigate whether they possess molecular mechanisms that store information about daytime temperatures to control stem growth during the night.
Findings: We observed that daytime temperatures affected the nighttime growth of the stem (hypocotyl). We analyzed the molecular dynamics of the key components of the signaling network and hypocotyl growth in mutants under different day/night temperatures. The short-term growth memory required ELF3, PIF4, COP1, and HY5, which carried daytime temperature information into the night. In response to increasing temperatures, ELF3 was grouped into condensates associated with reduced activity. However, upon cooling, these condensates barely changed. PIF4, controlled by ELF3, followed the same pattern. A system shows “hysteresis” when, as shown here, the response to an increasing or decreasing variable (such as temperature) shows different levels of sensitivity. We demonstrate that memorization of past conditions involving the formation of cellular condensates, which occurs in yeast and Drosophila, also occurs in plants.
Next steps: The natural environment of plants is complex due to the simultaneous exposure to several fluctuating cues. We aim to explore the molecular mechanisms involved in integrating this complex information.
Introduction
Warmer temperatures within the physiological range can selectively increase or decrease the growth of different plant organs. These changes lead to modifications in plant architecture, a process known as thermomorphogenesis (Quint et al., 2016; Casal and Balasubramanian, 2019). Thermomorphogenesis occurs in crop species, highlighting the need to further understand this process in the current context of global warming (Casal and Balasubramanian, 2019). A well-established model in thermomorphogenesis, which is useful for understanding the basic mechanisms, is the enhanced growth of the hypocotyl in Arabidopsis thaliana seedlings in response to nonstressful warm temperatures (Gray et al., 1998).
The photo-sensory receptor phytochrome B (phyB; Jung et al., 2016; Legris et al., 2016), the clock protein EARLY FLOWERING 3 (ELF3; Jung et al., 2020), and the transcription factor PHYTOCHROME-INTERACTING FACTOR 7 (PIF7; Chung et al., 2020) are the three plant temperature sensors involved in controlling hypocotyl growth identified so far. Warm temperatures accelerate the rate of thermal reversion of active phyB to its inactive conformer (Jung et al., 2016; Legris et al., 2016; Burgie et al., 2021). ELF3 is a component of the evening complex whose binding to target gene promoters is reduced by warm temperatures (Box et al., 2015; Ezer et al., 2017; Silva et al., 2020). Arabidopsis ELF3 contains a predicted prion-like domain with a polyglutamine repeat that is important for the reversible phase transition from the active to inactive state of ELF3 under warm conditions (Jung et al., 2020). According to Jung et al. (2020), warm temperatures can enhance the formation of nuclear speckles containing ELF3, but Ronald et al. (2021) reported the opposite pattern. Therefore, there is controversy about the link between these sub-nuclear bodies and ELF3 activity. Warmth-induced changes in the RNA hairpin present at the 5′-untranslated region of the PIF7 transcript favor its translation, increasing PIF7 protein abundance (Chung et al., 2020). phyB and ELF3 signaling converge on PIF4, as phyB physically interacts with PIF4 and negatively regulates its stability (Cheng et al., 2021), and the evening complex binds to the PIF4 promoter, reducing its expression during the early night (Nusinow et al., 2011). Therefore, both PIF4 expression (Koini et al., 2009; Stavang et al., 2009; Box et al., 2015) and PIF4 protein stability (Foreman et al., 2011) increase under elevated temperatures. In addition, ELF3 sequestrates PIF4 by a direct physical interaction, preventing it from binding to its transcriptional targets (Nieto et al., 2015).
CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), ELONGATED HYPOCOTYL 5 (HY5), and LONG HYPOCOTYL IN FAR‐RED (HFR1) are among the regulators of PIF4 activity. COP1 is a RING E3 ligase that is required for the hypocotyl growth response to warm temperatures (Delker et al., 2014). Warm temperatures increase the nuclear accumulation of COP1 (Park et al., 2017), thus enhancing the expression of PIF4 (Gangappa and Kumar, 2017). HY5 binds to the PIF4 promoter to negatively regulate its activity (Delker et al., 2014). HY5 also competes with PIF4 for binding to its target gene promoters (Toledo-Ortiz et al., 2014), while warm temperatures inhibit HY5 expression and can reduce HY5 protein stability (Catalá et al., 2011; Delker et al., 2014; Toledo-Ortiz et al., 2014; Park et al., 2017). HFR1 is stabilized under warm temperatures (Romero-Montepaone et al., 2021) and inhibits the binding of PIF4 and PIF7 to their targets via a direct physical interaction (Hornitschek et al., 2009; Paulišić et al., 2021). PIF4 (Crawford et al., 2012; Sun et al., 2012) and PIF7 (Fiorucci et al., 2020) bind to the promoters of auxin biosynthesis genes to increase auxin levels in the warmth. Auxin produced in the cotyledons travels down to the hypocotyl to promote growth (Bellstaedt et al., 2019).
In nature, temperatures typically fluctuate between the day and night. We recently observed that night temperature information stored in phyB affects hypocotyl growth during the subsequent photoperiod (Murcia et al., 2020). Here, we report that nighttime hypocotyl growth and gene expression depend not only on the temperature during the night itself but also on the former daytime temperature.
Results
Nighttime growth depends on daytime temperatures
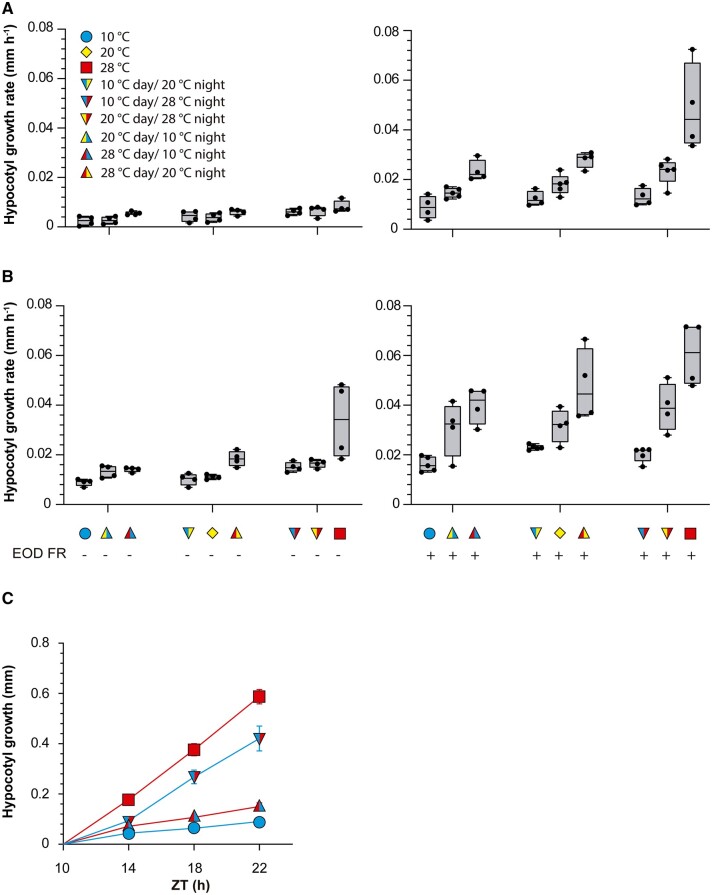
To investigate whether the growth of the hypocotyl during the night responds exclusively to the current temperature or is also affected by the conditions experienced during the previous photoperiod, we exposed wild-type (WT) Columbia-0 (Col-0) Arabidopsis plants to all possible combinations of three daytime temperatures and three nighttime temperatures (10°C, 20°C, and 28°C). Furthermore, to analyze whether the status of photo-sensory receptors modifies the influence of previous temperature, we exposed plants to simulated sunlight or shade conditions during the daytime and either a far-red light pulse (EOD FR; 10 min) or no light pulse at dusk. As expected, warmer nights accelerated nighttime growth (Figure 1, A and B; Supplemental File S1A). More importantly, for all nighttime conditions, warmer daytime temperatures caused faster nighttime growth (Figure 1, A and B; Supplemental File S1A). EOD FR or daytime shade accelerated hypocotyl growth during the night (Supplemental File S1A). Significant interactions indicate that the effect of daytime temperature was stronger when nighttime temperatures were warmer and in seedlings exposed to EOD FR (Supplemental File S1A). Taken together, these results indicate that nighttime growth depends not only on the nighttime temperature itself but also on prior daytime temperature, particularly under the conditions that elicit faster nighttime growth (warm night, daytime shade, and EOD FR). Based on these observations, in subsequent experiments, we used daytime shade and EOD FR to optimize the analysis. The phenomenon reported here should not be confused with the known effects of day/night temperature differentials, which actually affect daytime growth (Bours et al., 2013, 2015).
Figure 1.
Daytime temperatures affect nighttime hypocotyl growth. A and B, Hypocotyl growth rate measured in Col-0 seedlings during the night (ZT 10 h to ZT 24 h). The seedlings received all possible combinations of temperature during the preceding photoperiod (10°C, 20°C, or 28°C), nighttime temperature (10°C, 20°C, or 28°C), daytime light (A) or daytime shade (B) and either control or EOD FR. The left- and right-hand sides of the symbols indicate daytime and nighttime temperatures, respectively. Box plots show median, interquartile range 1–3 and the maximum–minimum interval of four replicate boxes with seedlings (see Supplemental File S1A for detailed statistics). C, Increase in hypocotyl length measured in Col-0 seedlings during the night (starting at ZT = 10 h), as affected by four combinations of daytime temperatures (10°C or 28°C) and nighttime temperatures (10°C or 28°C). All seedlings received shade during the day and EOD FR. Data are means ± se of nine replicate boxes of seedlings for each time point. The interaction between nighttime and daytime temperatures persists beyond ZT = 14 (see Supplemental File S1B for detailed statistics).
To obtain a detailed kinetics of nighttime growth, we exposed the seedlings to two different temperatures during the day (10°C and 28°C) and two different night temperatures (10°C and 28°C) in all four combinations. Hypocotyl growth rate responded to current and previous temperature conditions (Figure 1C; Supplemental File S1B). After the initial 4 h of the night, hypocotyl growth proceeded at a constant rate, indicating that the transition interval had ended by then. However, growth rate remained significantly affected by previous temperatures beyond that point (Figure 1C). In effect, according to a multiple regression analysis, the increase in hypocotyl length between zeitgeber time (ZT) = 14 h and ZT = 22 h depended on time (coefficient ± standard error (SE) 4.4 E-03 ± 1.4E-03, P = 0.0025), the interaction between time and night temperature (0.04 ± 1.4E-03, P < 0.0001), and the interaction with daytime temperature (0.01 ± 1.4E-03, P < 0.0001). These observations indicate that the daytime cue remains stored in the system, persistently affecting growth. In plants that experienced a cold day, a warm night did not increase hypocotyl growth to the rates exhibited by plants already exposed to warmth during the day. Conversely, in plants that experienced a warm day, a cold night did not decrease hypocotyl growth to the rates exhibited by plants already exposed to cold temperature during the day.
Nighttime gene expression depends on daytime temperatures
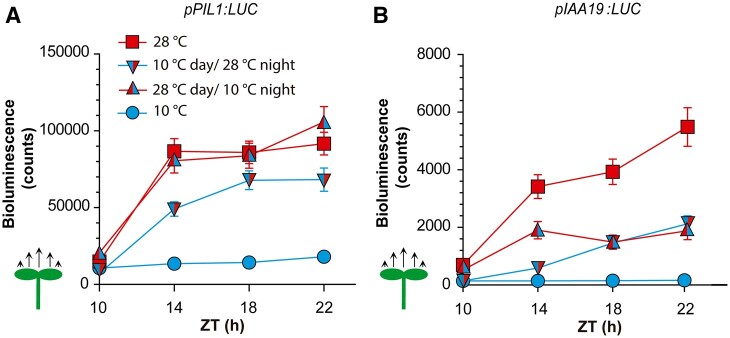
Prompted by the observed growth patterns, we investigated whether nighttime gene promoter activities also depended on daytime temperatures. We selected the PIF3-LIKE 1 (PIL1) and INDOLE-3-ACETIC ACID-INDUCIBLE 19 (IAA19) genes because warm daytime temperatures do not affect their expression at the end of the photoperiod (Romero-Montepaone et al., 2021), but night temperatures affected their expression in preliminary experiments. We grew transgenic lines bearing the pPIL1:LUC or pIAA19:LUC promoter–reporter fusions at two different temperatures during the day (10°C and 28°C) and two different night temperatures (10°C and 28°C) in all four combinations. For both promoters, the activities in seedlings transferred from 10°C to 28°C did not reach the levels observed in seedlings exposed to 28°C day and night. Moreover, in seedlings transferred from 28°C to 10°C, the promoter activities did not drop to the levels observed for plants exposed to 10°C day and night (Figure 2, A and B; Supplemental File S1, C and D). Notably, a cold night did not cause any reduction in the activity of the pPI1L:LUC promoter after a warm day. Therefore, not only growth but also gene expression showed effects of daytime temperature that persisted during the night.
Figure 2.
Daytime temperatures affect nighttime gene expression. A and B, Time course of LUC activity driven by pPIL1:LUC (A) or pIAA19:LUC (B). Luminescence was recorded during the night (starting at ZT = 10 h), as affected by four combinations of daytime temperature (10°C or 28°C) and nighttime temperature (10°C or 28°C). All seedlings received shade during the day and EOD FR. Data are means ± se of three plates with seedlings (see Supplemental File S1, C and D for detailed statistics).
Genetic requirements for the effects of daytime temperature on nighttime growth
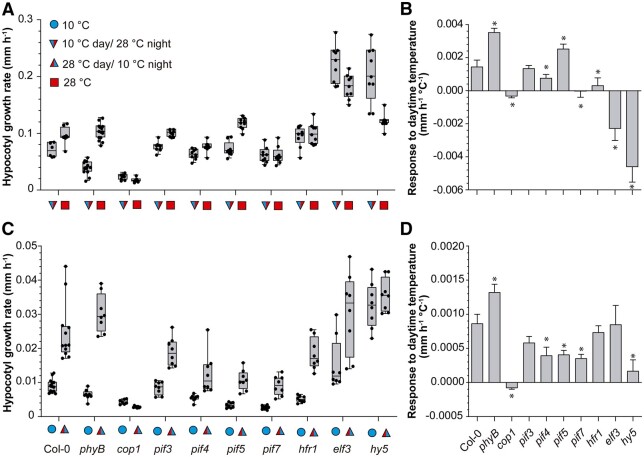
We compared the WT to different mutants to gain insight into the genetic requirements for the effects of daytime temperature on nighttime growth. We grew seedlings at 28°C or 10°C and exposed them to shade during daytime, followed by EOD FR. In a first set of experiments, we exposed all the seedlings to 28°C during the night. The WT showed faster growth during the night when subjected to the warmer temperature during daytime (Figure 3, A and B, Supplemental File S1E). The pif4, pif7, and hfr1 mutants showed a reduced response to daytime temperature and the cop1, elf3, and hy5 mutants showed an inverted response. The pif5 mutation did not reduce the effect of daytime temperature and actually partially rescued the defect of pif4 and pif7 (Supplemental Figure S1A; Supplemental File S1Z). In the second set of experiments, we exposed all the seedlings to 10°C during the night. The cop1, pif4, pif5, pif7, and hy5 mutations impaired the effects of daytime temperature on nighttime growth (Figure 3, C and D; Supplemental File S1F).
Figure 3.
Genetic requirements of the effects of daytime temperature on nighttime growth. A and C, Hypocotyl growth rate measured in seedlings of the indicated genotypes during the night (ZT 10 h to ZT 24 h), as affected by two different daytime temperatures (10°C or 28°C). B and D, Slope of the responses to daytime temperature (linear regression analysis of the data in (A) and (C), respectively). All seedlings received shade during the day and EOD FR. Night temperature was 28°C (A and B) or 10°C (C and D). See Supplemental Figure S1 for hypocotyl growth in multiple pif mutants. Box plots show median, interquartile range 1–3 and the maximum–minimum interval of 8–12 replicate boxes with seedlings (see Supplemental File S1, E and F for detailed statistics). In (B) and (D), the asterisks indicate significant differences from Col-0 according to t tests (*P < 0.05).
Nighttime memory of daytime temperature carried by signaling components
Since the pif4, pif7, hfr1, cop1, elf3, and hy5 mutants did not show the normal nighttime memory of daytime temperatures observed in the WT, we investigated whether the night status of these signaling components carried daytime information.
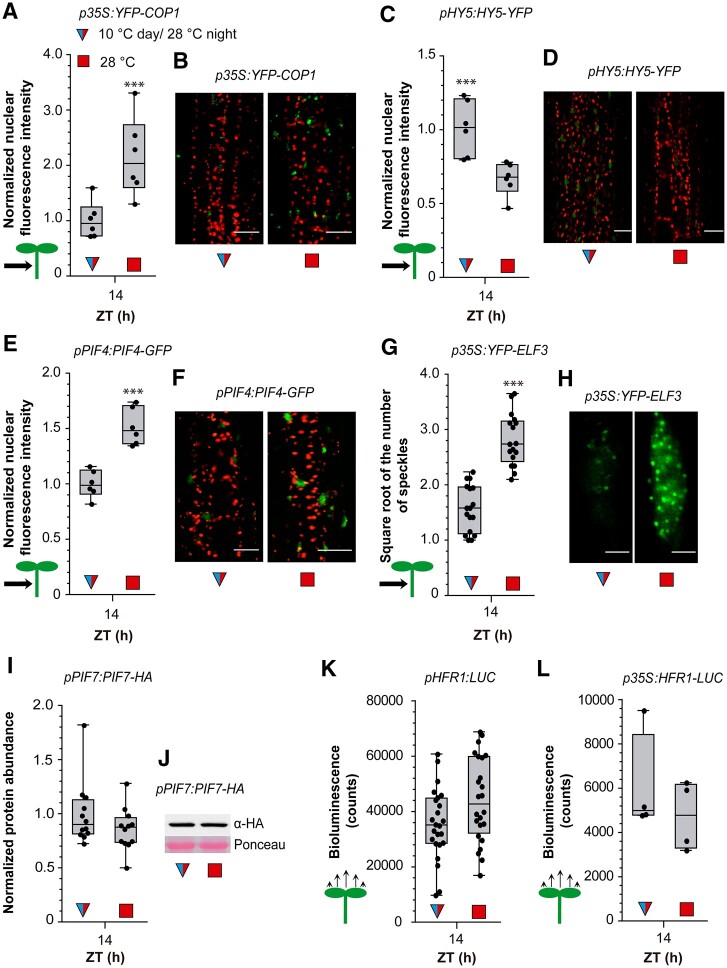
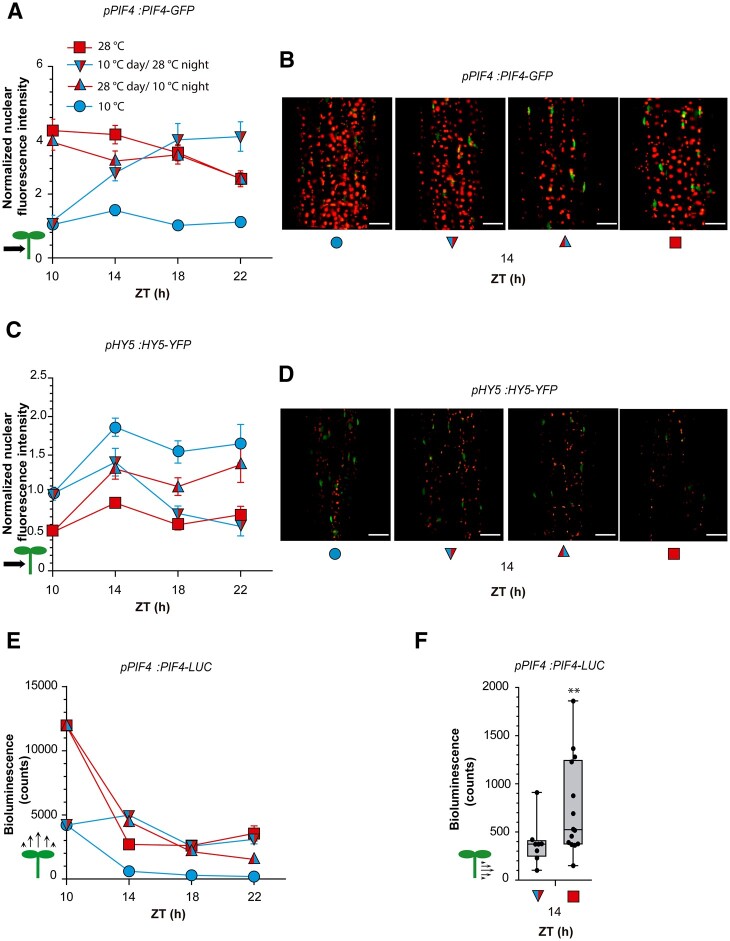
Warm temperatures increase PIF4 (Legris et al., 2017) and COP1 levels (Park et al., 2017) and decrease HY5 levels in the nucleus (Romero-Montepaone et al., 2021). We used confocal microscopy to analyze the nuclear fluorescence driven by pPIF4:PIF4-GFP, pHY5:HY5-YFP, and p35S:YFP-COP1 transgenic lines 4 h into the night (ZT = 14 h, 28°C) in seedlings exposed to 10°C and 28°C during the daytime. Compared to 10°C treatment, 28°C treatment during the day increased the nuclear abundance of PIF4 and COP1 and decreased the nuclear abundance of HY5 in hypocotyl cells (Figure 4, A–F).
Figure 4.
Daytime temperatures affect nighttime signaling status. A and B, Nuclear fluorescence driven in hypocotyl cells by p35S:YFP-COP1. C and D, Nuclear fluorescence driven in hypocotyl cells by pHY5:HY5-YFP. E and F, Nuclear fluorescence driven in hypocotyl cells by pPIF4:PIF4-GFP. G and H, Number of ELF3 nuclear speckles (square root-transformed data) in the hypocotyl cells of the line expressing the p35S:YFP-ELF3 transgene. I and J, Abundance of PIF7 in seedlings bearing pPIF7:PIF7-HA. K and L, LUC activity driven by pHFR1:LUC (K) or p35S:HFR1-LUC (L). The seedlings received shade and either 10°C or 28°C during the day, EOD FR and 28°C during the night. Box plots show median, interquartile range 1–3 and the maximum–minimum interval of 6 (A, C, and E), 16 (G), 12 (I), 25 (K), and 4 (L) replicate boxes or plates with seedlings. Representative confocal (B, D, F, and H) and protein blot (J) images are shown. Scale bar = 35 µm (B, D, and F) and 2.61 µm (H). Asterisks indicate significant differences in t tests (***P < 0.001).
Warm temperatures were previously shown to either increase (Jung et al., 2020) or decrease (Ronald et al., 2021) the formation of nuclear speckles containing ELF3. Since overexpression of ELF3 had no effect on temperature responsiveness compared to the WT (Thines and Harmon, 2010; Jung et al., 2020), we used the p35S:YFP-ELF3 line to facilitate the quantitative analysis of ELF3 speckles (the same line used by Ronald et al., 2021). We observed a significantly higher number of ELF3 speckles in seedlings exposed to 28°C versus 10°C during the day (Figure 4, G and H).
Warm temperatures increase PIF7 protein levels (Fiorucci et al., 2020; Chung et al., 2020) and enhance HFR1 protein stability (Romero-Montepaone et al., 2021). However, protein blot analysis of seedlings sampled at ZT 14 after 4 h of 28°C nighttime treatment did not reveal differences in PIF7 protein abundance caused by contrasting daytime temperatures (Figure 4, I and J). Similarly, in luminometer readings, we did not observe significant effects of daytime temperature on nighttime HFR1 promoter activity or HFR1 stability (Figure 4, K and L).
PIF4 and COP1 promote hypocotyl growth, whereas HY5 inhibits hypocotyl growth (e.g. Figure 3). Cold days decreased the nighttime activities of PIF4 and COP1 and increased the nighttime activities of HY5 (Figure 4, A–F), suggesting that PIF4, HY5, and COP1 convey daytime temperature information to nighttime growth. This would also be the case for ELF3 if enhanced speckle formation correlates with reduced activity, a link analyzed in further detail below. Conversely, the short-term memory of daytime temperature requires PIF7 and HFR1, but apparently, these factors do not carry prior temperature information because their levels during the night showed no influence of daytime conditions (Figure 4, I–L).
Nighttime PIF4 and HY5 kinetics account for growth responses
Since ELF3 and COP1 control hypocotyl growth partially via PIF4 and HY5 (Nusinow et al., 2011; Box et al., 2015; Gangappa and Kumar, 2017; Park et al., 2017;), we analyzed the nighttime kinetics of the nuclear abundance of these two transcription factors in further detail using confocal microscopy of hypocotyl cells. Seedlings exposed to 28°C during the day began the night with more PIF4 and less HY5 in the nuclei of their hypocotyl cells than plants exposed to 10°C during the day, and these differences persisted for at least several hours (Figure 5, A–D). Seedlings that were transferred from 10°C to 28°C at the beginning of the night reached similar nuclear levels of PIF4 or HY5 to seedlings kept at 28°C during the daytime by 8 h after the temperature shift (ZT = 18 h, Figure 5, A–D). In seedlings transferred from 28°C to 10°C, similar HY5 nuclear levels to seedlings already exposed to 10°C during the day were only observed 12 h later (ZT = 22 h, Figure 5, C and D). Notably, the 28°C to 10°C shift at the beginning of the night caused almost no PIF4 response (Figure 5, A and B). Taken together, these results indicate that the nighttime kinetics of PIF4 and HY5 depend on both nighttime and daytime temperatures (Supplemental File S1, G and H). Differences in their protein levels caused by contrasting temperatures during the day extended several hours into the night, either due to slower transitions between the levels typical for day and night temperatures (PIF4 from 10°C to 28°C and HY5 in both directions) or to a nearly complete insensitive response (PIF4 from 28°C to 10°C).
Figure 5.
Daytime temperature affects nuclear levels of PIF4 and HY5 during the night. A–D, Nighttime kinetics of nuclear fluorescence intensity driven in hypocotyl cells by pPIF4:PIF4-GFP (A and B) or pHY5:HY5-YFP (C and D). See Supplemental Figure S2 for nuclear fluorescence driven by pPIF4:PIF4-GFP in cotyledons. E, Nighttime kinetics of luminescence driven by pPIF4:PIF4-LUC in whole seedlings. F, Luminescence driven by pPIF4:PIF4-LUC in isolated hypocotyls harvested at ZT = 14 h. Confocal images or luminescence values were recorded during the night (starting at ZT = 10 h), as affected by four combinations of daytime temperature and nighttime temperature. All seedlings received shade during the day and EOD FR. (A, C, and E) Data are means ± se of three replicate boxes with seedlings. F, Box plots show median, interquartile range 1–3 and the maximum–minimum interval of 10–14 replicate boxes with seedlings. B and D, Representative confocal images at ZT = 14 h. Scale bar = 35 µm. See Supplemental File S1, G–I for detailed statistics of (A, C, and E). In (F), the asterisks indicates significant differences in t test (**P < 0.01).
Genetic studies indicated that the effects of daytime temperature depend on PIF4 and HY5 (Figure 3). Therefore, we explored the quantitative association between the kinetics of growth rate and that of PIF4 and HY5 nuclear levels using multiple regression analysis across three temporal phases (ZT 10–14 h, 14–18 h, and 18–22 h). For each temporal phase, we averaged the PIF4- or HY5-fluorescence values obtained by confocal microscopy at the beginning and end of the phase (Figure 5, A and C) as explanatory variables. The model accounted for 80% of the variability in growth rate (Supplemental File S1I). Both PIF4 and HY5 levels contributed significantly, indicating that the information provided by PIF4 and HY5 dynamics is important and not redundant.
We also investigated the abundance of PIF4 protein during the night using transgenic lines bearing pPIF4:PIF4-LUC. As observed by confocal microscopy analysis, there was little difference in PIF4 protein level if the plants continued at 28°C or were shifted from 28°C to 10°C at the beginning of the night (Figure 5E; Supplemental File S1J). Also resembling the hypocotyl pattern, PIF4 levels remained low throughout the night in plants grown at 10°C during the day and night and increased when the plants were transferred from 10°C to 28°C at the beginning of the night (Figure 5E), although this response was faster than in the confocal studies. The upward cotyledon signal dominates luminescence readings of whole seedlings; therefore, we used the line bearing the pPIF4:PIF4-GFP transgene to evaluate nuclear levels of PIF4 in cotyledon cells. The results confirmed elevated levels of PIF4 during the day (see also Stavang et al., 2009) and showed that these differences had already been inverted by 4 h after the beginning of night (ZT = 14; Supplemental Figure S2). Bioluminescence analysis of PIF4 levels in isolated hypocotyls of the pPIF4:PIF4-LUC line actually confirmed that the shift to 28°C at the beginning of the night was not enough to achieve the levels detected in this organ in seedlings grown at 28°C during the day (Figure 5F).
Given the faster change in PIF4 levels in cotyledons, we investigated whether the growth of this organ still responded to daytime temperatures in a PIF4-dependent manner. The cotyledon area increased more during the night at 28°C if the seedlings were exposed to 10°C during the daytime (mean ± se n = 40, 4.0 E-03 ± 4.0 E-04 mm2) than when the daytime temperature was 28°C (2.7 E-03 ± 2.1 E-04 mm2, P = 0.005). This short-term memory effect was absent in the pif4 mutant. Compared to the WT, pif4 showed an enhanced cotyledon expansion under a 28°C daytime temperature (4.3 E-03 ± 4.8 E-04 mm2, P = 0.0017), similar to that observed at 10°C (4.2 E-03 ± 4.8 E-04 mm2; Huq and Quail, 2002). Therefore, differences in PIF4 levels in cotyledons exposed to 28°C compared to 10°C during the day, despite showing shorter persistence during warm nights, convey daytime temperature information to nighttime growth of this organ.
PIF4 promoter activity accounts for PIF4 dynamics during the night
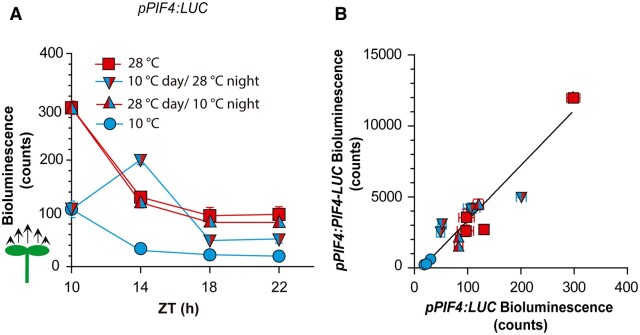
The nighttime kinetics of PIF4 promoter activity exhibited the key features observed for PIF4 protein kinetics: PIF4 promoter activity depended on nighttime and daytime activities and showed asymmetric responses to temperature shifts (Figure 6A; Supplemental File S1K). The stronger PIF4 promoter activity established by 28°C compared to 10°C treatment during the day was largely irreversible during the night, as transfer to 10°C did not reduce this activity below the levels of the 28°C controls (Figure 6A). Conversely, the transfer of seedlings from 10°C to 28°C at the beginning of the night did increase PIF4 promoter activity (Figure 6A). Luminescence readings driven by promoter and protein fusions showed a strong correlation (Figure 6B), supporting a major role of PIF4 promoter activity in controlling nighttime PIF4 levels.
Figure 6.
Changes in PIF4 promoter activity account for nighttime PIF4 protein dynamics. A, Nighttime kinetics of luminescence driven by pPIF4:LUC in whole seedlings. Luminescence was recorded during the night (starting at ZT = 10 h), as affected by four combinations of daytime temperature (10°C or 28°C) and nighttime temperature (10°C or 28°C). All seedlings received shade during the day and EOD FR. Data are means ± SE of three plates with seedlings. See Supplemental File S1, J and K for detailed statistics. B, Correlation between luminescence driven by pPIF4:PIF4-LUC (from 5E) and pPIF4:LUC (from A) in the Col-0 background. The correlation is significant at P < 0.0001.
The nighttime patterns of PIF4 protein and PIF4 promoter activities showed two features. The first feature is that daytime differences persisted at least several hours into the night. The second feature is the asymmetric response to temperature shifts in contrasting directions (increase compared to decrease in temperature). This asymmetric response to a variable when it either increases or decreases its values is typical of hysteretic systems (Davies, 2017; Jiang and Hao, 2021).
The effects of daytime temperatures on differences in PIF4 levels at night requires ELF3
The differences in PIF4 nuclear levels (Figure 5, A and C) and PIF4 promoter activity (Figure 6A) that persisted during the night were already present in cotyledons and hypocotyls at the end of the day. We therefore investigated the processes involved in their generation during the photoperiod. Luminescence readings revealed that the abundance of PIF4 protein (Figure 7A) and the activity of the PIF4 promoter (Figure 7B) abruptly increased early in the morning, when the seedlings were transferred from 20°C to 28°C, compared to 10°C. Initial changes induced by such contrasting temperatures slightly decreased during the course of the day but were still present at the beginning of the night. The early increase in the PIF4 protein signal in response to warm temperatures was more intense than that in PIF4 promoter activity (cf. Figure 7, A and B at 4 h), which is reflected by a higher protein/promoter activity ratio (Figure 7C). The protein blot results using a line bearing the p35S:PIF4-HA transgene are consistent with the higher stability of PIF4 at 28°C than at 10°C early in the morning (Supplemental Figure S3, A and B). However, later in the day, PIF4 protein levels increased in plants exposed to 10°C without a concomitant increase in PIF4 promoter activity, causing a higher protein/promoter activity ratio in plants exposed to cooler temperatures (Figure 7C). Therefore, the differences in PIF4 protein levels at the beginning of the night were largely due to temperature effects on PIF4 promoter activity, with only transient posttranscriptional effects.
Figure 7.
Effects of temperature on end-of-day PIF4 promoter activity and PIF4 protein abundance require ELF3. A and B, Time course of daytime LUC activity driven by pPIF4:PIF4-LUC (A) or pPIF4:LUC (B) in the Col-0 and elf3-8 background, as affected by 10°C or 28°C. C, Ratio between the LUC signals driven by pPIF4:PIF4-LUC and pPIF4:LUC. See Supplemental Figure S3 for protein blots supporting posttranscriptional effects of temperature on PIF4 abundance. D, Luminescence driven by pPIF4:LUC in isolated hypocotyls harvested from Col-0 and elf3-8 seedlings at ZT = 14 h. All seedlings received shade during the day and EOD FR. A and B, Data are means ± se of three plates with seedlings. D, Box plots show median, interquartile range 1–3 and the maximum–minimum interval of 24–30 replicate plates seedlings; asterisk indicates significant differences in t tests (*P < 0.05). See Supplemental File S1, L and M for detailed statistics of (A and B).
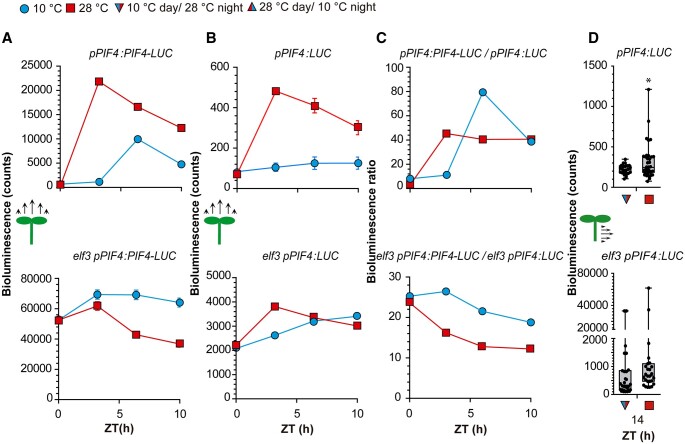
We investigated whether the differences in PIF4 levels observed at the end of the day require ELF3. Experiments using lines bearing the pPIF4:LUC transgene in the elf3 background indicated that, early in the photoperiod, warm temperature promoted PIF4 activity even in the absence of ELF3 (Figure 7B), pointing to the actions of other transcriptional regulators. However, temperature-induced changes in PIF4 promoter activity showed an absolute requirement for ELF3 during the rest of the photoperiod (Figure 7B). During warm afternoons, PIF4 promoter activity declined, even in the elf3 background (Figure 7B); this might reflect an effect of GIGANTEA, which reduces PIF4 transcript levels particularly during the afternoon (Anwer et al., 2020).
Since luminometer readings of whole seedlings are mainly affected by PIF4 promoter activity in cotyledons, we conducted experiments with isolated hypocotyls; that is, the organ where differences in PIF4 are more persistent during warm nights (compare Figure 5, C and E). The nighttime memory of daytime temperatures of the PIF4 promoter was also absent in hypocotyls in the elf3 background (Figure 7D).
Kinetics of ELF3
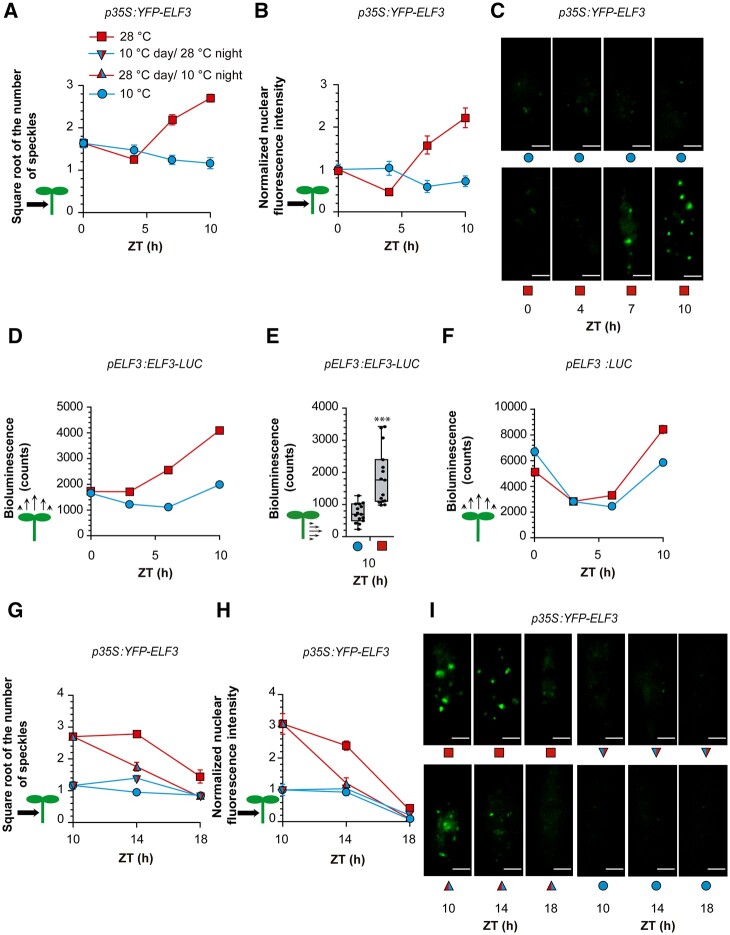
We investigated the dynamics of ELF3 under the conditions where it controlled PIF4 promoter activity. Under warm temperatures, the formation of speckles decreased during the morning and increased strongly during the afternoon (Figure 8, A and C). In addition to PIF4, ELF3 negatively regulates TIMING OF CAB EXPRESSION 1 (TOC1) and positively regulates LATE ELONGATED HYPOCOTYL (LHY), and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1; Thines and Harmon, 2010; Fehér et al., 2011; Herrero et al., 2012; Ezer et al., 2017), in all cases by directly binding to their promoters. TOC1 promoter activity increased whilst LHY and CCA1 promoter activity decreased in response to warm temperatures (Supplemental Figure S4). In particular, TOC1 did not show responses during the morning. These results, together with the expression pattern of PIF4, indicate that ELF3 speckle formation and ELF3 activity are negatively correlated.
Figure 8.
ELF3 dynamics under contrasting temperatures. A–C, Number of ELF3 nuclear speckles (A), nuclear fluorescence intensity (B), and representative confocal images (C) from hypocotyl cells of seedlings expressing p35S:YFP-ELF3. D, LUC activity driven by pELF3:ELF3-LUC in whole seedlings. E, LUC activity driven by pELF3:ELF3-LUC in isolated hypocotyls. F, LUC activity driven by pELF3:LUC in whole seedlings. G–I, Number of ELF3 nuclear speckles (square root-transformed data, G), nuclear fluorescence intensity (H) and representative confocal images (I) from hypocotyl cells of seedlings expressing p35S:YFP-ELF3. All seedlings received shade during the day and EOD FR. See Supplemental Figure S4 for the kinetics of TOC1, LHY, and CCA1 promoter activities functionally linked to ELF3. (A and B and G and H) Data are means ± se of 20 replicate plates with seedlings. C and I, Representative confocal images. Scale bar = 2.61 µm. D and F, Data are means ± se of three plates with seedlings. E, Box plots show median, interquartile range 1–3 and the maximum–minimum interval of 15 replicate plates with seedlings. See Supplemental File S1, N–S for detailed statistics of (A and B, D, F–H). In (E), asterisks indicate significant differences in t test (***P < 0.001).
During the morning, warm temperatures decreased ELF3 nuclear abundance (Figure 8B) and the bioluminescence signal driven by the pELF3:ELF3-LUC transgene (Figure 8D). During the afternoon, warm temperatures increased both features (Figure 8, B and D). Harvested hypocotyls also showed elevated nuclear levels of ELF3 at 28°C (Figure 8E). Conversely, bioluminescence driven by the pELF3:LUC transgene increased only slightly at 28°C (Figure 8F). These results point to posttranscriptional control of ELF3 protein levels, which might involve increased stability within the speckles.
As described above (Figure 4, G and H), during the night, the shift from 10°C to 28°C did not increase the number of ELF3 speckles to the levels observed in seedlings already exposed to 28°C during the day. Actually, the number of speckles decreased during the night under all temperature conditions (Figure 8, G and I). However, such a decrease does not reflect enhanced ELF3 activity because a steady drop in ELF3 protein levels accompanied the decrease in speckle number (Figure 8, H and I).
Hysteresis in PIF4 promoter activity
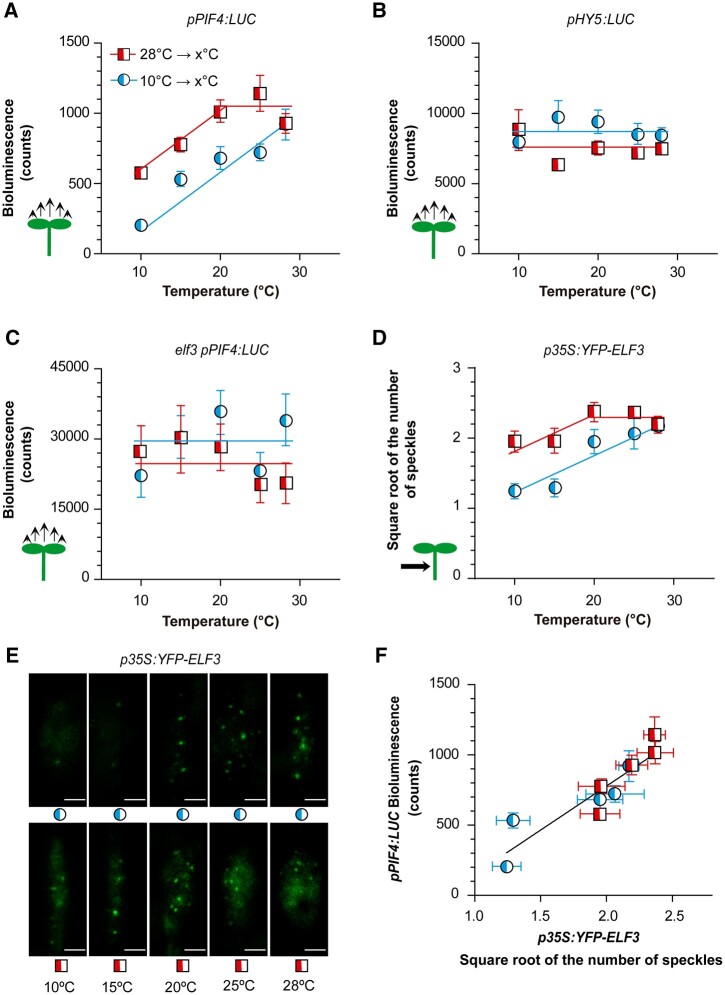
In all the experiments described above, temperature shifts coincided with end of the day. However, under natural conditions, temperature fluctuations can occur throughout the photoperiod. We, therefore, investigated whether the response pattern of the PIF4 promoter during the day was similar to the behavior observed during the night. For this purpose, we exposed seedlings to 10°C or 28°C and transferred them at ZT = 4 h either from 10°C to warmer temperatures (15°C, 20°C, 25°C, or 28°C) or from 28°C to cooler temperatures (25°C, 20°C, 15°C, or 10°C), whilst the controls remained at 10°C or 28°C. We harvested the seedlings 3 h after the temperature shift, that is, still within the photoperiod (ZT = 7 h). As shown in Figure 9A, a memory of the previous temperature was retained, because warmer morning temperatures yielded higher PIF4 promoter activities for most afternoon temperatures (X-axis). This short-term memory is entirely associated with hysteresis in promoter activity, as demonstrated by a shift in sensitivity on the way up compared to the way down.
Figure 9.
Hysteresis of ELF3 drives hysteresis in PIF4 promoter activity. A–C, Response of the luminescence driven by pPIF4:LUC (A and C) or pHY5:LUC (B) in the Col-0 (A and B) or elf3 (C) backgrounds to increasing or decreasing temperature. D and E, Response of the number of ELF3 nuclear speckles (square root-transformed data) from hypocotyl cells of seedlings expressing p35S:YFP-ELF3 to increasing or decreasing temperature. See Supplemental Figure S5 for information on the kinetics of ELF3 speckles and their response to nighttime temperature during the morning. F, Correlation between luminescence driven by pPIF4:LUC (from A) and the number of speckles driven by p35S:YFP-ELF3 (from D). The seedlings were exposed to white light at either 10°C or 28°C and 4 h after the beginning of the photoperiod were transferred to the temperature indicated on the x-axes (including controls that remained at 10°C or 28°C). Luminescence (A–C) or confocal images (D and E) were taken 3 h later. Data are means ± se of three plates with seedlings (A–C) or 15 replicate boxes with seedlings (D and E). E, Representative confocal images. Scale bar = 2.61 µm. In (F), the correlation is significant at P = 0.0002. See Supplemental File S1, T–X for detailed statistics.
We simultaneously analyzed the activity of the HY5 promoter, which remained largely unresponsive to the temperature shifts in either of the two directions (Figure 9B). This indicates that different mechanisms mediate the persistent effects of previous temperature on PIF4 and HY5 nuclear levels. Compared to the WT, the analysis of seedlings bearing pPIF4-LUC in the elf3 mutant background showed a completely distorted pattern of response to temperature (Figure 9C). This indicates that the hysteresis pattern depends on ELF3. We therefore investigated the response of ELF3 itself.
Hysteresis in ELF3 speckle formation
We analyzed the formation of ELF3 speckles in response to increasing or decreasing temperatures during the day, under the same conditions used to investigate the hysteresis of the PIF4 promoter. Seedlings exposed to 28°C showed more speckles than those exposed to 10°C (Figure 9D). Notably, the response curve showed a strong shift in the range of sensitivity. On the way up, the number of speckles increased between 10°C and 28°C. On the way down, the number of speckles decreased only ˂20°C. Therefore, the subnuclear localization of ELF3 itself showed the hysteretic pattern. The activity of the PIF4 promoter strongly correlated with the number of ELF3 speckles formed under the same conditions (Figure 9E), supporting a link between speckle formation and reduced ELF3 activity.
The kinetics of ELF3 speckle formation deserve particular consideration. Compared to 10°C, 28°C did not increase the number of speckles during the first 4 h of the day (Supplemental Figure S5, A and B, see also Figure 8A). Therefore, all the differences observed in Figure 9D originated during the afternoon (i.e. between 4 and 7 h; Supplemental Figure S5, A and B). This means that the speckles appeared rapidly during the afternoon, but not in the morning (Supplemental Figure S5, A and B), suggesting that other components required for their formation became available in the afternoon. Furthermore, the additional speckles formed between 4 and 7 h, even in seedlings exposed during the first 4 h to 28°C and then transferred to 10°C. This demonstrates that the induction of ELF3 speckle formation by warmth persisted in the cold until the additional putative components of the speckles became available. The persistence of a warmth-induced state of ELF3 is consistent with the occurrence of hysteresis.
Finally, since elf3 showed a distorted memory of nighttime temperatures in the control of daytime growth (Murcia et al., 2020), we investigated whether nighttime temperatures affect daytime ELF3 speckle formation. For this purpose, we transferred seedlings grown at 20°C to either 10°C or 28°C at the beginning of the night on Day 4. At the beginning of the photoperiod on Day 5, we transferred half of the seedlings of each group to the opposite temperature and obtained confocal images at ZT = 4 h. Confirming the above observations (Figure 8A; Supplemental Figure S5, A and B), warm morning temperatures did not enhance speckle formation in the morning. However, a warm night increased the number of ELF3 speckles (Supplemental Figure S5, C and D). Therefore, ELF3 also stores night temperature information for the control of daytime growth.
Discussion
The results reported here demonstrate that the control of hypocotyl growth and gene expression during the night respond not only to the current temperature environment but also to the temperature experienced during the preceding photoperiod (Figures 1 and 2). Therefore, plants have a nighttime memory of daytime temperature.
PIF4 and HY5 store the daytime temperature information that controls hypocotyl growth during the night. In fact, both HY5 and PIF4 showed daytime-induced differences in nuclear protein abundance, which persisted during at least part of the night (Figures 4 and 5), and the short-term memory of daytime temperatures that affect nighttime growth required HY5 and PIF4 (Figure 3). In plants exposed to different daytime and nighttime temperature combinations, the dynamics of the nuclear abundances of PIF4 and HY5 during the night jointly accounted for 80% of the variability in growth rates over the same period (Supplemental File S1I), indicating that the analysis presented here has identified the major components of the system. Two features of the kinetics of PIF4 and HY5 nuclear protein levels revealed a carryover of daytime information into the night. The first feature is the slow transitions (lasting 8–12 h) of PIF4 abundance in plants shifted from cool to warm temperature conditions, and of HY5 abundance in plants shifted in any of the two directions (Figure 5, A and B). At least in the case of PIF4, the slow transition was night-specific, because during the day, the transitions were rapid (<4 h; Murcia et al., 2020). The second, more striking feature of the nighttime memory of daytime temperatures is that in plants grown under warm daytime temperatures, PIF4 levels remained high and were nearly unresponsive to cold nights (Figure 5A).
Regarding the storage mechanisms of previous temperature information, in the case of HY5, temperature had little effect on HY5 promoter activity (Figure 9B), pointing to a posttranscriptional control, likely mediated by COP1 (Park et al., 2017), which itself stored daytime information into the night (Figure 4A). Conversely, in the case of PIF4, changes in PIF4 promoter activity largely accounted for the dynamics of nuclear protein levels, both during daytime (Figure 7, A and B) and at night (Figure 6, A and B). Apparent posttranscriptional effects were only transient (Figure 6C). In turn, the nighttime memory of daytime temperature exhibited by PIF4 promoter activity showed an absolute requirement for ELF3 (Figures 7, B, D and 9, C), which binds to this promoter via the evening complex (Box et al., 2015; Ezer et al., 2017; Silva et al., 2020). The tight association between ELF3 and PIF4 does not rule out other potential control mechanisms. For instance, TOC1 gating might contribute to the slow nighttime response of PIF4 to warmth (Zhu et al., 2016) and ELF3, as a clock component, might control the starch degradation required for nighttime growth (Graf et al., 2010; Pyl et al., 2012).
Warm temperatures reduce the transcriptional activity of the evening complex (Box et al., 2015; Ezer et al., 2017; Silva et al., 2020), but there is some controversy regarding the response to temperature of nuclear ELF3 speckle formation and the function of these sub-nuclear structures. According to Jung et al. (2020), warm temperatures induce the formation of speckles containing ELF3 in root cells, but according to Ronald et al. (2021), warm temperatures reduce the formation of speckles in hypocotyl and root cells. Here we showed that both patterns are not mutually exclusive because warm temperatures reduced speckle formation in hypocotyl cells during the morning and increased speckle formation during the afternoon (Figure 8A; Supplemental Figure S5, A and B). Therefore, the cellular context appears to affect ELF3 speckle formation. ELF4 does not mediate this time-of-day effect (Ronald et al., 2021). There is a negative association between ELF3 activity and PIF4 expression (Nusinow et al., 2011; Box et al., 2015; Raschke et al., 2015; Press et al., 2016). Under our conditions, ELF3 speckle formation correlated with enhanced PIF4 promoter activity (Figure 9F) and hence reduced ELF3 activity. Membrane-less compartments have been linked to changes in the stability of their components (Kim et al., 2021; Emenecker et al., 2021), and warm temperatures increased the abundance of ELF3 (Figure 8, B and D) (see also Ding et al., 2018; and time-dependent effects in Zhang et al., 2021). This observation suggests that condensation in speckles protects ELF3 from degradation, as is the case of phyB in nuclear bodies (Rausenberger et al., 2010; Van Buskirk et al., 2014).
We observed a strong asymmetry in the responses of the PIF4 protein and the PIF4 promoter. During the night, warming caused significant increases in PIF4 protein levels and PIF4 promoter activity, but cooling resulted in barely detectable changes (Figures 5, A and 6, A). The PIF4 target promoter PIL1 showed the same pattern (Figure 2A). During the afternoon, the PIF4 promoter did respond to cooling, but with poor sensitivity (Figure 9A). This pattern revealed strong hysteresis of PIF4 protein and PIF4 promoter activity; that is, the response to temperature followed one pattern in the forward direction but a different one in the return direction (Davies, 2017; Jiang and Hao, 2021).
The formation of speckles by ELF3 itself showed hysteresis, as revealed by a significant shift in sensitivity during cooling compared to warming (Figure 9D). Since the pattern of PIF4 promoter activity requires ELF3 and correlates with ELF3 speckle formation (Figure 9, A, C and F), we conclude that ELF3 achieves hysteresis and drives the PIF4 promoter into the same behavior. What mechanisms generate ELF3 hysteresis? Nuclear speckles are condensates, that is, membrane-less compartments that might be formed by liquid–liquid phase separation (Emenecker et al., 2021). ELF3 undergoes phase separation to form nuclear speckles under warm temperatures, and both in vitro phase separation and nuclear speckle formation depend on the intrinsically disordered prion-like domain of the protein (Jung et al., 2020). Hysteretic phase separation of a purified intrinsically disordered protein can emerge from intermolecular interactions that stabilize the aggregated phase (Quiroz et al., 2019). Although not discussed in the original report, purified ELF3 peptides showed hysteresis, because in response to increasing temperatures, they formed liquid droplets in vitro, and reversibility in response to decreasing temperature was shifted toward lower temperatures (Jung et al., 2020). Therefore, the hysteresis of in vivo ELF3 speckle formation observed here (Figure 9D) could have its origin in ELF3–ELF3 protein interactions. The so-called “mnemons” are proteins that oligomerize to form condensates and establish long-lasting signaling changes, encoding a memory of previous conditions (Reichert and Caudron, 2021). There is abundant evidence for the function of these assemblies in yeast and Drosophila (Reichert and Caudron, 2021).
In conclusion, temperature is a variable aspect of the environment. Here we showed that the history-dependent behavior of the nuclear levels of PIF4 and HY5 enables a short-term memory of past temperature conditions for the control of hypocotyl growth. ELF3 stores information about warm conditions, transmitting this information to the activity of the PIF4 promoter, and from there to PIF4 protein abundance. The ELF3–PIF4 module drives this memory in one direction, as cooling barely affected the status established by warmth. We speculate that in parallel, COP1 could transmit daytime temperature information to HY5 by regulating its stability.
Materials and methods
Plant material
We used A. thaliana in this study. The experiments where we measured hypocotyl growth or cotyledon expansion included the WT Col-0. The mutant alleles and transgenic reporter lines are listed in Supplemental Table S1.
Growth conditions
We used clear plastic boxes (4 × 3.5 × 2 cm3 height) for growth (14 seeds per genotype and box), microscopy (5 seeds per box), and protein blot analysis (80 seeds per box) and microtiter plates for bioluminescence experiments (one seed per well). The substrate was 1.5% agar-water. After sowing, we incubated the seeds for 4 days at 4°C in darkness and transferred the stratified seeds to white light at 90 μmol m−2 s−1 (400–700 nm), provided by a mixture of fluorescent and halogen lamps, with a red/far-red ratio typical of sunlight (1.1), a photoperiod of 10 h (short day), and 20°C for 4 days. At the beginning of the fourth photoperiod (ZT = 0 h), the seedlings received either white light or simulated shade (9 μmol m−2 s−1 between 400 and 700 nm with a red/far-red ratio of 0.1) at 10°C, 20°C, or 28°C. For simulated shade, we combined the white light source with two green acetate filters (LEE filters 089). The seedlings initiated the night (ZT = 10 h) with or without 10 min of far-red light at 7 μmol m−2 s−1 (EOD FR), provided by 150 W incandescent lamps (R95, Philips, Eindhoven, Netherlands) in combination with yellow, orange, and red acetate filters (LEE filters, Panavision, Los Angeles, CA, 101, 105, and 106, respectively) and six blue acrylic filters (Paolini 2031, Buenos Aires, Argentina). Night temperature was 10°C, 20°C, or 28°C. To investigate the occurrence of hysteresis without involving a light-to-dark transition, in some experiments, at 4 h after the beginning of the fourth photoperiod (ZT = 4 h), we introduced temperature shifts while the plants remained under white light.
Hypocotyl growth rate
We photographed the seedlings with a digital camera (PowerShot; Canon, Tokyo, Japan) at the beginning of the night (ZT = 10 h) of the fourth photoperiod and either at the end of the night (ZT = 24 h) or at intermediate times (ZT = 14, 18, and 22 h). We measured hypocotyl length increments using image processing software as described (Legris et al., 2016).
Bioluminescence
We detected luciferase (LUC) activity with a Centro LB 960 (Berthold, Stuttgart, Germany) luminometer by adding 20 μL of 0.2 mM D-luciferin per well 24 h before starting the measurements (Romero-Montepaone et al., 2021). To estimate the direct effect of temperature on the LUC reporter, we rapidly (5–7 min) cooled down or warmed up the plates with seedlings grown at 20°C to reach either 10°C or 28°C, respectively. The LUC activity values (±se) normalized to the 20°C control were 0.74 ± 0.04 (10°C), 1.00 ± 0.07 (20°C), and 1.38 ± 0.21 (28°C) for elf3-8 pPIF4:PIF4-LUC. This distortion applies for the comparison of different current temperatures but not for the short-term memory effect because in the latter case, the plants were already at the same temperature at the time of LUC measurements.
Confocal microscopy
We obtained confocal fluorescence images from the epidermis and the first sub-epidermal layers of either the upper third portion of the hypocotyl (PIF4, HY5, and COP1) or individual nuclei present in the same region (ELF3) with a LSM5 Pascal laser-scanning microscope (Zeiss, Jena, Germany). The water-immersion objective lens were C-Apochromat X40/1.2 or C-Apochromatic X63/1.2 (Zeiss), respectively. We used an argon laser (λ = 488 nm) for excitation of GFP or YFP and a BP 505-530 filter for fluorescence detection. We used a He-Ne laser (λ = 543 nm) for excitation of chlorophyll and a LP 560 filter for detection of its fluorescence and configured a transmitted light channel to visualize cellular structures. We performed image analysis in batch with an image segmentation program developed in Icy (http://icy.bioimageanalysis.org/; Sellaro et al., 2019).
Protein blots
We extracted total protein from 100 mg of seedlings by homogenizing plant material in extraction buffer containing 50-mM Tris–HCl (pH 7.5), 200-mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Tween-20, 1-mM PMSF and protease inhibitors (Roche), centrifuged the sample twice (20,000g, 15 min, at 4°C), and transferred the supernatant into fresh Eppendorf reaction tubes. The protein concentration in the supernatant was determined by the Bradford assay (Bio-Rad, Hercules, CA, USA). Protein samples were boiled (95°C, 5 min) in TMx4 loading buffer. We run 20 µg of protein in 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) followed by wet blotting (100-mM Tris/Glycine, 10% MeOH, 1.5 h). Homogeneous protein transfer to nitrocellulose membranes (Whatman, Cytiva, Kent, England) was confirmed by Ponceau red staining. Membranes were blocked (5% milk powder, 0.1% Tween-20 in TBS, 2 h) and incubated with primary antibody (anti-HA, 1:1,000; Sigma, St Louis, MO, USA) overnight, followed by incubation with anti-rabbit HRP-conjugated antibody (1:5,000; Sigma) for 2 h. For 35S:PIF4-HA, we used an anti-HA Peroxidase (Roche, Basel, Switzerland) antibody. We used ImageJ for quantification of the bands.
Statistical analysis
We used boxes or microtiter plates of seedlings (randomly assigned to the different conditions) as replicates in the analyses. Except for protein blots (where all the seedlings in the box were harvested as a single sample), data obtained per seedling were averaged per box to produce one replicate. We pooled replicates from independent experiments, which included all the conditions to avoid biases. We used factorial analysis of variance, t tests (two tails), correlation analysis, linear and multiple linear regression analysis as described in Supplemental File S1 or the figure legends. Representative images correspond to samples with fluorescence or band-intensity values similar to the average shown in the quantitative data display.
Accession numbers
Accession numbers based on The Arabidopsis Information Resource (https://www.arabidopsis.org) for all genes examined in this study are PIL1 (AT2G46970), IAA19 (AT3G15540), PHYB (AT2G18790), COP1 (AT2G32950), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), PIF7 (AT5G61270), HFR1 (AT1G02340), ELF3 (AT2G25930), HY5 (AT5G11260), TOC1 (AT5G61380), LHY (AT1G01060), and CCA1 (AT2G46830).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects of daytime temperature on nighttime growth in multiple pif mutants.
Supplemental Figure S2. Daytime temperature affects nuclear fluorescence driven by the pPIF4:PIF4-GFP transgene in the cotyledons.
Supplemental Figure S3. Warm temperatures posttranscriptionally enhance PIF4 abundance.
Supplemental Figure S4. Temperature effects on the time course of TOC1, LHY, and CCA1 promoter activities.
Supplemental Figure S5. Number of nuclear speckles in hypocotyl cells of the line expressing the p35S:YFP-ELF3 transgene.
Supplemental Table S1. Mutant and transgenic lines used in this study.
Supplemental File S1. Detailed statistical analysis of the data.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Christian Fankhauser (University of Lausanne) for providing seed samples of the lines pPIF7:PIF7-3HA-tPIF7 and p35S:PIF4-HA.
Funding
This work was supported by the Argentinian Agencia Nacional de Promoción Científica y Tecnológica (grants PICT-2016-1459 and PICT-2018-01695 to J.J.C.), Universidad de Buenos Aires (grant 20020170100505BA to J.J.C.) and the Spanish Ministerio de Ciencia e Innovación (grant BIO2017-90056-R to S.P.).
Conflict of interest statement. None declared.
Contributor Information
Germán Murcia, Fundación Instituto Leloir, Instituto de Investigaciones Bioquímicas de Buenos Aires-CONICET, Buenos Aires, Argentina.
Cristina Nieto, Department of Plant Molecular Genetics, CNB-CSIC, Madrid, 28049, Spain.
Romina Sellaro, Universidad de Buenos Aires, Consejo Nacional de Investigaciones Científicas y Técnicas, Instituto de Investigaciones Fisiológicas y Ecológicas Vinculadas a la Agricultura (IFEVA), Facultad de Agronomía, Buenos Aires, Argentina.
Salomé Prat, Department of Plant Molecular Genetics, CNB-CSIC, Madrid, 28049, Spain.
Jorge J Casal, Fundación Instituto Leloir, Instituto de Investigaciones Bioquímicas de Buenos Aires-CONICET, Buenos Aires, Argentina; Universidad de Buenos Aires, Consejo Nacional de Investigaciones Científicas y Técnicas, Instituto de Investigaciones Fisiológicas y Ecológicas Vinculadas a la Agricultura (IFEVA), Facultad de Agronomía, Buenos Aires, Argentina.
G.M. and J.J.C. conceived and designed the experiments. S.P. and C.N. provided insightful suggestions. G.M., C.N., and R.S. performed the experiments. S.P. provided new reporter lines. G.M., C.N., R.S., S.P., and J.J.C. analyzed the data. G.M. and J.J.C. wrote the paper with input from the other authors.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Jorge J. Casal (casal@ifeva.edu.ar) and Salomé Prat (salome.prat@cragenomica.es).
References
- Anwer MU, Davis A, Davis SJ, Quint M (2020) Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J 101: 1397–1410 [DOI] [PubMed] [Google Scholar]
- Bellstaedt J, Trenner J, Lippmann R, Poeschl Y, Zhang X, Friml J, Quint M, Delker C (2019) A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol 180: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours R, Kohlen W, Bouwmeester HJ, van der Krol A (2015) Thermoperiodic control of hypocotyl elongation depends on auxin-induced ethylene signaling that controls downstream PHYTOCHROME INTERACTING FACTOR3 activity. Plant Physiol 167: 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours R, van Zanten M, Pierik R, Bouwmeester H, van der Krol A (2013) Antiphase light and temperature cycles affect PHYTOCHROME B-controlled ethylene sensitivity and biosynthesis, limiting leaf movement and growth of Arabidopsis. Plant Physiol 163: 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Burgie ES, Gannam ZTK, McLoughlin KE, Sherman CD, Holehouse AS, Stankey RJ, Vierstra RD (2021) Differing biophysical properties underpin the unique signaling potentials within the plant phytochrome photoreceptor families. Proc Natl Acad Sci USA 118: e2105649118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk EK, Reddy AK, Nagatani A, Chen M (2014) Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol 165: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70: 321–346 [DOI] [PubMed] [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Kathare PK, Paik I, Huq E (2021) Phytochrome signaling networks. Annu Rev Plant Biol 72: 217–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA (2020) An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat Plants 6: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA (2012) High temperature exposure increases plant cooling capacity. Curr Biol 22: R396–7 [DOI] [PubMed] [Google Scholar]
- Davies J (2017) Using synthetic biology to explore principles of development. Development 144: 1146–1158 [DOI] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Ding L, Wang S, Song ZT, Jiang Y, Han JJ, Lu SJ, Li L, Liu JX (2018) Two B-box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep 25: 1718–1728.e4 [DOI] [PubMed] [Google Scholar]
- Emenecker RJ, Holehouse AS, Strader LC (2021) Biological phase separation and biomolecular condensates in plants. Annu Rev Plant Biol 72: 17–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, Zubieta C, et al. (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér B, Kozma-Bognár L, Kevei É, Hajdu A, Binkert M, Davis SJ, Schäfer E, Ulm R, Nagy F (2011) Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J 67: 37–48 [DOI] [PubMed] [Google Scholar]
- Fiorucci AS, Galvão VC, Ince YÇ, Boccaccini A, Goyal A, Allenbach Petrolati L, Trevisan M, Fankhauser C (2020) PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol 226: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Johansson H, Hornitschek P, Josse EMM, Fankhauser C, Halliday KJ (2011) Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J 65: 441–452 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Kumar SV (2017) DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep 18: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail P (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Hao N (2021) Memorizing environmental signals through feedback and feedforward loops. Curr Opin Cell Biol 69: 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585: 256–260 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HG, Seo PJ (2021) Get closer and make hotspots: liquid–liquid phase separation in plants. EMBO Rep 22: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie E, Costigliolo Rojas C, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Legris M, Nieto C, Sellaro R, Prat S, Casal JJ (2017) Perception and signalling of light and temperature cues in plants. Plant J 90: 683–697 [DOI] [PubMed] [Google Scholar]
- Murcia G, Enderle B, Hiltbrunner A, Casal JJ (2020) Phytochrome B and PCH1 protein dynamics store night temperature information. Plant J 105: 22–33 [DOI] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Ha JH, Kim JY, Park CM (2017) COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol 215: 269–280 [DOI] [PubMed] [Google Scholar]
- Paulišić S, Qin W, Harshul Arora Verasztó CT, Nogue B, Alary F, Tsiantis M, Hothorn M, Martínez-García JF (2021) Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J 40: e104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MO, Lanctot A, Queitsch C (2016) PIF4 and ELF3 act independently in Arabidopsis thaliana thermoresponsive flowering. PLoS One 11: 14–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyl ET, Piques M, Ivakov A, Schulze W, Ishihara H, Stitt M, Sulpice R (2012) Metabolism and growth in Arabidopsis depend on the daytime temperature but are temperature-compensated against cool nights. Plant Cell 24: 2443–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Quiroz FG, Li NK, Roberts S, Weber P, Dzuricky M, Weitzhandler I, Yingling YG, Chilkoti A (2019) Intrinsically disordered proteins access a range of hysteretic phase separation behaviors. Sci Adv 5: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke A, Ibañez C, Ullrich KK, Anwer MU, Becker S, Glöckner A, Trenner J, Denk K, Saal B, Sun X, et al. (2015) Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin responses. BMC Plant Biol 15: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J, Hussong A, Kircher S, Kirchenbauer D, Timmer J, Nagy F, Schäfer E, Fleck C (2010) An integrative model for phytochrome B mediated photomorphogenesis: from protein dynamics to physiology. PLoS One 5: e10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert P, Caudron F (2021) Mnemons and the memorization of past signaling events. Curr Opin Cell Biol 69: 127–135 [DOI] [PubMed] [Google Scholar]
- Romero-Montepaone S, Sellaro R, Hernando CE, Costigliolo-Rojas C, Bianchimano L, Ploschuk EL, Yanovsky MJ, Casal JJ (2021) Functional convergence of growth responses to shade and warmth in Arabidopsis. New Phytol 231: 1890–1905 [DOI] [PubMed] [Google Scholar]
- Ronald J, Wilkinson AJ, Davis SJ (2021) EARLY FLOWERING3 sub-nuclear localization responds to changes in ambient temperature. Plant Physiol 187: 2352–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Smith RW, Legris M, Fleck C, Casal JJ (2019) Phytochrome B dynamics departs from photoequilibrium in the field. Plant Cell Environ 42: 606–617 [DOI] [PubMed] [Google Scholar]
- Silva CS, Nayak A, Lai X, Hutin S, Hugouvieux V, Jung JH, López-Vidriero I, Franco-Zorrilla JM, Panigrahi KCS, Nanao MH, et al. (2020) Molecular mechanisms of Evening Complex activity in Arabidopsis. Proc Natl Acad Sci USA 117: 6901–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolomé J, Gómez MD, Yoshida S, Asami T, Olsen JE, García-Martínez JL, Alabadí D, Blázquez MA (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Harmon FG (2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LL, Li W, Tian YY, Davis SJ, Liu JX (2021) The E3 ligase XBAT35 mediates thermoresponsive hypocotyl growth by targeting ELF3 for degradation in Arabidopsis. J Integr Plant Biol 63: 1097–1103 [DOI] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY (2016) TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.